Abstract
Gestational diabetes mellitus (GDM) is a growing public health problem worldwide that threatens both maternal and fetal health. Identifying individuals at high risk for GDM and diabetes after GDM is particularly useful for early intervention and prevention of disease progression. In the last decades, a number of studies have used metabolomics, genomics, and proteomic approaches to investigate associations between biomolecules and GDM progression. These studies clearly demonstrate that various biomarkers reflect pathological changes in GDM. The established markers have potential use as screening and diagnostic tools in GDM and in postpartum diabetes research. In the present review, we summarize recent studies of metabolites, single-nucleotide polymorphisms, microRNAs, and proteins associated with GDM and its transition to postpartum diabetes, with a focus on their predictive value in screening and diagnosis.
Keywords: Gestational diabetes mellitus, Biomarkers, Metabolomics, Proteomics, microRNA, Single-nucleotide polymorphism
Introduction
Gestational diabetes mellitus (GDM), one of the most common pregnancy complications, is defined as diabetes diagnosed in the second or third trimester of pregnancy that is not clearly overt diabetes prior to gestation.[1] The prevalence of GDM varies widely depending on population characteristics and diagnostic criteria. In 2017, the International Diabetes Federation reported that around 21.3 million live births (16.2%) worldwide were affected by hyperglycemia in pregnancy, including 18.4 million cases involving GDM. Notably, estimates of the raw prevalence of hyperglycemia in pregnancy range from 9.5% in Africa to 26.6% in Southeast Asia.[2] Risk factors for GDM include pre-pregnancy body mass index (BMI), advanced maternal age, ethnicity, family history of diabetes, smoking, and perfluorochemicals during pregnancy.[3–5] GDM is a cause of morbidity and mortality in both mothers and infants. Patients with GDM have a high risk of pre-eclampsia, polyhydramnios, operative delivery, and birth canal lacerations, while short-term consequences for offspring include shoulder dystocia, macrosomia, neonatal hypoglycemia, jaundice, and perinatal mortality. While GDM usually resolves following delivery, it can have a long-term impact on both the mother and the infant, including increased risks of hyperglycemia, diabetes, obesity, and cardiovascular diseases in the future. This contributes to the transgenerational cycle of diabetes and cardiometabolic disorders.[6,7] Therefore, GDM is a considerable threat to the health of mothers and infants worldwide, and research on its pathogenesis, early diagnosis, and intervention is necessary.
Similar to diabetes, GDM development has multiple mechanisms, including β-cell dysfunction, insulin resistance, adipose tissue dysfunction, gluconeogenesis, gut microbiota dysbiosis, and oxidative stress.[8] Owing to this complexity, our current understanding of the pathogenesis of this disease remains limited.
Biomarkers are quantifiable indicators of the physiological and pathological status of an organism. They are clinically useful because they can be used to assess the risk of disease development in healthy individuals. In the subclinical phase of diseases, biomarkers serve as screening tools for diagnosis, the prevention of progression, and monitoring pharmacological responses to therapeutic interventions.[9] Many molecular biomarkers for GDM have been investigated, including metabolites, single-nucleotide polymorphisms (SNPs), microRNAs (miRNAs), and proteins. These biomarkers may complement existing clinical risk factors to identify women at high risk of developing GDM during pregnancy as well as type 2 diabetes mellitus (T2D) after delivery. Furthermore, they provide insights into the pathophysiology of the disease and reveal the molecular mechanisms underlying the development of GDM.[10–12] Hence, molecular biomarkers have contributed substantially to GDM research in recent decades.
Metabolomics is the comprehensive analysis of low-molecular-weight compounds, known as metabolites, in biofluids, cells, and tissues. As the end products of metabolic processes, metabolites can reflect the internal physiological status of an organism, gene expression, and changes in the response to environmental factors. Due to the high sensitivity, even subtle changes in metabolic networks can be detected. Moreover, metabolomics provides an integrated profile of the biological status, without considering the effects of individual factors.[13] The most frequently used experimental technologies in metabolomics are liquid or gas chromatography–mass spectrometry (MS) and proton nuclear magnetic resonance (1H NMR) spectroscopy. MS-based approaches provide high throughput, sensitivity, and versatility, while NMR-based methods offer an overview of structural information, dynamic process, and higher reproducibility.[14,15] Though MS is usually more sensitive than 1H NMR, the biological fluid in NMR does not require any physical or chemical treatment prior to the analysis, when compared with MS-based methods. Furthermore, NMR does not damage analytes, which is useful when exploring metabolites in tissues that should be used in further experiments.[13] Untargeted and targeted profiling are the main methodologies used for metabolomics. Untargeted metabolomics allows for studies of metabolites without a priori information and is therefore suitable for candidate biomarker discovery, hypothesis generation, and analyses of metabolic mechanisms. In contrast, targeted metabolomics is usually employed for the analysis of specific metabolites. This type of analysis can be used to validate biomarkers and study metabolic pathways of interest.[16] Since samples of biological fluids, such as blood or urine, can be collected fairly easily, metabolites and their biological responses to diseases can be easily studied in detail. Metabolic groups include branched-chain amino acids (BCAAs), aromatic amino acids, sulfur-containing amino acids, phospholipids, and other metabolites, which mainly participate in amino acid, lipid, and carbohydrate metabolism.[17–19]
SNPs refer to alterations in a single nucleotide in the genome sequence. While most SNPs do not alter the function or expression of genes, some influence gene expression and contribute to diseases. Comparisons of SNP frequencies between individuals with a disease and control individuals can reveal candidate loci associated with the disease.[20] One general approach for these studies is the candidate gene approach, which is used to investigate the association between a particular susceptibility gene and a phenotype.[21] Technologies such as TaqMan assay, SNaPshot, and PCR-restriction fragment length polymorphism methods are widely used.[22] For the reason that each SNP contains limited genetic information, a great deal of effort has been devoted to improving the throughput of genotyping. SNPlex, the Illumina BeadArray, the Sequenom MassARRAY iPLEX platform, and other SNP genotyping platforms are developed to serve different purposes.[22,23] SNPs in several genes, particularly genes responsible for lipid and glucose metabolism, insulin secretion, and insulin resistance, have been associated with GDM. These genetic variants are potential biomarkers; however, individually, they are likely to lack sensitivity.
miRNAs are specialized short non-coding RNAs approximately 22 nucleotides in length. Functionally, mature miRNAs guide the RNA-induced silencing complex to recognize target mRNAs at 3′ untranslated regions, thus regulating gene expression.[24–26] miRNAs are promising biomarkers of diseases owing to their high stability in body fluids and their enrichment in particular tissues. miRNAs have been associated with GDM and its health complications and may have diagnostic, prognostic, and predictive value.[27–29] Three common methods for miRNA analysis are miRNA sequencing, microarray, and quantitative real-time PCR (qRT-PCR).[30] miRNA sequencing utilizes next-generation sequencing for high-throughput analysis of miRNA. Though it has advantages of sensitivity, accuracy, and repeatability, it is also time consuming and costly.[31] Microarray is also a technology for multiplex analysis of miRNAs. However, the relatively low sensitivity and specificity of microarray could be a challenge.[32] qRT-PCR is currently the most widely used detection technology for miRNAs.[33] Sensitivity, specificity, and dynamic range of qRT-PCR are considered as excellent, but it is useable only for miRNAs that have been previously identified and not suitable for discovery studies.[34] Despite of its imperfections, qRT-PCR is the most common method for routine testing in the clinical laboratory because it is performed on standard equipment.[35]
Proteomics is the large-scale analysis of peptides or proteins in cells, tissues, and body fluids and has attracted substantial attention over the past few decades owing to the roles of proteins in almost all biological processes.[36] Four major approaches are involved in quantitative proteomics, including gel-based, stable isotope labeling, label free, and targeted proteomics. One-dimensional gel electrophoresis, two-dimensional polyacrylamide gel electrophoresis, and difference gel electrophoresis approaches have been utilized for protein separation in gel-based proteomics.[37,38] In labeling proteomics, stable isotope labeling by amino acids in cell culture, isobaric tag for relative and absolute quantitation, and tandem mass tag are commonly used.[39] Selected reaction monitoring and multiple reaction monitoring are MS-based techniques that act as reliable quantitation methods in targeted proteomics. A triple quadrupole mass spectrometer is employed to detect and analyze targeted proteins. Using this instrument, specific peptides representing each of the targeted proteins are selected based on their mass to charge (m/z) ratio, and they subsequently fragment into smaller ions for quantitation analysis.[40,41] Label-free proteomics is not restricted to labels and consequently reduces the overall experimental error. It also enables the comparison of protein expression among a great number of samples from various sources. MS has been invaluable for the revolution of proteomics, for example, stable isotope labeling coupled with MS-based techniques tremendously improved sensitivity and protein detection in comparison to classical approaches.[42,43] Several proteomics studies have identified potential biomarkers for GDM and its complications, although GDM is believed to share a similar mechanism with T2D. These protein biomarkers, such as transthyretin, serum retinol binding protein 4, and the apolipoprotein (Apo) superfamily, are related to insulin resistance, glycolipid metabolism, inflammatory pathways, and oxidative stress.[44,45]
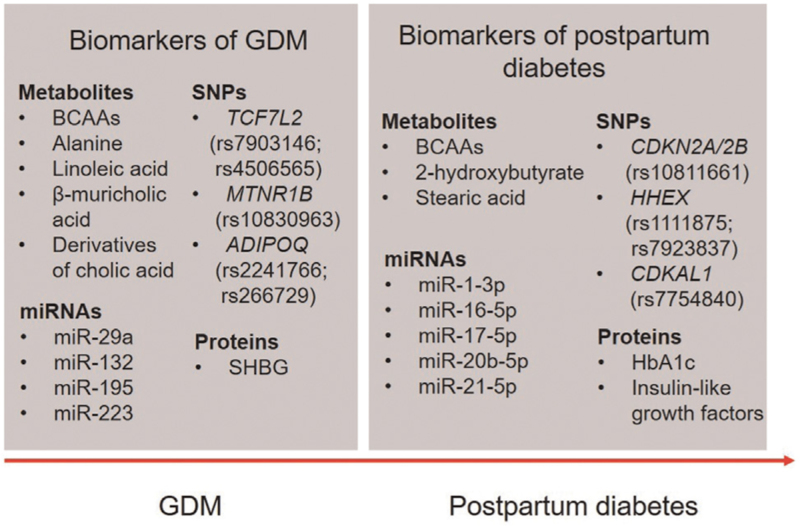
In this review, we searched for studies of metabolites, SNPs, miRNAs, and proteins in human subjects with GDM published between January 2010 and September 2021. PubMed was systematically searched using search terms “gestational diabetes mellitus” or “postpartum diabetes” combined with “metabolite,” “single-nucleotide polymorphism,” “microRNAs” or “protein” and corresponding synonyms and associated terms for each word. Articles published in languages other than English were excluded. We describe altered biomolecule profiles, summarize the mechanisms of action of specific biomolecules, and discuss their use as biomarkers in the occurrence and development of GDM and postpartum diabetes. The main findings are summarized in [Figure 1].
