Abstract
CD154 is necessary for mice to clear a Cryptosporidium parvum infection, but whether this ligand has to be expressed on T cells with specificity for C. parvum has not been determined. We infected DO11.10 (ovalbumin specific) T-cell receptor transgenic mice that had been bred to a RAG−/− background with C. parvum and found that the infection was cleared within 6 weeks, while RAG−/− controls were unable to clear C. parvum infection. Recovery was accompanied by an increase in the number of splenic T cells with the CD44high phenotype that characterizes memory cells. To determine whether a C. parvum-infected environment sufficed to activate transgenic T cells, we reconstituted C. parvum-infected BALB/c SCID mice with DO11.10 RAG−/− splenocytes. Fecal excretion of C. parvum antigen ceased in the 12 weeks following the adoptive transfer, unless the mice were also injected with tolerizing doses of ovalbumin. DO11.10 T cells were found in the submucosa of C. parvum-infected, but not uninfected, BALB/c SCID hosts within 48 h of injection. The transferred DO11.10 T cells divided and acquired a CD44high memory phenotype in C. parvum-infected, but not uninfected, recipients. DO11.10 splenocytes from CD154 knockout donors failed to clear a C. parvum infection, confirming a requirement for CD154 in recovery. In vitro, the DO11.10 cells did not proliferate in response to C. parvum antigen, and a tBlast GenBank search revealed no matches between the ovalbumin peptide and C. parvum DNA sequences. C. parvum-infected SCID mice given RAG−/− CD8+ T cells with a Listeria-specific transgene did not recover from C. parvum infection. Our data suggest that antigen-nonspecific CD4+ T-cell effector mechanisms in combination with the innate arm of the immune system are sufficient for the eradication of C. parvum infection.
Cryptosporidium parvum causes severe diarrhea and weight loss in immunodeficient subjects, including those with AIDS (13) and boys with CD154 gene mutations causing X-linked immunodeficiency with hyper-immunoglobulin M (IgM) (17, 20), but healthy adults have little if any symptomatology (9). T-cell-deficient mice fed C. parvum oocysts are infected first in the gut and some 6 weeks later in the intra- and extrahepatic biliary tree (29). Previous studies showed that C. parvum-infected SCID mice would clear the C. parvum infection if they were reconstituted with CD4 T cells (22, 24) and that recovery was impaired by antibody to gamma interferon (IFN-γ) (7, 30), interleukin 4 (IL-4) (2), and IL-12 (31). Along with a requirement for major histocompatibility complex (MHC) class II expression (1), it seems likely that CD4 responses are central to recovery from C. parvum infection. Whether the CD4 response is contingent on stimulation of C. parvum-specific T cells by a C. parvum peptide on a class II molecule, or whether bystander T cells of other specificities might suffice to clear a C. parvum infection, is not known. Our own results indicate that a T-cell activation molecule, CD154, is necessary for C. parvum defense, because mice lacking either CD154 or its cell surface receptor, CD40, do not clear C. parvum infection (8). CD40 is a tumor necrosis factor-like receptor found primarily on B cells, macrophages, and dendritic cells. It transduces signals promoting upregulation of Bcl-2 (3) and isotype switching in B lymphocytes (19), increased expression of adhesion and accessory (B7-1 and B7-2) molecules on macrophages (23), and apoptosis of cells whose synthesis of antiapoptotic proteins such as Bcl-x is inhibited (33). CD154 is transiently expressed on activated T cells. When ligated, CD154 may mediate an afferent signal through a tyrosine kinase (6), resulting in increased IFN-γ production (23).
Whether CD154-CD40 interactions contribute to immunity to C. parvum by promoting T-cell stimulation (15), by an action on macrophages (26), through effector function on infected cells (27), or by any combination of these pathways is not currently established. Our own in vitro data show that C. parvum-infected biliary epithelial cells detach and undergo apoptosis after ligation by a CD154 fusion protein (28). These results raise the possibility that any source of CD154 could suffice to eradicate a C. parvum infection in T-cell-deficient mice. Preliminary studies in which C. parvum-infected SCID mice were treated with a CD154-CD8 fusion protein suggested that infection in the liver might be affected; however, these experiments were not pursued, because it was not possible to measure the in vivo distribution or persistence of the fusion protein. To test the hypothesis that T cells of an unrelated specificity might suffice to clear a C. parvum infection, we infected RAG−/− knockout mice expressing the DO11.10 T-cell receptor (TCR) for ovalbumin as a transgene. Since these mice recovered from C. parvum infection within 8 weeks, we looked for evidence of T-cell activation and a requirement for CD154 expression in a transfer model in which DO11.10 cells were injected into C. parvum-infected SCID mice. Here, we show that adoptive transfer of non-C. parvum-specific DO11.10 RAG−/− cells into C. parvum-infected SCID mice is sufficient to eradicate the infection.
MATERIALS AND METHODS
Mice.
