Abstract
Claudin 18.2 (CLDN18.2) is an emerging target for the treatment of gastric cancers. We aim to develop tracers to image the expression of CLDN18.2. A humanized nanobody targeting CLDN18.2 (clone hu19V3) was produced and labeled with 68Ga, 64Cu, and 18F. The tracers were investigated in subcutaneous and metastatic models established using two different mouse types (nude and Balb/c mice) and two different cell lines (CHO-CLDN18.2 and CT26-CLDN18.2). Gastric cancer patient-derived xenograft (PDX) models were further established for validation experiments. Three novel CLDN18.2-targeted tracers (i.e., [68Ga]Ga-NOTA-hu19V3, [64Cu]Cu-NOTA-hu19V3, and [18F]F-hu19V3) were developed with good radiochemical yields and excellent radiochemical purities. [68Ga]Ga-NOTA-hu19V3 immuno-positron emission tomography (immunoPET) rapidly delineated subcutaneous CHO-CLDN18.2 lesions and CT26-CLDN18.2 tumors, as well as showing excellent diagnostic value in PDX models naturally expressing CLDN18.2. While [68Ga]Ga-NOTA-hu19V3 had high kidney accumulation, [64Cu]Cu-NOTA-hu19V3 showed reduced kidney accumulation and improved image contrast at late time points. Moreover, [18F]F-hu19V3 was developed via click chemistry reaction under mild conditions and precisely disseminated CHO-CLDN18.2 lesions in the lungs. Furthermore, region of interest analysis, biodistribution study, and histopathological staining results correlated well with the in vivo imaging results. Taken together, immunoPET imaging with the three tracers can reliably visualize CLDN18.2 expression.
Keywords: Claudin 18.2, immunoPET, nanobody, radiopharmaceuticals, companion diagnostics
Graphical abstract

Claudin 18.2 (CLDN18.2) is a biomarker for developing novel cancer therapeutics. There is an urgent need to develop approaches to map the dynamics of CLDN18.2. We developed three novel molecular imaging tracers noninvasively visualizing CLDN18.2. Clinical translation of the tracers may optimize the use of CLDN18.2-targeted antibody and cell therapies.
Introduction
Claudin 18.2 (CLDN18.2) is exclusively expressed in tight junctions of gastric mucosal cells, remaining inaccessible to intravenously administered antibodies.1 With the malignant transformation of gastric mucosal cells, CLDN18.2 is exposed and becomes available by therapeutic antibodies. Zolbetuximab (IMAB362) is a chimeric monoclonal antibody (mAb) and mediates specific killing of CLDN18.2-positive cells through antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity.2 Following initial studies reporting the manageable safety profiles and anti-tumor activity of zolbetuximab in patients with gastric or gastro-esophageal junction patients,3,4 a more recent randomized phase II study reported that the addition of zolbetuximab to the first-line EOX (epirubicin + oxaliplatin + capecitabine) significantly improved the progression-free survival and overall survival than that obtained in the EOX treatment group.5 The novel treatment option also improved the quality of life of the included patients.6 Ongoing phase III studies are investigating the therapeutic efficacy of zolbetuximab 800/600 mg/m2 in patients with moderate to strong CLDN18.2 expression (>70% of tumor cells). Currently, CLDN18.2 is among the most promising targets that can be leveraged to improve the theranostic landscape of gastric cancers. As far as we know, immunohistochemistry (IHC) staining with CLAUDETECT 18.2 is the only available option to determine CLDN18.2 expression. However, IHC is limited to examine the surgically resected tissue or the biopsied tissue, failing to provide the heterogeneous expression of CLDN18.2 across the whole tumor tissue or in the metastases. In this setting, novel techniques that can determine the expression level of Claudin18.2 and further diagnose CLDN18.2-expressing tumors are urgently needed.
Immuno-positron emission tomography, also known as immunoPET, is a rapidly expanding direction in the field of molecular imaging and in the era of precision medicine.7 ImmunoPET has a well-recognized role in guiding the early development of antibody therapeutics and is increasingly used in clinical practice to select patients for molecularly targeted therapies and assess the responses following the therapies. By labeling mAbs with radiometals (e.g., 64Cu or 89Zr),8,9,10,11,12 we have developed several novel immunoPET probes and characterized the diagnostic efficacies in preclinical settings. Admittedly, the clinical translational way of these probes is arduous and long. Hurdles impeding the clinical translation include high expenditure in producing humanized mAbs, scarcity of long-lived radiometals or radiohalogens, multiple cycles of imaging across a week, and the resultant high radiation exposure to patients and staff. To alleviate these concerns and facilitate same-day imaging, antibody derivatives and protein scaffold mimicking antibodies have been leveraged to develop next-generation molecular imaging probes.7,13 One such example is the variable region of the heavy chain of heavy chain-only antibody (VHH), which is also known as nanobody. Nanobodies are naturally occurring antigen recognition domains mainly found in alpacas, llamas, and camels.14 They are 12–15 kDa in size, highly stable in various conditions, display strong binding affinities, and can be expressed in bacterial, yeast, and mammalian expression systems with high yields. Nanobodies are the most promising candidates to substitute their full-size counterparts for at least diagnostic applications.15 Nanobody-derived immunoPET probes targeting human epidermal growth factor receptor 2 and programmed death ligand-1 have been successfully translated to the bedside.16,17,18 We have established screening technologies and produced a variety of nanobodies targeting pivotal biomarkers highly expressed in either hematological or solid tumors. In recent work, we designed nanobody-based CD38-targeted probes [68Ga]Ga-NOTA-Nb1053/[18F]F-Nb1053 and further reported the excellent diagnostic potencies in multiple myeloma models.19,20 We also developed [68Ga]Ga-NOTA-SNA006 to noninvasively map human CD8+ T cells.21 Nanobody-based probes are favored for clinical translation and routine applications in our view.
