Abstract
This article presents data concerning STX18-AS1, a long noncoding RNA gene identified from a Genome-wide association study of Atrial Septal Defect (ASD). The data describes its expression patterns in human tissues and functions in regulating cardiomyocyte differentiation in vitro. STX18-AS1 is a lncRNA with a higher abundance in developing tissues, including hearts. Its transcription distribution within the embryonic hearts during key heart septation stages supports STX18-AS1’s association with risk SNPs for ASD. The CRISPR stem cell pool in which STX18-AS1 was knocked down, showed reduced CM differentiation efficiency and lower expression of key cardiac transcriptional factors. This indicated its regulative role in supporting the lineage specification from cardiac mesoderm into cardiac progenitors and cardiomyocytes. These data can benefit the understanding of human embryonic heart developmental biology, and the time-course changes of cardiac transcriptional factors during in vitro cardiomyocyte differentiation from human embryonic stem cells.
Keywords: Long noncoding RNA, Human heart, Cardiomyocyte differentiation, CRISPR, Time-course, Cardiac development
Abbreviations: ASD, Atrial Septal Defect; SNP, Single Nucleotide Polymorphism; GWAS, Genome-Wide Association Study; eQTL, Expression Quantitative Trait Loci; hESC, human Embryonic Stem Cell; CM, Cardiomyocyte; lncRNA, long noncoding RNA
Specification Table
| Subject | Biological sciences |
| Specific subject area | Cardiology and Cardiovascular Medicine Functional genetics Stem cell technology |
| Type of data | Figure Video Excel |
| How the data were acquired | Images and videos were recorded with the microscope. Expressional and genotyping data were acquired with qPCR and recorded in .xlsx format; analysed data were shown as graphs. Genomic locations were derived from the UCSC genome browser with the following link: https://genome.ucsc.edu. |
| Data format | Raw Analysed |
| Description of data collection | Human RNA and DNA samples for eQTL analyses were from patients undergoing cardiac surgeries collected in a previous project [1]. Human embryonic heart samples for in situ hybridisation were provided by the HDBR group. Functional experimental data were generated with in vitro human embryonic stem cell-cardiomyocytes differentiation model (hESC-CM) at D0, D2, D4, D6, D8, and D15. The control cells for experiments were transduced with CRISPR vector without targeting sgRNA sequences. |
| Data source location | Institution: Division of Cardiovascular Sciences, University of Manchester City: Manchester Country: UK |
| Data accessibility | Analysed data and figures are available in the article. Raw data of eQTL are available in “Raw data for STX18-AS1”, Mendeley Data, V1, doi: 10.17632/rpzs6fcdhz.1 Supplementary videos are in “Video supplements”, Mendeley Data, V1, doi: 10.17632/rw949cx3y8.1 |
| Related research article | Yingjuan Liu, Mun-kit Choy, Sabu Abraham, Gennadiy Tenin, Graeme C. Black, Bernard D. Keavney. “Atrial Septal Defect (ASD) associated long noncoding RNA STX18-AS1 maintains time-course of in vitro cardiomyocyte differentiation” Genes and Diseases, DOI:10.1016/j.gendis.2022.07.010. |
Value of the Data
-
•
This data supports the functional study of a GWAS-identified long noncoding RNA gene, STX18-AS1. It describes the step-by-step investigations of a de novo ASD risk lncRNA gene, using online genetic tools, eQTL analyses, expressional quantifications, and gene functional evaluations with gene editing and in vitro hESC-CM differentiation model.
-
•
This data provides the genotyping results of top risk ASD SNPs, rs870142, rs16835979 and rs6864295, and expression of STX18-AS1 in human atrial appendage samples. Also, this data provides the time-course expression of key cardiac transcriptional factors during D0-D15, which will be useful reference information for researchers attempting to use human expressional data and in vitro hESC-CM model in their own projects.
-
•
This data provides the whole-mount stained human embryonic hearts and sections, which is of rare availability. This will be beneficial resource for the understanding of human heart morphology at very early developmental stages.
