Abstract
Preoperative neoadjuvant chemoradiotherapy, combined with total mesorectal excision, has become the standard treatment for advanced localized rectal cancer (RC). However, the biological complexity and heterogeneity of tumors may contribute to cancer recurrence and metastasis in patients with radiotherapy-resistant RC. The identification of factors leading to radioresistance and markers of radiosensitivity is critical to identify responsive patients and improve radiotherapy outcomes. MicroRNAs (miRNAs) are small, endogenous, and noncoding RNAs that affect various cellular and molecular targets. miRNAs have been shown to play important roles in multiple biological processes associated with RC. In this review, we summarized the signaling pathways of miRNAs, including apoptosis, autophagy, the cell cycle, DNA damage repair, proliferation, and metastasis during radiotherapy in patients with RC. Also, we evaluated the potential role of miRNAs as radiotherapeutic biomarkers for RC.
Keywords: MicroRNAs, Rectal cancer, Radiotherapy, Mechanisms
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related deaths globally, with >1.9 million new cases (10.0%) and 935,000 deaths (9.4%) reported in 2020.[1] According to the NCCN Guidelines (2020), the rectum was defined as below the virtual line from the promontory of the sacrum to the upper margin of the symphysis, as determined by magnetic resonance imaging, rectal cancer (RC) accounts for approximately 39% of CRC cases.[1,2] Although preoperative neoadjuvant chemoradiotherapy combined with total mesorectal excision has become the standard treatment for advanced localized RC,[2] previous studies have indicated that only approximately 15% to 20% of patients with RC achieved a complete pathological regression, whereas the remainders have incomplete response or no response.[3,4] Evidence from the previous studies suggests that radiotherapy resistance may be regulated through the mechanisms of apoptosis, autophagy, the cell cycle, and DNA damage repair.[5–8] Therefore, it is of great importance to explore the mechanisms of radiotherapy resistance in RC, to understand the pathways of tumor cell survival inhibition in patients undergoing radiotherapy, and to develop new therapeutic strategies targeting these mechanisms.
MicroRNAs (miRNAs), a family of small non-coding RNAs, are approximately 22 nucleotides in length and they exert their effects by binding to complementary sequences on the 3’-untranslated regions (3’-UTRs) or the open reading frames of target genes. miRNAs regulate gene expression at the post-transcriptional level, leading to the degradation of target mRNAs or the inhibition of mRNA translation.[9] In some cases, miRNAs interact with long noncoding RNAs to form a network that regulates tumorigenesis.[10–12] It is believed that up to 30% of human genes are regulated by miRNAs. They play key roles in the regulation of biological processes, such as apoptosis, cell differentiation, development, and cell proliferation.[13,14] miRNAs are secreted into bodily fluids with minimal degradation; they are highly stable during storage; and they are easy to quantify using quantitative polymerase chain reaction, microarray, bead array, or sequencing approaches.[15,16]
Findings from many previous studies have indicated that miRNAs may be markers of the response to cancer treatment, which may facilitate the development of new strategies to overcome therapeutic resistance. Recent studies have shown that miRNAs may be involved in the regulation of radiotherapy sensitivity in RC. Zhang et al[17] found that miR-124 increased the radiosensitivity of CRC cells by blocking the expression of paired related homeobox 1. The expression level of miR-15b has been shown to predict the degree of postoperative tumor degeneration and the sensitivity of CRC cells to chemo- therapy/radiotherapy. Double cortin-like kinase 1 (DCLK1) is a direct target gene of miR-15b, and its expression level is negatively correlated with the prognosis of patients with RC.[18] miR-149-3p sensitizes CRC cells to radiation by inhibiting WAP four-disulfide core protein 2 (WFDC2).[19] Previous studies have revealed various possible mechanisms by which miRNAs are involved in the response to radiation. Here, we summarize the findings related to miRNAs involved in RC radiotherapy. We focus on miRNAs involved in regulating apoptosis, autophagy, DNA damage repair, the cell cycle, cell proliferation, and metastasis in response to ionizing radiation (IR). Furthermore, we provide an overview of the roles of miRNAs and their target genes in the resistance to RC radiotherapy, and we discuss the value of using miRNAs as biomarkers of radiosensitivity and the potential of miRNAs as targets of improved treatment strategies.
miRNAs Affect the Response to RC Radiotherapy by Regulating Apoptosis
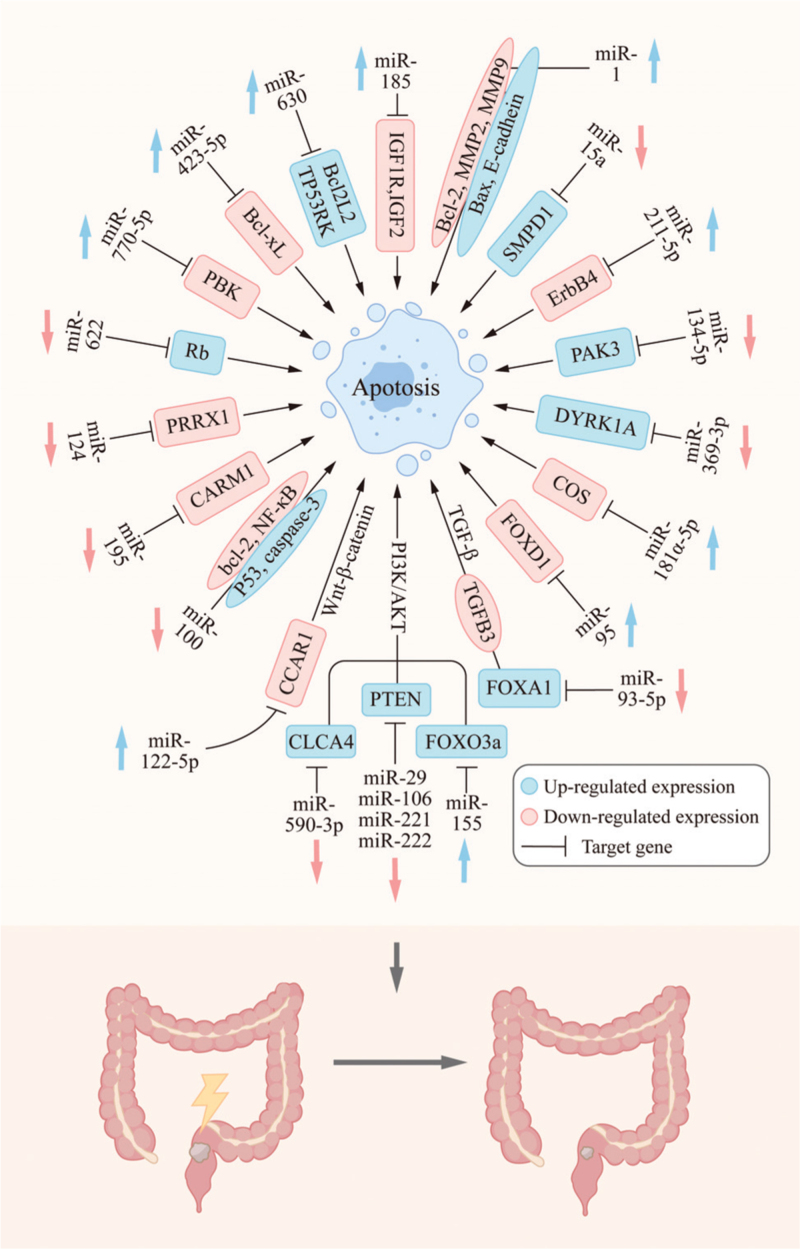
Apoptosis refers to programmed cell death that is initiated after the damage to DNA or cellular organelles, such as the mitochondria and endoplasmic reticulum.[20] Apoptosis usually occurs after the exposure of cells to stress conditions, such as oxidative stress, IR, chemotherapy drugs, hypoxia, or high temperature.[21] It is modulated via different signaling pathways that affect extrinsic or intrinsic mediators of apoptosis. Previous studies have shown that apoptosis is an indicator of cell radiosensitivity and an important prognostic factor for radiotherapy outcomes.[22,23] Moreover, miRNAs have been found to regulate apoptosis after IR. Here, we summarize the miRNAs involved in the regulation of apoptosis after IR [Table 1 and Figure 1].
