Abstract
The type b capsule of pathogenic Haemophilus influenzae is a critical factor for H. influenzae survival in the blood and the establishment of invasive infections. Other pathogenic factors associated with type b strains may also play a role in invasion and sustained bacteremia, leading to the seeding of deep tissues. The gene encoding haemocin is the only noncapsular gene found to be specific to type b strains until now. Here we report the discovery of an approximately 16-kb genetic locus, HiGI1, that is present primarily in type b strains. Pulsed-field gel electrophoresis and Southern hybridization were used to map this new locus between secG (HI0445) and fruA (HI0446), which are contiguous in Rd, a nonpathogenic derivative of a serotype d strain. It is inserted at the 3′ end of tRNA4Leu and has regions whose G+C content differs from the average genomic G+C content of H. influenzae. An integrase gene, which encodes a CP4-57 like integrase, is located downstream of tRNA4Leu. Hybridization probes based on the sequences within the HiGI1 locus have been used to screen 61 H. influenzae strains (2 type a, 22 type b, 2 type c, 1 type d, 3 type e, 7 type f, and 21 nontypeable H. influenzae [NTHi]) from our collection. This HiGI1 locus exists in all 22 type b strains and two NTHi strains and is likely to have been acquired by an ancestral type b strain.
Haemophilus influenzae causes a variety of human infections. Type b capsule, LOS, and pili have been shown to play important role in pathogenesis (13, 29). Encapsulated H. influenzae type b (Hib) strains cause invasive infections, including meningitis and septicemia, in infants and children, while H. influenzae of other capsule types (a, c, d, e, and f) rarely cause invasive infections. Encapsulated strains only occasionally colonize the upper respiratory tract, whereas nontypeable H. influenzae (NTHi) strains often colonize the respiratory tract and can cause a variety of respiratory infections, such as otitis media, sinusitis, bronchitis, and conjunctivitis (31).
The entire genomic DNA sequence of H. influenzae strain Rd, a nonencapsulated, nonpathogenic derivative of a serotype d strain, became available in 1995 (11, 45). The Rd genome is estimated to be 270 kb smaller than that of virulent type b strain Eagan (6). Capsule type b (24), pili (hif) (44), tryptophanase (tna) (27), and haemocin (hmc) (30) genes are present in Hib strains and are not found in Rd. The cap b, hif, and tna loci are each flanked by direct repeats. The cap b gene cluster, containing a duplication of two ∼18-kb segments, lies between direct repeats of IS1016 (21, 24). In each ∼18-kb segment, there is a central serotype-specific region II which has a substantially lower G+C content, 32% (43). The hif gene cluster is inserted between pepN (HI1614) and purE (HI1615). This cluster has a G+C ratio of 39%, typical of H. influenzae. Analysis of the regions flanking the pilus gene cluster of type b strain reveals a duplication of the 57-bp pur regulatory region (44). The tryptophan (tna) genes are situated between nlpD (HI0706) and mutS (HI0707), are found at the same map location of all indole-positive strains, and are missing from Rd, type d, and type e genomes. Most interestingly, this locus is flanked by 43-bp direct repeats of paired Haemophilus uptake signal sequences (USSs) (27). The hmc locus produces haemocin, a protein toxic to all non-Hib strains and one which appears to play a role in the onset of invasive type b disease in the infant rat model (26, 30).
Many bacterial pathogens contain virulence genes located on pathogenicity islands, which may be derived from integrated bacteriophages that are associated with tRNA or single-stranded RNA genes or, alternatively, might arise from the insertion sequence-mediated gene transfer (8, 16). Tizard et al. have proposed that loci similar to pathogenicity islands that either do not contain virulence genes or have not yet been shown to contain virulence genes be called genetic islands (42). Genetic islands may represent a class of genetic elements whose acquisition contribute to microbial evolution (40).
From a search for other potential virulence genes that might contribute to the ability of Hib strains to cause invasive diseases, we report here a ∼16-kb locus in strain Eagan that appears to be found primarily in type b strains. It is situated between secG (HI0445) and fruA (HI0446), is adjacent to the tRNA4Leu gene, is flanked by 23-bp direct repeats (DR1), has regions different in G+C content from the rest of the genome, and contains a phage-related integrase gene, suggesting it could be of bacteriophage origin. We propose to call this locus HiGI1 (for H. influenzae genetic island 1).
MATERIALS AND METHODS
Bacterial strains.
A total of 61 H. influenzae strains were used in this study: 22 Hib, 2 type a, 2 type c, 1 type d, 3 type e, 7 type f, and 24 NTHi. The majority of these isolates have been previously characterized (9, 15). Seventeen strains (one type a, nine type b, four type f, and three NTHi) were isolated from cerebrospinal fluid or blood. Hib strain Eagan, the source for the mapping, cloning, and sequencing procedures, was originally isolated from a child with meningitis (12). In contrast to the majority of Hib strains, which belong to multilocus enzyme phylogenetic division I, strain R9 (otherwise known as Rab) belongs to multilocus enzyme phylogenetic division II (32, 33). Strains AAr64 (14) and AAr117 (9) have lost type b capsules. NTHi strains 315-3 and 316-4 were isolated from blood of patients with immunodeficiency disease. H. influenzae biogroup aegyptius strain ATCC 49252 was isolated from blood of a Brazilian purpuric fever patient. Strain ATCC 11116 is a type strain of biogroup aegyptius (4). H. influenzae strains were grown on brain heart infusion plates, solidified with 1.2% agar and supplemented with 10% Levinthal base (28) and NAD (2 μg/ml), in a 35°C CO2 incubator.
