Graphical abstract
Abbreviations: GDT, global distance test; p-lDDT, predicted local distance difference test; SBDD, Structure-based drug design; TBM, template-based modeling or template-based model; RMSD, root-mean-squared deviation; AF, AlphaFold; DL, deep learning; GALD, Rosetta GA LigandDock; GD3, GalaxyDock3; MD, molecular dynamics; CAPRI, critical assessment of predicted interactions, DOF, Degree-of-freedom
Keywords: GPCR, Deep learning, Ligand docking, Protein structure prediction, Drug discovery
Abstract
While deep learning (DL) has brought a revolution in the protein structure prediction field, still an important question remains how the revolution can be transferred to advances in structure-based drug discovery. Because the lessons from the recent GPCR dock challenge were inconclusive primarily due to the size of the dataset, in this work we further elaborated on 70 diverse GPCR complexes bound to either small molecules or peptides to investigate the best-practice modeling and docking strategies for GPCR drug discovery. From our quantitative analysis, it is shown that substantial improvements in docking and virtual screening have been possible by the advance in DL-based protein structure predictions with respect to the expected results from the combination of best pre-DL tools. The success rate of docking on DL-based model structures approaches that of cross-docking on experimental structures, showing over 30% improvement from the best pre-DL protocols. This amount of performance could be achieved only when two modeling points were considered properly: 1) correct functional-state modeling of receptors and 2) receptor-flexible docking. Best-practice modeling strategies and the model confidence estimation metric suggested in this work may serve as a guideline for future computer-aided GPCR drug discovery scenarios.
1. Introduction
Deep learning (DL) has revolutionized protein structure prediction [1], [2] and started to have a large impact on structure-based drug design (SBDD) [3], [4]. Before the emergence of such methods, predicted structures played minor roles in SBDD. Prior to the breakthrough, template-based models (TBM) were regarded as the best protein structure models [5]. However, TBMs have sufficient accuracy for SBDD only at limited conditions when very close (e.g. sequence identity > 50 %) homolog structures are available. Unlike TBMs, the model accuracy of AlphaFold [1] reported in CASP14 (2020) was comparable to such close-homolog TBMs regardless of the existence of such homologous structures when enough related sequences were available. The paradigm has been shifted since then, and now a majority of proteins having reasonable homologous “sequences” (not necessarily structures) have their near-experiment accuracy models by DL and hence could serve as reasonable targets for SBDD, covering a far larger portion in the protein space than before. The key question has also shifted from “whether the protein of interest (or its homolog) has experimental structure” to “how to model protein–ligand interactions” using DL-based tools, and “what’s the best status we can reach now compared to the previous best modeling scenario and whether it is a meaningful progress”. Whether those DL tools would be advantageous to drug discovery-related problems is a crucial question [6], [7], but not many works have been published to date providing realistic guidelines or quantitative analyses.
In this work, we aim to find the best contemporary strategy for the structure-based and computer-aided G-protein-coupled receptor (GPCR) drug discovery. GPCRs are a group of related proteins sharing the same 7-transmembrane helical topology, therefore the shared activation mechanism for many members with some exceptions, being responsible for sensing various external signals (lights, chemicals, pressures, etc.) [8]. Because of their huge importance as drug targets, continuous efforts have been made by researchers to apply computer-aided drug discovery pipelines [7], [9], [10], [11]. Especially, the recent GPCRdock 2021 has shown an important step toward understanding the aforementioned key questions regarding the recent advance in DL structure prediction. The main lesson from the challenge was that, unlike previous rounds of the challenge held in the early 2010 s, now receptors and receptor-peptide complexes could be modeled with reasonable accuracy by many participants, majorly powered by the breakthrough in DL-based structure prediction. However, the lessons learned from the challenge were quite limited primarily due to the size of the dataset tested in the challenge, in which only two small-molecule binders and three peptide binders were included. To derive lessons that are as general as possible, we not only expand the dataset size but also test a variety of receptor families and ligand types including both small molecules and peptides. We first carefully collected 70 unique GPCR complexes with either small molecules or peptides. Then 5 receptor modeling strategies and a total of 8 docking strategies (4 for small molecules and 4 for peptides) are benchmarked to evaluate the best-practice receptor modeling and docking protocols.
2. Overview of the benchmark dataset
We tried to consider a large number of receptor types when curating the benchmark dataset. The dataset employed in this work covers 33 unique families in human GPCRs spanning classes A, B1, C, and F. We selected complex structures that were experimentally resolved in the bound form with one of 51 small molecules or 19 peptides, comprising a total of 38 active-state and 32 inactive-state complexes (Fig. 1). To fairly benchmark the effect of considering the activation state excluding the effect by the information underlined in the receptor types or sequences, we included the receptors that have PDB entries for both active and inactive states as many as possible. Resolutions of selected complex structures were lower than 3.5 and 3.0 Å for active and inactive complexes, respectively. A full list of complex PDB entries, their receptor names, and ligands is listed in Supplementary Table S1.
Fig. 1.
Dataset used in the study. A) The GPCR phylogenetic tree and 33 selected families, highlighted by blue texts. The figure is modified from the original figure in GPCRdb (downloaded from https://gpcrdb.org/structure/statistics) [12]. B) Number of experimental methods and active versus inactive complexes in the dataset.
