Abstract
Background:
MicroRNA-20a (miR-20a) is dysregulated in many types of malignancies, including human hepatocellular carcinoma (HCC), but its expression level and functional significance in HCC are still disputed. We aimed to study the role of miR-20a-5p in HCC and its downstream molecular mechanisms.
Methods:
We used real-time polymerase chain reaction to detect the expression of miR-20a-5p and runt-related transcription factor 3 (RUNX3) in HCC and paraneoplastic tissue, transfected Huh7 and highly metastatic human hepatocellular carcinoma (MHCC97H) cells. A live cell workstation was used to observe the proliferation and migration of transfected cells. The invasiveness of transfected cells was verified by Transwell assay. Cell apoptosis was detected by flow cytometry. The expression levels of proteins after transfection were measured using simple western immunoblot measurements. Gene expression profiles between HCC and normal samples were obtained from The Cancer Genome Atlas. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment results were processed by the database for annotation, visualization and integrated discovery. Potential target genes of miR-20a-5p were predicted to further investigate how miR-20a-5p regulates epithelial-mesenchymal transition (EMT) in HCC.
Results:
MiR-20a-5p was significantly highly expressed in HCC tissues, and overexpression of miR-20a-5p significantly promoted HCC cell proliferation, migration, and invasion and inhibited apoptosis in vitro. The protein expression of E-cadherin was decreased and that of vimentin was increased after overexpression of miR-20a-5p in HCC cells. We discovered the intersection of genes from miRDB, miR TarBase, and TargetScan, obtained 397 target genes and finally focused on RUNX3. RUNX3 was not only reduced in HCC specimens but also drastically reduced in HCC cells overexpressing miR-20a-5p. RUNX3 expression decreased with elevated miR-20a-5p, which activated downstream EMT signaling and promoted cell proliferation, migration, and invasion.
Conclusions:
Since RUNX3 is involved in EMT in HCC, as proven by previous research, our findings provide further evidence for a novel regulatory pathway comprising the miR-20a/RUNX3/EMT axis that upregulates EMT signaling and enhances the migration of HCC cells.
Keywords: Epithelial-mesenchymal transition, Hepatocellular carcinoma, miR-20a-5p, RUNX3
Introduction
Primary liver cancer is the sixth most common cancer in the world, and 80% of liver cancer is hepatocellular carcinoma (HCC),[1,2] which is the second leading cause of cancer-related death globally.[3] Although the majority of cases occur in developing countries, the incidence has recently increased in developed countries.[4] In recent years, studies on the molecular mechanism of HCC have provided more indications for its pathogenesis, diagnosis, and prognosis. Many microRNAs (miRNAs) associated with HCC, including miR-21, miR-221, and miR-224, have increased expression in HCC, while others, such as miR-122a, miR-199a, and miR-223, have decreased expression in HCC.[5–8] Recent reports have demonstrated that some specific miRNAs could play important roles in the tumor microenvironment during HCC development.[9] miRNAs can also regulate multiple target genes and signaling pathways in HCC, and miRNA-targeted cancer treatment is an attractive approach for the prevention of HCC development.[10] Various miRNAs, including miR-32a-5p, miR-122, miR-221, and miR-1468, have potential as prognostic biomarkers for HCC, and upregulation of these miRNAs indicates poor prognosis in HCC patients.[11–14]
miR-20a has significantly higher expression in ovarian cancer, lung cancer, colorectal cancer, cervical cancer, gastric cancer, and head and neck cancer according to The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) statistics, and it exhibits potential for clinical application as a novel diagnostic biomarker and therapeutic target. In liver cancer, some studies reported that miR-20a was expressed at low levels in HCC tissues and cell lines,[15,16] while another found that 52% of miR-20a was highly expressed in 39 pairs of HCC specimens.[17] Similar findings have been reported in breast cancer,[18,19] and these conflicting results prompted us to further study the TCGA database and HCC specimen analysis.
Recently, it was proven that overexpression of miR-20a-5p promoted the development of liver cancer,[17] breast cancer,[19] lung cancer,[20] and cervical cancer[21] by targeting RUNX3. In addition, it was reported that downregulation of RUNX3 promoted epithelial-mesen-chymal transition (EMT) in HCC.[22] However, it is still unclear whether and how miR-20a-5p affects the EMT process in HCC. In the present study, we investigated the expression of miR-20a-5p and RUNX3 in HCC cells and tissues and found that miR-20a-5p affected the EMT process by regulating the expression of RUNX3 in HCC cells. We identified the miR-20a/RUNX3/EMT regulatory axis, providing strong evidence for molecular targeted therapy of liver cancer.
Methods
Ethical approval
All patients or patients’ families signed the written informed consent, and the procedure of tissue collection was approved by the Ethics Review Committee of the Affiliated Hospital of Inner Mongolia Medical University (No. WZ2021043).
Tissue specimens
Six tumor tissue specimens along with six adjacent tissues were obtained from HCC patients from the Affiliated Hospital of Inner Mongolia Medical University. After resection of the tumor and adjacent tissues, the specimens were immediately frozen in liquid nitrogen and stored at − 80°C. All experiments strictly adhered to the code of ethics of the World Medical Association and were conducted following the guidelines of Inner Mongolia Medical University.
