Abstract
Rationale
Common genetic variants have been associated with idiopathic pulmonary fibrosis (IPF).
Objectives
To determine functional relevance of the 10 IPF-associated common genetic variants we previously identified.
Methods
We performed expression quantitative trait loci (eQTL) and methylation quantitative trait loci (mQTL) mapping, followed by co-localization of eQTL and mQTL with genetic association signals and functional validation by luciferase reporter assays. Illumina multi-ethnic genotyping arrays, mRNA sequencing, and Illumina 850k methylation arrays were performed on lung tissue of participants with IPF (234 RNA and 345 DNA samples) and non-diseased controls (188 RNA and 202 DNA samples).
Measurements and Main Results
Focusing on genetic variants within 10 IPF-associated genetic loci, we identified 27 eQTLs in controls and 24 eQTLs in cases (false-discovery-rate-adjusted P < 0.05). Among these signals, we identified associations of lead variants rs35705950 with expression of MUC5B and rs2076295 with expression of DSP in both cases and controls. mQTL analysis identified CpGs in gene bodies of MUC5B (cg17589883) and DSP (cg08964675) associated with the lead variants in these two loci. We also demonstrated strong co-localization of eQTL/mQTL and genetic signal in MUC5B (rs35705950) and DSP (rs2076295). Functional validation of the mQTL in MUC5B using luciferase reporter assays demonstrates that the CpG resides within a putative internal repressor element.
Conclusions
We have established a relationship of the common IPF genetic risk variants rs35705950 and rs2076295 with respective changes in MUC5B and DSP expression and methylation. These results provide additional evidence that both MUC5B and DSP are involved in the etiology of IPF.
Keywords: pulmonary fibrosis, functional genomics, common genetic variant, transcriptome, epigenome
At a Glance Commentary
Scientific Knowledge on the Subject
Common genetic variants have been associated with idiopathic pulmonary fibrosis (IPF), but their functional consequences have not been fully elucidated.
What This Study Adds to the Field
Common IPF genetic risk variants rs35705950 and rs2076295 are associated with changes in MUC5B and DSP expression and methylation, respectively. These results provide additional evidence that both MUC5B and DSP are involved in the etiology of IPF.
While environmental factors play a role in the development of idiopathic pulmonary fibrosis (IPF) (1, 2), genetic risk factors explain a large portion of attributable risk (3) and represent promising approaches to identify disease before irreversible scarring, understand further disease pathogenesis, and identify additional therapeutic targets for this complex and incurable disease (4). Rare variants in telomerase and surfactant gene families have been associated with familial forms of pulmonary fibrosis but are unusual in sporadic cases of IPF (4–8). Common variants in 17 genetic loci have demonstrated genome-wide evidence for association with IPF (9–14).
To develop an integrated understanding of the rare and common variants located in the 10 primary genome-wide association study (GWAS) loci (12), we performed deep targeted resequencing across all 10 loci (3.15 Mb of DNA) in a large population of IPF patients (N = 3,624) and unaffected control subjects (N = 4,442). In that study, we identified 10 common variants that represent the common independent signals in these IPF risk loci and in aggregate account for at least 40% of the risk of IPF (8). Among them, the MUC5B promoter variant, rs35705950, was the strongest genetic risk variant for IPF (8). Questions remain, however, as to which causal biological mechanisms underlie these genetic associations and how identifying these mechanisms can help us understand disease pathogenesis or modify our approach to disease diagnosis and treatment. Our previous work used targeted approaches to assess the effect of the rs35705950 variant and promoter methylation in the regulation of MUC5B gene expression (15). We have more recently shown that the region around the rs35705950 variant functions as a classically defined enhancer subject to epigenetic programming (16). However, more comprehensive analysis of the MUC5B locus and other IPF-associated genetic loci has not been performed.
The Genotype-Tissue Expression (GTEx) project has shown that the majority of genes have regulatory genetic variants (17) and that expression quantitative trait locus (eQTL) mapping combined with co-localization of genetic and eQTL signal is a powerful approach to identify potentially causal genetic risk variants underlying GWAS signals (18). eQTL and co-localization approaches have also been successfully applied to other complex traits using diseased tissue (19, 20). Recently, GTEx also performed a cell type-interaction eQTL (ieQTL) analysis to analyze cell type–specificity of genetic regulation of gene expression across human tissues (21). Because methylation plays a critical role in regulating gene expression, DNA methylation may alter the effect of genetic variants on gene expression through methylation quantitative trait loci (mQTLs) (22, 23). Integration of mQTL and genetic signal at the same locus has also proven successful in prioritizing potential causal variants (22, 23) and mQTLs have previously been co-localized with genetic signal in COPD (24).
To investigate the functional relevance of common genetic IPF risk variants, we have performed genome, transcriptome, and methylome analyses on lung tissue from IPF and control subjects. Here, we report the results of eQTL and mQTL mapping to comprehensively study the effect of genetic variants on local (cis) gene expression and DNA methylation. We also performed co-localization and mediation analysis of eQTL and mQTL with genetic loci to prioritize potential causal risk variants, as well as functional validation of a region in MUC5B containing a novel mQTL by luciferase reporter assays.
Methods
We highlight the key methods in this section. Full methods are available in the online supplement. Count-level transcriptome and DNA methylome datasets are available through the Gene Expression Omnibus under accession GSE175459.
Study Population
Human tissue was collected after appropriate ethical review for the protection of human subjects through the National Heart Lung and Blood Institute (NHLBI)-sponsored Lung Tissue Research Consortium, Interstitial Lung Disease programs at the University of Colorado, National Jewish Health, University of California San Francisco and Vanderbilt University, as well as Committee for Oversight of Research and Clinical Training Involving Decedents for the Lung Donor Program at the University of Pittsburgh.
Genetic Data Processing and Imputation
We applied standard quality checks to the multi-ethnic genotyping array data (8, 12). Ancestral principal components (PCs) were derived by merging overlapping SNPs between our data and 1,000 Genomes samples. Imputation was performed against the Haplotype Reference Consortium v1.1 panel on the Michigan Imputation Server. This resulted in the final set of 7,975,707 SNPs available for further analysis.
Transcriptome Data Processing
mRNA libraries were prepared with TruSeq stranded mRNA library preparation kits (illumina) and sequenced at the average depth of 80M reads on the NovaSeq 6,000 (illumina). RNA paired-end reads were aligned to Ensembl GrCh38 using Kallisto (25). After quality control and filtering, transcriptome data were available on 21,449 genes/58,288 transcripts in 422 samples.
DNA Methylome Data Processing
DNA was bisulfite treated, labeled and hybridized to Illumina Infinium Human MethylationEPIC BeadChip using standard protocols. Illumina signal intensity files were processed using SeSAMe; within-sample normalization with out-of-band probes and dye bias correction were performed (26). After QC and filtering, methylome data were available on 712,229 probes in 547 samples.
Expression and Methylation Quantitative Trait Locus Analyses
To remove the effects of known and unknown batch effects, we used Probabilistic Estimation of Expression Residuals (PEER) (27). PEER factors are surrogate variables that capture technical and demographic batch effects present in the data. PEER models were run adjusting for age, sex, four genetic principal components, and 30 PEER factors. Four PCs from the genetic data were included to appropriately adjust for observed population stratification (see Figure E1 in the online supplement). Comparison of the top 36 Principal component regression analysis and PEER factors (Figure E2) demonstrates that PEER successfully captured confounding attributed to observed technical and demographic batch effects as well as unobserved population stratification. Because PEER adjusted for some of the effects of diagnosis, we ran eQTL and mQTL models separately in controls and IPF cases. eQTL and mQTL permutation models, in which the most significant SNP is identified for each transcript/CpG, were run using a distance cutoff between each SNP-gene/CpG pair of 1 Mb (2 Mb for the FAM13A locus because of the longer distance between the lead SNP and FAM13A gene [8]) in FastQTL (28). β-distributed P values from FastQTL output were adjusted to a 5% false discovery rate (FDR) using the Benjamini-Hochberg procedure (29). We performed confirmatory mQTL analysis using nominal model testing of all CpGs within 1 Mb (2 Mb for the FAM13A locus) of the lead SNPs in each IPF genetic locus. We examined quantile–quantile plots for all eQTL/mQTL models to determine the extent of genomic inflation; nominal mQTL models did not exhibit any inflation while some inflation was present in all permutation-based models (Figure E3). This is expected due to the nature of permutation-based testing in which case the most significant transcript/CpG-SNP pair is selected for each transcript/CpG tested. In general, residual inflation has been observed in studies of DNA methylation on Illumina arrays (30, 31).