3. Modeling in functional state-specific context results in the most accurate binding site structure prediction
We sought to benchmark several receptor modeling strategies to address two questions: what is the best DL-based modeling strategy to obtain the most accurate structure models, and what is the extent of model accuracy improvement compared to the previous state-of-the-art non-DL methods (i.e. template-based modeling). With this aim, four types of Alphafold protocol for active-state modeling and two types for inactive-state were employed, mainly varying in their inputs to take into account the functional state in different fashions (details provided in Methods). For the best-practice TBM for comparison, receptor models at inactive states were brought from the GPCR TBM database (https://github.com/benderb1/rosettagpcr) prepared by Bender et al [13] (only inactive states are available in the database). In this database, all TBMs were built using RosettaCM [14], known to be the best-performing template-based method in the previous CASP [15]. The method builds models by recombining multiple templates with a sequence identity of less than 40 % (more description of the database in Methods) and showed improved model accuracy over five other GPCR model databases using other methods [13], [15]. Because no close homologs were included as templates, TBM from this database may correspond to the best available models for “non-trivial modeling” rather than “trivial modeling” scenarios (e.g. TBM of 5HT1R using 5HT2R). However, given that non-trivial modeling is required for the majority (>80 %) of GPCRs, according to Bender et al, we thought benchmarking against TBMs built at such generic scenarios would be more informative to judge its coverage in the entire GPCR family tree. We also tested whether detailed AlphaFold parameters affect the AlphaFold model accuracy (e.g. 5 wt parameter set indices, number of recycles, AlphaFold versus AlphaFold_multimer when modeling more than a single chain, and so on), and found that changes in the parameters had minimal effects (Supplementary Fig. S1). Therefore, these parameters were fixed to default values (see Methods) throughout the work.
Receptor model qualities were measured by two metrics: 1) overall receptor TM-score [16] focusing on global model quality and 2) binding site backbone RMSD (bbRMSD) focusing on local model quality around the receptor-ligand interface. To support the conclusion derived by using these metrics, different measures, receptor global distance test (GDT) values [17], and binding site χ angle accuracy were also used, as presented in Supplementary Fig. S2.
In Fig. 2, receptor modeling results are presented in the two model quality metrics, and a few clear conclusions could be derived from the results. First, DL structure prediction outperformed TBM in both global and interface accuracy (Fig. 2A and 2B). Note that only inactive state models are available in the TBM database, therefore comparison is made only for inactive state models. Also, because AlphaFold models used templates without a sequence identity threshold, unlike TBMs, we further explored to what extent the AlphaFold model differs with the choice of templates (i.e. the same condition as TBM). Shown in Supplementary Fig. S3, we observe little difference in model quality with template choices, indicating AF modeling is little affected by the template quality as long as sequence identity is reasonable (>20 %), consistent observation with Heo et al [7]. Second, the active state model accuracy depends quite a lot on the modeling strategies (for instance, the difference in the active-state binding site accuracy of “AF,as-is” and those of the rest AF strategies is about 20 %, dark green bars in Fig. 2B); modeling together at the correct active-state context (“AF,G-pro”, “AF,Gɑ”, or “AF,bias”) equally outperformed receptor modeling without any functional state consideration (“AF,as-is”). For the modeling of inactive states, this gap is smaller but still, template-biasing (“AF,bias”) is slightly better than non-biasing (“AF,as-is”).
Fig. 2.
Receptor model accuracy upon modeling strategies. Different modeling strategies considered are as follows: “TBM”, template-based modeling; “AF,as-is”, AlphaFold without any tweak for the functional state, “AF,bias”, AlphaFold with a biased template set matching the functional state [7]; “AF,Gɑ”, AlphaFold_multimer modeling of receptor + alpha unit of G-protein; “AF,Gpro”, AlphaFold_multimer modeling of receptor + whole G-protein. Modeling details are described in Methods. A) Global receptor model accuracy measured by TM-score [16]. B) Binding site accuracy measured by the fraction of models with the binding site backbone RMSD < 0.5 Å and < 1.0 Å shown in dark and light bars, respectively. Backbone RMSD refers to RMSD between the backbone atoms of the model and the corresponding atoms in the experimental structure. Model accuracy measured by alternative metrics (GDT [17] and side-chain χ angle accuracy) is reported in Supplementary Fig. S2. C) Distribution of binding site accuracy for AlphaFold models (top), TBMs (middle), and experimental structures with no ligand or bound to other ligand molecules (“cross”, bottom). The number of receptors at each RMSD bin is shown on the y-axis. D) Examples of receptor models showing large differences, with the following color scheme: pink, experimental structure; blue, best-practice AF model; gray, “AF,as-is” for the three left panels and TBM for the right-end panel; orange, native ligand structures. Major conformational differences are highlighted by arrows. From left to right: GLR, PDBID 6wpw) The extracellular orientation was corrected when modeled with the G alpha subunit together. OX2R, PDBID 7l1u) The best-practice AF model showed the correct TM6 orientation. AA2AR, PDBID 5 nm4) AF-bias correctly modeled the extracellular loop alpha helix fragment, which contains lysine interacting with the ligand, while AF as-is failed with the unfolded loop. PE2R4, PDBID 5ywy) The TM helix I and II modeled by TBM intruded into the ligand binding pocket, while the AF-bias model showed a structure similar to the native.