Figure 1.
Biomarkers for GDM and postpartum diabetes. BCAAs: Branched-chain amino acids; GDM: Gestational diabetes mellitus; HbA1c: Hemoglobin A1c; miRNAs: microRNAs; SHBG: Sex hormone-binding globulin; SNPs: Single-nucleotide polymorphisms.
Metabolomics in GDM
The identification of GDM-related metabolic biomarkers is important for early intervention and may improve our understanding of the underlying mechanisms. Blood (plasma and serum), urine, and hair are frequently utilized in metabolomic studies of GDM. In particular, blood samples are the most commonly used biofluids because blood is abundant and highly dynamic; however, sampling requires invasive procedures.[46] Elevated levels of linoleic acid, alanine, leucine, lysophosphatidylcholine, tyrosine, phenylalanine, carnitine, and derivatives of cholic acid have been consistently reported among patients with GDM.[47–57] In contrast, decreases in serine,[47,58,59] glutamine,[58–60] and methionine[59,60] have been observed.
Most studies have reported that levels of circulating BCAAs, which consist of valine, leucine, and isoleucine, were increased in patients with GDM.[47,48,56] Increased levels of ketone bodies in GDM inhibit proteolysis and reduce the oxidation of BCAAs in skeletal muscle; thus, they are released at low rates from skeletal muscle and are mostly catabolized in the liver.[61] The activities of BCAA catabolic enzymes in the liver and adipose tissue are repressed, contributing to the high blood concentration of BCAAs.[62] Valine levels were also higher in infants of mothers with GDM than in infants of healthy mothers.[63] Among children with GDM, daily intake of BCAAs was associated with elevated risks of overweight and insulin resistance; however, this association was not fully independent of daily energy intake in children.[64] BCAAs are predictive of GDM and are associated with insulin resistance.[65,66] It has been proposed that BCAA metabolism may contribute to insulin resistance because the accumulation of toxic BCAA metabolites could result in β-cell mitochondrial dysfunction and high susceptibility to insulin resistance.[67] Another explanation for the association was that BCAAs activated the mammalian target of rapamycin pathway and phosphorylated insulin receptor substrate 1, thereby interfering with insulin signaling.[68] BCAAs can also participate in glucose uptake by increasing the translocation of the glucose transporter 1 (GLUT1) and GLUT4 to the cell surface.[69]
The area under the curve (AUC) is a typical parameter used to assess the predictive efficiency of potential biomarkers. In analyses of blood samples, a β-muricholic acid-based model had the best performance for the prediction of GDM, with the highest AUC (>0.95) and Youden index (>0.80) and a sensitivity of 92.1% and specificity of 96.3%.[55] β-Muricholic acid is an intermediate in the metabolism of cholesterol to tauro-β-muricholic acid in the liver. Tauro-β-muricholic acid is then exported to the intestine and hydrolyzed to β-muricholic acid by bile salt hydrolase.[70,71] As a potent farnesoid X receptor antagonist, tauro-β-muricholic acid inhibits intestinal farnesoid X receptor signaling, contributing to improved hepatic steatosis, insulin sensitivity, and glucose tolerance.[72,73]
While blood provides a snapshot of the metabolic status of individuals at the time of sampling, urine represents a summation of process occurring in the hours prior to sampling.[46] The acquisition of urine samples is non-invasive; however, the composition and volume depend on dietary habits and other factors. Women with GDM showed significant increases in urine levels of serotonin,[19,74] tryptophan,[19,54,75] glucuronide derivatives,[75,76] and phenylalanine.[54,77] Studies have also identified decreases in metabolites, including ethanolamine, lanthionine, 5-methoxytryptamin, threonine, methionine, methionine sulfoxide, acadesine, carnitine, argininate, and melatonin.[19,54,60,76] The AUC values for these factors ranged from 0.718[60] to 0.993,[76] demonstrating good predictive performance. In one of the studies with the highest predictive performance, untargeted and targeted metabolomics approaches were used to explore plasma and urine sample metabolites, yielding an AUC for GDM prediction (combined with BMI) of 0.99.[19] Another recent study of urinary metabolites in early pregnancy involving 46 women with GDM and 46 age-matched individuals without GDM applied a classification tree analysis based on saccharopine, dihydroorotate, nicotinate ribonucleoside, 7,8-dihydroneopterin, phenylglucuronide, lanthionine, and arginine and the AUC for the prediction of progression to GDM was 0.993.[76]
Hair is easy to obtain in a non-invasive manner and highly stable for retaining long-term information; thus, it is a useful source of biomarkers.[78] Analyses of hair samples are limited; however, increases in 2-aminobutyric acid and adipic acid have been detected.[79,80] Further studies of hair samples are needed to confirm these findings and to identify more differentially expressed metabolites in women with GDM.
Approximately, 50%–60% of patients with GDM develop dysglycemia (T2D and prediabetes) after delivery;[81,82] accordingly, a predictive test for this transition may improve patient management. Increases in BCAAs are predictors of the incident T2D risk after pregnancy complicated by GDM.[83–85] Six years after GDM, levels of BCAAs and the valine metabolite 3-hydroxyisobyturate were higher in women who developed T2DM, and there was a tendency for BCAA levels to increase in the impaired glucose tolerance group.[85] Dudzik et al[86] found that 2-hydroxybutyrate and stearic acid showed the best discriminative power (AUC of 0.90) for postpartum diabetes. Elevated levels of 2-hydroxybutyrate, which result from enhanced lipid oxidation, glutathione synthesis, and a redox imbalance,[86] are associated with insulin resistance and reduced insulin secretion.[87] These metabolite changes may be considered prognostic biomarkers for the prediction of postpartum diabetes among women with prior GDM.
Genomic Analysis of GDM
Since GDM shares pathophysiological similarities with T2D, some T2D-related genes are also important in the GDM process. The TCF7L2 gene, which is highly associated with T2D predisposition, encodes a transcription factor that operates at the end of the Wnt signaling cascade not only in β cells but also in other organs, including the liver.[88] rs7903146, rs4506565, rs7901695, rs12255372, and rs12243326 in the TCF7L2 gene are associated with GDM. As the most frequently reported polymorphism in TCF7L2, rs7903146 was associated with an increased risk of GDM.[89–95] In addition, patients with GDM harboring the T risk allele in rs7903146 were more likely to have early postprandial glycemic control failure and require insulin therapy during pregnancy.[96] rs7903146 is located in islet-selective open chromatin, indicating that the chromatin state at rs7903146 is more open in chromosomes carrying the T allele, allowing the binding of regulatory proteins to this locus in human islet cells.[97] Previous studies have shown that it was related to impaired insulin secretion, decreased incretin effects, and hepatic insulin resistance.[98,99] Pilgaard et al[99] found that risk allele carriers showed an increased proinsulin/insulin ratio, reduced insulinotropic effects of incretin hormones, and impaired β-cell responsiveness to glucose. Cropano et al[100] also revealed that the T allele of rs7903146 contributed to the development of hyperglycemia by altering the proinsulin secretory efficiency and reducing the ability of insulin to suppress hepatic endogenous glucose production. The T allele of rs4506565 was also a risk allele for GDM.[92,93,101] Pagan et al[101] found that TCF7L2 rs4506565 was associated with a two-fold increase in the risk of developing GDM. Pregnant individuals carrying this risk allele had elevated levels of resistin, a member of the adipokine family, in the plasma and cord blood. Additionally, the allele was associated with increased interleukin (IL)-6 levels, suggesting that it had an effect on GDM via inflammation.
MTNR1B encodes the melatonin MT2 receptor, also known as melatonin receptor type 1B. As a member of the G-protein-coupled receptor superfamily, it affects glucose intolerance and insulin resistance.[102] Four SNPs in the MTNR1B gene were frequently reported, including rs10830963. Most studies of rs10830963 have shown that the G allele was the risk allele and was correlated with increased fasting glucose, increased hemoglobin A1c (HbA1c), and an impaired early insulin response to glucose.[103–105] Glucokinase (GK) is a tissue-specific enzyme mainly present in the liver and pancreatic islets,[106] where glucose is phosphorylated by GK into glucose-6-phosphate for further glycometabolism.[107] Individuals carrying the G allele showed higher expression of MTNR1B via increased FOXA2-bound enhancer activity and neuronal differentiation 1 (NEUROD1) binding in islet cells,[108] and melatonin lowered intra-cellular cyclic adenosine monophosphate (cAMP) levels, leading to the downregulation of GK expression and impaired insulin secretion.[105,109] In an analysis of individuals with a high GDM risk (i.e., with a history of GDM and/or BMI ≥30 kg/m2), only non-carriers of the risk allele (i.e., G) benefited from lifestyle interventions, suggesting that the MTNR1B rs10830963 variant could modify the efficacy of lifestyle interventions.[110] Moreover, for patients with GDM with a pre-pregnancy BMI ≥29 kg/m2 carrying the rs10830963 G risk allele, a study reported that despite the medical nutrition therapy and lifestyle intervention, endogenous insulin secretion was insufficient to meet the increased insulin demand, and antenatal insulin therapy initiation may be necessary.[111]
The ADIPOQ gene spans 17 kb on chromosomal region 3q27 and encodes human adiponectin, which is secreted mainly by adipose tissue. This adipocyte-derived plasma protein can reverse insulin resistance and increase insulin efficacy in glucose metabolism.[112] It is related to obesity, T2D, and metabolic syndrome.[113] rs1501299, rs2241766, and rs266729 are the most frequent SNPs. Although studies of rs1501299 have not detected an association with GDM,[114,115] most studies of rs2241766 have shown that the G allele was associated with an increased risk of GDM.[116–119] Several studies have indicated that subjects with the rs2241766 G allele showed a decrease in adiponectin levels and an increase in the insulin resistance index.[120,121] Considering that rs2241766 is a silent polymorphism with no impact on the sequence of amino acids, it is possible that this SNP inactivates the gene by influencing transcript activity, such as the splicing accuracy or efficiency.[122] The association between rs266729 and GDM remains inconclusive.[114,115,123,124] Conflicting results can be explained by differences in environmental factors and lifestyle among populations. In addition, inconsistent detection methods and sample sizes might have contributed to the differences.