BALB/c SCID mice were purchased from Jackson Laboratories, and BALB/c RAG−/− mice were supplied by R. Gill. The C57BL/6 CD154 knockout mice (14) were supplied by R. A. Flavell, and the C57BL/6 CD40 knockout mice were supplied by H. Kikutani (19). These and the C57BL/6 SCID mice were bred in our specific-pathogen-free mouse facility. The DO11.10 CD154−/− mice were supplied by A. Abbas. All were housed in microisolater cages and received sterile food, water, and bedding. Eight milliliters of trimethoprim-sulfa antibiotic solution was added to drinking water on alternate weeks for Pneumocystis carinii prophylaxis. DO11.10 RAG−/− (Tg) mice were derived by back-crossing B6 DO11.10 Tg animals with BALB/cByJ RAG−/− animals through 11 generations, selecting for DO11.10 Tg animals that were also RAG−/−. Their CD4+ cells have a TCR with specificity for an ovalbumin peptide in the context of H-2D (34), and the cells express CD154 when activated. The C10.4 animals were generously donated by U. D. Staerz and R. E. Berg. Their CD8 T cells express a transgene with specificity for a Listeria peptide (5, 25) in the context of H2-M3. The conditions for animal care and experimentation were approved by the Institutional Animal Care and Use Committee.
Infection and testing for C. parvum.
For infection with C. parvum, animals were transferred to a biohazard facility, where they remained until they were killed. C. parvum oocysts were obtained from McKesson Bioservices (catalog no. 1372) through the National Institute of Allergy and Infectious Diseases (NIAID) AIDS Research and Reference Reagent Program of the NIAID, National Institutes of Health. These oocysts were obtained from C. R. Sterling (4). They were washed in phosphate-buffered saline (PBS) to remove potassium dichromate buffer, then in sodium hypochlorite to reduce bacterial contaminants, and finally in PBS to remove bleach. Over 50% of these oocysts excyst after being washed in bleach and after incubation for 4 h at 37°C. Animals were infected by gavage once with 106 oocysts in 0.1 ml of Hanks balanced salt solution (HBSS). Feces were collected weekly and stored at −70°C until infection status was determined by fecal C. parvum antigen load by using a commercial C. parvum antigen enzyme-linked immunosorbent assay (ELISA) kit (Prospect-T; Alexon, Ramsay, Minn.). Feces were resuspended in the homogenization buffer supplied by the manufacturer overnight at 4°C before testing according to the manufacturer's instructions. Positive and negative controls were included with each ELISA run. C. parvum-infected SCID mice given ovalbumin received 0.5 mg in 100 μl of PBS injected into the peritoneum (i.p.) three times on alternate days starting on the day following their reconstitution with DO11.10 cells. This dose is known to affect the survival of these cells following adoptive transfer (21). Animals were euthanized by CO2 inhalation (i) when they lost 15% of body weight or (ii) at 12 to 13 weeks after adoptive transfer of T cells. Tissues obtained at necropsy for routine histology were fixed in 10% buffered formalin and processed for paraffin-embedded sectioning and hematoxylin and eosin (H&E) staining. Tissues for the identification of cells labeled with 5 and 6)-carboxyfluorescein succinimidyl ester [5(6)-FAM, SE] (CFSE) were frozen for sectioning.
Cell suspensions: preparation and transfers.
Spleens were removed from euthanized animals immediately, and cell suspensions were prepared with HBSS. Clumps were removed with nylon mesh, and the cell suspension was adjusted to 108/ml. One hundred microliters (107 cells) was injected where indicated into the peritoneal cavity. For tracking purposes, donor cells were fluorescence labeled before transfer. CFSE (catalog no. C-1311; Molecular Probes, Eugene, Oreg.), stored as a 10 mM stock solution in dimethyl sulfoxide, was diluted to 4 μM in 10 ml of saline at 37°C, and 4 × 107 lymphocytes were added in 10 ml of saline. After 10 min at 37°C, the cells were pelleted by centrifugation, resuspended in sterile HBSS, and injected i.p.
Dividing cells were identified by bromodeoxyuridine (BUdR) (Sigma, St. Louis, Mo. [catalog no. 9285]) incorporation. Mice were injected with 1 mg of BUdR i.p. 36 h after adoptive transfer of T cells and euthanized 12 h later. Gut tissue was fixed in 70% ethanol in water overnight and embedded in paraffin for sectioning. BUdR was localized by staining with antiserum followed by staining with a horseradish peroxidase- and diaminobenzidine-conjugated anti-goat antibody kit from Vector Labs (Burlingame, Calif. [catalog no. SK4100]).
Staining.
For fluorescence-activated cell sorter (FACS) analysis, 106 spleen cells were spun down and resuspended in the presence of fluorochrome-conjugated antibodies to CD4 or CD8 (Caltag, Burlingame, Calif.) or biotinylated CD44 antibodies (Pharmingen, San Diego, Calif.) and incubated on ice for 30 min. Biotinylated antibodies were followed by addition of phycoerythrin-avidin (Molecular Probes). The cells were washed twice and fixed in 1% paraformaldehyde before being viewed on an EPICS Elite cytofluorograph. CFSE-stained cells were identified in frozen sections by fluorescence microscopy with a Leitz microscope with incident UV light and transmitted phase-contrast optics. Images were captured with a Spot camera (model 1.3.0; Diagnostic Instruments, Sterling Heights, Mich.) and processed with Adobe PhotoShop software.
In vitro lymphocyte stimulation.
Spleen cells suspended in RPMI 1640 with 10% fetal calf serum were cultured at 106 cells/ml in 0.2-ml aliquots in triplicates either alone (as an unstimulated control), with 1 μg of concanavalin A (ConA) per ml, or with 105 C. parvum sporozoites excysted and filtered as described previously (12). Cultures were pulsed for 8 h with 1 μCi [3H]thymidine on day 3 (for ConA) or day 5 (for antigen) of stimulation.
RESULTS
Recovery of DO11.10 RAG−/− Tg mice from C. parvum infection.