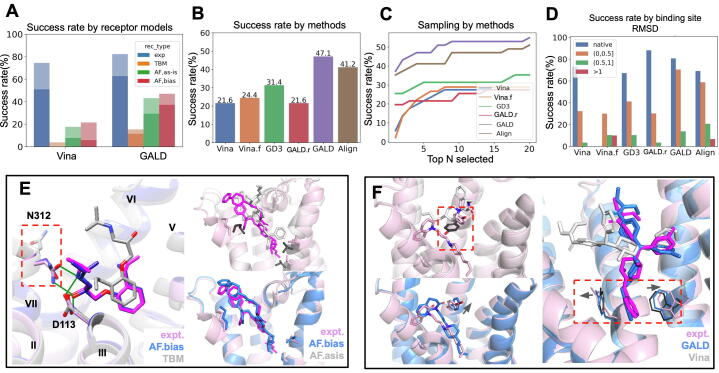
One of the surprising receptor modeling differences found was for class B1 (and a few other peptide-binding class A) GPCRs known as peptide-binders. Unlike others, these receptors possess large extracellular domains whose interfaces with the transmembrane domains often form binding sites for natural agonist peptides (the leftmost panel in Fig. 2D). We observe that the orientation of this domain could be modeled accurately only when modeling was performed with the G-alpha subunit using AlphaFold_multimer, in contrast to the failure observed when modeling receptor-only using AlphaFold (the monomeric version) [6]. By further analyzing 6 targets with bulky extracellular domains (ECDs) (Supplementary Fig. S4 and Table S2), we observed no clear dependence of ECD orientation on the templates provided as inputs. Therefore, we speculate a more relevant origin for the difference might be the difference in the network training procedures between AlphaFold and its multimer version (e.g. cropping size, training data, etc), which is beyond what could be addressed in this work. Two other examples shown on the right panels of Fig. 2D highlight the structural differences in small-molecule binding site loops or helical orientations originating from variations in modeling strategies.
How is the expected docking performance for the best-practice DL models compared to that for experimentally resolved receptor structures? To address this question, we compared the distribution of binding site accuracy statistics for the AlphaFold models versus experimental structures in “cross-docking” scenarios, which may correspond to the best possible receptor structures in real practice drug design situations. Here, cross-docking refers to docking on experimental structures determined at an unbound state or bound state with other ligands. We find that the AlphaFold model accuracy judged by the binding site backbone RMSD is far better than TBM, closely approaching the level of experimental structures for “cross-docking” (Fig. 2C).
To investigate if model accuracy depends on whether the target receptor was included in the training process of AlphaFold, statistics was examined separately by the deposited date of PDB entries. We see no distinction in model accuracy distributions (Supplementary Fig. S5), indicating the modeling protocol is robust throughout different GPCRs.
Best-practice protocol: In summary, we choose “AF,bias” as the best practice receptor modeling protocol for small-molecule docking because of its modeling convenience among strategies considering activation states. For peptide docking, “AF,Gɑ” is chosen as the best practice protocol which can optimally capture peptide binding sites.
4. Receptor modeling strategy and consideration of receptor flexibility are critical for small-molecule docking and virtual screening
From this point, we evaluate the overall docking performance across various small-molecule and peptide docking protocols. Details of docking protocols are reported in Methods. To reduce the complexity of benchmarking, we ran docking simulations on four selected types of receptor models: 1) TBM, 2) “AF,as-is”, 3) “AF,bias”, and 4) experimental structures. This setup would reveal the benefit of using DL models over TBMs as well as the carefully curated AlphaFold models over those naively modeled in docking scenarios.
Looking at small-molecule docking results first, the receptor modeling strategy largely determines the performances of all docking protocols (Fig. 3A). A successful prediction was defined as that with ligand RMSD < 2.5 Å for the best of the top 5 (top N refers to N top-scored ones) unless specified otherwise. While docking on TBM shows high rates of failure, using the AlphaFold models gives quite improved success rates over TBM by a maximum of 30 % depending on the docking tools (Fig. 3A). Of the various AF receptor models, docking on state-specific AF models (“AF,bias”) led to improved results over using the naive receptor models (“AF,as-is”), especially when measured by top 1 models (8 % difference). Examples in which inaccurate side-chain modeling by TBM (on the left) or “AF,as-is” (on the right) resulted in inaccurate docking results are shown in Fig. 3E.
Fig. 3.
Small-molecule pose prediction benchmark on various receptor models using different docking methods. Success rates are shown using the criterion of the best of top 5 unless specified; top N refers to N top-ranked ones by the tool. Tested methods: “Vina”: Autodock Vina (default) [18];”Vina.f”: Autodock Vina with flexible side chain option [20]; “GD3”: GalaxyDock3 [19]; “GALD”: Rosetta GALigandDock (default receptor-flexible mode); “GALD.r”: GALigandDock with rigid side chain option; and “Align”: CSAlign-Dock [21]. A) Docking performance by different receptor models, shown for the top 1 ligand pose (darker colors) or the best of top 5 (lighter colors) having ligand RMSD < 2.5 Å. Here “exp” corresponds to self-docking, meaning an artificial docking case to the experimental receptor structure bound to the same ligand. Per-receptor model type results for other tools (GD3 and Align) are reported in Supplementary Fig. S6. B) Docking results for various docking tools on the best-practice AlphaFold receptor models. C) Conformational sampling performance of docking tools. D) Dependence of docking results on binding site accuracy. E-F) Examples highlighting differences originating from receptor models or docking tools. Color schemes are shown in the inset. Incorrectly modeled side-chains are highlighted by dark gray colors. E) Examples showing the importance of receptor modeling strategy, ADRB2 (PDBID 6 ps2) on the left and AA2AR (PDBID 5wf5) on the right. F) Examples showing the importance of receptor-flexible docking, ACM1 (PDBID 6zfz) on the left and OPRD (PDBID 6pt3) on the right.
When the best-practice receptor models were used, the main determinant of the docking performance was the implementation of receptor flexibility in docking methods (Fig. 3B). GalaxyDock3 [19] was run with ligand flexibility but without receptor flexibility (no such option on it); Autodock Vina [18] and GALigandDock [20] can take into account receptor flexibility and both versions were run (detailed comparison of the tools in Methods). The best success rate obtained was 47 % by Rosetta GALigandDock with receptor flexibility, compared to 24 % by Autodock Vina (Vina.f, receptor flexible version) and 32 % by the GalaxyDock3. Large-scale side-chain rotations were required (left panel in Fig. 3F) sometimes, but in other cases, slight side-chain movements were sufficient to let ligands move into the correct poses (right panel in Fig. 3F). This discovery is further supported by comparing against GALigandDock without receptor flexibility option (GALD.r), as shown in Fig. 3B (the gap between red and purple bars), in which docking performance significantly drops without receptor flexibility. A reference ligand-guided docking method, CSAlign-Dock [21], which uses the same machinery as GalaxyDock3 but differs in the additional guidance by structure alignment to a reference ligand pose, performed better than GalaxyDock3 but not exceeded GALigandDock. This is likely because the information on the binding pose of a reference ligand helps tolerate receptor structure deviations, but to a limited degree due to the lack of explicit consideration of receptor flexibility.
