Abstract
Cyclophilin is known to act as a molecular chaperone in the endoplasmic reticulum. Recent studies have reported that the expression of cyclophilin B (CypB) is increased in ob/ob mice and its inhibitor suppresses adipocyte differentiation. However, the mechanism of action of CypB in adipocytes remains to be elucidated. The present study investigated the role of CypB in 3T3-L1 adipocyte differentiation. It showed that the expression level of CypB was increased during 3T3-L1 adipocyte differentiation by reverse transcription-quantitative PCR and western blotting analysis. CypB knockdown using short interfering RNA delayed cell cycle progression from the G1/S to G2/M phase through the mammalian target of rapamycin (mTOR) signaling pathway and inhibited the expression levels of adipogenic transcription factors including peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT-enhancer binding protein (C/EBP)α. Additionally, the accumulation of lipid droplets was inhibited by CypB knockdown. Conversely, overexpression of CypB promoted cell cycle progression from the G1/S to G2/M phase by the mTOR signaling pathway and enhanced the expression levels of adipogenic transcription factors including PPARγ and C/EBPα. Finally, the present study showed that CypB downregulated the expression of CHOP, a well-known negative regulator of adipogenesis. Taken together, the data suggested that CypB might serve important physiological regulatory roles in 3T3-L1 adipocyte differentiation.
Keywords: cyclophilin B, mitotic clonal expansion, adipogenesis, obesity, AKT/mTOR signaling pathway, 3T3-L1 adipocyte
Introduction
Obesity is a disease in which fat accumulates in the body due to an imbalance between food intake and energy expenditure. It is known to increase the risk of metabolic disorders such as type 2 diabetes, arteriosclerosis, hypertension and various cancers (1-3). Global research on obesity prevention and treatment is underway and various methods such as diet, exercise, surgery and drug therapy are being proposed (4-6). However, it is not easy to lose weight with behavioral therapy alone and drugs and surgery cause side effects such as mental disorders and depression (7). Obesity is induced by the accumulation of triglyceride in cells through adipogenesis, in which preadipocytes differentiate into mature adipocytes. Methods for preventing and treating obesity by regulating adipogenesis, one of the causes of obesity, are being studied (8,9).
The standard adipogenesis process of 3T3-L1 preadipocytes consists of three steps: Contact inhibition, mitotic clonal expansion (MCE) and terminal adipogenic differentiation (10). After contact inhibition and growth arrest, 3T3-L1 preadipocytes reenter the cell cycle using a differentiation medium, MDI [dexamethasone, 3-isobutyl-1-methylxanthine (IBMX), insulin]. Then, the cells undergo a few mitotic cycles during MCE, which is an essential step in the early stage of adipocyte differentiation (11). Therefore, anti-obesity therapies that inhibit adipocyte differentiation through MCE regulation are being studied (12-15). Transcription factors that regulate adipogenesis include peroxisome proliferator-activated receptors (PPARs) and CCAAT/enhancer-binding proteins (C/EBPs). During MCE, early adipogenic factors, C/EBPβ and C/EBPδ are temporally expressed and this event stimulates late adipogenic factors such as C/EBPα and peroxisome proliferator-activated receptor γ (PPARγ). C/EBPα and PPARγ coordinately induce terminal differentiation into a mature adipocyte phenotype by activating adipocyte-specific genes including adipocyte protein 2 (FABP4), a terminal marker of adipogenesis (16-18). In addition, the expression of the regulators has been reported to be associated with the expression of cyclin-dependent kinase inhibitors, including p27 (19,20).
CypB is a protein present in the endoplasmic reticulum. It has prolyl isomerase activity (PPIase) and the ability to inhibit endoplasmic reticulum (ER) stress and oxidative stress. It is involved in protein folding and acts as a chaperone to prevent or inhibit protein folding (21). CypB expression has been reported to increase in the serum of patients with metabolic disorders more than in healthy subjects and in various organs of ob/ob mice (obesity-induced mice) (22). Sanglifehrin-based cyclophilin inhibitor suppresses the carcinoma formation and progression of non-alcoholic fatty liver (23). Similar to CypB, CypA, which is known as a member of the PPIase family, has been reported as a novel adipogenic factor (24). Therefore, it is fascinating to study the role of cyclophilin family proteins in the progression of adipogenesis.
CHOP has been reported as a negative regulator that inhibits adipocyte differentiation (25-27). It has been reported that eIF2α phosphorylation increases CHOP production to repress adipocyte differentiation in response to ER stress in vitro and in vivo (28). It has been proposed that an elevated rate of protein synthesis would in turn increase phosphorylation of eIF2α in differentiation and later induce downstream target molecules such as CHOP. Therefore, inhibiting ER stress activation may be the key to reducing metabolic syndrome events such as adipogenesis (28). Further studies are needed to dissect the cellular and molecular mechanisms involved in the regulation of adipogenesis and ER stress.
The mammalian target of rapamycin (mTOR) is a phosphoinositide 3-kinase (PI3K)-like serine/threonine-protein kinase that controls protein and lipid synthesis, cell size, proliferation, differentiation, autophagy and metabolism according to intracellular and extracellular cues (29). mTOR is the catalytic core of two distinct multiprotein complexes, mTOR complex 1 (mTORC1) and 2 (mTORC2), which differ in their components, regulation, function and sensitivity to rapamycin (30). Insulin and insulin-like growth factors (IGFs) bind to the insulin receptor (IR) or IGF receptor (IGF-IR), thereby activating the receptor tyrosine kinase (31). Then, activated IR substrate proteins trigger PI3K/AKT signaling and promote cell proliferation and lipid synthesis (32). PI3K/AKT/mTOR signaling pathway and its downstream, ribosomal protein S6 kinase (P70S6K) are reported to be critical regulators of adipogenesis (33). In particular, mTOR complex 1 (mTORC1) phosphorylates P70S6K and upregulates the adipogenic transcription factors PPARγ and C/EBPα (34-36).
The present study, for the first time to the best of the authors' knowledge, investigated the molecular mechanism of CypB in 3T3-L1 adipocyte differentiation, with a focus on the early-stage MCE process of adipogenesis and the AKT/mTOR signaling pathway.
