Abstract
Coronary 18F-sodium-fluoride (18F-NaF) positron emission tomography (PET) showed promise in imaging coronary artery disease activity. Currently image processing remains subjective due to the need for manual registration of PET and computed tomography (CT) angiography data. We aimed to develop a novel fully automated method to register coronary 18F-NaF PET to CT angiography using pseudo-CT generated from non-attenuation corrected (NAC) PET by generative adversarial networks (GAN). Non-rigid registration was used to register pseudo-CT to CT angiography and the resulting transformation was subsequently used to align PET with CT angiography.
A total of 169 patients, 139 in the training and 30 in the testing sets were considered. We compared translations at the location of plaques, maximal standard uptake value (SUVmax) and target to background ratio (TBRmax), obtained after observer and automated alignment. Automatic end-to-end registration was performed for 30 patients with 88 coronary vessels and took 95 seconds per patient. Difference in displacement motion vectors between GAN-based and observer-based registration in the x, y and z directions was 0.8 ± 3.0 mm, 0.7 ± 3.0 mm, and 1.7 ± 3.9 mm respectively. TBRmax had a coefficient of repeatability (CR) of 0.31, mean bias of 0.03 and narrow limits of agreement (LOA) (95% LOA: −0.29 to 0.33). SUVmax had CR of 0.26, mean bias of 0 and narrow LOA (95% LOA: −0.26 to 0.26).
In conclusion, pseudo-CT generated by GAN from PET, which are perfectly aligned with PET, can be used to facilitate quick and fully automated registration of PET and CT angiography.
Keywords: PET, CT, Image analysis, Image Reconstruction, Multimodality
INTRODUCTION
Coronary positron emission tomography (PET) has shown promise for the non-invasive assessment of atherosclerotic plaque(1). By targeting processes directly involved in plaque progression and rupture (including inflammation and microcalcification) PET has broadened our understanding of plaque biology(2). Importantly, it has recently been demonstrated that beyond pathophysiological insights 18F-sodium fluoride (18F-NaF) provides prognostic implications with the coronary microcalcification activity (CMA) acting as a strong independent predictor of myocardial infarction(3, 4).
While 18F-NaF emerged as a promising tool for risk stratification in CAD patients, wider adoption of this imaging modality remains challenging(2). 18F-NaF PET requires co-registered computed tomography (CT) angiography images for precise anatomic localization of 18F-NaF activity within coronary plaques. Although typically the CT angiography is acquired on a hybrid PET/CT scanner during the same imaging session as PET, in order to allow for patient repositioning and the respiratory phase at which the CT angiography is acquired the reading physician has to carefully co-register both datasets(5). This important step is necessary for precise quantification of PET activity, which needs to be guided by the anatomical information derived from CT angiography(6, 7). Currently this step is time consuming, subjective and requires great operator expertise, adding to the complexity of coronary PET protocols. In view of the already existing tools for 18F-NaF PET quantification, CT angiography and PET co-registration emerges as the final obstacle for near-full automation of post-acquisition data processing and analysis which could facilitate widespread use of this promising imaging modality.
In the current study we aimed to develop and evaluate a novel, fully automated method for co-registering coronary PET and CT angiography datasets using a conditional generative adversarial network (GAN)(8, 9) and a diffeomorphic nonlinear registration algorithm(10, 11). GANs are a type of deep learning algorithms where two neural networks trained simultaneously, one responsible for generating images and the other classifying whether the generated images are realistic. The two networks are trained against each other, thus “adversarial”, to generate new realistic data. They are prominently used for generation and translation of image data with the objective of learning the underlying distribution of source domain to generate indistinguishable target realistic data samples. In medical imaging, GANs are often used for denoising(12), low-dose to high-dose translation(13) and increasing samples in medical imaging training datasets(14). In our study we employ GAN to generate “pseudo-CT” from corresponding non-attenuation corrected (NAC) PET data. The generated pseudo-CT is perfectly aligned to PET as it is derived from the PET image, unlike the actual non-contrast CT, which is acquired during the same imaging session but is prone to misalignment due to patient motion. The pseudo-CT is then non-linearly registered to CT angiography using diffeomorphic registration algorithm called demons which iteratively computes the displacement for each voxel in a computationally efficient manner. The resulting transformation is used for the final PET to CT angiography registration.
METHODS
Patient population
169 patients with established coronary artery disease undergoing hybrid coronary 18F-NaF PET and contrast CT angiography at the Edinburgh Heart Centre within the investigator-initiated, double-blind, randomized, parallel-group, placebo-controlled DIAMOND (Dual Antiplatelet Therapy to Reduce Myocardial Injury) trial (NCT02110303) were included in the current study(15). All patients underwent a comprehensive baseline clinical assessment and hybrid 18F-NaF PET imaging alongside coronary CT calcium scoring and coronary CT angiography. The study was approved by the local institutional review board, the Scottish Research Ethics Committee (REC reference: 14/SS/0089 and it was performed in accordance with the Declaration of Helsinki. All patients provided written informed consent before any study procedures were initiated.
Acquisition
All patients underwent 18F-NaF PET on a hybrid PET/CT scanner (128-slice Biograph mCT, Siemens Medical Systems, Knoxville, Tennessee) 60 min after intravenous 18F-NaF administration. During a single imaging session, we acquired a non-gated non-contrast CT attenuation correction (AC) scan for attenuation correction purposes, followed by a 30-min PET emission scan in list mode. The electrocardiogram (ECG)-gated list mode dataset was reconstructed using a standard ordered expectation maximization algorithm with time-of-flight and point-spread-function correction. Using 4 cardiac gates, the data were reconstructed on a 256 × 256 matrix (with 75 slices using 2 iterations, 21 subsets, and 5-mm Gaussian smoothing). After the PET scan, based on standard protocol, a gated low-dose non-contrast ECG-gated CT for calculation of the coronary artery calcium score was performed. Subsequently, a contrast-enhanced, ECG-gated coronary CT angiogram was obtained in mid-diastole on the same PET/CT system without repositioning the patient. The ECG-gated non-contrast and contrast CT were not used in automatic registration method.
Manual image registration
We used a dedicated software package for coronary PET image analysis (FusionQuant, Cedars-Sinai Medical Center, Los Angeles, California)(16). PET and CT angiography reconstructions were reoriented, fused, and systematically co-registered in 3 orthogonal planes(5). 18F-NaF uptake in the sternum, vertebrae, blood pool in the left, and right ventricle served as key points of reference. Subsequently, PET activity in the ascending aorta and aortic arch was aligned with the non-contrast CT AC and final refinement of co-registration was performed according to landmarks around the coronary arteries as well as the aortic and mitral valves (Figure 1).