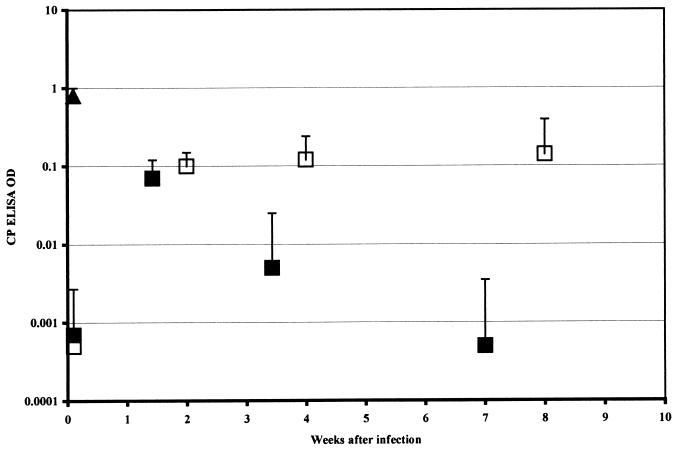
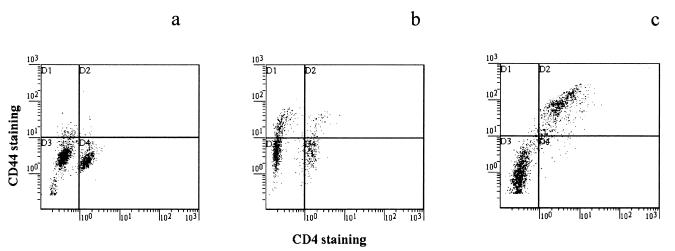
Five DO11.10 RAG−/− Tg mice were infected by gavage once with 106 C. parvum oocysts. Six BALB/c RAG−/− mice were similarly infected as controls. The increased stool C. parvum ELISA optical density (OD) of these animals 10 days later confirmed that infection had occurred (Fig. 1). The subsequent fall in stool C. parvum ELISA OD in the DO11.10 RAG−/− Tg animals indicates that they cleared the C. parvum infection. Eradication of infection was confirmed by histologic examination of the small and large intestine (not shown). Spleen cells were recovered at necropsy and stained for CD4 and CD44. The results (Fig. 2b) show an increase in CD44high cells in the C. parvum-infected mice.
FIG. 1.
ELISA ODs for fecal C. parvum (CP) antigen for DO11.10 RAG−/− Tg mice (■ [n = 5]) and RAG−/− controls (□ [n = 6]) following infection with 107 oocysts. ▴, positive ELISA control. Data points represent the mean + 1 standard deviation.
FIG. 2.
CD44 and CD4 staining of spleen cells from an uninfected DO11.10 RAG−/− Tg mouse (a), a DO11.10 RAG−/− Tg mouse 4 weeks after recovery from C. parvum infection (b), and a BALB/c SCID mouse recovered from C. parvum infection 12 weeks after reconstitution with DO11.10 RAG−/− cells (c).
Further analysis of the response of intact DO11.10 RAG−/− Tg mice to C. parvum infection was not attempted, because the overlap between the time courses of infection and recovery made them difficult to analyze as independent variables. Instead, we used an adoptive transfer model in which spleen cells from the DO11.10 RAG−/− Tg mice were transferred into SCID mice, whose infection by C. parvum could be established in advance.
Excretion of C. parvum antigen by SCID mice following reconstitution with T cells.
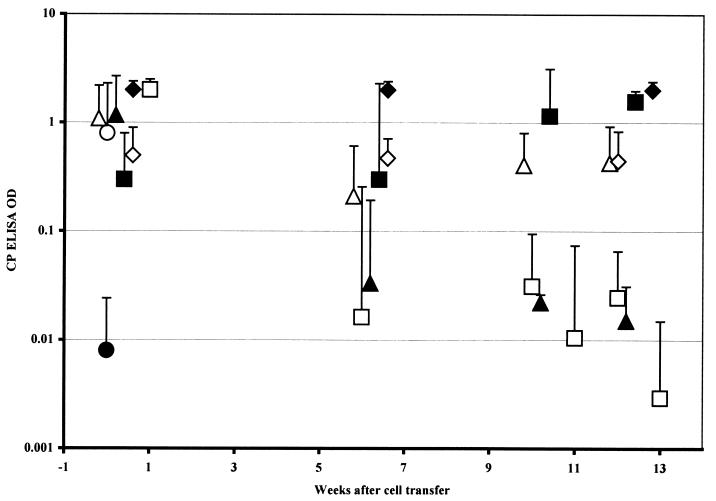
SCID mice became positive for C. parvum infection by stool ELISA between 7 and 14 days after gavage with 106 C. parvum oocysts. To determine the effect of adoptively transferred T cells on this infection, groups of six to eight mice were injected with 107 spleen cells from wild-type control or DO11.10 RAG−/− Tg donors. C. parvum-infected SCID mice reconstituted with the wild-type control, MHC-matched T cells and the DO11.10 RAG−/− cells became free of infection after 10 weeks (Fig. 3). The final OD readings for DO11.10 RAG−/− cell recipients at 12 weeks were <0.001, 0.021, <0.001, <0.001, 0.001, 0.003, and 0.001. C. parvum-infected SCID mice injected with ovalbumin for 1 week following transfer of the DO11.10 cells and the eight C. parvum-infected SCID mice given the C10.4 Tg cells were uniformly infected 6 weeks after cell transfer (mean ELISA OD, 1.5). Six of the C10.4 recipients died while infected, and two were still infected when killed 12 weeks after the adoptive transfer (final ODs of 2, 0.37, 0.13, 2, 0.7, 0.01, and 2). The difference between the ODs for the mice injected with DO11.10 and C10.4 cells is statistically significant, with P < 0.01 by two-tailed Wilcoxon test. Differences between the “recovered” DO11.10 RAG−/− and wild-type spleen cell recipients and all of the other control groups (whether injected with DO11.10 cells plus ovalbumin, DO11.10 CD154−/− cells, or ovalbumin alone [Fig. 3]) are significant at P < 0.05 by two-tailed Wilcoxon test at the 12-week point.
