Abstract
Since the arrival of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in December 2019, its characterization as a novel human pathogen, and the resulting coronavirus disease 2019 (COVID-19) pandemic, over 6.5 million people have died worldwide—a stark and sobering reminder of the fundamental and nonredundant roles of the innate and adaptive immune systems in host defense against emerging pathogens. Inborn errors of immunity (IEI) are caused by germline variants, typically in single genes. IEI are characterized by defects in development and/or function of cells involved in immunity and host defense, rendering individuals highly susceptible to severe, recurrent, and sometimes fatal infections, as well as immune dysregulatory conditions such as autoinflammation, autoimmunity, and allergy. The study of IEI has revealed key insights into the molecular and cellular requirements for immune-mediated protection against infectious diseases. Indeed, this has been exemplified by assessing the impact of SARS-CoV-2 infection in individuals with previously diagnosed IEI, as well as analyzing rare cases of severe COVID-19 in otherwise healthy individuals. This approach has defined fundamental aspects of mechanisms of disease pathogenesis, immunopathology in the context of infection with a novel pathogen, and therapeutic options to mitigate severe disease. This review summarizes these findings and illustrates how the study of these rare experiments of nature can inform key features of human immunology, which can then be leveraged to improve therapies for treating emerging and established infectious diseases.
Key words: SARS-CoV-2, COVID-19, inborn errors of immunity, primary immune deficiencies, immune dysregulation, type I IFN signaling, cytokine storm
Inborn errors of immunity (IEI) are diseases caused by germline pathogenic variants, typically in single genes.1, 2, 3, 4 IEI have an incidence of ∼1 per 5,000 to 10,000 individuals.1, 2, 3, 4, 5 Currently, pathogenic variants in more than 480 genes have been identified that cause IEI. These variants can lead to loss of expression, complete (null) or partial (hypomorphic) loss of function, gain of function (GOF; hypermorphic), haploinsufficiency, or dominant negative function of the encoded protein. IEI can present as autosomal dominant (AD; heterozygous variants), autosomal recessive (AR; homozygous/compound heterozygous variants), or X-linked (XL) recessive (hemizygous in male subjects; homozygous or heterozygous with skewed X inactivation in female subjects) conditions.4 , 6 However, some IEI have incomplete penetrance, with a significant proportion of individuals carrying some pathogenic variants compromising protein function remaining unaffected.7 The mechanism or mechanisms underlying incomplete penetrance remain unclear but may involve epistatic effects of modifier genes, epigenetics, and/or variants in additional genes.7 It is also worth noting that a monogenic cause for the most common IEI—common variable immunodeficiency (CVID)—has only been determined for ∼20-30% of affected individuals,8 thus suggesting that most cases of CVID are likely to be oligo- or polygenic.
IEI are characterized by defects in immune cell development, and/or impaired innate and adaptive immune function of hematopoietic and nonhematopoietic cells. Consequently, affected individuals are highly susceptible to severe, recurrent, and sometimes fatal infections.4 , 6 As a result of this immunodeficient state, vaccine efficacy can also be compromised in IEI, resulting in affected individuals having modest, if any, vaccine-induced immunity against infectious diseases. Thus, IEI patients continue to be susceptible to infection as well as being vulnerable to disease as a result of live-attenuated vaccines.9
Although historically considered to be immune deficiencies manifesting as infections, the clinical spectrum of IEI is extremely broad, with autoimmunity, autoinflammatory diseases, allergy, bone marrow failure, and/or malignancy also being common maladies of patients.1 , 3 , 4 , 6 , 10 , 11 Although most are individually rare, IEI are collectively common5 and have enabled the delineation of fundamental roles of individual genes, proteins, signaling pathways, and cell types in immune cell development; immune homeostasis and regulation; antitumor immunity; and host defense against infectious diseases.1, 2, 3 Thus, IEI provide insights into the molecular pathogenesis of more common diseases and have led to the development of targeted therapies for various immune dyscrasias.1, 2, 3 , 12
SARS-CoV-2 and the COVID-19 pandemic
Coronaviruses have caused pandemics in the human population for decades.13 Certainly we would have a short memory if we failed to recall the deadly toll of the original SARS coronavirus outbreak in 2002-3.13 In December 2019, the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged from Wuhan, China, and then spread rapidly to cause a catastrophic global pandemic.14 At the time of writing, more than 650 million people have been infected and at least 6.6 million people have died from SARS-CoV-2 infection (www.covid19.who.int/, www.worldometers.info/coronavirus/). The clinical spectrum of coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 infection ranges from asymptomatic to life-threatening disease. The global case fatality rate (CFR) due to SARS-CoV-2 infection is currently ∼1.1%, but this varies widely across different countries, ranging from 0.1% to 5%, and even up to 10% to 15% for some regions (www.ourworldindata.org/grapher/deaths-covid-19-vs-case-fatality-rate). Importantly, early during the pandemic, when viral screening was restricted to symptomatic individuals and vaccines were still 12 to 18 months away, the average global CFR was 5% to 7%, and as high as 10% to 20% in the United Kingdom and some European countries15 , 16 (www.ourworldindata.org/grapher/deaths-covid-19-vs-case-fatality-rate).
Several risk factors have been identified for developing severe disease, as defined by the World Health Organization. These include primarily age, with the frequency of severe cases/death escalating with each decade of increasing age. For example, the mortality rate for people aged <50 years was <1.0%; for individuals aged 60-80 or more years, the mortality rate was ∼4% to 25%. Male sex as well as comorbidities such as cardiovascular/pulmonary disease, obesity, diabetes, and liver/kidney dysfunction also have an impact, albeit less than age.16, 17, 18, 19 Correlates of severe disease and mortality include lymphopenia, increased levels of inflammatory mediators, cytokines, chemokines,18 , 20, 21, 22, 23, 24, 25, 26 and complement components,27, 28, 29 which indicate the intense immune activation and inflammation that can lead to severe and potentially fatal SARS-CoV-2–induced cytokine storm and consequent tissue pathology.
In healthy individuals, SARS-CoV-2 infection induces functional CD4+ and CD8+ T cells and memory B cells specific for viral epitopes, as well as neutralizing antibodies.30, 31, 32, 33, 34, 35, 36, 37, 38, 39 These correlates of protective immunity are detectable 1 or 2 weeks after infection and persist at peak levels for 3 to 4 months. However, in most cases, levels of neutralizing IgG and of SARS-CoV-2–specific memory B cells and T cells dramatically wane 8 to 12 months after infection,32, 33, 34 , 36, 37, 38, 39, 40 potentially compromising host defense against subsequent infections. Furthermore, several SARS-CoV-2 variants that have acquired mutations in the immunodominant spike domain, thus rendering these variants less susceptible to antibody-mediated neutralization, have emerged.41 Waning of acquired immunity after natural infection, combined with immune-escape variants, are a significant challenge in controlling SARS-CoV-2 infection, resulting in COVID-19 continuing to represent a significant global health risk.
SARS-CoV-2 infection, COVID-19, and IEI
Since the beginning of the pandemic, it was recognized that people diagnosed with an IEI were potentially at risk of developing severe COVID-19. Over the past 2 years, outcomes of SARS-CoV-2 infection have been reported for ∼1330 individuals with IEI. These studies range from reports of single cases or small numbers of patients42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88 to cohort studies from Iran,89, 90, 91 Turkey,92, 93, 94 Brazil,95 , 96 Israel,97 Italy,98, 99, 100, 101 Spain,102 the United Kingdom,15 , 103 , 104 Mexico,105 Denmark,106 , 107 Poland,108 the Czech Republic,109 France,110 and the United States,111, 112, 113, 114 as well as an international survey of 94 patients followed in 12 countries.115 These studies have revealed key outcomes of SARS-CoV-2 infection in IEI and defined fundamental requirements for host defense against infection.
Patients with IEI infected with SARS-CoV-2
Affected patients have been found to represent most, if not all, categories of IEI as defined by the International Union of Immunological Societies Committee (Table I ).6 Of the ∼1330 patients reported so far, approximately 60% have antibody deficiencies, consistent with antibody deficiency being the most common IEI.6 , 8 This includes CVID, hypogammaglobulinemia, and specific antibody and immunoglobulin subclass deficiencies due to unknown genetic causes8 (∼600 cases), as well as XL (BTK pathogenic variants) and AR (eg, TCF3 pathogenic variants) agammaglobulinemia (∼110 cases) and a series of patients with pathogenic variants in single genes known to disrupt B-cell function and humoral immunity, such as NFKB1, NFKB2, PIK3CD, or PIK3R1 (Table I). Outcomes of SARS-CoV-2 infections have also been reported for patients with the following:
-
•
Severe combined (JAK3, RAG, IL7RA, DCLRE1C) or combined (CD40LG, RASGRP1, RELB, STK4, WAS, ICOS, ATM, IKBKG, STAT3 DN, PGM3) immunodeficiencies.
-
•
Immune dysregulatory disorders (STAT3 GOF, AIRE, CTLA4, CD70, LRBA, RAB27A, SH2D1A, XIAP, RLTPR/CARML2, CD137, STXBP2, ALPS).
-
•
Phagocytic defects (chronic granulomatous disease, GATA2).
-
•
Innate immune defects (IFNGR1, IFNGR2, IFNAR1, IFNAR2, IL12RB1, IRAK4, MYD88, STAT1 GOF, CXCR4, TBK1, TLR3, TLR7, IRF3, IRF7, IRF9).
-
•
Autoinflammatory disorders (MEFV, TNFAIP3, IL36R, ADA2).
-
•
Complement deficiencies.
-
•
Phenocopies of IEI.
Table I.
SARS-CoV-2 infection in defined IEI
| Type of IEI | Gene defect/IEI | Approximate no. of patients | Study or studies |
|---|---|---|---|
| Severe combined immunodeficiency (n = 25) | JAK3 | 1 | 70 |
| RAG | 3 | 92, 97, 115 | |
| IL7RA | 1 | 91, 94 | |
| DCLRE1C | 1 | 49 | |
| IL2RG | 4 | 77, 95, 115 | |
| CD3D | 1 | 105 | |
| Not specified | 15 | 95, 99, 108 | |
| Combined immunodeficiency (n = 91) | STAT3 DN | 7 | 103, 109, 115, 176 |
| PGM3 | 1 | 102, 115 | |
| ARPC1B | 1 | 47, 105, 115 | |
| WAS | 8 | 47, 48, 95, 99, 100, 103, 105, 108, 109, 115 | |
| ZAP70 | 1 | 115 | |
| CD40L | 9 | 94, 95, 97, 103, 109, 111, 116, 143 | |
| RASGRP1 | 1 | 92 | |
| CARD11 | 1 | 92, 103 | |
| RELB | 3 | 97, 116 | |
| STK4 | 1 | 89 | |
| DNMT3B/NBS1 | 4 | 89, 91, 94 | |
| ICOS | 1 | 15, 103 | |
| IKBKG (NEMO) | 3 | 72, 78, 94 | |
| ATM | 11 | 91, 92, 94, 99, 100, 102, 103, 108 | |
| Di George syndrome | 16 | 99, 100, 108 | |
| Not specified | 23 | 89, 92, 94, 95, 99, 103, 108 | |
| Predominantly antibody deficient (n = 714) | CVID∗ | 589 | 51, 52, 58, 71, 75, 83, 92, 94, 95, 97, 98, 99, 100, 102, 103, 104, 105, 106, 107, 108, 109, 111, 112, 113, 114, 115, 143 |
| BTK | 98 | 15, 46, 51, 53, 55, 60, 61, 66, 73, 85, 86, 91, 92, 94, 95, 97, 98, 99, 100, 102, 103, 104, 105, 108, 109, 111, 115, 116, 139, 140, 143 | |
| AR agammaglobulinemia | 9 | 99, 100, 115 | |
| PIK3R1/PIK3CD GOF | 7 | 64, 82, 91, 95, 99, 100, 115 | |
| NFKB1 | 4 | 15, 91, 103, 111, 115 | |
| NFKB2 | 3 | 43, 103, 115, 143 | |
| IKZF1 | 1 | 91 | |
| Immune dysregulation (n = 64) | AIRE (APS1/APECED) | 29 | 57, 84, 94, 118, 122, 149 |
| CTLA4 | 7 | 15, 97, 103, 115, 177 | |
| LRBA | 3 | 92, 97, 115 | |
| SOCS1 | 1 | 76 | |
| STAT3 GOF | 1 | 111 | |
| RAB27A | 1 | 89 | |
| CD70 | 1 | 89 | |
| ALPS | 5 | 95, 99, 102, 108 | |
| XLP (XIAP, SH2D1A) | 4 | 63, 95, 108, 109, 115 | |
| PRKCD | 1 | 115 | |
| RLTPR/CARMIL2 | 2 | 94 | |
| CD137 | 1 | 94 | |
| STXBP2 | 2 | 88, 94 | |
| Not specified/other | 6 | 92, 99, 105, 108 | |
| Phagocytic defects, bone marrow failure (n = 36) | Chronic granulomatous disease (CYBB; NCF2) | 28 | 15, 59, 89, 95, 97, 102, 103, 105, 108, 115 |
| GATA2 | 2 | 15, 103, 115 | |
| DNAJC21 | 1 | 115 | |
| Not specified/other | 5 | 92, 99 | |
| Innate immune defects (n = 75) | TLR3/UNC93B/TRIF/IRF3/IRF7/IRF9/TBK1 | 23 | 65, 68, 69, 120, 123 |
| IFNAR1/2 | 7 | 42, 56, 87, 126 | |
| STAT1/TYK2 | 2 | 126 | |
| TLR7 | 22 | 90, 124, 125, 126 | |
| MYD88/IRAK4 | 8 | 45, 81, 95, 99, 102 | |
| IFNGR1/IFNGR2/IL12RB1 | 5 | 54, 79, 95, 111, 115 | |
| STAT1 GOF | 6 | 50, 92, 95, 102, 109, 115 | |
| CXCR4 GOF | 2 | 94, 95 | |
| Autoinflammatory disorders (n = 96) | MEFV | 68 | 93, 95, 110, 115 |
| IL1RN | 1 | 89 | |
| Aicardi-Goutières syndrome (RNASEH2B, SAMHD1) | 5 | 15, 99, 100, 115 | |
| TNFAIP3 | 1 | 15 | |
| NLRP1, NLRP3, NLRP12 | 3 | 91, 95 | |
| IL36RN | 1 | 74 | |
| ADA2 | 1 | 94 | |
| Not specified/other | 16 | 95, 108 | |
| Complement deficiencies (n = 55) | Hereditary angioedema (pathogenic SERPING variants), C3 deficiency, other | 55 | 15, 91, 95, 96, 109 |
| Phenocopies of IEI | Good syndrome | 13 | 83, 100, 103, 105, 109 |
| Autoantibodies to type I IFNs | Many! | 128, 129, 130, 131, 132, 133, 134, 135, 136 |
Including hypogamma, immunoglobulin subclass deficiency, and specific antibody deficiency.