Although the best docking result obtained (47 %) on the best-practice models outperforms that on TBM, the result is still far off from the artificial self-docking case on co-crystallized receptor structures (82 %). We sought to identify the factors that could have potentially reduced this gap. The first is a more aggressive modeling of receptor flexibility during ligand docking. In Fig. 3D, the dependence of docking results on binding site structure accuracy is shown. The performance of GALigandDock approaches that of self-docking when binding site RMSD is less than 0.5 Å but rapidly drops as the difference gets larger. Note that here we call it “difference” not “error”, because this amount of difference can readily occur even when experimental structures are used (i.e. in cross-docking scenarios), and thus should be taken into account at the docking stage than at the receptor modeling stage. One could insist a full receptor backbone flexible docking could rescue these failures in principle; however, developing a robust but practically affordable method (MD-based methods can be too expensive [22]) remains another big challenge. A full receptor-flexible method, Galaxy-7TM [23], has shown promise in this direction, but we observed worse performance than other tools when tested in this dataset (Supplementary Fig. S7), primarily because Galaxy-7TM was developed in the TBM context. We expect similar developmental efforts can be made that take more accurate DL-based models as inputs. The second is regarding how to select the docked models. When the selection criterion is further relieved to include the top 20 structures, this adds in about 10 % of more success (Fig. 3C), suggesting an extra model selection step can rescue a large portion of failures. Based on this finding, developing a complex model selection or discrimination tool could be a useful addition to the community.
Best-practice protocol: In summary, for small-molecule docking, we designate GALigandDock (flexible-receptor mode) as the best-practice protocol among the tools tested in this work. Other docking tools supporting robust receptor flexibility may be a good substituent. We expect that combining the reference ligand-guided docking idea (used in CSAlign-Dock) with carefully curated receptor flexible docking may push the accuracy limits.
Another important drug-discovery-related problem is virtual screening, in which a huge library of compounds is screened against a receptor to search for its potential hit binder compounds. We performed virtual screening on 10 different GPCR targets with 3 different receptor conformations for each target (experimental, AF, and TBM), leading to a total of 30 virtual screening tasks. For each screening task, 3 docking tools (Vina, Vina-RF [24], and GALigandDock) were compared. For 5 receptors (AA2AR, ADRB1, ADRB2, CXCR4, DRD3), their binders, and non-binder “decoy” compounds were brought from the DUD-E dataset [25]. For the rest (AGTR1, CRFR1, GRM2, OPRD, S1PR1), binders were brought from GPCRdb and decoys from the DUD-E web server (https://dude.docking.org/generate). Because the majority of binders in the set are inhibitors (antagonist or inverse agonists), we simply employed “AF,as-is” for the AlphaFold models in virtual screening tests. As shown in Fig. 4A-B, virtual screening on AlphaFold models showed somewhat worse performance than that on native experimental structures, but was superior to the results on TBMs, consistently through the docking tools (per-target breakdown analyses in Supplementary Fig. S8). The results again prove the improved performance of the DL-predicted receptor models for drug discovery. Meanwhile, GALD consistently outperformed Vina and VIna-RF throughout the receptor structure types and target receptors (full ROC curves in Supplementary Fig. S9), supporting successful pose prediction can be important for virtual screening performance. Despite the GALD’s improved performance over Vina and Vina-RF on GPCRs, the virtual screening result here needs to be carefully taken until it is benchmarked on a broader set of receptors along with many other docking tools.
Fig. 4.
Virtual screening results. 3 receptor structures (native, TBM, and AF,as-is) for each of 10 GPCRs (AA2AR, ADRB1, ADRB2, CXCR4, DRD3, AGTR1, CRFR1, GRM2, OPRD, S1PR1) are tested for virtual screening using 3 docking tools: Vina, Vina reranked with ML [24] (Vina-RF), and GALigandDock (GALD). Mean values over 10 targets are reported. A) Mean AUC values from Receiver operator characteristics (ROC) analysis are shown. B) Enrichment ratios of active molecules at a false positive ratio of 1% are shown. Detailed analyses can be found in Supplementary Figs. S8-9.
5. Deep-learning-based protein-peptide complex modeling outperforms previous peptide docking tools
Peptides are an important class of ligands for GPCRs. Class B1 GPCRs take peptides as their natural agonists, and also many class A GPCRs adopt peptides either as agonists or antagonists. Here we explore how we can benefit from using DL methods for GPCR-peptide complex structure prediction.
One clear advantage of DL methods over many other peptide docking tools is that they can predict the whole complex structure at once. With this advantage, both receptor and peptide ligand flexibility are naturally considered. Here, the AlphaFold multimer modeling strategy is compared against using three other available peptide docking tools [26], [27], [28] to dock peptides on pre-generated AlphaFold receptor models.
In Fig. 5A, peptide docking benchmark results are presented. We took the CAPRI [30] metric to measure success, considering a prediction with “the best of top 5 ligand poses being at least at acceptable accuracy” (measured by a combined evaluation of contacts and orientations, details in Methods) as a success. As shown in the figure, AlphaFold complex modeling outperformed all other methods using peptide docking tools. Compared to the best pre-DL scenario (applying the best pre-DL docking tool, i.e., MdockPep2, on TBM), the performance difference is 60 %. This result is consistent with the observation from small-molecule docking, that the consideration of receptor flexibility is critical for success which should be naturally captured during simultaneous modeling. In Fig. 5D, examples of three peptide-binding complexes, two corresponding to class A and one to class B1 are shown.