Cell culture and transfection
The human HCC cell line MHCC97H was cultured in Dulbecco's Modified Eagle's Medium replenished with 10% fetal bovine serum (FBS, Gibco, Life Technologies Limited, Paisley, UK). The human HCC cell line Human hepatocellular carcinoma-7 was cultured in Modified Eagle's Medium containing 10% FBS. All cells were cultured at 37°C in a humidified 5% CO2 incubator. The synthesized miR-20a-5p mimics and the random negative control RNAs (control mimic and inhibitor) were obtained from GenePharma (Shanghai, China). When the cells in the plate reached a fusion degree of 70% to 80% per well, the overexpression of miR-20a was performed by transfecting a miR-20a mimic with a synthesized double-stranded RNA oligonucleotide imitating the miR-20a precursor. The plasmids were transfected into the cells by an electroporation system (Bio-Rad, USA) based on the protocols.
Quantitative real-time (qRT) polymerase chain reaction (PCR)
TRIZOL reagent (Invitrogen, CA, USA) was used to isolate total RNA from frozen tumor tissues and cells. We used the (cDNA) Synthesis Kit (Takara, Tokyo, Japan) for the synthesis of cDNA. The qRT-PCR for miRNAs and (mRNA) was conducted with specific primers following the instructions of the manufacturer (Life Technologies, Carlsbad, CA, USA). qPCR results of miRNA and mRNA were expressed corresponding to Rnu6, gene ID 19862 snRNA or Glyceral-dehyde-3-phosphate dehydrogenase (GAPDH) mRNA, respectively.
Cell proliferation assay
The cells after transient transfection were seeded onto 96-well plates at 1 × 104 cells/well. After 0, 6, 12, 18, 24, 36, and 72 h of culture, the cells were captured in a living cell workstation (IncuCyte Zoom, Essen BioScience, USA).
Cell apoptosis assay
The Annexin V-fluorescein isothiocyanate (FITC)/PI staining kit (Becton, Dickinson and Company [BD] Biosciences, San Diego, CA, USA) was used for cell apoptosis. MHCC97H and Human hepatocellular carcinoma-7 cells were seeded in 6-well plates with a density of 106 cells/mL. After transfecting for 24 h, Annexin V-FITC was used to label the cells for 20 min in the darkness. In total, 50 μg/mL of PI was added and further incubated for 30 min. The situation of cell apoptosis was detected to calculate the percentage of cell death based on flow cytometry with the FACScan flow cytometer (BD Biosciences). All tests were performed in triplicate.
Wound healing assay
The transfected cells were cultured on 96-well plates forming a single cell layer, and allowed to grow to 100% confluence. A straight wound line was made in the middle of the cell layer. The closure of the wound was observed by taking pictures at different time points by the IncuCyte (IncuCyte Zoom, Essen BioScience) for 24 h.
Cell invasion assay
Matrigel (Fisher Scientific, USA) invasion chambers (8.0 μm pore size, BD Biosciences, Franklin Lakes, NJ, USA) were used to investigate the invasion capability of transfected cells. Relative cells (5 × 105 cells/mL) were seeded in a serum-free medium into the upper chamber, and the bottom chamber added 10% FBS as chemo-attractant inducing cells invading toward the bottom chamber. After 24 h, the cells invaded through the membrane and adhered to the underside of the membrane were stained by 0.05% crystal violet and counted under a microscope (Olympus, USA). The number of cells that migrated through the transwell pores was quantified using the Image J (NIH, USA) procedure. The reported data represent the average of three independent experiments performed in triplicate. Each experiment was repeated three times.
Western blot analysis
Cells were seeded in 6-well plates with a density of 4 × 105 cells/well and were treated with miR-20-5p mimics and inhibitor for 48 h. Followed by treatment, cells were lysed with radioimmunoprecipitation assay (RIPA) Lysis buffer (Solarbio, Beijing, China) and the protein concentration was quantified using the bicincho-ninic acid (BCA) protein assay kit (Applygen Technologies, China). The simple western immunoblots were performed on a Wes (Protein Simple Santa Clara, San Jose, CA, USA) using the Size Separation Master Kit with Split Buffer (12– 230 D) according to the manufacturer's standard instruction. The antibodies used included the following: anti-RUNX3 (ab40278, abcam, UK), anti-Vimentin (10366-1-AP, Proteintech, USA), anti-E-cadherin (20874-1-AP, Proteintech, USA), and anti-GAPDH (10494-1-AP, Proteintech, USA).
Data preprocessing and enrichment analysis
Gene expression profiles between HCC samples and normal samples were obtained from TCGA. Three publicly available databases, including miRDB (http://www.mirdb.org/mirdb/ontology.html), miRTarBase (https://mirtarbase.cuhk.edu.cn/), and TargetScan (http://www.targetscan.org/vert_72/), were used to predict the candidate targets of miR-20a. A functional enrichment analysis was performed to examine the enrichment of annotated terms. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted using the database for annotation, visualization and integrated discovery (DAVID) (https://david.abcc.ncifcrf.gov; version 6.7) with a threshold of P < 0.05.
Statistical analysis
Values were expressed as mean ± standard error of mean. The quantitative results presented were obtained from at least three independent experiments. The Student t test was used to evaluate the significance of differences between the two groups. Differences with P value <0.05 were considered to be statistically significant.