The host Escherichia coli strains used in the cloning experiments were INVaF [F endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169 λ−], TOP10 [F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacZX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG], both obtained from Invitrogen, Carlsbad, Calif., and DH5α [F-f80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 λ− gyrA96 Δ(lacZYA-argF)U169].
Pulsed-field gel electrophoresis (PFGE).
The protocol for preparation of Haemophilus genomic DNA in InCert agarose plugs was adapted from the manufacturer (FMC BioProducts, Rockland, Maine). After digestion with restriction enzymes, DNA fragments were resolved by contour-clamped homogeneous electric field (CHEF) electrophoresis using a CHEF-DRIII apparatus (Bio-Rad Laboratories, Richmond, Calif.) with an electric field of 6 V cm−1 and an angle of 120°. DNA fragment migration was performed in 1% SeaKem HGT agarose (FMC) and in 0.5× Tris-borate-EDTA buffer at 14°C. Pulsed time was ramped from 1 to 15 s or from 10 to 30 s over 8 to 20 h, according to the size of DNA fragment to be resolved.
DNA techniques.
Isolation of genomic DNA was performed using a Wizard genomic DNA purification kit (Promega, Madison, Wis.). Plasmid extractions were carried out as specified for the Wizard Plus minipreps DNA purification system (Promega). The DNA preparations were quantitated on ethidium bromide-stained gels by applying GibcoBRL DNA quantitation standards (Life Technologies, Gaithersburg, Md.).
PCR.
Low annealing temperature (40°C) and a relatively high concentration (up to 1 mg/100 μl) were used with the degenerate PCR primers, LVIED and GADDY, which were based on two regions of conserved amino acids shared by response regulators from a variety of bacteria (39) (Table 1). A 100-μl reaction mixture consisted of 10 μl of 10× reaction buffer, 4 μl of MgCl2 (2 mM, final concentration), 2 μl of dimethyl sulfoxide, 10 μl of deoxynucleoside triphosphate (dNTP) mix (2 mM), 1 μl of forward primer (50 μM), 1 μl of reverse primer (50 μM), 0.5 μl of Taq DNA polymerase (5 U/ml), 10 μl of genomic DNA template (>100 μg per reaction), and 61.5 μl of H2O (7, 46). Long PCR amplification was performed by one of the two following methods. The first long PCR used Taq DNA polymerase (Promega). A 50-μl reaction mixture consisted of 5 μl of reaction buffer, 3 μl of MgCl2 (25 mM), 2 μl of dNTP mix (10 mM), 1 μl of forward primer (20 mM), 1 μl of primer (20 mM), 1 μl of Taq DNA polymerase (5 U/ml), 1 μl of genomic DNA template (>200 ng), and 36 μl of H2O. One cycle of preamplification DNA denature at 94°C for 30 s, 25 cycles of denaturing at 94°C for 10 s, annealing at 55°C for 1 min, and extension at 72°C for 5 min, and one cycle of final extension at 72°C for 10 min were done on thermal cycler (MJ Research, Watertown, Mass.). Long PCR products (5 kb) were cloned into the original pCR2.1 vector (Invitrogen). The second long PCR amplification was performed using the TaqPlus Long polymerase mixture (Stratagene, La Jolla, Calif.). A 50-μl reaction mixture consisted of 5 μl of 10× TaqPlus Long low-salt buffer, 2 μl of dNTP mix (10 mM), 1 μl of forward primer (20 mM), 1 μl of backward primer (20 mM), 1 μl of TaqPlus Long polymerase mixture (5 U/ml), 1 μl of genomic DNA template (>200 ng) and 39 μl of H2O. One cycle of preamplification denaturing at 94°C for 30 s, 25 cycles of denaturing at 94°C for 10 s, annealing at 55°C for 1 min, and extension at 72°C for 10 min, and one cycle of final extension at 72°C for 15 min were done on a thermal cycler (MJ Research). Long PCR products were visualized and purified by agarose gel electrophoresis using crystal violet (35). Gel-purified long PCR products (11 kb) were cloned into pCR-XL-TOPO vector (Invitrogen). Subsequently, a 5-kb ClaI fragment, FC5, was subcloned into the ClaI-linearized pGEM7 vector.
TABLE 1.