A complete reference listing is provided in Table I.
Clinical features in IEI after SARS-CoV-2 infection
The clinical presentation of SARS-CoV-2 infection in patients with IEI resembled that of the general population16 , 19 inasmuch that symptoms frequently include fever, cough, headache, upper respiratory symptoms, fatigue, and dyspnea.15 , 89 , 91 , 92 , 94 , 95 , 97 , 99 , 102 , 103 , 105 , 106 , 108 , 109 , 115 Similarly, risk factors for hospital/intensive care unit (ICU) admission and developing severe and/or fatal disease were also consistent with those determined from studies of the general population. Thus, the most severe disease was observed in older patients with IEI as well as those with pre-existing comorbidities, such as previous infection; lung, kidney, heart, or gut disease, diabetes, and obesity; or after solid organ or hematopoietic stem cell transplantation.43 , 58 , 89 , 91 , 92 , 94 , 95 , 98 , 103 , 105 , 106 , 108 , 111 , 112 , 115 Other predictors of severe disease in IEI patients included leukopenia (reduced numbers of B, CD4+ T, and natural killer cells) and hypogammaglobulinemia/low IgG trough levels before infection, and increased levels of markers of systemic inflammation after infection.43 , 46 , 58 , 66 , 103 , 109 , 111 , 114 Interestingly, and similar to the general population, ∼10% to 20% of infected IEI patients were asymptomatic, and up to another ∼30% to 50% developed only mild disease.15 , 89 , 90 , 92, 93, 94, 95 , 97, 98, 99, 100, 101, 102, 103, 104, 105, 106 , 108, 109, 110, 111, 112, 113, 114, 115
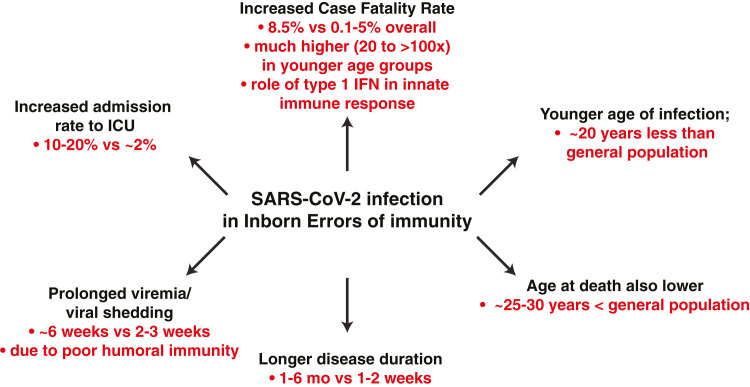
Despite such similarities in disease presentation and risk factors for the general population and IEI patients, there were notable differences. First, the age of affected IEI patients was markedly younger than the general population (∼28 years vs ∼50-plus years).16 , 44 , 79 , 81 , 84 , 89 , 91 , 93, 94, 95 , 100 , 102, 103, 104 , 108, 109, 110 , 115 There were also differences in age at infection for different IEI. Thus, SARS-CoV-2–infected patients with CVID, periodic fevers, or complement defects were generally older, and patients with defects in innate immune cell signaling due to pathogenic variants in IRAK4, MYD88, or IFNAR1/IFNAR2 were generally younger, than the entire cohort of published IEI patients (Fig 1 , A). Second, the proportion of IEI patients admitted to ICU—including younger individuals—was substantially higher than the general population (10-30% vs 2-5%).15 , 16 , 44 , 81 , 91 , 92 , 94 , 102 , 105 , 109 , 111 , 115 Third, duration of disease—likely a result of prolonged viremia and virus shedding—was longer (1-6 months vs 1-2 weeks), and the likelihood of reinfection was greater, than observed for the general population.46 , 55 , 59 , 77 , 83 , 84 , 92 , 99 , 100 , 104 , 106 , 107 , 116 Thus, COVID-19 generally manifests clinically at a younger age, runs a more protracted course, and has a more severe outcome requiring hospitalization and/or ICU admission in many individuals with IEI compared to the epidemiology of SARS-CoV-2 infection in the general population (Fig 2 ).16 , 44 This is reminiscent of findings for SARS-CoV-2 infection in patients with cystic fibrosis. Here, it was found that many cystic fibrosis patients had mild disease and common risk factors such as diabetes and previous solid organ transplantation, but subgroups of patients exhibited increased hospitalization rates and younger age at presentation relative to the general population.117
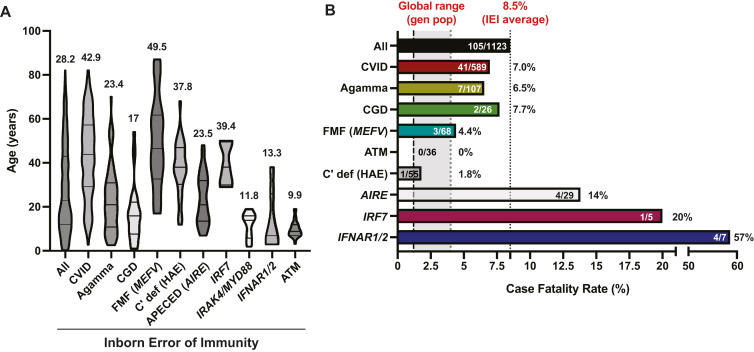
Fig 1.
Features of cohorts of patients with IEI and SARS-CoV-2 infection. (A) Age of patients with the indicated IEI. Data are shown as median ages and quartiles for each patient group. Values above each data set represent the mean ages of patients with the indicated IEI. (B) CFR for all IEI patients, as well as range for the CFR in the general population (www.covid19.who.int/, www.worldometers.info/coronavirus/). Values in each patient group represent the number of deaths/total number of patients with the indicated IEI. Agamma, Agammaglobulinemia; AIRE, patients with APECED; ATM, ataxia telangiectasia; C’ def, complement deficiency; CGD, chronic granulomatous disease; FMF, familial Mediterranean fever; IFNAR1/2, patients with pathogenic variants in type I IFN receptors; IRF7, MYD88/IRAK4, patients with pathogenic variants in IRF7 or MYD88/IRAK4 that disrupt type I IFN signaling.
Fig 2.
Consequences and outcomes of SARS-CoV-2 infection in patients with IEI.
Mortality due to SARS-CoV-2 infection in IEI
Depending on the country or region where different studies have been performed, as well as the size of the cohort being investigated, the CFR after SARS-CoV-2 infection in patients with IEI is highly variable, being 0,97 , 102 , 106 , 113 , 114 2% to 5%,99 , 101 , 107, 108, 109 5% to 10%,92 , 95 , 103 , 104 15% to 20%,105 , 112 20% to 30%,94 , 111 , 118 and >30%.15 , 89 , 91 From all available published studies, 113 of 1328 patients with IEI died after SARS-CoV-2 infection, resulting in an overall CFR of 8.5% (Fig 1, B). Remarkably, this is highly similar to the CFR reported by Meyts et al115 for an international survey of 94 patients with a broad range of IEI recruited from 12 countries (9.4%). The significant variability in CFR reported for many studies likely reflects the type of cohort being analyzed (eg, children vs adults; predominantly CVID due to unknown genetic defects vs severe combined immunodeficiency/combined immunodeficiency),108 the predominant SARS-CoV-2 variant at the time of study,41 the burden of SARS-CoV-2 infection in different countries and the relative impact this had on the respective health care systems, and the differences in screening for SARS-CoV-2 infection across the population. It is also important to note that the ∼500 IEI described exhibit enormous diversity6—so much so that it is challenging to draw conclusions when assessing these patient cohorts with limited granularity. It is also likely that some IEI will result in greater predisposition to severe COVID-19, while others may even be protective,119 thereby obscuring the overall severity of some IEI.
While it is difficult to make a direct comparison between CFR for IEI and the general population, this has been addressed for some countries. In Brazil,95 Italy,95 , 99, 100, 101 and the United Kingdom,103 the CFR in IEI was ∼2- to 4-fold greater than the general population. More strikingly, though, were findings from Iran, Italy, the United Kingdom, and an international study that the CFR for IEI patients aged 20-60 years or 60-75 years was 20-50 times or 2.5-5 times greater, respectively, than the general population.91 , 99 , 103 , 115 Furthermore, while the absolute number of patients analyzed is relatively small, the CFR for IEI patients aged 0-19 years is also much greater—possibly up to 100 times—than this age group in the general population.91 , 99 , 103 , 115 Consequently, the overall average age at death due to SARS-CoV-2 infection in IEI patients is much younger than the general population (Fig 2; ∼50 years vs ∼80 years).16 , 44 , 79 , 81 , 84 , 89 , 91 , 93, 94, 95 , 100 , 102, 103, 104 , 109 , 110 , 115 Thus, in addition to IEI patients’ generally presenting with COVID-19 at a younger age and a greater proportion requiring admission to ICU than the general population, the mortality rate of SARS-CoV-2 infection is greater in IEI, especially at ages where SARS-CoV-2 has a very low—even negligible—CFR in the general population (Fig 2).16 , 91 , 99 , 103 , 115
Innate immune defects predispose to severe and fatal SARS-CoV-2 infection
When comparing different IEI, there was often no correlation between the type of IEI and severity of disease/death after SARS-CoV-2 infection. For instance, the CFR for CVID, agammaglobulinemia, or chronic granulomatous disease were 7.2%, 6.2%, and 7.7%, respectively, compared to 8.5% for all IEI patients reported to date (Fig 1, B). However, there were several striking exceptions. First, although only few individuals have been identified, AR-pathogenic variants in IFNAR1 or IFNAR2, encoding individual receptor subunits for type I IFNs, resulted in lethal COVID-19 in 4 (57%) of 7 patients (Fig 1, B) and an average age at death of 11.8 years.42 , 56 , 87 , 120 Second, SARS-CoV-2 infection was severe in most patients with autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) as a result of biallelic pathogenic AIRE variants. These individuals develop neutralizing autoantibodies against a range of cytokines, including type I IFN.121 In the setting of SARS-CoV-2 infection of APECED patients, rates of hospitalization (72%, 21/29), ICU admission (59%, 17/29), and death (13.8%, 4/29)57 , 84 , 115 , 118 , 122 were higher than all IEI patients as well as the general population (Fig 1, B).16 , 19 Third, patients with biallelic pathogenic variants in MYD88, IRAK, or IRF7—which function downstream of virus-sensing Toll-like receptors to induce production of type I IFNs by dendritic cells—experience severe COVID-19, with 5 of 8 MYD88/IRAK-deficient and all 5 IRF7-deficient SARS-CoV-2–infected individuals developing COVID-19 pneumonia, requiring hospitalization and/or admission to ICU; 1 of 5 IRF7-deficient patient died (Fig 1, B).45 , 81 , 95 , 99 , 102 , 120 , 123 Thus, genetic lesions or autoantibodies that compromise innate immunity by disrupting production or function of type I IFNs underpin severe, life-threatening, and often fatal SARS-CoV-2 infection (Fig 3 ).