Materials and methods
Reagents
The 3T3-L1 cell was purchased from the Korean Cell Line Bank. Dulbecco's modified Eagle's medium (DMEM) and calf serum (CS) used for cell culture were purchased from HyClone (Cytiva) and penicillin/streptomycin was purchased from Corning, Inc. MDI used for differentiation was purchased from MilliporeSigma, fetal bovine serum (FBS) HyClone (Cytiva). Cyclosporine A (CsA) used as an inhibitor of CypB was purchased from Cell Signaling Technology, Inc. and Rapamycin used as an mTOR inhibitor was purchased from MilliporeSigma. RNA extraction was performed using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA synthesis was performed using a kit (Thermo Fisher Scientific, Inc.). Reverse transcription PCR was performed using a 7500 Reverse transcription PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and with Power SYBR® Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). All reagents used for Oil Red O Staining were purchased from MilliporeSigma. For cell cycle analysis, propidium iodide solution and RNase A solution were purchased from MilliporeSigma. Cell proliferation assay was performed using Chromo-CK cell viability assay reagent (Monobio). Antibodies used in the present study included mouse anti-β-actin; cat. no. sc-47778, mouse anti-vinculin; cat. no. sc-25336, mouse anti-C/EBPβ; cat. no. sc-7962, mouse anti-C/EBPα; cat. no. sc-166258, goat anti-FABP4; cat. no. sc-18661, rabbit anti-Adiponectin; cat. no. PA1-054 (Thermo Fisher Scientific, Inc.), mouse anti-hemagglutinin (HA); cat. no. sc-7392, mouse anti-CyclinD; cat. no. sc-8396, mouse anti-CyclineE; cat. no. sc247, mouse anti-CyclinA; cat. no. sc-271645, rabbit anti-AKT; cat. no. sc-8312 (Santa Cruz Biotechnology, Inc.), rabbit anti-PPAR; cat. no. 2443s, rabbit anti-CDK4; cat. no. 12790s, rabbit anti-p27; cat. no. 2552s, rabbit anti-pRB; cat. no. 8516s, rabbit anti-pAKT; cat. no. 9271s, rabbit anti-pmTOR; cat. no. 5536s, rabbit anti-mTOR; cat. no. 2983s, rabbit anti-phosphorylated (p-) p70S6K; cat. no. 9204L, rabbit anti-p70S6K; cat. no. 9202L (Cell Signaling Technology, Inc.), rabbit anti-CypB; cat. no. ab16045 (Abcam). Secondary antibodies used were goat anti-rabbit IgG; cat. no. 31460, goat anti-mouse IgG; cat. no. M32607 (Invitrogen; Thermo Fisher Scientific, Inc.) and rabbit anti goat IgG; cat. no. SA007-500 (GenDEPOT, LLC).
Cell culture and differentiation
3T3-Ll cells were cultured in DMEM supplemented with 10% CS and 1% P/S at 37°C in a 5% CO2 atmosphere. For 3T3-L1 cell differentiation, the cells when confluent were incubated for 2 more days (day 0) and then treated with MDI (500 µM IBMX, 0.25 µM dexamethasone, 10 µg/ml insulin) for 48 h at 37°C to induce differentiation. At 48 h after initiating 3T3-L1 cell differentiation, the DMEM medium (10% FBS, 1% P/S) was supplemented with 10 µg/ml insulin for 2 days (day 4). Day 4 after 3T3-L1 cell differentiation, DMEM medium (10% FBS, 1% P/S) was supplemented for 3 days (day 7). On day 7 of differentiation, differentiating cells were observed by capturing five different random fields of views for each well using a light microscopy at ×40 magnification (model IX73; Olympus Corporation). CsA (a CypB inhibitor) and mTOR-specific inhibitor rapamycin were dissolved in DMSO and added into the medium at 5 µg/ml (CsA) and 0.5 µM (Rapamycin), respectively at 37°C.
Plasmid and short interfering (si)RNAs transfection
The 3T3-L1 cells were seeded at a density of 5.5×105 cells/ml in a 60 mm dish or 12×105 cells/ml in a 100 mm dish and then incubated for 24 h at 37°C in a 5% CO2 atmosphere. Afterward, plasmid or siRNA transfection was performed using X-tremeGene® HP DNA Transfection Reagent (MilliporeSigma) or X-tremeGENE siRNA Transfection Reagent (MilliporeSigma) according to the manufacturer's instructions and treated for 48 h at 37°C in a 5% CO2 atmosphere. Subsequent experimentation was then performed 2 days later. CypB was HA-tagged at its 5' end and subcloned into a pcDNA vector (HA tag; 5′-TAC CCA TAC GAC GTC CCA GAC TAC GCT-3′). CypB-specific siRNA (5′GGA AAG ACU GUU CCA AAA A3′; D-001136-01-20) and scrambled siRNA (5′UAA GGC UAU GAA GAG AUA C3′) were purchased from Dharmacon.
Western blot analysis
Cells were lysed in RIPA buffer (50 mM Tris-HCl; pH 7.6; 150 mM NaCl; 1% Triton X-100; 1% Sodium deoxycholate). Protein concentrations was determined by BCA assay. Equal amounts of protein (20 µg) were separated with 6-15% SDS-PAGE gel and transferred to BioTrace NT nitro-cellulose membrane (Pall Life Sciences) and incubated with 5% blocking solution [(BSA; cat. no. A0100-010; GenDEPOT, LLC) or skimmed milk (cat. no. MB-S1667; MB cell)] for 1 h at room temperature. Afterward, the membrane was incubated with primary antibody overnight at 4°C. Next, the reaction was performed at room temperature for 1 h using a secondary antibody matching the primary antibody. The primary antibodies and secondary antibodies concentration used in the experiment were 1:1,000 and 1:10,000, respectively. The bands were detected with the enhanced chemiluminescence (Clarity™ Western ECL Substrate; cat. no. 170-5061; Bio-Rad Laboratories, Inc.), and the density and size of the bands were quantified using ImageJ 1.50i software (National Institutes of Health) by normalizing to β-actin and Vinculin.
Reverse transcription-quantitative (RT-q) PCR
The 3T3-L1 cells were seeded at a density of 5.5×105 cells/ml in a 60 mm dish. Total RNAs were extracted by TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized from 0.2 µg total RNA using RevertAid First Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). Reverse transcription PCR was performed using 7500 Real Time PCR system with SYBR-Green PCR Master Mix according to the manufacturer's instructions. The Real Time PCR cycling conditions consisted of an initial activation step of 95°C for 15 min, denaturation of 95°C for 15 sec, annealing of 60°C for 30 sec, extension for 72°C for 30 sec; for 40 cycles. The primer sequences used are shown in Table I. The data were quantified using the 2−ΔΔCq method and were normalized against the levels of GAPDH (37). These experiments were performed in triplicate and repeated five times with similar results.
Table I.
A list of primer sequences used in current study.
| Gene | Direction | Sequence |
|---|---|---|
| CypB | Forward | TATGAAGGTGCTCTTCGCCG |
| Reverse | AGTATACCTTGACTGTGACTTTAGG | |
| GAPDH | Forward | ATGGTGAAGGTCGGTGTGAA |
| Reverse | TGGAAGATGGTGATGGGCTT | |
| C/EBPβ | Forward | AGCTGAGCGACGAGTACAAG |
| Reverse | AGCTGCTCCACCTTCTTCTG | |
| C/EBPα | Forward | GCCATGTGGTAGGAGACAGA |
| Reverse | CAAGTTCCTTCAGCAACAGC | |
| PPARγ | Forward | ATCTTAACTGCCGGATCCAC |
| Reverse | TGGTGATTTGTCCGTTGTCT | |
| Adiponectin | Forward | TGTTCCTCTTAATCCTGCCCA |
| Reverse | CCAACCTGCACAAGTTCCTT | |
| FABP4 | Forward | AAAGAAGTGGGAGTGGGC |
| Reverse | CTGTCGTCTGCGGTGATT |
CypB, cyclophilin B; C/EBP, CCAAT-enhancer binding protein; PPARγ, peroxisome proliferator-activated receptor γ; FABP4, fatty acid binding protein 4.
Oil Red O staining
After day 7, differentiated 3T3-L1 cells were washed three times with phosphate-buffered saline (PBS) and then fixed with 3.7% paraformaldehyde for 1 h at room temperature and washed thrice again with PBS. The cells were stained with 0.5% Oil Red O in isopropanol for 1 h at room temperature. Stained cells were washed three times with distilled water. Then, the stained cells were observed by capturing five different random fields of views for each well using a light microscopy at ×100 magnification (model IX73; Olympus Corporation) and lipid droplets were measured with a microplate reader (Molecular Devices, LLC.) at 510 nm.