Figure 1: Methodology for manual co-registration of PET and CT angiography.
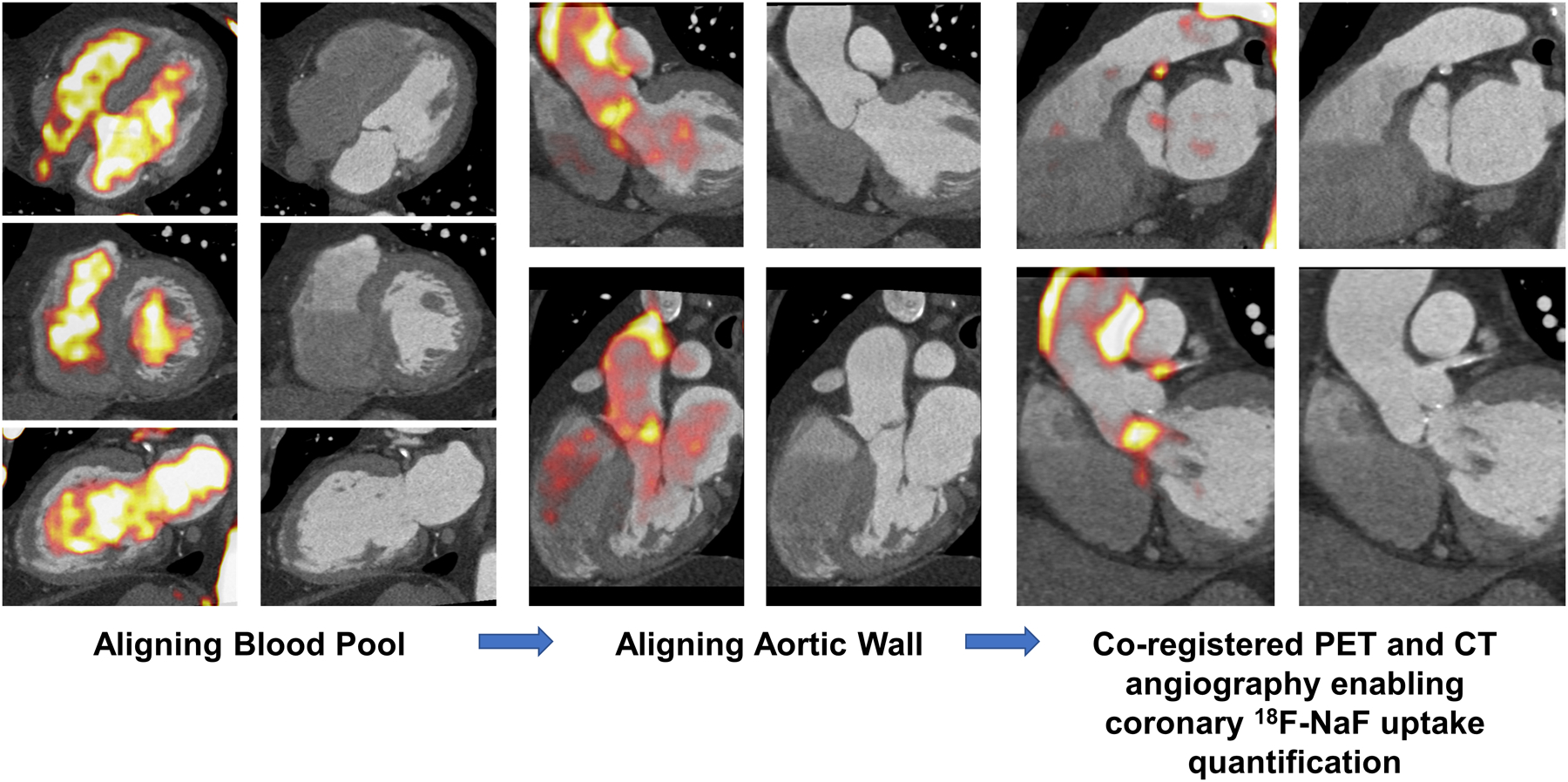
PET and CT angiography were manually co-registered by first aligning the blood pool (left) using key points of reference. The PET activity was subsequently aligned in the aorta to the CT angiography (middle). The final refinement of co-registration was performed according to landmarks around the coronary arteries as well as the aortic and mitral valves.
Automatic deep learning- based registration
We developed a fully automated PET and CT angiography registration using conditional generative adversarial network (GAN)(9) and nonlinear diffeomorphic (demons) registration (10, 11). In the first step, we generated CT images (pseudo-CT) from NAC PET images (Figure 2). We choose NAC PET for our registration (pipeline) as it is immune to potential misregistration of emission data and non-contrast CT AC images which can affect PET attenuation correction (AC) reconstructions. These pseudo-CTs are in perfect alignment with the original PET images. In the second step, these pseudo-CTs were used to register PET to CT angiography (Figure 3).
Figure 2: Generation of pseudo-CT from NAC PET.