Luciferase Assays in Cultured Cells
MUC5B reporter constructs were amplified (Table E1) from genomic DNA by PCR, cloned into pCR2.1-TOPO (Invitrogen), and subsequently ligated into the pGL3-Promoter vector (Invitrogen). A549 cells were transfected with reporter constructs using Lipofectamine 2,000 (Life Technologies) as instructed by the manufacturer. Approximately 24 hours later, luciferase activity was assayed using the Dual-Luciferase Reporter Assay System from Promega as described previously (32). Firefly luciferase activity was normalized to that of a Renilla luciferase internal control (pRL-SV40; Promega).
Results
Expression Quantitative Trait Loci (eQTLs)
As expected, the IPF subjects were older, more often male, and have a more extensive cigarette smoking history than healthy controls (Table 1). We did not observe significant differences in self-reported race and ethnicity between the groups. However, we did observe differences in the method used to obtain lung tissue between IPF subjects and controls (Table 1).
Table 1.
Cohort Characteristics for Controls and IPF Cases Included in the Gene Expression and DNA Methylation Datasets
| Gene Expression Dataset | Control (N = 188) | IPF (N = 234) | P Value |
|---|---|---|---|
| Age | 55.3 ± 16.8 | 61.4 ± 7.5 | 9.92 × 10−6 |
| Sex (M/F) | 110/78 (59/41%) | 184/50 (79/21%) | 1.11 × 10−5 |
| Self-reported race, White (yes/no) | 155/33 (82/18%) | 200/34 (85/15%) | 0.42 |
| Self-reported ethnicity, non-Hispanic (yes/no/unknown) | 162/6/20 (86/3/11%) | 197/20/16 (84/9/7%) | 0.20 |
| Cigarette smoke (ever/never/unknown) | 86/89/13 (46/47/7%) | 140/75/19 (60/32/8%) | 5.7 × 10−3 |
| Tissue sampling method (biopsy/explant/autopsy/unknown) | 4/184/0/0 (3/73/0/1%) | 18/145/3/68 (8/62/1/29%) | 2.2 × 10−16 |
| DNA Methylation Dataset | Control (N = 202) | IPF (N = 345) | P Value |
|---|---|---|---|
| Age | 54.8 ± 16.3 | 61.6 ± 7.8 | 7.22 × 10−8 |
| Sex (M/F) | 123/79 (61/39%) | 263/82 (76/24%) | 2.09 × 10−4 |
| Self-reported race, White (yes/no) | 168/34 (83/17%) | 295/50 (85/15%) | 0.46 |
| Self-reported ethnicity, non-Hispanic (yes/no/unknown) | 174/8/20 (86/4/10%) | 288/25/32 (84/7/9%) | 0.30 |
| Cigarette smoke (ever/never/unknown) | 90/98/14 (45/48/7%) | 209/106/30 (60/31/9%) | 1.90 × 10−4 |
| Tissue sampling method (biopsy/explant/autopsy/unknown) | 4/193/0/5 (2/96/0/2%) | 37/185/2/121 (11/54/1/35%) | 2.2 × 10−16 |
Definition of abbreviation: IPF = idiopathic pulmonary fibrosis.
P values were determined by two tailed Student’s t test for age and Fisher Exact test for all other variables
Using residuals from the PEER model, we ran permutation-based transcriptome-wide cis-eQTL analysis in controls and IPF cases separately. This analysis identified 4,745 transcripts with significant association of genetic variants and gene expression (eQTLs) in controls and 6,047 in cases (FDR-adjusted P < 0.05; Table E2). Of the 20,704 tested transcripts, 16,685 also appeared in GTEx lung eQTLs. Concordance of our results with GTEx, estimated via a calculation, suggested broad replication of our eQTLs in the GTEx dataset ( = 0.58). Concordance was particularly high for highly significant eQTLs (q < 5 × 10−4) as demonstrated by the high correlation coefficient of the effect size (slope from the linear model) in our data compared with GTEx lung eQTL data (Pearson r = 0.90, Figure E4).
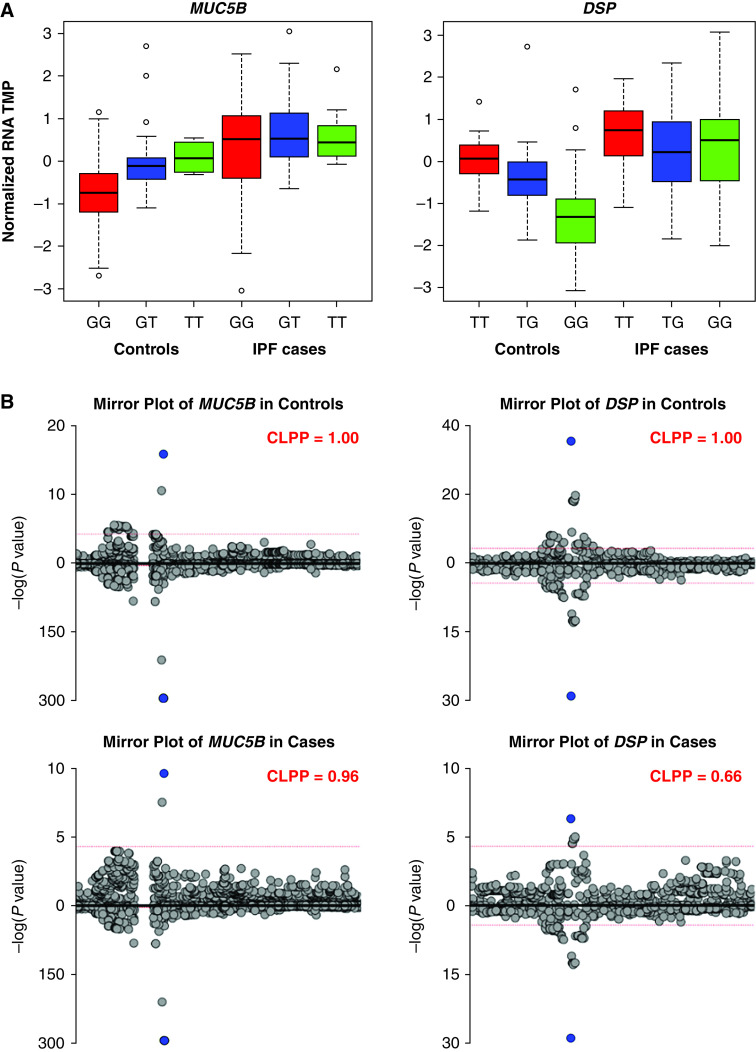
We next focused on cis-eQTLs in the 10 IPF genetic loci analyzed in our recent resequencing study (8). Of the 161 transcripts within the boundaries of the 10 IPF genetic loci, defined in Moore and colleagues (8), 27 eQTLs were significant in controls and 24 in cases (FDR-adjusted bpval < 0.05; Table 2). Among these eQTLs, only expression of MUC5B and DSP mRNA was associated with the lead genetic variants identified in the two loci (11p15/MUC5B and 6p24/DSP, respectively). In the MUC5B locus, the presence of the alternate allele (T) compared with the major allele (G) at rs35705950 was associated with higher expression of MUC5B mRNA in controls (Figure 1A, left panel). The effect is not apparent in cases with transcript level without adjustment for PEER factors plotted (Figure 1A, left panel) but it is statistically significant although both significance level and the effect size of the eQTL are diminished in cases compared with controls (Table 2). When PEER-normalized transcript levels are plotted, the effect of rs35705950 on MUC5B expression is visually apparent (Figure E5). In the DSP locus, the presence of the alternate allele (G) compared with major allele (T) at rs2076295 was associated with lower expression of DSP mRNA in controls with diminished effect in cases (Figure 1A, right panel and Figure E5).
Table 2.