Data Description
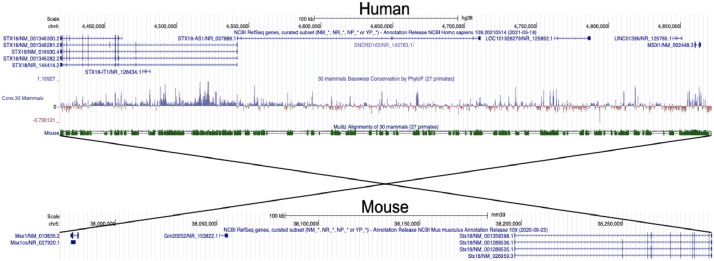
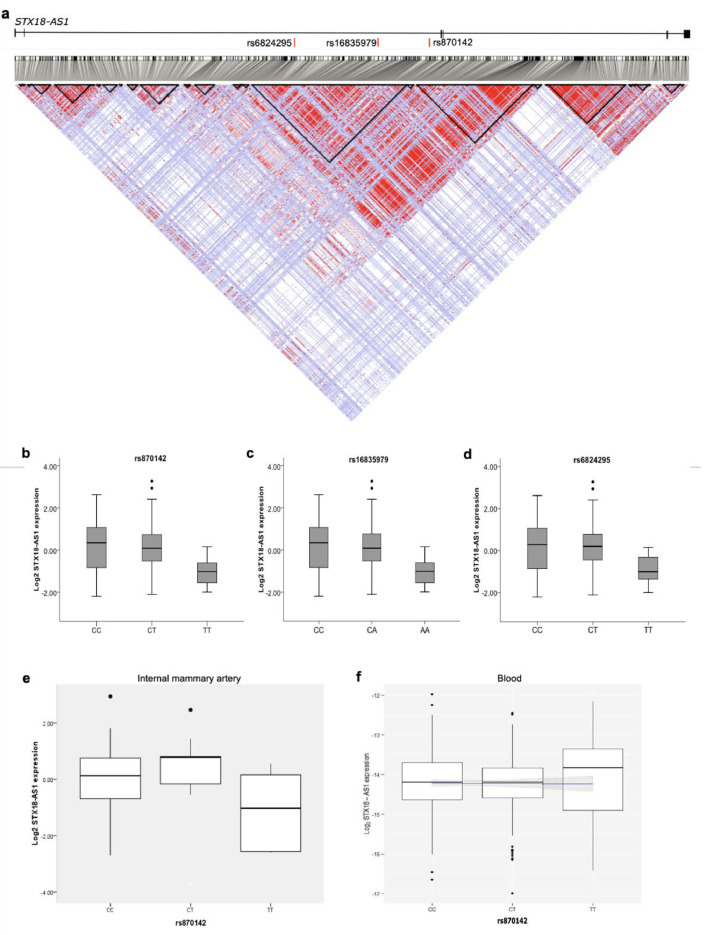
STX18-AS1 is the long noncoding RNA harbouring three top risk SNPs (rs870142, rs16835979, and rs6824295) for Atrial Septal Defect identified from a Genome-wide association study (GWAS). This gene is located at Chr4p16 neighbouring STX18 and MSX1. The first exon of STX18-AS1 overlaps with a rare transcript of STX18 but no other common isoforms. STX18-AS1 in the sequence is not a conserved lncRNA gene across species, like the mouse (Fig. 1). The homologues of this gene can only be identified in primates. The three SNPs are highly correlated and fall in the same linkage disequilibrium block (Fig. 2a). With 108 human atrial appendage samples, all three SNPs were eQTL for STX18-AS1, whereby the minor alleles produced less transcription of STX18-AS1 (Fig. 2b-d). However, the correlation between SNPs genotypes and STX18-AS1 transcriptions was not found in internal mammary artery samples or blood samples (Fig. 2e-f), indicating the SNPs are tissue type-specific eQTLs for STX18-AS1.
Fig. 1.
Genomic location and conservation scores of STX18-AS1 and surrounding genes between Human and Mouse genome. STX18 and MSX1 are the two neighbouring genes of STX18-AS1 in humans. No annotated genes at a similar location between STX18 and MSX1 are identified in the mouse genome. Phylop score shows the conservation score spare species. The sequences across STX18-AS1 have lower Phylop scores than neighbouring genes.