Oligonucleotide primer sequences used for PCR
| Oligonucleotide | Sequencea |
|---|---|
| Degenerate primers | |
| LVIED | CCC CTC TAG ACT NGT NAT NGA NGA |
| GADDY | CCC CGC ATC CGT AAT CAT CNG CGC C |
| DNA probes (mapping) | |
| HI0401 (omp1) | TCG TTG CGC CAG TGA ATG ATA A |
| GCC CCT AAT GCA ACA CGA GAG T | |
| HI0406 (accA) | ATC GCC CAC GCC AAT AGC |
| ATC GGT CAT CAA AAA GGT CGT TCT | |
| HI0410 (tyrR) | GTG CGG GTT TGC CTG ATG |
| ACG AAC AAG CGC GAT AAA GAG T | |
| HI0429 (glmS) | TGC CGC CAT ATA GCC CTT TTC |
| GTG TTA CCC GCC GTT TTA TCT TTT | |
| HI0444 (topB) | AGC GGA TGT GGC AAG AGG AAT AAA |
| TGC GGC GAT AAT GAT GTG GTA ACT | |
| HI0445 (secG) | CAT CAG GTA CAA TGT TTG GCT CTG |
| TGT CTT TCG CTG GAG CTG CTT | |
| HI0446 (fruA) | CCC GCA TCG CAT TGG CTA AC |
| TAA TGC GGG AAC GAA AGA AGA AAG | |
| HI0448 (fruB) | GCC CCG CAT TAA TCG CAA CTA |
| ATG GCA TCG CTA TTC CTC ACG | |
| HI0457 (pabC) | TTC TTG CAC CGC TTT GTT ATG TT |
| TTG GCG AAA AGA TCT TGA AAA TG | |
| HI0465 (serA) | AAT AAA TCC GCG ACT GGC TCT CA |
| CGG GCT CAA CGG GGA ATA CA | |
| DNA probes (screening) | |
| Region I | CGG TAA ATG CGG AAT GGT CA |
| GCC ACT CTT TGA CAA ATG GTT GAG | |
| Region II | GTG CCA CCT TTC TAA TTG TTG CTG |
| GAA CGA TAG CAC GCC TTT TAA CC | |
| Region III | TTC GCT TGT TCT CTC CAC GC |
| GAC CGC ACT TTT TAC CTT TGT CA | |
| Long PCR primers | |
| F5E | CTT TGA CTT GTG CGC AAT AAG TCG |
| TAA TGC GGG AAC GAA AGA AGA AAG | |
| F10G | CAT CAG GTA CAA TGT TTG GCT CTG |
| GCA TTA CGC AGC TTT CGT ATC GT |
Underlined region in LVIED is XbaI site; underlined region in GADDY is BamHI site.
Nucleotide sequencing and analysis.
DNA sequencing of clone F2 was carried out by the dideoxy-chain terminating method (37) with a Sequenase 2.0 sequencing kit (U.S. Biochemical, Cleveland, Ohio) in conjunction with 35S (Sigma Chemical. St. Louis, Mo.). Double-stranded DNAs from three other overlapping clones (F5E, FC5, and F10G) were subjected to automated sequencing run by the DNA Sequencing Core, University of Michigan, Ann Arbor, with reagents from a dye terminator kit (Applied Biosystems). MacVector sequencing analysis software (version 5.0; Oxford Molecular Group) was used to analyze the DNA sequencing for identification of open reading frames, restriction sites, base composition, and codon frequency. The nucleotide sequences were used in searches against GenBank, EMBL, DDBJ, and PDB databases. The predicted amino acid sequences of each open reading frame were used in searches against the GenBank CDS translation, PDB, SwissProt, PIR, and PRF databases, using the BLAST2.0 program (1). The codon letter G+C content of genes in each region was calculated and compared with those of H. influenzae Rd, accessed from CUTG (codon usage tabulated from GenBank) database at the Kazusa DNA Research Institute web site (www.kazusa.or.jp) (34).
DNA probes and hybridization.
Probes for mapping and probes (I, II, and III) for screening were PCR amplified. Probe IVa was a KpnI/SacI-digested fragment from clone F2. Probe IVb was a HincII-digested fragment from clone F5E. DNA was labeled with digoxigenin-11-dUTP by the random-primed method as specified by the according manufacturer (Boehringer Mannheim, Indianapolis, Ind.). Restriction digested DNAs were electrophoresed in SeaKem HGT agarose gels and then transferred to a positively charged nylon membrane (Boehringer Mannheim). For analysis of distribution of HiGI1 among different strains, bacterial genomic DNA was denatured by adding dot blot-denaturing solution (4 M NaOH, 100 mM EDTA) and was pipetted onto a positively charged nylon membrane (Boehringer Mannheim). Hybridizations were performed under stringent conditions: at 65°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1.0% (wt/vol) blocking reagent–0.1% N-lauroylsarcosine–0.02% sodium dodecyl sulfate, and the membranes were washed at 65°C in 0.5× SSC containing 0.1% sodium dodecyl sulfate.
Oligonucleotide sequences.
Oligonucleotide sequences for degenerate PCR primers, long PCR primers, and probe preparations are listed in Table 1.
Nucleotide sequence accession number.
The nucleotide sequence of HiGI1 has been submitted to GenBank and assigned accession no. AF198256.
RESULTS
Discovery of a locus present in Eagan and other Hib isolates.