To the best of our knowledge, there are no reliable molecular imaging tracers specific for CLDN18.2 so far. In this work, we aim to develop a series of nanobody-based tracers to noninvasively visualize CLDN18.2 expression and diagnose CLDN18.2-expressing tumors. Such tracers may also help optimize the preclinical development and clinical use of CLDN18.2-targeted therapies.
Results
Validation of CLDN18.2 expression in normal mice tissues
Human and mouse CLDN18.2 have identical amino acid sequences.1 From the literature report and public database (https://www.proteinatlas.org/ENSG00000066405-CLDN18/tissue), we can know that the expression of CLDN18.2 is strictly restricted to the stomach. We first stained archived normal nude mouse tissues and validated the highest expression of CLDN18.2 in the stomach (Figure 1). Negligible expression was found in the liver, kidney, lung, pancreas, or spleen. Under normal conditions, CLDN18.2 is located in gastric mucosal cells and is isolated from the external environment. Malignant transformation of the gastric mucosal cell leads to exposure of CLDN18.2 epitopes and opens the therapeutic window.22 With the above information in hand, we can better develop molecular imaging tracers and interpret the imaging results.
Figure 1.
Expression of CLDN18.2 in normal mouse tissues
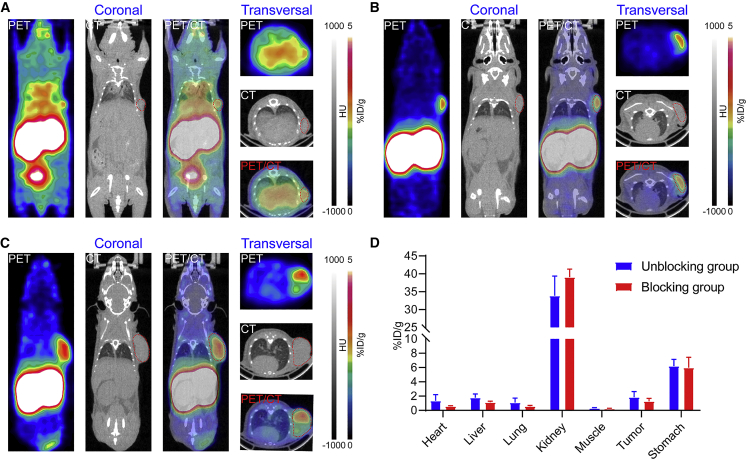
[68Ga]Ga-NOTA-hu19V3 immunoPET imaging of subcutaneous CHO-CLDN18.2
In our efforts to develop molecular imaging tracers targeting CLDN18.2, [68Ga]Ga-NOTA-hu19V3 was first synthesized. The RCP of the radiopharmaceutical meets the requirement for in vivo imaging (Figure S1). Two groups of mice subcutaneously inoculated with CHO-CLDN18.2 cells were used for imaging experiments, where one group of mice was pre-injected with an excess dose of hu19V3 (1 mg/mouse). [68Ga]Ga-NOTA-hu19V3 immunoPET imaging not only detected tiny subcutaneous CHO-CLDN18.2 lesions (Figure 2A), but it also rapidly detected CHO-CLDN18.2 lesions of relatively larger volumes (Figures 2B and 2C). In another two groups of tumor-bearing mice, premedication with hu19V3 reduced the tumor and stomach uptake (Figure S2). But the region of interest (ROI) analysis showed no statistical significance (Figure 2D). The tumor uptake reduced from 1.85% ± 0.79% ID/g to 1.27% ± 0.42% ID/g (n = 5, P = 0.229), while the stomach uptake reduced from 6.20% ± 0.95% ID/g to 5.98% ± 1.46% ID/g (n = 5, P = 0.787). Although the rapid clearance of hu19V3 from the circulation led to less thorough blocking effect, the preferential uptake of the tracer in the stomach demonstrated high specificity of the tracer.
Figure 2.
[68Ga]Ga-NOTA-hu19V3 immunoPET imaging precisely detected CHO-CLDN18.2 lesions
(A–C) [68Ga]Ga-NOTA-hu19V3 immunoPET imaging of CHO-CLDN18.2 lesions of different volumes 1 hour after injection of the tracer. The tumors are indicated by red circles on PET and PET/CT images. (D) Region of interest analysis of the immunoPET imaging data.
In-depth analysis of the biodistribution data confirmed the comparable uptake of the tracer in the tumors and major tissues in the two groups (Figure 3A). Premedication with excess hu19V3 reduced the uptake of [68Ga]Ga-NOTA-hu19V3 in the stomach and tumor, two organs where CLDN18.2-targeted agents are supposed to accumulate at high levels. Based on the biodistribution data, the stomach uptake dropped from 2.77% ± 1.02% ID/g to 1.71% ± 0.58% ID/g (n = 5, P = 0.556), and the corresponding tumor uptake dropped from 3.62% ± 0.83% ID/g to 2.49% ± 0.66% ID/g (n = 5, P = 0.389). The results demonstrated the specificity of [68Ga]Ga-NOTA-hu19V3 in visualizing CLDN18.2 expression in CHO-CLDN18.2 models. Use of hu19V3 reduced uptake of [68Ga]Ga-NOTA-hu19V3 in the stomach and tumor without statistical significance, assumably due to the short half-life of the single valent hu19V3. Histopathological staining studies showed positive expression of CLDN18.2 on the CHO cells, reflecting the stable expression of the protein in in vivo conditions (Figures 3B and 3C). The above data together indicated the in vivo targeting ability and specificity of [68Ga]Ga-NOTA-hu19V3.
Figure 3.