Results
miR-20a-5p is upregulated in HCC tissues
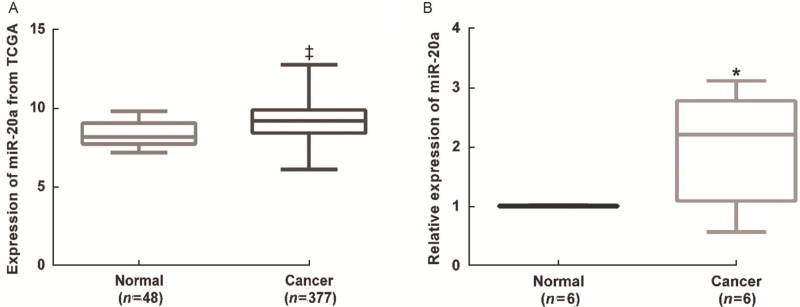
miR-20a is among the most frequently altered miRNAs in human cancers. To investigate the expression of miR-20a-5p in HCC, we collected data on miR-20a-5p expression in a large number of HCC patients from the TCGA database. Through TCGA analysis, we found miR-20a-5p was significantly increased in HCC tissues compared with normal liver tissues (P < 0.001) [Figure 1A]. miR-20a-5p expression was further measured in six pairs of tumors and adjacent normal tissues of patients with primary HCC in our hospital and was significantly increased in HCC tissues compared with adjacent normal tissues (P < 0.05) [Figure 1B].
Figure 1.
Analysis of miR-20a-5p expression by TCGA database (A) and qRT-PCR (B) in HCC tissues. (A) Data from TCGA database showed increased expression of miRNA-20a in HCC tumor samples (n = 377) compared with normal liver tissues (n = 48). (B) The relative expression level of miR-20a-5p was significantly increased in HCC tissues compared with adjacent normal tissues (n = 6). ∗P < 0.05, ‡P < 0.001 vs. Normal. HCC: Hepatocellular carcinoma; qRT-PCR: quantitative real-time PCR; TCGA: The Cancer Genome Atlas.
Overexpression of miR-20a-5p promotes the proliferation and inhibits apoptosis of HCC cells
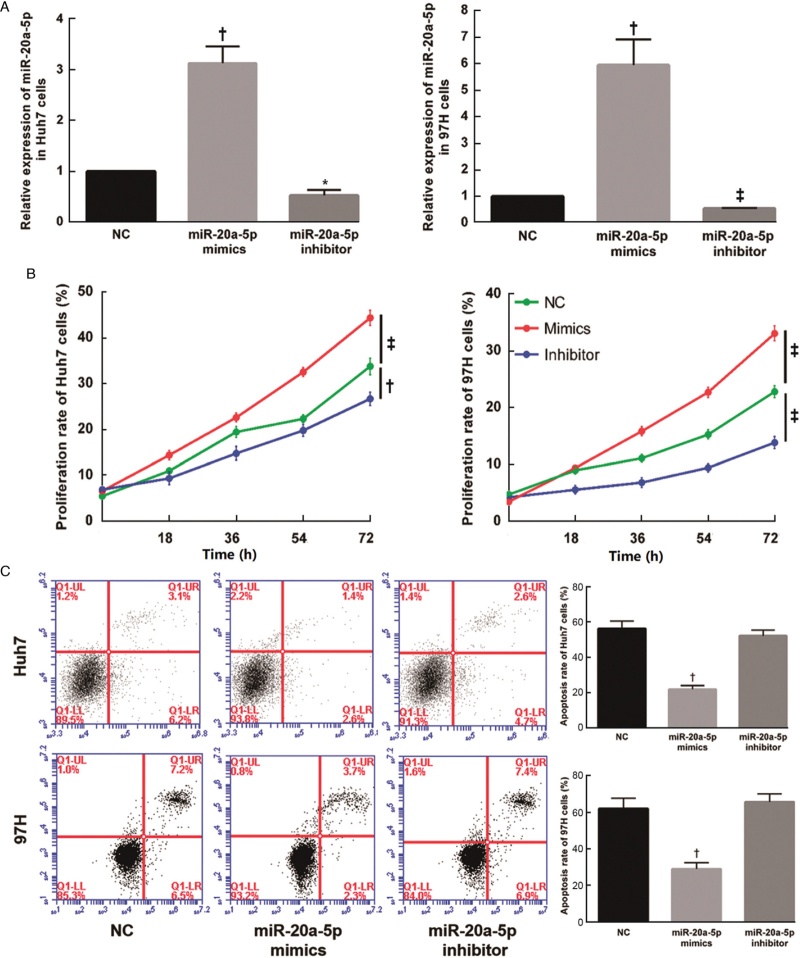
To investigate the effect of miR-20a-5p on cell proliferation and apoptosis, we overexpressed both miR-20a-5p mimics and a miR-20a-5p inhibitor in Huh7 and MHCC97H cells. The efficiency of transfection was confirmed by qRT-PCR. At 48 h posttransfection with miR-20a-5p mimics, the expression of miR-20a-5p was increased ∼3- to 5-fold in the two HCC cell lines (P < 0.01). The expression of miR-20a-5p was reduced after transfection with the inhibitor [Figure 2A]. The data from a live cell workstation showed a significant increase in proliferation in cells after transfection with miR-20a-5p mimics (P < 0.001) [Figure 2B]. After Annexin V-FITC/PI staining, flow cytometry analysis showed that transfection of miR-20a-5p mimics resulted in a significant reduction in the apoptotic rate (P < 0.01), while the opposite effect was observed upon treatment with the miR-20a-5p inhibitor [Figure 2C].