A pair of degenerate PCR primers, LVIED and GADDY, were used to identify potential response regulators of two-component regulatory systems in H. influenzae strain Eagan (7). One 300-bp PCR fragment, f-1, was found in strain Eagan but not in the published sequences of Rd (11). In a preliminary screen, hybridization analysis using f-1 as a probe indicated that f-1 sequences were present uniformly in 21 tested Hib isolates but not in H. influenzae with other capsular types (one type a, one type c, one type d, two type e, and seven type f) or in 15 NTHi strains (7). A larger clone, F2, a 1,837-bp TaqI-digested fragment, was isolated from strain Eagan using the f-1 probe, and its entire sequence was also absent in Rd. We then proceeded to map this region on strain Eagan and delineate its complete size and sequence.
Location and boundary sequence analysis of unique type b locus.
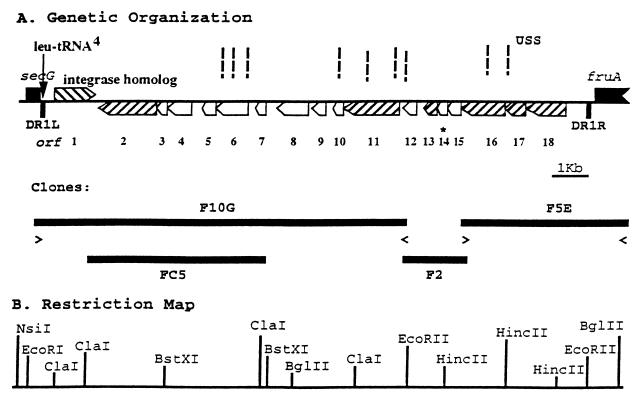
PFGE was used in combination with Southern hybridization to position the F2 region onto an existing large-scale restriction map of strain Eagan for the enzymes EagI, NaeI, RsrI, and SmaI (6). This location was further refined by using PCR probes based on Rd sequences, looking for DNA fragments that hybridized to a given PCR probe and to probe IVa (Table 1; Fig. 1). This dual approach localized the strain Eagan-specific DNA to a region located between secG (HI0445, protein translocation protein) and fruA (HI0446, fructose-permease IIBC component), which are contiguous in the Rd genome (11). Using sequences within clone F2 and the flanking known genes, long PCR was used to isolate clones containing the rest of the region (Materials and Methods). The sequence of the region was determined, and two maps of the region are shown in Fig. 1. For reasons described below, we have decided to name this region H. influenzae genetic island 1 (HiGI1). The left boundary of HiGI1 is the end of the tRNA4Leu gene. The HiGI1 sequence is flanked by direct repeats. The left-most member of the first direct repeat (DR1L) is 23 bp in length (5′-ttcaagtctcgcccagagcacca-3′) and is almost completely contained within the 3′ end of tRNA4Leu gene. The right-most member of the first direct repeat (DR1R) is 22 bp in length (5′-ttcacttctcgcccag_gcacca-3′), and 20 of 23 bases are identical between DR1L and DR1R (differences are underlined). DR2L starts 162 bp on the right of DR1L, is 22 bp in length, and is perfect match to DR2R (DR2, 5′-ttagtaaccaaaatagtaacca-3′). Each repeat consists of two 9-bp identical units (underlined). DR2R is located 49 bp to the left of DR1R. Of these strain Eagan sequences, only DR1L is retained in Rd. A stretch of six 10-bp short direct repeats (5′-gtctttaatt-3′) also can be found between DR1L and DR2L. In strain Eagan, inverted repeat 1 is located downstream of tRNA4Leu coding sequences (Fig. 2).
FIG. 1.
(A) Genetic organization of the HiGI1 locus and positions of clones. Open reading frames homologous to genes found in a variety of phage are indicated by diagonal lines in boxes. Dashed vertical lines, USS; < and >, long PCR primers; *orf14, containing the 300-bp f-1 sequences. (B) Partial restriction map. Only the restriction enzymes used to position the clone F2 between secG and fruA are shown.
FIG. 2.
Nucleotide sequences at the boundaries of the HiGI1 locus. Both ends of HiGI1 contain a single copy of DR1 (boldface). (DR1L on the left boundary and DR1R on the right boundary). Rd lacks the whole 15,660-bp sequence of HiGI1 but retains one copy of the 23-bp sequence DR1L. Another pair of repeats, DR2L and DR2R, are underlined. SDR, a stretch of six 10-bp short direct repeats; IR, inverted repeats.
Identification of genes in this locus.
The locus consists of 18 open reading frames between two direct repeats (Fig. 1). Just downstream of the tRNA4Leu coding region, orf1 shows a high degree of amino acid sequence similarity (52%) to E. coli prophage CP4-57 integrase, SlpA. The predicted amino acid sequence of the next open reading frame (orf2) shares significant similarity (score; 291; similarity, 53%) with phage phi-R73 primase. The predicted amino acid sequence of orf1 1 are homologous to phage D3 terminase. The predicted amino acid sequences of orf13 and orf16 are similar to those of phage phi-105 holin and ORF25, respectively. The predicted amino acid sequences of the last two open reading frames, orf17 and orf18, show low-level (BLAST score, <80) similarity to gp35 and gp36 of Streptomyces temperate phage phi-C31, respectively (Table 2). This is consistent with the recent conclusion about the evolutionary relationships among prophages that all double-stranded DNA phage genomes are mosaics in nature and capable of horizontal exchange (20).
TABLE 2.