Significant eQTLs within the Boundaries of IPF Genetic Loci in Controls and Cases
| Controls | ||||||
|---|---|---|---|---|---|---|
| IPF Locus (Chr: Region) | Gene Name | SNP ID | Distance, bp | Slope | bpval | adj bpval |
| 3:169723712-169910112 | LRRC31 | rs9290376 | 27278 | −0.41834 | 0.006212 | 0.019114 |
| 4:88684849-89177649 | NAP1L5 | rs2860500 | 30003 | 0.304358 | 0.017387 | 0.044866 |
| 6:7493767-7687767 | DSP | rs2076295 | 21423 | −0.66886 | 2.18E-26 | 3.49E-25 |
| 6:7493767-7687767 | AL031058.1 | rs2076295 | 22547 | −0.43322 | 5.25E-08 | 2.71E-07 |
| 7:99889391-100167139 | MBLAC1 | rs139788064 | 381983 | 0.910552 | 1.89E-05 | 8.17E-05 |
| 7:99889391-100167139 | TRIM4 | rs2527925 | 5056 | 0.651133 | 5.06E-28 | 1.09E-26 |
| 7:99889391-100167139 | AZGP1 | rs2527889 | −15304 | −0.56105 | 7.53E-08 | 3.76E-07 |
| 7:99889391-100167139 | AP4M1 | rs9649220 | −59212 | −0.28104 | 0.000245 | 0.000853 |
| 7:99889391-100167139 | LAMTOR4 | rs77712869 | −66934 | −0.64989 | 9.85E-05 | 0.000358 |
| 7:99889391-100167139 | TAF6 | rs1050542 | 133 | −0.17539 | 6.03E-05 | 0.000235 |
| 7:99889391-100167139 | CNPY4 | rs144305120 | 28348 | −0.38747 | 0.010134 | 0.028445 |
| 7:99889391-100167139 | ZSCAN21 | rs3736591 | 103619 | −0.26315 | 0.010567 | 0.029149 |
| 10:103837373-104069233 | SH3PXD2A | rs11191773 | 106615 | −0.21871 | 0.005672 | 0.017795 |
| 10:103837373-104069233 | SLK | rs2864020 | 572109 | −0.15223 | 0.007512 | 0.021464 |
| 11:1008000-1737755 | MUC5B | rs35705950 | −3076 | 0.560129 | 2.97E-11 | 1.76E-10 |
| 11:1008000-1737755 | KRTAP5-AS1 | rs1809668 | 27323 | −0.38391 | 1.87E-05 | 8.17E-05 |
| 11:1008000-1737755 | AC068580.4 | rs8839 | 18084 | −0.51894 | 9.62E-05 | 0.000358 |
| 11:1008000-1737755 | IFITM10 | rs72843946 | 136930 | −0.43939 | 0.007374 | 0.021452 |
| 11:1008000-1737755 | LINC02688 | rs1127800 | −179414 | 0.652602 | 0.0167 | 0.044479 |
| 13:112648686-113136861 | AL356740.1 | rs4907571 | −654 | 0.312598 | 0.002177 | 0.006966 |
| 15:40293756-40521315 | INAFM2 | rs934935 | 19311 | −0.27894 | 0.001665 | 0.00555 |
| 15:40293756-40521315 | KNSTRN | rs76277863 | −25060 | 1.02178 | 1.59E-25 | 2.32E-24 |
| 15:40293756-40521315 | BAHD1 | rs6492945 | −41976 | −0.19031 | 0.014081 | 0.038186 |
| 15:40293756-40521315 | IVD | rs8033938 | 5119 | −0.62869 | 8.64E-12 | 5.32E-11 |
| 15:40293756-40521315 | DISP2 | rs11636147 | 70693 | 0.94725 | 1.07E-10 | 6.14E-10 |
| 19:4635088-4766379 | MYDGF | rs72620526 | 51891 | 0.211868 | 0.016958 | 0.044479 |
| 19:4635088-4766379 | DPP9 | rs62115083 | 548680 | 0.215823 | 0.017666 | 0.044866 |
| Cases | ||||||
|---|---|---|---|---|---|---|
| IPF Locus (Chr: Region) | Gene Name | SNP ID | Distance | Slope | bpval | adj bpval |
| 3:169723712-169910112 | LRRC31 | rs2276718 | 30773 | −0.31134 | 0.007675 | 0.020813 |
| 4:88684849-89177649 | NAP1L5 | rs8605 | 320 | 0.233498 | 0.003354 | 0.009937 |
| 6:7493767-7687767 | DSP | rs2076295 | 21423 | −0.15647 | 0.005279 | 0.015081 |
| 7:99889391-100167139 | TRIM4 | rs2527926 | 2942 | 0.627493 | 4.29E-48 | 2.29E-46 |
| 7:99889391-100167139 | AZGP1 | rs2527898 | −30314 | −0.32512 | 0.018054 | 0.045136 |
| 7:99889391-100167139 | ZSCAN21 | rs4729571 | 869 | −0.32981 | 3.43E-07 | 1.57E-06 |
| 7:99889391-100167139 | CNPY4 | rs114198920 | −54504 | −0.61142 | 8.66E-06 | 3.08E-05 |
| 7:99889391-100167139 | AP4M1 | rs13236456 | −23615 | −0.23271 | 1.58E-05 | 5.38E-05 |
| 7:99889391-100167139 | TAF6 | rs2293481 | −5068 | −0.22596 | 4.13E-06 | 1.57E-05 |
| 7:99889391-100167139 | MBLAC1 | rs146579476 | 6792 | 0.970627 | 2.41E-08 | 1.13E-07 |
| 11:1008000-1737755 | MUC5B | rs35705950 | −3076 | 0.307958 | 8.32E-06 | 3.03E-05 |
| 11:1008000-1737755 | TOLLIP-AS1 | rs80158222 | −85073 | −0.43748 | 1.62E-06 | 6.33E-06 |
| 11:1008000-1737755 | BRSK2 | rs36064646 | −935 | −0.26786 | 0.0198 | 0.048739 |
| 11:1008000-1737755 | KRTAP5-AS1 | rs34995599 | 9984 | −0.42908 | 2.21E-11 | 1.26E-10 |
| 11:1008000-1737755 | IFITM10 | rs111693235 | 16583 | −0.27613 | 0.010576 | 0.02774 |
| 11:1008000-1737755 | AC068580.4 | rs111693235 | 14172 | −0.26365 | 0.014982 | 0.038049 |
| 13:112648686-113136861 | AL139384.1 | rs2274774 | 2122 | −0.26887 | 5.40E-06 | 2.01E-05 |
| 13:112648686-113136861 | AL356740.1 | rs4907571 | −654 | 0.428474 | 1.81E-11 | 1.07E-10 |
| 15:40293756-40521315 | CHST14 | rs28521889 | −249578 | 0.492508 | 0.005897 | 0.016554 |
| 15:40293756-40521315 | INAFM2 | rs7171143 | −195 | −0.3379 | 1.09E-05 | 3.79E-05 |
| 15:40293756-40521315 | DISP2 | rs56221586 | −35 | 0.975899 | 3.55E-18 | 2.71E-17 |
| 15:40293756-40521315 | KNSTRN | rs17671194 | −7384 | 1.13297 | 1.25E-39 | 3.99E-38 |
| 15:40293756-40521315 | IVD | rs8033938 | 5119 | −0.52893 | 2.52E-12 | 1.55E-11 |
| 19:4635088-4766379 | DPP9 | rs12462642 | 39779 | −0.22049 | 0.003044 | 0.009188 |
Definition of abbreviations: adj bpval = Benjamini-Hochberg adjusted bpval; bp = base pairs; bpval = P value of association adjusted for the number of variants tested in cis given by the fitted β distribution; Chr = chromosome; eQTLs = expression quantitative trait loci; IPF = idiopathic pulmonary fibrosis.
Highlighted are eQTLs associated with the lead variants from the IPF resequencing study by Moore et al. (8)
Figure 1.
eQTLs at MUC5B and DSP loci co-localize with genetic signal. (A) Box plots for MUC5B and DSP eQTLs. Normalized RNA TMP refers to transcripts per million after trimmed mean of M values (TMM) normalization across samples and inverse normal transformation on a per-gene basis, as has been done in GTEx. eQTL analysis was performed in 188 controls and 234 cases with the following genotype breakdown. MUC5B rs35705950: 141 GG, 42 GT, and 5 TT in controls and 105 GG, 114 GT, and 15 TT in IPF. DSP rs2076295: 54 TT, 90 GT, and 44 GG in controls and 54 TT, 101 GT, and 79 GG in IPF. (B) Mirror plots for co-localization of eQTL (top) with genetic signal (bottom) at MUC5B and DSP loci. CLPP = co-localization posterior probability; eQTL = expression quantitative trait loci; IPF = idiopathic pulmonary fibrosis.