FIG. 3.
ELISA ODs for fecal C. parvum antigen in SCID mice in the weeks following transfer of 107 spleen cells from DO11.10 RAG−/− donors (□ [n = 8]), DO11.10 RAG−/− donors followed by 0.5-mg ovalbumin injections (▵ [n = 5]), C10.4 Tg donors (⧫ [n = 5]), CD154−/− DO11.10 donors (◊ [n = 4]), or C57BL/6 wild-type donors (▴ [n = 6]). ■, six controls injected with ovalbumin but no cells; ●, OD for six uninfected controls; ○, positive control supplied by the kit manufacturer. The ODs at ≥10 weeks for the animals receiving DO11.10 RAG−/− cells without ovalbumin and the wild-type controls differ (P < 0.05) from those for the other four C. parvum-infected groups. Data points represent the mean + 1 standard deviation.
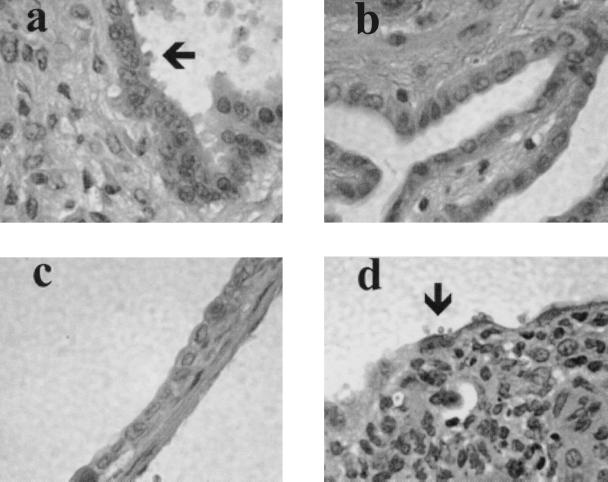
H&E-stained sections of gut and gall bladder were examined to confirm that C. parvum was eliminated from the bile ducts of the animals whose ELISAs became negative. Examples of the histology show organisms on the gall bladder epithelium of unreconstituted SCID mice (Fig. 4a). Mice given either control BALB/c spleen cells (Fig. 4b) or DO11.10 cells (Fig. 4c) are free of C. parvum. Gall bladder epithelium of mice given the C10.4 Tg cells remained infected (Fig. 4d), as did the DO11.10 reconstituted SCID animals that were injected with ovalbumin (data not shown).
FIG. 4.
Gall bladder epithelium of SCID mice 16 weeks after infection with C. parvum. (a) Without spleen cell reconstitution. (b) Twelve weeks after reconstitution with control BALB/c spleen cells. (c) Twelve weeks after reconstitution with DO11.10 Tg spleen cells (d) Twelve weeks after reconstitution with C10.4 Tg cells. C. parvum sporozoites in panels a and d are shown by arrows.
Thymidine uptake proliferation assays were used to determine whether C. parvum could stimulate DO11.10 RAG−/− spleen cells. The results show that Tg cells obtained from DO11.10 mice that had recovered from C. parvum infection did not proliferate in response to C. parvum sporozoites, whereas they did respond to ovalbumin and to ConA (Table 1). Spleen cells from a control mouse that had recovered from a C. parvum infection proliferated in response to C. parvum sporozoites. This suggests that there is no specific cross-priming of the ovalbumin-specific DO11.10 RAG−/− T cells by an antigen present in C. parvum sporozoites. A database search using advanced tBlast (ncbi.nlm.nih.gov) showed no similarities between the no. 324-339 ovalbumin peptide sequence and any apicomplexan nucleotide sequences.
TABLE 1.
Thymidine uptake by spleen cells from DO11.10 RAG−/− mice and SCID recipients of DO11.10 cells
| Spleen cells tested | Thymidine uptake (cpm) with antigen stimulusa:
|
|||
|---|---|---|---|---|
| None | Ovalbumin | C. parvum | ConA | |
| Tg DO11.10 | 625 ± 140 | 34,602 ± 6,670 | 584 ± 270 | 10,954 ± 3,290 |
| SCID | 305 ± 110 | 380 ± 170 | 288 ± 80 | 380 ± 170 |
| DO11.10 (reconstituted SCID) | 428 ± 166 | 6,600 ± 2,270 | 316 ± 70 | 4,100 ± 220 |
| Control B6 mouse recovered from C. parvum infection | 271 ± 90 | 1,660 ± 180 | 3,765 ± 900 | 79,175 ± 22,100 |
Results are the mean cpm ± 1 standard deviation of triplicate wells for groups of two to four mice cultured for 3 days with 1 μg of ConA per ml or for 5 days with 1 mg of ovalbumin per ml or 104 freshly excysted C. parvum sporozoites.