Table 1.
Mechanisms and targets of miRNAs involvement in radiotherapy of RC.
| MiRNA | Up/ Down | Mechanism | Target | Up/Down | Sensitivity/ Resistance | Sample type | Reference |
| miR-15a | Down | Apoptosis | SMPD1 | Up | Sensitivity | Cell lines | [38] |
| miR-29a | Up | Apoptosis | PTEN | Down | Resistance | Cell lines | [26] |
| miR-95 | Down | Apoptosis | FOXD1 | Down | Sensitivity | Cell lines, CDX | [39] |
| miR-134-5p | Down | Apoptosis | PAK3 | Up | Resistance | Cell lines | [31] |
| miR-145 | Down | Apoptosis | — | — | Resistance | Tissue, cell lines, PDX, CDX | [75,77] |
| miR-155 | Up | Apoptosis | FOXO3a | Up | Resistance | Cell lines | [27] |
| miR-181a-p | Up | Apoptosis | COS | Down | Sensitivity | Cell lines | [11] |
| miR-205-3p | Up | Apoptosis | — | — | Sensitivity | Cell lines | [78] |
| miR-211-5p | Up | Apoptosis | ErbB4 | Down | Sensitivity | Tissue, cell lines, CDX | [43] |
| miR-221 | Down | Apoptosis | PTEN | Up | Sensitivity | Cell lines | [28] |
| miR-222 | Up | Apoptosis | PTEN | Down | Resistance | Cell lines | [27] |
| miR-338-3p | Down | Apoptosis | — | — | Resistance | Cell lines | [79] |
| miR-369-3p | Down | Apoptosis | DYRK1A | Up | Sensitivity | Cell lines | [12] |
| miR-423-5P | Up | Apoptosis | bcl-xL | Down | Sensitivity | Tissue, cell lines | [34] |
| miR-630 | Up | Apoptosis | BCL2L2, TP53RK | Up | Sensitivity | Cell lines | [35] |
| miR-888 | Down | Apoptosis | — | — | Sensitivity | Tissue | [80] |
| miR-106b | Up | Apoptosis, cell cycle | PTEN, P21 | Down | Resistance | Tissue, cell lines, CDX | [24] |
| miR-100 | Up | Apoptosis, DNA damage | — | — | Sensitivity | Tissue, cell lines | [7] |
| miR-122-5p | Up | Apoptosis, DNA damage | CCAR1 | Down | Sensitivity | Plasma, cell lines, C57BL/6 mouse | [40] |
| miR-124 | Down | Apoptosis, DNA damage | PRRX1 | Down | Sensitivity | Tissue, cell lines, CDX | [17] |
| miR-185 | Up | Apoptosis, DNA damage | IGF1R, IGF2 | Down | Sensitivity | Cell lines | [41] |
| miR-195 | Down | Apoptosis, DNA damage | CARM1 | Down | Sensitivity | Cell lines, CDX | [33] |
| miR-590-3p | Up | Apoptosis, DNA damage | CLCA4 | Down | Resistance | Tissue, cell lines, CDX | [30] |
| miR-622 | Up | Apoptosis, DNA damage | Rb | Down | Resistance | Tissue, cell lines | [37] |
| miR-1 | Up | Apoptosis, migration, invasion | — | — | Sensitivity | Tissue, cell lines | [73] |
| miR-101-3p | Up | Apoptosis, migration, invasion | — | — | Sensitivity | Cell lines | [10] |
| miR-93-5p | Up | Apoptosis, proliferation | FOXA1 | Down | Resistance | Tissue, cell lines, CDX | [36] |
| miR-770-5p | Up | Apoptosis, proliferation | PBK | Down | Sensitivity | Cell lines, CDX | [44] |
| miR-296-5p | Up | Apoptosis, proliferation, cell cycle | IGF1R | Down | Sensitivity | Tissue, cell lines, CDX | [42] |
| miR-18a | Up | Autophagy | ATM | Up | Sensitivity | Cell lines | [48] |
| miR-93 | Up | Autophagy | ATG12 | Down | Sensitivity | Tissue, cell lines, CDX, plasma | [52] |
| miR-129-5p | Up | Autophagy | beclin-1 | Down | Sensitivity | Cell lines, CDX | [50] |
| miR-183-5p | Down | Autophagy | ATG5 | Up | Sensitivity | Tissue, cell lines, CDX | [51] |
| miR-210 | Down | Autophagy | Bcl-2 | Down | Sensitivity | Cell lines | [55] |
| miR-31 | Up | Autophagy, apoptosis | — | — | Sensitivity | Tissue, cell lines | [49] |
| miR-214 | Up | Autophagy, apoptosis | ATG12 | Down | Sensitivity | Tissue, cell lines, CDX, plasma | [5] |
| let-7e | Up | Cell cycle, apoptosis | IGF-1R | Down | Sensitivity | Cell lines | [58] |
| miR-31 | Up | DNA damage | STK40 | Down | Sensitivity | Tissue, cell lines | [66] |
| miR-31-5p | Down | DNA damage | hMLH1 | Up | Resistance | Cell lines, (Cre; Apc) mouse | [68] |
| miR-130a | Up | DNA damage, invasion | SOX4 | Down | Sensitivity | Cell lines, CDX | [64] |
| miR-15b | Up | Migration, invasion, proliferation | DCLK1 | Down | Sensitivity | Tissue, cell lines, CDX | [18] |
| miR-32-5p | Down | Migration, invasion | TOB1 | Down | Sensitivity | Tissue, cell lines | [74] |
| miR-140-5p | Up | Proliferation | — | — | Sensitivity | Plasma, cell lines | [70] |
| miR-451a-5p | Up | Proliferation | CAB39, EMSY | Down | Sensitivity | Tissue, cell lines | [69] |
| miR-506-3p | Up | Proliferation | — | — | Sensitivity | Plasma, cell lines | [70] |
| miR-149-3p | Up | — | WFDC2 | Down | Sensitivity | Tissue, cell lines, CDX | [19] |
CDX: Cell line derived xenograft; DNA: deoxyribonucleic acid; PDX: Patient-derived xenograft; miRNAs: MicroRNAs; RC: Rectal cancer; –: Not applicable.