Relatively few studies have evaluated the genetic risk of postpartum diabetes among women with GDM. Notably, rs10811661 of CDKN2A/2B and rs1111875 and rs7923837 of HHEX increased the risk of early conversion (2 months), whereas rs7754840 of CDKAL1 increased the risk of late conversion (more than a year).[125] Some SNPs associated with postpartum glycemic traits have been identified in women with a history of GDM. MTNR1B rs10830963 was genotyped in 1025 women with previous GDM, revealing its relationship with postpartum fasting glucose levels. In a stratified analysis, the MTNR1B genotype was related to postpartum changes in the 2-hour oral glucose tolerance test (OGTT) across categories of inadequate, adequate, and excessive gestational weight gain.[126] Similarly, rs10830963 and rs1387153 of MTNR1B were associated with elevated fasting glucose levels in another study, with highly significant P values.[90] In a study of 1208 women with prior GDM, MC4R rs6567160 was not significantly associated with postpartum fasting glucose but was positively associated with 2-hour OGTT glucose concentrations and increased HbA1c.[127]
The genetic risk score (GRS) is computed as the sum of unweighted or weighted risk variants. This parameter combines genetic information for multiple variants and thereby has high predictive power. Given the relatively small effect sizes of individual risk loci, the GRS includes SNPs at independent loci to predict the genetic risk of diseases. A GRS based on 48 genetic variants showed good prediction performance for T2D in women with GDM; women who developed diabetes after GDM pregnancy had a higher unweighted and weighted GRS (wGRS) than those who did not meet the diabetes diagnostic criteria. The risk of diabetes increased as the quartiles of wGRS increased. The hazard for diabetes incidence was 5.52 times higher in the highest wGRS quartile than in the lowest wGRS quartile. In a complex clinical model for the prediction of future diabetes, the c-statistic increased from 0.741 without the wGRS to 0.775 with the wGRS and the net reclassification improvement index was 0.430.[128] Another GRS model based on 36 SNPs was also predictive of pre-diabetes and T2D in women with previous GDM; when the explained-variance GRS was added to a model including age and BMI, the AUC increased from 0.6269 to 0.6672, indicating an improved predictive value.[129]
miRNAs in GDM
There is substantial interest in the roles of miRNAs in the pathogenesis of GDM. In the last few years, a number of miRNAs that are upregulated in women with GDM have been identified, including miR-16-5p,[130–132] miR-19a,[132,133] miR-19b,[132,133] miR-101,[134,135] miR-137,[136,137] miR-195,[134,138–140] miR-223,[141,142] miR-330-3p,[28,143,144] miR-342-3p,[134,145] and miR-657.[146,147] Like many other miRNAs, miR-223 has been implicated in various physiological and pathological conditions, including inflammatory disorders,[148,149] infection,[150,151] and cancer.[152,153] It was downregulated in T2D[154] but was upregulated in the insulin-resistant heart of patients with T2D.[155] Independent of phosphoinositide 3-kinase signaling or AMP kinase activity, miR-223 increased glucose uptake via GLUT4 protein expression in cardiomyocytes.[155] It was speculated that the overexpression of miR-223 in the insulin-resistant heart is a compensatory mechanism for the systemic reduction of miR-223.[156] However, miR-223 was upregulated in GDM, and a prediction model based on miR-223 alone had a better accuracy than that of a model including the differentially expressed miRNAs miR-223 and miR-23a, with an AUC value of 0.94 and accuracy of 0.90.[141] Another model based on miR-223 showed a similar predictive value (AUC = 0.92).[142] Serum expression of miR-29a was significantly downregulated in pregnant women with GDM compared with healthy women, and its expression decreased ahead of the elevation of serum glucose.[157] However, the AUC for miR-29a varied greatly, from 0.658[157] to 0.829.[158] The knockdown of miR-29a increases the expression of Insig1 gene and subsequently enhances the level of phosphoenolpyruvate carboxy kinase 2. As a key enzyme in gluconeogenesis and glycolysis, phosphoenolpyruvate carboxy kinase 2 expression may lead to increased glucose level.[157] miR-29a is not only a potential regulator of serum glucose but also a negative regulator of cannabinoid type 1 receptor. Overexpressing miR-29a inhibits the expression of proinflammatory and profibrogenic mediators, preventing diabetic glomeruli damage caused by fibrosis.[159]
Other miRNA-related mechanisms contributing to the pathogenesis of GDM have been discovered. For example, miR-657 could promote macrophage proliferation, migration, and polarization toward the M1 phenotype by downregulating family with sequence similarity 46 member C, thus effecting macrophage-mediated immunity and inflammation in GDM.[147] The downregulation of miR-770-5p could enhance pancreatic β-cell proliferation, promote insulin secretion, and suppress cell apoptosis via the TP53 regulated inhibitor of apoptosis 1 (TRIAP1)/apoptotic peptidase activating factor 1 (APAF1) pathway; hence, the miRNA plays a protective role in GDM.[160] miR-96 also contributes to β-cell proliferation and function by targeting and downregulating p21-activated kinase 1.[161] The downregulation of miR-29b may be partially related to the development of GDM by increasing the expression of hypoxia-inducible factor 3A, promoting trophoblast cell activity.[162] Serum aberrant expression of miR-132 in GDM is observed prior to glucose abnormality,[157] and miR-132 may exert a protective role against GDM through abrogating the inhibiting effects of high glucose on trophoblast cell proliferation.[163] The dysfunction of vascular endothelial cells could account for the high risk of cardiovascular diseases in patients with GDM and their offspring. The overexpression of miR-137 induces human umbilical vein endothelial cell dysfunction under high glucose conditions by promoting the secretion of CC chemokine ligand-2 (CCL2)/monocyte chemoattractant protein-1 (MCP-1), upregulating the levels of IL-6, intercellular cell adhesion molecule-1, vascular cell adhesion molecule-1, and E-selectin, and downregulating IL-8 and vascular endothelial growth factor.[136] By the inhibition of the target gene peroxisome proliferator-activated receptor-α, miR-518d may disrupt the homeostatic balance between cellular fatty acid and glucose metabolism and increase resistance to insulin.[164] These findings clearly establish the importance of miRNAs in the development of GDM, suggesting that further investigations of miRNAs may shed light on the pathophysiology of the disease.
In addition to associations between miRNAs and the risk of GDM, some investigators have evaluated trimester-specific miRNA changes in cases and controls. Herrera-Van Oostdam et al[130] found that in the second trimester, miR-517-3p and miR-518-5p levels were higher in cases than in controls, while the opposite pattern was observed in the third trimester. Lamadrid-Romero et al[137] also observed that the level of miR-125b-5p increased in the first trimester and decreased during the second trimester. Additionally, levels of miR-200b-3p and miR-183-5p, which were upregulated in the first and second trimester, respectively, were downregulated in the third trimester. These findings may be helpful in tracking disease progression and clarifying the pathophysiology.
Other studies of the risk of developing diabetes after delivery among women with a history of GDM based on miRNAs are lacking. However, Hromadnikova et al[165] assessed the risk of later development of diabetes mellitus and cardiovascular and cerebrovascular diseases in 111 women with GDM and 89 age-matched women after normal pregnancies. They selected a series of miRNAs involved in the pathogenesis of diabetes mellitus and cardiovascular and cerebrovascular diseases. The expression levels of these miRNAs were higher in women with a history of GDM than in the control group. Among these miRNAs, miR-1-3p, miR-16-5p, miR-17-5p, miR-20b-5p, miR-21-5p, miR-23a-3p, miR-26a-5p, miR-29a-3p, miR-103a-3p, miR-133a-3p, miR-146a-5p, miR-181a-5p, miR-195-5p, miR-199a-5p, miR-221-3p, and miR-499a-5p showed good sensitivity at a 10.0% false positive rate and high accuracy for the identification of women previously affected with GDM at a higher risk of postpartum diabetes mellitus and cardiovascular and cerebrovascular diseases. These miRNAs were highly discriminative, with an AUC of 0.900, sensitivity of 77.48%, and specificity of 93.26%.
Proteomics in GDM
Protein biomarkers in women with GDM included immune molecules, hormones, enzymes, polypeptides, and glycoproteins. Sex hormone-binding globulin (SHBG) is a glycoprotein synthesized mainly in the liver that binds to and regulates sex steroids with high affinity and specificity.[166] Low concentrations of SHBG are a biomarker for early GDM.[167,168] Even pre-pregnancy SHBG levels are a predictor of subsequent GDM; women with SHBG levels below the median appeared to have a 2.6-fold increased risk for GDM. A greater effect of low SHBG levels was observed in women who were overweight or obese (BMI ≥25.0 kg/m2), with a 5.3-fold increased risk of GDM compared with that of normal weight women with high SHBG concentrations.[169] Despite controversies regarding the inhibitory effects of insulin on SHBG production,[170–172] there is evidence for an association between SHBG levels and the development of insulin resistance.[173,174] Some studies have attempted to explain the inverse association. For instance, Wang et al[175] speculated that SHBG may exert its biological effects by inhibiting the extra-cellular signal-regulated kinase (ERK) pathway, thus influencing insulin secretion and participating in the onset of insulin resistance and GDM. Disruption of the ERK isoform ERK1 in mice resulted in resistance to high-fat diet-induced obesity and improved insulin sensitivity.[176] Activation of the ERK pathway by IL-1β could decrease insulin-induced glucose transport mainly by inhibiting insulin receptor substrate 1 expression at the transcriptional level.[177] In addition, Chi et al[178] found that SHBG in the placenta may regulate the expression of GLUT1 via the activation of the cAMP/protein kinase A (PKA)/cAMP responsive element binding protein 1 (CREB1) pathway, thereby affecting glucose metabolism and improving insulin resistance.