C. parvum-dependent activation of DO11.10 RAG−/− cells in adoptive hosts.
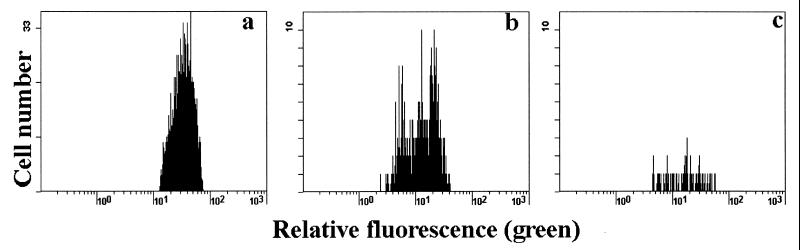
Spleens were recovered from mice at the time of necropsy. Detection of CD4+ T cells by FACS showed that the Tg cells persisted after adoptive transfer (Table 2). FACS analysis demonstrated that, ex vivo, DO11.10 Tg CD4+ spleen cells from uninfected animals do not stain for the memory cell marker CD44, also referred to as Pgp-1 (Fig. 2a). Spleen cells from DO11.10 mice that have recovered from a C. parvum infection have increased levels of CD44 on both CD4+ and CD4− cells (Fig. 2b). Substantially higher expression of CD44 was seen on DO11.10 CD4+ T cells from the spleens of reconstituted SCID animals that have recovered from a C. parvum infection (Fig. 2c), while DO11.10 cells injected into uninfected SCID mice retained a CD44low phenotype (not shown). Additional evidence for activation of DO11.10 cells in C. parvum-infected mice came from cell membrane fluorescence labeling. Few CD4+ cells labeled with CFSE were recovered from the spleen and lymph nodes obtained from uninfected recipients 6 days after transfer of labeled DO11.10 spleen cells (Fig. 5c), and their fluorescence profile does not separate into individual peaks. Four to eight times more lymphocytes were recovered 6 days after transfer from the spleen and lymph nodes of C. parvum-infected mice (Fig. 5b). The fluorescence profile of these CFSE-labeled cells shows several peaks that are of reduced intensity compared with those of the cells before transfer (Fig. 5a). This separation into peaks indicates that the CFSE-labeled spleen and lymph node cells had gone through one to two cycles of cell division.
TABLE 2.
Percentage of CD4+ cells in spleens of C. parvum-infected SCID mice injected with DO11.10 cellsa
| Group | Treatment | % of CD4+ cells in spleen |
|---|---|---|
| 1 | Ovalbumin injected alone | <1 |
| 2 | 107 DO11.10 cells | 35 ± 7 |
| 3 | 107 DO11.10 cells + ovalbumin | 21 ± 6 |
| 4 | 107 BALB/c cells | 38 ± 5 |
Groups of six BALB/c SCID mice were infected with C. parvum. Four weeks later, groups 2 to 4 were given 107 DO11.10 or control spleen cells by i.p. injection. The percentage of CD4+ cells in spleen ± 1 standard deviation was determined after necropsy by FACS analysis, 12 weeks after cell transfer. Groups 2 and 4 do not differ significantly, while group 3 is significantly lower than group 2 or 4 (P < 0.04).
FIG. 5.
CFSE fluorescence profiles for DO11.10 RAG−/− cells recovered from spleen and lymph node 6 days after transfer into BALB/c SCID mice. (a) Input cells (before transfer). (b) C. parvum-infected recipient. (c) Uninfected recipient. Results are representative for groups of six uninfected and six infected mice.
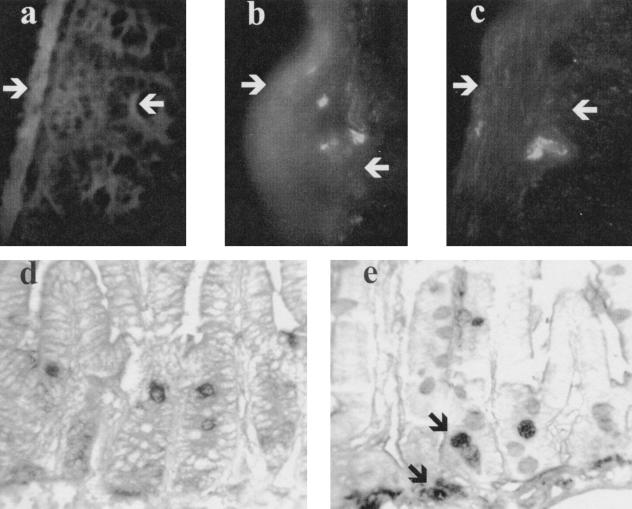
Proliferation studies using CFSE- and BUdR-labeled cells were employed to determine whether or not the adoptively transferred DO11.10 RAG−/− T cells were detectable in the gut. Thirty-six hours after CFSE-labeled DO11.10 RAG−/− splenocytes were injected into infected or control SCID recipients, the recipients were injected with BUdR. The mice were necropsied 12 h later. Frozen sections of small bowel from uninfected control recipients did not contain CFSE-labeled cells (Fig. 6a). However, CFSE-positive cells were found in the small intestinal submucosa of C. parvum-infected recipients (examples from two mice are shown in Fig. 6b and c). When paraffin-embedded sections from uninfected mice were stained with anti-BUdR antibody, only epithelial cells in the crypts became labeled (Fig. 6d). BUdR staining of gut from infected animals showed an additional population of labeled mononuclear cells beneath the crypts in the C. parvum-infected recipients (Fig. 6e). These labeled cells have the same location as the CSFE-labeled cells identified by fluorescence microscopy.
FIG. 6.