Fig. 2.
Risk SNPs for ASD and eQTL analyses between SNPs and STX18-AS1. a, Linkage disequilibrium (LD) across STX18-AS1 with SNP data from CEU population of 1000 Genomes Project. The LD map was generated with HaploView. The LD of SNP pairs is represented as r2 in white to red (0-1). The locations of the three risk SNPs identified in GWAS are labelled in red bars at the top of the figure. The black triangles represent haplotype blocks (defining with confidence intervals according to Gabriel's method). b-d, eQTL analyses with 108 human heart right atrial appendage (RAA) samples using the qPCR method for three top SNPs identified from GWAS, rs870142 (b), rs16835979(c), and rs6824295(d). The significant association between SNPs and STX18-AS1 transcription was tested using the linear regression model, with P values between 0.038-0.039. e-f, The eQTL analyses of rs870142 and STX18-AS1 transcription in internal mammary artery (IMA) samples (e) and human blood samples (f). The linear regressions are not significant for both IMA and blood eQTL analyses.
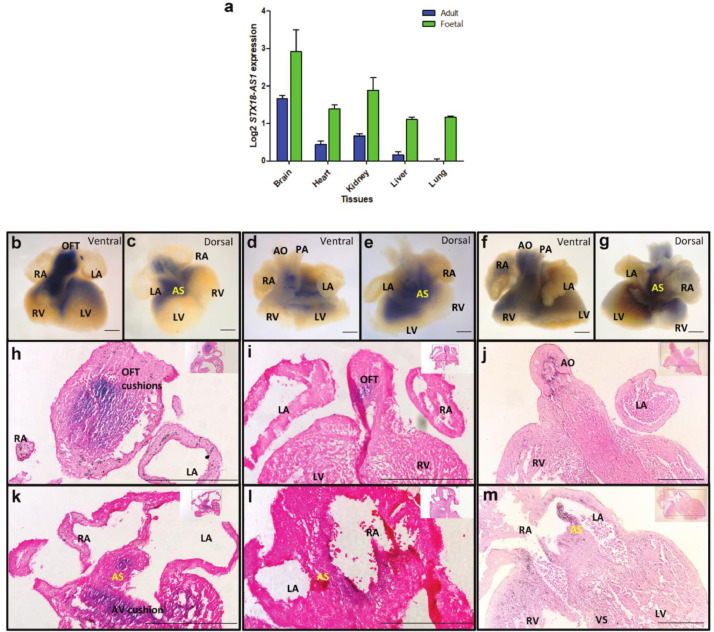
In alignment with the tissue dependent eQTLs, the distribution of STX18-AS1 expression is also relevant to developmental stages and tissue structures. Among all five available human tissues samples with both adult and foetal stages, STX18-AS1 transcripts were detected in all tissue types with higher levels in developing tissues (foetal) than adult tissues (Fig. 3a). Though cardiac STX18-AS1 is not a cardiac-specific gene, its transcription in developing hearts is prominently detected with in situ hybridisation with an antisense STX18-AS1 probe. From human embryonic whole heart samples of Carnegie stage 17, 18, and 19 (Fig. 3b-g), the transcripts of STX18-AS1 are mainly centred at the out-flow tract (OFT) (Fig. 3h-j), atrial septum (AS), atrio-ventricular cushions (AVC), and part of ventricles (Fig. 3k-m), accompanying with the key stages of heart septation into four chambers. Along with the development stages of hearts, the distribution of STX18-AS1 transcripts is more restricted to developing septum and valves but less in ventricles (Fig. 3b-g).
Fig. 3.
STX18-AS1 expression in human tissues and developing hearts. a, comparison of STX18-AS1 transcription in Adult and Foetal tissues (cDNA from Human MTCTM Panel I and Human Foetal MTCTM Panel). b-g, whole-mount ISH of human embryonic hearts (b-c, CS17; d-e, CS18; f-g, CS19) from ventral and dorsal views. Blue colour indicates the STX18-AS1 expression in hearts. h-j, the section of all three ISH hearts (h, CS17 heart; i, CS18 heart; j, CS19 heart) with cuts at the view of OFT. k-m, section of all three ISH hearts (k, CS17 heart; l, CS18 heart; m, CS19 heart) with cuts at the view of AS. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; OFT, out-flow tract; AS, atrial septum (yellow); AO, aorta; PA, pulmonary artery; VS, ventricular septum. STX18-AS1 transcripts are stained blue and counterstained with Eosin.; scale bars are 200µm.