In addition to differences in protein expression levels between patients with GDM and healthy women, longitudinal changes in differentially expressed proteins have been investigated. In a prospective cohort study, serum proteins were screened in the early stage (12–16 weeks) and middle stage (24–28 weeks) of pregnancy in 60 participants (30 GDM cases and 30 healthy controls). In total, 31 and 27 proteins were differentially expressed between GDM cases and controls in the early and middle stages, respectively. When compared with the early stage, 38 and 28 proteins were altered in the middle stage in healthy controls and patients with GDM, respectively, and these proteins may be associated with the progression of normal pregnancy and GDM.[179] Among these proteins, beta-ala-his dipeptidase was highly discriminative (AUC = 0.98),[180] whereas Apo E was less discriminative (AUC = 0.965). The combination of Apo E, coagulation factor IX, fibrinogen alpha chain, and insulin-like growth factor-binding protein 5 increased the AUC to 0.985, with 80% sensitivity and 95% specificity.[181]
An increasing number of studies have focused on the clinical value of HbA1c levels in predicting the development of postpartum diabetes among patients with GDM. A logistic regression analysis indicated that HbA1c ≥ 5.4% was associated with a 5.5-fold increased risk of postpartum diabetes.[182] In another study, at the optimal HbA1c cutoff value of 5.55%, the AUC was 0.846, with 78.6% sensitivity and 72.5% specificity.[183] Coetzee et al[184] investigated the value of HbA1c levels at the time of GDM diagnosis (t1) and in the 4 weeks preceding delivery (t2) for the prediction of postpartum diabetes. The receiver operating characteristic curve analysis indicated that HbA1c at GDM diagnosis performed well (AUC = 0.90). At a cutoff of 6.2%, the sensitivity and specificity were 95% and 62%, respectively. The optimal cutoff value for HbA1c in the 4 weeks preceding delivery was 6.2%, with an AUC value of 0.81, and patients with GDM and HbA1c ≥6.2% had a four-fold (at t1) or five-fold (at t2) increased risk of diabetes, respectively. Thus, HbA1c could be used as a tool to predict postpartum diabetes in women with GDM.
Insulin-like growth factors and their binding proteins are correlated with the development of T2D. Lappas et al[185] further showed that postpartum insulin-like growth factor-binding protein-2 levels were significantly and negatively associated with the development of T2D among women with previous GDM, even after adjusting for age and BMI. In contrast, insulin-like growth factor I levels were positively associated with postpartum T2D. When the above two factors were added to a base model (including age, BMI, fasting glucose, and postnatal fasting glucose), the predictive model identified individuals with postpartum diabetes with an AUC of 0.892. Similarly, in 2019, Lappas et al[185] explored the relationship between postpartum diabetes and Apo species based on their roles in T2D. Apo CIII levels as well as the Apo CIII/Apo AI, Apo CIII/Apo AII, Apo CIII/Apo CII, Apo CIII/Apo E, and Apo E/Apo CIII ratios were positively associated with the development of diabetes; when these parameters were added to the base model, the accuracy (83.2% to 86.3%), sensitivity (30% to 40%), and AUC values (0.782–0.824) were improved.[186] Thus, these variables were identified as risk factors for the prediction of T2D in women with prior GDM.
Challenges and Future Directions
Several biomolecules have recently emerged as biomarkers for GDM and postpartum diabetes and can offer a basis for early diagnosis and targeted treatment. Biomarkers, including metabolites, SNPs, miRNAs, and proteins, are involved in glucolipid metabolism, insulin resistance, and inflammation. Thus, a complex network linking these factors should exist to integrate the biological information. However, the results of some GDM studies are highly inconsistent and difficult to replicate. Sample size and demographic factors are major bottlenecks in identifying credible diagnostic and prognostic biomarkers of GDM. In addition, most studies are case-control studies; these retrospective analyses do not provide insight into prevalence or incidence. Therefore, prospective studies are needed to explore the associations between biomarkers and GDM in a more powerful way. More importantly, while most biological samples are collected in the second trimester, at the time of GDM diagnosis, samples collected prior to the existence of GDM or at its onset are more critical for early prediction and diagnosis. The widely accepted approach for GDM diagnosis is 75 g OGTT performed during 24–28 gestational weeks, which seems a little late for intervention and prevention. In this sense, biomarkers released in the first trimester or ahead of glucose abnormality may help identify women at high risk of GDM and early GDM. However, relatively few studies target this area and no single molecule currently performs sufficiently well to be an established screening tool for early prediction and diagnosis of GDM. Though women with GDM are recommended to screen for T2D after pregnancy,[187] the compliance among this group is relatively low probably due to lack of awareness of the need for screening and the time-consuming nature of the tests.[188] Since some pathophysiological changes occur long before the elevation of blood glucose, it is possible to build up a more accurate and acceptable test based on biomolecules for the prediction of T2D following GDM pregnancy. However, existing studies are mostly limited to certain racial groups and short-term follow-up.
Hence, biological samples obtained at different stages of pregnancy would be of great use in exploring the etiology and prognosis of GDM. Biomolecules, especially those released in the early stage, need to be tested on larger and more diverse populations to assess their predictive value. Further research with a much longer follow-up period may help explore their potential utility as screening tools, considering sensitivity, specificity, cost, and patient acceptability. Since the mechanism underlying the pathological progression of GDM is not entirely clear, the integration of data from various omics-based approaches is crucial, although numerous challenges remain.
Conflicts of interest
None.
Footnotes
How to cite this article: Lu W, Hu C. Molecular biomarkers for gestational diabetes mellitus and postpartum diabetes. Chin Med J 2022;135:1940–1951. doi: 10.1097/CM9.0000000000002160
Contributor Information
Wenqian Lu, Email: 635481691@qq.com.
Cheng Hu, Email: alfredhc@sjtu.edu.cn.
References
- 1.American Diabetes Association. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care 2020; 43:S14–S31. doi: 10.2337/dc20-S002. [DOI] [PubMed] [Google Scholar]
- 2.Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, et al. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract 2018; 138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 3.Mutsaerts MA, Groen H, Buiter-Van der Meer A, Sijtsma A, Sauer PJ, Land JA, et al. Effects of paternal and maternal lifestyle factors on pregnancy complications and perinatal outcome. A population-based birth-cohort study: The GECKO Drenthe cohort. Hum Reprod 2014; 29:824–834. doi: 10.1093/humrep/deu006. [DOI] [PubMed] [Google Scholar]
- 4.Mattsson K, Källen K, Longnecker MP, Rignell-Hydbom A, Rylander L. Maternal smoking during pregnancy and daughters’ risk of gestational diabetes and obesity. Diabetologia 2013; 56:1689–1695. doi: 10.1007/s00125-013-2936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Sundaram R, Maisog J, Calafat AM, Barr DB, Buck Louis GM. A prospective study of prepregnancy serum concentrations of perfluorochemicals and the risk of gestational diabetes. Fertil Steril 2015; 103:184–189. doi: 10.1016/j.fertnstert.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McIntyre HD, Catalano P, Zhang C, Desoye G, Mathiesen ER, Damm P. Gestational diabetes mellitus. Nat Rev Dis Primers 2019; 5:47.doi: 10.1038/s41572-019-0098-8. [DOI] [PubMed] [Google Scholar]
- 7.Mitanchez D, Yzydorczyk C, Siddeek B, Boubred F, Benahmed M, Simeoni U. The offspring of the diabetic mother - short- and long-term implications. Best Pract Res Clin Obstet Gynaecol 2015; 29:256–269. doi: 10.1016/j.bpobgyn.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Plows JF, Stanley JL, Baker PN, Reynolds CM, Vickers MH. The pathophysiology of gestational diabetes mellitus. Int J Mol Sci 2018; 19:3342.doi: 10.3390/ijms19113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001; 69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 10.Smirnakis KV, Plati A, Wolf M, Thadhani R, Ecker JL. Predicting gestational diabetes: choosing the optimal early serum marker. Am J Obstet Gynecol 2007; 196:e1–e6. doi: 10.1016/j.ajog.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, Choi SH, et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 2012; 61:531–541. doi: 10.2337/db11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirier C, Desgagne V, Guerin R, Bouchard L. MicroRNAs in pregnancy and gestational diabetes mellitus: emerging role in maternal metabolic regulation. Curr Diab Rep 2017; 17:35.doi: 10.1007/s11892-017-0856-5. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature 2008; 455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 14.Mao X, Chen X, Chen C, Zhang H, Law KP. Metabolomics in gestational diabetes. Clin Chim Acta 2017; 475:116–127. doi: 10.1016/j.cca.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Francis E, Hu G, Chen L. Metabolomic profiling of women with gestational diabetes mellitus and their offspring: Review of metabolomics studies. J Diabetes Complications 2018; 32:512–523. doi: 10.1016/j.jdiacomp.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016; 17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Li M, Rahman ML, Hinkle SN, Wu J, Weir NL, et al. Plasma phospholipid n-3 and n-6 polyunsaturated fatty acids in relation to cardiometabolic markers and gestational diabetes: a longitudinal study within the prospective NICHD Fetal Growth Studies. PLoS Med 2019; 16:e1002910.doi: 10.1371/journal.pmed.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cetin I, de Santis MS, Taricco E, Radaelli T, Teng C, Ronzoni S, et al. Maternal and fetal amino acid concentrations in normal pregnancies and in pregnancies with gestational diabetes mellitus. Am J Obstet Gynecol 2005; 192:610–617. doi: 10.1016/j.ajog.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Leitner M, Fragner L, Danner S, Holeschofsky N, Leitner K, Tischler S, et al. Combined metabolomic analysis of plasma and urine reveals AHBA, tryptophan and serotonin metabolism as potential risk factors in gestational diabetes mellitus (GDM). Front Mol Biosci 2017; 4:84.doi: 10.3389/fmolb.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, Rotter JI. Genome-wide association studies. JAMA 2019; 322:1705–1706. doi: 10.1001/jama.2019.16479. [DOI] [PubMed] [Google Scholar]