Sections of small bowel from SCID recipients 48 h after injection of CFSE-labeled spleen cells from DO11.10 RAG−/− Tg mice. The mucosa of uninfected (control) recipients does not contain CFSE-labeled cells (a), while portions of labeled cells are seen in the submucosa of C. parvum-infected recipients (b and c). White arrows mark the serosal surface and the base of the crypts. (d and e) Paraffin-embedded sections from uninfected (d) and infected (e) mice injected with BUdR 36 h after cell transfer and harvested 48 h after transfer. In panel d, only dividing epithelial cells in the crypts are labeled, while the BUdR staining of gut from infected animals shows an additional population of labeled mononuclear cells beneath the crypts (black arrows in panel e). These labeled cells have the same location as the CSFE-labeled cells identified by fluorescence microscopy.
DISCUSSION
SCID mice are unable to rearrange antigen receptors on B and T cells, so they are unable to make specific immune responses. They generally do not clear C. parvum infections (as with the six controls injected with ovalbumin only) unless they are reconstituted with normal T lymphocytes (22, 24). The general persistence of C. parvum infection in SCID mice implicates T cells in the elimination of C. parvum. Whether recovery requires the development of a population of C. parvum-specific effectors, or whether an antigen nonspecific effector function mediated by T cells is sufficient to clear the infection, has not previously been explored. Our observation that both boys with mutated CD154 and CD154-deficient mice had difficulty clearing C. parvum infections (8) prompted the speculation that cell surface expression of CD154 was required for an appropriate immune response to C. parvum infection. In vitro results, in which a soluble CD154 fusion protein reduced the infection of a cultured cell line by C. parvum (17), suggested that T cells might clear C. parvum infection by virtue of CD154 expression without C. parvum antigen-specific T cells. T cells expressing an ovalbumin TCR transgene provide a means to test this possibility, provided that the Tg cells come from an animal with a RAG−/− background so alternative TCR-α chain rearrangements are prevented.
The DO11.10 RAG−/− Tg mice we infected cleared C. parvum within 8 weeks. These results were confirmed in adoptive transfer experiments, in which DO11.10 RAG−/− Tg T cells cleared a C. parvum infection from SCID mice in the 10 weeks following their adoptive transfer. This result suggests that DO11.10 cells are as efficient in mediating a response to C. parvum as the heterogeneous populations of CD4 T cells that are present in a mouse spleen. Although the adoptively transferred spleen cells were heterogeneous, it is likely that it was the T-cell component that was critical for the recovery of SCID mice from C. parvum infection. This conclusion comes from our finding that those SCID mice injected with ovalbumin to impair the function of the DO11.10 cells after their adoptive transfer did not eliminate the C. parvum infection. While formal evidence for tolerization of the transferred DO11.10 cells was not sought, the ovalbumin injections significantly reduced the number of T cells found in the spleen at necropsy. Antigen (ovalbumin) injections would be expected to affect only the number and function of the ovalbumin-specific CD4+ T cells in the adoptive hosts. Current models would suggest that the ovalbumin injections may have stimulated activation-induced cell death (21). Clearance of C. parvum infections by unreconstituted SCID mice has been reported, with diet as one variable (15), so negative controls are important for the interpretation of our study. These negative controls included the adoptive transfer of DO11.10 CD154−/− spleen cells. This population of T cells was tested because a requirement for CD154 to clear C. parvum infections had previously been established (8). We also tested C10.4 CD8+ Tg mouse T cells (5, 25), because these, in common with CD8 cells in general, do not express CD154 following activation. The persistence of C. parvum infection in the recipients of these cells and in C. parvum-infected SCID mice that were not reconstituted makes it unlikely that the handling of the mice or dietary factors were responsible for clearance of the infection. Furthermore, the evidence for clearing of the infection by the DO11.10 RAG−/− cells seems secure in that the ELISA is sensitive and was corroborated by histologic examination of gut and bile ducts.
Survival of the DO11.10 RAG−/− cells in the SCID recipients was confirmed by phenotyping and by the identification of CFSE-labeled DO11.10 cells in the submucosa of the small intestine. DO11.10 cells retained their CD44low phenotype when injected into uninfected mice, whereas they acquired the CD44high phenotype of memory cells (10) in C. parvum-infected SCID recipients. Given the diversity of the T-cell repertoire, the chance that there might be some cross-reactivity between C. parvum antigens and the ovalbumin peptide recognized by DO11.10 cells seems low. A GenBank database search revealed no similarities between the ovalbumin peptide sequence and any apicomplexan nucleotide sequences. The absence of an in vitro proliferative response by the DO11.10 cells (recovered from SCID mice after their C. parvum infection had cleared) to C. parvum sporozoites (when the response to ovalbumin was positive) also argues against a fortuitous cross-reaction or a superantigen-like effect. Perhaps C. parvum infection sufficiently alters the environment in the intestinal wall or gut-associated lymphoid tissue to activate costimulatory pathways that would allow even the low affinity that the TCR has for self-MHC to trigger a response. Alternatively, Tg T cells might be activated through one of the antigen-independent pathways that have been identified in vitro. Stimulation through pairs of CD2 antibodies is a familiar example (11), but whether these in vitro phenomena have in vivo counterparts is not known. Whatever the mechanism for activation, the acquisition of the CD44high phenotype by DO11.10 cells in the spleens of DO11.10 mice that had recovered from C. parvum infection and in C. parvum-infected adoptive transfer recipients suggests that C. parvum influences the behavior of T cells of unrelated specificities in the intestine. Perhaps DO11.10 T cells can traffic into the intestinal submucosa of infected animals. The uptake of BUdR by mononuclear cells in the submucosa of C. parvum-infected, but not uninfected, recipients is compatible with activation of the transferred cells to a level that resulted in proliferation. The accompanying reduction in CFSE staining per cell and the increase in CD44 expression are both consistent with the interpretation that the DO11.10 cells completed one or two cycles of cell division in the 6 days following transfer into C. parvum-infected SCID recipients. The C. parvum infection is likely to have contributed to this division, because the few adoptively transferred cells that were recovered from uninfected recipients had not divided. The mechanism for C. parvum elimination from the C. parvum-infected SCID mice is unknown, but it was likely to have involved CD154 expression by the DO11.10 cells. In this context, our previous studies have shown that adoptively transferred T cells need to be able to express CD154 to eradicate a C. parvum infection from SCID recipients (8).