Figure 1.
miRNAs regulate apoptosis through the corresponding target genes and affect the radiation sensitivity of RC. miRNAs: MicroRNAs; RC: Rectal cancer.
The phosphatidylinositol 3-kinase/AKT (PI3K/AKT) path way is known to regulate cell survival and increase radiation resistance.[24,25] Previous studies have shown that phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a natural inhibitor of PI3K, negatively regulates the PI3K/AKT pathway, resulting in apoptosis. There is evidence suggesting that miR-29a, miR-106b, and miR-222 decrease the radiosensitivity of RC by negatively regulating PTEN expression, to activate the PI3K/AKT signaling pathway.[24,26,27] It has also been shown that an miR-221 antisense oligonucleotide enhances IR sensitivity by mediating the upregulation of PTEN.[28] Furthermore, forkhead box O3 (FOXO3a), a member of the FOXO family, is a downstream effector of the PI3K/AKT pathway.[29] Khoshinani et al[27] showed that FOXO3a is involved in the regulation of the radiation resistance of RC as a direct target gene of miR-155. Moreover, Chen et al[30] demonstrated that exosomal miR-590-3p derived from cancer-associated fibroblasts (CAFs) increases the radiation resistance of RC by positively regulating the chloride channel accessory 4-dependent PI3K/AKT signaling pathway in vivo and in vitro. Another study showed that, in vitro, the lncRNA, TTN antisense RNA 1, promotes the radiotherapy resistance of CRC cells by negatively regulating miR-134-5p and increasing the expression levels of p21-activated kinase 3,[31] which may be associated with the P21 and AKT/glycogen synthase kinase-3β/β-catenin pathways.
Several miRNAs also affect apoptosis through other signaling pathways and participate in the regulation of the radiotherapy sensitivity of RC. Some members of the Bcl-2 protein family, such as Bad, Bid, and Bax, promote apoptosis, whereas others, such as Bcl-2, Bcl-x, and Bcl-w, prevent apoptosis.[32] Yang et al[7] reported that miR-100 promotes the X-ray-induced apoptosis of CCL-244 cells and regulates the expression of apoptosis-related proteins, including increasing the expression levels of the pro- apoptotic proteins, P53 and caspase-3, and decreasing the levels of the anti-apoptotic proteins, Bcl-2 and NF-κB, further increasing the radiosensitivity of CCL-244 cells. The forced expression of miR-195 has been shown to induce apoptosis, upregulate Bax and γ-H2AX levels, inhibit the expression of Bcl-2, and increase the radiosensitivity of CRC cells by downregulating coactivator associated arginine methyltransferase 1 in vivo and in vitro.[33] Data from the study reported by Shang et al[34] implied that overexpression of miR-423-5p promotes radiation-induced apoptosis through downregulation of Bcl-XL, ultimately increasing the sensitivity of HCT116 and RKO cells to IR. The expression level of miR-630 in CRC cells is positively correlated with radiosensitivity and the induction of apoptosis.[35] BCL2L2 and TP53 regulating kinase (TP53RK) have been identified as target genes of miR-630. BCL2L2, also known as Bcl-w, belongs to the Bcl-2 protein family. The kinase, TP53RK, inhibits apoptosis after mitotic stress. Zhang et al[35] reported that cAMP-response element-binding protein/miR-630/BCL2L2 and TP53RK constitute a novel signaling cascade that modulates the radiosensitivity of CRC cell lines by inducing apoptosis.
Other studies have also reported findings related to the mechanism of apoptosis. CAF-derived exosomes upregulate the level of miR-93-5p and downregulate the level of forkhead box A1, the target gene of miR-93-5p, further inhibiting apoptosis and promoting the resistance of RC to radiation in vivo and in vitro.[36] Ma et al[37] found that the overexpression of miR-622 induces radiation resistance in vitro. During the radiation response, the retinoblastoma (Rb)-E2F1-P/CAF complex transcriptionally activates pro-apoptotic genes. Rb overexpression has also been shown to reverse radiation resistance induced by miR-622 in vitro.[37] The results reported by Rana et al[38] suggest that the activation of acid sphingolipids (sphingomyelin phosphodiesterase 1, SMPD1) and the production of ceramides are key processes in the regulation of apoptosis in response to cellular stress, including radiation. Low expression levels of miR-15a upregulate SMPD1 levels, inducing apoptosis and increasing radiosensitivity. Low expression levels of miR-95 have been shown to increase the radiosensitivity of LoVo cells and promote apoptosis, which may be related to the inhibition of forkhead box D1 protein activity.[39] It has been noted that miR-122-5p significantly inhibits cell survival and increases radiotherapy sensitivity and apoptosis by silencing the apoptosis regulator, cell division cycle, and apoptosis regulator 1.[40] Antisense noncoding RNA in the INK4 locus negatively regulates radiosensitivity induced by chitooligosaccharides in CRC cells by sponging miR-181a-5p.[11] miR-185 and miR-296-5p trigger apoptosis through the downstream IGF1R signaling pathway and enhance the radiosensitivity of CRC cells.[41,42] Li et al[43] found that eosinophil granule ontogeny transcript (EGOT) expression levels are upregulated in RC tissues and cells, and that its level of expression is related to the pathological stage. The downregulation of EGOT may inhibit the growth of Colo320 cells by regulating the miR-211-5p/receptor tyrosine-protein kinase erbB-4 axis, inducing the apoptosis of cancer cells, and enhancing the effects of radiotherapy for RC in vivo and in vitro.[43] Apoptosis induced by miR-214 significantly increases the radiosensitivity of CRC cells.[5] When miR-369-3p expression is downregulated, the downstream target gene, dual-specificity tyrosine-phosphorylation-regulation kinase 1A, is upregulated, promoting apoptosis and increasing the radiosensitivity of CRC cells.[12] When miR-770-5p is overexpressed, the apoptosis of MCF7 and A549 cells increases, leading to a decrease in the relative cell number. Moreover, miR-770-5p has been shown to negatively regulate PDZ-bound kinase, increasing apoptosis and the sensitivity of tumors to radiation.[44]
Relationship Between miRNAs, Autophagy, and Radiotherapy in RC
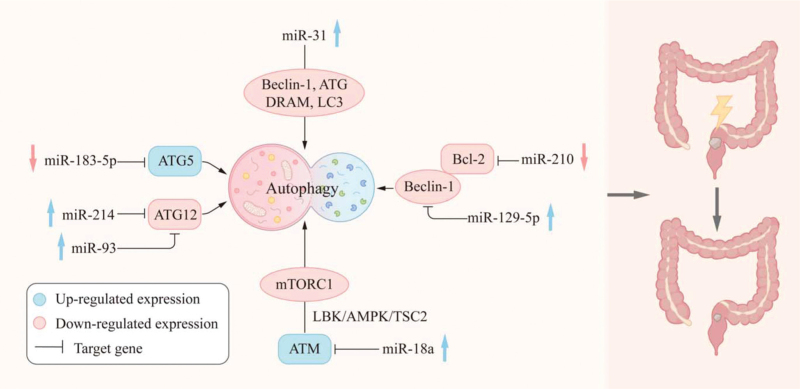
Autophagy is a highly conserved critical regulatory process in which cells degrade aging proteins and damaged organelles through lysosomes, resulting in the circulation of cellular material and the maintenance of homeostasis.[45] The formation of autophagosomes is regulated by autophagy-related genes (ATGs), such as ATG12, ATG5, and microtubule-associated protein light chain 3 (LC3). ATG12 and ATG5 a conjugated complex that plays an important role in autophagosome expansion.[46] Various stimuli, such as starvation, hypoxia, DNA damage, and IR, may activate autophagy. Previous studies have demon strated that autophagy is tightly linked to various cellular functions and that dysfunctional autophagy leads to various diseases, including cancer.[47] In recent years, there have been many investigations of the mechanism whereby autophagy affects the radiosensitivity of cancer cells [Table 1 and Figure 2].