Biodistribution and histopathological staining results
(A) Biodistribution data showing detailed uptake of [68Ga]Ga-NOTA-hu19V3 in various organs/tissues with or without hu19V3 premedication. (B) H&E staining of a collected CHO-CLDN18.2 lesion. (C) Immunohistochemistry staining of CLDN18.2 expression in the CHO-CLDN18.2 lesion (left). CLDN18.2 is preferentially expressed at the edge (red arrowhead, middle) but not at the necrotic areas or center of the lesion (black arrowhead, right).
[64Cu]Cu-NOTA-hu19V3 immunoPET imaging of disseminated CHO-CLDN18.2
One phenomenon of [68Ga]Ga-NOTA-hu19V3 is the compelling accumulation in the kidneys. Currently, there is limited evidence labeling nanobodies with long-lived radionuclides. We wondered if the kidney accumulation of the radiometal-labeled nanobody could decrease in a relatively long imaging period. For this purpose, we developed [64Cu]Cu-NOTA-hu19V3 and investigated the pharmacokinetic and pharmacodynamic traits in nude mice models, in which CHO-CLDN18.2 cells were intravenously injected 18 days before tracer administration. [64Cu]Cu-NOTA-hu19V3 was developed with RCY of 61.54% and RCP of 100% (Figure S3). [64Cu]Cu-NOTA-hu19V3 immunoPET imaging at three time points (1 h, 24 h, and 48 h; Figures 4A–4C) showed rapid clearance of the tracer from the circulation. The high kidney accumulation decreased time dependently. Specifically, the initial uptake of 13.73% ± 2.16% ID/g at 1 h post-injection substantially decreased to 0.11% ± 0.05% ID/g at 48 h post-injection (p = 0.0021, n = 4; Figure 4E). In general, [64Cu]Cu-NOTA-hu19V3 showed excellent imaging contrast as the kidney accumulation reduced to the background level (Figures 4C and 4E). Apart from the lower kidney accumulation, [64Cu]Cu-NOTA-hu19V3 produced images of improved contrast. Although [64Cu]Cu-NOTA-hu19V3 immunoPET delineated stomach (Figure 4D), it did not detect any disseminated CLDN18.2-expressing CHO lesions, probably due to the limited detection efficacy at the early stages or failed growth and formation of lesions of the intravenously injected CHO-CLDN18.2 cells.
Figure 4.
[64Cu]Cu-NOTA-hu19V3 showed favorable pharmacokinetics
(A–C) [64Cu]Cu-NOTA-hu19V3 immunoPET imaging at different time points (1 h, 24 h, and 48 h) post-injection of the tracer. [64Cu]Cu-NOTA-hu19V3 demonstrated improved image contrast and reduced kidney accumulation. D [64Cu]Cu-NOTA-hu19V3 accumulated in the stomach (shown by red circles on CT and PET/CT images), indicating the in vivo affinity and specificity of the tracer. (E) Region of interest analysis showing the dynamic change of [64Cu]Cu-NOTA-hu19V3 in different organs/tissues over time.
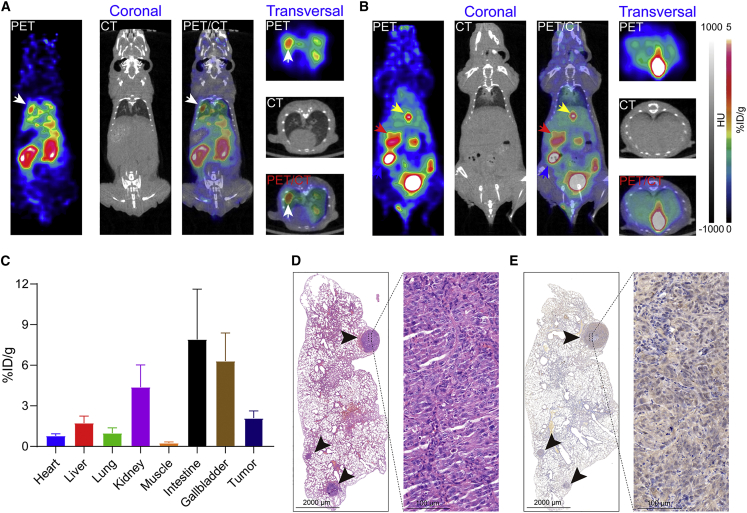
[18F]F-hu19V3 immunoPET imaging of disseminated CHO-CLDN18.2
By taking advantage of click chemistry,23 we further developed the third CLDN18.2-targeted tracer [18F]F-hu19V3 with RCP of 100% and non-decay corrected RCY of 5.72% (Figure S4). Another group of tumor-bearing mice models was used to evaluate the diagnostic efficacy of [18F]F-hu19V3, with a time lag of 28 days between tumor cell inoculation and PET imaging. As shown in Figure 5A, immunoPET imaging with [18F]F-hu19V3 delineated multiple signals in the lungs. Different from [68Ga]Ga-NOTA-hu19V3 and [64Cu]Cu-NOTA-hu19V3, which largely underwent renal clearance, [18F]F-hu19V3 was cleared from the urinary and hepatobiliary systems (Figure 5B). The uptake in the kidney, gallbladder, and intestine was 4.38% ± 1.65% ID/g (n = 4), 6.30% ± 2.08% ID/g (n = 4), and 7.90% ± 3.72% ID/g (n = 4), respectively. The average uptake of [18F]F-hu19V3 in the disseminated CHO-CLDN18.2 lesions was 2.10% ± 0.53% ID/g (n = 4) (Figure 5C). After termination of the imaging, major organs (heart, liver, lung, kidney, spleen, pancreas, and stomach) were collected and used for histopathological studies. Hematoxylin and eosin (H&E) staining showed disseminated CHO lesions in the left lung (Figure 5D). IHC staining further revealed intensive CLDN18.2 staining in the specific CHO lesions (Figure 5E). These results demonstrate that [18F]F-hu19V3 immunoPET can visualize the expression of disseminated CLDN18.2-expressing CHO lesions and may facilitate precise diagnosis of CLDN18.2-expressing metastases.