Flow cytometric cell cycle analysis
The cells were harvested after MDI treatment for 48 h. Harvested cells were fixed 70% cold ethanol overnight at -20°C and washed with cold PBS. The fixed cells were centrifuged at 500 × g for 10 min at room temperature. The centrifuged cell pellet was resuspended in 10 µg/ml of RNase A at 37°C for 1 h. Then, 40 µg/ml propidium iodine solution was added at 37°C for 10 min in the dark. Cells were analyzed using a FACSCalibur system (BD Biosciences) according to the manufacturer's instructions. The results were averaged, and the percentage of cell cycle progression was calculated using the BD CellQuest™ Pro software (version 5.1; BD Biosciences).
Statistical analysis
Results were presented as the mean ± standard deviation. Error bars represent the mean ± standard deviation of three or five independent experiments performed in triplicate. Depending on the design of the experiment, data were analyzed one-way ANOVA with post hoc multicomparison analysis (Tukey's test) and comparisons between the two groups were performed using an unpaired two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of CypB during adipogenesis in 3T3-L1 cells
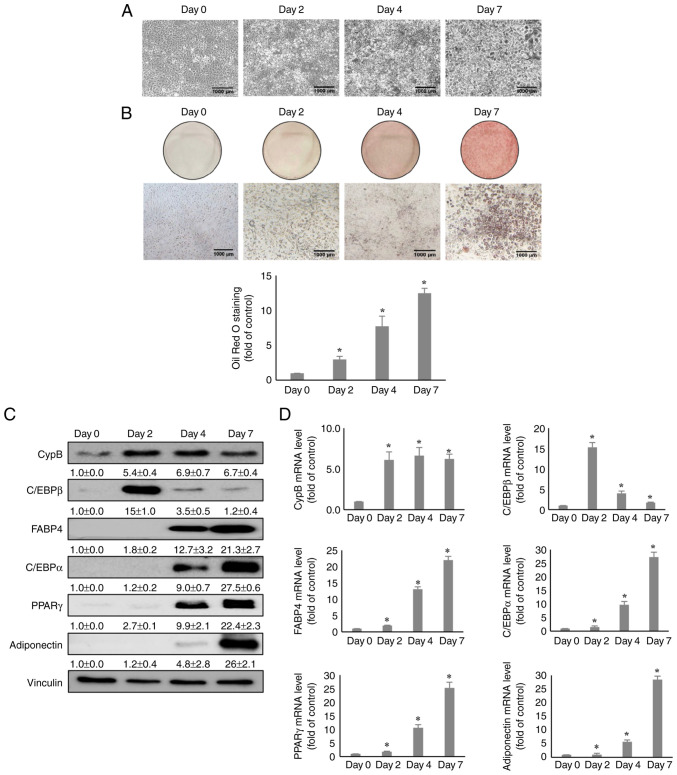
Post confluent 3T3-L1 pre-adipocytes were treated with MDI, an adipogenic inducer and were observed for 7 days to determine the expression level of CypB during adipocyte differentiation and adipogenesis. Fig. 1A shows the morphology image of 3T3-L1 cells during the differentiation period under a micro-scope and Fig. 1B shows that the lipid droplets were stained with Oil Red O staining assay for each day, confirming that the differentiation was normal. For reference, the expression levels of key adipogenic markers such as C/EBPβ, PPARγ, C/EBPα, FABP4 and adiponectin were monitored. The C/EBPβ gene is expressed in the early stage of adipogenesis and at the middle and later stages, PPARγ, C/EBPα, FABP4 and Adiponectin are expressed (38). As shown in Fig. 1C, CypB and C/EBPβ were transiently induced on day 2 and the protein expression levels of PPARγ, C/EBPα, FABP4 and Adiponectin were significantly increased after day 4. Notably, CypB protein was upregulated by 5-6 fold after day 2. mRNA levels of CypB were measured by RT-qPCR during adipocyte differentiation (Fig. 1D). Consistent with previous reports (16,38), the mRNA levels of adipogenic genes including C/EBPβ, PPARγ, C/EBPα and FABP4 were increased after treatment with MDI. The mRNA level of CypB was also significantly upregulated during MDI-induced adipocyte differentiation, consistent with the CypB protein expression data.
Figure 1.
Expression of CypB during 3T3-L1 cell adipogenesis. (A) The morphology images of 3T3-L1 cells during the adipogenesis period. (B) During adipogenesis, cells (Day 0, 2, 4 and 7) were stained with lipid droplets through Oil Red O staining assay. Quantification of the stained lipid droplets was performed by measuring the absorbance at 510 nm. (C) At 0, 2, 4 and 7 days after induction of 3T3-L1 cell adipogenesis, protein levels of CypB and adipogenic markers (C/EBPβ, PPARγ, C/EBPα, FABP4 and adiponectin) during 3T3-L1 cell adipogenesis was measured by western blotting. Vinculin was used as a loading control. (D) The mRNA levels of CypB and adipogenic marker genes (C/EBPβ, PPARγ, C/EBPα, FABP4 and adiponectin) were measured by reverse transcription-quantitative PCR. The values were shown as the mean ± standard deviation of three independent experiments. *P<0.001 vs. the data of Day 0. CypB, cyclophilin B; C/EBP, CCAAT-enhancer binding protein; PPARγ, peroxisome proliferator-activated receptor γ; FABP4, fatty acid binding protein 4.
The influence of CypB knockdown on adipocyte differentiation
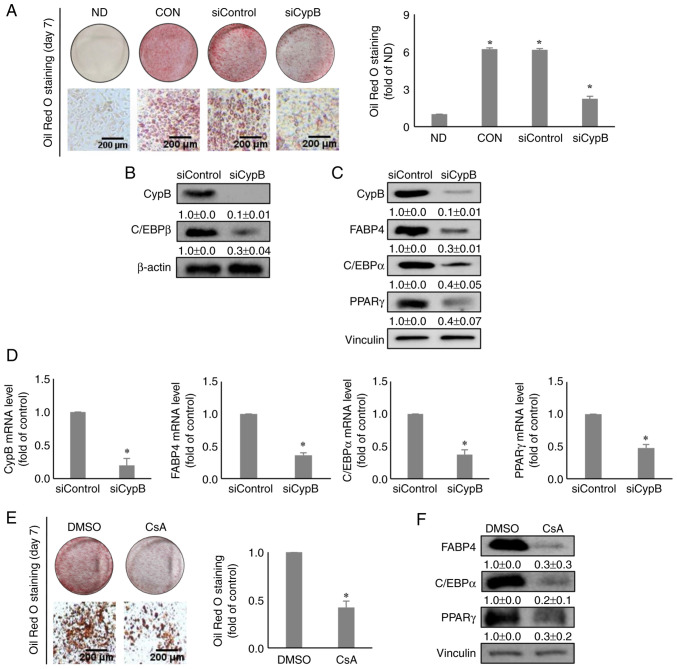
To investigate the functional roles of CypB in 3T3-L1 cell adipogenesis, the effects of CypB gene silencing were monitored using siRNA on adipocyte differentiation. Following CypB-knockdown cells were treated with MDI for 48 h to induce differentiation, Oil Red O staining assay was performed to evaluate the effects of CypB knockdown on lipid accumulation during adipocyte differentiation. The results showed that lipid accumulation was reduced by >50% in siCypB, compared to siControl (Fig. 2A). Next, western blot analysis was performed to monitor a key early-stage adipogenic marker, C/EBPβ. As shown in Fig. 2B, both CypB and C/EBPβ were significantly suppressed in CypB knockdown, compared with siControl. Other key adipogenic factors such as FABP4, C/EBPα and PPARγ were significantly downregulated in CypB knockdown, compared with siControl (Fig. 2C). Likewise, mRNA levels of key adipogenic markers such as FABP4, C/EBPα and PPARγ were significantly decreased in siCypB, compared to siControl (Fig. 2D). To confirm these results, CsA was used. Consistent with the CypB knockdown data, treatment with CsA inhibited lipid accumulation by >50%, compared to the DMSO (Fig. 2E). In addition, the CsA treatment led to a reduction the expression levels of key adipogenic markers such as FABP4, C/EBPα and PPARγ (Fig. 2F). These data suggested that CypB was required for adipogenesis in 3T3-L1 cells.