Figure 2.
miRNAs regulate autophagy through the corresponding target genes and affect the radiation sensitivity of RC. miRNAs: MicroRNAs; RC: Rectal cancer.
Heterotopic overexpression of miR-18a in HCT116 cells enhances IR-induced autophagy.[48] In addition, evidence suggests that miR-18a overexpression results in upregulation of the expression of the autophagy activator, ataxia telangiectasia mutated, and the inhibition of the mammalian target of rapamycin compound 1 activity. Yang et al[49] found that the treatment of CAFs with an miR-31 mimic inhibited the expression of ATGs Beclin-1, ATG, DRAM, and LC3, thus increasing the radiosensitivity of CRC cells co-cultured with CAFs. Xu et al[50] indicated that the overexpression of miR-129-5p inhibits Beclin-1, a key autophagy-related gene, and inhibits autophagy. However, the overexpression of Beclin-1 eliminates the effect of miR-129-5p. These results suggest that miR-129-5p significantly enhances the radiosensitivity of CRC cells by inhibiting Beclin-1-mediated autophagy. Zheng et al[51] reported that the upregulation of miR-183-5p expression levels and the downregulation of ATG5 expression levels are associated with the poor prognosis of patients with RC. In vitro and in vivo experiments have also shown that miR-183-5p knockdown increases the radiosensitivity of CRC cells by directly targeting ATG5. Liu et al[52] reported that the overexpression of miR-93 inhibits IR-induced autophagy and enhances the radiosensitivity of RC by down regulating its target gene, ATG12. miR-214 has also been found to significantly increase the radiosensitivity of CRC by targeting ATG12, to inhibit autophagy and induce apoptosis.[5] Furthermore, the combination of Bcl-2 and Beclin-1 may prevent the inappropriate activation of autophagy, while the disruption of this interaction may induce autophagy.[53,54] Autophagy has been shown to contribute to the decrease in radiosensitivity in hypoxic environments. This finding suggests that, under hypoxic conditions, hypoxia-inducible factor-1a induces miR-210 to downregulate the expression of Bcl-2 in CRC cells, thereby increasing autophagy and reducing radiosensitivity.[55]
miRNAs are Involved in the Radiotherapy of RC by Regulating the Cell Cycle
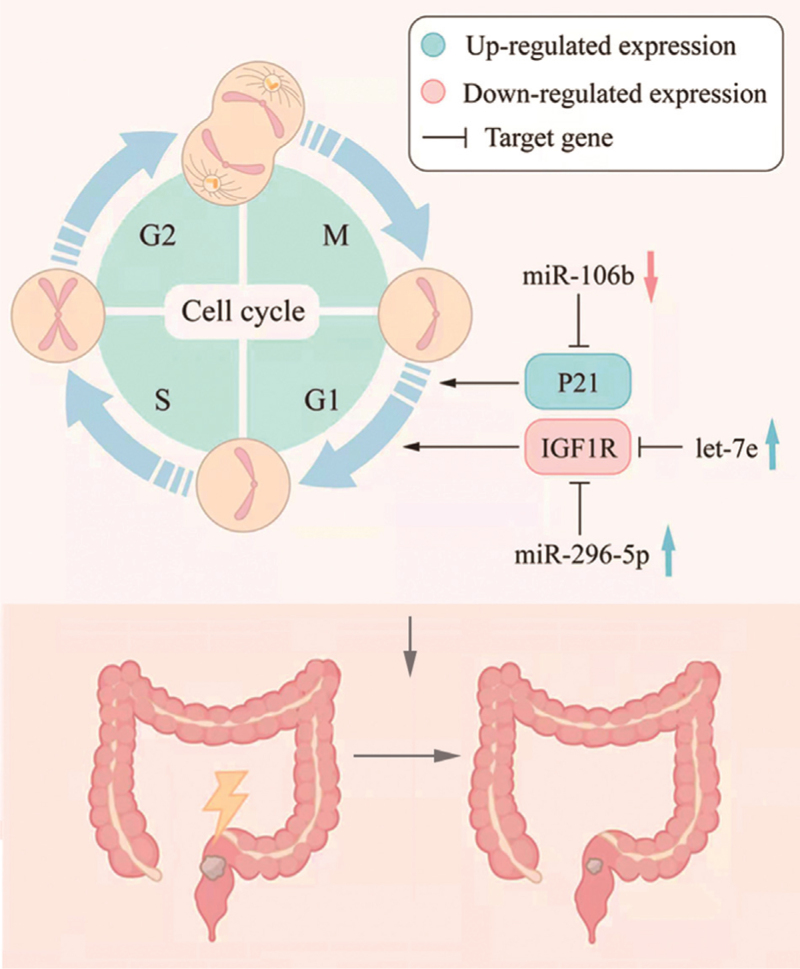
In normal cells, IR delays entry into the G1, S, and G2 phases of the cell cycle to allow DNA repair and prevent the accumulation of harmful genomic damage. The phosphorylation of P53 by ATM induces the expression and phosphorylation of the cyclin-dependent kinase inhibitor, P21. This leads to the inhibition of CDK4/6- cyclin D and CDK1-cyclin B, causing the cell cycle to arrest at G1 and G2, respectively.[56] In addition, ATM- and ATR-activated signal transducers, CHK1 and CHK2, promote the degradation of CDC25, leading to the inhibition of CDK2-cyclin E and CDK1-cyclin B, thereby promoting cell cycle arrest at G1 and G2, respectively.[57] The efficient induction of cell cycle arrest promotes radiosensitivity, suggesting that cell cycle progression after IR contributes to the radiation resistance of tumors.[6] Several miRNAs have been shown to play a role in cell cycle regulation after IR in CRC [Table 1 and Figure 3].
Figure 3.
miRNAs regulate cell cycle through the corresponding target genes and affect the radiation sensitivity of RC. miRNAs: MicroRNAs; RC: Rectal cancer.