Summary of BLAST search and G+C content for each open reading frame
| HiGI1 | Similar to source | % GC | Score | Identity | E value |
|---|---|---|---|---|---|
| Region I | |||||
| orf1 | Integrase/phage CP4-57 | 34.9 | 254 | 149/391 | 1e-66 |
| orf2 | Primase/phage phi-R73 | 37.3 | 291 | 161/425 | 1e-77 |
| Region II | |||||
| orf3 | No significant match | 42.8 | |||
| orf4 | Intracellular hyaluronic acid | 41.8 | 36.7 | 52/187 | 0.15 |
| Binding protein/Mus musculus | |||||
| orf5 | Retrotransposon-like protein/Arabidopsis thaliana | 39.5 | 33.6 | 24/61 | 1.6 |
| orf6 | Hypothetical protein/Thermotoga maritima | 41.1 | 41 | 27/110 | 0.016 |
| orf7 | Putative DNA binding protein/satellite phage P4 | 43.4 | 34 | 14/52 | 2.0 |
| Region III | |||||
| orf8 | Hypothetical protein/Plasmodium falciparum | 29.3 | 41 | 40/160 | 0.012 |
| orf9 | No significant match | 32.8 | |||
| orf10 | Heat shock protein HTPG/Mycobacterium tuberculosis | 35.8 | 29 | 12/37 | 7.1 |
| Region IV | |||||
| orf11 | Putative terminase/phage D3 | 45.7 | 244 | 165/493 | 2e-63 |
| orf12 | Hypothetical protein Rv1578c/M. tuberculosis | 43.5 | 54 | 28/97 | 5e-07 |
| orf13 | Holin/phage phi-105 | 47.1 | 105 | 56/110 | 2e-22 |
| orf14 | Synuclein, alpha interacting protein/Homo sapiens | 46.2 | 30 | 18/55 | 6.8 |
| orf15 | Gene16/phage SPP1 | 44.9 | 34.8 | 20/73 | 0.27 |
| orf16 | ORF25/phage phi-105 | 45.8 | 164 | 103/371 | 2e-39 |
| orf17 | gp35/phage phi-C31 | 45.3 | 68 | 44/108 | 4e-1 |
| orf18 | gp36/phage phi-C31 | 45.1 | 72 | 68/210 | 2e-11 |
The predicted amino acid sequences of the remaining 11 open reading frames, however, show very low level (BLAST score, <50) of similarities to genes from diverse origins, such as Mycobacterium tuberculosis and Plasmodium falciparum (Table 2).
Base composition of DNA and codon letter G+C content.
The average G+C content of HiGI1 is approximately 41%, slightly higher than the genomewide average of approximately 38%, but the distribution of G+C is uneven. The base composition of a region that contains prophage CP4-57 integrase homologue and phi-R73 primase homologue, designated region I, is 36.3% G+C. The G+C content of region II, which contains orf3 to orf7, 7, is 41.6%. Region III, containing orf8 to orf10, has a low G+C content (31.2%). An 8-kb fragment designated region IV, which contains several phage-related genes and others, has a 45.4% G+C content (Fig. 3A; Table 2). The bias for A- or T-ending codons in the four different G+C regions reflects its base composition using G- or C-ending codons: 29.0, 40.1, 24.9, and 46.4%, respectively (Table 3).
FIG. 3.
(A) G+C content of each region of HiGI1 and region covered in each probe. (B) Hybridization of HiGI1 locus DNA probes to chromosomal DNA of various H. influenzae strains. The presence or absence of hybridization is indicated by + or −, respectively. ∗, cerebrospinal fluid or blood isolate.
TABLE 3.
Codon letter G+C content
| Genome or region | % G+C content
|
|||
|---|---|---|---|---|
| Arg | 1st letter | 2nd letter | 3rd letter | |
| Rd (1,709 genes) | 38.8 | 51.0 | 36.2 | 29.1 |
| Region I | 36.3 | 45.6 | 34.3 | 29.0 |
| Region II | 41.6 | 48.9 | 35.3 | 40.1 |
| Region III | 31.2 | 41.0 | 27.6 | 24.9 |
| Region IV | 45.4 | 50.9 | 38.6 | 46.4 |
Distribution of the HiGI1 among H. influenzae strains.
Five probes, corresponding to regions of HiGI1 differing in G+C content, were used to determine whether homologous sequences were present in the genomic DNA preparations of 61 H. influenzae strains from our collection. Using probes I, II, III, IVa and IVb, hybridizations occurred in all 22 type b strains and 2 NTHi strains. Among Hib strains are 9 strains isolated from patients with invasive diseases and 13 isolates from the upper respiratory tract, including 2 strains that have lost expression of type b capsules. Thus, this genetic island not only associates with Hib strains causing invasive diseases but also exists in Hib strains isolated from the upper respiratory tract. It is noteworthy, however, that two NTHi strains, AAr176 and Mr31, also hybridized to probe II. The same hybridization analysis also indicated that the entire locus is absent from 2 type a, 2 type c, 1 type d, 3 type e, 7 type f, and 10 other NTHi strains, including the invasive nontypeable strains 315-3 and 316-4 (Fig. 3B).
USS.