We also performed a subgroup analysis of cis-eQTLs in the 10 IPF genetic loci in 140 controls and 140 cases frequency-matched on age, sex, and smoking (Table E3). Given the reduction in sample size, it is not surprising that eQTLs in MUC5B and DSP remained highly significant in controls but did not reach significance in cases. However, the effect size measured by the slope in the linear model remained similar. For MUC5B, slopes are 0.560 in the full cohort and 0.553 in the subgroup analysis in controls, and 0.308 versus 0.247 in full cohort versus subgroup analysis in cases. For DSP, slopes are -0.669 in the full cohort and -0.695 in the subgroup analysis in controls, and −0.156 versus −0.166 in full versus subgroup analysis in cases. We have previously observed less pronounced effects of the MUC5B genetic variant on lung gene expression in cases than in controls (15, 33) and the reduced effect size in cases is in line with these observations. We believe that extensive remodeling of IPF lung that is associated with changes in expression of many genes may be responsible for masking some of the effects of the genetic variants compared with what we observed in controls. Overall, however, this subgroup analysis demonstrates that age, sex and smoking do not significantly influence our key eQTL findings.
Co-localization of eQTLs and Genetic Loci
To assess potential causality of the genetic variants that act as eQTLs within the IPF loci, we performed a co-localization analysis of genetic (from the resequencing study; exclusively non-Hispanic White [8]) and eQTL (from the current study; >80% non-Hispanic White) data. Using eCAVIAR co-localization posterior probability (CLPP) scores, we demonstrate almost perfect co-localization of genetic and eQTL signal for the MUC5B promoter variant rs35705950 (CLPP = 1.00 in controls and 0.96 in cases; Figure 1B, left panel). We also observed a strong co-localization of genetic and eQTL signal for the DSP risk variant rs2076295 (CLPP = 1.00 in controls and 0.66 in cases; Figure 1B, right panel). Examination of all CLPP scores (Table E4) revealed only a few other co-localization results in the chr11 and chr6 loci. rs12802931 and MUC5B had CLPP of 0.93 in controls and 0.50 in cases; this variant is in linkage disequilibrium with rs35705950 (D’ = 1.00, R2 = 0.54 in the EUR population), indicating that rs12802931 does not represent an independent signal. Moreover, conditional testing at rs35705950 indicated that rs12802931 does not represent an independent signal from that of rs35705950 (P = 0.27 after conditional testing). On chr6, rs2076295 had a CLPP of 0.99 in controls with lncRNA AL031058.1, which is antisense to DSP and located in the promoter of DSP. However, we observed no evidence of co-localization in cases (CLPP = 0.040). Similarly, rs55938083 has a CLPP of 0.89 with DSP and 0.44 with lncRNA AL031058.1 in controls but this co-localization was not present in cases (CLPP = 6.3 × 10−4 for DSP and 9.4 × 10−3 for lncRNA AL031058.1). This variant is in linkage disequilibrium with rs2076295 (D’ = 1.00, R2 = 0.54 in the EUR population) and therefore not an independent signal. All other CLPP scores in controls and cases were low (CLPP < 0.10) and provided no evidence for co-localization of genetic and eQTL signals in the remaining IPF loci.
Cell Type Interaction eQTLs (ieQTLs)
To address the issue of cell type-specificity in complex tissue, we performed cell type–interaction eQTL (ieQTL) analysis restricted to IPF loci in seven cell populations that are enriched in lung tissue (Figure E6A). This ieQTL analysis revealed 81 significant associations in total, ranging between 1 and 7 significant ieQTLs in each of the 14 models (7 cell types, controls and IPF cases; Table E5). Only one of these associations is related to an IPF locus lead variant, rs2076295 with DSP in smooth muscle cells. We observed decreased expression of DSP with the alternate allele, as expected from our eQTL analysis and previous work (12), when enrichment for smooth muscle cells is low; the effect of the allele on expression diminishes with increasing enrichment of smooth muscle cells (Figure E6B). eCAVIAR analysis demonstrated co-localization of this smooth muscle cell ieQTL in controls with the genetic signal (Figure E6B; CLPP = 0.87). As ieQTLs may capture compensatory cell abundance changes, we asked whether this anticorrelation with smooth muscle cell enrichment may be driven by a compensatory increased abundance in another cell type. Because of the interest in DSP expression in epithelial cells in IPF (34), we also visually examined the effect in epithelial cells, even though no significant interaction was observed; opposite of smooth muscle results, the alternate allele is associated with a decrease in DSP expression in controls when enrichment for epithelial cells is high. These data suggest that the effect of the genetic variant on DSP expression in control tissue is potentially attributable to epithelial cells. However, epithelial cell populations in lung tissue are complex and these preliminary results need further validation. We did not observe any ieQTL effects in IPF lung tissue, likely because of the widespread aberrant expression of DSP in diseased lung, including epithelial cells (Figure E7).
DNA Methylation Quantitative Trait Loci (mQTLs) and Co-localization of mQTLs and Genetic Loci
We next focused on the possibility that IPF genetic variants may influence DNA methylation. We performed both a permutation-based analysis of all SNP-CpG pairs within the boundaries of the risk loci (analogous to the analysis we performed for the eQTLs) and nominal analysis of only the 10 lead risk variants (as previously published mQTL analyses used nominal models [35–40]). Permutation analysis revealed on the order of 10 to a few hundred significant associations of the genetic variants with DNA methylation levels (cis-mQTLs) in each genetic locus (Table 3 and Table E6); of these, seven mQTLs in controls and four mQTLs in cases were associated with lead genetic variants, and only two (rs35705950-cg17589883 in the MUC5B locus and rs2076295-cg08964675 in the DSP locus) were identified in both controls and cases (Table 4). More conventional nominal mQTL models focusing on only lead genetic variants confirmed the association of these two SNP-CpG pairs (Table E7). We also performed subgroup permutation and nominal mQTL analyses in the same set of 140 controls and 140 cases frequency-matched on age, sex, and smoking that were used in the subgroup eQTL analysis. Not surprisingly, this reduction in cohort size substantially reduced the number of significant loci both in permutation and nominal models (Table E8). Importantly, however, the effect sizes remain similar. For rs35705950-cg17589883 mQTL in the MUC5B locus, slopes are 0.201 in the full cohort and 0.205 in the subgroup analysis in controls, and 0.142 versus 0.166 in full versus subgroup analysis in cases. For rs2076295-cg08964675 mQTL in the DSP locus, slopes are 0.223 in the full cohort and 0.196 in the subgroup analysis in controls, and 0.152 versus 0.149 in full versus subgroup analysis in cases. These results demonstrate that age, sex, and smoking do not significantly influence our key mQTL results.
Table 3.
Significant mQTLs within the Boundaries of IPF Genetic Loci. Summary of Significant mQTLs in Permutation-based and Nominal Analyses
| IPF Locus (Chr: Region) | Number of CpGs Tested | Significant mQTLs in Controls | Significant mQTLs in Cases |
|---|---|---|---|
| 3:169723712-169910112 | 609 | 15 | 20 |
| 4:88684849-89177649 | 816 | 33 | 38 |
| 5:1213146-1385099 | 2047 | 37 | 80 |
| 6:7493767-7687767 | 618 | 31 | 31 |
| 7:99889391-100167139 | 1687 | 51 | 95 |
| 10:103837373-104069233 | 875 | 41 | 60 |
| 11:1008000-1737755 | 3347 | 221 | 348 |
| 13:112648686-113136861 | 2256 | 178 | 244 |
| 15:40293756-40521315 | 1272 | 70 | 103 |
| 19:4635088-4766379 | 1753 | 12 | 15 |
Definition of abbreviations: Chr = chromosome; IPF = idiopathic pulmonary fibrosis; mQTLs = methylation quantitative trait loci.
Table 4.