Specificity is one of the hallmarks of adaptive immunity, and in the context of the response to most pathogens, prior infection or immunization is necessary to stimulate the expansion of antigen-specific lymphocytes. Nevertheless, the susceptibility of human immunodeficiency virus-infected subjects to C. parvum appears to correlate with the total number of CD4+ cells in the blood. While our results do not negate the possibility that this correlation is primarily with the number of apicomplexan-specific T cells, we do show that, in mice, T cells of unrelated antigen specificity can adoptively transfer the capacity to eliminate this opportunistic infection.
ACKNOWLEDGMENTS
This work was supported in part by grants from the March of Dimes (6-0266) and the National Institutes of Health (AI 41075 and 40870).
We thank Mike Arrowood and Giovanni Widmer for helpful discussions and Leslie Bloomquist for histologic processing.
REFERENCES
- 1.Aguirre S A, Mason P H, Perryman L E. Susceptibility of major histocompatibility complex (MHC) class I- and MHC class II-deficient mice to Cryptosporidium parvum infection. Infect Immun. 1994;62:697–699. doi: 10.1128/iai.62.2.697-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguirre S A, Perryman L E, Davis W C, McGuire T C. IL-4 protects adult C57BL/6 mice from prolonged Cryptosporidium parvum infection: analysis of CD4+alpha beta+IFN− gamma+ and CD4+alpha beta+IL-4+ lymphocytes in gut-associated lymphoid tissue during resolution of infection. J Immunol. 1998;161:1891. [PubMed] [Google Scholar]
- 3.Akifusa S, Ohguchi M, Koseki T, Nara K, Semba I, Yamato K, Okahashi N, Merino R, Nunez G, Hanada N, Takehara T, Nishihara T. Increase in Bcl-2 level promoted by CD40 ligation correlates with inhibition of B cell apoptosis induced by vacuolar type H(+)-ATPase inhibitor. Exp Cell Res. 1998;238:82–89. doi: 10.1006/excr.1997.3848. [DOI] [PubMed] [Google Scholar]
- 4.Arrowood M J, Sterling C R. Isolation of cryptosporidium oocysts sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol. 1987;73:314–319. [PubMed] [Google Scholar]
- 5.Berg R E, Princiotta M F, Irion S, Moticka J A, Dahl K R, Staerz U D. Positive selection of an H2M3 restricted T cell receptor. Immunity. 1999;11:33–43. doi: 10.1016/s1074-7613(00)80079-5. [DOI] [PubMed] [Google Scholar]
- 6.Brenner B, Koppenhoeffer U, Lepple-Wienhaues A, Grassme H, Muller C, Speer C-P, Lang F, Gulbins E. The CD40 ligand directly activates T-lymphocytes via tyrosine phosphorylation dependent PKC activation. Biochem Biophys Res Commun. 1997;239:11–17. doi: 10.1006/bbrc.1997.7415. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Harp J A, Harmsen A G, Havell E A. Gamma interferon functions in resistance to Cryptosporidium parvum infection in severe combined immunodeficient mice. Infect Immun. 1993;61:3548–3551. doi: 10.1128/iai.61.8.3548-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosyns M, Tsirkin S, Jones M, Flavell R, Kikutani H, Hayward A R. Requirement for CD40-CD40 ligand interaction for elimination of Cryptosporidium parvum from mice. Infect Immun. 1998;66:603–607. doi: 10.1128/iai.66.2.603-607.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuPont H L, Chappell C L, Sterling C R, Okhuysen P C, Rose J B, Jakubowski W. The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med. 1995;332:855–859. doi: 10.1056/NEJM199503303321304. [DOI] [PubMed] [Google Scholar]
- 10.Dutton R W, Bradley L M, Swain S L. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 11.Fox D A, Hussey R E, Fitzgerald K A, Bensussan A, Daley J F, Schlossman S F, Reinherz E L. Activation of human thymocytes via the 50 kD T11 sheep erythrocyte binding protein induces the expression of interleukin 2 receptors on both T3+ and T3− populations. J Immunol. 1985;144:2851–2859. [PubMed] [Google Scholar]
- 12.Gomez Morales M A, Ausiello C M, Urbani F, Pozio E. Crude extract and recombinant protein of Cryptosporidium parvum oocysts induce proliferation of human peripheral blood mononuclear cells in vitro. J Infect Dis. 1995;172:211–216. doi: 10.1093/infdis/172.1.211. [DOI] [PubMed] [Google Scholar]
- 13.Goodgame R W, Kimball K, Ou C N, White A C, Genta R M, Lifschitz C H, Chappell C L. Intestinal function and injury in acquired immunodeficiency syndrome-related cryptosporidiosis. Gastroenterology. 1995;108:1075–1082. doi: 10.1016/0016-5085(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 14.Grewal I S, Xu J, Flavell R A. Impairment of antigen-specific T-cell priming in mice lacking CD40 ligand. Nature. 1995;378:617–620. doi: 10.1038/378617a0. [DOI] [PubMed] [Google Scholar]