Analysis of sequences between two direct repeats (DR1) reveals nine USS sites (Fig. 1), four in the plus orientation (5′-AAGTGCGGT) and five in the minus orientation (5′-ACCGCACTT). None of the sites are in inverted repeat pairs. The mean distance between sites was 1,050 bp, with the range of 333 to 2833 bp. This is comparable to the genomewide mean distance between sites of 1,248 bp, with a range of 50 bp to 8 kb (38). Three USS sites fall into region II and four are located in region III, with the remaining two in region IV. All nine USS sites are located in open reading frames. In Rd, only 65% of 1,465 copies of USS sites are found in open reading frames, while about 86% of the genome is coding sequence (38).
DISCUSSION
We have characterized a ∼16-kb locus from H. influenzae strain Eagan that appears primarily in type b strains. This locus has several features characteristic of a genetic island (16). It contains 18 open reading frames differing in G+C content from the average H. influenzae genome (Table 2), is adjacent to tRNA4Leu gene, is bracketed by two 23-bp and two 22-bp direct repeats, and possesses a prophage CP4-57 integrase homologue. Genetic islands are thought to arise when a large region of foreign DNA is inserted into a bacterial genome. We are thus naming this locus H. influenzae genetic island 1 (HiGI1). While the HiGI1 locus differs in G+C content from other H. influenzae regions, it does contain nine H. influenzae uptake sequences (38).
tRNA loci are often targets for the integration of bacteriophage and pathogenicity islands into the chromosomes of various bacterial species, such as Pseudomonas aeruginosa (19), Vibrio cholerae (22), Yersinia pseudotuberculosis (5), and E. coli (3). In H. influenzae, leucine tRNA loci seem to be the most favored sites for phage integration. Two cryptic prophages, Mu-like phage (11) and φflu (20), are found in Rd. There are no clear boundary sequences around the proposed Mu-like phage, while φflu is found to integrate into tRNA4Leu. The temperate phage Hp1c1 (17) is capable of integrating into tRNA4Leu. However, Mu-like phage, φflu, and phage HP1c1 are not known to play any role in virulence (18). HiGI1 is located at the 3′ end of tRNA4Leu gene, a rare tRNA gene as opposed to more abundant tRNA1 and tRNA2. This rare tRNA4Leu gene might also act as a regulator for genes that frequently use this leucine-specific codon. In uropathogenic E. coli strain 536, pathogenicity island II was found to be inserted into the leuX locus, which encoded the rare tRNA4Leu, and deleted at a frequency of 10−3 to 10−4 per cell per generation (23). The deletion event also distorted the leuX locus and was shown to affect the expression of several virulence properties, such as type 1 fimbriae, flagella, serum resistance (36), and uropathogenesis (41).
There are two sets of direct repeats (DR1 and DR2) in the flanking regions of HiGI1 locus. The first set of repeats, DR1L and DR1R, were probably created during HiGI1 integration into tRNA4Leu. The direct repeats that flank the genetic islands play important role in their integration or excision (16). The excision of pathogenicity islands I and II from uropathogenic E. coli 536 occurs due to recombination within repeating sequences within tRNA coding sequences (3). However, we do not know whether the excision of HiGI1 can occur. The second set of direct repeats are internal to the HiGI1 locus. The role (if any) and origin of these repeats are not known, nor is it known if they facilitate rearrangement or deletion of genetic elements in HiGI1 locus. Whether this locus has gone through rearrangement or deletion in different strains, particularly in the three NTHi strains that possess only region II of HiGI1 locus (Fig. 3), remains to be explored.
HiGI1 is present in all Hib strains and two NTHi strains in our collection. Two possible mechanisms might have contributed to the distribution of HiGI1 in type b strains. HiGI1 could have been of bacteriophage origin, “HiGI1φ,” which might have played important role in the distribution of HiGi1 within H. influenzae. However, we do not know whether this putative HiGI1φ was transferable between different strains. A more likely scenario is that a type b ancestral strain acquired the HiGi1 locus before it diverged into different type b strains. As for the HiGI1-possessing nontypeable strains, they might have acquired HiGI1, or part of it, by horizontal uptake of DNA and homologous recombination, because HiGi1 evolved to contain several H. influenzae USS sites. The HiGI1 locus could also have been acquired by an ancestral nontypeable strain, and subsequent recombination between two direct repeats resulted in the loss of all or part of the HiGI1 locus from most nontypeable strains.
The G+C contents of region I (36.3%) and region II (41.6%) do not differ very much from the genome average (38%). This indicates that they might have been acquired from species with G+C content similar to that of H. influenzae or that the base composition of such acquired DNA has gradually adapted to the host genome over time (25). However, the G+C contents of region III (31.2%) and region IV (45.4%) vary substantially from the genomewide average. In A+T-rich H. influenzae, the average G+C content of the third codon letter is only 29.1% in 1,709 genes of Rd (34). In G+C-rich M. tuberculosis (∼65% G+C), there is a strong bias toward G- or C-ending codons for every amino acid; the G+C content at the third position of codons is 83% (2). The G+C usage in the third codon position of regions III and IV (24.9 and 46.4%, respectively) show strong bias toward each region's G+C content. These observations support the evidence discussed above that the HiGI1 locus might have been acquired by phage-mediated gene transfer; furthermore, the original element transferred in might have been composed of at least four different elements, from different sources.