The let-7 miRNA family of tumor suppressors is down- regulated in different types of human malignancies, including CRC. It has been shown that increasing let-7e levels reduces IGF-1R protein levels and inhibits the downstream signaling pathway, resulting in cell cycle arrest at G1, and significantly reducing CRC cell proliferation, survival, and radiation resistance.[58] In addition, miR-296-5p overexpression inhibits cell proliferation and cell cycle progression and promotes apoptosis and radiosensitivity by down-regulating insulin-like growth factor I receptor (IGF1R).[42] Zheng et al[24] showed that the overexpression of miR-106b decreased the expression levels of the direct targets of PTEN and P21. Meanwhile, the restoration of PTEN or P21 expression in cells stably overexpressing miR-106b reestablishes the effect of miR-106b on CRC cell radioresistance. These findings indicate that miR-106b mediates G1 growth arrest and cellular senescence by targeting P21.[24] In addition, this process is accompanied by enhanced tumor promotion, suggesting that miR-106b may be related to the resistance of RC to radiation therapy.
Relationship Between miRNAs, DNA Damage Repair, and Radiotherapy in RC
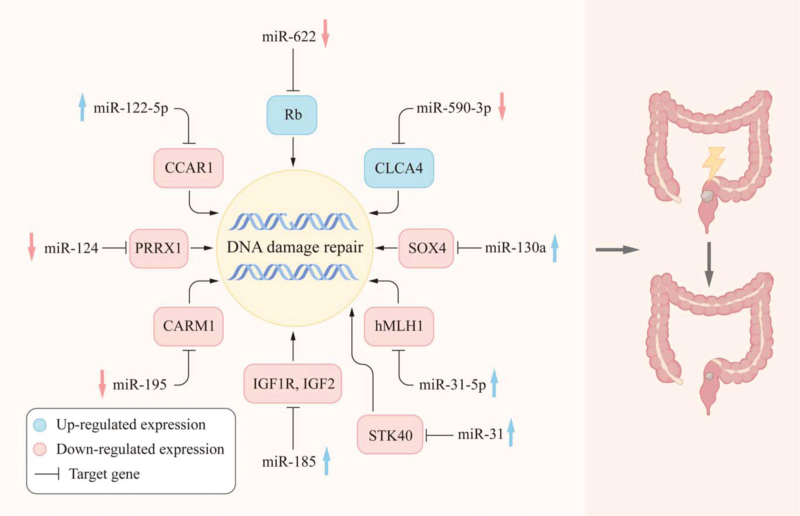
DNA double-strand breaks (DSBs) are the most deleterious type of DNA damage as they may initiate genomic instability, ultimately leading to cancer.[59] Two pathways are specifically dedicated to the repair of DSBs: nonhomologous end-joining (NHEJ) and homologous recombination (HR) repair.[60,61] The repression of these efficient repair systems permits the accumulation of DNA damage in rapidly dividing cells (such as cancer cells), thus inducing apoptosis. This mechanism plays an important role in the radiotherapy of RC. Recently, the role of miRNAs in DNA damage repair has been recognized,[62] suggesting that miRNAs control the DNA damage response by interacting with DNA repair genes. Here, we summarize the miRNAs involved in DNA damage repair following IR [Table 1 and Figure 4].
Figure 4.
miRNAs regulate DNA damage repair through the corresponding target genes and affect the radiation sensitivity of RC. IGF: Insulin-like growth factor; miRNAs: MicroRNAs; RC: Rectal cancer.
There is evidence that changes in miRNA-binding sites in the 3’-UTR of base-resected repair genes may modulate the prognosis and treatment response in patients with RC, due to effects on DNA damage repair.[63] Overexpression of miR-130a has been shown to inhibit the repair of cells after radiation-induced DNA damage.[64] Moreover, there is evidence that SRY-Box transcription factor 4, which is a direct target of miR-130a, increases the activation of ATM signals by increasing the expression level of NBS1 and promoting the interaction between NBS1 and p-ATM, thereby mediating DNA damage repair.[64] In a study of the relationship between miR-100 and radiotherapy for RC, miR-100 was found to be involved in radiation-induced apoptosis and to modulate the sensitization of CCL-244 cells to radiation by enhancing radiation-induced DNA damage repair.[7] miR-122-5p overexpression or CCAR1 silencing, combined with IR, significantly reduces the levels of p-CHK2 and enhances radiosensitivity, indicating that CHK2 is an important regulatory kinase in the DNA damage response.[40,65] Zhang et al[66]reported that miR-31 suppresses NF-κB signaling pathways by targeting serine/threonine kinase 40, thereby improving RC radiotherapy sensitivity. Moreover, it is possible to improve sensitivity to radiation by promoting DNA damage after radiotherapy. miR-185 regulates radiation-induced HR and NHEJ by targeting IGF1R and IGF2 genes.[41] CAF-derived exosomal miR-590-3p induces radiotherapy resistance in RC through the CLCA4/PI3K/AKT axis, thereby inhibiting the DNA damage response.[30] Overexpression of miR-124 and miR-195, combined with radiotherapy, increases the levels of phosphorylated γ-H2AX.[33,67] miR-31-5p inhibitors may modulate radiosensitivity through IR-induced DNA damage repair.[68]
Relationship Between miRNAs, Other Mechanisms, and Radiotherapy in RC
Unrestricted proliferation is the basis of cancer development. In addition, the invasion and metastasis of tumor cells are the major causes of RC-related deaths. Many studies have shown that miRNAs play important roles in pathways affecting tumor cell proliferation, invasion, and metastasis, thus influencing the efficacy of radiotherapy.
Ruhl et al[69] found higher expression levels of miR-451a and lower levels of calcium-binding protein 39 (CAB39) and EMSY transcriptional repressor, BRCA2 interacting (EMSY) in patients who responded to radiotherapy, compared with patients who did not respond to radiotherapy. Further exploration revealed that miR-451a was induced by radiation, and it may affect the proliferation of RC through the CAB39 and EMSY pathways. A series of studies have shown that overexpression of let-7e, miR-93-5p, miR-140-5p, miR-296-5p, miR-506-3p, and miR-770-5p significantly reduces the proliferation of CRC cells after radiotherapy and improves radiotherapy sensitivity.[36,42,44,58,70] Recent studies have suggested that miR-1 may play an important role in RC. The downregulation of miR-1 and metastasis-associated in colon cancer-1 increases MET expression levels and promotes RC metastasis.[71] Furukawa et al[72] found that the miR-1-notch receptor 3-Asef pathway plays an important role in CRC cell migration. Another study reported that miR-1 mimics promote the expression of Bax and E-cadherin and decreased the expression levels of Bcl-2, MMP2, and MMP9, significantly inhibiting the invasion and migration of CRC cells in conjunction with radiotherapy.[73] miR-1 is thought to increase the radiosensitivity of CRC cells by inducing apoptosis and synergistically inhibiting invasive phenotypes. Ji et al[18] showed that overexpression of miR-15b inhibits the proliferation, invasion, and metastasis of CRC cells. Liang et al[74] reported that miR-32-5p may be a prognostic tool and therapeutic target for RC. miR-32-5p was found to directly decrease the levels of transducer of ERBB2,1 mRNA by binding to its 3’-UTR, thus sensitizing RC to the effects of radiotherapy and inhibiting metastasis.