Fig. 5.
Peptide docking benchmark on different receptor models using various docking methods. The success rate is measured by the fraction of predictions with CAPRI accuracy of “acceptable” quality (see Methods). Tested docking tools: HPEPDOCK [26]; MDockPeP2 [27]; GalaxyPepDock [28]; AF_multimer, complex modeling by AlphaFold_multimer [29]. A) Success rate of docking onto either TBM or AF receptor models compared against AlphaFold_multimer. B) Sampling performance of MDockPeP2 when docked on AF models, TBM models, and bound experimental structures, compared to the AlphaFold_multimer top5 (green). C) Dependence of docking tools on peptide length. D) Examples of docking results for different methods. Peptide structures by modeling tools are colored by the scheme shown on the right bottom. Receptor type (PDB ID) from left: CCKAR (7ezm), NPY1R (7vgx), and GLR (6wpw).
We also found that the DL method decreased the dependency of the modeling performance on the peptide length. Interestingly, AlphaFold multimer modeling was not influenced much by the size of peptides (Fig. 5C) while other peptide docking tools showed a strong dependency. When six large peptides (>14 amino acids) were tested, none of the flexible peptide docking tools was able to recover the native poses while AlphaFold succeeded for four of them. Enabling peptide docking to be independent of peptide length can be regarded as one of the major triumphs achieved by DL-based peptide complex modeling.
AlphaFold generates only a limited number of solutions, but peptide docking tools can offer more diverse conformation samples some of which are more accurate than the best AlphaFold models. In Fig. 5B, sampling performances of a docking method MDockPeP2 are shown. While AlphaFold outperformed the rest at the top 1 or the best of the top 5 metric, a non-DL tool, MDockPeP2 exceeded AlphaFold in the self-docking scenario when 10 docked models were considered. This reinforces the previous findings from the small-molecule complex benchmark, in that external docking tools can be integrated for a “meta approach” to gain additional success as suggested in a recent work [31].
Best-practice protocol: The best performing protocol was to run AlphaFold_multimer for the complex modeling of the peptide and receptor only (without G protein) with proper recycle steps of 10 (parameter sweep reported in Supplementary Fig. S10). Although including G protein was helpful for the modeling of the receptor alone, it deteriorated the results when peptides were added. We attribute this performance degradation to some unknown artifact within AlphaFold multimer when more than two chains (GPCR, G-protein, and peptide) are provided as input altogether, however, there is a great chance that an improved protocol is developed.
6. Modeling and docking confidences can be estimated from the predicted receptor structure accuracy
It is very important in real practice to evaluate the reliability of a predicted model structure and docked conformations. Here, we propose a guideline to estimate the likelihood of successful docking given an AlphaFold receptor prediction. In Fig. 6, docking success rates are binned by the predicted accuracy (p-lDDT) of receptor binding site residues. For small-molecule complexes, chances to get accurate docked poses become 70 % when the binding site p-lDDT is larger than 0.95 but drops to 0 % when it is smaller than 0.85. Similarly for peptides, when the binding site p-lDDT is larger than 0.85 predictions are always accurate, but when lower than 0.85, the chance drops to 40 %. Because p-lDDT can be obtained without any knowledge of the experimental structure, these borderlines may help evaluate the prediction results.
Fig. 6.
Dependence of the docking performance on the estimated binding site accuracy, measured by AlphaFold binding site p-lDDT [1], [2]. Left) small-molecule complexes, right) peptide complexes. p-lDDT ranges are shown on the x-axis. No peptide complex had a binding site p-lDDT above 0.95. The numbers on top of the bars represent the number of samples in each bin.
7. Discussion
In this work, we have shown the most up-to-date status of GPCR docking coupled with recent DL-based protein structure predictions. We aimed to demonstrate realistic expectations in docking results to the researchers employing the recent DL modeling and to provide the best-practice protocols to our knowledge. Quantitative analyses shown in this work revealed that the protocols integrating DL-based modeling excel in docking or screening GPCRs compared to the best pre-DL, TBM modeling. We also showed that this advance can be maximally delivered to the prediction results only if the modeling and docking options are chosen appropriately. We encourage the readers to perceive the best practice guideline provided at the end of every section after understanding the different parameters that we have tested for the benchmark studies. We also presented how to estimate the prediction confidence, which will be valuable in practical docking scenarios. Finally, we stressed that considering larger receptor flexibility efficiently (binding site RMSD > 0.5 Å) during docking remains a major challenge in GPCR docking.
A few limitations to this study have to be pointed out. First, our guidelines can be temporary in this fast-changing era. There are many ongoing types of research that can substantially improve results over all the protocols tested in this study. This work would still be valuable because the quantitative analysis we collected can serve as a sound baseline for future research. Second, our guidelines may not always provide reliable solutions to GPCR docking practices. As it was insisted throughout the manuscript, the expected performance of the best-practice protocol is far from perfect yet. We, therefore, suggest users carefully judge their complex models based on the confidence estimation suggested in the last section. We believe that subsequent efforts by many researchers targeting more real-world problems, as demonstrated in the recent GPCRDock 2021 challenge, may help fill this gap.