- 21.Lee JE. High-throughput genotyping. Forum Nutr 2007; 60:97–101. doi: 10.1159/000107078. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng 2007; 9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- 23.Ding C, Jin S. High-throughput methods for SNP genotyping. Methods Mol Biol 2009; 578:245–254. doi: 10.1007/978-1-60327-411-1_16. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 25.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol 2013; 48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 2002; 21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wander PL, Boyko EJ, Hevner K, Parikh VJ, Tadesse MG, Sorensen TK, et al. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res Clin Pract 2017; 132:1–9. doi: 10.1016/j.diabres.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebastiani G, Guarino E, Grieco GE, Formichi C, Delli Poggi C, Ceccarelli E, et al. Circulating microRNA (miRNA) expression profiling in plasma of patients with gestational diabetes mellitus reveals upregulation of miRNA miR-330-3p. Front Endocrinol (Lausanne) 2017; 8:345.doi: 10.3389/fendo.2017.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pheiffer C, Dias S, Rheeder P, Adam S. Decreased expression of circulating miR-20a-5p in South African women with gestational diabetes mellitus. Mol Diagn Ther 2018; 22:345–352. doi: 10.1007/s40291-018-0325-0. [DOI] [PubMed] [Google Scholar]
- 30.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019; 234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012; 13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Dong L, Zhang J, Zhao Y, Li Z. Recent advances in microRNA detection. Analyst 2018; 143:1758–1774. doi: 10.1039/C7AN02001E. [DOI] [PubMed] [Google Scholar]
- 33.Zöllner H, Hahn SA, Maghnouj A. Quantitative RT-PCR specific for precursor and mature miRNAs. Methods Mol Biol 2014; 1095:121–134. doi: 10.1007/978-1-62703-703-7_10. [DOI] [PubMed] [Google Scholar]
- 34.Kappel A, Keller A. miRNA assays in the clinical laboratory: workflow, detection technologies and automation aspects. Clin Chem Lab Med 2017; 55:636–647. doi: 10.1515/cclm-2016-0467. [DOI] [PubMed] [Google Scholar]
- 35.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 2010; 50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 36.Patterson SD, Aebersold RH. Proteomics: the first decade and beyond. Nat Genet 2003; 33: (Suppl): 311–323. doi: 10.1038/ng1106. [DOI] [PubMed] [Google Scholar]
- 37.Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: past, present and future. J Proteomics 2010; 73:2064–2077. doi: 10.1016/j.jprot.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 38.Tannu NS, Hemby SE. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat Protoc 2006; 1:1732–1742. doi: 10.1038/nprot.2006.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002; 1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 40.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 2012; 9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 41.Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, et al. Mass spectrometry based targeted protein quantification: methods and applications. J Proteome Res 2009; 8:787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng 2009; 11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 43.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature 2003; 422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 44.Kim SM, Park JS, Norwitz ER, Lee SM, Kim BJ, Park CW, et al. Identification of proteomic biomarkers in maternal plasma in the early second trimester that predict the subsequent development of gestational diabetes. Reprod Sci 2012; 19:202–209. doi: 10.1177/1933719111417889. [DOI] [PubMed] [Google Scholar]
- 45.Fruscalzo A, Londero AP, Driul L, Henze A, Tonutti L, Ceraudo M, et al. First trimester concentrations of the TTR-RBP4-retinol complex components as early markers of insulin-treated gestational diabetes mellitus. Clin Chem Lab Med 2015; 53:1643–1651. doi: 10.1515/cclm-2014-0929. [DOI] [PubMed] [Google Scholar]
- 46.Gika H, Theodoridis G. Sample preparation prior to the LC-MS-based metabolomics/metabonomics of blood-derived samples. Bioanalysis 2011; 3:1647–1661. doi: 10.4155/bio.11.122. [DOI] [PubMed] [Google Scholar]
- 47.Enquobahrie DA, Denis M, Tadesse MG, Gelaye B, Ressom HW, Williams MA. Maternal early pregnancy serum metabolites and risk of gestational diabetes mellitus. J Clin Endocrinol Metab 2015; 100:4348–4356. doi: 10.1210/jc.2015-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy C, Tremblay PY, Anassour-Laouan-Sidi E, Lucas M, Forest JC, Giguere Y, et al. Risk of gestational diabetes mellitus in relation to plasma concentrations of amino acids and acylcarnitines: a nested case-control study. Diabetes Res Clin Pract 2018; 140:183–190. doi: 10.1016/j.diabres.2018.03.058. [DOI] [PubMed] [Google Scholar]
- 49.Li G, Gao W, Xu Y, Xie M, Tang S, Yin P, et al. Serum metabonomics study of pregnant women with gestational diabetes mellitus based on LC-MS. Saudi J Biol Sci 2019; 26:2057–2063. doi: 10.1016/j.sjbs.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Li H, Chung ACK, Xiang L, Li X, Zheng Y, et al. Large-scale longitudinal metabolomics study reveals different trimester-specific alterations of metabolites in relation to gestational diabetes mellitus. J Proteome Res 2019; 18:292–300. doi: 10.1021/acs.jproteome.8b00602. [DOI] [PubMed] [Google Scholar]
- 51.Hou W, Meng X, Zhao A, Zhao W, Pan J, Tang J, et al. Development of multimarker diagnostic models from metabolomics analysis for gestational diabetes mellitus (GDM). Mol Cell Proteomics 2018; 17:431–441. doi: 10.1074/mcp.RA117.000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T, Li J, Xu F, Wang M, Ding S, Xu H, et al. Comprehensive analysis of serum metabolites in gestational diabetes mellitus by UPLC/Q-TOF-MS. Anal Bioanal Chem 2016; 408:1125–1135. doi: 10.1007/s00216-015-9211-3. [DOI] [PubMed] [Google Scholar]
- 53.Hajduk J, Klupczynska A, Derezinski P, Matysiak J, Kokot P, Nowak DM, et al. A combined metabolomic and proteomic analysis of gestational diabetes mellitus. Int J Mol Sci 2015; 16:30034–30045. doi: 10.3390/ijms161226133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dudzik D, Zorawski M, Skotnicki M, Zarzycki W, Kozlowska G, Bibik-Malinowska K, et al. Metabolic fingerprint of gestational diabetes mellitus. J Proteomics 2014; 103:57–71. doi: 10.1016/j.jprot.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 55.Gao J, Xu B, Zhang X, Cui Y, Deng L, Shi Z, et al. Association between serum bile acid profiles and gestational diabetes mellitus: a targeted metabolomics study. Clin Chim Acta 2016; 459:63–72. doi: 10.1016/j.cca.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 56.Mokkala K, Vahlberg T, Houttu N, Koivuniemi E, Laitinen K. Distinct metabolomic profile because of gestational diabetes and its treatment mode in women with overweight and obesity. Obesity (Silver Spring) 2020; 28:1637–1644. doi: 10.1002/oby.22882. [DOI] [PubMed] [Google Scholar]
- 57.Jiang R, Wu S, Fang C, Wang C, Yang Y, Liu C, et al. Amino acids levels in early pregnancy predict subsequent gestational diabetes. J Diabetes 2020; 12:503–511. doi: 10.1111/1753-0407.13018. [DOI] [PubMed] [Google Scholar]
- 58.Walejko JM, Chelliah A, Keller-Wood M, Wasserfall C, Atkinson M, Gregg A, et al. Diabetes leads to alterations in normal metabolic transitions of pregnancy as revealed by time-course metabolomics. Metabolites 2020; 10:350.doi: 10.3390/metabo10090350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burzynska-Pedziwiatr I, Jankowski A, Kowalski K, Sendys P, Zieleniak A, Cypryk K, et al. Associations of arginine with gestational diabetes mellitus in a follow-up study. Int J Mol Sci 2020; 21:7811.doi: 10.3390/ijms21217811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakurai K, Eguchi A, Watanabe M, Yamamoto M, Ishikawa K, Mori C. Exploration of predictive metabolic factors for gestational diabetes mellitus in Japanese women using metabolomic analysis. J Diabetes Investig 2019; 10:513–520. doi: 10.1111/jdi.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pappa KI, Vlachos G, Theodora M, Roubelaki M, Angelidou K, Antsaklis A. Intermediate metabolism in association with the amino acid profile during the third trimester of normal pregnancy and diet-controlled gestational diabetes. Am J Obstet Gynecol 2007; 196:e1–e5. doi: 10.1016/j.ajog.2006.06.094. [DOI] [PubMed] [Google Scholar]
- 62.She P, Van Horn C, Reid T, Hutson SM, Cooney RN, Lynch CJ. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metab 2007; 293:E1552–E1563. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dani C, Bresci C, Berti E, Ottanelli S, Mello G, Mecacci F, et al. Metabolomic profile of term infants of gestational diabetic mothers. J Matern Fetal Neonatal Med 2014; 27:537–542. doi: 10.3109/14767058.2013.823941. [DOI] [PubMed] [Google Scholar]
- 64.Lu J, Gu Y, Liu H, Wang L, Li W, Li W, et al. Daily branched-chain amino acid intake and risks of obesity and insulin resistance in children: a cross-sectional study. Obesity (Silver Spring) 2020; 28:1310–1316. doi: 10.1002/oby.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandler V, Reisetter AC, Bain JR, Muehlbauer MJ, Nodzenski M, Stevens RD, et al. Associations of maternal BMI and insulin resistance with the maternal metabolome and newborn outcomes. Diabetologia 2017; 60:518–530. doi: 10.1007/s00125-016-4182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Connell TM. The complex role of branched chain amino acids in diabetes and cancer. Metabolites 2013; 3:931–945. doi: 10.3390/metabo3040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 2014; 10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab 2006; 3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 69.Nishitani S, Takehana K, Fujitani S, Sonaka I. Branched-chain amino acids improve glucose metabolism in rats with liver cirrhosis. Am J Physiol Gastrointest Liver Physiol 2005; 288:G1292–G1300. doi: 10.1152/ajpgi.00510.2003. [DOI] [PubMed] [Google Scholar]