Figure 5.
18FF-hu19V3 immunoPET rapidly detected disseminated CHO-CLDN18.2 lesions
(A) 18FF-hu19V3 immunoPET detected a disseminated CHO-CLDN18.2 lesion in the left lung 1 hour after injection of the tracer. The CHO-CLDN18.2 lesion was indicated by the white arrowhead. The left panel shows coronal images, and the right panel shows transversal images. (B) 18FF-hu19V3 also accumulated in the stomach (red arrowhead) with the remaining excreted from the urinary and hepatobiliary system. The gallbladders and intestines are indicated by yellow and blue arrowheads, respectively. The left panel are coronal images, and the right panel are transversal images. (C) Region of interest analysis showing the detailed uptake of 18FF-hu19V3 in different organs/tissues. (D) H&E staining showed multiple metastases in the left lung. (E) Immunohistochemistry further showed positive expression of CLDN18.2 in the metastases, which were indicated by black arrowheads.
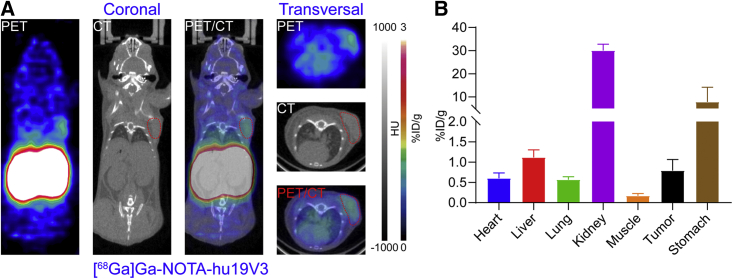
[68Ga]Ga-NOTA-hu19V3 immunoPET imaging of murine colorectal cancers
We further established CLDN18.2-overexpressing CT26 cell line (denoted as CLDN18.2-CT26) and Balb/c tumor models with the cell line. As expected, [68Ga]Ga-NOTA-hu19V3 immunoPET rapidly delineated the subcutaneous CLDN18.2-CT26 tumor and stomach (Figure 6A), with the latter naturally expressing CLDN18.2 at a higher level and consequently exhibiting a higher uptake of the tracer. ROI analysis results correlated well with the imaging findings and showed preferential uptake of [68Ga]Ga-NOTA-hu19V3 and staining of CLDN18.2 in the tumor and stomach (Figures 6B and S5), with the corresponding uptake value of 0.79% ± 0.28% ID/g and 7.80% ± 6.37% ID/g (n = 4), respectively. As expected, the tracer accumulated at the highest level at the kidneys (30.0% ± 2.81% ID/g, n = 4).
Figure 6.
[68Ga]Ga-NOTA-hu19V3 immunoPET imaging of murine colorectal cancers
(A) [68Ga]Ga-NOTA-hu19V3 immunoPET detected CLDN18.2-CT26 tumors (yellow circles) 1 hour after injection of the tracer. (B) Region of interest analysis showing the detailed uptake of [68Ga]Ga-NOTA-hu19V3 in different organs/tissues. The tumor is indicated by red circles on PET and PET/CT images.
[68Ga]Ga-NOTA-hu19V3 immunoPET imaging in PDX models
We further established and screened 12 patient-derived xenograft (PDX) models in our lab. Positive expression of CLDN18.2 was found in the No. 144 PDX model, a gastric cancer model maintaining glandular structures (Figure 7A). A low to negative expression of CLDN18.2 was found in the No. 490 PDX model (Figure 7D). These two gastric cancer models can be prepared with excellent reproducibility. In addition, CLDN18.2 expressed heterogeneously in other gastric cancer PDX models (Figure S6). Based on the above staining results of PDX models, we established No. 144 PDX models and validated the diagnostic value of [68Ga]Ga-NOTA-hu19V3 immunoPET in the models. The results demonstrated the potency of [68Ga]Ga-NOTA-hu19V3 in mapping CLDN18.2 and outlining the tumors (Figure 7C). ROI analysis data showed the detailed accumulation of the tracer in major organs/tissues (Figure 7D). Notably, tumor uptake of [68Ga]Ga-NOTA-hu19V3 was statistically higher in No. 144 PDX models than in CLDN18.2-CT26 models (2.53% ± 0.75% ID/g [n = 3] vs. 0.79% ± 0.28% ID/g [n = 4, P = 0.0168]). These results together demonstrate that [68Ga]Ga-NOTA-hu19V3 can detect CLDN18.2 expression in various models, but No. 144 PDX models were the best because of the natural and high expression of CLDN18.2.
Figure 7.
[68Ga]Ga-NOTA-hu19V3 immunoPET imaging in patient-derived xenograft (PDX) models
(A) No.144 was a representative gastric cancer PDX model showing positive expression of CLDN18.2. (B) Another representative gastric cancer PDX model showing negative expression of CLDN18.2. (C) [68Ga]Ga-NOTA-hu19V3 immunoPET imaging in No. 144 PDX models 1 hour after injection of the tracer. The tumor is indicated by red circles on CT and PET/CT images. (D) Region of interest analysis showing the detailed uptake of [68Ga]Ga-NOTA-hu19V3 in different organs/tissues.