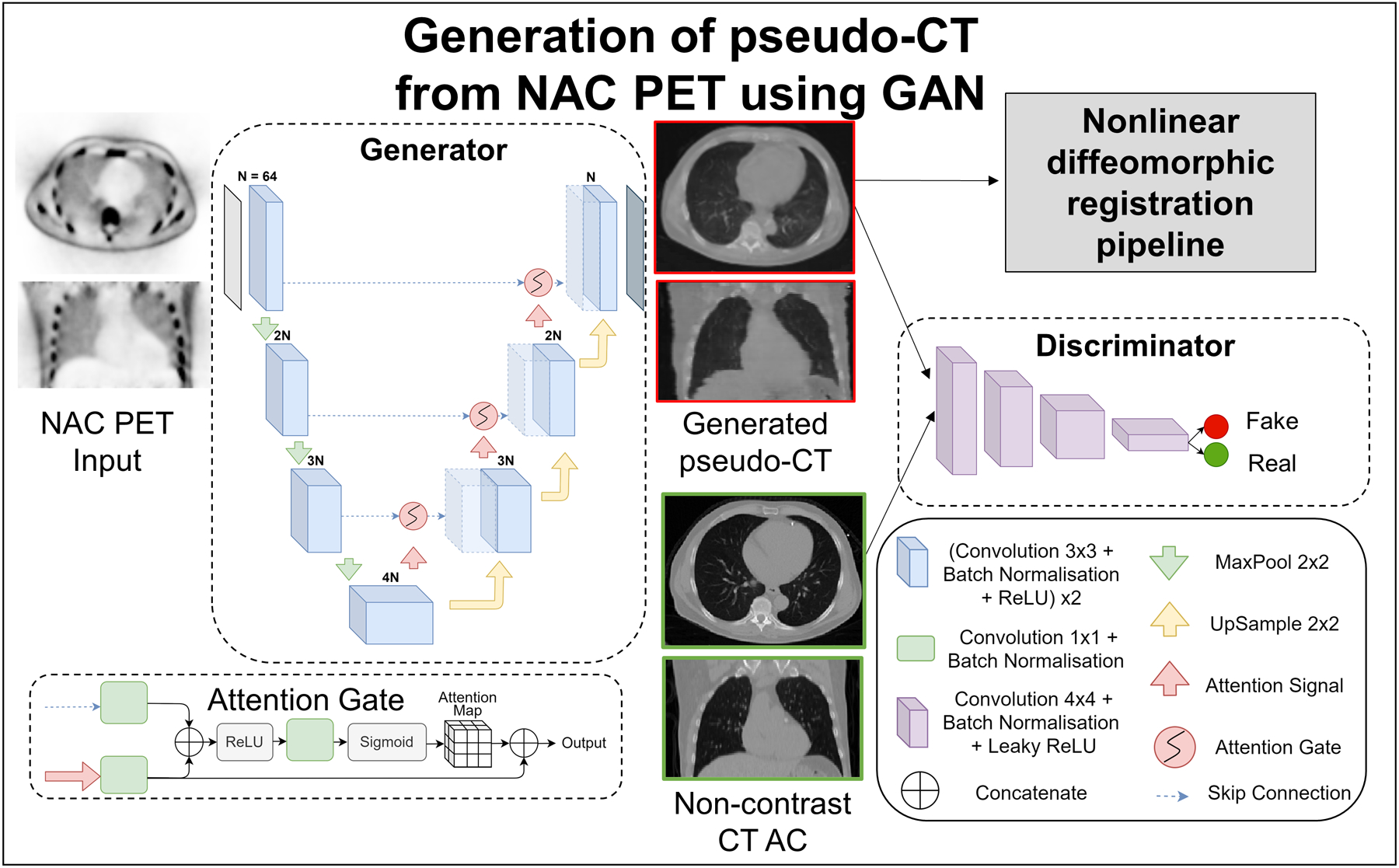
The GAN consisted of two deep learning networks, the generator and discriminator which were trained together to generate realistic pseudo-CT images. The input to the UNet based generator was a 2D NAC PET slice and the output was the corresponding pseudo-CT slice. The encoder part of the generator consisted of 4 contracting convolution blocks each. Each convolution block was followed by a 2×2 max pool operation for dimension reduction. The decoder block consisted of 3 repeated upsampling blocks each doubling the number of feature channels. Batch normalization normalized inputs from a layer and stabilized the learning process. ReLU is used to set all negative inputs to zero and pass all positive inputs to introduce non-linearity in the network. Skip connections were used with attention gates connecting the encoder to the decoder. The discriminator with convolution blocks had inputs of generated pseudo-CT and corresponding real CT AC slice. The output of the network was an averaged similarity between the two inputs. The generated pseudo-CT slices were consolidated per patient and input to the diffeomorphic registration pipeline.
AC: attenuation correction, CT: computed tomography, NAC: non-attenuation corrected, PET: positron emission tomography, ReLU: rectified linear unit
Figure 3: Overview of nonlinear diffeomorphic registration pipeline.
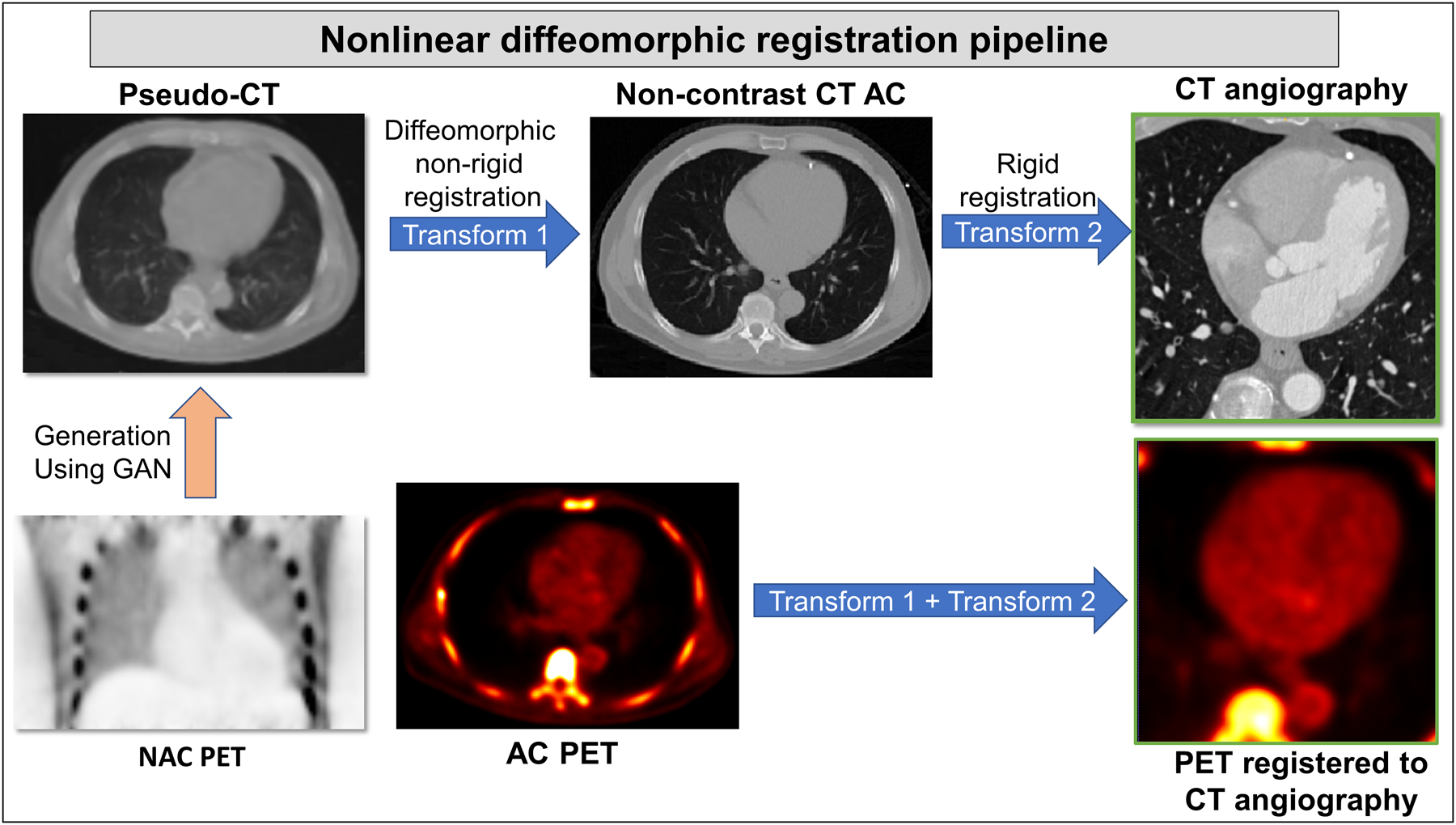
The GAN generated pseudo-CT from NAC PET was registered to the corresponding non-contrast CT attenuation correction image using diffeomorphic (demons) non-rigid registration (Transform 1). The non-contrast CT AC was then registered rigidly to CT angiography image of the same patient (Transform 2). These two transforms (Transforms 1 and 2) were applied to AC PET image, registering PET to CT angiography automatically.