Figure 2.
Overexpression of miR-20a-5p promotes the proliferation and inhibits apoptosis of HCC cells. (A) The relative expression level of miR-20a-5p was examined by qRT-PCR in Huh7 and MHCC97H cells transfected with NC, miR-20a-5p mimics and inhibitor. The expression of miR-20a-5p was increased after transfected with miR-20a-5p mimics. Otherwise reduced after transfection with the inhibitor. (B) Cell proliferation was examined by IncuCyte after transfection with NC, miR-20a-5p mimics and inhibitor. A significant increase in proliferation was detected after transfection with miR-20a-5p mimics. (C) The cell apoptosis rate of two cell lines transfected with NC, miR-20a-5p mimics and inhibitor was measured by flow cytometry. Transfection of miR-20a-5p mimics resulted in a significant reduction in the apoptotic rate. ∗P < 0.05, †P < 0.01, ‡P < 0.001 vs. NC. HCC: Hepatocellular carcinoma; NC: Negative control; qRT-PCR: Quantitative real-time PCR.
Overexpression of miR-20a-5p promotes the migration and invasion of HCC cells
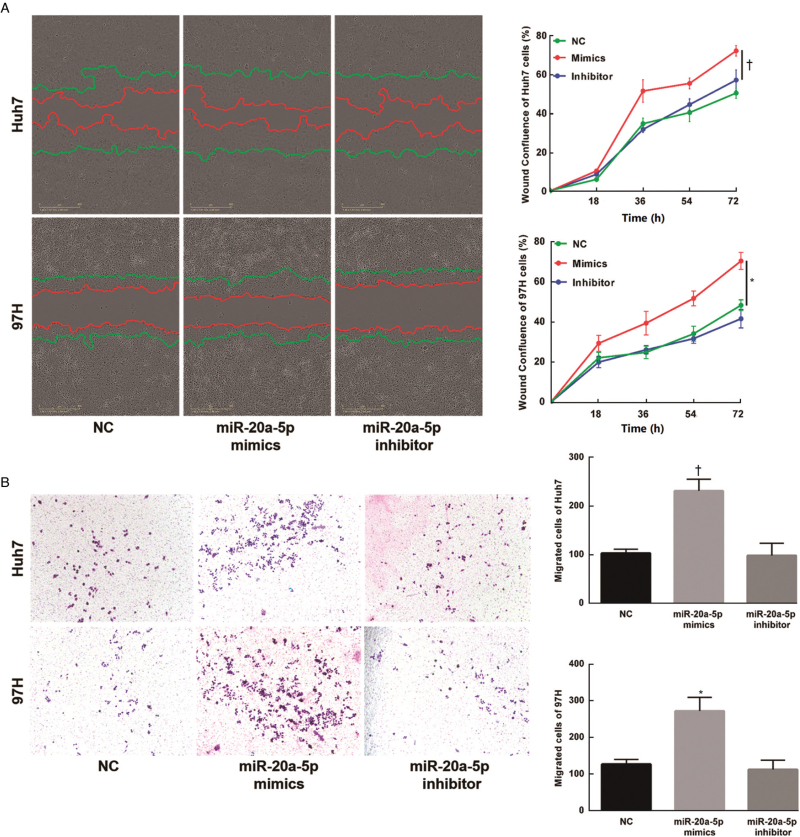
The wound healing assay showed that miR-20a-5p overexpression also enhanced Huh7 cell (P < 0.01) and MHCC-97H cell (P < 0.05) migration ability in vitro [Figure 3A]. Accordingly, treatment with the miR-20a-5p inhibitor caused the opposite effects. The Transwell assay results showed a significant increase in the number of invasive cells after transfection with miR-20a-5p mimics (P < 0.01), which was greatly reduced by the miR-20a-5p inhibitor [Figure 3B]. These data indicated that HCC cell migration and invasion could be modulated by the expression of miR-20a-5p.
Figure 3.
Overexpression of miR-20a-5p promotes the migration and invasion of HCC cells. (A) The effect of transfection of NC, miR-20a-5p mimics and inhibitor on cell migration was observed by wound healing assays. The wound healing was accelerated after transfection with miR-20a-5p mimics. (B) Transwell assays revealed cell invasion after transfection with NC, miR-20a-5p mimics and the inhibitor. The cells were stained by 0.05% crystal violet. The number of Invasive cells after transfection with miR-20a-5p mimics was significantly increased. ∗P < 0.05, †P < 0.01 vs. NC. HCC: Hepatocellular carcinoma; NC: Negative control.