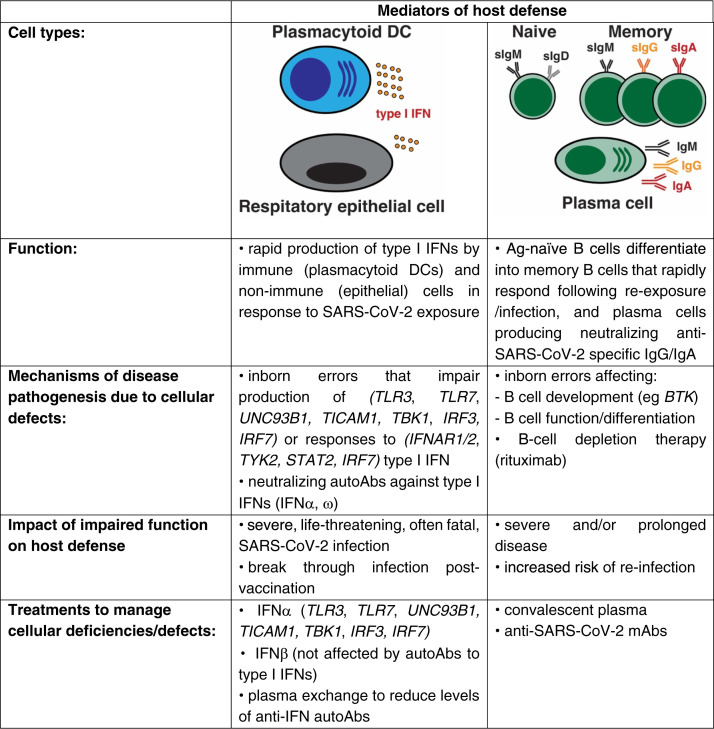
Fig 3.
Critical roles of innate and adaptive immune cells in host defense against SARS-CoV-2 infection and disease pathogenesis.
These findings have been validated by a forward genetics approach. Whole-exome and -genome sequencing of adults and children who developed severe and/or life-threatening SARS-CoV-2 infection/COVID-19 identified pathogenic variants in genes involved in type I IFN signaling. These include genes required for the production of (TLR3, TLR7, UNC93B1, TICAM1, TBK1, IRF3, IRF7) or responses to (IFNAR1, IFNAR2, TYK2, STAT2, IRF7) type I IFN produced by plasmacytoid dendritic cells or respiratory epithelial cells after viral infection.65 , 90 , 120 , 123, 124, 125, 126 Overall, genetic variants in the type I IFN signaling pathway were the cause of severe COVID-19 in ∼3% of adults and ∼10% of children (Fig 3).120 , 126 , 127
Parallel to these genetic studies was the discovery that neutralizing autoantibodies specific for type I IFNs cause severe COVID-19 in 10% to 20% of otherwise healthy individuals infected with SARS-CoV-2.128, 129, 130, 131, 132, 133, 134, 135, 136, 137 Interestingly, these autoantibodies were: (1) predominantly directed against IFN-α and IFN-ω but not IFN-β; (2) found in increasing proportions of affected patients with each decade of life; (3) associated with disease severity, prolonged virus clearance, and admission to ICU; (4) inversely related to serum levels of type I IFNs and interferon-stimulated gene signatures in myeloid cells;78 , 128, 129, 130, 131, 132, 133, 134, 135, 136, 137 and (5) enriched in affected male subjects compared to female subjects across different age intervals. This, together with XL TLR7 deficiency, may contribute to the increased incidence of hospitalization and severe COVID-19 in male versus female subjects. These genetic and serologic studies unequivocally identified a fundamental nonredundant role for type I IFN–dependent immunity against SARS-CoV-2 infection, with 20% to 25% of cases of severe and life-threatening COVID-19 resulting from defective type I IFN production or function (Fig 3).
Additional anecdotal data have also linked impaired type I IFN–dependent immunity with susceptibility to SARS-CoV-2 infection. First, the CFR for autoinflammatory conditions such as Aicardi-Goutières syndrome or familial Mediterranean fever was lower than that for all reported cases of IEI (4.4% vs 8.5%; Fig 1, B).15 , 93 , 95 , 99 , 110 , 115 Thus, increased basal type I IFN signaling in these conditions may enable prompt host defense against SARS-CoV-2. Second, a recent study of patients with systemic lupus erythematosus, which is characterized by overproduction of type I IFNs, found that a subset of these patients also produced autoantibodies against type I IFNs. Remarkably, while these autoantibody-positive patients were less likely to develop active lupus disease, members of this same group were at increased risk of severe viral infections and sequelae including COVID-19 pneumonia.138
B cells and protective IgG in host defense against SARS-CoV-2
The study of COVID-19 in IEI provides an elegant opportunity to define redundant and nonredundant requirements for host defense against SARS-CoV-2. Initial studies found that patients with congenital B-cell deficiency and agammaglobulinemia had relatively mild disease and prompt recovery after SARS-CoV-2 infection.51 , 73 , 92 , 97 , 98 This led to a suggestion that B cells and neutralizing IgG may not be necessary for controlling SARS-CoV-2 infection and preventing severe COVID-19.98 Consistent with this, the CFR for XL/AR agammaglobulinemia patients is lower than all IEI patients (6.2%, 6/97, vs 8.5%, Fig 1, B). However, COVID-19 and SARS-CoV-2 viremia/virus shedding are prolonged in many B-cell–deficient/agammaglobulinemia patients, resulting in pneumonia requiring extended or multiple hospital stays, as well as numerous treatments to control viral infection.46 , 55 , 60 , 61 , 85 , 86 , 99 , 100 , 104 , 116 , 139 There have also been reports of chronic and/or repeated infections with worse outcomes than primary infection before vaccination, as well as breakthrough infections after vaccination in some XL agammaglobulinemia (XLA) patients.100 , 104 , 109 , 116 , 140 Similar observations in terms of relapsing COVID-19, as well as reinfection and/or sustained infection with SARS-CoV-2, have been made for patients with primary antibody deficiencies,82 , 104 , 116 further underscoring an important role for secreted immunoglobulin in controlling and clearing viral infection and attenuating disease. These findings from analysis of SARS-CoV-2 infection in individuals with congenital B-cell deficiency are also supported by studies of patients with rheumatic/musculoskeletal autoimmune diseases (rheumatoid arthritis, vasculitis, Sjögren syndrome, systemic lupus erythematosus) who are treated with B-cell–depleting therapies such as rituximab. In these cases, therapeutic B-cell depletion can result in high rates of hospital admissions, severe COVID-19 including protracted pneumonia and acute respiratory distress syndrome, and death after SARS-CoV-2 infection.141 , 142 Thus, the inability to generate specific IgG responses to novel antigens as a result of a lack of naive B cells can have dire consequences in the setting of SARS-CoV-2 infection (Fig 3).
This apparent paradox of prolonged illness and viremia but often-milder disease and lower CFR in XLA patients who completely lack B cells may be explained by the nature of the genetic defect. On the one hand, agammaglobulinemia in these patients highlights a key role for specific immunoglobulins in controlling and clearing viral infection, even when responses of innate immune cells and CD4+ and CD8+ T cells are intact.46 , 83 Indeed, administration of convalescent plasma isolated from previously infected healthy donors or anti–SARS-CoV-2–specific monoclonal antibodies (mAbs) led to rapid reductions in virus load and recovery in XLA—more so than observed with antiviral treatments alone (Fig 3).46 , 53 , 55 , 61 , 85 , 86 , 104 , 139 , 143 Although convalescent plasma or anti–SARS-CoV-2 mAbs are a logical treatment for XLA patients, similar results have also been reported for other IEI patients who have near-normal B cells and serum immunoglobulin levels but defects in generating functional and protective IgG-dependent humoral immunity. For instance, passive IgG therapy led to dramatic improvements in the clinical course of SARS-CoV-2 infection in patients with pathogenic variants in NFKB2,43 IL2RG,77 IKBKG (NEMO),72 and PIK3CD GOF,82 as well as many cases of CVID.83 , 103 , 104 , 109 , 143 In fact, anti–SARS-CoV-2 mAb or convalescent plasma greatly improved virus clearance and disease outcomes when combined with antivirals (eg, remdesivir).43 , 104 , 143 Thus, while type I IFN–mediated innate immunity is indispensable for containing acute SARS-CoV-2 infection, antibodies are necessary to mitigate prolonged viral infection, minimize disease, and prevent reinfections (Fig 3).
On the other hand, Bruton tyrosine kinase (BTK) deficiency—the genetic cause of XLA—compromises production of inflammatory cytokines by myeloid cells.144 Thus, relatively mild pulmonary disease in XLA may result from a lessened cytokine storm after SARS-CoV-2–induced activation of BTK-deficient myeloid cells. This is consistent with findings that some SARS-CoV-2–infected XLA patients have lower serum IL-6 levels than infected individuals in the general population,111 observations of mild COVID-19 in patients with B-cell malignancies who were treated with BTK inhibitors,145 and rapid clinical improvement in COVID-19 patients treated with a BTK inhibitor as a therapeutic intervention.146 These findings reveal dual roles for BTK in host defense and tissue pathology after SARS-CoV-2 infection. First, B cells and virus-specific antibodies are important for controlling prolonged infection. Second, BTK in myeloid cells may drive the SARS-CoV-2–induced cytokine storm characteristic of severe COVID-19. These findings provide a rationale for the use of passive immunoglobulin serotherapy (intravenous immunoglobulin, mAbs) to expedite virus clearance in IEI characterized by impaired humoral immunity, as well as of BTK inhibitors, Janus kinase (JAK) inhibitors, and tocilizumab (anti–IL-6R)146, 147, 148 to quell SARS-CoV-2–induced production of inflammatory cytokines by myeloid cells. However, it needs to be emphasized that timing of the delivery of these treatments can also influence outcome and efficacy. For instance, if administered too early, JAK inhibits may attenuate the protective effect of type I IFNs, while delayed treatment with tocilizumab may be ineffectual. Similarly, these interventions may be better suited for some specific types of IEI, particularly as results from clinical trials of these inhibitors in the general population have been variable.
Gene-directed therapies for COVID-19 in some IEI
Delineation of the genetic and serologic causes of severe COVID-19 has led to the implementation of specific therapies in some IEI. For instance, the discovery that inborn errors in type I IFN signaling are a risk factor for severe COVID-19 inspired the use of IFN-α2a or IFN-β, anti–SARS-CoV-2 mAbs, or convalescent plasma to treat SARS-CoV-2 infection in individuals with pathogenic variants in TLR3, IRF3, IRF7, or IRF9,68 , 69 , 123 which genetically disrupt type I IFN function, or patients with pathogenic AIRE variants or incontinentia pigmenti due to pathogenic IKBKG variants that result in production of neutralizing anti–type I IFN autoantibodies.78 , 84 , 149 However, convalescent plasma has also been found to contain neutralizing anti–type I IFN autoantibodies,137 which obviously could impact the efficacy of this treatment.