Figure 2.
Effect of knockdown of CypB on adipogenesis in 3T3-L1 cells. 3T3-L1 cells at 70-80% confluence were transfected with scrambled siRNA (siControl) or CypB siRNA (siCypB at 37°C for 48 h. Then the fully confluent cells were differentiated for 2 to 7 days. (A) The differentiated cells (ND, CON, siControl and siCypB) were stained with Oil Red O to detect cytoplasmic lipid droplets. Quantification of the stained lipid droplets was performed by measuring the absorbance at 510 nm. CypB knockdown efficiency was monitored by western blotting and reverse transcription-quantitative PCR. β-actin and vinculin were used as loading controls. (B) The expression of C/EBPβ, an early adipogenic factor was analyzed by western blotting 2 days after induction of adipogenesis. β-actin was used as a loading control. (C) The expressions of late adipogenic factors (C/EBPα, PPARγ and FABP4) were analyzed by western blotting 7 days after induction of adipogenesis. Vinculin was used as a loading control. (D) At 7 days after induction of adipogenesis, the mRNA levels of CypB and adipogenic marker genes (C/EBPβ, PPARγ, C/EBPα and FABP4) were measured by reverse transcription-quantitative PCR. The values were shown as the mean ± standard deviation of five independent experiments. *P<0.001 vs. siControl. (E) The CsA-treated differentiated cells were stained with Oil Red O to detect cytoplasmic lipid droplets. Quantification of the stained lipid droplets was performed as aforementioned. (F) After treatment with 5 µg/ml CsA (dissolved in DMSO), 3T3-L1 cells were induced to differentiate into adipocytes. The expressions of late adipogenic factors (C/EBPα, PPARγ and FABP4) were analyzed by western blotting 7 days after induction of adipogenesis. Vinculin was used as a loading control. The control was treated with DMSO. The values were shown as the mean ± standard deviation of five independent experiments. *P<0.001 vs. DMSO. CypB, cyclophilin B; si, short interfering; ND, non-differentiation; CON, untreated differentiation control; C/EBP, CCAAT-enhancer binding protein; PPARγ, peroxisome proliferator-activated receptor γ; FABP4, fatty acid binding protein 4.
The influence of CypB overexpression during 3T3-L1 differentiation
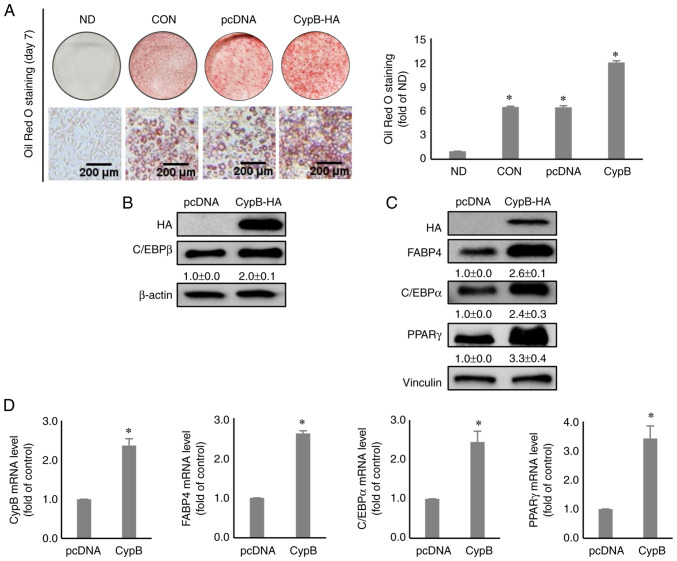
To investigate the functional role of CypB in adipogenesis, 3T3-L1 pre-adipocytes were transfected with a CypB expression vector. After the pre-adipocytes were treated with MDI for 2 days, Oil Red O staining assay was performed to evaluate the effects of overexpression of CypB on lipid accumulation during adipocyte differentiation. The results showed that overexpression of CypB increased lipid accumulation by almost two-fold, compared to pcDNA (Fig. 3A). Next, western blot analysis was performed to monitor the levels of a key early-stage adipogenic marker, C/EBPβ. As shown in Fig. 3B, C/EBPβ was significantly increased during CypB-overexpression, compared with pcDNA. Other key adipogenic factors such as FABP4, C/EBPα and PPARγ were significantly upregulated under the conditions of CypB-overexpression on day 7, compared with pcDNA (Fig. 3C). Likewise, CypB overexpression significantly increased mRNA levels of key adipogenic markers such as FABP4, C/EBPα and PPARγ, compared to pcDNA (Fig. 3D).
Figure 3.
Effect of CypB overexpression on adipogenesis in 3T3-L1 cells. 3T3-L1 cells at 70-80% confluence were transfected with pcDNA or CypB-HA at 37°C for 48 h. Then the fully confluent cells were differentiated for 2 to 7 days. (A) The CypB-overexpressed and differentiated cells were stained with Oil Red O to detect cytoplasmic lipid droplets. Quantification of the stained lipid droplets was performed as before. CypB overexpression efficiency was monitored by western blotting. (B) The expression of C/EBPβ, an early adipogenic factor was analyzed by western blotting 2 days after induction of adipogenesis. β-actin was used as a loading control. (C) The expressions of late adipogenic factors (C/EBPα, PPARγ and FABP4) were analyzed by western blotting 7 days after induction of adipogenesis. Vinculin was used as a loading control. (D) At 7 days after induction of adipogenesis, the mRNA levels of CypB and adipogenic marker genes (C/EBPβ, PPARγ, C/EBPα and FABP4) were measured by reverse transcription-quantitative PCR. The values were shown as the mean ± standard deviation of five independent experiments. *P<0.001 vs. pcDNA. CypB, cyclophilin B; si, short interfering; ND, non-differentiation; CON, untreated differentiation control; C/EBP, CCAAT-enhancer binding protein; PPARγ, peroxisome proliferator-activated receptor γ; FABP4, fatty acid binding protein 4.