Discussion
Since the expression of CLDN18.2 protein is highly conserved, an ideal CLDN18.2-targeted molecular imaging approach may identify CLDN18.2 expression across a variety of species including mouse, rat, rabbit, dog, monkey, and human.1,24 In this work, we have successfully developed three state-of-the-art CLDN18.2-targeted molecular imaging tracers. [68Ga]Ga-NOTA-hu19V3 immunoPET visualized CLDN18.2 expression in the stomach of mice, rats, and rabbits (data not shown). More importantly, [68Ga]Ga-NOTA-hu19V3 immunoPET delineated subcutaneously inoculated CHO-CLDN18.2 lesions and patient-derived tumors. Meanwhile, [18F]F-hu19V3 immunoPET detected intravenously injected CHO-CLDN18.2 cells, demonstrating the potency and accuracy of the novel imaging strategies. We also developed [64Cu]Cu-NOTA-hu19V3 to detect disseminated CLDN18.2 because delayed imaging at 24 h and 48 h post-injection of the tracer allowed improved image contrast. Admittedly, radiometals like 64Cu are relatively scarce and expensive. While [68Ga]Ga-NOTA-hu19V3 can only be produced on site for small-scale clinical use, [18F]F-hu19V3 can be produced in large amount and even shipped to other medical centers for multi-center clinical trials.25 Therefore, we believe that [68Ga]Ga-NOTA-hu19V3 and [18F]F-hu19V3 will have complementary roles in visualizing CLDN18.2 expression upon clinical translation.
Gastric and gastroesophageal junction adenocarcinomas are on the top list for CLDN18-targeted theranostics. However, accumulating evidence indicates that several other types of cancers may also express CLDN18.2. Previous studies reported CLDN18.2 expression in 60–90% of pancreatic ductal adenocarcinoma (PDAC). A more recent study reported that over 50% of primary and metastatic PDACs highly express CLDN18.2,26 adding to the validity of the previous evidence. Aberrant activation or expression of Claudin18.2 was also found in non-small-cell lung cancer and colitis-associated colorectal adenocarcinomas.27,28 These results indicate that CLDN18.2-targeted diagnostic or therapeutic agents would be in principle eligible for a considerable number of tumors.29 With close collaboration with oncologists at the hospital, we can investigate the diagnostic accuracies of the reported tracers in broad types of tumors in the future.
There are several limitations of the current work. Firstly, the diagnostic efficacy of the tracer was only validated in a CLDN18.2-expressing CT26 cancer cell line, due to a lack of other CLDN18.2-expressing cell lines.2 We have screened the expression of CLDN18.2 in a variety of gastric cancer PDX models. Our ongoing work is exploring the diagnostic and predictive value of [68Ga]Ga-NOTA-hu19V3 in the course of CLDN18.2-targeted therapies. Secondly, nanobody is rapidly cleared through kidneys due to the small size, resulting in high kidney accumulation. The undesirable kidney accumulation may cause nephrotoxicity and compromise the diagnostic value for tumor lesions located near the kidneys. To develop next-generation theranostic platforms, the short half-life of nanobodies can be overcome via fusion to an albumin-binding domain or the Fc fragment of IgG.30,31 We have produced fusion proteins of prolonged circulation and are currently determining the theranostic potential of the radiolabeled derivatives. More specifically, 64Cu/89Zr-labeled versions are used for immunoPET imaging, and 177Lu/131I-labeled cousins were used for radioimmunotherapy. Moreover, nanobody drug conjugates using the described nanobody and its derivatives are under development in our lab. With the development and clinical translation of CLDN18.2-targeted molecular imaging techniques, we can better guide the development and use of CLDN18.2-targeted therapeutics.
Conclusions
We synthesized three CLDN18.2-targeted tracers ([68Ga]Ga-NOTA-hu19V3, [18F]F-hu19V3, and [64Cu]Cu-NOTA-hu19V3) and reported the excellent diagnostic efficacy in preclinical models. Translational studies are warranted to investigate the clinical value.
Materials and methods
Nanobody, cell lines, and tumor models
A healthy vicugna pacos was immunized with CHO-CLDN18.2 cells three times, followed by RNA extraction, phage library construction and display, next-generation sequencing, and recombinant expression of the selected clones in E. coli or mammalian expression systems. The details will be reported elsewhere, and protocols are available from the authors upon request. A specific clone hu19V3 was produced at large scale and used for constructing molecular imaging tracers in the current work. CHO-CLDN18.2 and CT26-CLDN18.2 cell lines were prepared to establish tumor models following standard protocols. All animal experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee (Renji Hospital, School of Medicine, Shanghai Jiao Tong University). Nude mice and Balb/c mice aged 4–5 weeks were purchased from GemPharmatech to establish CHO-CLDN18.2-bearing and CT26-CLDN18.2-bearing tumor models, respectively. For subcutaneous tumor models, 2 × 106 CHO-CLDN18.2 or CT26-CLDN18.2 cells were suspended in 100–150 μL of Matrigel and PBS mixture and inoculated. PDX models were prepared with NOD-Prkdcem26Cd52Il2rgem26Cd22/Nju mice (NCG, GemPharmatech). The models were used for immunoPET imaging 2–4 weeks after inoculation of the cells, assessing the diagnostic efficacies of the developed tracers at different disease stages. For metastatic models, 50 × 104 CHO-CLDN18.2 cells in sterile PBS were injected intravenously.