AC: attenuation corrected, CT: computed tomography, NAC: non-attenuation corrected, PET: positron emission tomography
Pseudo-CT generation
GANs(8) are a type of deep learning networks which use generative modelling to synthesize realistic images from a given input, consisting of two key components generator and discriminator. The generator learns the mapping between two types of images, in our case source (PET) and target (non-contrast CT AC), generated on the condition of the target image corresponding to input source image(9) (Figure 2). The second component is the discriminator which tries to classify the generated images as real or fake (generated from source). These two networks were trained in an adversarial fashion(8), competing with one another, until the discriminator network was unable to distinguish between the real and generated cases of CT.
In our implementation, the generator used was a modified UNet(17) with skip connections and attention gates(18). The UNet is a popular encoder decoder network commonly used in biomedical imaging to learn the information present in the input image and encapsulate it. The input to the generator was a 2D NAC PET slice and the output was the generated pseudo-CT slice. The encoder part consisted of repeated convolution layers each followed by a batch normalization(19) and a rectified linear unit (ReLU) (20) followed by a maxpool operation for dimension reduction. The decoder part upsampled the information learnt in a meaningful representation based on the condition of same slice of real CT AC image. Skip connections were used with attention gates to connect layers of encoder with corresponding layers of decoder to localize high-level salient features present in PET, often lost in downsampling. Attention gates help to suppress irrelevant background noise without the need of segmenting heart regions and preserve features relevant for CT generation. The output of attention gates was concatenated with the corresponding upsampling block.
The discriminator used was a deep convolutional neural network (CNN) which had two inputs: the pseudo-CT which was the output of the generator, and the corresponding CT AC image slice. The network took a 70×70 patch of both the inputs and estimated a similarity metric. The discriminator repeated this process for the entire slice, averaging the similarities for each patch, to provide a single probability of whether the pseudo-CT slice was real or fake when compared to the CT AC image slice.
Image preprocessing
CT images were resampled and resized to PET dimensions, per patient. For generation of pseudo-CT, each slice of PET and CT were normalized by subtracting with mean and dividing with the voxel range of gaussian smoothed patient data(21). The images were cropped using the largest CT slice by automatically obtaining the bounding box using the boundary of CT scan. The same bounding box was applied to the corresponding PET scan. Slices of output pseudo-CT were combined and overlayed with the real CT AC image to obtain the background.
Training of pseudo-CT generation
The patient population was randomly divided into train and test sets of 139 and 30 patients. The generator model was trained using a combination of adversarial loss for discriminator and a mean squared loss between input PET and generated pseudo-CT slice. The adversarial loss was optimized to ensure the generator produces realistic pseudo-CT slices and the discriminator was unable to distinguish between real and pseudo-CT slices. The conditional GAN was trained for 250 epochs with a learning rate of 0.0001, an input batch size of 8 NAC PET slices of 256×256 voxels. The output pseudo-CT slices were consolidated per patient and used as reference for the final registration.
Nonlinear diffeomorphic registration
The pipeline for the end-to-end registration of PET to CT angiography is shown in Figure 3. We first registered the generated pseudo-CT scan to the non-contrast CT AC using a nonlinear diffeomorphic registration algorithm called demons(10, 11). The non-contrast CT AC is subsequently registered rigidly to CT angiography. Both these transforms were applied by integrating the 2 motion vector fields from the first step with the second step to PET thus registering PET to CT angiography using the generated pseudo-CT.
Image quantification
On co-registered PET and CT angiography images, 18F-NaF uptake within coronary plaque we measured activity using automatically extracted whole-vessel tubular and tortuous 3-dimensional volumes of interest from CT angiography data sets using FusionQuant(6, 7, 16). These encompass all the main native epicardial coronary vessels and their immediate surroundings (4-mm radius), enabling both per vessel and per patient uptake quantification. Within these volumes of interest, we measured maximum standardized uptake values (SUVmax) (Figure 4) and target to background (TBR) values - calculated by dividing the coronary SUVmax by the blood pool activity measured in the right atrium (mean SUV in cylindrical volumes of interest at the level of the right coronary artery ostium: radius 10 mm and thickness 5 mm)(6, 7). We also measured the coronary microcalcification activity (CMA) which quantifies 18F-NaF activity across the entire coronary vasculature, is highly reproducible, and acts as an independent predictor of myocardial infarction(3). CMA was defined as the integrated activity in the region where standardized uptake value exceeded the corrected background blood-pool mean standardized uptake value +2 SDs. The per patient CMA was defined as the sum of the per vessel CMA values. The same analysis was repeated after automatic co-registration utilizing the same region of interests. Translation vectors in each of the 3 directions for observer and automatic registered PET were exported from observer marked vessels per patient using FusionQuant.
Figure 4: Case example of original, expert observer and GAN-based registration of PET and CT angiography with SUVmax values.