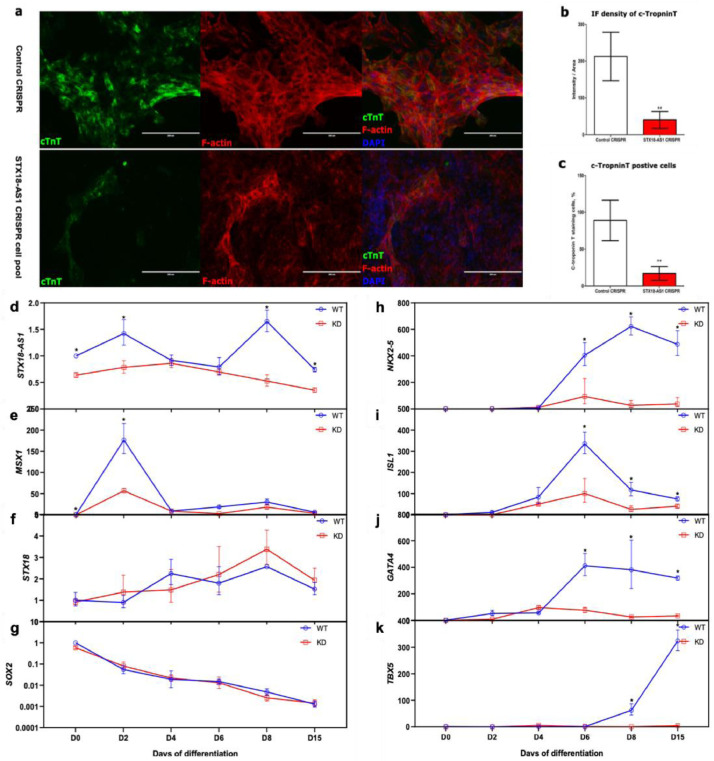
STX18-AS1 is expressed in hESC-derived cardiomyocytes with an appropriate peaking transcription level surrounding Day8 and Day2 of in vitro hESC-CM differentiation (Fig. 4d). The STX18-AS1 transcripts are comparatively higher in early-stage cardiomyocytes (Day8) than relative mature stage (Day15), commensurate with its expression pattern in human tissues. Using CRISPR knockdown, STX18-AS1 transcription in the mixed cell pool of STX18-AS1 CRISPR (which are CRISPR transduced cells following puromycine selection, containing STX18-AS1 deleted and non-deleted cells) is reduced by about 40-70% across the hESC-CM differentiation periods (Fig. 4d). Along with the haploinsufficiency of STX18-AS1 in hESCs, the differentiation into CM was interrupted with fewer beating cell colonies observed after 8 days of differentiation (Supplementary videos, doi: 10.17632/rw949cx3y8.1). Cardiomyocytes derived from the STX18-AS1 CRISPR cell pool were fewer in cTroponin-positive cell numbers and cTroponin expression levels (Fig. 4a-c). In accordance with the reduced CM differentiation efficiency, transcriptions of key cardiac transcriptional factors, including NKX2-5, ISL1, GATA4, and TBX5 during the hESC-CM differentiation, are also decreased at the stages of Day6-Day15, the period of cardiac lineage specification from cardiac mesoderm and cardiac progenitor into cardiomyocytes (Fig. 4h-k). No apparent difference in the time-course of pluripotency marker SOX2 and neighbouring gene STX18 was identified across the differentiation procedures (Fig. 4f-g). The reduction of another neighbouring gene, MSX1, at Day2 (Fig. 4e) does not explain the inhibited CM differentiation efficiency, according to a previous report that MSX1 deficiency will minimise the requirement of WNT inhibition for CM differentiation and promote the CM differentiation rate [2].
Fig. 4.