Similarly, plasma exchange was effective at reducing serum levels of neutralizing anti–type I IFN autoantibodies in an APECED patient.57 While it is difficult to draw specific conclusions regarding possible therapies for SARS-CoV-2 infection in IEI from these anecdotal investigations, most treated patients exhibited mild disease, experienced rapid resolution of symptoms, and made a full recovery.57 , 68 , 69 , 78 , 84 , 149 This contrasts with those IEI patients who did not receive specific treatments and experienced severe and even fatal COVID-19.120 , 126 , 127 Thus, early provision of type I IFN or antibody against SARS-CoV-2 may represent an immunotherapeutic approach to prevent critical pneumonia in patients who are most vulnerable to severe SARS-CoV-2 infection due to disrupted type I IFN–mediated immunity. Furthermore, because anti–type I IFN autoantibodies are mostly directed against IFN-α and IFN-ω, IFN-β can still be used therapeutically for severe COVID-19 in individuals who develop these neutralizing autoantibodies.
Vaccines against SARS-CoV-2
The global rollout of several different SARS-CoV-2 vaccines (mRNA, adenoviral based, inactivated virus, viral proteins) has dramatically attenuated COVID-19–associated mortality.150 These vaccines induce SARS-CoV-2–specific CD4+ and CD8+ T cells, memory B cells, and neutralizing serum IgG in >95% of healthy donors. Readouts of vaccine-induced immunity generally peaked 2 or 3 weeks after receipt of the second vaccine dose and then either significantly declined (specific IgG titers, CD8+ T cells), plateaued (CD4+ T cells), or even increased (memory B cells).150, 151, 152 Regardless of these trajectories, SARS-CoV-2–specific adaptive cellular and humoral immunity remained detectable ∼6 months after vaccination.150, 151, 152 The magnitude of these vaccine-induced correlates of immunity in healthy individuals was generally comparable to or greater than those observed in convalescent individuals recovering from natural SARS-CoV-2 infection.150, 151, 152
While these findings are encouraging, several challenges remain in controlling SARS-CoV-2. First, vaccine efficacy declines from 85-95% at 2 to 4 weeks after full vaccination to 20-50% 6 months later, thus revealing an inability to completely resist future infection and highlighting the need for vaccine boosters.150 , 153 , 154 Second, while successfully reducing disease severity, hospital admissions, and mortality, current vaccines do not effectively prevent SARS-CoV-2 transmission.150 , 155 Third, the emergence of variants of concern—which can arise in immunocompromised individuals156—compromise vaccine efficacy, with vaccine-induced immunity being significantly reduced against several SARS-CoV-2 variants.41 , 154 , 157 Thus, COVID-19 continues to represent a significant health risk despite the availability of several SARS-CoV-2 vaccines. Furthermore, findings from studies of IEI have established the importance of SARS-CoV-2–specific neutralizing IgG in preventing severe and prolonged disease as well as reinfection, so it is critical to continue encouraging vaccine and booster uptake in the general population.
Efficacy of SARS-CoV-2 vaccines in IEI patients
Many studies have initially assessed the immunogenicity and effectiveness of SARS-CoV-2 vaccines in IEI. The general findings from these studies were that (1) fewer patients mounted SARS-CoV-2–specific IgG (30-75%) and T-cell responses (∼50-70%) compared to healthy donors (∼95-100%), (2) titers of SARS-CoV-2–specific IgG, efficacy of virus neutralization, and magnitude of T-cell responses were reduced in patients compared to healthy donors, and (3) poor vaccine-induced immunity in patients correlated with reduced numbers of CD4+ T cells or memory B cells, low serum IgG and IgA, and older age.80 , 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173 Importantly, IEI that disrupt type I IFN–mediated immunity or autoantibodies against type I IFN do not impair humoral immune responses to RNA vaccines.174 Furthermore, despite normal levels of neutralizing IgG, some patients with anti–type I IFN autoantibodies develop breakthrough COVID-19 pneumonia.175
Overall, these studies established that SARS-CoV-2 vaccines are safe and well tolerated in people with IEI, and that they can induce specific adaptive immune responses, albeit at reduced levels compared to the general population. However, several significant unknowns remain. First, most studies assessed immune responses 2 to 8 weeks after the second vaccine dose. Thus, sustained durability of vaccine-induced immunity in IEI patients against SARS-CoV-2 has not been determined. Second, while almost all vaccine studies measured SARS-CoV-2–specific IgG, only a few determined virus neutralization. Thus, it is unknown whether vaccine-induced immunoglobulin in IEI patients can neutralize the original SARS-CoV-2 strain and emerging variants. Third, specific CD4+ and CD8+ T-cell responses in vaccinated IEI patients were not assessed in most studies. The paucity of data relating to responses of T-cell subsets impacts our ability to predict vulnerability of individuals with intrinsic T-cell defects to SARS-CoV-2 infection. Fourth, how waning immunity and SARS-CoV-2 variants impact host defense, as well as the capacity of vaccine booster doses to amplify immunity, in IEI patients is unexplored. Fifth, ∼80% of all IEI patients assessed in these studies did not have a molecular diagnosis; most had CVID. Thus, it is difficult to (1) delineate cellular and molecular mechanisms underlying impaired immunity in IEI patients, (2) extrapolate these findings from predominantly CVID and antibody-deficient patients to IEI in general, (3) identify which pathways are necessary to elicit robust and long-lived immune responses, and (4) leverage these findings to develop methods to target specific key molecules/pathways to improve host defense against infectious diseases induced by next-generation vaccines. These are issues that need to be addressed in ongoing and future studies.
Conclusion
Analysis of individuals with single-gene defects that result in immune dysregulation have defined the fundamental requirements for immune homeostasis and host defense against a broad range of infectious agents. It was upon this foundation that the fields of genetics/genomics, basic and clinical immunology, and infectious diseases combined to make profound advances in unraveling the complexity of SARS-CoV-2 infection and severe COVID-19. Indeed, some of the key discoveries over the past 2 or 3 years have arisen from studying severe COVID-19 in otherwise healthy individuals, as well as in individuals with IEI. These studies established the framework to further define host factors necessary for early innate and sustained adaptive immune-mediated protection against SARS-CoV-2 infection and the establishment of immunologic memory, as well as mechanisms of severe disease and identifying opportunities for therapeutic intervention to manage COVID-19. Despite these breakthrough findings, there remains significant uncertainty regarding SARS-CoV-2 and IEI patients. These include the impact of standard treatments for IEI on immunity against SARS-CoV-2 infection and vaccination (eg, JAK inhibitors, TNF inhibitors, abatacept, rapamycin), long-term effects of SARS-CoV-2 infection/reinfection on IEI patients with autoimmunity and/or malignancy, whether long COVID and neurologic impacts are more prevalent in IEI compared to the general population, and the protective effect of neutralizing antibodies that are accumulating in donor blood products used for immunoglobulin replacement therapy. However, with the rapid pace of the advances already made since we first became aware of SARS-CoV-2, there is no doubt that answers to these questions—and more—will be delivered as we move into the third year (and, I hope, the last frontier) of this pandemic.
Acknowledgments
I would like to acknowledge all the clinicians, nurses, caregivers, parents, and patient advocacy groups who have ensured the safety and well-being of patients with IEI during the COVID-19 pandemic. I also want to thank Jean-Laurent Casanova and Helen Su for their leadership of the COVID-19 Human Genetic Effort consortium (www.covidhge.com), as well as their insightful discussions and inspiration since the onset of the pandemic; and Isabelle Meyts and many members of the COVID-19 Human Genetic Effort for their constructive feedback.
Members of the COVID Human Genetic Effort consortium are as follows: Laurent Abel (INSERM U1163, University of Paris, Imagine Institute, Paris, France); Salah Al-Muhsen (Immunology Research Lab, Department of Pediatrics, College of Medicine, King Saud University, Riyadh, Saudi Arabia); Alessandro Aiuti (San Raffaele Telethon Institute for Gene Therapy, IRCCS Ospedale San Raffaele, and Vita Salute San Raffaele University, Milan, Italy); Saleh Al-Muhsen (Immunology Research Laboratory, Department of Pediatrics, College of Medicine and King Saud University Medical City, King Saud University, Riyadh, Saudi Arabia); Fahd Al-Mulla (Dasman Diabetes Institute, Department of Genetics and Bioinformatics, Dasman, Kuwait); Mark S. Anderson (Diabetes Center, University of California, San Francisco, Calif); Evangelos Andreakos (Biomedical Research Foundation of the Academy of Athens, Athens, Greece); Antonio Novelli (Laboratory of Medical Genetics, IRCCS Bambino Gesù Children’s Hospital, Rome, Italy); Andrés A. Arias (Group of Primary Immunodeficiencies, University of Antioquia UdeA, Medellin, Colombia); Hagit Baris Feldman (The Genetics Institute, Tel Aviv Sourasky Medical Center and Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel); Alexandre Belot (Pediatric Nephrology, Rheumatology, Dermatology, HFME, Hospices Civils de Lyon, National Referee Centre RAISE, and INSERM U1111, Université de Lyon, Lyon, France); Catherine M. Biggs (Department of Pediatrics, British Columbia Children’s Hospital, University of British Columbia, Vancouver, British Columbia, Canada); Ahmed A. Bousfiha (Clinical Immunology Unit, Department of Pediatric Infectious Disease, CHU Ibn Rushd, and LICIA, Laboratoire d’Immunologie Clinique, Inflammation et Allergie, Faculty of Medicine and Pharmacy, Hassan II University, Casablanca, Morocco); Petter Brodin (SciLifeLab, Department of Women’s and Children’s Health, Karolinska Institutet, Stockholm, Sweden); John Christodoulou (Murdoch Children’s Research Institute and Department of Paediatrics, University of Melbourne, Melbourne, Australia); Antonio Condino-Neto (Department of Immunology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil); Clifton L. Dalgard (Department of Anatomy, Physiology, and Genetics, Uniformed Services University of the Health Sciences, Bethesda, Md); Sara Espinosa-Padilla (National Institute of Pediatrics, Mexico City, Mexico); Jacques Fellay (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne; Precision Medicine Unit, Lausanne University Hospital; and University of Lausanne, Lausanne, Switzerland); Carlos Flores (Genomics Division, Instituto Tecnológico y de Energías Renovables [ITER], Santa Cruz de Tenerife; Research Unit, Hospital Universitario NS de Candelaria, Santa Cruz de Tenerife; and CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid, Spain); José Luis Franco (Group of Primary Immunodeficiencies, University of Antioquia UdeA, Medellin, Colombia); Antoine Froidure (Pulmonology Department, Cliniques Universitaires Saint-Luc, and Institut de Recherche Expérimentale et Clinique [IREC], Université Catholique de Louvain, Brussels, Belgium); Filomeen Haerynck (Department of Paediatric Immunology and Pulmonology, Centre for Primary Immunodeficiency Ghent [CPIG], PID Research Laboratory, Jeffrey Modell Diagnosis and Research Centre, Ghent University Hospital, Ghent, Belgium); Rabih Halwani (Sharjah Institute of Medical Research, College of Medicine, University of Sharjah, Sharjah, United Arab Emirates [UAE]); Lennart Hammarström (Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden); Sarah E. Henrickson (Department of Pediatrics, Division of Allergy Immunology, Children’s Hospital of Philadelphia; and Department of Microbiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pa); Elena W. Y. Hsieh (Departments of Pediatrics, Immunology, and Microbiology, University of Colorado, School of Medicine, Aurora, Colorado); Yuval Itan (Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, and Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY); Timokratis Karamitros (Bioinformatics and Applied Genomics Unit, Hellenic Pasteur Institute, Athens, Greece); Yu-Lung Lau (Department of Paediatrics and Adolescent Medicine, University of Hong Kong, Hong Kong); Davood Mansouri (Department