8. Methods & materials
8.1. Dataset curation
First, 33 receptor families having complex structures for both agonists and antagonists were selected according to the information from the GPCRdb website (https://gpcrdb.org) [12]. Because their ligands could be either small molecules or peptides or both, counting peptide complexes separately resulted in 51 small-molecule complexes and 19 peptide complexes in the end. The total number of active and inactive state complexes is 38 and 32, respectively. When more than one structure exists for a complex type for a given complex (e.g. small-molecule complex in the active state), the PDB entry with the highest resolution was chosen. Non-canonical amino acids in peptide ligands were converted to their counterpart canonical amino acids as closely as possible if the modification was not judged to affect their interactions by human inspection. Because AlphaFold can be only run with canonical amino acids, those peptide ligands possessing at least one residue that could not be converted to canonical amino acid were dropped from the benchmark. Sequences for receptors and G-proteins were taken from the UniProt database [32].
8.2. Receptor modeling
8.2.1. AlphaFold modeling
AlphaFold and AlphaFold_multimer version 2.28 was used throughout the study [1], [29]. Templates were searched from PDB70 (version Apr 2020) using the HHsearch tool in HH-suite 3 [33], but only with the entries deposited before the query. Sequences were searched against the UniRef90 (version 2022) [34], BFD (version Mar 2019) [35], and Mgnify (version Dec 2018) [36] metagenomics databases using jackhmmer (version 3.3.2) [37].
Various receptor modeling options were tested within AlphaFold, and a default parameter set was chosen that consistently performs near optimally (Supplementary Fig. S1). These default parameters for the receptor modeling are: i) model_1 of 5 AlphaFold parameter indices, ii) recycle steps of 3, and iii) AMBER force field relaxation at the end. For any instance of modeling more than a single chain (AF,Gpro, AF,Ga), raw MSAs for each protein (or peptide) chain were combined to an unpaired MSA, and the AlphaFold_multimer version was applied. Template-biased models were downloaded from Heo et al. [7] if a model does not exist for the receptor, otherwise, they were modeled using the script provided by the authors.
Receptor models with biased activation states include the modeling chains as follows.
AF,Gpro: GPCR and whole chains in G-protein, no template-biasing
AF,Ga: GPCR and alpha chain in G-protein, no template-biasing
AF,bias: GPCR only, models directly brought from Heo et al. [7]
For the GPCR-peptide complex modeling, all 5 model indices from AlphaFold_multimer at recycling steps of 10 were used for evaluation.
8.2.2. Template-based models
Template-based models (TBMs) deposited in Bender et al. [13] were generated only for inactive state conformations using RosettaCM [14], a method hybridizing multiple templates’ secondary structure chunks along with fragment insertions at unaligned regions. Models were relaxed by using the Rosetta energy function at the end to ensure physical likeliness (e.g. no Ramachandran or rotameric outliers, clash, etc.). Structures deposited as of June 2020 were used as templates (April 2020 for the AlphaFold_multimer template database for comparison). In this work, multiple templates with a sequence identity of less than 40 % were intentionally selected as input templates to mimic medium-to-hard homology modeling scenarios which correspond to the modeling of >80 % GPCRs. They reported their protocol showed better performance than I-Tasser, which is known as one of the best template-based modeling protocols.
8.3. Evaluation metrics
Evaluation metrics for protein structures and docked poses used in this study follow the standards taken in the related works. The binding site is defined for all receptor residues having at least one atom closer than 6 Å to their native ligand. Small-molecule docking accuracy is measured by ligand RMSD and considered successful if the best of the top 5 values is less than 2.5 Å. Peptide docking accuracy is measured by the CAPRI metric [30] (modified following Ref. [38]) on the best of the top 5 models, and having at least “acceptable” quality is considered a success. The CAPRI “acceptable” quality corresponds to contact accuracy (fnat) above 0.2 and either ligand RMSD below 4.0 or interface RMSD below 2.0. Acceptable quality amounts to the cases in which the predictions can be used to guide site-directed mutagenesis or other biochemical experiments to identify the correct contacts.
8.4. Ligand preparation for small molecules
Ligand structures extracted from the PDB files were converted to SMILES by OpenBabel (version 3.1.0) [39] and then reconverted to 3D structures by CORINA (version 4.4.0) [40] to erase any experimental information on the ligand structures. We do see almost no difference in the results when docking simulations are run using the experimental ligands instead (Supplementary Fig. S11).
For Rosetta GALigandDock, inputs were generated following the input preparation guideline (https://www.rosettacommons.org/docs/latest/scripting_documentation/RosettaScripts/Movers/GALigandDock); hydrogens were attached at pH 7 and partial charges were assigned using the MMFF94 force field [41] by OpenBabel. For input ligands of GalaxyDock3, CSAlign-Dock, Galaxy7TM, and AutoDock Vina, Chimera [42] was used for protonation and Gasteiger partial charge [43] calculation.
8.5. Docking tools
For small-molecule docking, we tested various tools covering different degrees of freedom (DOFs). DL-based docking tools were not included in this study because their improvements were rather marginal. Moreover, the current dataset might have been included in their training set, and most critically, the reported results were not reproduced by our own runs. Below are comparisons of the docking methods tested in this work, mainly focusing on their receptor flexibility considerations (also summarized in Supplementary Table S3).
AutoDock Vina[18] Ligand translation, rotation, and torsion angles are varied.
AutoDock Vina with flexible sidechain[15] In addition to the original DOFs considered in Vina, receptor sidechain torsions are additionally varied (the method “Vina.f” in the manuscript). Flexible residues are assigned when two criteria are satisfied together: 1) any of the side-chain atoms are within 3 Å of any ligand atoms, 2) Any χ angle is different from that of the experimental structure by more than 40°. Two strategies are tested: 1) All side-chains with the criteria, leading to 1 ∼ 21 flexible sidechains, and 2) capping up to two closest residues passing the criteria above [7]. The former led to a better result and was reported in the manuscript.
GalaxyDock3[19] In addition to the DOFs considered in Vina, ligand ring flexibility is considered by discrete sampling within a crystal ring structure library.