Radiolabeling and quality control
NOTA-derivatized hu19V3 was labeled with 68Ga according to our previously reported protocol.19 Briefly, hu19V3 was conjugated with bifunctional chelator 2-S-(4-Isothiocyanatobenzyl)-1,4,7-triazacyclononane-1,4,7-triacetic acid (p-SCN-Bn-NOTA; Macrocyclics) in the first place. Briefly, 1 mg of hu19V3 in the PBS was prepared, and the pH of the solution was adjusted to 9.0–10 by adding 0.1 M Na2CO3 buffer. NOTA was freshly dissolved in dimethyl sulfoxide (DMSO) and immediately added to the nanobody solution with a chelator/nanobody ratio of 10:1. The reaction was incubated at room temperature for 2 hours, followed by purification with equilibrated PD-10 desalting columns (GE Healthcare) and concentration with Amicon Ultra Centrifugal Filter (10 kDa, Merck). NOTA-hu19V3 was stored at 4°C for subsequent use in the short term. For 68Ga-labeling, 370–555 MBq of freshly eluted 68Ga in 0.1 M hydrogen chloride (pH = 1) was mixed with equal volume of 1 M sodium acetate (pH = 5). The radiometal solution with a final volume of 2 mL (pH = 4.0–4.5) was added to 100–200 μg of NOTA-hu19V3, followed by incubation of the mixture at 37°C for 5–10 min under constant shaking (600 rpm). Copper-64 was freshly produced on site at our department. For 64Cu-labeling, 44.92 MBq of the activity in 160 μL of 2 M HCl was added to 800 μL of 1 M sodium acetate (pH = 5), resulting in a final pH = 5 ± 0.5 of the reaction. The labeling reaction was incubated at 37°C for 30 min under constant shaking.
To facilitate click chemistry-mediated 18F-labeling, the hu19V3 was first modified with DBCO-NHS ester (MeloPEG). Briefly, 2 mg of hu19V3 was prepared, and the pH of the solution was adjusted to ∼10 by adding 0.1 M Na2CO3 buffer. DBCO-NHS ester was freshly dissolved in DMSO and added to the hu19V3 solution with a molar ratio of 10:1. The reaction was incubated at room temperature for 2 hours, followed by purification and concentration as previously stated. The conjugated DBCO-hu19V3 was stored at 4°C for subsequent use. The 18F-labeling precursor (denoted as RJDJ01) was synthesized following prior publications.20,32 [18F]F-RJDJ01 was synthesized in an automatic module. Briefly, 39.9 GBq of 18F-fluoride was produced and eluted with potassium carbonate (3 mg) and 4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane (15 mg) in 1.4 mL of acetonitrile. The solvent was dried, and to this mixture was added RJDJ01 (2.3 mg) in 1 mL of dimethyl sulfoxide. This mixed solution was heated at 120°C for 14 min, followed by dilution with 3 mL of water, and transferred onto a Luna C18 (250 · 10 mm) HPLC. The prosthetic group [18F]F-RJDJ01 was purified using a mobile phase mixture of 32% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 4.6 mL/min. [18F]F-RJDJ01 was collected into a 100-mL flask that contained 25 mL of water, and its contents were delivered to a C18 cartridge. [18F]F-RJDJ01 was released from the cartridge with 3 mL of ethanol, evaporated to dryness, and reconstituted into 0.1 mL of PBS. To 15 mCi of [18F]F-RJDJ01 was added 166 μg of DBCO-hu19V3 in PBS. The reaction mixture was gently mixed for 45 min at 45°C. The final radiopharmaceuticals were collected after purification with equilibrated PD-10 columns. The radiochemical purities of the final products were assessed by instant thin-layer chromatography (iTLC, Eckert & Ziegler Radiopharma).
PET imaging and data analysis
The averaged injection dose for [68Ga]Ga-NOTA-hu19V3, [18F]F-hu19V3, and [64Cu]Cu-NOTA-hu19V3 was 6.7 ± 1.3 MBq (n = 10), 1.7 ± 0.56 MBq (n = 4), and 5.5 ± 1.7 MBq (n = 4), respectively. For the blocking studies, the hu19V3 (1 mg/mouse) was administered to the mice 1 hour before tracer administration. The mice were anesthetized and placed in the prone position in the scanning bed 1 hour after tracer administration. PET/CT data were acquired in sequence with an IRIS PET/CT system (Inviscan Imaging Systems). The spatial resolution of the PET is ∼1 mm (CFOV), 1.3 mm (50 mm off-center). The spatial resolution of the CT is ∼70 μm @ 10% MTF. In the series studies, we used a 10-min acquisition protocol, 9 min for the PET acquisition, and 1 min for the CT acquisition. PET data were reconstructed using a non-scatter-corrected 3D-ordered subset expectation optimization/maximum a posteriori (OSEM3D/MAP) algorithm. The data were analyzed using OsiriX Lite software (Pixmeo SARL) and Inveon Research Workplace (Siemens Preclinical Solutions).
Biodistribution and histopathological studies
The mice were sacrificed after PET imaging at the last time point. Samples including blood were collected and wet-weighed, and the radioactivity of the samples was counted using an automated γ-counter (PerkinElmer). The uptake value in terms of percent ID/g (mean ± SD) was calculated and given for major organs or tissues. H&E and IHC staining of the fixed tissues was carried out to evaluate the expression pattern of CLDN18.2. Briefly, sections of 10 μm were cut and stained for H&E and CLDN18.2 (EPR19202, ab222512, Abcam) following the standard protocols. The clone used for IHC reacts with mouse, rat, and human CLDN18.2 and the dilution ratio of 1/200. All the stained tissues were scanned for subsequent analysis.
Statistics
Statistical analyses were performed using the GraphPad software. Data were presented as means ± SD, and P < 0.05 was considered statistically significant.
Acknowledgments
We thank our colleagues for the helpful discussions. This work was supported in part by the National Key Research and Development Program of China (Grant No. 2020YFA0909000 and 2021YFA0910000), the National Natural Science Foundation of China (Grant No. 82171972 and 82001878), and the Shanghai Rising-Star Program (Grant No. 20QA1406100).