STX18-AS1 haploinsufficiency restricted in vitro hESC-cardiomyocyte differentiation. a, Immunostaining of cTNT (red fluorescence, referencing to DAPI in blue) at Day 8 of differentiation in both Control CRISPR hESC-CMs and STX18-AS1 CRISPR cell pool (KD in following gragh) derived CMs. Scale bars represent 200µm. b-c, the quantitative analyses of cTNT immunostaining on fluorescence density (b) and cTNT-positive cells (c). **, P< 0.01, using t-test. d, Time-course expression of STX18-AS1 during the CM differentiation from Control CRISPR and STX18-AS1 CRISPR cell pool. e-k, the time-course transcription changes of neighbouring genes [MSX1 (e) and STX18 (f)], pluripotency marker [SOX2(g)], and key cardiac transcriptional factors [NKX2-5(h), ISL1 (i), GATA4 (j), and TBX5 (k)] during D0-D15 of hESC-CM differentiation. *, P< 0.05. Two-way ANOVA test with Bonferroni adjustment is applied for generating the P values for comparisons at each time point (n>=3).
Experimental Design, Materials and Methods
The genomic structures, sequences and conservation scores of STX18-AS1 are acquired using online genome browser UCSC (https://genome-euro.ucsc.edu) with the human genome of hg38 and mouse genome mm10.
Expression data for human tissues were generated with quantitative PCR using TaqMan probes (Table 1) and pooled cDNA samples of human foetal tissues (Fetal MTC Panel, 636747, Lot#1308272A) and adult tissues (MTC panel I, 636742, Lot#1303120A) sourced from Clontech. Human RNA and DNA samples for eQTL analyses were from patients undergoing cardiac surgeries collected in our previous project [1].
Table 1.
TaqMan assay probes for qPCR.
| Target | Purpose | Assay ID |
|---|---|---|
| rs870142 | Genotyping | C___8840003_10 |
| rs16835979 | Genotyping | C__34027652_20 |
| rs6824295 | Genotyping | C__29284797_10 |
| STX18-AS1 | Gene expression | Hs00416742_m1 |
| NKX2-5 | Gene expression | Hs00231763_m1 |
| GATA4 | Gene expression | Hs00171403_m1 |
| MSX1 | Gene expression | MSXPROB design from [7] |
| TBX5 | Gene expression | Hs00361155_m1 |
| ISL1 | Gene expression | Hs00158126_m1 |
| STX18 | Gene expression | Hs00560288_g1 |
| SOX2 | Gene expression | Hs01053049_s1 |
| IPO8 | Gene expression | Hs00183533_m1 |
In situ hybridisation. The whole human embryonic hearts for in situ hybridisation were fixed in 4% PFA and obtained from the MRC-Wellcome HDBR. Whole-mount in situ hybridisation (ISH) was conducted as previously described [3]. Using the human foetal heart cDNA as a template, DIG-labeled STX18-AS1 RNA probes antisense to the first three exons of STX18-AS1 (514bp) were produced via in vitro RNA syntheses. Primers for RNA probe synthesis: Forward primer (GCGAGCTCTTCTGTGTCTGT) and reverse primer (TGCTGGAAGACACAGGCTTT) tagged by T3 sequence (AATTAACCCTCACTAAAGGG), which is the recognisable start site for in vitro transcription with T3 polymerase (Promega) and DIG RNA probe labelling kit (Roche). The antisense RNA probes are used for ISH of three whole embryonic hearts of CS17, CS18, AND CS19, the stages of atrial septation with peaks of STX18-AS1 expression. Hearts following ISH were dehydrated with 50% Ethanol, 75% Ethanol, 100% Ethanol gradient (30min each) and 100% Ethanol overnight. After histological clearing with successive incubation (30min) in 50% HistoClear (with Ethanol, RT), 100% HistoClear (RT), and 100% HistoClear (65℃), the hearts were stabilised in paraffin (65℃) overnight and subjected to embedding and Microtome sectioning (Leica RM2145). Sections were counterstained with Eosin and then mounted with DPX Mounting Media (Merck) for observation.
SNP eQTL analyses. DNA (blood-derived) and RNA from 108 right atrial appendage (RAA) samples were extracted as previously described [1]. Three SNPs were genotyped with Taqman genotyping probes using qPCR, while the transcription of STX18-AS1 and housekeeping control gene IPO8 were quantified with Taqman expression probes. The correlation between genotypes and transcription levels was analysed with the linear model using SPSS14.0.