CSAlign-Dock[21] DOFs are identical to GalaxyDock3. A shape score measuring the similarity of the query ligand to a reference ligand is added to the docking score to guide docking. GalaxySite [44] was used to search three reference molecules with the highest Tanimoto coefficients.
Rosetta GALigandDock[20] Receptor flexibility is allowed for a set of side-chains plus their backbones automatically detected around the input ligand. On average 9.8 residues are detected as flexible. Unlike other tools, the docking process is repeated 15 times following the guidelines for receptor-flexible docking for increased convergence. For the rigid receptor option (“GALD.r” in the manuscript), no side-chain and backbone optimization is performed.
Galaxy7TM[23] All residues' backbones and side-chains are fully allowed to move. The method first presamples receptor conformational ensemble and runs docking on the ensemble.
For peptide docking, three tools were tested including two global docking tools, MDockPeP2, HPEPDOCK, and a template-based docking tool, GalaxyPepDock.
HPEPDOCK[21] An ensemble of peptide conformations generated by MODPEP [45] is docked and selected by a hierarchical search guided by a knowledge-based potential.
MDockPeP2[22] Peptide conformations are searched on-the-fly by assembling fragments collected from PDB along with global rigid sampling and local flexible minimization. A hybrid scoring function combining the Vina score and PC_score (physicochemical similarity score) is applied for model ranking.
GalaxyPepDock[23] Initial peptide conformations are brought from a template search based on protein structure similarity and protein-peptide interaction similarity. Models are further refined by the protein and peptide adjustment of the backbone and side-chain inspired by GalaxyRefine [42] and GalaxyRefineComplex [43].
8.6. Virtual screening
10 GPCRs were selected for the benchmark with a sufficient number of known ligands considering protein family diversity. The overview of the procedure is described in Supplementary Fig. S12. Each receptor had a set of molecules to screen with at least 49 binders and 40 times more decoys. The number of binders and decoys is summarized in Supplementary Table S4.
For 5 receptors included in the DUD-E benchmark set (AA2AR, ADRB1, ADRB2, CXCR4, and DRD3), 3D conformation files of ligands and decoys in mol2 format were downloaded from the website. If there were multiple protonated forms, we selected one state for ligands of the same ChEMBL ID. For general comparison with other results, original protonation states were preserved except for CXCR4 ligands, in which an important key proton was missing and remodeled using Chimera.
5 receptors (AGTR1, CFCR1, GRM2, OPRD, and S1PR1) were selected additionally to cover other classes of GPCRs (class A, B1, and C). Ligands were collected from GPCRdb and curated with the following steps: 1) filtered by the number of heavy atoms (<70) and experimental affinity higher than 10 nM; 2) removed duplicates in SMILES and ChEMBL IDs; 3) clustered by scaffolds and picked the highest affinity ligand as representative. Decoys were generated from the DUD-E server (https://dude.docking.org/generate). The generation method predicts the ligand’s protonation states in pH 6 ∼ 8 and then selects a set of molecules from the ZINC database with identical molecular properties but different scaffolds to ligands.
We used the Autodock Vina, a random forest estimator using Vina results [24], and finally, Rosetta GALigandDock (GALD) for benchmarking virtual screening at its VSH mode (https://www.rosettacommons.org/docs/latest/scripting_documentation/RosettaScripts/Movers/GALigandDock). Although its performance for virtual screening has not been published yet, our in-house benchmark showed the GALD’s AUROC (area under receiver-operator characteristics curve) of 0.79 on the original DUD set [46], superior to many other docking tools (GLIDE, GOLD, GalaxyDock, Vina, etc.).
CRediT authorship contribution statement
Sumin Lee: Software, Visualization. Seeun Kim: Software, Visualization. Gyu Rie Lee: Software, Visualization. Sohee Kwon: Software. Hyeonuk Woo: Software. Chaok Seok: Conceptualization, Supervision. Hahnbeom Park: Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by National Research Foundation of Korea (NRF) grants (No. 2022R1C1C1007817 to HP and No. 2020M3A9G7103933 to CS), Institute of Information & communications Technology Planning & Evaluation (IITP) grant funded by the Korean government (MSIT) (No. 2021-0-02068, Artificial Intelligence Innovation Hub) (No.2021-0-01343, Artificial Intelligence Graduate School Program (Seoul National University)) (to CS), and KIST Institutional grant 2E31523 (to HP). We thank Nuri Jung at Seoul National University for technical support and Dr. Jinsol Yang at Galux Inc. for helpful discussions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.11.057.
Contributor Information
Chaok Seok, Email: chaok@snu.ac.kr.
Hahnbeom Park, Email: hahnbeom@kist.re.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seok C, Baek M, Steinegger M, Park H, Lee GR, Won J. Accurate protein structure prediction: what comes next? Biodesign. 2021. pp. 47–50. 10.34184/kssb.2021.9.3.47.