- 70.Gonzalez FJ, Jiang C, Patterson AD. An intestinal microbiota-farnesoid X receptor axis modulates metabolic disease. Gastroenterology 2016; 151:845–859. doi: 10.1053/j.gastro.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun L, Pang Y, Wang X, Wu Q, Liu H, Liu B, et al. Ablation of gut microbiota alleviates obesity-induced hepatic steatosis and glucose intolerance by modulating bile acid metabolism in hamsters. Acta Pharm Sin B 2019; 9:702–710. doi: 10.1016/j.apsb.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest 2015; 125:386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L, et al. An intestinal farnesoid X receptor-ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes 2017; 66:613–626. doi: 10.2337/db16-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Law KP, Han TL, Mao X, Zhang H. Tryptophan and purine metabolites are consistently upregulated in the urinary metabolome of patients diagnosed with gestational diabetes mellitus throughout pregnancy: a longitudinal metabolomics study of Chinese pregnant women part 2. Clin Chim Acta 2017; 468:126–139. doi: 10.1016/j.cca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 75.López-Hernández Y, Herrera-Van Oostdam AS, Toro-Ortiz JC, López JA, Salgado-Bustamante M, Murgu M, et al. Urinary metabolites altered during the third trimester in pregnancies complicated by gestational diabetes mellitus: relationship with potential upcoming metabolic disorders. Int J Mol Sci 2019; 20:1186.doi: 10.3390/ijms20051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koos BJ, Gornbein JA. Early pregnancy metabolites predict gestational diabetes mellitus: implications for fetal programming. Am J Obstet Gynecol 2020; 224:215.e1–215.e7. doi: 10.1016/j.ajog.2020.07.050. [DOI] [PubMed] [Google Scholar]
- 77.Lorenzo MP, Dudzik D, Varas E, Gibellini M, Skotnicki M, Zorawski M, et al. Optimization and validation of a chiral GC-MS method for the determination of free D-amino acids ratio in human urine: application to a gestational diabetes mellitus study. J Pharm Biomed Anal 2015; 107:480–487. doi: 10.1016/j.jpba.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 78.Sulek K, Han TL, Villas-Boas SG, Wishart DS, Soh SE, Kwek K, et al. Hair metabolomics: identification of fetal compromise provides proof of concept for biomarker discovery. Theranostics 2014; 4:953–959. doi: 10.7150/thno.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, de Seymour JV, Han TL, Xia Y, Chen C, Zhang T, et al. Metabolomic biomarkers and novel dietary factors associated with gestational diabetes in China. Metabolomics 2018; 14:149.doi: 10.1007/s11306-018-1445-6. [DOI] [PubMed] [Google Scholar]
- 80.He X, de Seymour JV, Sulek K, Qi H, Zhang H, Han TL, et al. Maternal hair metabolome analysis identifies a potential marker of lipid peroxidation in gestational diabetes mellitus. Acta Diabetol 2016; 53:119–122. doi: 10.1007/s00592-015-0737-9. [DOI] [PubMed] [Google Scholar]
- 81.Goyal A, Gupta Y, Kalaivani M, Sankar MJ, Kachhawa G, Bhatla N, et al. Long term ((1year) postpartum glucose tolerance status among Indian women with history of gestational diabetes mellitus (GDM) diagnosed by IADPSG criteria. Diabetes Res Clin Pract 2018; 142:154–161. doi: 10.1016/j.diabres.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 82.Lowe WL, Jr, Scholtens DM, Lowe LP, Kuang A, Nodzenski M, Talbot O, et al. Association of gestational diabetes with maternal disorders of glucose metabolism and childhood adiposity. JAMA 2018; 320:1005–1016. doi: 10.1001/jama.2018.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allalou A, Nalla A, Prentice KJ, Liu Y, Zhang M, Dai FF, et al. A predictive metabolic signature for the transition from gestational diabetes mellitus to type 2 diabetes. Diabetes 2016; 65:2529–2539. doi: 10.2337/db15-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai M, Liu Y, Ronnett GV, Wu A, Cox BJ, Dai FF, et al. Amino acid and lipid metabolism in post-gestational diabetes and progression to type 2 diabetes: a metabolic profiling study. PLoS Med 2020; 17:e1003112.doi: 10.1371/journal.pmed.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andersson-Hall U, Gustavsson C, Pedersen A, Malmodin D, Joelsson L, Holmang A. Higher concentrations of BCAAs and 3-HIB are associated with insulin resistance in the transition from gestational diabetes to type 2 diabetes. J Diabetes Res 2018; 2018:4207067.doi: 10.1155/2018/4207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dudzik D, Zorawski M, Skotnicki M, Zarzycki W, García A, Angulo S, et al. GC-MS based gestational diabetes mellitus longitudinal study: identification of 2-and 3-hydroxybutyrate as potential prognostic biomarkers. J Pharm Biomed Anal 2017; 144:90–98. doi: 10.1016/j.jpba.2017.02.056. [DOI] [PubMed] [Google Scholar]
- 87.Vangipurapu J, Fernandes Silva L, Kuulasmaa T, Smith U, Laakso M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care 2020; 43:1319–1325. doi: 10.2337/dc19-2533. [DOI] [PubMed] [Google Scholar]
- 88.Grant SFA. The TCF7L2 locus: a genetic window into the pathogenesis of type 1 and type 2 diabetes. Diabetes Care 2019; 42:1624–1629. doi: 10.2337/dci19-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pappa KI, Gazouli M, Economou K, Daskalakis G, Anastasiou E, Anagnou NP, et al. Gestational diabetes mellitus shares polymorphisms of genes associated with insulin resistance and type 2 diabetes in the Greek population. Gynecol Endocrinol 2011; 27:267–272. doi: 10.3109/09513590.2010.490609. [DOI] [PubMed] [Google Scholar]
- 90.Huopio H, Cederberg H, Vangipurapu J, Hakkarainen H, Pääkkönen M, Kuulasmaa T, et al. Association of risk variants for type 2 diabetes and hyperglycemia with gestational diabetes. Eur J Endocrinol 2013; 169:291–297. doi: 10.1530/EJE-13-0286. [DOI] [PubMed] [Google Scholar]
- 91.Franzago M, Fraticelli F, Marchetti D, Celentano C, Liberati M, Stuppia L, et al. Nutrigenetic variants and cardio-metabolic risk in women with or without gestational diabetes. Diabetes Res Clin Pract 2018; 137:64–71. doi: 10.1016/j.diabres.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Ding M, Chavarro J, Olsen S, Lin Y, Ley SH, Bao W, et al. Genetic variants of gestational diabetes mellitus: a study of 112 SNPs among 8722 women in two independent populations. Diabetologia 2018; 61:1758–1768. doi: 10.1007/s00125-018-4637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huerta-Chagoya A, Vázquez-Cárdenas P, Moreno-Macías H, Tapia-Maruri L, Rodríguez-Guillén R, López-Vite E, et al. Genetic determinants for gestational diabetes mellitus and related metabolic traits in Mexican women. PLoS One 2015; 10:e0126408.doi: 10.1371/journal.pone.0126408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franzago M, Fraticelli F, Nicolucci A, Celentano C, Liberati M, Stuppia L, et al. Molecular analysis of a genetic variants panel related to nutrients and metabolism: association with susceptibility to gestational diabetes and cardiometabolic risk in affected women. J Diabetes Res 2017; 2017:4612623.doi: 10.1155/2017/4612623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papadopoulou A, Lynch KF, Shaat N, Håkansson R, Ivarsson SA, Berntorp K, et al. Gestational diabetes mellitus is associated with TCF7L2 gene polymorphisms independent of HLA-DQB1∗0602 genotypes and islet cell autoantibodies. Diabet Med 2011; 28:1018–1027. doi: 10.1111/j.1464-5491.2011.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Potasso L, Perakakis N, Lamprinou A, Polyzou E, Kassanos D, Peter A, et al. Clinical impact of the TCF7L2 gene rs7903146 type 2 diabetes mellitus risk polymorphism in women with gestational diabetes mellitus: impaired glycemic control and increased need of insulin therapy. Exp Clin Endocrinol Diabetes 2020; 128:663–666. doi: 10.1055/a-1008-9223. [DOI] [PubMed] [Google Scholar]
- 97.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, et al. A map of open chromatin in human pancreatic islets. Nat Genet 2010; 42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 2007; 117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pilgaard K, Jensen CB, Schou JH, Lyssenko V, Wegner L, Brøns C, et al. The T allele of rs7903146 TCF7L2 is associated with impaired insulinotropic action of incretin hormones, reduced 24 h profiles of plasma insulin and glucagon, and increased hepatic glucose production in young healthy men. Diabetologia 2009; 52:1298–1307. doi: 10.1007/s00125-009-1307-x. [DOI] [PubMed] [Google Scholar]
- 100.Cropano C, Santoro N, Groop L, Dalla Man C, Cobelli C, Galderisi A, et al. The rs7903146 variant in the TCF7L2 gene increases the risk of prediabetes/type 2 diabetes in obese adolescents by impairing beta-cell function and hepatic insulin sensitivity. Diabetes Care 2017; 40:1082–1089. doi: 10.2337/dc17-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pagán A, Sabater-Molina M, Olza J, Prieto-Sánchez MT, Blanco-Carnero JE, Parrilla JJ, et al. A gene variant in the transcription factor 7-like 2 (TCF7L2) is associated with an increased risk of gestational diabetes mellitus. Eur J Obstet Gynecol Reprod Biol 2014; 180:77–82. doi: 10.1016/j.ejogrb.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 102.Karamitri A, Jockers R. Melatonin in type 2 diabetes mellitus and obesity. Nat Rev Endocrinol 2019; 15:105–125. doi: 10.1038/s41574-018-0130-1. [DOI] [PubMed] [Google Scholar]
- 103.Song JY, Wang HJ, Ma J, Xu ZY, Hinney A, Hebebrand J, et al. Association of the rs10830963 polymorphism in MTNR1B with fasting glucose levels in Chinese children and adolescents. Obes Facts 2011; 4:197–203. doi: 10.1159/000329306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Langenberg C, Pascoe L, Mari A, Tura A, Laakso M, Frayling TM, et al. Common genetic variation in the melatonin receptor 1B gene (MTNR1B) is associated with decreased early-phase insulin response. Diabetologia 2009; 52:1537–1542. doi: 10.1007/s00125-009-1392-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spégel P, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 2009; 41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bedoya FJ, Matschinsky FM, Shimizu T, O’Neil JJ, Appel MC. Differential regulation of glucokinase activity in pancreatic islets and liver of the rat. J Biol Chem 1986; 261:10760–10764. doi: 10.1016/S0021-9258(18)67451-4. [PubMed] [Google Scholar]