Cell culture. HEK293T cells were maintained in DMEM complete medium (DMEM [Gibco] supplemented with 10% FBS and 100 UI of Penicillin/Streptomycin). H9 human embryonic stem cell line [WiCell] was maintained with mTesR1 plus medium (STEMCELL Technologies) supplemented with 10µM ROCK inhibitor (Millipore) in Matrigel-coated plates.
CRISPR/Cas9 design. In deleting a region of about 3-4kb, sequences of ∼1kb at both ends of the targeted region were extracted and used as templates for sgRNA designs. The CRISPR sgRNAs were designed with an online tool developed by Zhang's Lab [4] (http://crispr.mit.edu/) and ATUM grna design tool (https://www.atum.bio/eCommerce/cas9/input). Designs with high scores in both tools were selected for CRISPR construction. AACCGCCCGGTCTCAGTGAGGG is the upstream sgRNA, and CAGCAGCAACACCTATGCAAGG is the downstream sgRNA, with underlining PAMs. The pairs of sgRNAs intermediated by the human U6 (hU6) promoter were cloned into plentiCRISPR_V2 plasmid (Addgene, #52961) following the original hU6 promoter sequence according to Vidigal et al.’s protocol [5]. The primers designed for the detection of deletions are located outsides the targeted regions with CGGAATAGCAGCGTGATGTC as the forward primer and TGTCCTTGGTTGGCTATGCT as the reverse primer, showing a product of 600bp for deletions and 3.4kb for non-deletions.
Lentivirus construction and transduction. The plasmids were packaged into lentivirus by co-transfecting HEK293T cells with pPAX2 (Addgene, #35002) and pMD2.G (Addgene, #12259) in a proportion of 4:3:2 using Lipofectamine 2000 (ThermoFisher). After transfection, the medium of the first 12 hours was discarded and changed into fresh DMEM complete medium. The lentivirus was collected and filtered (PVDF 0.45µm) after additional 48 hours for immediate use in transduction or aliquoted and stored at -80℃ for future use. Triple lentivirus transductions of H9 cells (Passage 35-40) were conducted for getting the CRISPR cell pool before puromycine selection. The first transduction started at the confluency of 40-60%. For each transduction, cells were permeabilised first with 1ml/well (6-well plate) mTESR1 plus medium with 8µg/ml polybrene (Millipore) for 15 min (37℃). Afterwards, 2ml lentivirus soup with 8µg/ml polybrene was added (24 hours). The transduction procedures were conducted successively for three days. After the 3rd transduction, cells were maintained in mTesR1 plus medium for 1-2 days before puromycine selection (0.8µg/ml puromycine [ThermoFisher]). Transduced cells were maintained in 0.8µg/ml puromycine at least 7 days before subjected to CM differentiation and other analyses.
Cardiomyocyte differentiation. Cardiomyocyte differentiation was performed as previously described [6] and summarised as follows. The differentiation was started on Day 0 with confluency of 90-100% hESCs (H9 line). For the first two days, hESCs were treated with 6µM Chir99021 (GST inhibitor [Millipore]) in B27- medium (RPMI 1640 medium [Invitrogen] supplemented with 1× B27 minus insulin [50×, Gibco]). On Day 2-4, cells were changed to 2µM C59 (WNT antagonist [Abcam]) in B27- medium. Day 4-8, cells were maintained in B27- medium. All mediums were refreshed every day. From Day 8, cells were maintained with B27+ (1×B27 [50×, Gibco] supplemented RPMI 1640 medium [Invitrogen]) till Day15. Beating cells were typically started to be seen from Day 6-8.
Quantitative polymerase chain reaction (PCR). RNA from hESCs and derived cells were extracted with Trizol (Invitrogen) following the manufacturer's protocol. The first-strand cDNA was synthesised using M-MLV Reverse Transcriptase (Promega) with both random hexamer and oligo(dT) primers (Promega) after the DNase (Promega) treatment. Gene expression was quantified by TaqMan assays (ThermoFisher, Table 1) using a ViiA7 qPCR system (Life Technologies). For each qPCR reaction, ∼40ng cDNA templates were used with 2.5ul TaqMan™ Gene Expression Master Mix (2×) and 0.25ul TaqMan assay probes (20×). Three replicates were done for each reaction with the program of 95℃×10min, 40× (95℃×30s, 60℃×1min).