- 15.Grewal I S, Borrow P, Pamer E G, Oldstone M B A, Flavell R A. The CD40-CD154 system in anti-infective host defense. Curr Opin Immunol. 1997;9:491–497. doi: 10.1016/s0952-7915(97)80100-8. [DOI] [PubMed] [Google Scholar]
- 16.Harp J A, Chen W, Harmsen A G. Resistance of severe combined immunodeficient mice to infection with Cryptosporidium parvum: the importance of intestinal microflora. Infect Immun. 1992;60:3509–3512. doi: 10.1128/iai.60.9.3509-3512.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward A R, Levy L, Facchetti F, Notarangelo L, Ochs H D, Etzioni A, Weinberg A. Cholangiopathy and tumors of the pancreas, liver and biliary tree in boys with X-linked immunodeficiency with hyper-IgM (XHIM) J Immunol. 1995;158:977–983. [PubMed] [Google Scholar]
- 18.Karmann K, Hughes C C, Schechner J, Fanslow W C, Pober J S. CD40 on human endothelial cells: inducibility by cytokines and functional regulation of adhesion molecule expression. Proc Natl Acad Sci USA. 1995;92:4342–4346. doi: 10.1073/pnas.92.10.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 20.Kroczek R A, Graf D, Brugnoni D, Giliani S, Korthauer U, Ugazio A, Senger G, Mages H W, Villa A, Notarangelo L D. Defective expression of CD40 ligand on T cells causes “X linked immunodeficiency with hyper IgM (XHIM).”. Immunol Rev. 1994;138:39–59. doi: 10.1111/j.1600-065x.1994.tb00846.x. [DOI] [PubMed] [Google Scholar]
- 21.London C A, Perez V L, Abbas A. Functional characteristics and survival requirements of memory CD4+ T lymphocytes in vivo. J Immunol. 1998;162:766. [PubMed] [Google Scholar]
- 22.McDonald V, Bancroft G J. Mechanisms of innate and acquired resistance to Cryptosporidium parvum infection in SCID mice. Parasite Immunol. 1994;16:315–320. doi: 10.1111/j.1365-3024.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 23.McDyer J F, Goletz T J, Thomas E, June C H, Seder R A. CD40 ligand/CD40 stimulation regulates the production of IFN-gamma from human peripheral blood mononuclear cells in an IL-12- and/or CD28-dependent manner. J Immunol. 1998;160:1701–1707. [PubMed] [Google Scholar]
- 24.Perryman L E, Mason P H, Chrisp C E. Effect of spleen cell populations on resolution of Cryptosporidium parvum infection in SCID mice. Infect Immun. 1994;62:1474–1477. doi: 10.1128/iai.62.4.1474-1477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Princiotta M F, Lenz L L, Bevan M J, Staerz U D. H2-M3 restricted presentation of a Listeria-derived leader peptide. J Exp Med. 1998;187:1711–1719. doi: 10.1084/jem.187.10.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridge J P, DiRosa F, Matzinger P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature. 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 27.Ruby J, Bluethman H, Auguet M, Ramshaw I A. CD40 ligand has potent antiviral activity. Nat Med. 1995;1:437. doi: 10.1038/nm0595-437. [DOI] [PubMed] [Google Scholar]
- 28.Stephens J, Cosyns M, Jones M, Hayward A. Liver and bile duct pathology following Cryptosporidium parvum infection of immunodeficient mice. Hepatology. 1999;30:27–35. doi: 10.1002/hep.510300138. [DOI] [PubMed] [Google Scholar]
- 29.Tzipori S, Rand W, Theodos C. Evaluation of a two-phase scid mouse model preconditioned with anti-interferon-gamma monoclonal antibody for drug testing against Cryptosporidium parvum. J Infect Dis. 1995;172:1160–1164. doi: 10.1093/infdis/172.4.1160. [DOI] [PubMed] [Google Scholar]
- 30.Ungar B L, Kao T C, Burris J A, Finkelman F D. Cryptosporidium infection in an adult mouse model. Independent roles for IFN-γ and CD4+ T lymphocytes in protective immunity. J Immunol. 1991;147:1014–1022. [PubMed] [Google Scholar]
- 31.Urban J F, Fayer R, Chen S J, Gause W C, Gately M K, Finkelman F D. IL-12 protects immunocompetent and immunodeficient neonatal mice against infection with Cryptosporidium parvum. J Immunol. 1996;156:263–268. [PubMed] [Google Scholar]
- 32.Van Gool S W, Vandenberghe P, de Boer M, Ceuppens J L. CD80, CD86 and CD40 provide accessory signals in a multiple-step T-cell activation model. Immunol Rev. 1996;153:47–83. doi: 10.1111/j.1600-065x.1996.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Karras J G, Howard R G, Rothstein T L. Induction of bcl-x by CD40 engagement rescues sIg-induced apoptosis in murine B cells. J Immunol. 1995;155:3722. [PubMed] [Google Scholar]
- 34.White J, Haskins K M, Marrack P, Kappler J. Use of I-region restricted, antigen-specific, T cell hybridomas to produce idiotypically specific anti-receptor antibodies. J Immunol. 1983;130:1033–1043. [PubMed] [Google Scholar]