- 4.Ourmazd A., Moffat K., Lattman E.E. Structural biology is solved — now what? Nat Methods. 2022:24–26. doi: 10.1038/s41592-021-01357-3. [DOI] [PubMed] [Google Scholar]
- 5.Moult J., Fidelis K., Kryshtafovych A., Schwede T., Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins. 2018;86(Suppl 1):7–15. doi: 10.1002/prot.25415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X.-H., You C.-Z., Jiang H.-L., Jiang Y., Eric X.H., Cheng X. AlphaFold2 versus experimental structures: evaluation on G protein-coupled receptors. Acta Pharmacol Sin. 2022 doi: 10.1038/s41401-022-00938-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo L., Feig M. Multi-state modeling of G-protein coupled receptors at experimental accuracy. Proteins. 2022 doi: 10.1002/prot.26382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trzaskowski B., Latek D., Yuan S., Ghoshdastider U., Debinski A., Filipek S. Action of molecular switches in GPCRs–theoretical and experimental studies. Curr Med Chem. 2012;19:1090–1109. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster S.R., Hauser A.S., Vedel L., Strachan R.T., Huang X.-P., Gavin A.C., et al. Discovery of human signaling systems: pairing peptides to G protein-coupled receptors. Cell. 2019:895–908.e21. doi: 10.1016/j.cell.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein R.M., Kang H.J., McCorvy J.D., Glatfelter G.C., Jones A.J., Che T., et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature. 2020;579:609–614. doi: 10.1038/s41586-020-2027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Sun Y., Song Y., Dai D., Zhao Z., Zhang Q., et al. Fragment-based computational method for designing GPCR ligands. J Chem Inf Model. 2020;60:4339–4349. doi: 10.1021/acs.jcim.9b00699. [DOI] [PubMed] [Google Scholar]
- 12.Kooistra A.J., Mordalski S., Pándy-Szekeres G., Esguerra M., Mamyrbekov A., Munk C., et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 2021;49:D335–D343. doi: 10.1093/nar/gkaa1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender B.J., Marlow B., Meiler J. Improving homology modeling from low-sequence identity templates in Rosetta: A case study in GPCRs. PLoS Comput Biol. 2020:e1007597. doi: 10.1371/journal.pcbi.1007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y., DiMaio F., Wang R.Y.-R., Kim D., Miles C., Brunette T., et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryshtafovych A., Moult J., Bales P., Bazan J.F., Biasini M., Burgin A., et al. Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10. Proteins. 2014;82(Suppl 2):26–42. doi: 10.1002/prot.24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 17.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J., Baek M., Seok C. GalaxyDock3: Protein–ligand docking that considers the full ligand conformational flexibility. J Comput Chem. 2019:2739–2748. doi: 10.1002/jcc.26050. [DOI] [PubMed] [Google Scholar]
- 20.Park H., Zhou G., Baek M., Baker D., DiMaio F. Force field optimization guided by small molecule crystal lattice data enables consistent sub-angstrom protein-ligand docking. J Chem Theory Comput. 2021:2000–2010. doi: 10.1021/acs.jctc.0c01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Kwon S., Bae S.-H., Park K.M., Yoon C., Lee J.-H., et al. GalaxySagittarius: structure- and similarity-based prediction of protein targets for druglike compounds. J Chem Inf Model. 2020;60:3246–3254. doi: 10.1021/acs.jcim.0c00104. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Vass M, Shi D, Abualrous E, Chambers J, Chopra N, et al. Benchmarking refined and unrefined AlphaFold2 structures for hit discovery. 10.26434/chemrxiv-2022-kcn0d. [DOI] [PubMed]
- 23.Lee G.R., Seok C. Galaxy7TM: flexible GPCR-ligand docking by structure refinement. Nucleic Acids Res. 2016;44:W502–W506. doi: 10.1093/nar/gkw360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C., Zhang Y. Improving scoring-docking-screening powers of protein-ligand scoring functions using random forest. J Comput Chem. 2017;38:169–177. doi: 10.1002/jcc.24667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P., Jin B., Li H., Huang S.-Y. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018;46:W443–W450. doi: 10.1093/nar/gky357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X., Zou X. Predicting protein-peptide complex structures by accounting for peptide flexibility and the physicochemical environment. J Chem Inf Model. 2022;62:27–39. doi: 10.1021/acs.jcim.1c00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H., Heo L., Lee M.S., Seok C. GalaxyPepDock: a protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 2015;43:W431–W435. doi: 10.1093/nar/gkv495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lensink M.F., Velankar S., Wodak S.J. Modeling protein-protein and protein-peptide complexes: CAPRI. Proteins. 2017;85:359–377. doi: 10.1002/prot.25215. [DOI] [PubMed] [Google Scholar]
- 31.Tsaban T., Varga J.K., Avraham O., Ben-Aharon Z., Khramushin A., Schueler-Furman O. Harnessing protein folding neural networks for peptide–protein docking. Nat Commun. 2022 doi: 10.1038/s41467-021-27838-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinegger M., Meier M., Mirdita M., Vöhringer H., Haunsberger S.J., Söding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinf. 2019;20:473. doi: 10.1186/s12859-019-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt Consortium UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinegger M., Mirdita M., Söding J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat Methods. 2019;16:603–606. doi: 10.1038/s41592-019-0437-4. [DOI] [PubMed] [Google Scholar]
- 36.Forster S.C., Kumar N., Anonye B.O., Almeida A., Viciani E., Stares M.D., et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khramushin A., Marcu O., Alam N., Shimony O., Padhorny D., Brini E., et al. Modeling beta-sheet peptide-protein interactions: Rosetta FlexPepDock in CAPRI rounds 38–45. Proteins. 2020;88:1037–1049. doi: 10.1002/prot.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadowski J., Gasteiger J., Klebe G. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J Chem Inf Comput Sci. 1994:1000–1008. doi: 10.1021/ci00020a039. [DOI] [Google Scholar]
- 41.Halgren T.A. Force Fields: MMFF94. Encyclopedia of Computational Chemistry. 2002 doi: 10.1002/0470845015.cma012m. [DOI] [Google Scholar]
- 42.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron. 1980:3219–3228. doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- 44.Heo L., Shin W.-H., Lee M.S., Seok C. GalaxySite: ligand-binding-site prediction by using molecular docking. Nucleic Acids Res. 2014;42:W210–W214. doi: 10.1093/nar/gku321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y., Zhang D., Huang S.-Y. Efficient conformational ensemble generation of protein-bound peptides. J Cheminform. 2017;9:59. doi: 10.1186/s13321-017-0246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang N., Shoichet B.K., Irwin J.J. Benchmarking Sets for Molecular Docking. J Med Chem. 2006:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.