Effect of CypB on the regulation of the AKT/mTOR signaling pathway during 3T3-L1 differentiation
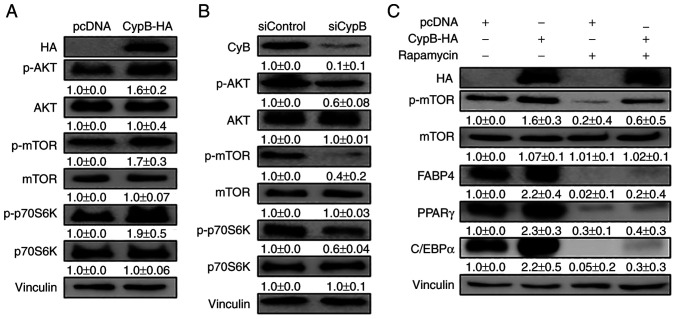
AKT/mTOR plays an important role in adipocyte differentiation, which affects lipid metabolism through the insulin pathway (39). To examine whether CypB affects the AKT/mTOR pathway in adipocyte differentiation, CypB-HA was overexpressed in 3T3-L1 preadipocytes before inducing the differentiation of 3T3-L1 cells with MDI, followed by western blotting. As shown in Fig. 4A, CypB overexpression activated the AKT/mTOR pathway. P70S6 kinase (P70S6K) is a mitogen-activated Ser/Thr protein kinase that is required for cell growth and G1 cell cycle progression. CypB overexpression increased the expression of p-P70S6K. Next, the effects of CypB gene silencing were tested using siRNA on the AKT/mTOR pathway by western blotting (Fig. 4B). The expression levels of p-AKT, p-mTOR and p-P70S6K were significantly reduced in CypB knockdown. To confirm these results, rapamycin was used in the assay. Treatment with rapamycin negated the effects of overexpressed CypB on the AKT/mTOR pathway (Fig. 4C). These results suggested that CypB plays an important role in the AKT/mTOR pathway involved in adipocyte differentiation.
Figure 4.
CypB regulates the AKT/mTOR signaling pathway in 3T3-L1 adipocytes. 3T3-L1 cells at 70-80% confluence were transfected with plasmids or siRNA at 37°C for 48 h. Then the fully confluent cells were differentiated with MDI treatment for 24 h. CypB overexpression and CypB knockdown efficiency were monitored by western blotting. The protein band intensities were quantified by ImageJ and were normalized to the expression of vinculin. (A) Following transfection with CypB-HA or pcDNA, 3T3-L1 preadipocytes were differentiated into adipocytes for 24 h. The expression of AKT/mTOR signaling pathway molecules (p-AKT, p-mTOR and p-p70S6K) was monitored by Western blotting. (B) After gene knockdown by siCypB or siControl, 3T3-L1 preadipocytes were differentiated into adipocytes for 24 h. AKT/mTOR signaling pathway molecules (p-AKT, p-mTOR and p-p70S6K) were monitored by western blotting. (C) Following transfection with CypB-HA or pcDNA, 3T3-L1 preadipocytes were differentiated into adipocytes in the presence or absence of rapamycin and mTOR inhibitor for 7 days and subjected to western blotting with antibodies against late adipogenic factors such as C/EBPα, PPARγ and FABP4. Vinculin was used as a loading control. The control was pcDNA (-rapamycin). The values were shown as the mean ± standard deviation of three independent experiments. CypB, cyclophilin B; AKT, protein kinase B; mTOR, mammalian target of rapamycin; p-phosphorylated; si, short interfering; C/EBP, CCAAT-enhancer binding protein; PPARγ, peroxisome proliferator-activated receptor γ; FABP4, fatty acid binding protein 4; ND, non-differentiation; CON, untreated differentiation control.
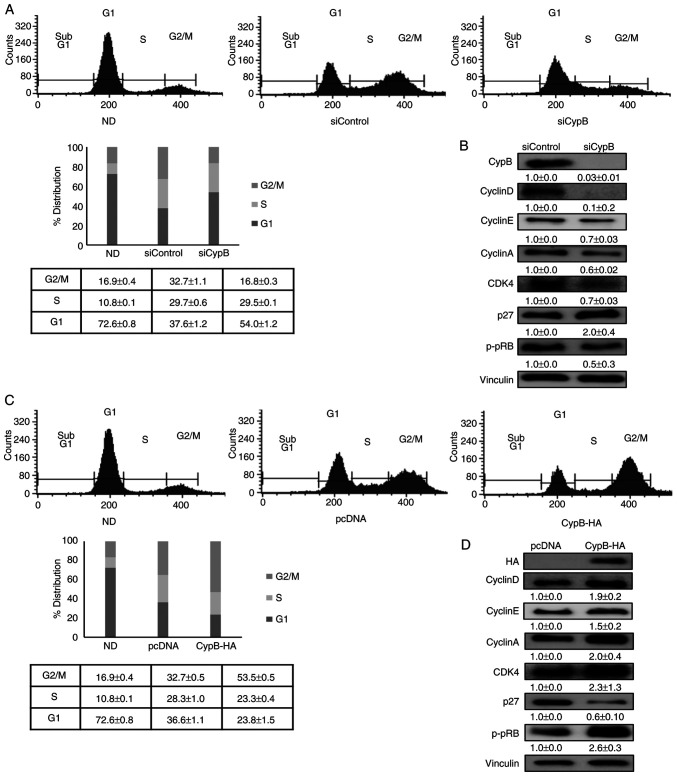
CypB promotes mitotic clonal expansion in adipogenesis
Next, how cell cycle events during mitotic clonal expansion are affected by CypB were examined. G1-arrested 3T3-L1 cells synchronously re-enter the cell cycle and undergo mitotic clonal expansion during adipogenesis (11). As shown in Fig. 5A, CypB knockdown (siCypB) reduced the G2/M population during adipocyte differentiation, compared to siControl. The DNA content of the cells in the G1 phase in the siControl and siCypB groups was 37.6 and 54.0%, respectively, while that of the cells in the G2/M phase in the two groups was 32.7 and 16.8%, respectively. Next, the expression of cell cycle-associated genes such as CyclinD, CylclinE, CyclinA, CDK4, p27 and pRB in CypB knockdown were monitored (Fig. 5B). Compared to siControl, expression levels of CyclinD, CylclinE, CyclinA, CDK4 and pRB were significantly reduced in siCypB, while p27 expression level was increased in siCypB by two-fold. These results suggested that the cell cycle transition from G1 into the G2/M phase was inhibited in CypB knockdown. As shown in Fig. 5C, overexpression of CypB increased the G2/M population during adipocyte differentiation, compared to pcDNA control. The DNA content of the cells in the G1 phase in pcDNA and CypB-HA groups was 36.6 and 23.8%, respectively, while that of the cells in the G2/M phase in the two groups was 35.7 and 53.5%, respectively. Compared to pcDNA, expression levels of CyclinD, CylclinE, CyclinA, CDK4 and pRB were significantly increased by CypB overexpression, while p27 expression level was significantly decreased in CypB overexpression. These results suggested that the cell cycle transition from G1 into the G2/M phase was facilitated by CypB overexpression. Altogether, the data suggested that CypB regulates adipocyte differentiation by modulation of cell cycle progression.
Figure 5.