of Clinical Immunology and Infectious Diseases, National Research Institute of Tuberculosis and Lung Diseases, Clinical Tuberculosis and Epidemiology Research Center, National Research Institute of Tuberculosis and Lung Diseases [NRITLD], Masih Daneshvari Hospital, Shahid Beheshti, University of Medical Sciences, Tehran, Iran); Isabelle Meyts (Department of Pediatrics, University Hospitals Leuven, Department of Microbiology, Immunology, and Transplantation, and Laboratory for Inborn Errors of Immunity, KU Leuven, Leuven, Belgium); Trine H. Mogensen (Department of Biomedicine, Aarhus University, Aarhus, Denmark); Tomohiro Morio (Tokyo Medical and Dental University Hospital, Tokyo, Japan); Lisa F. P. Ng (A∗STAR Infectious Disease Labs, Agency for Science, Technology and Research; and Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore); Luigi D. Notarangelo (National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH], Bethesda, Md); Giuseppe Novelli (Department of Biomedicine and Prevention, Tor Vergata University of Rome, Rome, Italy); Satoshi Okada (Department of Pediatrics, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan); Tayfun Ozcelik (Department of Molecular Biology and Genetics, Bilkent University, Bilkent, Ankara, Turkey); Qiang Pan-Hammarström (Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden); Rebeca Perez de Diego (Laboratory of Immunogenetics of Human Diseases, Innate Immunity Group, IdiPAZ Institute for Health Research, La Paz Hospital, Madrid, Spain); Carolina Prando (Faculdades Pequeno Príncipe, Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil); Aurora Pujol (Neurometabolic Diseases Laboratory, Bellvitge Biomedical Research Institute [IDIBELL]), L’Hospitalet de Llobregat; Catalan Institution of Research and Advanced Studies [ICREA]; and Center for Biomedical Research on Rare Diseases [CIBERER], ISCIII, Barcelona, Spain); Laurent Renia (A∗STAR Infectious Disease Labs, Agency for Science, Technology and Research; and Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore); Igor Resnick (Department of Medical Genetics, Medical University; and Department of Hematology and BMT, University Hospital St Marina, Varna, Bulgaria); Carlos Rodríguez-Gallego (Department of Immunology, University Hospital of Gran Canaria Dr Negrín, Canarian Health System; and Department of Clinical Sciences, University Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain); Vanessa Sancho-Shimizu (Department of Paediatric Infectious Diseases and Virology, Imperial College London; and Centre for Paediatrics and Child Health, Faculty of Medicine, Imperial College London, London, United Kingdom); Mikko R. J. Seppänen (Adult Immunodeficiency Unit, Infectious Diseases, Inflammation Center, University of Helsinki and Helsinki University Hospital; Rare Diseases Center and Pediatric Research Center, Children’s Hospital, University of Helsinki; and Helsinki University Hospital, Helsinki, Finland); Anna Shcherbina (Department of Immunology, Dmitry Rogachev National Medical Research Center of Pediatric Hematology, Oncology, and Immunology, Moscow, Russia); Andrew L. Snow (Department of Pharmacology and Molecular Therapeutics, Uniformed Services University of the Health Sciences, Bethesda, Md); Pere Soler-Palacín (Pediatric Infectious Diseases and Immunodeficiencies Unit, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain); András N. Spaan (St Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, New York, NY; and Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, The Netherlands); Ivan Tancevski (Department of Internal Medicine II, Medical University of Innsbruck, Innsbruck, Austria); Stuart G. Tangye (Garvan Institute of Medical Research, Darlinghurst; and St Vincent’s Clinical School, Faculty of Medicine, University of New South Wales Sydney, Sydney, Australia); Ahmad Abou Tayoun (Al Jalila Children’s Hospital, Dubai, UAE); Sehime G. Temel (Bursa Uludag University, Medical Faculty, Department of Medical Genetics, Bursa, Turkey); Stuart E. Turvey (BC Children’s Hospital, The University of British Columbia, Vancouver, Canada); Mohammed J. Uddin (College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, UAE; and Cellular Intelligence [CI] Lab, GenomeArc Inc, Toronto, Ontario, Canada); Donald C. Vinh (Department of Medicine, Division of Infectious Diseases, McGill University Health Centre; and Infectious Disease Susceptibility Program, Research Institute, McGill University Health Centre, Montreal, Quebec, Canada); Mayana Zatz (Biosciences Institute, University of São Paulo, São Paulo, Brazil); Keisuke Okamoto (Tokyo Medical and Dental University, Tokyo, Japan); David S. Pelin (Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ); Graziano Pesole (Department of Biosciences, Biotechnology, and Biopharmaceutics, University of Bari A. Moro, Bari, Italy); Diederik van de Beek (Department of Neurology, Amsterdam Neuroscience, Amsterdam University Medical Center, University of Amsterdam, Amsterdam, The Netherlands); Roger Colobran (Hospital Universitari Vall d’Hebron, Barcelona, Spain); Joost Wauters (Department of General Internal Medicine, Medical Intensive Care Unit, University Hospitals Leuven, Leuven, Belgium); Helen C. Su (NIAID, NIH, Bethesda, Md); Jean-Laurent Casanova (Rockefeller University and Howard Hughes Medical Institute, New York, NY; and Necker Hospital for Sick Children and INSERM, Paris, France).
Footnotes
S.G.T. is supported by an Investigator Grant awarded by the National Health and Medical Research Council of Australia, the Allergy & Immunology Foundation of Australia, the Jeffrey Modell Foundation, and a University of New South Wales Sydney COVID Rapid Response Initiative grant. H.C.S. and L.D.N. (listed under the COVID Human Genetic Effort consortium) are supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH. J.L.C. (listed under the COVID Human Genetic Effort consortium) is supported by the NIH (R01AI088364, R01AI163029, UL1TR001866), Fisher Center for Alzheimer’s Research Foundation, Meyer Foundation, JPB Foundation, French National Research Agency (ANR-10-IAHU-01, ANR-10-LABX-62-IBEID, ANR-20-CE93-003, ANR-20-CO11-0001, French Foundation for Medical Research (EQU201903007798), the ANRS-COV05, European Union’s Horizon 2020 Research and Innovation Program (824110; EASI-genomics), HORIZON-HLTH-2021-DISEASE-04 Program (01057100; UNDINE), the ANR-RHU COVIFERON Program (ANR-21-RHUS-08), the Square Foundation, Grandir - Fonds de solidarité pour l’enfance, the Fondation du Souffle, the SCOR Corporate Foundation for Science, and French Ministry of Higher Education, Research, and Innovation (MESRI-COVID-19).
Disclosure of potential conflict of interest: The author declares no relevant conflicts of interest.
Contributor Information
COVID Human Genetic Effort consortium:
Laurent Abel, Salah Al-Muhsen, Alessandro Aiuti, Saleh Al-Muhsen, Fahd Al-Mulla, Mark S. Anderson, Evangelos Andreakos, Antonio Novelli, Andrés A. Arias, Hagit Baris Feldman, Alexandre Belot, Catherine M. Biggs, Ahmed A. Bousfiha, Petter Brodin, John Christodoulou, Antonio Condino-Neto, Clifton L. Dalgard, Sara Espinosa-Padilla, Jacques Fellay, Carlos Flores, José Luis Franco, Antoine Froidure, Filomeen Haerynck, Rabih Halwani, Lennart Hammarström, Sarah E. Henrickson, Elena W.Y. Hsieh, Yuval Itan, Timokratis Karamitros, Yu-Lung Lau, Davood Mansouri, Isabelle Meyts, Trine H. Mogensen, Tomohiro Morio, Lisa F.P. Ng, Luigi D. Notarangelo, Giuseppe Novelli, Satoshi Okada, Tayfun Ozcelik, Qiang Pan-Hammarström, Rebeca Perez de Diego, Carolina Prando, Aurora Pujol, Laurent Renia, Igor Resnick, Carlos Rodríguez-Gallego, Vanessa Sancho-Shimizu, Mikko R.J. Seppänen, Anna Shcherbina, Andrew L. Snow, Pere Soler-Palacín, András N. Spaan, Ivan Tancevski, Stuart G. Tangye, Ahmad Abou Tayoun, Sehime G. Temel, Stuart E. Turvey, Mohammed J. Uddin, Donald C. Vinh, Mayana Zatz, Keisuke Okamoto, David S. Pelin, Graziano Pesole, Diederik van de Beek, Roger Colobran, Joost Wauters, Helen C. Su, and Jean-Laurent Casanova
References
- 1.Casanova J.L., Abel L. Human genetics of infectious diseases: unique insights into immunological redundancy. Semin Immunol. 2018;36:1–12. doi: 10.1016/j.smim.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova J.L., Abel L. From rare disorders of immunity to common determinants of infection: following the mechanistic thread. Cell. 2022;185:3086–3103. doi: 10.1016/j.cell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer A., Rausell A. What do primary immunodeficiencies tell us about the essentiality/redundancy of immune responses? Semin Immunol. 2018;36:13–16. doi: 10.1016/j.smim.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Notarangelo L.D., Bacchetta R., Casanova J.L., Su H.C. Human inborn errors of immunity: an expanding universe. Sci Immunol. 2020 doi: 10.1126/sciimmunol.abb1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q., Frange P., Blanche S., Casanova J.L. Pathogenesis of infections in HIV-infected individuals: insights from primary immunodeficiencies. Curr Opin Immunol. 2017;48:122–133. doi: 10.1016/j.coi.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tangye S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., et al. Human inborn errors of immunity: 2022 update on the classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieux-Laucat F., Casanova J.L. Immunology. Autoimmunity by haploinsufficiency. Science. 2014;345:1560–1561. doi: 10.1126/science.1260791. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez N.J., Posadas-Cantera S., Caballero-Oteyza A., Camacho-Ordonez N., Grimbacher B. There is no gene for CVID—novel monogenetic causes for primary antibody deficiency. Curr Opin Immunol. 2021;72:176–185. doi: 10.1016/j.coi.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Poyhonen L., Bustamante J., Casanova J.L., Jouanguy E., Zhang Q. Life-threatening infections due to live-attenuated vaccines: early manifestations of inborn errors of immunity. J Clin Immunol. 2019;39:376–390. doi: 10.1007/s10875-019-00642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thalhammer J., Kindle G., Nieters A., Rusch S., Seppanen M.R.J., Fischer A., et al. Initial presenting manifestations in 16,486 patients with inborn errors of immunity include infections and noninfectious manifestations. J Allergy Clin Immunol. 2021;148:1332–1341.e5. doi: 10.1016/j.jaci.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Fischer A., Provot J., Jais J.P., Alcais A., Mahlaoui N. Autoimmune and inflammatory manifestations occur frequently in patients with primary immunodeficiencies. J Allergy Clin Immunol. 2017;140:1388–1393.e8. doi: 10.1016/j.jaci.2016.12.978. [DOI] [PubMed] [Google Scholar]
- 12.Leiding J.W., Ballow M. Redefining precision medicine in disorders of immune dysregulation. J Allergy Clin Immunol Pract. 2019;7:2801–2803. doi: 10.1016/j.jaip.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 13.Weiss S.R. Forty years with coronaviruses. J Exp Med. 2020 doi: 10.1084/jem.20200537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shields A.M., Burns S.O., Savic S., Richter A.G., UK PIN COVID-19 Consortium COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J Allergy Clin Immunol. 2021;147:870–875.e1. doi: 10.1016/j.jaci.2020.12.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stokes E.K., Zambrano L.D., Anderson K.N., Marder E.P., Raz K.M., El Burai Felix S., et al. Coronavirus disease 2019 case surveillance—United States, January 22-May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Xu S., Yu M., Wang K., Tao Y., Zhou Y., et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146:110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell T.W., Hellewell J., Jarvis C.I., van Zandvoort K., Abbott S., Ratnayake R., et al. Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.12.2000256. February 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abers M.S., Delmonte O.M., Ricotta E.E., Fintzi J., Fink D.L., de Jesus A.A.A., et al. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6 doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold T., Jurinovic V., Arnreich C., Lipworth B.J., Hellmuth J.C., von Bergwelt-Baildon M., et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. 2020;146:128–136.e4. doi: 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quartuccio L., Sonaglia A., Pecori D., Peghin M., Fabris M., Tascini C., et al. Higher levels of IL-6 early after tocilizumab distinguish survivors from nonsurvivors in COVID-19 pneumonia: a possible indication for deeper targeting of IL-6. J Med Virol. 2020;92:2852–2856. doi: 10.1002/jmv.26149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc Natl Acad Sci U S A. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou H., Zhang Y., Tang G., Luo Y., Liu W., Cheng C., et al. Immunologic memory to SARS-CoV-2 in convalescent COVID-19 patients at 1 year postinfection. J Allergy Clin Immunol. 2021;148:1481–1492.e2. doi: 10.1016/j.jaci.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breton G., Mendoza P., Hagglof T., Oliveira T.Y., Schaefer-Babajew D., Gaebler C., et al. Persistent cellular immunity to SARS-CoV-2 infection. J Exp Med. 2021 doi: 10.1084/jem.20202515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bilich T., Nelde A., Heitmann J.S., Maringer Y., Roerden M., Bauer J., et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci Transl Med. 2021 doi: 10.1126/scitranslmed.abf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriyama S., Adachi Y., Sato T., Tonouchi K., Sun L., Fukushi S., et al. Temporal maturation of neutralizing antibodies in COVID-19 convalescent individuals improves potency and breadth to circulating SARS-CoV-2 variants. Immunity. 2021;54:1841–1852.e4. doi: 10.1016/j.immuni.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherina N., Piralla A., Du L., Wan H., Kumagai-Braesch M., Andrell J., et al. Persistence of SARS-CoV-2–specific B and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med (N Y) 2021;2:281–295.e4. doi: 10.1016/j.medj.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balachandran H., Phetsouphanh C., Agapiou D., Adhikari A., Rodrigo C., Hammoud M., et al. Maintenance of broad neutralizing antibodies and memory B cells 1 year post-infection is predicted by SARS-CoV-2–specific CD4+ T cell responses. Cell Rep. 2022;38:110345. doi: 10.1016/j.celrep.2022.110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abayasingam A., Balachandran H., Agapiou D., Hammoud M., Rodrigo C., Keoshkerian E., et al. Long-term persistence of RBD+ memory B cells encoding neutralizing antibodies in SARS-CoV-2 infection. Cell Rep Med. 2021;2:100228. doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chia W.N., Zhu F., Ong S.W.X., Young B.E., Fong S.W., Le Bert N., et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. 2021;2:e240–e249. doi: 10.1016/S2666-5247(21)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirabara S.M., Serdan T.D.A., Gorjao R., Masi L.N., Pithon-Curi T.C., Covas D.T., et al. SARS-COV-2 variants: differences and potential of immune evasion. Front Cell Infect Microbiol. 2021;11:781429. doi: 10.3389/fcimb.2021.781429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abolhassani H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., et al. Inherited IFNAR1 deficiency in a child with both critical COVID-19 pneumonia and multisystem inflammatory syndrome. J Clin Immunol. 2022;42:471–483. doi: 10.1007/s10875-022-01215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham R.S., Marshall J.M., Kuehn H.S., Rueda C.M., Gibbs A., Guider W., et al. Severe SARS-CoV-2 disease in the context of a NF-kappaB2 loss-of-function pathogenic variant. J Allergy Clin Immunol. 2021;147:532–544.e1. doi: 10.1016/j.jaci.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beccuti G., Ghizzoni L., Cambria V., Codullo V., Sacchi P., Lovati E., et al. A COVID-19 pneumonia case report of autoimmune polyendocrine syndrome type 1 in Lombardy, Italy: letter to the editor. J Endocrinol Invest. 2020;43:1175–1177. doi: 10.1007/s40618-020-01323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bucciol G., Moens L., Corveleyn A., Dreesman A., Meyts I. A novel kindred with MyD88 deficiency. J Clin Immunol. 2022;42:885–888. doi: 10.1007/s10875-022-01240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckland M.S., Galloway J.B., Fhogartaigh C.N., Meredith L., Provine N.M., Bloor S., et al. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat Commun. 2020;11:6385. doi: 10.1038/s41467-020-19761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castano-Jaramillo L.M., Yamazaki-Nakashimada M.A., Scheffler Mendoza S.C., Bustamante-Ogando J.C., Espinosa-Padilla S.E., Lugo Reyes S.O. A male infant with COVID-19 in the context of ARPC1B deficiency. Pediatr Allergy Immunol. 2021;32:199–201. doi: 10.1111/pai.13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cenciarelli S., Calbi V., Barzaghi F., Bernardo M.E., Oltolini C., Migliavacca M., et al. Mild SARS-CoV-2 infection after gene therapy in a child with Wiskott-Aldrich syndrome: a case report. Front Immunol. 2020;11:603428. doi: 10.3389/fimmu.2020.603428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabryszewski S.J., England R.N., Sun D., Gentile T.L., Hochgertel W., Jyonouchi S., et al. Self-limited COVID-19 in a patient with artemis hypomorphic SCID. J Clin Immunol. 2021 doi: 10.1007/s10875-021-01093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guisado Hernandez P., Blanco Lobo P., Villaoslada I., de Felipe B., Lucena J.M., Martin Gutierrez G., et al. SARS-CoV-2 infection in a pediatrics STAT1 GOF patient under ruxolitinib therapy—a matter of balance? J Clin Immunol. 2021 doi: 10.1007/s10875-021-01081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta S., Agrawal S., Sandoval A., Su H., Tran M., Demirdag Y. SARS-CoV-2–specific and functional cytotoxic CD8 cells in primary antibody deficiency: natural infection and response to vaccine. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta S., Su H., Narsai T., Agrawal S. SARS-CoV-2–associated T-cell responses in the presence of humoral immunodeficiency. Int Arch Allergy Immunol. 2021;182:195–209. doi: 10.1159/000514193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iaboni A., Wong N., Betschel S.D. A patient with X-linked agammaglobulinemia and COVID-19 infection treated with remdesivir and convalescent plasma. J Clin Immunol. 2021;41:923–925. doi: 10.1007/s10875-021-00983-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin H., Moss R., Reed J.C., Hertzberg E., Cruz M.R., Akkoyun E., et al. IFN-gamma receptor 2 deficiency initial mimicry of multisystem inflammatory syndrome in children (MIS-C) J Allergy Clin Immunol Pract. 2021;9:989–992.e1. doi: 10.1016/j.jaip.2020.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin H., Reed J.C., Liu S.T.H., Ho H.E., Lopes J.P., Ramsey N.B., et al. Three patients with X-linked agammaglobulinemia hospitalized for COVID-19 improved with convalescent plasma. J Allergy Clin Immunol Pract. 2020;8:3594–3596.e3. doi: 10.1016/j.jaip.2020.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khanmohammadi S., Rezaei N., Khazaei M., Shirkani A. A case of autosomal recessive interferon alpha/beta receptor alpha chain (IFNAR1) deficiency with severe COVID-19. J Clin Immunol. 2022;42:19–24. doi: 10.1007/s10875-021-01166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemarquis A., Campbell T., Aranda-Guillen M., Hennings V., Brodin P., Kampe O., et al. Severe COVID-19 in an APS1 patient with interferon autoantibodies treated with plasmapheresis. J Allergy Clin Immunol. 2021;148:96–98. doi: 10.1016/j.jaci.2021.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.London J., Boutboul D., Lacombe K., Pirenne F., Heym B., Zeller V., et al. Severe COVID-19 in patients with B cell alymphocytosis and response to convalescent plasma therapy. J Clin Immunol. 2021;41:356–361. doi: 10.1007/s10875-020-00904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mantravadi V., Nguyen S.T., Morley S.C., Bednarski J.J., Kitcharoensakkul M., Cooper M.A. Recovery from COVID-19 in a child with chronic granulomatous disease and T cell lymphopenia. J Clin Immunol. 2021;41:23–25. doi: 10.1007/s10875-020-00896-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milosevic I., Jovanovic J., Stevanovic O. Atypical course of COVID-19 in patient with Bruton agammaglobulinemia. J Infect Dev Ctries. 2020;14:1248–1251. doi: 10.3855/jidc.13840. [DOI] [PubMed] [Google Scholar]
- 61.Mira E., Yarce O.A., Ortega C., Fernandez S., Pascual N.M., Gomez C., et al. Rapid recovery of a SARS-CoV-2–infected X-linked agammaglobulinemia patient after infusion of COVID-19 convalescent plasma. J Allergy Clin Immunol Pract. 2020;8:2793–2795. doi: 10.1016/j.jaip.2020.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullur J., Wang A., Feldweg A. A fatal case of coronavirus disease 2019 in a patient with common variable immunodeficiency. Ann Allergy Asthma Immunol. 2021;126:90–92. doi: 10.1016/j.anai.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narahari P.G., Gebbia J., Alperstein W., Kleiner G., Gans M. Post–SARS-CoV-2 atypical inflammatory syndrome in a toddler with X-linked inhibitor of apoptosis deficiency after stem cell transplant. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01316-3. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez Clemente N., Penner J., Breuer J., Ip W., Booth C. Case report: a severe paediatric presentation of COVID-19 in APDS2 immunodeficiency. Front Immunol. 2022;13:881259. doi: 10.3389/fimmu.2022.881259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt A., Peters S., Knaus A., Sabir H., Hamsen F., Maj C., et al. TBK1 and TNFRSF13B mutations and an autoinflammatory disease in a child with lethal COVID-19. NPJ Genom Med. 2021;6:55. doi: 10.1038/s41525-021-00220-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Foca E., et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vignesh P., Mondal S., Sudhakar M., Sharma Y.K., Bansal A., Singh M., et al. SARS-CoV-2 infection in a child with severe congenital neutropenia. J Clin Immunol. 2021;41:1165–1168. doi: 10.1007/s10875-021-01054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levy R., Bastard P., Lanternier F., Lecuit M., Zhang S.Y., Casanova J.L. IFN-alpha2a therapy in two patients with inborn errors of TLR3 and IRF3 infected with SARS-CoV-2. J Clin Immunol. 2021;41:26–27. doi: 10.1007/s10875-020-00933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levy R., Zhang P., Bastard P., Dorgham K., Melki I., Hadchouel A., et al. Monoclonal antibody–mediated neutralization of SARS-CoV-2 in an IRF9-deficient child. Proc Natl Acad Sci U S A. 2021 doi: 10.1073/pnas.2114390118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al-Saud B., Hazzazi K.M., Mohammed R., Al Najjar A., Al Hazmi T., Monies D., et al. SARS-CoV-2–related acute respiratory distress syndrome uncovers a patient with severe combined immunodeficiency disease. J Clin Immunol. 2021 doi: 10.1007/s10875-021-01063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aljaberi R., Wishah K. Positive outcome in a patient with coronavirus disease 2019 and common variable immunodeficiency after intravenous immunoglobulin. Ann Allergy Asthma Immunol. 2020;125:349–350. doi: 10.1016/j.anai.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alkan G., Artac H., Oz S.K.T., Emiroglu M. Management of COVID-19 pneumonia in a child with NEMO deficiency. Immunol Res. 2021;69:391–393. doi: 10.1007/s12026-021-09184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Almontasheri A., Al-Husayni F., Alsuraihi A.K., Binyahib H., Albanna A.S. The clinical course of COVID-19 pneumonia in a 19-year-old man on intravenous immunoglobulin replacement therapy for X-linked agammaglobulinemia. Am J Case Rep. 2021;22 doi: 10.12659/AJCR.929447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bozonnat A., Assan F., LeGoff J., Bourrat E., Bachelez H. SARS-CoV-2 infection inducing severe flare up of deficiency of interleukin thirty-six (IL-36) receptor antagonist (DITRA) resulting from a mutation invalidating the activating cleavage site of the IL-36 receptor antagonist. J Clin Immunol. 2021 doi: 10.1007/s10875-021-01076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fill L., Hadney L., Graven K., Persaud R., Hostoffer R. The clinical observation of a patient with common variable immunodeficiency diagnosed as having coronavirus disease 2019. Ann Allergy Asthma Immunol. 2020;125:112–114. doi: 10.1016/j.anai.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee P.Y., Platt C.D., Weeks S., Grace R.F., Maher G., Gauthier K., et al. Immune dysregulation and multisystem inflammatory syndrome in children (MIS-C) in individuals with haploinsufficiency of SOCS1. J Allergy Clin Immunol. 2020;146:1194–1200.e1. doi: 10.1016/j.jaci.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Oers N.S.C., Hanners N.W., Sue P.K., Aquino V., Li Q.Z., Schoggins J.W., et al. SARS-CoV-2 infection associated with hepatitis in an infant with X-linked severe combined immunodeficiency. Clin Immunol. 2021;224:108662. doi: 10.1016/j.clim.2020.108662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bastard P., Levy R., Henriquez S., Bodemer C., Szwebel T.A., Casanova J.L. Interferon-beta therapy in a patient with incontinentia pigmenti and autoantibodies against type I IFNs infected with SARS-CoV-2. J Clin Immunol. 2021;41:931–933. doi: 10.1007/s10875-021-01023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Penafiel Vicuna A.K., Yamazaki Nakashimada M., Leon Lara X., Mendieta Flores E., Nunez Nunez M.E., Lona-Reyes J.C., et al. Mendelian susceptibility to mycobacterial disease: retrospective clinical and genetic study in Mexico. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bloomfield M., Parackova Z., Hanzlikova J., Lastovicka J., Sediva A. Immunogenicity and safety of COVID-19 mRNA vaccine in STAT1 GOF patients. J Clin Immunol. 2022;42:266–269. doi: 10.1007/s10875-021-01163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahmood H.Z., Madhavarapu S., Almuqamam M. Varying Illness severity in patients with MyD88 deficiency infected with coronavirus SARS-CoV-2. Pediatrics. 2021;147:453–454. [Google Scholar]