In Rd, a cryptic Mu-like phage (11) with relatively high G+C content (∼50%), is located in the interval from 1.56 to 1.59 Mb on the genome. Two regions of 14,441 and 8,239 bp in this area contain no USS site. However, there are USSs in cryptic prophage φflu (11) and phage HP1c1 (10). The distribution of USSs in the Rd genome is not entirely random and is overrepresented in the intergenic regions. Most USS sequences in the H. influenzae genome appear as inverted-repeat pairs just beyond the 3′ ends of genes (38). In contrast, the USS sequences in HiGI1 are single and are found within coding regions. So far, the only similarity between the newly identified HiGI1 locus and the rest of genome is that they all contain USS sites.
Our results demonstrate that the HiGI1 locus might have resulted from a phage-mediated transfer, as evidenced by its being flanked by the tRNA4Leu gene and harboring a prophage CP4-57 integrase gene homologue just downstream of tRNA gene. The G+C content and codon usage of HiGI1 are different from the rest of host genome. To date, there is no experimental evidence to indicate that HiGi1 is a pathogenicity island. It is, however, conserved in Hib strains, which are responsible for most invasive diseases, and is absent from the majority of other strains studied. These facts raise the potential that it might be a virulence-associated region. As we continue our studies on the HiGI1 locus, we will dissect its structure among different H. influenzae strains and evaluate its possible role in the virulence of pathogenic strains.
ACKNOWLEDGMENT
This work was supported in part by Public Health Service grant RO1 AI25630 from the National Institute of Allergy and Infectious Diseases to J.R.G.
REFERENCES
- 1.Altschul S F, Madden T L, Achaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson S G E, Sharp P M. Codon usage in the Mycobacterium tuberculosis complex. Microbiology. 1996;142:915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- 3.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner D J, Mayer L W, Carlone G M, Harrison L H, Bibb W F, Brandileone M C, Sottnek F O, Irino K, Reeves M W, Swenson J M, Birkness K A, Weyant R S, Berkley S F, Woods T C, Steigerwalt A G, Grimont P A D, McKinney R M, Fleming D W, Gheesling L L, Cooksey R C, Arko R J, Broome C V The Brazilian Purpuric Fever Study Group. Biochemical, genetic, and epidemiologic characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazilian purpuric fever. J Clin Microbiol. 1988;26:1524–1534. doi: 10.1128/jcm.26.8.1524-1534.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn-tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 6.Butler P D, Moxon E R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990;136:2333–2342. doi: 10.1099/00221287-136-12-2333. [DOI] [PubMed] [Google Scholar]
- 7.Chang C-C. Ph.D. thesis. Ann Arbor: University of Michigan; 1999. [Google Scholar]
- 8.Cheetham B F, Katz M E. A role for bacteriophage in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 9.Clemens D L, Marrs C F, Patel M, Duncan M, Gilsdorf J R. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect Immun. 1998;66:656–663. doi: 10.1128/iai.66.2.656-663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzmaurice W P, Benjamin R C, Huang P C, Scocca J J. Characterization of sites on DNA segments from bacteriophage HP1c1 which interact with specific DNA recognition system of transformable Haemophilus influenzae Rd. Gene. 1984;31:187–196. doi: 10.1016/0378-1119(84)90209-9. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J F, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Forney L J, Marrs C F, Bektesh S L, Gilsdorf J R. Comparison and analysis of the nucleotide sequences of pilin genes from Haemophilus influenzae type b strains Eagan and M43. Infect Immun. 1991;59:1991–1996. doi: 10.1128/iai.59.6.1991-1996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilsdorf J R, McCrea K W, Marrs C F. Role of pili in Haemophilus influenzae adherence and colonization. Infect Immun. 1997;65:2997–3002. doi: 10.1128/iai.65.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilsdorf J R, McCrea K W, Forney L J. Conserved and nonconserved epitopes among Haemophilus influenzae type b pili. Infect Immun. 1990;58:2252–2257. doi: 10.1128/iai.58.7.2252-2257.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilsdorf J R, Chang H Y, McCrea K W, Bakaletz L O. Comparison of hemagglutinating pili of Haemophilus influenzae type b with similar structures of nontypeable H. influenzae. Infect Immun. 1992;60:374–379. doi: 10.1128/iai.60.2.374-379.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function, and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 17.Hauser M A, Scocca J J. Location of the host attachment site for phage HP1 within a cluster of Haemophilus influenzae tRNA genes. Nucleic Acids Res. 1990;18:5305. doi: 10.1093/nar/18.17.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser M A, Scocca J J. Site-specific integration of the Haemophilus influenzae bacteriophage HP1: location of the boundaries of the phage attachment site. J Bacteriol. 1992;174:6674–6677. doi: 10.1128/jb.174.20.6674-6677.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi T, Matsumoto H, Ohnishi M, Terawaki Y. Molecular analysis of a cytotoxin-converting phage, φCTX, of Pseudomonas aeruginosa: structure of attP-cos-ctx region and integration into the serine tRNA gene. Mol Microbiol. 