Effect of CypB on cell cycle progression during 3T3-L1 cell differentiation. (A) 3T3-L1 cells at 70-80% confluence were transfected with scrambled siRNA (siControl) or CypB siRNA (siCypB). Then the fully confluent cells were differentiated into adipocyte with MDI treatment for 24 h. Cell cycle progression was monitored with flow cytometric analysis. 3T3-L1 cells were stained with propidium iodide nuclear dye under the indicated conditions. Cellular DNA content was analyzed by flow cytometric analysis and cells were distributed in the three phases of the cycle (G1. S and G2/M). Results were presented in percentage for each plot and are representative of three independent experiments. (B) The expressions of cell cycle-associated genes (CyclinD, CylclinE, CyclinA, CDK4, p27 and pRB) were analyzed in CypB knockdown by western blotting. Vinculin was used as a loading control. (C) 3T3-L1 cells transfected with pcDNA or CypB-HA were induced to differentiate into adipocytes. Following CypB overexpression, cell cycle progression was monitored as before. (D) Following overexpression of CypB, the expression of cell cycle-associated proteins (CyclinD, CylclinE, CyclinA, CDK4, p27 and pRB) was analyzed by western blotting. The values were shown as the mean ± standard deviation of three independent experiments. CypB, cyclophilin B; si, short interfering; p-phosphorylated.
CypB modulates the expression of CHOP, a negative regulator in adipogenesis
Our previous study reported that CypB interacts with p300 to degrade CHOP in tumor cells (40). It has also been reported that CHOP inhibits adipogenesis and p300 is the transcriptional co-activator of C/EBPα (25,41). Therefore, whether CypB regulates the expression of CHOP in 3T3-L1 cells was tested by western blotting. CypB knockdown decreased the expression level of C/EBPβ and increased the expression level of CHOP (Fig. 6A). Compared with pc-DNA, overexpression of CypB increased the expression level of C/EBPβ and significantly decreased the expression level of CHOP (Fig. 6B). By contrast, CypB knockdown decreased the expression level of C/EBPβ and increased the expression level of CHOP (Fig. 6B). As expected, these data suggested that CypB regulates adipogenesis by regulating CHOP.
Figure 6.
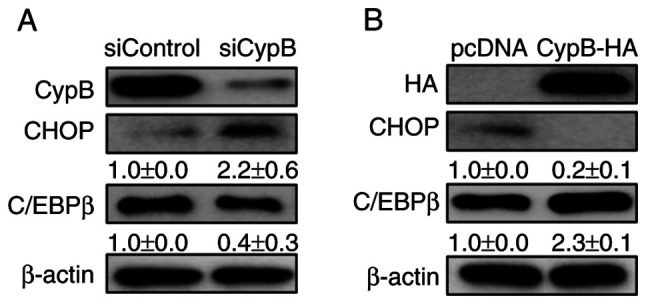
CypB modulates the expression of CHOP, a negative adipocyte regulator. (A) Two days after the induction of adipogenesis, the expression levels of CHOP and C/EBPβ were monitored in CypB knockdown by western blotting. (B) After overexpression of CypB, the expression level of CHOP and C/EBPβ was monitored by western blotting. β-actin was used as a loading control. The values were shown as the mean ± standard deviation of three independent experiments. CypB, cyclophilin B; CHOP, C/EBP homologous protein; si, short interfering.
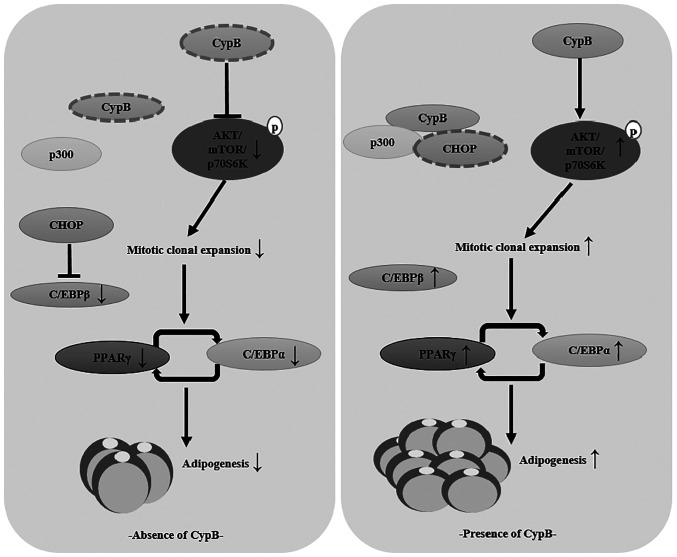
Schematic model of the role played by CypB in 3T3-L1 adipocyte differentiation
The present study showed that CypB regulated adipocyte differentiation through modulation of mitotic clonal expansion and AKT/mTOR signaling pathway (Fig. 7).
Figure 7.
Schematic diagram of the mechanism by which CypB regulates adipogenesis. CypB, cyclophilin B; CHOP, C/EBP homologous protein; AKT, protein kinase B; mTOR, mammalian target of rapamycin; PPARγ, peroxisome proliferator-activated receptor γ.
Discussion
The present study described that CypB regulated adipogenesis in 3T3-L1 cells. At present, CypB is known to be related to cell collagen formation and the growth of various cancer cells (42,43). However, it has been reported that CypA, which has similar properties to CypB, serves an important role in adipocyte differentiation (24). Also, it has been reported that the expression level of CypB was only enhanced in the liver, visceral and subcutaneous adipose tissue rather than in the lung and kidney of ob/ob mice. Moreover, serum CypB expression levels were more associated with hypertriglyceridemia and decreased HDL cholesterol levels than with hypertension and DM or hyperglycemia, among all components of metabolic syndrome, further suggesting the relationship between CypB and lipid metabolism (22). Although cyclophilin family proteins have been speculated to be involved in adipogenesis and lipogenesis, the present study is the first one to show that CypB plays important role in adipocyte differentiation, to the best of the authors' knowledge.
CHOP is generally expressed when ER stress occurs in cells and is known to induce cell death through apoptosis (44,45). It has also been reported as a negative regulator of adipogenic factors such as C/EBPβ and C/EBPα (25,27,45). Previously, we have shown that CypB cooperates with p300 to degrade CHOP (40). Consistently, the data showed that the expression of CHOP was also suppressed by CypB overexpression or increased by CypB knockdown in adipocytes (Fig. 6). It is suggestive that the molecular mechanism of CypB function in CHOP degradation is good evidence for the role of CypB in adipogenesis.
Several signaling pathways are involved in adipogenesis (46-49). AKT/mTOR serves an important role in cell proliferation and adipogenesis. The present study showed that CypB regulated adipocyte differentiation via the AKT/mTOR/p70S6K pathway (Fig. 4). In particular, mTOR is known to regulate cell cycle progression by P70S6K. STAT3 knockdown and kaempferol treatment suppress adipogenesis by inhibiting or delaying cell cycle progression during adipocyte differentiation (50,51). In the present study, CypB knockdown inhibited the cell cycle transition from G1 to G2/M phase, thereby suppressing adipogenesis. A previous report demonstrated that p27 protein levels decrease during phase transitions of 3T3-L1 clonal expansion by the 26S proteasome (52). Consistently, the present study showed that the expression level of p27 was reduced by CypB overexpression (Fig. 5D). Increased p27 in CypB knockdown appeared to inhibit the CDK4/Cyclin D, CDK2/Cyclin E and CDK2/Cyclin A complexes, thereby leading to reduced levels of phospho-pRB (Fig. 5B).