MiR-20a-5p promotes EMT of HCC cells
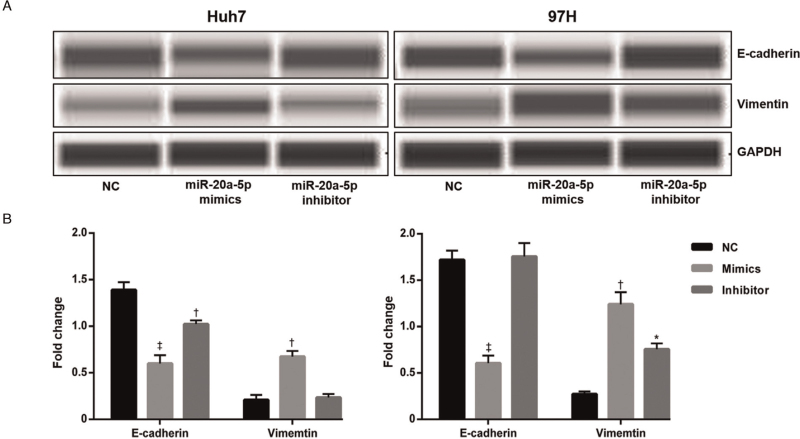
The invasion and migration of tumors involve many biological processes (BP), among which EMT plays a very important role.[23] To determine how miR-20a-5p promotes the migration and invasion of HCC cells, we determined whether miR-20a-5p could regulate the EMT process. The results revealed that the expression of E-cadherin, a growth and invasion suppressor of the EMT pathway, was significantly decreased after overexpression of miR-20a-5p (P < 0.001). In addition, the expression of Vimentin, a marker correlated with the EMT phenotype, was increased markedly upon miR-20a-5p overexpression (P < 0.01). The inhibition of miR-20a-5p expression showed the opposite result [Figure 4].
Figure 4.
E-cadherin and Vimentin protein expression was analyzed by western blotting (A and B). The expression of E-cadherin was significantly decreased and Vimentin was increased remarkably upon after overexpression of miR-20a-5p. ∗P < 0.05, †P < 0.01, ‡P < 0.001 vs. NC. NC: Negative control.
The miR-20a/RUNX3/EMT axis was established via enrichment analysis and validated
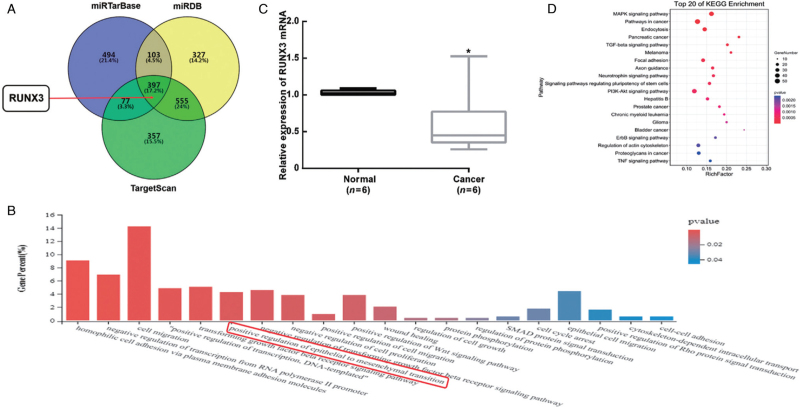
It was proven that miR-20a-5p could regulate the EMT process and then promote the migration and invasion of HCC cells. To investigate the miR-20a-5p target that regulates the EMT process, miRDB, miRTarBase, and TargetScan were used to predict potential mRNAs that could be targeted by miR-20a-5p. After the intersection of the three databases, we obtained 397 target genes, except for the enrichment of “the positive regulation of epithelial to mesenchymal transition,” and we finally found that RUNX3 was one of the common predicted target genes [Figure 5A and 5B]. Compared with the adjacent normal tissues, the expression of RUNX3 mRNA significantly decreased in HCC specimens with high miR-20a-5p expression (P < 0.05) [Figure 5C]. In addition, the enriched pathways of the 397 target genes by KEGG analysis were associated with “cell migration” and “cell adhesion” [Figure 5D].
Figure 5.
MiR-20a-5p targets the mRNA RUNX3, and GO functional terms are enriched in the positive regulation of EMT. (A) total 397 mRNAs targeted by miR-20a-5p were intersected from miRDB, miRTarBase and TargetScan databases, and RUNX3 was one of the predicted target genes. (B) GO BP for genomic drivers determined in this study. The “positive regulation of epithelial to mesenchymal transition” in biological processes was enriched in 397 target genes. (C) The relative expression level of RUNX3 mRNA was evaluated in six pairs of frozen HCC specimens by qRT-PCR. The expression of RUNX3 mRNA significantly decreased in HCC specimens with high miR-20a-5p expression. ∗P < 0.05 vs. Normal. (D) The bubble diagram indicated the top 20 enriched KEGG pathways including “cell migration” and “cell adhesion”. BP: Biological processes; EMT: Epithelial-mesenchymal transition; GO: Gene ontology; HCC: Hepatocellular carcinoma; KEGG: Kyoto Encyclopedia of Genes and Genomes; qRT-PCR: Quantitative real-time PCR.