1993;7:657–667. doi: 10.1111/j.1365-2958.1993.tb01157.x. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix R W, Smith M C M, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoiseth S K, Moxon E R, Silver R P. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc Natl Acad Sci USA. 1986;83:1106–1110. doi: 10.1073/pnas.83.4.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knapp S, Hacker J, Jarchau T, Goebel W. Large unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli strain 536. J Bacteriol. 1986;168:22–30. doi: 10.1128/jb.168.1.22-30.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroll J S, Loynds B M, Moxon E R. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol. 1991;5:1549–1560. doi: 10.1111/j.1365-2958.1991.tb00802.x. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence J G, Ochman H. Amelioration of bacterial genome: rates of change and exchange. J Mol Evol. 1996;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 26.LiPuma J J, Richman H, Stull T L. Haemocin, the bacteriocin produced by Haemophilus influenzae: species distribution and role in colonization. Infect Immun. 1990;58:1600–1605. doi: 10.1128/iai.58.6.1600-1605.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin K, Morlin G, Smith A, Nordyke A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaels R H, Stonebraker F E, Robbins J B. Use of antiserum agar for detection of Haemophilus influenzae type b in the pharynx. Pediatr Res. 1975;9:513–516. doi: 10.1203/00006450-197505000-00010. [DOI] [PubMed] [Google Scholar]
- 29.Moxon E R. Molecular basis of Haemophilus influenzae type b disease. J Infect Dis. 1992;165:s77–s81. doi: 10.1093/infdis/165-supplement_1-s77. [DOI] [PubMed] [Google Scholar]
- 30.Murley Y M, Edlind T D, Plett P A, LiPuma J J. Cloning of haemocin locus of Haemophilus influenzae type b and assessment of the role of haemocin in virulence. Microbiology. 1998;144:2531–2538. doi: 10.1099/00221287-144-9-2531. [DOI] [PubMed] [Google Scholar]
- 31.Murphy T F, Apicella M A. Nontypable Haemophilus influenzae: a review of clinical aspects, surface antigens, and the human immune response to infection. Rev Infect Dis. 1987;9:1–15. doi: 10.1093/clinids/9.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Musser J M, Kroll J S, Granoff D M, Moxon E R, Brodeur B R, Campos J, Dabernat H, Frederiksen W, Hamel J, Hammond G, Hoiby E A, Jonsdottir K E, Kabeer M, Kallings I, Khan W N, Kilian M, Knowles K, Koornhof H J, Law B, Li K I, Montgomery J, Pattison P E, Piffaretti J-D, Takala A K, Thong M E, Wall R A, Ward J I, Selander R K. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis. 1990;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 33.Musser J M, Kroll J S, Moxon E R, Selander R K. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci USA. 1988;85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequences databases; its status 1999. Nucleic Acids Res. 1999;27:292. doi: 10.1093/nar/27.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rand, K. N. 1996, posting date. Crystal violet can be used to visualize DNA bands during gel electrophoresis and to improve cloning efficiency. Elsevier Trends Journals Technical Tips Online. http://biomednet.com.
- 36.Ritter A, Blum G, Emody L, Kerenyi M, Bock A, Neuhierl B, Rabsch W, Scheutz F, Hacker J. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic E. coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5436–5464. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith H O, Tomb J-F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 39.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan J T, Ronson C W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Susa M, Kreft B, Wasenauer G, Ritter A, Hacker J, Marre R. Influence of cloned tRNA genes from a uropathogenic Escherichia coli strain on adherence to primary human renal tubular epithelial cells and nephropathogenicity in rats. Infect Immun. 1996;64:5390–5394. doi: 10.1128/iai.64.12.5390-5394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tizard M, Bull T, Millar D, Doran T, Martin H, Sumar N, Ford J, Hermon-Taylor J. A low G+C content genetic island in Mycobacterium avium subsp. Paratuberculosis and M. avium subsp. silvaticum with homologous genes in Mycobacterium tuberculosis. Microbiology. 1998;144:3413–3423. doi: 10.1099/00221287-144-12-3413. [DOI] [PubMed] [Google Scholar]
- 43.van Eldere J, Brophy L, Loynds B, Celis P, Hancock I, Carman S, Kroll J S, Moxon E R. Region II of the Haemophilus influenzae type b capsulation locus involved in serotype-specific polysaccharide synthesis. Mol Microbiol. 1995;15:107–118. doi: 10.1111/j.1365-2958.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 44.van Ham S M, van Alphen L, Mool F R, van Putten J P M. The fimbrial gene cluster of Haemophilus influenzae type b. Mol Microbiol. 1994;13:673–684. doi: 10.1111/j.1365-2958.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 45.Wilcox K W, Smith H O. Isolation and characterization of mutants of Haemophilus influenzae deficient in an adenosine 5′-triphosphate deoxyribonuclease activity. J Bacteriol. 1975;122:443–453. doi: 10.1128/jb.122.2.443-453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wren B W, Colby S M, Cubberley R R, Pallen M J. Degenerate PCR primers for the amplification of fragments from genes encoding response regulators from a range of pathogenic bacteria. FEMS Microbiol Lett. 1992;99:287–292. doi: 10.1016/0378-1097(92)90042-m. [DOI] [PubMed] [Google Scholar]