Immunofluorescence (IF). Cells in the plate were fixed with 4% PFA for 15min at room temperature (RT). After three PBT (PBS with 0.5% Triton-100) washes, cells were blocked with blocking buffer (PBT with 10% sheep serum) for 30min at RT. Cells were then incubated with primary antibody c-TroponinT (rabbit, Abcam, 1:200 diluted in PBT with 1% sheep serum) overnight at 4℃. The next day, cells were incubated with fluorophore-conjugated secondary antibodies goat-anti-rabbit H&L FITC (Cohesion Biosciences, 1:200, 1 hour at RT in the dark) after PBT washes. Following additional washes, cells were incubated with DAPI (1:1000 [Invitrogen] in PBS) and AF680-Phalloidin (F-actin, 1:1000 [Invitrogen]) for 20min at RT. After three PBT washes, cells were photographed using a fluorescence microscope (EVOS FL). The intensity of the fluorescent signal was analysed with ImageJ [8].
Ethics Statements
Human foetal materials obtained from the MRC/Wellcome Human Developmental Biology Resource were under NHS Research Ethics references 18/LO/0822 and 18/NE/0290. All participants providing atrial and blood samples used for eQTL analyses provided informed consent, and samples were collected under NHS Research Ethics reference 12/NE/0072. The study conforms to the principles of the Declaration of Helsinki.
CRediT authorship contribution statement
Yingjuan Liu: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft. Mun-kit Choy: Methodology, Resources, Visualization. Sabu Abraham: Methodology, Resources, Visualization. Gennadiy Tenin: Methodology, Resources, Visualization. Graeme C. Black: Supervision. Bernard D. Keavney: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by The University of Manchester-Peking University Health Science Centre Alliance, China Scholarships Council, and BHF Progamme Grant RG/15/12/31616 and RG/F/21/110050. BK holds a British Heart Foundation Personal Chair and the BHF Accelerator Award AA/18/4/34221. We are thankful to Dr Ruairidh Martin for the collection of the samples for eQTL analyses and the data of peripheral blood. We thank HDBR group for supplying human hearts samples for expression analyses.
Contributor Information
Yingjuan Liu, Email: yingjuan.liu@manchester.ac.uk.
Bernard D. Keavney, Email: bernard.keavney@manchester.ac.uk.
Data Availability
Raw data for STX18-AS1 (Original data) (Mendeley Data).
Video supplements (Original data) (Mendeley Data).
References
- 1.Martin R.I., Babaei M.S., Choy M.K., Owens W.A., Chico T.J., Keenan D., Yonan N., Koref M.S., Keavney B.D. Genetic variants associated with risk of atrial fibrillation regulate expression of PITX2, CAV1, MYOZ1, C9orf3 and FANCC. J. Mol. Cell Cardiol. 2015;85:207–214. doi: 10.1016/j.yjmcc.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Rao J., Pfeiffer M.J., Frank S., Adachi K., Piccini I., Quaranta R., Arauzo-Bravo M., Schwarz J., Schade D., Leidel S., Scholer H.R., Seebohm G., Greber B. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016;18(3):341–353. doi: 10.1016/j.stem.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Henrique D., Adam J., Myat A., Chitnis A., Lewis J., Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375(6534):787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 4.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vidigal J.A., Ventura A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat. Commun. 2015;6(1):8083. doi: 10.1038/ncomms9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venza I., Visalli M., Parrillo L., De Felice M., Teti D., Venza M. MSX1 and TGF-β3 are novel target genes functionally regulated by FOXE1. Hum. Mol. Genet. 2010;20(5):1016–1025. doi: 10.1093/hmg/ddq547. [DOI] [PubMed] [Google Scholar]
- 8.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data for STX18-AS1 (Original data) (Mendeley Data).
Video supplements (Original data) (Mendeley Data).