- 82.Rivalta B., Amodio D., Giancotta C., Santilli V., Pacillo L., Zangari P., et al. Case report: successful treatment with monoclonal antibodies in one apds patient with prolonged SARS-CoV-2 infection not responsive to previous lines of treatment. Front Immunol. 2022;13:891274. doi: 10.3389/fimmu.2022.891274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steiner S., Schwarz T., Corman V.M., Gebert L., Kleinschmidt M.C., Wald A., et al. SARS-CoV-2 T cell response in severe and fatal COVID-19 in primary antibody deficiency patients without specific humoral immunity. Front Immunol. 2022;13:840126. doi: 10.3389/fimmu.2022.840126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schidlowski L., Iwamura A.P.D., Covid S.U.D., Condino-Neto A., Prando C. Diagnosis of APS-1 in two siblings following life-threatening COVID-19 pneumonia. J Clin Immunol. 2022;42:749–752. doi: 10.1007/s10875-022-01245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palomba E., Carrabba M., Zuglian G., Alagna L., Saltini P., Fortina V., et al. Treatment of SARS-CoV-2 relapse with remdesivir and neutralizing antibodies cocktail in a patient with X-linked agammaglobulinaemia. Int J Infect Dis. 2021;110:338–340. doi: 10.1016/j.ijid.2021.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hovey J.G., Tolbert D., Howell D. Bruton’s agammaglobulinemia and COVID-19. Cureus. 2020;12 doi: 10.7759/cureus.11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duncan C.J.A., Skouboe M.K., Howarth S., Hollensen A.K., Chen R., Borresen M.L., et al. Life-threatening viral disease in a novel form of autosomal recessive IFNAR2 deficiency in the Arctic. J Exp Med. 2022:219. doi: 10.1084/jem.20212427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reiff D.D., Zhang M., Smitherman E.A., Mannion M.L., Stoll M.L., Weiser P., et al. A rare STXBP2 mutation in severe COVID-19 and secondary cytokine storm syndrome. Life (Basel) 2022;12 doi: 10.3390/life12020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delavari S., Abolhassani H., Abolnezhadian F., Babaha F., Iranparast S., Ahanchian H., et al. Impact of SARS-CoV-2 pandemic on patients with primary immunodeficiency. J Clin Immunol. 2021;41:345–355. doi: 10.1007/s10875-020-00928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abolhassani H., Vosughimotlagh A., Asano T., Landegren N., Boisson B., Delavari S., et al. X-linked TLR7 deficiency underlies critical COVID-19 pneumonia in a male patient with ataxia-telangiectasia. J Clin Immunol. 2022;42:1–9. doi: 10.1007/s10875-021-01151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abolhassani H., Delavari S., Landegren N., Shokri S., Bastard P., Du L., et al. Genetic and immunological evaluation of children with inborn errors of immunity and severe or critical COVID-19. J Allergy Clin Immunol. 2022 doi: 10.1016/j.jaci.2022.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Esenboga S., Ocak M., Akarsu A., Bildik H.N., Cagdas D., Iskit A.T., et al. COVID-19 in patients with primary immunodeficiency. J Clin Immunol. 2021 doi: 10.1007/s10875-021-01065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guven S.C., Erden A., Karakas O., Armagan B., Usul E., Omma A., et al. COVID-19 outcomes in patients with familial Mediterranean fever: a retrospective cohort study. Rheumatol Int. 2021;41:715–719. doi: 10.1007/s00296-021-04812-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karakoc Aydiner E., Bilgic Eltan S., Babayeva R., Aydiner O., Kepenekli E., Kolukisa B., et al. Adverse COVID-19 outcomes in immune deficiencies: inequality exists between subclasses. Allergy. 2022;77:282–295. doi: 10.1111/all.15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goudouris E.S., Pinto-Mariz F., Mendonca L.O., Aranda C.S., Guimaraes R.R., Kokron C., et al. Outcome of SARS-CoV-2 infection in 121 patients with inborn errors of immunity: a cross-sectional study. J Clin Immunol. 2021 doi: 10.1007/s10875-021-01066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grumach A.S., Goudouris E., Dortas Junior S., Marcelino F.C., Alonso M.L.O., Martins R.O., et al. COVID-19 affecting hereditary angioedema patients with and without C1 inhibitor deficiency. J Allergy Clin Immunol Pract. 2021;9:508–510. doi: 10.1016/j.jaip.2020.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marcus N., Frizinsky S., Hagin D., Ovadia A., Hanna S., Farkash M., et al. Minor clinical impact of COVID-19 pandemic on patients with primary immunodeficiency in Israel. Front Immunol. 2020;11:614086. doi: 10.3389/fimmu.2020.614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quinti I., Lougaris V., Milito C., Cinetto F., Pecoraro A., Mezzaroma I., et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. 2020;146:211–223.e4. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Milito C., Lougaris V., Giardino G., Punziano A., Vultaggio A., Carrabba M., et al. Clinical outcome, incidence, and SARS-CoV-2 infection-fatality rates in Italian patients with inborn errors of immunity. J Allergy Clin Immunol Pract. 2021;9:2904–2906.e2. doi: 10.1016/j.jaip.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giardino G., Milito C., Lougaris V., Punziano A., Carrabba M., Cinetto F., et al. The impact of SARS-CoV-2 infection in patients with inborn errors of immunity: the experience of the Italian Primary Immunodeficiencies Network (IPINet) J Clin Immunol. 2022 doi: 10.1007/s10875-022-01264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Milito C., Cinetto F., Palladino A., Garzi G., Punziano A., Lagnese G., et al. Biomedicines; 2022. Mortality in severe antibody deficiencies patients during the first two years of the COVID-19 pandemic: vaccination and monoclonal antibodies efficacy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deya-Martinez A., Garcia-Garcia A., Gonzalez-Navarro E.A., Yiyi L., Vlagea A., Jordan I., et al. COVID-19 in children and young adults with moderate/severe inborn errors of immunity in a high burden area in pre-vaccine era. Clin Immunol. 2021;230:108821. doi: 10.1016/j.clim.2021.108821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shields A.M., Anantharachagan A., Arumugakani G., Baker K., Bahal S., Baxendale H., et al. Outcomes following SARS-CoV-2 infection in patients with primary and secondary immunodeficiency in the UK. Clin Exp Immunol. 2022 doi: 10.1093/cei/uxac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown L.K., Moran E., Goodman A., Baxendale H., Bermingham W., Buckland M., et al. Treatment of chronic or relapsing COVID-19 in immunodeficiency. J Allergy Clin Immunol. 2022;149:557–561.e1. doi: 10.1016/j.jaci.2021.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castano-Jaramillo L.M., Yamazaki-Nakashimada M.A., O’Farrill-Romanillos P.M., Muzquiz Zermeno D., Scheffler Mendoza S.C., Venegas Montoya E., et al. COVID-19 in the context of inborn errors of immunity: a case series of 31 patients from Mexico. J Clin Immunol. 2021 doi: 10.1007/s10875-021-01077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Drabe C.H., Hansen A.E., Rasmussen L.D., Larsen O.D., Moller A., Mogensen T.H., et al. Low morbidity in Danish patients with common variable immunodeficiency disorder infected with severe acute respiratory syndrome coronavirus 2. Infect Dis (Lond) 2021;53:953–958. doi: 10.1080/23744235.2021.1957144. [DOI] [PubMed] [Google Scholar]
- 107.Katzenstein T.L., Rasmussen L.D., Drabe C.H., Larsen C.S., Hansen A.E., Staerkind M., et al. Outcome of SARS-CoV-2 infection among patients with common variable immunodeficiency and a matched control group: a Danish nationwide cohort study. Front Immunol. 2022;13:994253. doi: 10.3389/fimmu.2022.994253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koltan S., Zietkiewicz M., Grzesk E., Becht R., Berdej-Szczot E., Cienkusz M., et al. COVID-19 in unvaccinated patients with inborn errors of immunity-polish experience. Front Immunol. 2022;13:953700. doi: 10.3389/fimmu.2022.953700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Milota T., Sobotkova M., Smetanova J., Bloomfield M., Vydlakova J., Chovancova Z., et al. Risk factors for severe COVID-19 and hospital admission in patients with inborn errors of immunity—results from a multicenter nationwide study. Front Immunol. 2022;13:835770. doi: 10.3389/fimmu.2022.835770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bourguiba R., Delplanque M., Vinit C., Ackermann F., Savey L., Grateau G., et al. Clinical course of COVID-19 in a cohort of 342 familial Mediterranean fever patients with a long-term treatment by colchicine in a French endemic area. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-218707. [DOI] [PubMed] [Google Scholar]
- 111.Ho H.E., Mathew S., Peluso M.J., Cunningham-Rundles C. Clinical outcomes and features of COVID-19 in patients with primary immunodeficiencies in New York City. J Allergy Clin Immunol Pract. 2021;9:490–493.e2. doi: 10.1016/j.jaip.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cohen B., Rubinstein R., Gans M.D., Deng L., Rubinstein A., Eisenberg R. COVID-19 infection in 10 common variable immunodeficiency patients in New York City. J Allergy Clin Immunol Pract. 2021;9:504–507.e1. doi: 10.1016/j.jaip.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Greenmyer J.R., Joshi A.Y. COVID-19 in CVID: a case series of 17 patients. J Clin Immunol. 2022;42:29–31. doi: 10.1007/s10875-021-01150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuster J.K., Unlu S., Makin T.A., Par-Young J., Simonov M., Shafi S., et al. Low IgG trough and lymphocyte subset counts are associated with hospitalization for COVID-19 in patients with primary antibody deficiency. J Allergy Clin Immunol Pract. 2022;10:633–636.e3. doi: 10.1016/j.jaip.2021.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marcus N., Ashkenazi-Hoffnung L., Ovadia A., Dalal I., Yoffe S., Kropach N., et al. SARS-CoV-2 symptomatic reinfection among patients with primary antibody deficiency. J Allergy Clin Immunol Pract. 2022;10:1907–1909. doi: 10.1016/j.jaip.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Terlizzi V., Motisi M.A., Pellegrino R., Padoan R., Chiappini E. Risk factors for severe COVID-19 in people with cystic fibrosis: a systematic review. Front Pediatr. 2022;10:958658. doi: 10.3389/fped.2022.958658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bastard P., Orlova E., Sozaeva L., Levy R., James A., Schmitt M.M., et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med. 2021:218. doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tangye S.G., Bucciol G., Meyts I. Mechanisms underlying host defense and disease pathology in response to severe acute respiratory syndrome (SARS)-CoV2 infection: insights from inborn errors of immunity. Curr Opin Allergy Clin Immunol. 2021;21:515–524. doi: 10.1097/ACI.0000000000000786. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Puel A., Bastard P., Bustamante J., Casanova J.L. Human autoantibodies underlying infectious diseases. J Exp Med. 2022 doi: 10.1084/jem.20211387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meisel C., Akbil B., Meyer T., Lankes E., Corman V.M., Staudacher O., et al. Mild COVID-19 despite autoantibodies against type I IFNs in autoimmune polyendocrine syndrome type 1. J Clin Invest. 2021 doi: 10.1172/JCI150867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Campbell T.M., Liu Z., Zhang Q., Moncada-Velez M., Covill L.E., Zhang P., et al. Respiratory viral infections in otherwise healthy humans with inherited IRF7 deficiency. J Exp Med. 2022 doi: 10.1084/jem.20220202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci Immunol. 2021 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Q., Matuozzo D., Le Pen J., Lee D., Moens L., Asano T., et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J Exp Med. 2022 doi: 10.1084/jem.20220131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Q., Bastard P., Effort C.H.G., Cobat A., Casanova J.L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022 doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Troya J., Bastard P., Planas-Serra L., Ryan P., Ruiz M., de Carranza M., et al. Neutralizing autoantibodies to type I IFNs in >10% of patients with severe COVID-19 pneumonia hospitalized in Madrid, Spain. J Clin Immunol. 