- 107.Matschinsky FM. Banting Lecture 1995. A lesson in metabolic regulation inspired by the glucokinase glucose sensor paradigm. Diabetes 1996; 45:223–241. doi: 10.2337/diab.45.2.223. [DOI] [PubMed] [Google Scholar]
- 108.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Mägi R, Reschen ME, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 2015; 47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fernandez-Mejia C, Vega-Allende J, Rojas-Ochoa A, Rodriguez-Dorantes M, Romero-Navarro G, Matschinsky FM, et al. Cyclic adenosine 3′,5′-monophosphate increases pancreatic glucokinase activity and gene expression. Endocrinology 2001; 142:1448–1452. doi: 10.1210/endo.142.4.8100. [DOI] [PubMed] [Google Scholar]
- 110.Grotenfelt NE, Wasenius NS, Rönö K, Laivuori H, Stach-Lempinen B, Orho-Melander M, et al. Interaction between rs10830963 polymorphism in MTNR1B and lifestyle intervention on occurrence of gestational diabetes. Diabetologia 2016; 59:1655–1658. doi: 10.1007/s00125-016-3989-1. [DOI] [PubMed] [Google Scholar]
- 111.Firneisz G, Rosta K, Al-Aissa Z, Hadarits O, Harreiter J, Nádasdi Á, et al. The MTNR1B rs10830963 variant in interaction with pre-pregnancy BMI is a pharmacogenetic marker for the initiation of antenatal insulin therapy in gestational diabetes mellitus. Int J Mol Sci 2018; 19:3734.doi: 10.3390/ijms19123734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med 2001; 7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 113.Achari AE, Jain SK. Adiponectin, a therapeutic target for obesity, diabetes, and endothelial dysfunction. Int J Mol Sci 2017; 18:1321.doi: 10.3390/ijms18061321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pawlik A, Teler J, Maciejewska A, Sawczuk M, Safranow K, Dziedziejko V. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J Assist Reprod Genet 2017; 34:511–516. doi: 10.1007/s10815-016-0866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beltcheva O, Boyadzhieva M, Angelova O, Mitev V, Kaneva R, Atanasova I. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch Gynecol Obstet 2014; 289:743–748. doi: 10.1007/s00404-013-3029-z. [DOI] [PubMed] [Google Scholar]
- 116.Feng Y, Jiang CD, Chang AM, Shi Y, Gao J, Zhu L, et al. Interactions among insulin resistance, inflammation factors, obesity-related gene polymorphisms, environmental risk factors, and diet in the development of gestational diabetes mellitus. J Matern Fetal Neonatal Med 2019; 32:339–347. doi: 10.1080/14767058.2018.1446207. [DOI] [PubMed] [Google Scholar]
- 117.Han Y, Zheng YL, Fan YP, Liu MH, Lu XY, Tao Q. Association of adiponectin gene polymorphism 45TG with gestational diabetes mellitus diagnosed on the new IADPSG criteria, plasma adiponectin levels and adverse pregnancy outcomes. Clin Exp Med 2015; 15:47–53. doi: 10.1007/s10238-014-0275-8. [DOI] [PubMed] [Google Scholar]
- 118.Low CF, Mohd Tohit ER, Chong PP, Idris F. Adiponectin SNP45TG is associated with gestational diabetes mellitus. Arch Gynecol Obstet 2011; 283:1255–1260. doi: 10.1007/s00404-010-1548-4. [DOI] [PubMed] [Google Scholar]
- 119.Takhshid MA, Haem Z, Aboualizadeh F. The association of circulating adiponectin and + 45 T/G polymorphism of adiponectin gene with gestational diabetes mellitus in Iranian population. J Diabetes Metab Disord 2015; 14:30.doi: 10.1186/s40200-015-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakatani K, Noma K, Nishioka J, Kasai Y, Morioka K, Katsuki A, et al. Adiponectin gene variation associates with the increasing risk of type 2 diabetes in non-diabetic Japanese subjects. Int J Mol Med 2005; 15:173–177. [PubMed] [Google Scholar]
- 121.Matharoo K, Arora P, Bhanwer AJ. Association of adiponectin (AdipoQ) and sulphonylurea receptor (ABCC8) gene polymorphisms with Type 2 Diabetes in North Indian population of Punjab. Gene 2013; 527:228–234. doi: 10.1016/j.gene.2013.05.075. [DOI] [PubMed] [Google Scholar]
- 122.Choe EY, Wang HJ, Kwon O, Kim KJ, Kim BS, Lee BW, et al. Variants of the adiponectin gene and diabetic microvascular complications in patients with type 2 diabetes. Metabolism 2013; 62:677–685. doi: 10.1016/j.metabol.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 123.Dziedziejko V, Safranow K, Tarnowski M, Pawlik A. Common type 2 diabetes genetic risk variants improve the prediction of gestational diabetes. Horm Metab Res 2019; 51:655–660. doi: 10.1055/a-0945-0328. [DOI] [PubMed] [Google Scholar]
- 124.Kasuga Y, Hata K, Tajima A, Ochiai D, Saisho Y, Matsumoto T, et al. Association of common polymorphisms with gestational diabetes mellitus in Japanese women: a case-control study. Endocr J 2017; 64:463–475. doi: 10.1507/endocrj.EJ16-0431. [DOI] [PubMed] [Google Scholar]
- 125.Kwak SH, Choi SH, Jung HS, Cho YM, Lim S, Cho NH, et al. Clinical and genetic risk factors for type 2 diabetes at early or late post partum after gestational diabetes mellitus. J Clin Endocrinol Metab 2013; 98:E744–E752. doi: 10.1210/jc.2012-3324. [DOI] [PubMed] [Google Scholar]
- 126.Nisa H, Qi KHT, Leng J, Zhou T, Liu H, Li W, et al. The circadian rhythm-related MTNR1B genotype, gestational weight gain, and postpartum glycemic changes. J Clin Endocrinol Metab 2018; 103:2284–2290. doi: 10.1210/jc.2018-00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.de Carvalho AM, Shao P, Liu H, Cheng HL, Zheng Y, Leng J, et al. The MC4R genotype is associated with postpartum weight reduction and glycemic changes among women with prior gestational diabetes: longitudinal analysis. Sci Rep 2017; 7:9654.doi: 10.1038/s41598-017-10101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kwak SH, Choi SH, Kim K, Jung HS, Cho YM, Lim S, et al. Prediction of type 2 diabetes in women with a history of gestational diabetes using a genetic risk score. Diabetologia 2013; 56:2556–2563. doi: 10.1007/s00125-013-3059-x. [DOI] [PubMed] [Google Scholar]
- 129.Cormier H, Vigneault J, Garneau V, Tchernof A, Vohl MC, Weisnagel SJ, et al. An explained variance-based genetic risk score associated with gestational diabetes antecedent and with progression to pre-diabetes and type 2 diabetes: a cohort study. BJOG 2015; 122:411–419. doi: 10.1111/1471-0528.12937. [DOI] [PubMed] [Google Scholar]
- 130.Herrera-Van Oostdam AS, Toro-Ortíz JC, López JA, Noyola DE, García-López DA, Durán-Figueroa NV, et al. Placental exosomes isolated from urine of patients with gestational diabetes exhibit a differential profile expression of microRNAs across gestation. Int J Mol Med 2020; 46:546–560. doi: 10.3892/ijmm.2020.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cao YL, Jia YJ, Xing BH, Shi DD, Dong XJ. Plasma microRNA-16-5p, -17-5p and -20a-5p: novel diagnostic biomarkers for gestational diabetes mellitus. J Obstet Gynaecol Res 2017; 43:974–981. doi: 10.1111/jog.13317. [DOI] [PubMed] [Google Scholar]
- 132.Zhu Y, Tian F, Li H, Zhou Y, Lu J, Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynaecol Obstet 2015; 130:49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 133.Wang F, Zhang X, Zhou H. Role of cell free microRNA-19a and microRNA-19b in gestational diabetes mellitus patients. 3 Biotech 2019; 9:406.doi: 10.1007/s13205-019-1952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tagoma A, Alnek K, Kirss A, Uibo R, Haller-Kikkatalo K. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene 2018; 672:137–142. doi: 10.1016/j.gene.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 135.Floris I, Descamps B, Vardeu A, Mitic T, Posadino AM, Shantikumar S, et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler Thromb Vasc Biol 2015; 35:664–674. doi: 10.1161/ATVBAHA.114.304730. [DOI] [PubMed] [Google Scholar]
- 136.Peng HY, Li HP, Li MQ. High glucose induces dysfunction of human umbilical vein endothelial cells by upregulating miR-137 in gestational diabetes mellitus. Microvasc Res 2018; 118:90–100. doi: 10.1016/j.mvr.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 137.Lamadrid-Romero M, Solís KH, Cruz-Reséndiz MS, Pérez JE, Díaz NF, Flores-Herrera H, et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci Res 2018; 130:8–22. doi: 10.1016/j.neures.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 138.Liao X, Zhou Z, Zhang X. Effects of miR1955p on cell proliferation and apoptosis in gestational diabetes mellitus via targeting EZH2. Mol Med Rep 2020; 22:803–809. doi: 10.3892/mmr.2020.11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang J, Pan Y, Dai F, Wang F, Qiu H, Huang X. Serum miR-195-5p is upregulated in gestational diabetes mellitus. J Clin Lab Anal 2020; 34:e23325.doi: 10.1002/jcla.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tang L, Li P, Li L. Whole transcriptome expression profiles in placenta samples from women with gestational diabetes mellitus. J Diabetes Investig 2020; 11:1307–1317. doi: 10.1111/jdi.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yoffe L, Polsky A, Gilam A, Raff C, Mecacci F, Ognibene A, et al. Early diagnosis of gestational diabetes mellitus using circulating microRNAs. Eur J Endocrinol 2019; 181:565–577. doi: 10.1530/EJE-19-0206. [DOI] [PubMed] [Google Scholar]
- 142.Abdeltawab A, Zaki ME, Abdeldayem Y, Mohamed AA, Zaied SM. Circulating micro RNA-223 and angiopoietin-like protein 8 as biomarkers of gestational diabetes mellitus. Br J Biomed Sci 2021; 78:12–17. doi: 10.1080/09674845.2020.1764211. [DOI] [PubMed] [Google Scholar]
- 143.Martínez-Ibarra A, Martínez-Razo LD, Vázquez-Martínez ER, Martínez-Cruz N, Flores-Ramírez R, García-Gómez E, et al. Unhealthy levels of phthalates and bisphenol a in Mexican pregnant women with gestational diabetes and its association to altered expression of mirnas involved with metabolic disease. Int J Mol Sci 2019; 20:3343.doi: 10.3390/ijms20133343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pfeiffer S, Sánchez-Lechuga B, Donovan P, Halang L, Prehn JHM, Campos-Caro A, et al. Circulating miR-330-3p in late pregnancy is associated with pregnancy outcomes among lean women with GDM. Sci Rep 2020; 10:908.doi: 10.1038/s41598-020-57838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gillet V, Ouellet A, Stepanov Y, Rodosthenous RS, Croft EK, Brennan K, et al. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab 2019; 104:5157–5169. doi: 10.1210/jc.2018-02693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang P, Wang H, Li C, Zhang X, Xiu X, Teng P, et al. Dysregulation of microRNA-657 influences inflammatory response via targeting interleukin-37 in gestational diabetes mellitus. J Cell Physiol 2019; 234:7141–7148. doi: 10.1002/jcp.27468. [DOI] [PubMed] [Google Scholar]