The present study discovered a novel function of CypB in adipocytes and clearly showed that the reduction of CypB inhibited adipogenesis. CsA, a well-known inhibitor of CypB, has been shown to inhibit adipogenic differentiation through the reduced expression of phosphorylation levels of PPARγ and C/EBPα, which is consistent with the present study (53). Our previous study reported that honokiol, a natural compound, inhibited the migration of human hepatocellular carcinoma by decreasing CypB expression in hepatocarcinoma cells (54). Consistently, in a recent study, it was reported that HNK inhibits adipogenesis and promotes the browning of white adipose tissues (55). However, further studies are needed to determine whether adipogenesis is indeed suppressed by HNK-induced CypB reduction.
Accumulated reports have demonstrated that ER stress can induce dysregulation of metabolism in adipocytes. A total of two FDA-approved chaperones, 4-phenylbutyric acid (4-PBA) and taurine-conjugated ursodeoxycholic acid have been investigated in adipocytes and β-cells for their ability to reduce ER-stress induced dysfunction (56). The two chaperones have been reported to reduce activation of ER stress in obese mice (57). However, further studies are needed to elucidate the mechanism of intertwining ER stress and adipogenesis.
Taken together, it was proved that CypB plays an important role as a novel adipogenesis regulator in 3T3-L1 cells and this is expected to be helpful in obesity treatment studies.
Acknowledgments
Not applicable.
Abbreviations
- CypB
cyclophilin B
- C/EBP
CCAAT-enhancer binding protein
- CHOP
C/EBP homologous protein
- PPARγ
peroxisome proliferator-activated receptor γ
- p300
histone acetyltransferase p300
- FABP4
fatty acid binding protein 4
- mTOR
mammalian target of rapamycin
- p70S6K
p70S6 kinase
- pRB
retinoblastoma protein
- MCE
mitotic clonal expansion
- IBMX
3-Isobutyl-1-methylxanthine
- PPIase
prolyl isomerase
- ob/ob mice
obesity-induced mice
- ER
endoplasmic reticulum
- PI3K
phosphoinositide 3-kinases
- mTORc1
mammalian target of rapamycin complex 1
- CS
calf serum
- FBS
fetal bovine serum
- CypA
cyclophilin A
- CsA
cyclosporine
- AKT
protein kinase B, HA, hemagglutinin
Funding Statement
The present study was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education of the Korean government (grant no. NRF-2018R1A6A1A030525124).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JY and WC participated in study design, performed all experiments, collected data, and drafted the manuscript. IK, JH, SK, and WC analyzed and interpreted the data and revised the manuscript. JY and WC confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS, Marks JS. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA. 2003;289:76–79. doi: 10.1001/jama.289.1.76. [DOI] [PubMed] [Google Scholar]
- 2.Smith PD, O'Halloran P, Hahn DL, Grasmick M, Radant L. Screening for obesity: Clinical tools in evolution, a WREN study. WMJ. 2010;109:274–278. [PMC free article] [PubMed] [Google Scholar]
- 3.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/S0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 4.Tak YJ, Lee SY. Anti-Obesity Drugs: Long-Term efficacy and safety: An updated review. World J Mens Health. 2021;39:208–221. doi: 10.5534/wjmh.200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman E, Golden J, Roberts M, Egan A, Weaver J, Rosebraugh C. The FDA's assessment of two drugs for chronic weight management. N Engl J Med. 2012;367:1577–1579. doi: 10.1056/NEJMp1211277. [DOI] [PubMed] [Google Scholar]
- 6.Schauer PR, Bhatt DL, Kirwan JP, Wolski K, Aminian A, Brethauer SA, Navaneethan SD, Singh RP, Pothier CE, Nissen SE, et al. Bariatric surgery versus intensive medical therapy for diabetes-5-year outcomes. N Engl J Med. 2017;376:641–651. doi: 10.1056/NEJMoa1600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Every-Palmer S, Romans SE, Stubbs R, Tomlinson A, Gandhi S, Huthwaite M. Experiences of weight-loss surgery in people with serious mental Illness: A qualitative study. Front Psychiatry. 2020;11:419. doi: 10.3389/fpsyt.2020.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: Tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Chang E, Kim CY. Natural products and obesity: A focus on the regulation of mitotic clonal expansion during adipogenesis. Molecules. 2019;24:1157. doi: 10.3390/molecules24061157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitterberger MC, Zwerschke W. Mechanisms of resveratrol-induced inhibition of clonal expansion and terminal adipogenic differentiation in 3T3-L1 preadipocytes. J Gerontol A Biol Sci Med Sci. 2013;68:1356–1376. doi: 10.1093/gerona/glt019. [DOI] [PubMed] [Google Scholar]
- 11.Tang QQ, Otto TC, Lane MD. Mitotic clonal expansion: A synchronous process required for adipogenesis. Proc Natl Acad Sci USA. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang MK, Yun YR, Kim JH, Park MH, Jung MH. Gomisin N inhibits adipogenesis and prevents high-fat diet-induced obesity. Sci Rep. 2017;7:40345. doi: 10.1038/srep40345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson BS, Nam H, Morrison RF. Curcumin Inhibits 3T3-L1 preadipocyte proliferation by mechanisms involving post-transcriptional p27 regulation. Biochem Biophys Rep. 2016;5:16–21. doi: 10.1016/j.bbrep.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maki C, Funakoshi-Tago M, Aoyagi R, Ueda F, Kimura M, Kobata K, Tago K, Tamura H. Coffee extract inhibits adipogenesis in 3T3-L1 preadipocyes by interrupting insulin signaling through the downregulation of IRS1. PLoS One. 2017;12:e0173264. doi: 10.1371/journal.pone.0173264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang HJ, Seo HA, Go Y, Oh CJ, Jeoung NH, Park KG, Lee IK. Dimethylfumarate suppresses adipogenic differentiation in 3T3-L1 preadipocytes through inhibition of STAT3 activity. PLoS One. 2013;8:e61411. doi: 10.1371/journal.pone.0061411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/S1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee JE, Schmidt H, Lai B, Ge K. Transcriptional and epigenomic regulation of adipogenesis. Mol Cell Biol. 2019;39:e00601–18. doi: 10.1128/MCB.00601-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 19.Muller C, Calkhoven CF, Sha X, Leutz A. The CCAAT enhancer-binding protein alpha (C/EBPalpha) requires a SWI/SNF complex for proliferation arrest. J Biol Chem. 2004;279:7353–7358. doi: 10.1074/jbc.M312709200. [DOI] [PubMed] [Google Scholar]
- 20.Morrison RF, Farmer SR. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem. 1999;274:17088–17097. doi: 10.1074/jbc.274.24.17088. [DOI] [PubMed] [Google Scholar]
- 21.Price ER, Zydowsky LD, Jin MJ, Baker CH, McKeon FD, Walsh CT. Human cyclophilin B: A second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc Natl Acad Sci USA. 1991;88:1903–1907. doi: 10.1073/pnas.88.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Fan Q, Xie H, Lu L, Tao R, Wang F, Xi R, Hu J, Chen Q, Shen W, et al. Elevated serum cyclophilin B levels are associated with the prevalence and severity of metabolic syndrome. Front Endocrinol (Lausanne) 2017;8:360. doi: 10.3389/fendo.2017.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo J, Serrano SS, Gronberg A, Massoumi R, Hansson MJ, Gallay P. Cyclophilin inhibitor NV556 reduces fibrosis and hepatocellular carcinoma development in mice with Non-alcoholic steatohepatitis. Front Pharmacol. 2019;10:1129. doi: 10.3389/fphar.2019.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Li Z, Zhang B, He H, Bai Y. PPIA is a novel adipogenic factor implicated in obesity. Obesity (Silver Spring) 2015;23:2093–2100. doi: 10.1002/oby.21208. [DOI] [PubMed] [Google Scholar]