Significant mQTLs Associated with Lead Variants in IPF Loci from the IPF Resequencing Study by Moore et al. (8)
| SNP | CpG | Gene Name | CpG Gene Relation | Distance, bp | Slope | bpval | Adjusted bpval |
|---|---|---|---|---|---|---|---|
| Controls | |||||||
| rs2609260 | cg25055244 | FAM13A | Body | 48245 | −0.13281 | 0.00292 | 0.044863 |
| rs2076295 | cg08964675 | DSP | Body | 423 | 0.222649 | 3.02E-05 | 0.000989 |
| rs2076295 | cg12817734 | DSP | Body | 9397 | −0.07546 | 8.18E-08 | 5.05E-06 |
| rs35705950 | cg03298405 | MUC5B | Promoter | −1206 | 0.133362 | 0.000937 | 0.017711 |
| rs35705950 | cg16842717 | MUC5B | Body | −11050 | 0.329679 | 0.000101 | 0.002513 |
| rs35705950 | cg17589883 | MUC5B | Body | −18978 | 0.200628 | 4.44E-07 | 1.60E-05 |
| rs35705950 | cg19488922 | MUC5B | Body | −8223 | 0.237018 | 0.000579 | 0.011594 |
| IPF cases | |||||||
| rs2076295 | cg03818715 | SNRNP48 | Body | −28116 | 0.091787 | 0.000559 | 0.01329 |
| rs2076295 | cg08964675 | DSP | Body | 423 | 0.151517 | 0.000201 | 0.005579 |
| rs35705950 | cg02522041 | MUC5B | Body | −7787 | 0.07022 | 0.000376 | 0.004949 |
| rs35705950 | cg17589883 | MUC5B | Body | −18978 | 0.142382 | 6.92E-08 | 1.48E-06 |
Definition of abbreviations: adj bpval = Benjamini-Hochberg adjusted bpval; bp = base pairs; IPF = idiopathic pulmonary fibrosis; mQTLs = methylation quantitative trait loci.
Highlighted are eQTLs associated with the lead variants from the IPF resequencing study by Moore et al. (8)
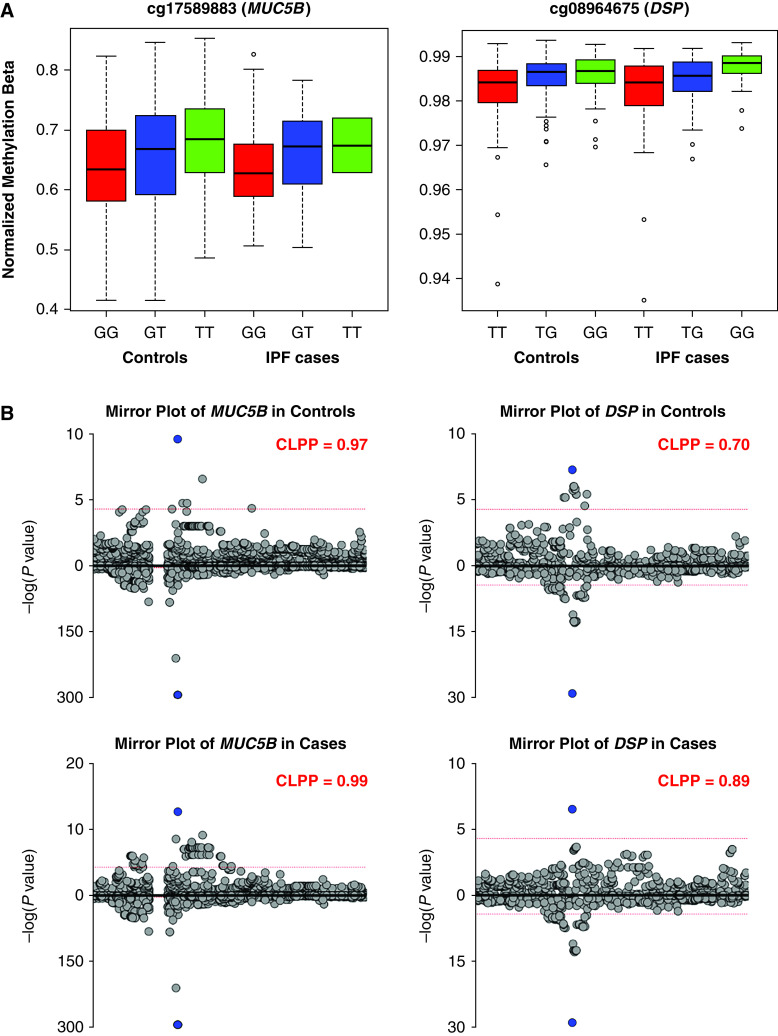
The presence of the alternate allele (T) compared with the major allele (G) at rs35705950 was associated with higher methylation at the cg17589883 CpG within the gene body (exon 26) of MUC5B in both controls and cases (Figures 2A and E8, left panel). Similarly, the presence of the alternate allele (G) compared with major allele (T) at rs2076295 was associated with higher methylation at the cg08964675 CpG within the gene body (intron 4) of DSP in both controls and cases (Figures 2A and E8, right panel). Both effects were more pronounced in controls than cases, analogous to our eQTL results. Similar to the eQTL co-localization, we observed almost perfect co-localization of genetic and mQTL loci for rs35705950 and cg17589883 (CLPP of 0.97 in controls and 0.99 in cases; Figure 2B, left panel). We also observed a strong co-localization of genetic and mQTL loci for rs2076295 and cg08964675 (CLPP of 0.70 in controls and 0.89 in cases; Figure 2B, right panel).
Figure 2.
mQTLs at MUC5B and DSP loci co-localize with genetic signal. (A) Box plots for MUC5B (cg17589883) and DSP (cg08964675) mQTLs. Normalized methylation β refers to β values of DNA methylation level measured on Illumina arrays (scale 0–1) after SeSame data preprocessing and normalization. mQTL analysis was performed in 202 controls and 345 cases with the following genotype breakdown. MUC5B rs35705950: 165 GG, 35 GT, and 2 TT in controls and 160 GG, 164 GT, and 21 TT in IPF. DSP rs2076295: 61 TT, 89 GT, and 52 GG in controls and 88 TT, 149 GT, and 108 GG in IPF. (B) Mirror plots for co-localization of mQTL (top) with genetic signal (bottom) at MUC5B and DSP loci. CLPP = co-localization posterior probability; IPF = idiopathic pulmonary fibrosis; mQTL = methylation quantitative trait loci.
Mediation Analysis
Given the evidence for potential causality at the MUC5B and DSP genetic risk loci from co-localization analyses, we performed a mediation analysis by running a series of regression models, while adjusting for sex and four genetic PCs (results of all models summarized in Table E9). The association of rs35705950 with disease risk was attenuated from an odds ratio (OR) of 3.08 (95% confidence interval [CI], 1.96–4.85; P = 1.14 × 10−6) to 1.95 (95% CI, 1.18–3.21; P = 8.74 × 10−3) after adjusting for MUC5B transcript expression (36.7% reduction in OR; 95% CI, −49.55% to −21.99%). Conversely, the association of rs2076295 with disease risk was enhanced from an OR of 1.14 (95% CI, 0.84–4.27; P = 0.39) to 1.77 (95% CI, 1.22–8.54; P = 2.85 × 10−3) when DSP transcript expression is included in the model (54.8% increase in OR; 95% CI, 26.04–107.58%). Because the presence of the alternate allele at rs35705950 is positively associated with both MUC5B expression and disease risk, the attenuation of the association between rs35705950 and IPF is suggestive of partial mediation of the effect of the genetic variant on disease risk. Because the alternate (G) allele at rs2076295 is negatively associated with DSP expression but positively associated with disease risk, the observed enhancement of the effect of the G allele on disease risk after adjustment for expression is consistent with expression being a negative confounder in the relationship between the allele and disease risk. We have noted this complex relationship in the DSP locus previously (12). We did not observe any evidence of mediation through DNA methylation (Table E10).
Transcriptional Activity of the MUC5B Locus
Examination of genomic functional element annotation provided by The Encyclopedia of DNA Elements (ENCODE) Consortium indicates that both CpGs reside in regions enriched for transcription factor binding, sensitivity to nuclease digestion, and histone marks associated with open/active chromatin (Figure E9), and therefore may be important in gene regulation. To determine whether the region in the MUC5B locus harboring the cg17589883 CpG has a functional role, we generated a luciferase reporter containing this CpG and several hundred base pairs of flanking DNA (referred to as “Reg 4”; see Figure 3). We also generated reporters from several additional putative regulatory regions of the MUC5B locus based on ENCODE data. We tested these reporters along with a previously described construct spanning the MUC5B rs35705950 site that exhibits enhancer activity (“Reg 1”) (16). In this assay, the cg17589883 CpG region repressed reporter activity in the “Reg 4” construct, whereas a region near the 3′ UTR in the “Reg 6” construct functioned as an enhancer (Figure 3). The other tested regions had minimal effect. These data provide further support for a functional role of the CpG-containing region in regulating MUC5B expression. Future work will need to determine whether this repressor region within the MUC5B gene interacts with the enhancer containing rs35705950.