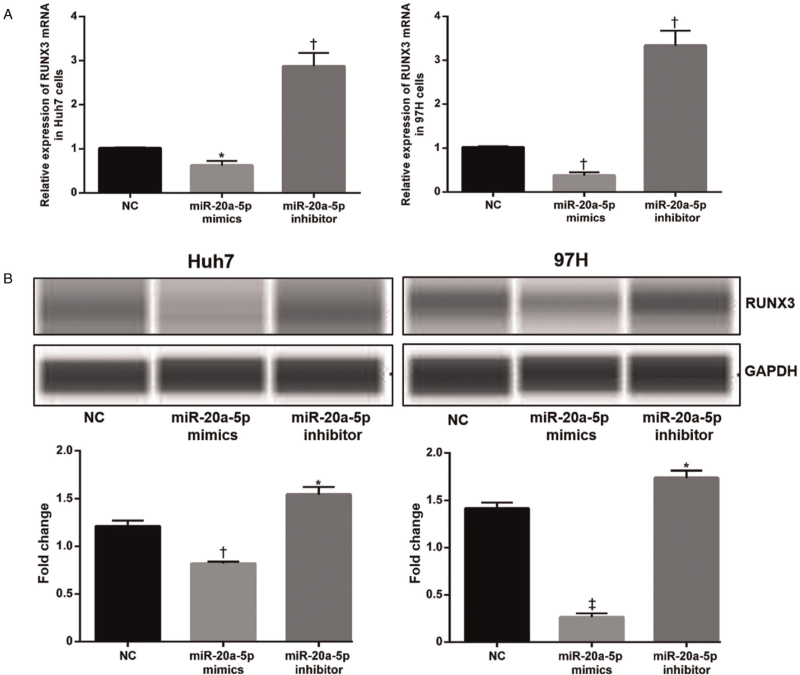
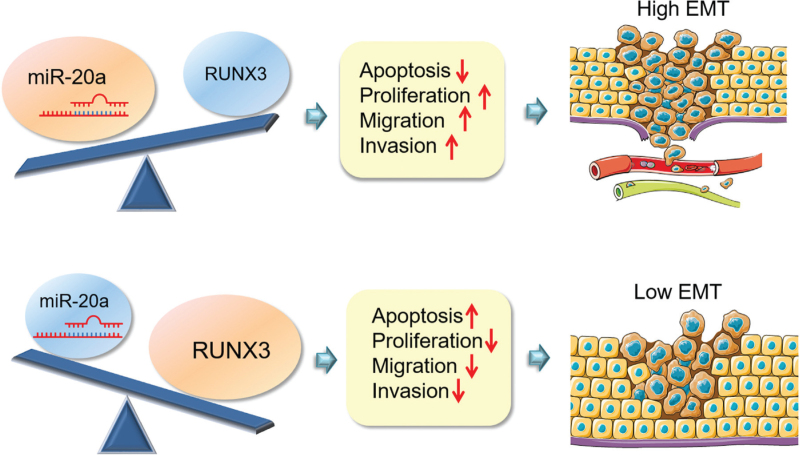
Further analysis was carried out to determine whether miR-20a-5p can regulate the target gene RUNX3 in vitro. The qRT-PCR result showed that overexpression of miR-20a-5p led to decreased levels of RUNX3 mRNA [Figure 6A]. Accordingly, transfection of the miR-20a-5p inhibitor increased the level of RUNX3 protein [Figure 6B]. RUNX3 expression decreased with increasing miR-20a-5p, which in turn promoted cell proliferation, migration, and invasion and further activated downstream EMT signaling. These changes contributed to the proliferation and migration of HCC cells, ultimately increasing tumor formation and metastasis [Figure 7].
Figure 6.
MiR-20a-5p downregulates the protein level of RUNX3. (A) The relative expression level of RUNX3 mRNA was measured after transfection with miR-20-5p mimics and inhibitor by qRT-PCR. (B)The RUNX3 protein level was measured after transfection of miR-20-5p mimics and inhibitor by western blotting. mRNA and protein expression levels of RUNX3 decreased after miR-20a-5p mimics transfection in Huh7 and MHCC97H cells. ∗P < 0.05, †P < 0.01, ‡P < 0.001 vs. NC. NC: Negative control; qRT-PCR: Quantitative real-time PCR.
Figure 7.
A working model describing the interaction among miR-20a/RUWX3/EMT during HCC metastasis. miR-20a directly regulates the expression of the target gene RUNX3. RUNX3 expression decreases with elevated miR-20a, which in turn inhibits cell apoptosis, promotes migration and invasion, and further stimulates downstream EMT signaling. Vice versa, RUNX3 expression increases with inhibited miR-20a-5p, which in turn promotes cell apoptosis, inhibits migration and invasion, and further attenuates downstream EMT signaling. EMT: Epithelial-mesenchymal transition; HCC: Hepatocellular carcinoma.
Discussion
miRNAs affect HCC tumor growth and metastasis at multiple stages of tumor development, and dysregulation of miRNAs has been related to poor outcome of patients with HCC. To date, 55 upregulated miRNAs, including miR-17–92 polycistron and miR-21, as well as 96 downregulated miRNAs, including miR-29 and miR-122, have been found in liver cancer.[24] For miR-20a-5p, the different expression trends have made it an ongoing hot topic in HCC research. Some studies have suggested that the levels of miR-20a-5p are elevated in HCC cell lines and tissues,[15,25] while others have concluded that miR-20a-5p expression is lower in primary HCC than that in normal liver tissue.[26] We used primary HCC tissue to verify that the expression of miR-20a-5p was higher than that in adjacent normal tissue. Moreover, we found that overexpression of miR-20a-5p significantly promoted cell proliferation, migration, and invasion and inhibited apoptosis.
To identify the target genes of miR-20a-5p and their functions, we first predicted the target genes of miR-20a-5p using public databases: miRTarBase, miRDB, and TargetScan. A total of 397 target genes of miR-20a-5p were obtained after the intersection of the above three databases, and the DAVID website was used for KEGG pathway and GO functional enrichment analyses. The KEGG analysis results showed that a portion of the enriched pathways was associated with “cell migration” and “cell adhesion”. The BP in GO functional analysis was enriched in the “ positive regulation of epithelial to mesenchymal transition”. Overexpression of miR-20a-5p could inhibit the expression of E-cadherin and stimulate the expression of Vimentin, an invasion suppressor and promoter of the EMT pathway, respectively.