Author contributions
W.W., G.H., and J.L. collaboratively conceived and designed the project. W.W. and D.Z. performed the experiments and wrote most of the manuscript. Y.H. and J.D. produced the proteins used in the study. H.Z., C.W., and C.L. produced the radionuclides. L.L., Y.J., S.A., and Y.Z. contributed to the characterization of the radiotracers. W.W., G.H., and J.L. supervised the study and revised and finalized the manuscript.
Declaration of interests
W.W., D.Z., and J.L. are co-inventors of a patent encompassing the technologies reported in the work.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.11.003.
Contributor Information
Weijun Wei, Email: wwei@shsmu.edu.cn.
Gang Huang, Email: huanggang@sumhs.edu.cn.
Jianjun Liu, Email: nuclearj@163.com.
Supplemental information
Data availability
All data involved in the work can be obtained from Prof. W. Wei upon reasonable request.
References
- 1.Türeci O., Koslowski M., Helftenbein G., Castle J., Rohde C., Dhaene K., Seitz G., Sahin U. Claudin-18 gene structure, regulation, and expression is evolutionary conserved in mammals. Gene. 2011;481:83–92. doi: 10.1016/j.gene.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Türeci Ӧ., Mitnacht-Kraus R., Wöll S., Yamada T., Sahin U. Characterization of zolbetuximab in pancreatic cancer models. Oncoimmunology. 2019;8:e1523096. doi: 10.1080/2162402X.2018.1523096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahin U., Schuler M., Richly H., Bauer S., Krilova A., Dechow T., Jerling M., Utsch M., Rohde C., Dhaene K., et al. A phase I dose-escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro-oesophageal junction cancer. Eur. J. Cancer. 2018;100:17–26. doi: 10.1016/j.ejca.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Türeci O., Sahin U., Schulze-Bergkamen H., Zvirbule Z., Lordick F., Koeberle D., Thuss-Patience P., Ettrich T., Arnold D., Bassermann F., et al. A multicentre, phase IIa study of zolbetuximab as a single agent in patients with recurrent or refractory advanced adenocarcinoma of the stomach or lower oesophagus: the MONO study. Ann. Oncol. 2019;30:1487–1495. doi: 10.1093/annonc/mdz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin U., Türeci Ö., Manikhas G., Lordick F., Rusyn A., Vynnychenko I., Dudov A., Bazin I., Bondarenko I., Melichar B., et al. FAST: a randomised phase II study of zolbetuximab (IMAB362) plus EOX versus EOX alone for first-line treatment of advanced CLDN18.2-positive gastric and gastro-oesophageal adenocarcinoma. Ann. Oncol. 2021;32:609–619. doi: 10.1016/j.annonc.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Lordick F., Al-Batran S.E., Ganguli A., Morlock R., Sahin U., Türeci Ö. Patient-reported outcomes from the phase II FAST trial of zolbetuximab plus EOX compared to EOX alone as first-line treatment of patients with metastatic CLDN18.2+ gastroesophageal adenocarcinoma. Gastric Cancer. 2021;24:721–730. doi: 10.1007/s10120-020-01153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei W., Rosenkrans Z.T., Liu J., Huang G., Luo Q.Y., Cai W. ImmunoPET: concept, design, and applications. Chem. Rev. 2020;120:3787–3851. doi: 10.1021/acs.chemrev.9b00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei W., Jiang D., Lee H.J., Engle J.W., Akiba H., Liu J., Cai W. ImmunoPET imaging of TIM-3 in murine melanoma models. Adv. Ther. 2020;3:2000018. doi: 10.1002/adtp.202000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W., Jiang D., Lee H.J., Li M., Kutyreff C.J., Engle J.W., Liu J., Cai W. Development and characterization of CD54-targeted immunoPET imaging in solid tumors. Eur. J. Nucl. Med. Mol. Imaging. 2020;47:2765–2775. doi: 10.1007/s00259-020-04784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W., Liu Q., Jiang D., Zhao H., Kutyreff C.J., Engle J.W., Liu J., Cai W. Tissue factor-targeted ImmunoPET imaging and radioimmunotherapy of anaplastic thyroid cancer. Adv. Sci. 2020;7:1903595. doi: 10.1002/advs.201903595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei W., Jiang D., Ehlerding E.B., Barnhart T.E., Yang Y., Engle J.W., Luo Q.Y., Huang P., Cai W. CD146-Targeted multimodal image-guided photoimmunotherapy of melanoma. Adv. Sci. 2019;6:1801237. doi: 10.1002/advs.201801237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., Wei W., Barnhart T.E., Jiang D., Cao T., Fan K., Engle J.W., Liu J., Chen W., Cai W. ImmunoPET/NIRF/Cerenkov multimodality imaging of ICAM-1 in pancreatic ductal adenocarcinoma. Eur. J. Nucl. Med. Mol. Imaging. 2021;48:2737–2748. doi: 10.1007/s00259-021-05216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W., Ni D., Ehlerding E.B., Luo Q.Y., Cai W. PET imaging of receptor tyrosine kinases in cancer. Mol. Cancer Ther. 2018;17:1625–1636. doi: 10.1158/1535-7163.MCT-18-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 15.Ingram J.R., Schmidt F.I., Ploegh H.L. Exploiting nanobodies' singular traits. Annu. Rev. Immunol. 2018;36:695–715. doi: 10.1146/annurev-immunol-042617-053327. [DOI] [PubMed] [Google Scholar]