- 147.Wang P, Wang Z, Liu G, Jin C, Zhang Q, Man S, et al. miR-657 promotes macrophage polarization toward M1 by targeting FAM46C in gestational diabetes mellitus. Mediators Inflamm 2019; 2019:4851214.doi: 10.1155/2019/4851214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol 2010; 71:206–211. doi: 10.1016/j.humimm.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 149.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. J Immunol 2011; 187:5834–5841. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 2011; 50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 151.Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 2009; 113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 2011; 9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 153.Li S, Li Z, Guo F, Qin X, Liu B, Lei Z, et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci 2011; 18:24.doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010; 107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 155.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 2010; 86:410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 156.Haneklaus M, Gerlic M, O’Neill LAJ. Masters SL. miR-223: infection, inflammation and cancer. J Intern Med 2013; 274:215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu Y, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One 2011; 6:e23925.doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deng L, Huang Y, Li L, Chen H, Su J. Serum miR-29a/b expression in gestational diabetes mellitus and its influence on prognosis evaluation. J Int Med Res 2020; 48:300060520954763.doi: 10.1177/0300060520954763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tung CW, Ho C, Hsu YC, Huang SC, Shih YH, Lin CL. MicroRNA-29a attenuates diabetic glomerular injury through modulating cannabinoid receptor 1 signaling. Molecules 2019; 24:264.doi: 10.3390/molecules24020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang YL, Chen XQ. Dysregulation of microRNA-770-5p influences pancreatic-beta-cell function by targeting TP53 regulated inhibitor of apoptosis 1 in gestational diabetes mellitus. Eur Rev Med Pharmacol Sci 2020; 24:793–801. doi: 10.26355/eurrev_202001_20062. [DOI] [PubMed] [Google Scholar]
- 161.Li L, Wang S, Li H, Wan J, Zhou Q, Zhou Y, et al. microRNA-96 protects pancreatic beta-cell function by targeting PAK1 in gestational diabetes mellitus. Biofactors 2018; 44:539–547. doi: 10.1002/biof.1461. [DOI] [PubMed] [Google Scholar]
- 162.Sun DG, Tian S, Zhang L, Hu Y, Guan CY, Ma X, et al. The miRNA-29b is downregulated in placenta during gestational diabetes mellitus and may alter placenta development by regulating trophoblast migration and invasion through a HIF3A-dependent mechanism. Front Endocrinol (Lausanne) 2020; 11:169.doi: 10.3389/fendo.2020.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou X, Xiang C. Zheng X. miR-132 serves as a diagnostic biomarker in gestational diabetes mellitus and its regulatory effect on trophoblast cell viability. Diagn Pathol 2019; 14:119.doi: 10.1186/s13000-019-0899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao C, Zhang T, Shi Z, Ding H, Ling X. MicroRNA-518d regulates PPARalpha protein expression in the placentas of females with gestational diabetes mellitus. Mol Med Rep 2014; 9:2085–2090. doi: 10.3892/mmr.2014.2058. [DOI] [PubMed] [Google Scholar]
- 165.Hromadnikova I, Kotlabova K, Dvorakova L, Krofta L. Diabetes mellitus and cardiovascular risk assessment in mothers with a history of gestational diabetes mellitus based on postpartal expression profile of micrornas associated with diabetes mellitus and cardiovascular and cerebrovascular diseases. Int J Mol Sci 2020; 21:2437.doi: 10.3390/ijms21072437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee IR, Dawson SA, Wetherall JD, Hahnel R. Sex hormone-binding globulin secretion by human hepatocarcinoma cells is increased by both estrogens and androgens. J Clin Endocrinol Metab 1987; 64:825–831. doi: 10.1210/jcem-64-4-825. [DOI] [PubMed] [Google Scholar]
- 167.Maged AM, Moety GA, Mostafa WA, Hamed DA. Comparative study between different biomarkers for early prediction of gestational diabetes mellitus. J Matern Fetal Neonatal Med 2014; 27:1108–1112. doi: 10.3109/14767058.2013.850489. [DOI] [PubMed] [Google Scholar]
- 168.Zhang T, Du T, Li W, Yang S, Liang W. Sex hormone-binding globulin levels during the first trimester may predict gestational diabetes mellitus development. Biomark Med 2018; 12:239–244. doi: 10.2217/bmm-2016-0030. [DOI] [PubMed] [Google Scholar]
- 169.Hedderson MM, Xu F, Darbinian JA, Quesenberry CP, Sridhar S, Kim C, et al. Prepregnancy SHBG concentrations and risk for subsequently developing gestational diabetes mellitus. Diabetes Care 2014; 37:1296–1303. doi: 10.2337/dc13-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Haffner SM, Katz MS, Stern MP, Dunn JF. The relationship of sex hormones to hyperinsulinemia and hyperglycemia. Metabolism 1988; 37:683–688. doi: 10.1016/0026-0495(88)90091-1. [DOI] [PubMed] [Google Scholar]
- 171.Plymate SR, Matej LA, Jones RE, Friedl KE. Inhibition of sex hormone-binding globulin production in the human hepatoma (Hep G2) cell line by insulin and prolactin. J Clin Endocrinol Metab 1988; 67:460–464. doi: 10.1210/jcem-67-3-460. [DOI] [PubMed] [Google Scholar]
- 172.Selva DM, Hogeveen KN, Innis SM, Hammond GL. Monosaccharide-induced lipogenesis regulates the human hepatic sex hormone-binding globulin gene. J Clin Invest 2007; 117:3979–3987. doi: 10.1172/JCI32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yasui T, Tomita J, Miyatani Y, Yamada M, Uemura H, Irahara M, et al. Associations of adiponectin with sex hormone-binding globulin levels in aging male and female populations. Clin Chim Acta 2007; 386:69–75. doi: 10.1016/j.cca.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 174.Akin F, Bastemir M, Alkis E. Effect of insulin sensitivity on SHBG levels in premenopausal versus postmenopausal obese women. Adv Ther 2007; 24:1210–1220. doi: 10.1007/BF02877767. [DOI] [PubMed] [Google Scholar]
- 175.Wang X, Chi X, Feng C, Zhang X, Jin Z. Sex hormone-binding globulin regulates the activity of the ERK pathway in the placentas of patients with gestational diabetes mellitus. Biochem Biophys Res Commun 2020; 532:613–619. doi: 10.1016/j.bbrc.2020.08.100. [DOI] [PubMed] [Google Scholar]
- 176.Bost F, Aouadi M, Caron L, Even P, Belmonte N, Prot M, et al. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes 2005; 54:402–411. doi: 10.2337/diabetes.54.2.402. [DOI] [PubMed] [Google Scholar]
- 177.Jager J, Gremeaux T, Cormont M, Le Marchand-Brustel Y, Tanti JF. Interleukin-1beta-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology 2007; 148:241–251. doi: 10.1210/en.2006-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chi X, Feng C, Wang X, Jin Z. Sex hormone-binding globulin regulates glucose metabolism in human placental trophoblasts via cAMP/PKA/CREB1. J Obstet Gynaecol Res 2020; 46:2340–2346. doi: 10.1111/jog.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shen L, Zhao D, Chen Y, Zhang K, Chen X, Lin J, et al. Comparative proteomics analysis of serum proteins in gestational diabetes during early and middle stages of pregnancy. Proteomics Clin Appl 2019; 13:e1800060.doi: 10.1002/prca.201800060. [DOI] [PubMed] [Google Scholar]
- 180.Mavreli D, Evangelinakis N, Papantoniou N, Kolialexi A. Quantitative comparative proteomics reveals candidate biomarkers for the early prediction of gestational diabetes mellitus: a preliminary study. In Vivo 2020; 34:517–525. doi: 10.21873/invivo.11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhao D, Shen L, Wei Y, Xie J, Chen S, Liang Y, et al. Identification of candidate biomarkers for the prediction of gestational diabetes mellitus in the early stages of pregnancy using iTRAQ quantitative proteomics. Proteomics Clin Appl 2017; 11.doi: 10.1002/prca.201600152. [DOI] [PubMed] [Google Scholar]
- 182.Claesson R, Ignell C, Shaat N, Berntorp K. HbA1c as a predictor of diabetes after gestational diabetes mellitus. Prim Care Diabetes 2017; 11:46–51. doi: 10.1016/j.pcd.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 183.Kwon SS, Kwon JY, Park YW, Kim YH, Lim JB. HbA1c for diagnosis and prognosis of gestational diabetes mellitus. Diabetes Res Clin Pract 2015; 110:38–43. doi: 10.1016/j.diabres.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 184.Coetzee A, Mason D, Hall DR, Hoffmann M, Conradie M. Evidence for the utility of antenatal HbA1c to predict early postpartum diabetes after gestational diabetes in South Africa. Diabetes Res Clin Pract 2018; 143:50–55. doi: 10.1016/j.diabres.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 185.Lappas M, Jinks D, Shub A, Willcox JC, Georgiou HM, Permezel M. Postpartum IGF-I and IGFBP-2 levels are prospectively associated with the development of type 2 diabetes in women with previous gestational diabetes mellitus. Diabetes Metab 2016; 42:442–447. doi: 10.1016/j.diabet.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 186.Lappas M, Georgiou HM, Velagic A, Willcox JC, Permezel M, Shub A. Do postpartum levels of apolipoproteins prospectively predict the development of type 2 diabetes in women with previous gestational diabetes mellitus? Exp Clin Endocrinol Diabetes 2019; 127:353–358. doi: 10.1055/a-0577-7700. [DOI] [PubMed] [Google Scholar]
- 187.Draznin B, Aroda VR, Bakris G, Benson G, et al. American Diabetes Association Professional Practice Committee, American Diabetes Association Professional Practice Committee. 15. management of diabetes in pregnancy: standards of medical care in diabetes-2022. Diabetes Care 2022; 45:S232–S243. doi: 10.2337/dc22-S015. [DOI] [PubMed] [Google Scholar]
- 188.Blatt AJ, Nakamoto JM, Kaufman HW. Gaps in diabetes screening during pregnancy and postpartum. Obstet Gynecol 2011; 117:61–68. doi: 10.1097/AOG.0b013e3181fe424b. [DOI] [PubMed] [Google Scholar]