Figure 3.
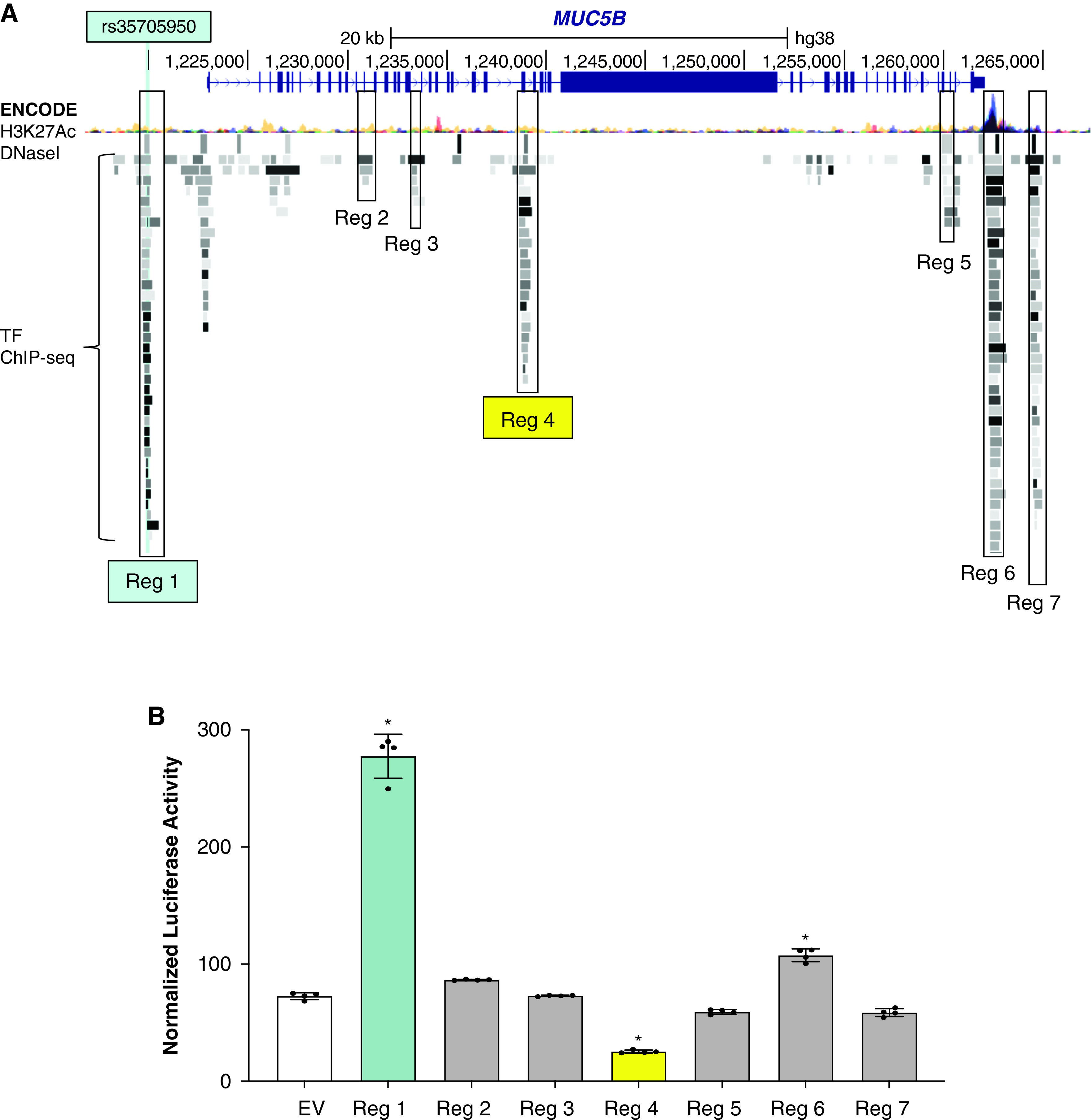
Reporter assays ascribe selective regulatory function to cg17589883 region in MUC5B. (A) UCSC Genome Browser screenshot of the MUC5B locus with H3K27Ac Mark, DNase I Hypersensitivity Peak, and Transcription Factor (TF) ChIP-seq Cluster tracks from The Encyclopedia of DNA Elements Consortium enabled and used to identify regions with putative regulatory function for cloning into luciferase reporters, as indicated. (B) Mean ± SD normalized luciferase activity of indicated reporter constructs and empty vector (EV) control in A549 cells under basal culture conditions (n = 4 per group and *P < 0.0001 versus EV via one-way ANOVA with Bonferroni correction). ANOVA = Analysis of Variance; Reg = region.
Discussion
Our findings lead us to conclude that the IPF risk variants rs35705950 and rs2076295 likely perturb the expression and methylation of MUC5B and DSP, respectively, and are involved in the etiology of IPF. More specifically, our findings demonstrate strong co-localization of rs35705950 and rs2076295 with both gene expression and DNA methylation marks. Moreover, mQTL results led to identification of a putative internal repressor element within MUC5B. Collectively, these results provide additional evidence that both MUC5B and DSP are involved in the etiology of IPF, and that the expression of these genes is regulated by both genetic and epigenetic factors. However, using transcriptional and epigenetic approaches, we were unable to identify potential regulatory roles for the other eight common IPF risk variants (8) and further work will be necessary to disentangle the causal variants and perturbed biological pathways at these loci.
Several lines of investigation, in addition to the findings presented in this manuscript, suggest that MUC5B is involved in the etiology of pulmonary fibrosis. First, rs35705950 (15, 33) is the dominant risk factor for IPF and is present in >50% of affected patients (33); this finding has been validated in at least 10 independent studies (8, 9, 12, 14, 33, 41–47). Second, MUC5B is normally not expressed in the terminal bronchioles (48–50). However, MUC5B is expressed in the bronchiolar epithelia, epithelial cells lining honeycomb cysts, and co-expressed with surfactant protein C (SFTPC) in alveolar type II cells in IPF (33, 51–54), indicating that cell types involved in lung fibrosis in the distal airspace express MUC5B. In addition, the variant allele is specifically associated with increased expression of MUC5B in the terminal bronchiole (52). Third, overexpression of Muc5b in bronchoalveolar epithelia in mice is directly related to the extent and persistence of bleomycin-induced lung fibrosis, honeycomb metaplasia, and mortality (53, 55). Fourth, we recently demonstrated that the MUC5B variant rs35705950 resides within an enhancer that is subject to epigenetic remodeling and contributes to pathologic misexpression in IPF (16). Findings presented in the current study demonstrate further the importance of the MUC5B promoter variant on both risk and MUC5B expression and identify the regulatory importance of methylation marks in repressing the effects of the MUC5B promoter variant.
Accumulating evidence suggests that DSP, part of the desmosome, is also involved in the etiology of IPF. DSP is critical to cell–cell adhesion, wound repair, and epithelial barrier function. While rs2076295 has been repeatedly found to be associated with IPF (8, 34), other variants in DSP have also been associated with cardiac fibrosis (56), right ventricular dysplasia (57), and keratodermas (58), suggesting that loss of cell–cell adhesion may result in a number of conditions involving injury, tissue remodeling, and fibrosis. Deletion of the DNA region spanning rs2076295 using CRISPR/Cas9 editing leads to reduced expression of DSP and an edited G allele at rs2076295 results in lower expression of DSP compared with the wild-type T allele in a human bronchial epithelial cell line (59). This is consistent with the results of our eQTL analysis that show a decrease in DSP gene expression with the presence of the alternate allele. Moreover, loss of DSP enhanced ECM-related gene expression and promoted cell migration, potentially contributing functionally to the pathogenesis of IPF (59). A recent study also showed that stiff matrix induces DSP gene expression in lung epithelial cells and that this induction is regulated by DNA methylation of a conserved region in the proximal DSP promoter (60). In aggregate, these findings suggest that reduced expression of DSP could adversely affect wound healing and promote fibroproliferation.