- 16.Xing Y., Chand G., Liu C., Cook G.J.R., O'Doherty J., Zhao L., Wong N.C.L., Meszaros L.K., Ting H.H., Zhao J. Early phase I study of a (99m)Tc-labeled anti-programmed death ligand-1 (PD-L1) single-domain antibody in SPECT/CT assessment of PD-L1 expression in non-small cell lung cancer. J. Nucl. Med. 2019;60:1213–1220. doi: 10.2967/jnumed.118.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keyaerts M., Xavier C., Heemskerk J., Devoogdt N., Everaert H., Ackaert C., Vanhoeij M., Duhoux F.P., Gevaert T., Simon P., et al. Phase I study of 68Ga-HER2-Nanobody for PET/CT assessment of HER2 expression in breast carcinoma. J. Nucl. Med. 2016;57:27–33. doi: 10.2967/jnumed.115.162024. [DOI] [PubMed] [Google Scholar]
- 18.D'Huyvetter M., Vos J.D., Caveliers V., Vaneycken I., Heemskerk J., Duhoux F.P., Fontaine C., Vanhoeij M., Windhorst A.D., Aa F.v.d., et al. Phase I trial of (131)I-GMIB-Anti-HER2-VHH1, a new promising candidate for HER2-targeted radionuclide therapy in breast cancer patients. J. Nucl. Med. 2021;62:1097–1105. doi: 10.2967/jnumed.120.255679. [DOI] [PubMed] [Google Scholar]
- 19.Wang C., Chen Y., Hou Y.N., Liu Q., Zhang D., Zhao H., Zhang Y., An S., Li L., Hou J., et al. ImmunoPET imaging of multiple myeloma with [(68)Ga]Ga-NOTA-Nb1053. Eur. J. Nucl. Med. Mol. Imaging. 2021;48:2749–2760. doi: 10.1007/s00259-021-05218-1. [DOI] [PubMed] [Google Scholar]
- 20.Wei W., Zhang D., Wang C., Zhang Y., An S., Chen Y., Huang G., Liu J. Annotating CD38 expression in multiple myeloma with [(18)F]F-Nb1053. Mol. Pharm. 2022;19:3502–3510. doi: 10.1021/acs.molpharmaceut.1c00733. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H., Wang C., Yang Y., Sun Y., Wei W., Wang C., Wan L., Zhu C., Li L., Huang G., Liu J. ImmunoPET imaging of human CD8(+) T cells with novel (68)Ga-labeled nanobody companion diagnostic agents. J. Nanobiotechnology. 2021;19:42. doi: 10.1186/s12951-021-00785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqui A.Z., Almhanna K. Beyond chemotherapy, PD-1, and HER-2: novel targets for gastric and esophageal cancer. Cancers (Basel) 2021;13:4322. doi: 10.3390/cancers13174322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huisman M.C., Niemeijer A.L.N., Windhorst A.D., Schuit R.C., Leung D., Hayes W., Poot A., Bahce I., Radonic T., Oprea-Lager D.E., et al. Quantification of PD-L1 expression with (18)F-BMS-986192 PET/CT in patients with advanced-stage non-small cell lung cancer. J. Nucl. Med. 2020;61:1455–1460. doi: 10.2967/jnumed.119.240895. [DOI] [PubMed] [Google Scholar]
- 24.Zhu G., Foletti D., Liu X., Ding S., Melton Witt J., Hasa-Moreno A., Rickert M., Holz C., Aschenbrenner L., Yang A.H., et al. Targeting CLDN18.2 by CD3 bispecific and ADC modalities for the treatments of gastric and pancreatic cancer. Sci. Rep. 2019;9:8420. doi: 10.1038/s41598-019-44874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei W., Younis M.H., Lan X., Liu J., Cai W. Single-domain antibody theranostics on the horizon. J. Nucl. Med. 2022;63:1475–1479. doi: 10.2967/jnumed.122.263907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wöll S., Schlitter A.M., Dhaene K., Roller M., Esposito I., Sahin U., Türeci Ö. Claudin 18.2 is a target for IMAB362 antibody in pancreatic neoplasms. Int. J. Cancer. 2014;134:731–739. doi: 10.1002/ijc.28400. [DOI] [PubMed] [Google Scholar]
- 27.Micke P., Mattsson J.S.M., Edlund K., Lohr M., Jirström K., Berglund A., Botling J., Rahnenfuehrer J., Marincevic M., Pontén F., et al. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int. J. Cancer. 2014;135:2206–2214. doi: 10.1002/ijc.28857. [DOI] [PubMed] [Google Scholar]
- 28.Iwaya M., Hayashi H., Nakajima T., Matsuda K., Kinugawa Y., Tobe Y., Tateishi Y., Iwaya Y., Uehara T., Ota H. Colitis-associated colorectal adenocarcinomas frequently express claudin 18 isoform 2: implications for claudin 18.2 monoclonal antibody therapy. Histopathology. 2021;79:227–237. doi: 10.1111/his.14358. [DOI] [PubMed] [Google Scholar]
- 29.Sahin U., Koslowski M., Dhaene K., Usener D., Brandenburg G., Seitz G., Huber C., Türeci O. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin. Cancer Res. 2008;14:7624–7634. doi: 10.1158/1078-0432.CCR-08-1547. [DOI] [PubMed] [Google Scholar]
- 30.Pymm P., Adair A., Chan L.J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O'Neill M.T., Tan L.L., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101918118. e2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laursen N.S., Friesen R.H.E., Zhu X., Jongeneelen M., Blokland S., Vermond J., van Eijgen A., Tang C., van Diepen H., Obmolova G., et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362:598–602. doi: 10.1126/science.aaq0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donnelly D.J., Smith R.A., Morin P., Lipovšek D., Gokemeijer J., Cohen D., Lafont V., Tran T., Cole E.L., Wright M., et al. Synthesis and biologic evaluation of a novel (18)F-labeled adnectin as a PET radioligand for imaging PD-L1 expression. J. Nucl. Med. 2018;59:529–535. doi: 10.2967/jnumed.117.199596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data involved in the work can be obtained from Prof. W. Wei upon reasonable request.