2021;41:914–922. doi: 10.1007/s10875-021-01036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years and account for ∼20% of COVID-19 deaths. Sci Immunol. 2021 doi: 10.1126/sciimmunol.abl4340. al e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.de Prost N., Bastard P., Arrestier R., Fourati S., Mahevas M., Burrel S., et al. Plasma exchange to rescue patients with autoantibodies against type I interferons and life-threatening COVID-19 pneumonia. J Clin Immunol. 2021;41:536–544. doi: 10.1007/s10875-021-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abers M.S., Rosen L.B., Delmonte O.M., Shaw E., Bastard P., Imberti L., et al. Neutralizing type-I interferon autoantibodies are associated with delayed viral clearance and intensive care unit admission in patients with COVID-19. Immunol Cell Biol. 2021 doi: 10.1111/imcb.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Manry J., Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., et al. The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2200413119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eto S., Nukui Y., Tsumura M., Nakagama Y., Kashimada K., Mizoguchi Y., et al. Neutralizing type I Interferon autoantibodies in Japanese patients with severe COVID-19. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R., et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021;13:eabh2624. doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Goncalves D., Mezidi M., Bastard P., Perret M., Saker K., Fabien N., et al. Antibodies against type I interferon: detection and association with severe clinical outcome in COVID-19 patients. Clin Transl Immunology. 2021;10:e1327. doi: 10.1002/cti2.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Raadsen M.P., Gharbharan A., Jordans C.C.E., Mykytyn A.Z., Lamers M.M., van den Doel P.B., et al. Interferon-alpha2 auto-antibodies in convalescent plasma therapy for COVID-19. J Clin Immunol. 2022;42:232–239. doi: 10.1007/s10875-021-01168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mathian A., Breillat P., Dorgham K., Bastard P., Charre C., Lhote R., et al. Lower disease activity but higher risk of severe COVID-19 and herpes zoster in patients with systemic lupus erythematosus with pre-existing autoantibodies neutralising IFN-alpha. Ann Rheum Dis. 2022 doi: 10.1136/ard-2022-222549. [DOI] [PubMed] [Google Scholar]
- 139.Nguyen H., Salkeld J., Agarwal S., Goodman A. Compassionate use of REGN-COV2 in the treatment of COVID-19 in a patient with impaired humoral immunity. Clin Infect Pract. 2021;12:100089. doi: 10.1016/j.clinpr.2021.100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Loh S.Y., Bassett J., Hoodless E.J., Walshaw M. Possible COVID-19 reinfection in a patient with X-linked agammaglobulinaemia. BMJ Case Rep. 2021 doi: 10.1136/bcr-2020-240765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Daoussis D., Leonidou L., Kalogeropoulou C., Paliogianni F., Tzouvelekis A. Protracted severe COVID-19 pneumonia following rituximab treatment: caution needed. Rheumatol Int. 2021;41:1839–1843. doi: 10.1007/s00296-021-04969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loarce-Martos J., Garcia-Fernandez A., Lopez-Gutierrez F., Garcia-Garcia V., Calvo-Sanz L., Del Bosque-Granero I., et al. High rates of severe disease and death due to SARS-CoV-2 infection in rheumatic disease patients treated with rituximab: a descriptive study. Rheumatol Int. 2020;40:2015–2021. doi: 10.1007/s00296-020-04699-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lang-Meli J., Fuchs J., Mathe P., Ho H.E., Kern L., Jaki L., et al. Case series: convalescent plasma therapy for patients with COVID-19 and primary antibody deficiency. J Clin Immunol. 2022;42:253–265. doi: 10.1007/s10875-021-01193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lougaris V., Baronio M., Vitali M., Tampella G., Cattalini M., Tassone L., et al. Bruton tyrosine kinase mediates TLR9-dependent human dendritic cell activation. J Allergy Clin Immunol. 2014;133:1644–1650.e4. doi: 10.1016/j.jaci.2013.12.1085. [DOI] [PubMed] [Google Scholar]
- 145.Treon S.P., Castillo J.J., Skarbnik A.P., Soumerai J.D., Ghobrial I.M., Guerrera M.L., et al. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID-19–infected patients. Blood. 2020;135:1912–1915. doi: 10.1182/blood.2020006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roschewski M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., et al. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci Immunol. 2020:5. doi: 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Roumier M., Paule R., Vallee A., Rohmer J., Ballester M., Brun A.L., et al. Tocilizumab for severe worsening COVID-19 pneumonia: a propensity score analysis. J Clin Immunol. 2021;41:303–314. doi: 10.1007/s10875-020-00911-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jamilloux Y., Henry T., Belot A., Viel S., Fauter M., El Jammal T., et al. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun Rev. 2020;19 doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ferré E.N.N., Schmitt M.M., Ochoa S., Rosen L.B., Shaw E.R., Burbelo P.D., et al. SARS-CoV-2 spike protein–directed monoclonal antibodies may ameliorate COVID-19 complications in APECED patients. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.720205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Barouch D.H. COVID-19 vaccines—immunity, variants, boosters. N Engl J Med. 2022;387:1011–1020. doi: 10.1056/NEJMra2206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sette A., Crotty S. Immunological memory to SARS-CoV-2 infection and COVID-19 vaccines. Immunol Rev. 2022;310:27–46. doi: 10.1111/imr.13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Galvez R.I., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–2451.e17. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chemaitelly H., Tang P., Hasan M.R., AlMukdad S., Yassine H.M., Benslimane F.M., et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N Engl J Med. 2021;385:e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N Engl J Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Siddle K.J., Krasilnikova L.A., Moreno G.K., Schaffner S.F., Vostok J., Fitzgerald N.A., et al. Transmission from vaccinated individuals in a large SARS-CoV-2 Delta variant outbreak. Cell. 2022;185:485–492.e10. doi: 10.1016/j.cell.2021.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wilkinson S.A.J., Richter A., Casey A., Osman H., Mirza J.D., Stockton J., et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 2022;8:veac050. doi: 10.1093/ve/veac050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Amodio D., Ruggiero A., Sgrulletti M., Pighi C., Cotugno N., Medri C., et al. Humoral and cellular response following vaccination with the BNT162b2 mRNA COVID-19 vaccine in patients affected by primary immunodeficiencies. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.727850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Antoli A., Rocamora-Blanch G., Framil M., Mas-Bosch V., Navarro S., Bermudez C., et al. Evaluation of humoral and cellular immune responses to the SARS-CoV-2 vaccine in patients with common variable immunodeficiency phenotype and patient receiving B-cell depletion therapy. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.895209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Arroyo-Sanchez D., Cabrera-Marante O., Laguna-Goya R., Almendro-Vazquez P., Carretero O., Gil-Etayo F.J., et al. Immunogenicity of anti–SARS-CoV-2 vaccines in common variable immunodeficiency. J Clin Immunol. 2022;42:240–252. doi: 10.1007/s10875-021-01174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barmettler S., DiGiacomo D.V., Yang N.J., Lam T., Naranbhai V., Dighe A.S., et al. Response to severe acute respiratory syndrome coronavirus 2 initial series and additional dose vaccine in patients with predominant antibody deficiency. J Allergy Clin Immunol Pract. 2022;10:1622–1634.e4. doi: 10.1016/j.jaip.2022.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bergman P., Blennow O., Hansson L., Mielke S., Nowak P., Chen P., et al. Safety and efficacy of the mRNA BNT162b2 vaccine against SARS-CoV-2 in five groups of immunocompromised patients and healthy controls in a prospective open-label clinical trial. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bergman P., Wullimann D., Gao Y., Wahren Borgstrom E., Norlin A.C., Lind Enoksson S., et al. Elevated CD21low B cell frequency is a marker of poor immunity to Pfizer-BioNTech BNT162b2 mRNA vaccine against SARS-CoV-2 in patients with common variable immunodeficiency. J Clin Immunol. 2022;42:716–727. doi: 10.1007/s10875-022-01244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carrabba M., Baselli L.A., Consonni D., Ceriotti F., Fabio G. Responses to SARS-CoV-2 vaccines of patients with common variable immune deficiencies and X-linked agammaglobulinemia. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01251-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Delmonte O.M., Bergerson J.R.E., Burbelo P.D., Durkee-Shock J.R., Dobbs K., Bosticardo M., et al. Antibody responses to the SARS-CoV-2 vaccine in individuals with various inborn errors of immunity. J Allergy Clin Immunol. 2021;148:1192–1197. doi: 10.1016/j.jaci.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hagin D., Freund T., Navon M., Halperin T., Adir D., Marom R., et al. Immunogenicity of Pfizer-BioNTech COVID-19 vaccine in patients with inborn errors of immunity. J Allergy Clin Immunol. 2021 doi: 10.1016/j.jaci.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pham M.N., Murugesan K., Banaei N., Pinsky B.A., Tang M., Hoyte E., et al. Immunogenicity and tolerability of COVID-19 messenger RNA vaccines in primary immunodeficiency patients with functional B-cell defects. J Allergy Clin Immunol. 2022;149:907–911.e3. doi: 10.1016/j.jaci.2021.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ponsford M.J., Evans K., Carne E.M., Jolles S. COVID-19 vaccine uptake and efficacy in a national immunodeficiency cohort. J Clin Immunol. 2022;42:728–731. doi: 10.1007/s10875-022-01223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pulvirenti F., Fernandez Salinas A., Milito C., Terreri S., Piano Mortari E., Quintarelli C., et al. Cells; 2021. B cell response induced by SARS-CoV-2 infection is boosted by the BNT162b2 vaccine in primary antibody deficiencies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Romano C., Esposito S., Donnarumma G., Marrone A. Detection of neutralizing anti-severe acute respiratory syndrome coronavirus 2 antibodies in patients with common variable immunodeficiency after immunization with messenger RNA vaccines. Ann Allergy Asthma Immunol. 2021;127:499–501. doi: 10.1016/j.anai.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Salinas A.F., Mortari E.P., Terreri S., Quintarelli C., Pulvirenti F., Di Cecca S., et al. SARS-CoV-2 vaccine induced atypical immune responses in antibody defects: everybody does their best. J Clin Immunol. 2021;41:1709–1722. doi: 10.1007/s10875-021-01133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shields A.M., Faustini S.E., Hill H.J., Al-Taei S., Tanner C., Ashford F., et al. SARS-CoV-2 vaccine responses in individuals with antibody deficiency: findings from the COV-AD study. J Clin Immunol. 2022 doi: 10.1007/s10875-022-01231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van Leeuwen L.P.M., GeurtsvanKessel C.H., Ellerbroek P.M., de Bree G.J., Potjewijd J., Rutgers A., et al. Immunogenicity of the mRNA-1273 COVID-19 vaccine in adult patients with inborn errors of immunity. J Allergy Clin Immunol. 2022;149:1949–1957. doi: 10.1016/j.jaci.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sokal A., Bastard P., Chappert P., Barba-Spaeth G., Fourati S., Vanderberghe A., et al. Human type I IFN deficiency does not impair B cell response to SARS-CoV-2 mRNA vaccination. J Exp Med. 2022 doi: 10.1084/jem.20220258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bastard P., Vazquez S., Liu J., Laurie M.T., Wang C.Y., Gervais A., et al. Vaccine breakthrough hypoxemic COVID-19 pneumonia in patients with auto-Abs neutralizing type I IFNs. Sci Immunol. 2022 doi: 10.1126/sciimmunol.abp8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Khalid M., Urban A., Darnel D., Freeman A. Clinical outcomes of SARS-CoV2 infection in STAT3 deficiency. J Clin Immunol. 2021;41 abstract 83. [Google Scholar]
- 177.Ochoa S., Rosen L., Lionakis M., Uzel G., Suez D. COVID-19 in 3 patients with CTLA4 haploinsufficiency and absence of autoantibodies to type 1 interferons. J Clin Immunol. 2021 [Google Scholar]