Our findings underscore the complex etiology of IPF. The presence of both genetic and epigenetic changes associated with disease-defining transcriptional changes in the IPF lung strongly suggest that lung fibrosis is driven by gene-by-environment interactions. Although the MUC5B promoter variant is the dominant genetic risk variant for the development of IPF (33), chronic hypersensitivity pneumonitis (61), rheumatoid arthritis–associated interstitial lung disease (62), and asbestosis (63), only a small portion of individuals with these common genetic variants go on to develop lung fibrosis, raising the possibility that environmental factors such as microscopic lung injury caused by inhaled particles or toxins are inadequately cleared and/or cause excessive lung injury in genetically susceptible hosts. In fact, several genetic studies (33, 64–67) indicate that a specific gene variant or locus may cause different types of lung fibrosis within the same family. This supports our hypothesis that sharing the same genetic variant does not necessarily result in the same disease pattern and that other influences, including specific environmental exposures or possibly other genetic variants are pivotal to the final phenotype that emerges.
It is particularly intriguing that of the 10 IPF common risk loci, only MUC5B and DSP co-localized with gene expression and methylation marks. Importantly, variants in telomerase genes may exert their effects through circulating cells or at different stages of lung fibrosis; TERT and TERC have undetectable expression in lung tissue and therefore were not included in the analysis and lead genetic variants at the OBFC1/STN1 locus had no eQTLs. Genetic variants in telomerase genes are much more likely to function by telomere shortening (68) or cell senescence in alveolar cells (69). It is also important to note that this study focused on cis-eQTLs in the 10 IPF common risk loci identified by our group but that we report genome-wide cis-eQTL results for all genotype-expression pairs and that these data can be mined by others for additional IPF common variant loci.
The main limitation of the current study is the use of whole lung tissue. Cell heterogeneity in whole lung tissue is likely the reason we observe small effect sizes in our mQTL results. We partially addressed the issue of cell heterogeneity by performing cell type-interaction eQTL analysis, as has been done by GTEx (21). However, a major limitation of this analysis is that deconvolution of whole lung tissue gene expression data does not work well on specialized cell types in the lung such as MUC5B-producing secretory cells. Future single cell eQTL/mQTL studies (70) will be necessary to fully address cell specificity of the relationship of genetic variant to gene expression and DNA methylation. Another limitation that is inherent to using bisulfite conversion for DNA methylation measurements is inability to distinguish 5-methylcytosine from 5-hydroxymethylcytosine. However, given that the relative amount of 5-hydroxymethylcytosine is small compared with 5-methylcytosine, this is only a minor limitation and likely does not influence the key findings. Lastly, sample size and power were limited in our subgroup analysis of age-, sex-, and smoking-matched cases and controls. Despite these limitations, our study provides substantial evidence for roles of rs35705950 (MUC5B) and rs2076295 (DSP) genetic variants in the development of IPF. Future studies should also use additional assays to assess chromatin accessibility (ATAC-seq, for example) and tools to assess the functionality of the identified eQTL/mQTLs (CRISPR-Cas9, for example) in primary cells.
Footnotes
Supported by the NIH-NHLBI (R01HL097163 and P01HL092870) and Vertex Pharmaceuticals. R.B. was supported by “la bourse du college des enseignants de pneumologie.”
Author Contributions: J.B.R., A.N.G., T.E.F., N.S., S.L.P., Z.Z., D.A.S., and I.V.Y. conceived and designed the study. R.B., S.K.S., F.G., E.D., and A.W. collected the data. R.B., J.C., I.R.K., C.M.M., W.Z., and T.N. analyzed the data. M.R., P.J.W., K.K.B., and T.S.B. performed clinical phenotyping of the subjects. J.P. and J.B. enrolled the study participants and collected the samples. R.B., I.R.K., S.L.P., D.A.S., and I.V.Y. wrote the manuscript. All authors edited and approved the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202110-2308OC on July 11, 2022.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med . 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 2. Martinez FJ, Collard HR, Pardo A, Raghu G, Richeldi L, Selman M, et al. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers . 2017;3:17074. doi: 10.1038/nrdp.2017.74. [DOI] [PubMed] [Google Scholar]
- 3. Leavy OC, Ma SF, Molyneaux PL, Maher TM, Oldham JM, Flores C, et al. Proportion of idiopathic pulmonary fibrosis risk explained by known common genetic loci in European populations. Am J Respir Crit Care Med . 2021;203:775–778. doi: 10.1164/rccm.202008-3211LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mathai SK, Newton CA, Schwartz DA, Garcia CK. Pulmonary fibrosis in the era of stratified medicine. Thorax . 2016;71:1154–1160. doi: 10.1136/thoraxjnl-2016-209172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet . 2015;47:512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dressen A, Abbas AR, Cabanski C, Reeder J, Ramalingam TR, Neighbors M, et al. Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study. Lancet Respir Med . 2018;6:603–614. doi: 10.1016/S2213-2600(18)30135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borie R, Le Guen P, Ghanem M, Taillé C, Dupin C, Dieudé P, et al. The genetics of interstitial lung diseases. Eur Respir Rev . 2019;28:190053. doi: 10.1183/16000617.0053-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moore C, Blumhagen RZ, Yang IV, Walts A, Powers J, Walker T, et al. Resequencing study confirms that host defense and cell senescence gene variants contribute to the risk of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2019;200:199–208. doi: 10.1164/rccm.201810-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allen RJ, Guillen-Guio B, Oldham JM, Ma SF, Dressen A, Paynton ML, et al. Genome-wide association study of susceptibility to idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2020;201:564–574. doi: 10.1164/rccm.201905-1017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allen RJ, Porte J, Braybrooke R, Flores C, Fingerlin TE, Oldham JM, et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med . 2017;5:869–880. doi: 10.1016/S2213-2600(17)30387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fingerlin TE, Zhang W, Yang IV, Ainsworth HC, Russell PH, Blumhagen RZ, et al. Genome-wide imputation study identifies novel HLA locus for pulmonary fibrosis and potential role for auto-immunity in fibrotic idiopathic interstitial pneumonia. BMC Genet . 2016;17:74. doi: 10.1186/s12863-016-0377-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet . 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mushiroda T, Wattanapokayakit S, Takahashi A, Nukiwa T, Kudoh S, Ogura T, et al. Pirfenidone Clinical Study Group A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet . 2008;45:654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 14. Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med . 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Helling BA, Gerber AN, Kadiyala V, Sasse SK, Pedersen BS, Sparks L, et al. Regulation of MUC5B expression in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol . 2017;57:91–99. doi: 10.1165/rcmb.2017-0046OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gally F, Sasse SK, Kurche J, Gruca MA, Cardwell JH, Okamoto T, et al. The MUC5B-associated variant, rs35705950, resides within an enhancer subject to lineage- and disease-dependent epigenetic remodeling. JCI Insight . 2021;6:e144294. doi: 10.1172/jci.insight.144294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. GTEx Consortium. Genetic effects on gene expression across human tissues. Nature . 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet . 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu Y, Broadaway KA, Raulerson CK, Scott LJ, Pan C, Ko A, et al. Colocalization of GWAS and eQTL signals at loci with multiple signals identifies additional candidate genes for body fat distribution. Hum Mol Genet . 2019;28:4161–4172. doi: 10.1093/hmg/ddz263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franceschini N, Giambartolomei C, de Vries PS, Finan C, Bis JC, Huntley RP, et al. MEGASTROKE Consortium GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat Commun . 2018;9:5141. doi: 10.1038/s41467-018-07340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim-Hellmuth S, Aguet F, Oliva M, Muñoz-Aguirre M, Kasela S, Wucher V, et al. GTEx Consortium Cell type-specific genetic regulation of gene expression across human tissues. Science . 2020;369:eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol . 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McRae AF, Marioni RE, Shah S, Yang J, Powell JE, Harris SE, et al. Identification of 55,000 replicated DNA methylation QTL. Sci Rep . 2018;8:17605. doi: 10.1038/s41598-018-35871-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morrow JD, Glass K, Cho MH, Hersh CP, Pinto-Plata V, Celli B, et al. Human lung DNA methylation quantitative trait loci colocalize with chronic obstructive pulmonary disease genome-wide association loci. Am J Respir Crit Care Med . 2018;197:1275–1284. doi: 10.1164/rccm.201707-1434OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol . 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 26. Zhou W, Triche TJ, Jr, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res . 2018;46:e123. doi: 10.1093/nar/gky691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc . 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics . 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B . 1995;57:289–300. [Google Scholar]