Conclusions
With the development of biotechnologies, such as high- throughput sequencing, bioinformatics analysis, genome modification, and mouse models of disease, functional studies can provide new insights into the anticancer activity of miRNAs. By identifying downstream targets, many studies have shown that miRNAs regulate various signaling pathways (such as PI3K/AKT and Wnt/β-catenin) that play roles in a series of various processes, such as RC proliferation, metastasis, autophagy, and apoptosis, and affect the efficacy of radiotherapy for RC. Moreover, miRNAs are often involved in multiple mechanisms to regulate the activity of cancer cells, thus inducing radiotherapy sensitivity or resistance. These studies provide a new theoretical basis for the study of radiosensitivity and the identification of effective biomarkers and promising therapeutic targets for RC. However, many challenges remain, and the targets and downstream pathways of some miRNAs are still being explored.[75,76] In addition, most studies in this field are still at the basic stage, with little progress in in-vivo studies and translational direction. Moreover, most studies have been limited to specific tumor types or treatment modalities. Future studies should focus on exploring these questions. First, for related miRNAs, there is a need to further understand and evaluate genomic and functional approaches for basic and translational research, to help select appropriate and specific targets from a large number of candidates. More importantly, for suitable candidate genes that have been identified and stable delivery vectors that have been developed, there should be a greater focus on clinical studies to assess patients’ responses to miRNA-related therapies. This will improve our understanding of the long-term effects and adverse reactions associated with these therapies and help us to achieve translational results in cancer research.
Funding
This study was supported by the Basic Research for Application of Sichuan Province, China (No. 2020YJ0066), the Key Research and Development Projects of Sichuan Province, China (No. 2020YFS0430), and the Key Research and Development Projects of Sichuan Province, China (No. 2018SZ0136).
Conflicts of interest
The authors declare no competing financial interests.
Footnotes
How to cite this article: Zhu L, Wang M, Chen N, Zhang Y, Xu T, Zhuang W, Xiao S, Dai L. Mechanisms of microRNA action in rectal cancer radiotherapy. Chin Med J 2022;135:2017–2025. doi: 10.1097/CM9.0000000000002139
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. NCCN guidelines insights: rectal cancer, version 6.2020. J Natl Compr Canc Netw 2020; 18:806–815. doi: 10.6004/jnccn.2020.0032. [DOI] [PubMed] [Google Scholar]
- 3.Martin ST, Heneghan HM, Winter DC. Systematic review and meta analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br J Surg 2012; 99:918–928. doi: 10.1002/bjs.8702. [DOI] [PubMed] [Google Scholar]
- 4.de Campos-Lobato LF, Stocchi L, da Luz Moreira A, Geisler D, Dietz DW, Lavery IC, et al. Pathologic complete response after neoadjuvant treatment for rectal cancer decreases distant recurrence and could eradicate local recurrence. Ann Surg Oncol 2011; 18:1590–1598. doi: 10.1245/s10434-010-1506-1. [DOI] [PubMed] [Google Scholar]
- 5.Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y, et al. Inhibition of ATGl2-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis 2018; 7:16.doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys 2004; 59:928–942. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG, Ru G, et al. Role of miR-100 in the radioresistance of colorectal cancer cells. Am J Cancer Res 2015; 5:545–559. [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, Tie Y, Lv G, Zhu J, Fu H, Zheng X. Transcriptional activation of miR-320a by ATF2, ELK1 and YY1 induces cancer cell apoptosis under ionizing radiation conditions. Int J Oncol 2018; 53:1691–1702. doi: 10.3892/ijo.2018.4497. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Guo J, Ding Y, Yang H, Guo H, Zhou X, Chen X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp Mol Pathol 2020; 115:104448.doi: 10.1016/j.yexmp.2020.104448. [DOI] [PubMed] [Google Scholar]
- 11.Sun C, Shen C, Zhang Y, Hu C. LncRNA ANRIL negatively regulated chitooligosaccharide-induced radiosensitivity in colon cancer cells by sponging miR-181a-5p. Adv Clin Exp Med 2021; 30:55–65. doi: 10.17219/acem/128370. [DOI] [PubMed] [Google Scholar]
- 12.Zou Y, Yao S, Chen X, Liu D, Wang J, Yuan X, et al. LncRNA OIP5- AS1 regulates radioresistance by targeting DYRK1A through miR- 369-3p in colorectal cancer cells. Eur J Cell Biol 2018; 97:369–378. doi: 10.1016/j.ejcb.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer 2009; 8:102.doi: 10.1186/1476-4598-8 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agostini M, Pucciarelli S, Calore F, Bedin C, Enzo M, Nitti D. MiRNAs in colon and rectal cancer: a consensus for their true clinical value. Clin Chim Acta 2010; 411:1181–1186. doi: 10.1016/j. cca.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Eriksen AH, Andersen RF, Pallisgaard N, Sørensen FB, Jakobsen A, Hansen TF. MicroRNA expression profiling to identify and validate reference genes for the relative quantification of microRNA in rectal cancer. PLoS One 2016; 11:e0150593.doi: 10.1371/journal. pone.0150593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaedcke J, Grade M, Camps J, Søkilde R, Kaczkowski B, Schetter AJ, et al. The rectal cancer microRNAome - microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res 2012; 18:4919–4930. doi: 10.1158/1078-0432.Ccr-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, et al. MiR-124 radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One 2014; 9:e93917.doi: 10.1371/journal.pone.0093917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji D, Zhan T, Li M, Yao Y, Jia J, Yi H, et al. Enhancement of sensitivity to chemo/radiation therapy by using miR-15b against DCLK1 in colorectal cancer. Stem Cell Rep 2018; 11:1506–1522. doi: 10.1016/j.stemcr.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi LP, Guo HL, Su YB, Zheng ZH, Liu JR, Lai SH. MicroRNA-149 sensitizes colorectal cancer to radiotherapy by downregulating human epididymis protein 4. Am J Cancer Res 2018; 8:30–38. [PMC free article] [PubMed] [Google Scholar]
- 20.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moeller BJ, Richardson RA, Dewhirst MW. Hypoxia and radiother apy: opportunities for improved outcomes in cancer treatment. Cancer Metastasis Rev 2007; 26:241–248. doi: 10.1007/s10555-007-9056-0. [DOI] [PubMed] [Google Scholar]