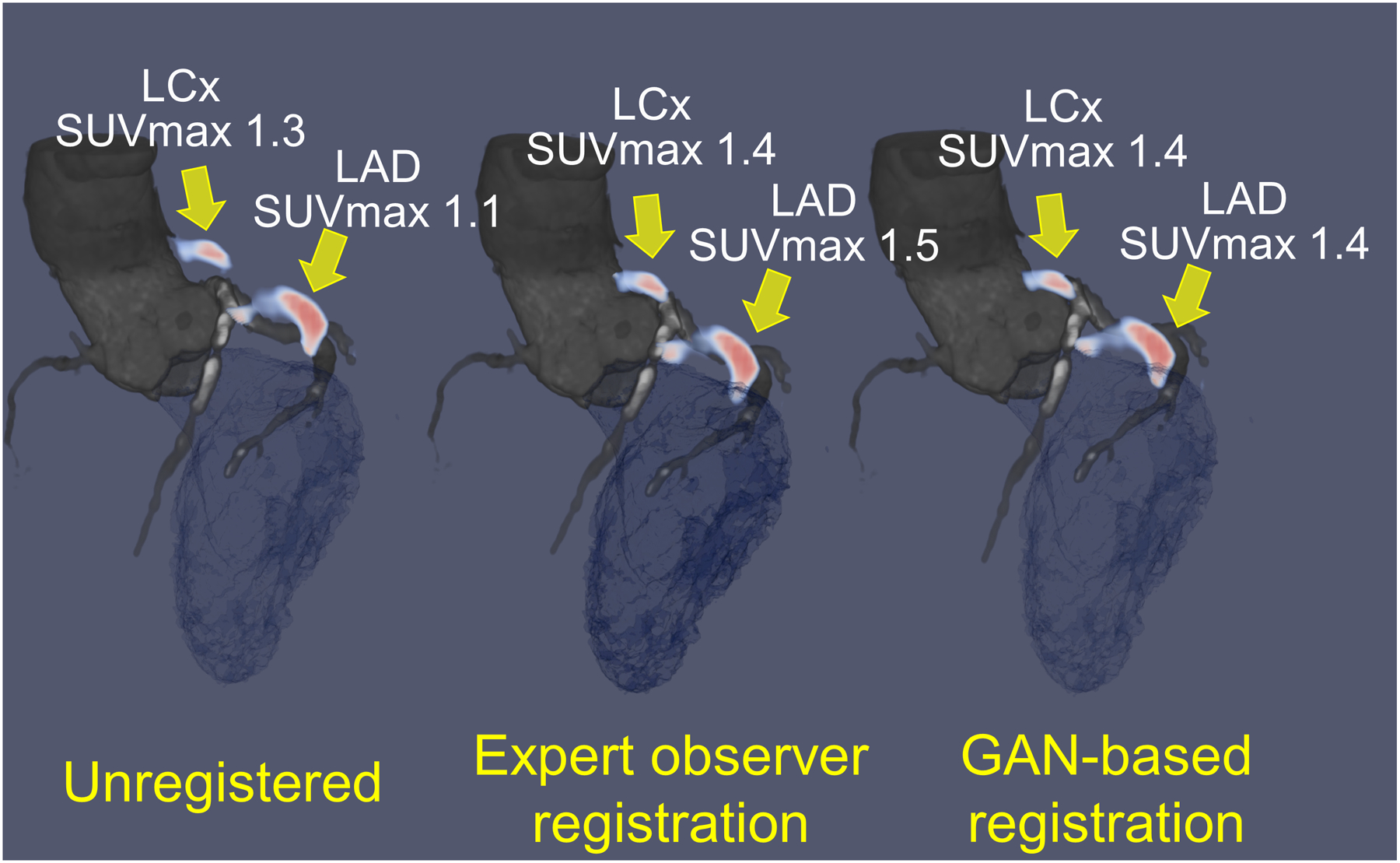
Hybrid CT angiography (grey blue) and 18F-NaF PET (red) of the left anterior descending (LAD) and left circumflex (LCx) coronary arteries of a 52-year-old man; unregistered (left), registered by expert observer (center) and by GAN-based nonlinear diffeomorphic registration (right). Manual and GAN-based registration have similar increase in standard uptake value (SUVmax) compared to unregistered PET/CT angiography.
Statistical Analysis
Categorical variables are presented as frequencies (percentages) and continuous variables as medians (interquartile range). Variables were compared using a Pearson χ2 statistic for categorical variables and a Wilcoxon rank-sum or Kruskal-Wallis test for continuous variables. We assessed the distribution of data with the Shapiro-Wilk test.
The performance of the proposed method was evaluated through the coefficients of reproducibility (CR) with observer manual registration using Bland-Altman plots with 95% limits of agreement (LOA) and difference in displacement vector fields.
RESULTS
The study population comprised 169 patients (81.1% men; mean age: 65.3± 8.4 years) with 139 used for training and 30 for testing. All participants had advanced stable coronary atherosclerosis with a high burden of cardiovascular risk factors (hypertension: n=101; 60%; hyperlipidemia: n=148; 88%; tobacco use: n=113; 67%) and were on guideline recommended therapy (statin: n=151; 90%; antiplatelet therapy: n=155; 92%; angiotensin-converting enzyme inhibitor or angiotensin receptor blockers: n=113; 67%) and had high rates of prior revascularization (n=137; 81%). Baseline study population characteristics of train and test sets are presented in Table 1.
Table 1:
Patient characteristics of training and testing sets
| Characteristics | Training set (n = 139) |
Testing set (n = 30) |
|---|---|---|
| Age (years) | 65.3 ± 8.2 | 65.2 ± 9.4 |
| Gender (males) | 104 (74.8) | 25 (83.3) |
| Body-mass index (kg/m2) | 29.6 ± 5.4 | 29.1 ± 4.8 |
| Cardiovascular risk factors | ||
| Diabetes mellitus (type II) | 25 (18) | 3 (10) |
| Current smoker | 22 (15.8) | 2 (6.7) |
| Hypertension | 74 (53.2) | 14 (46.7) |
| Hyperlipidemia | 128 (92.1) | 28 (93.3) |
| Agatston Calcium Score (AU) | 299 [90.8 – 685] | 565 [183 – 1201] |
Continuous variables reported as mean ± SD or median and interquartile range [IQR]; categorical variables reported as n (%), Continuous variables reported as mean ± SD or median and interquartile range [IQR]; categorical variables reported as n (%).
Twenty-five (83%) of the 30 patients in the testing set had poor registration between NAC PET and non-contrast CT AC, according to expert observer. To achieve perfect alignment of the PET and CT images all datasets in the training dataset required adjustments made by the interpreting physician. These were most prominent in the z axis reflecting the discordance in the diaphragm position – which is a result of breathing (while CT AC data can be acquired during a breath-hold the PET scan last for 30 minutes).
A case example of GAN-based nonlinear diffeomorphic registration in Figure 4 shows similar registration and SUVmax in the coronary arteries compared to expert observer. Activity of 88 vessels in the 30 patients, was assessed using TBRmax, SUVmax and CMA. TBRmax had a coefficient of repeatability (CR) of 0.31, mean bias of 0.92 and narrow limits of agreement (LOA) (95% LOA – 0.29 to 0.33) (Figure 5A). SUVmax had excellent CR of 0.26, mean bias of 0 and narrow limits of agreement (95% LOA – 0.26 to 0.26) (Figure 5B). Between observer and GAN based registration, CMA had CR of 0.57, mean bias of 0.07 and narrow limits of agreement (95% LOA – 0.54 to 0.60) (Figure 6). Difference in displacement motion vectors between GAN and observer-based registration (Figure 7) was 0.8 ± 3.02 mm in the x direction, 0.68 ± 2.98 mm in the y direction and 1.66 ± 3.94 mm in the z direction. The overall time for the GAN-based registration with analysis was 95 seconds on a standard CPU workstation. The overall registration and analysis time for the experienced observer was 15±2.5 minutes.