- 30. Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet . 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics . 2019;20:366. doi: 10.1186/s12864-019-5761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasse SK, Mailloux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, et al. The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol . 2013;33:2104–2115. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med . 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mathai SK, Pedersen BS, Smith K, Russell P, Schwarz MI, Brown KK, et al. Desmoplakin variants are associated with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2016;193:1151–1160. doi: 10.1164/rccm.201509-1863OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor DL, Jackson AU, Narisu N, Hemani G, Erdos MR, Chines PS, et al. Integrative analysis of gene expression, DNA methylation, physiological traits, and genetic variation in human skeletal muscle. Proc Natl Acad Sci USA . 2019;116:10883–10888. doi: 10.1073/pnas.1814263116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun . 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McClay JL, Shabalin AA, Dozmorov MG, Adkins DE, Kumar G, Nerella S, et al. Swedish Schizophrenia Consortium High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biol . 2015;16:291. doi: 10.1186/s13059-015-0842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun . 2019;10:4267. doi: 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hannon E, Gorrie-Stone TJ, Smart MC, Burrage J, Hughes A, Bao Y, et al. Leveraging DNA-methylation quantitative-trait loci to characterize the relationship between methylomic variation, gene expression, and complex traits. Am J Hum Genet . 2018;103:654–665. doi: 10.1016/j.ajhg.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet . 2014;10:e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y, Noth I, Garcia JG, Kaminski N. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med . 2011;364:1576–1577. doi: 10.1056/NEJMc1013504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stock CJ, Sato H, Fonseca C, Banya WA, Molyneaux PL, Adamali H, et al. Mucin 5B promoter polymorphism is associated with idiopathic pulmonary fibrosis but not with development of lung fibrosis in systemic sclerosis or sarcoidosis. Thorax . 2013;68:436–441. doi: 10.1136/thoraxjnl-2012-201786. [DOI] [PubMed] [Google Scholar]
- 43. Borie R, Crestani B, Dieude P, Nunes H, Allanore Y, Kannengiesser C, et al. The MUC5B variant is associated with idiopathic pulmonary fibrosis but not with systemic sclerosis interstitial lung disease in the European Caucasian population. PLoS One . 2013;8:e70621. doi: 10.1371/journal.pone.0070621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei R, Li C, Zhang M, Jones-Hall YL, Myers JL, Noth I, et al. Association between MUC5B and TERT polymorphisms and different interstitial lung disease phenotypes. Transl Res . 2014;163:494–502. doi: 10.1016/j.trsl.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horimasu Y, Ohshimo S, Bonella F, Tanaka S, Ishikawa N, Hattori N, et al. MUC5B promoter polymorphism in Japanese patients with idiopathic pulmonary fibrosis. Respirology . 2015;20:439–444. doi: 10.1111/resp.12466. [DOI] [PubMed] [Google Scholar]
- 46. Peljto AL, Selman M, Kim DS, Murphy E, Tucker L, Pardo A, et al. The MUC5B promoter polymorphism is associated with idiopathic pulmonary fibrosis in a Mexican cohort but is rare among Asian ancestries. Chest . 2015;147:460–464. doi: 10.1378/chest.14-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van der Vis JJ, Snetselaar R, Kazemier KM, ten Klooster L, Grutters JC, van Moorsel CH. Effect of MUC5B promoter polymorphism on disease predisposition and survival in idiopathic interstitial pneumonias. Respirology . 2016;21:712–717. doi: 10.1111/resp.12728. [DOI] [PubMed] [Google Scholar]
- 48. Fahy JV, Dickey BF. Airway mucus function and dysfunction. N Engl J Med . 2010;363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dickey BF, Whitsett JA. Understanding interstitial lung disease: it’s in the mucus. Am J Respir Cell Mol Biol . 2017;57:12–14. doi: 10.1165/rcmb.2017-0116ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, et al. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am J Respir Crit Care Med . 2019;199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One . 2013;8:e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakano Y, Yang IV, Walts AD, Watson AM, Helling BA, Fletcher AA, et al. MUC5B promoter variant rs35705950 affects MUC5B expression in the distal airways in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2016;193:464–466. doi: 10.1164/rccm.201509-1872LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hancock LA, Hennessy CE, Solomon GM, Dobrinskikh E, Estrella A, Hara N, et al. MUC5B overexpression causes mucociliary dysfunction and enhances lung fibrosis in mice. Nat Commun . 2018;9:5363. doi: 10.1038/s41467-018-07768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Conti C, Montero-Fernandez A, Borg E, Osadolor T, Viola P, De Lauretis A, et al. Mucins MUC5B and MUC5AC in distal airways and honeycomb spaces: comparison among idiopathic pulmonary fibrosis/usual interstitial pneumonia, fibrotic nonspecific interstitial pneumonitis, and control lungs. Am J Respir Crit Care Med . 2016;193:462–464. doi: 10.1164/rccm.201507-1322LE. [DOI] [PubMed] [Google Scholar]
- 55. Kurche JS, Dobrinskikh E, Hennessy CE, Huber J, Estrella A, Hancock LA, et al. Muc5b enhances murine honeycomb-like cyst formation. Am J Respir Cell Mol Biol . 2019;61:544–546. doi: 10.1165/rcmb.2019-0138LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Z, Bowles NE, Scherer SE, Taylor MD, Kearney DL, Ge S, et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Res . 2006;99:646–655. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 57. Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med . 2008;5:258–267. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Armstrong DK, McKenna KE, Purkis PE, Green KJ, Eady RA, Leigh IM, et al. Haploinsufficiency of desmoplakin causes a striate subtype of palmoplantar keratoderma. Hum Mol Genet . 1999;8:143–148. doi: 10.1093/hmg/8.1.143. [DOI] [PubMed] [Google Scholar]
- 59. Hao Y, Bates S, Mou H, Yun JH, Pham B, Liu J, et al. Genome-wide association study: functional variant rs2076295 regulates desmoplakin expression in airway epithelial cells. Am J Respir Crit Care Med . 2020;202:1225–1236. doi: 10.1164/rccm.201910-1958OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Qu J, Zhu L, Zhou Z, Chen P, Liu S, Locy ML, et al. Reversing mechanoinductive DSP expression by CRISPR/dCas9-mediated epigenome editing. Am J Respir Crit Care Med . 2018;198:599–609. doi: 10.1164/rccm.201711-2242OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ley B, Newton CA, Arnould I, Elicker BM, Henry TS, Vittinghoff E, et al. The MUC5B promoter polymorphism and telomere length in patients with chronic hypersensitivity pneumonitis: an observational cohort-control study. Lancet Respir Med . 2017;5:639–647. doi: 10.1016/S2213-2600(17)30216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Juge PA, Lee JS, Ebstein E, Furukawa H, Dobrinskikh E, Gazal S, et al. MUC5B promoter variant and rheumatoid arthritis with interstitial lung disease. N Engl J Med . 2018;379:2209–2219. doi: 10.1056/NEJMoa1801562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Platenburg MGJP, Wiertz IA, van der Vis JJ, Crestani B, Borie R, Dieude P, et al. The MUC5B promoter risk allele for idiopathic pulmonary fibrosis predisposes to asbestosis. Eur Respir J . 2020;55:1902361. doi: 10.1183/13993003.02361-2019. [DOI] [PubMed] [Google Scholar]
- 64. Thomas AQ, Lane K, Phillips J, III, Prince M, Markin C, Speer M, et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med . 2002;165:1322–1328. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- 65. Nogee LM, Dunbar AE, III, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med . 2001;344:573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 66. Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med . 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 67. Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA . 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Courtwright AM, El-Chemaly S. Telomeres in interstitial lung disease: the short and the long of it. Ann Am Thorac Soc . 2019;16:175–181. doi: 10.1513/AnnalsATS.201808-508CME. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yao C, Guan X, Carraro G, Parimon T, Liu X, Huang G, et al. Senescence of alveolar type 2 cells drives progressive pulmonary fibrosis. Am J Respir Crit Care Med . 2021;203:707–717. doi: 10.1164/rccm.202004-1274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. van der Wijst M, de Vries DH, Groot HE, Trynka G, Hon CC, Bonder MJ, et al. The single-cell eQTLGen consortium. eLife . 2020;9:e52155. doi: 10.7554/eLife.52155. [DOI] [PMC free article] [PubMed] [Google Scholar]