- 22.Borràs-Fresneda M, Barquinero JF, Gomolka M, Hornhardt S, Rössler U, Armengol G, et al. Differences in DNA repair capacity, cell death and transcriptional response after irradiation between a radiosensitive and a radioresistant cell line. Sci Rep 2016; 6:27043.doi: 10.1038/srep27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell 2019; 179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y, Yuan L, et al. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med 2015; 13:252.doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao G, Cai C, Yang T, Qiu X, Liao B, Li W, et al. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One 2013; 8:e53906.doi: 10.1371/journal.pone.0053906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Xu J, Fu J, Yuan D, Guo F, Zhou C, et al. MiR-29a regulates radiosensitivity in human intestinal cells by targeting PTEN gene. Radiat Res 2016; 186:292–301. doi: 10.1667/rr14428.1. [DOI] [PubMed] [Google Scholar]
- 27.Khoshinani HM, Afshar S, Pashaki AS, Mahdavinezhad A, Nikzad S, Najafi R, et al. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Jpn J Radiol 2017; 35:664–672. doi: 10.1007/s11604-017-0679-y. [DOI] [PubMed] [Google Scholar]
- 28.Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ, Li GX. Anti-miRNA-221 sensitizes human colorectal carcinoma cells to radiation by upregulating PTEN. World J Gastroenterol 2013; 19:9307–9317. doi: 10.3748/wjg.v19.i48.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández de Mattos S, Villalonga P, Clardy J, Lam EW. FOXO3a mediates the cytotoxic effects of cisplatin in colon cancer cells. Mol Cancer Ther 2008; 7:3237–3246. doi: 10.1158/1535-7163.Mct-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Liu Y, Zhang Q, Liu B, Cheng Y, Zhang Y, et al. Exosomal miR-590-3p derived from cancer-associated fibroblasts confers radioresistance in colorectal cancer. Mol Ther Nucleic Acids 2020; 24:113–126. doi: 10.1016/j.omtn.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo Z, Ji S, He L, Zhang Y, Peng Z, Han J. LncRNA TTN-AS1/miR- 134-5p/PAK3 axis regulates the radiosensitivity of human large intestine cancer cells through the P21 pathway and AKT/GSK-3β/β-catenin pathway. Cell Biol Int 2020; 44:2284–2292. doi: 10.1002/cbin.11436. [DOI] [PubMed] [Google Scholar]
- 32.Basu A. The interplay between apoptosis and cellular senescence: Bcl- 2 family proteins as targets for cancer therapy. Pharmacol Ther 2021; 230:107943.doi: 10.1016/j.pharmthera.2021.107943. [DOI] [PubMed] [Google Scholar]
- 33.Zheng L, Chen J, Zhou Z, He Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. Onco Targets Ther 2017; 10:1027–1038. doi: 10.2147/ott.S125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang Y, Wang L, Zhu Z, Gao W, Li D, Zhou Z, et al. Downregulation of miR-423-5p contributes to the radioresistance in colorectal cancer cells. Front Oncol 2020; 10:582239.doi: 10.3389/fonc.2020.582239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Yu J, Liu H, Ma W, Yan L, Wang J, et al. Novel epigenetic CREB-miR-630 signaling axis regulates radiosensitivity in colorectal cancer. PLoS One 2015; 10:e0133870.doi: 10.1371/journal. pone.0133870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Liu J, Zhang Q, Liu B, Cheng Y, Zhang Y, et al. Exosome- mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J Exp Clin Cancer Res 2020; 39:65.doi: 10.1186/s13046-019-1507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma W, Yu J, Qi X, Liang L, Zhang Y, Ding Y, et al. Radiation- induced microRNA-622 causes radioresistance in colorectal cancer cells by down-regulating Rb. Oncotarget 2015; 6:15984–15994. doi: 10.18632/oncotarget.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rana S, Espinosa-Diez C, Ruhl R, Chatterjee N, Hudson C, Fraile- Bethencourt E, et al. Differential regulation of microRNA-15a by radiation affects angiogenesis and tumor growth via modulation of acid sphingomyelinase. Sci Rep 2020; 10:5581.doi: 10.1038/s41598-020-62621-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang WL, Xiao TB, Yuan F, Cao YB, Yan DG. Effects of microRNA95 targeting FOXD1 gene on radiosensitivity of colorectal cancer loVo cells and its mechanism (in Chinese). Chin J Appl Phys 2020; 36:148–151. doi: 10.12047/j.cjap.5864.2020.033. [DOI] [PubMed] [Google Scholar]
- 40.Ge Y, Tu W, Li J, Chen X, Chen Y, Xu Y, et al. MiR-122-5p increases radiosensitivity and aggravates radiation-induced rectal injury through CCAR1. Toxicol Appl Pharmacol 2020; 399:115054.doi: 10.1016/j.taap.2020.115054. [DOI] [PubMed] [Google Scholar]
- 41.Afshar S, Najafi R, Sedighi Pashaki A, Sharifi M, Nikzad S, Gholami MH, et al. MiR-185 enhances radiosensitivity of colorectal cancer cells by targeting IGF1R and IGF2. Biomed Pharmacother 2018; 106:763–769. doi: 10.1016/j.biopha.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Sun Y, Yang Y, Chen Y, Liu H. Circ_0067835 knockdown enhances the radiosensitivity of colorectal cancer by miR-296-5p/IGF1R axis. Onco Targets Ther 2021; 14:491–502. doi: 10.2147/ott. S281011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C, Liu H, Wei R, Liu Z, Chen H, Guan X, et al. LncRNA EGOT/miR-211-5p affected radiosensitivity of rectal cancer by competitively regulating ErbB4. Onco Targets Ther 2021; 14:2867–2878. doi: 10.2147/ott.S256989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HC, Her NG, Kang D, Jung SH, Shin J, Lee M, et al. Radiation inducible miR-770-5p sensitizes tumors to radiation through direct targeting of PDZ-binding kinase. Cell Death Dis 2017; 8:e2693.doi: 10.1038/cddis.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Islam Khan MZ, Tam SY, Law HKW. Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular events in cancer. Front Genet 2018; 9:750.doi: 10.3389/fgene.2018. 00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunanopparat A, Kimkong I, Palaga T, Tangkijvanich P, Sirichin-dakul B, Hirankarn N. Increased ATG5-ATG12 in hepatitis B virus- associated hepatocellular carcinoma and their role in apoptosis. WorldJ Gastroenterol 2016; 22:8361–8374. doi: 10.3748/wjg.v22. i37.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Liu WL, Su L, Lu ZC, He XS. The role of autophagy in cancer radiotherapy. Curr Mol Pharmacol 2020; 13:31–40. doi: 10.2174/1874467212666190809154518. [DOI] [PubMed] [Google Scholar]
- 48.Qased AB, Yi H, Liang N, Ma S, Qiao S, Liu X. MicroRNA-18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells. Mol Med Rep 2012; 7:559–564. doi: 10.3892/mmr.2012.1214. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, Xu X, Zhu J, Zhang S, Wu Y, Wu Y, et al. miR-31 affects colorectal cancer cells by inhibiting autophagy in cancer-associated fibroblasts. Oncotarget 2016; 7:79617–79628. doi: 10.18632/onco- target.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu JY, Zheng FC, Yu WS. MicroRNA-129-5p-mediated inhibition of autophagy enhanced the radiosensitivity of human colon cancer cells. Int J Clin Exp Pathol 2016; 9:12179–12187. [Google Scholar]