Figure 5: Expert observer and GAN-based registration: Agreement of TBRmax and SUVmax.
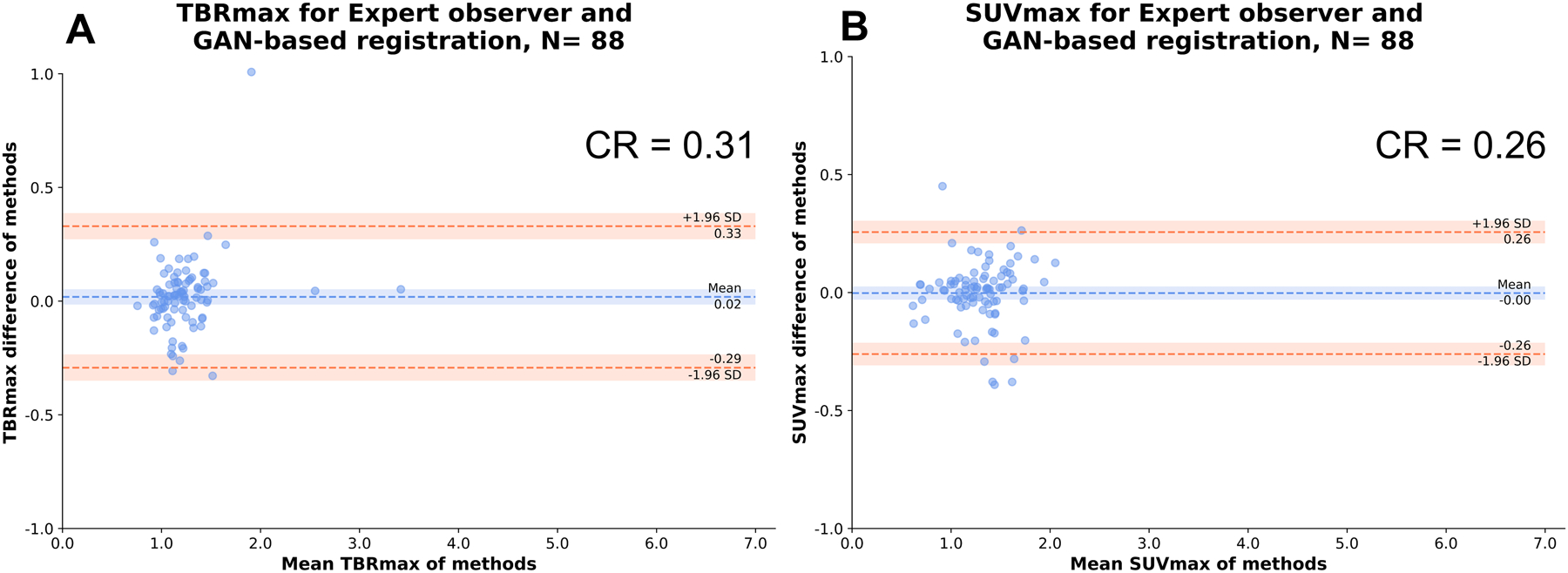
Bland Altman plots of observer and GAN-based nonlinear diffeomorphic registration measurements of maximal target to background ratio (TBRmax) (A) and standard uptake value [SUVmax] (B) at vessel level.
CR: coefficient of repeatability, SD: standard deviation
Figure 6: Expert observer and GAN-based registration: Agreement of CMA.
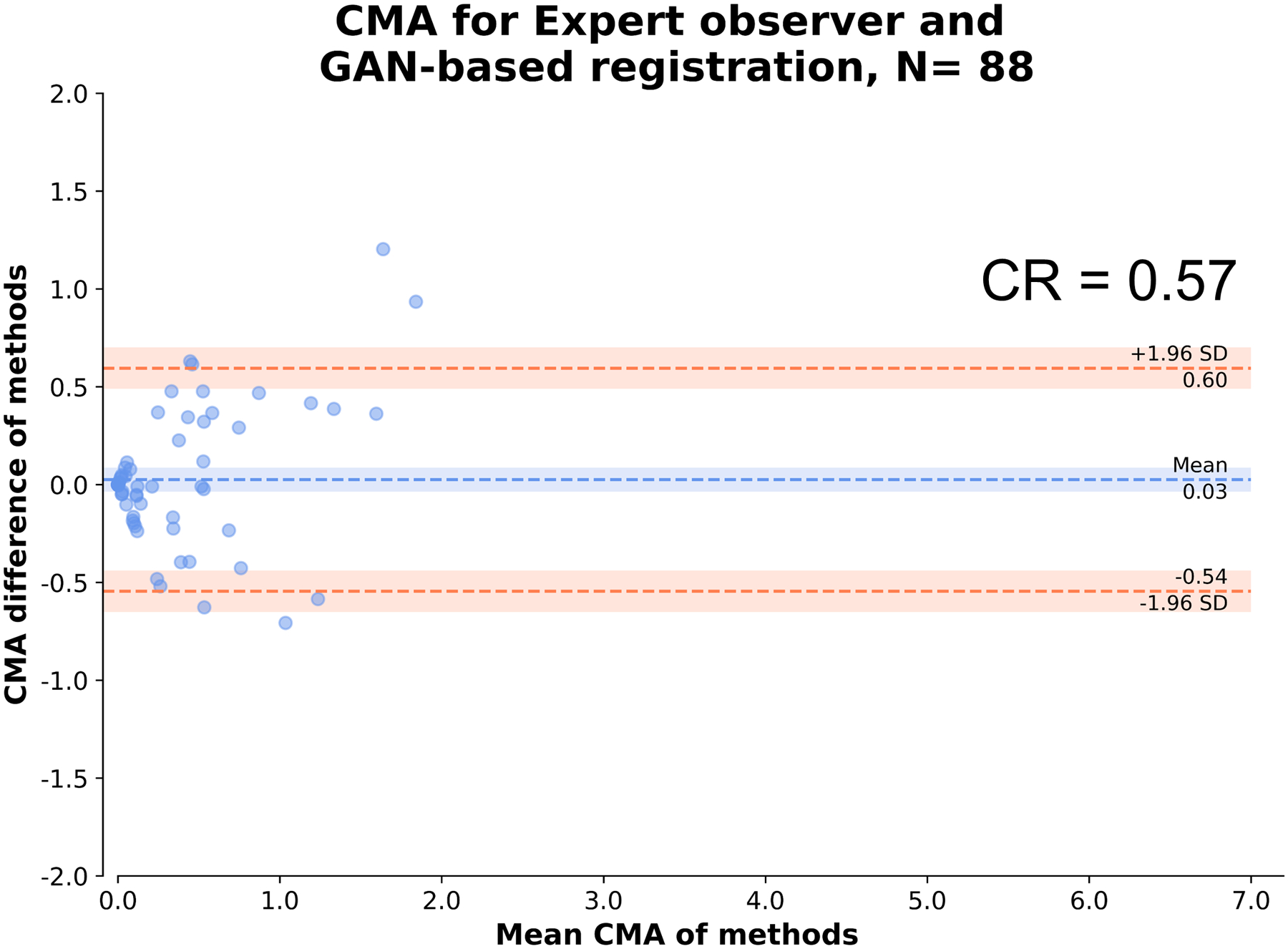
Bland Altman plot of observer and GAN-based nonlinear diffeomorphic registration measurements of coronary microcalcification activity (CMA) at vessel level.