- 25.Tang QQ, Lane MD. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc Natl Acad Sci USA. 2000;97:12446–12450. doi: 10.1073/pnas.220425597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batchvarova N, Wang XZ, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153) EMBO J. 1995;14:4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X, Ordemann J, Muller JM, Dubiel W. The COP9 signalosome, cullin 3 and Keap1 supercomplex regulates CHOP stability and adipogenesis. Biol Open. 2012;1:705–710. doi: 10.1242/bio.20121875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J, Murthy R, Wood B, Song B, Wang S, Sun B, Malhi H, Kaufman RJ. ER stress signalling through eIF2α and CHOP, but not IRE1α, attenuates adipogenesis in mice. Diabetologia. 2013;56:911–924. doi: 10.1007/s00125-012-2809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Bracho-Valdes I, Moreno-Alvarez P, Valencia-Martinez I, Robles-Molina E, Chavez-Vargas L, Vazquez-Prado J. mTORC1- and mTORC2-interacting proteins keep their multifunctional partners focused. IUBMB Life. 2011;63:896–914. doi: 10.1002/iub.558. [DOI] [PubMed] [Google Scholar]
- 31.Nissley SP, Haskell JF, Sasaki N, De Vroede MA, Rechler MM. Insulin-like growth factor receptors. J Cell Sci Suppl. 1985;3:39–51. doi: 10.1242/jcs.1985.Supplement_3.5. [DOI] [PubMed] [Google Scholar]
- 32.Wong RH, Sul HS. Insulin signaling in fatty acid and fat synthesis: A transcriptional perspective. Curr Opin Pharmacol. 2010;10:684–691. doi: 10.1016/j.coph.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Crawford R, Chen C, Xiao Y. The key regulatory roles of the PI3K/Akt signaling pathway in the functionalities of mesenchymal stem cells and applications in tissue regeneration. Tissue Eng Part B Rev. 2013;19:516–528. doi: 10.1089/ten.teb.2012.0672. [DOI] [PubMed] [Google Scholar]
- 34.Shockley KR, Rosen CJ, Churchill GA, Lecka-Czernik B. PPARgamma2 regulates a molecular signature of marrow mesenchymal stem cells. PPAR Res. 2007;2007:81219. doi: 10.1155/2007/81219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieberthal W, Levine JS. The role of the mammalian target of rapamycin (mTOR) in renal disease. J Am Soc Nephrol. 2009;20:2493–2502. doi: 10.1681/ASN.2008111186. [DOI] [PubMed] [Google Scholar]
- 36.Showkat M, Beigh MA, Andrabi KI. mTOR signaling in protein translation regulation: Implications in cancer genesis and therapeutic interventions. Mol Biol Int. 2014;2014:686984. doi: 10.1155/2014/686984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong K, Kim H, Kim K, Kim SJ, Hahn BS, Jahng GH, Yoon KS, Kim SS, Ha J, Kang I, Choe W. Cyclophilin B is involved in p300-mediated degradation of CHOP in tumor cell adaptation to hypoxia. Cell Death Differ. 2014;21:438–450. doi: 10.1038/cdd.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erickson RL, Hemati N, Ross SE, MacDougald OA. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem. 2001;276:16348–16355. doi: 10.1074/jbc.M100128200. [DOI] [PubMed] [Google Scholar]
- 42.Fang F, Zheng J, Galbaugh TL, Fiorillo AA, Hjort EE, Zeng X, Clevenger CV. Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells. J Mol Endocrinol. 2010;44:319–329. doi: 10.1677/JME-09-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR, III, Hefferan TE, Yaszemski MJ, Bram RJ. Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet. 2009;5:e1000750. doi: 10.1371/journal.pgen.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu H, Tian M, Ding C, Yu S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front Immunol. 2019;9:3083. doi: 10.3389/fimmu.2018.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei Y, Wang S, Ren B, Wang J, Chen J, Lu J, Zhan S, Fu Y, Huang L, Tan J. CHOP favors endoplasmic reticulum stress-induced apoptosis in hepatocellular carcinoma cells via inhibition of autophagy. PLoS One. 2017;12:e0183680. doi: 10.1371/journal.pone.0183680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Chaar D, Gagnon A, Sorisky A. Inhibition of insulin signaling and adipogenesis by rapamycin: Effect on phosphorylation of p70 S6 kinase vs eIF4E-BP1. Int J Obes Relat Metab Disord. 2004;28:191–198. doi: 10.1038/sj.ijo.0802554. [DOI] [PubMed] [Google Scholar]
- 47.Ahmad B, Serpell CJ, Fong IL, Wong EH. Molecular mechanisms of adipogenesis: The Anti-adipogenic role of AMP-Activated protein kinase. Front Mol Biosci. 2020;7:76. doi: 10.3389/fmolb.2020.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang MJ, Kim KK, Son BY, Nam SW, Shin PG, Kim GD. The anti-adipogenic activity of a new cultivar, pleurotus eryngii var. ferulae 'Beesan No. 2', through Down-Regulation of PPAR ү and C/EBP α in 3T3-L1 Cells. J Microbiol Biotechnol. 2016;26:1836–1844. doi: 10.4014/jmb.1606.06049. [DOI] [PubMed] [Google Scholar]
- 49.Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol Metab. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YJ, Choi HS, Seo MJ, Jeon HJ, Kim KJ, Lee BY. Kaempferol suppresses lipid accumulation by inhibiting early adipogenesis in 3T3-L1 cells and zebrafish. Food Funct. 2015;6:2824–2833. doi: 10.1039/C5FO00481K. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Y, Xi Y, Chen J, Zhu P, Kang J, Zou Z, Wang F, Bu S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem Biophys Res Commun. 2017;482:1360–1366. doi: 10.1016/j.bbrc.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 52.Auld CA, Fernandes KM, Morrison RF. Skp2-mediated p27(Kip1) degradation during S/G2 phase progression of adipocyte hyperplasia. J Cell Physiol. 2007;211:101–111. doi: 10.1002/jcp.20915. [DOI] [PubMed] [Google Scholar]
- 53.Qu Y, Lin Q, Yuan Y, Sun Z, Li P, Wang F, Jiang H, Chen T. Cyclosporin A inhibits adipogenic differentiation and regulates immunomodulatory functions of murine mesenchymal stem cells. Biochem Biophys Res Commun. 2018;498:516–522. doi: 10.1016/j.bbrc.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 54.Lee YS, Jeong S, Kim KY, Yoon JS, Kim S, Yoon KS, Ha J, Kang I, Choe W. Honokiol inhibits hepatoma carcinoma cell migration through downregulated Cyclophilin B expression. Biochem Biophys Res Commun. 2021;552:44–51. doi: 10.1016/j.bbrc.2021.03.011. [DOI] [PubMed] [Google Scholar]
- 55.Ding Y, Zhang L, Yao X, Zhang H, He X, Fan Z, Song Z. Honokiol alleviates high-fat diet-induced obesity of mice by inhibiting adipogenesis and promoting white adipose tissue browning. Animals (Basel) 2021;11:1493. doi: 10.3390/ani11061493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, Ogawa S, Hori M, Yamasaki Y, Matsuhisa M. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 57.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Görgün CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.