- 51.Zheng S, Zhong YF, Tan DM, Xu Y, Chen HX, Wang D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J Biosci 2019; 44:92. [PubMed] [Google Scholar]
- 52.Liu Y, Chen X, Chen X, Liu J, Gu H, Fan R, et al. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis 2020; 11:175.doi: 10.1038/s41419-020-2268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 2007; 3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 55.Sun Y, Xing X, Liu Q, Wang Z, Xin Y, Zhang P, et al. Hypoxia- induced autophagy reduces radiosensitivity by the HIF-1α/miR-210/Bcl-2 pathway in colon cancer cells. Int J Oncol 2015; 46:750–756. doi: 10.3892/ijo.2014.2745. [DOI] [PubMed] [Google Scholar]
- 56.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet 2012; 28:128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maier P, Hartmann L, Wenz F, Herskind C. Cellular pathways in response to ionizing radiation and their targetability for tumor radiosensitization. Int J Mol Sci 2016; 17:102.doi: 10.3390/ijms17010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samadi P, Afshar S, Amini R, Najafi R, Mahdavinezhad A, Sedighi Pashaki A, et al. Let-7e enhances the radiosensitivity of colorectal cancer cells by directly targeting insulin-like growth factor 1 receptor. J Cell Physiol 2018; 234:10718–10725. doi: 10.1002/jcp.27742. [DOI] [PubMed] [Google Scholar]
- 59.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet 2007; 8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 60.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev Genet 2006; 40:363–383. doi: 10.1146/annurev. genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 61.Jekimovs C, Bolderson E, Suraweera A, Adams M, O’Byrne KJ, Richard DJ. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Front Oncol 2014; 4:86.doi: 10.3389/fonc.2014. 00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thapar R. Regulation of DNA double-strand break repair by non coding RNAs. Molecules (Basel, Switzerland) 2018; 23:2789.doi: 10.3390/molecules23112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pardini B, Rosa F, Barone E, Di Gaetano C, Slyskova J, Novotny J, et al. Variation within 3’-UTRs of base excision repair genes and response to therapy in colorectal cancer patients: a potential modulation of microRNAs binding. Clin Cancer Res 2013; 19:6044–6056. doi: 10.1158/1078-0432.Ccr-13-0314. [DOI] [PubMed] [Google Scholar]
- 64.Ha Thi HT, Kim HY, Kim YM, Hong S. MicroRNA-130a modulates a radiosensitivity of rectal cancer by targeting SOX4. Neoplasia 2019; 21:882–892. doi: 10.1016/j.neo.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan Z, Ma J, Meng X, Chen N, Shen M. Chk2 deficiency alleviates irradiation-induced taste dysfunction by inhibiting p53-dependent apoptosis. Oral Dis 2018; 24:856–863. doi: 10.1111/odi.12822. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W, Zhu Y, Zhou Y, Wang J, Jiang P, Xue L. miRNA-31 increases radiosensitivity through targeting STK40 in colorectal cancer cells. Asia Pac J Clin Oncol 2021; Online ahead of print. doi: 10.1111/ajco.13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin SM, Xia Q, Zhang YQ, Sun AM, Shi YS, Zheng L, et al. miR-124 regulates radiosensitivity of colorectal cancer cells by targeting PRRX1 (in Chinese). Nan Fang Yi Ke Da Xue Xue Bao 2016; 36:1110–1116. [PubMed] [Google Scholar]
- 68.Kim SB, Zhang L, Barron S, Shay JW. Inhibition of microRNA-31-5p protects human colonic epithelial cells against ionizing radiation. Life Sci Space Res 2014; 1:67–73. doi: 10.1016/j.lssr.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 69.Ruhl R, Rana S, Kelley K, Espinosa-Diez C, Hudson C, Lanciault C, et al. microRNA-451a regulates colorectal cancer proliferation in response to radiation. BMC Cancer 2018; 18:517.doi: 10.1186/s12885-018-4370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao F, Chen X, Peng P, Dong W. RWR-algorithm-based dissection of microRNA-506-3p and microRNA-140-5p as radiosensitive biomarkers in colorectal cancer. Aging 2020; 12:20512–20522. doi: 10.18632/aging.103907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Migliore C, Martin V, Leoni VP, Restivo A, Atzori L, Petrelli A, et al. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin Cancer Res 2011; 18:737–747. doi: 10.1158/1078-0432.Ccr-11-1699. [DOI] [PubMed] [Google Scholar]
- 72.Furukawa S, Kawasaki Y, Miyamoto M, Hiyoshi M, Kitayama J, Akiyama T. The miR-1-NOTCH3-asef pathway is important for colorectal tumor cell migration. PLoS One 2013; 8:e80609.doi: 10.1371/journal.pone.0080609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y, Pu N, Su W, Yang X, Xing C. Downregulation of miR-1 in colorectal cancer promotes radioresistance and aggressive pheno types. J Cancer 2020; 11:4832–4840. doi: 10.7150/jca.44753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang H, Tang Y, Zhang H, Zhang C. MiR-32-5p regulates radiosensitization, migration and invasion of colorectal cancer cells by targeting TOB1 gene. Onco Targets Ther 2019; 12:9651–9661. doi: 10.2147/OTT.S228995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang P, Yang Y, An W, Xu J, Zhang G, Jie J, et al. The long noncoding RNA-ROR promotes the resistance of radiotherapy for human colorectal cancer cells by targeting the p53/miR-145 pathway. J Gastroenterol Hepatol 2017; 32:837–845. doi: 10.1111/jgh.13606. [DOI] [PubMed] [Google Scholar]
- 76.Saleh AD, Savage JE, Cao L, Soule BP, Ly D, DeGraff W, et al. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS One 2011; 6:e24429.doi: 10.1371/journal.pone.0024429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, et al. miR-145 antagonizes SNAI1-mediated stemness and radiation resistance in colorectal cancer. Mol Ther 2018; 26:744–754. doi: 10.1016/j.ymthe.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andaur R, Tapia JC, Moreno J, Soto L, Armisen R, Marcelain K. Differential miRNA expression profiling reveals miR-205-3p to be a potential radiosensitizer for low- dose ionizing radiation in DLD-1 cells. Oncotarget 2018; 9:26387–26405. doi: 10.18632/oncotar- get.25405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Peng X, Lu X, Wei Q, Chen M, Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio sensitivity of colon cancer cells via tumor suppressor miR-338-3p: effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol Res Pract 2018; 215:689–696. doi: 10.1016/j.prp.2018.12.032. [DOI] [PubMed] [Google Scholar]
- 80.Meng WJ, Pathak S, Zhang X, Adell G, Jarlsfelt I, Holmlund B, et al. Expressions of miR-302a, miR-105, and miR-888 play critical roles in pathogenesis, radiotherapy, and prognosis on rectal cancer patients: a study from rectal cancer patients in a Swedish rectal sancer trial of preoperative radiotherapy to big database analyses. Front Oncol 2020; 10:567042.doi: 10.3389/fonc.2020.567042. [DOI] [PMC free article] [PubMed] [Google Scholar]