CR: coefficient of repeatability, SD: standard deviation
Figure 7: Expert observer and GAN-based registration displacement differences in x, y and z directions.
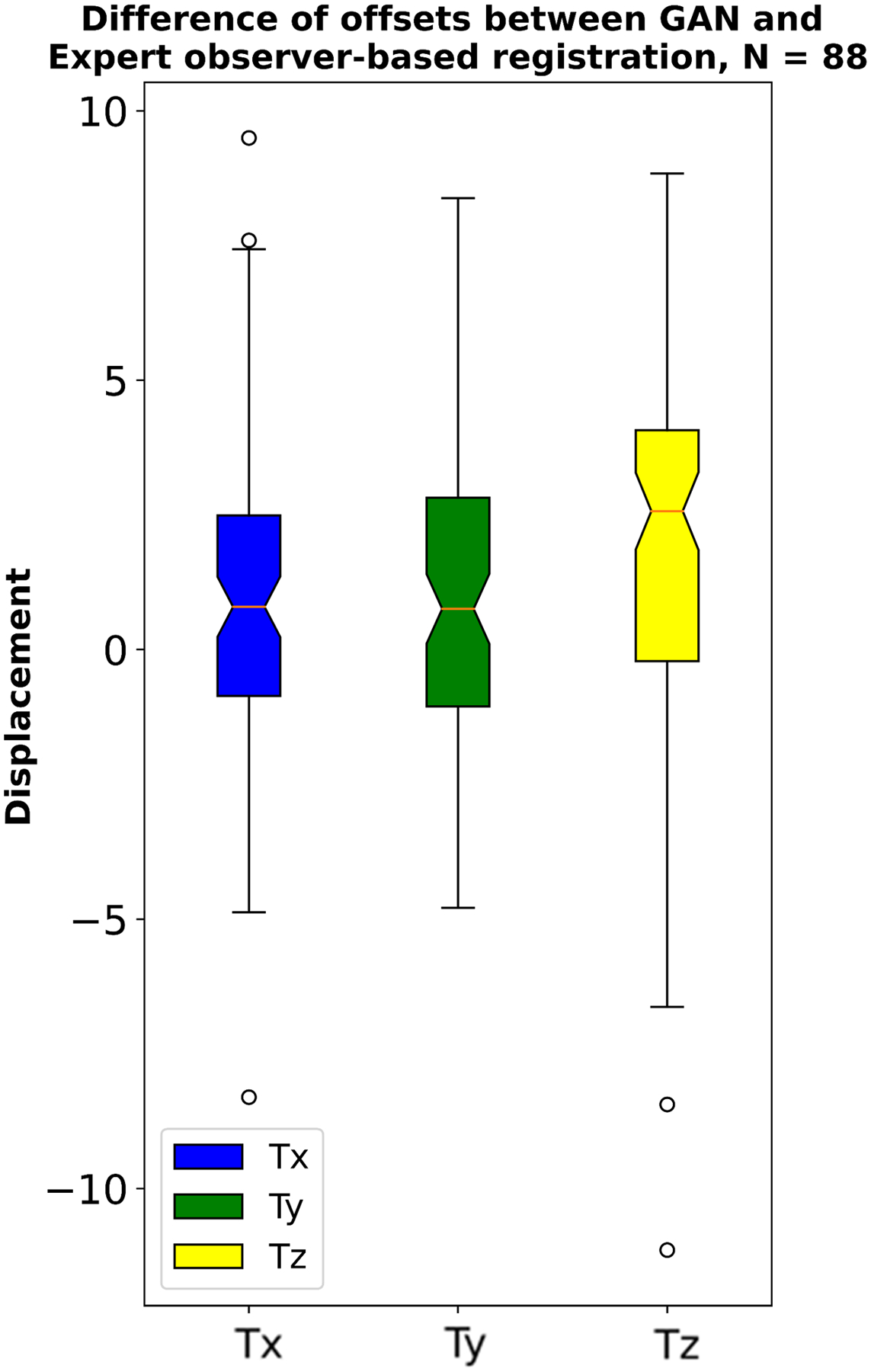
Expert observer and GAN-based nonlinear diffeomorphic registration displacement difference at vessel level in mm, in the location of the quantified vessels.
DISCUSSION
We propose a fully automated deep learning-based framework to register 18F-NaF PET to CT angiography images. A conditional GAN was used to synthesize pseudo-CT from coronary NAC PET images. The perfectly registered pseudo-CT provides an input to a non-linear registration pipeline which transforms PET alignment to match CT angiography. We trained the proposed method with 139 pairs of coronary PET/CT angiography images. The evaluation in a separate cohort of 30 patients demonstrated excellent correlation of TBRmax, SUVmax and CMA between observer and our method. The proposed method runs automatically in 95 seconds, approximately 10 times faster than expert observer, and has great potential to streamline time consuming manual rigid registration which is necessary for coronary PET/CT angiography data. Our approach is the first to use pseudo-CT generated from GAN for nonlinear diffeomorphic registration of coronary PET and CT angiography images. This development paves the way for more widespread utilization of coronary PET. Since acquisition protocols have been already validated across multiple centers and image analysis has been automated, by providing automatic co-registration an 18F-NaF CMA uptake value can become available as soon as image reconstruction is completed, and CT angiography coronary centerlines are available.
18F-NaF PET has emerged as a promising noninvasive imaging tool for assessment of active calcification processes across a wide range of cardiovascular conditions(16, 22–27). In coronary artery disease 18F-NaF uptake provides an assessment of disease activity and prediction of subsequent disease progression and clinical events(1, 3). Over the past years multiple technical refinements for 18F-NaF PET have been introduced(28–31). Motion correction techniques address heart contractions, tidal breathing and patient repositioning during the prolonged PET acquisitions(32–34). Novel uptake measures such as CMA enable a patient level assessment of disease activity which is guided by centerlines derived from contract enhanced CT angiography(6, 7). Dedicated software tools, optimized timing of the acquisition and dedicated reconstruction algorithms further streamline 18F-NaF coronary imaging(16, 35). In view of all these refinements to date, the need for manual co-registration of the CT angiography and PET datasets remains the last step, which still requires advanced cardiac imaging expertise for image quantification and is associated with subjectivity. Our current study addresses this important aspect of 18F-NaF coronary PET imaging. By leveraging AI, we were able to develop and test a fully automated tool aligning the CT angiography and PET datasets.