To further investigate the miR-20a-5p target involved in regulating EMT, we also examined the intersection of the three predicted results by Venn diagram and found that RUNX3 was one of the common predicted target genes. Previous research proved that miR-20a-5p directly targets RUNX3 by using a luciferase assay.[17] Here, by performing western blot experiments, we further proved that miR-20a-5p can directly downregulate the protein level of RUNX3. RUNX3 is frequently lost in heterozygosity in HCC,[27] and homozygous deletions are found in 35% to 40% of HCC cases, resulting in the absence of RUNX3 expression.[28,29] This clinical evidence indicates that loss of RUNX3 causes the development and progression of HCC.
The role of RUNX3 in HCC EMT has been proven in previous research. For example, Tanaka[22] proved that E-cadherin expression was induced by exogenous RUNX3 protein in low-EMT cells, and Gou[30] further found that RUNX3 increased E-cadherin expression by repressing microRNA-186. In this study, we examined the expression of RUNX3 and classic EMT proteins simultaneously by up- or downregulation of miR-20a expression. An increase in miR-20a-5p expression was followed by a decrease in RUNX3 protein expression, a significant decrease in E-cadherin expression, and an increase in Vimentin expression. When miR-20a-5p expression was inhibited, the opposite result was obtained. RUNX3 expression decreased with elevated miR-20a-5p, which activated downstream EMT signaling and further promoted cell proliferation, migration, and invasion.
Since EMT plays an important role in tumor invasion and metastasis, we surmised that RUNX3 may prevent HCC cell metastasis by modulating EMT. Studies have shown that the expression levels of E-cadherin were increased and those of Snail and N-cadherin were decreased in the RUNX3-overexpressing group compared with the vector control group.[31]RUNX3 inhibits the expression of claudin, another cell−cell junction protein involved in the development of EMT. As both E-cadherin and claudin are negatively regulated in EMT, RUNX3 could be an EMT regulator.[32] In addition, RUNX3 controls Notch signaling, which is closely linked to cancer stem cells.[33] Ectopic RUNX3 expression deactivated Notch signaling by decreasing jagged-1 expression in HCC.[34] These are the possible molecular mechanisms by which RUNX3 promotes the EMT signaling pathway and migration in HCC cells.
In conclusion, our findings provide further evidence for a novel regulatory pathway comprising the miR-20a/ RUNX3/EMT axis in HCC. miR-20a-5p can promote EMT and then enhance migration by targeting RUNX3. We believe that these findings significantly improve our understanding of HCC metastasis. These results suggest that miR-20a-5p may be an effective target for HCC and is worth investigating through in vivo experiments.
Funding
This work was supported by the Inner Mongolia Natural Science Foundation (No. 2015MS0827), a Major Project of the Affiliated Hospital of Inner Mongolia Medical University (No. NYFY ZD017), and Scientific Research Projects in Higher Education of Inner Mongolia (No. NJZY20139).
Conflicts of interest
None.
Footnotes
How to cite this article: Wang X, Wei P, Yang L, Liu F, Tong X, Yang X, Su L. MicroRNA-20a-5p regulates the epithelial-mesenchymal transition of human hepatocellular carcinoma by targeting RUNX3. Chin Med J 2022;135:2089–2097. doi: 10.1097/CM9.0000000000001975
Xianjue Wang and Ping Wei contributed equally to the work.
References
- 1.Aly A, Ronnebaum S, Patel D, Doleh Y, Benavente F. Epidemiologic, humanistic and economic burden of hepatocellular carcinoma in the USA: a systematic literature review. Hepat Oncol 2020; 7:HE27.doi: 10.2217/hep-2020-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J 2021; 134:783–791. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trevisani F, Frigerio M, Santi V, Grignaschi A, Bernardi M. Hepatocellular carcinoma in non-cirrhotic liver: a reappraisal. Dig Liver Dis 2010; 42:341–347. doi: 10.1016/j.dld.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007; 67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 7.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006; 25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 8.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 2008; 135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 2012; 22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung HJ, Choi YE, Kim ES, Han YH, Park MJ, Bae IH. miR-29b attenuates tumorigenicity and stemness maintenance in human glioblastoma multiforme by directly targeting BCL2L2. Oncotarget 2015; 6:18429–18444. doi: 10.18632/oncotarget.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res 2018; 37:52.doi: 10.1186/s13046-018-0677-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med 2018; 7:1670–1679. doi: 10.1002/cam4.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F, Li XF, Fu DS, Huang JG, Yang SE. Clinical potential of miRNA-221 as a novel prognostic biomarker for hepatocellular carcinoma. Cancer Biomark 2017; 18:209–214. doi: 10.3233/CBM-161671. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L, et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res 2018; 37:49.doi: 10.1186/s13046-018-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Liu DL, Lu LL, Dong LL, Liu Y, Bian XY, Lian BF, et al. miR-17-5p and miR-20a-5p suppress postoperative metastasis of hepatocellular carcinoma via blocking HGF/ERBB3-NF-kappaB positive feedback loop. Theranostics 2020; 10:3668–3683. doi: 10.7150/thno.41365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tipanee J, Di Matteo M, Tulalamba W, Samara-Kuko E, Keirsse J, Van Ginderachter JA, et al. Validation of miR-20a as a tumor suppressor gene in liver carcinoma using hepatocyte-specific hyperactive piggyBac transposons. Mol Ther Nucleic Acids 2020; 19:1309–1329. doi: 10.1016/j.omtn.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Wang X, Cheng J, Wang Z, Jiang T, Hou N, et al. MicroRNA-20a-5p targets RUNX3 to regulate proliferation and migration of human hepatocellular cancer cells. Oncol Rep 2016; 36:3379–3386. doi: 10.3892/or.2016.5144. [DOI] [PubMed] [Google Scholar]
- 18.Si W, Shen J, Du C, Chen D, Gu X, Li C, et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Differ 2018; 25:406–420. doi: 10.1038/cdd.2017.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai X, Han G, Liu Y, Jiang H, He Q. MiRNA-20a-5p promotes the growth of triple-negative breast cancer cells through targeting RUNX3. Biomed Pharmacother 2018; 103:1482–1489. doi: 10.1016/j.biopha.2018.04.165. [DOI] [PubMed] [Google Scholar]
- 20.Qin X, Wang XY, Fei JW, Li FH, Han J, Wang HX. MiR-20a promotes lung tumorigenesis by targeting RUNX3 via TGF-ß signaling pathway. J Biol Regul Homeost Agents 2020; 34.doi: 10.23812/20-12a. [DOI] [PubMed] [Google Scholar]
- 21.Qin X, Zhou M, Lv H, Mao X, Li X, Guo H, et al. Long noncoding RNA LINC00657 inhibits cervical cancer development by sponging miR-20a-5p and targeting RUNX3. Cancer Lett 2021; 498:130–141. doi: 10.1016/j.canlet.2020.10.044. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka S, Shiraha H, Nakanishi Y, Nishina S, Matsubara M, Horiguchi S, et al. Runt-related transcription factor 3 reverses epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Cancer 2012; 131:2537–2546. doi: 10.1002/ijc.27575. [DOI] [PubMed] [Google Scholar]
- 23.Tang KW, Guo ZX, Wu ZH, Zhou C, Sun J, Wang X, et al. Circ_0049447 acts as a tumor suppressor in gastric cancer through reducing proliferation, migration, invasion, and epithelial-mesenchy-mal transition. Chin Med J (Engl) 2021; 134:1345–1355. doi: 10.1097/cm9.0000000000001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghafouri-Fard S, Tamizkar KH, Hussen BM, Taheri M. MicroRNA signature in liver cancer. Pathol Res Pract 2021; 219:153369.doi: 10.1016/j.prp.2021.153369. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Zheng L, Ding Y, Li Q, Wang R, Liu T, et al. MiR-20a induces cell radioresistance by activating the PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Int J Radiat Oncol Biol Phys 2015; 92:1132–1140. doi: 10.1016/j.ijrobp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J Exp Clin Cancer Res 2013; 32:21.doi: 10.1186/1756-9966-32-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol 2000; 10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 28.Xiao WH, Liu WW. Hemizygous deletion and hypermethylation of RUNX3 gene in hepatocellular carcinoma. World J Gastroenterol 2004; 10:376–380. doi: 10.3748/wjg.v10.i3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori T, Nomoto S, Koshikawa K, Fujii T, Sakai M, Nishikawa Y, et al. Decreased expression and frequent allelic inactivation of the RUNX3 gene at 1p36 in human hepatocellular carcinoma. Liver Int 2005; 25:380–388. doi: 10.1111/j.1478-3231.2005.1059.x. [DOI] [PubMed] [Google Scholar]
- 30.Gou Y, Zhai F, Zhang L. CuiL.RUNX3 regulates hepatocellular carcinoma cell metastasis via targeting miR-186/E-cadherin/EMT pathway. Oncotarget 2017; 8:61475–61486. doi: 10.18632/oncotarget.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Z, Tian Y, Jia Y, Shen Q, Jiang W, Chen G, et al. RUNX3 inhibits the invasion and migration of esophageal squamous cell carcinoma by reversing the epithelialmesenchymal transition through TGFbeta/Smad signaling. Oncol Rep 2020; 43:1289–1299. doi: 10.3892/or.2020.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang TL, Ito K, Ko TK, Liu Q, Salto-Tellez M, Yeoh KG, et al. Claudin-1 has tumor suppressive activity and is a direct target of RUNX3 in gastric epithelial cells. Gastroenterology 2010; 138:255–265. e1–3. doi: 10.1053/j.gastro.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, Chen Y, Wu KC, Liu J, Zhao YQ, Pan YL, et al. RUNX3 directly interacts with intracellular domain of Notch1 and suppresses Notch signaling in hepatocellular carcinoma cells. Exp Cell Res 2010; 316:149–157. doi: 10.1016/j.yexcr.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 34.Nishina S, Shiraha H, Nakanishi Y, Tanaka S, Matsubara M, Takaoka N, et al. Restored expression of the tumor suppressor gene RUNX3 reduces cancer stem cells in hepatocellular carcinoma by suppressing Jagged1-Notch signaling. Oncol Rep 2011; 26:523–531. doi: 10.3892/or.2011.1336. [DOI] [PubMed] [Google Scholar]