GAN AI methods(8, 9) have recently become popular in medical imaging for image-to-image translation tasks(36). By learning the mapping from one type of the image to the to another, they are often used for denoising(13, 37, 38), segmentation(39, 40), low-dose to high-dose reconstruction(41) and registration(42–44) tasks. Dong et al.(45) have used GANs for generation of CT from PET has been used for attenuation correction. In this study we propose to use GANs for the challenging task of fully automated coronary PET and CT angiography image registration. Automatic registration of PET and CT angiography is difficult to accomplish due to nonlinear respiratory and cardiac motion displacement between the two modalities and limited anatomical information provided by coronary PET. We leverage the fact that the generated pseudo-CT is perfectly registered to PET, which is the input to GAN, unlike the non-contrast attenuation correction image acquired in the same imaging session. The latter misalignment occurs due patient motion and the lengthy PET acquisition. By registering the pseudo-CT to the actual CT and then subsequently to CT angiography with nonlinear diffeomorphic registration, these issues are overcome, and we obtain the transform to register PET to CT angiography.
Automatic GAN-based nonlinear diffeomorphic registration of PET and CT angiography can be employed for accurate alignment of images. This method facilitates automated analysis of 18F-NaF coronary uptake with the user input limited to careful inspection whether extra-coronary activity does not corrupt the coronary 18F-NaF uptake measurements. In view of the already available tools for quantification of 18F-NaF activity on a per vessel and per patient level(6, 7), by developing automatic PET to CT angiography registration we have paved the way for more widespread use of coronary PET. This development could simplify further the complex processing protocols needed for clinical application of coronary PET imaging.
Several studies have attempted automatic cardiac PET to CT registration. In previous studies, Nakazato et al.(46) have evaluated a non-AI based method to register myocardial perfusion 13N-ammonia PET to CT angiography. Yu et al.(47) proposed an AI based framework using convolution neural networks to non-rigidly register PET/CT images. However, 18F-NaF coronary PET imaging does not visualize the myocardium and so direct PET to CT angiography registration is not feasible. Our proposed solution overcomes the difficulties in image registration of images with different visual appearance.
Our study has limitations. It is a post-hoc analysis of single-center data and was acquired on one hybrid PET/CT scanner. The observer registration was performed rigidly using a summed PET scan registered to the diastolic gate, however the pseudo-CT was generated using summed NAC PET data and CT angiography was registered non-rigidly using corresponding AC PET scan. The CT AC misregistration may occur between the PET and CT affecting the quality of the PET images. This causes difference in observer registration, which was performed using potentially incorrect attenuation corrected PET, and automatic registration, which was performed with NAC pet images. Further studies could be performed utilizing the generation of pseudo-CT for optimizing attenuation correction of the PET signal. However, to date no clinical studies have utilized such corrections. The user correction was limited to rigid translation of the vessel in contrast to automatic nonlinear alignment by our method. Nevertheless, there is currently no other suitable standard to evaluate the misalignment and the vessel based rigid co-registration is the basis of the current clinical analysis.
CONCLUSIONS
Pseudo-CT generated by GAN from PET, which are perfectly aligned with PET, can be used to facilitate rapid and fully automated nonlinear diffeomorphic registration of PET and CT angiography. We show that our method has excellent agreement and is approximately 10 times faster as compared to expert observer registration.
ACKNOWLEDGEMENTS
We would like to thank all the individuals involved in collecting and acquiring data.
SOURCES OF FUNDING
This research was supported in part by grant R01HL135557 from the National Heart, Lung, and Blood Institute/ National Institutes of Health (NHLBI/NIH) (PI: Piotr Slomka). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DISCLOSURES
Dr. Slomka and Mr. Cadet participate in software royalties for cardiac imaging software at Cedars-Sinai Medical Center. Dr. Slomka has received research grant support from Siemens Medical Systems. D.E.N. (CH/09/002, RG/16/10/32375, RE/18/5/34216), and M.R.D. (FS/14/78/31020) are supported by the British Heart Foundation. MCW (FS/ICRF/20/26002) is supported by the British Heart Foundation. MCW has given talks for Canon Medical Systems and Siemens Healthineers. D.E.N. is the recipient of a Wellcome Trust Senior Investigator Award (WT103782AIA). M.R.D. is supported by the Sir Jules Thorn Biomedical Research Award 2015 (15/JTA). The remaining authors have no relevant disclosures.
Abbreviations and Acronyms
- AI
artificial intelligence
- CMA
coronary macrocalcification activity
- CR
coefficient of repeatability
- CT
computed tomography
- GAN
generative adversarial networks
- PET
positron emission tomography
- SUV
standard uptake value
- TBR
target to background
REFERENCES
- (1).Joshi NV, Vesey AT, Williams MC, Shah AS, Calvert PA, Craighead FH et al. 18F-fluoride positron emission tomography for identification of ruptured and high-risk coronary atherosclerotic plaques: a prospective clinical trial. The Lancet 2014;383:705–13. [DOI] [PubMed] [Google Scholar]
- (2).Kwiecinski J, Slomka PJ, Dweck MR, Newby DE, Berman DS. Vulnerable plaque imaging using 18F-sodium fluoride positron emission tomography. The British Journal of Radiology 2020;93:20190797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kwiecinski J, Tzolos E, Cadet S, Adamson PD, Moss A, Joshi NV et al. 18F-sodium fluoride coronary uptake predicts myocardial infarctions in patients with known coronary artery disease. Journal of the American College of Cardiology 2020;75:3667–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kwiecinski J, Tzolos E, Meah MN, Cadet S, Adamson PD, Grodecki K et al. Machine learning with 18F-sodium fluoride PET and quantitative plaque analysis on CT angiography for the future risk of myocardial infarction. Journal of Nuclear Medicine 2022;63:158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kwiecinski J, Adamson PD, Lassen ML, Doris MK, Moss AJ, Cadet S et al. Feasibility of coronary 18F-sodium fluoride positron-emission tomography assessment with the utilization of previously acquired computed tomography angiography. Circulation: Cardiovascular Imaging 2018;11:e008325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tzolos E, Kwiecinski J, Lassen ML, Cadet S, Adamson PD, Moss AJ et al. Observer repeatability and interscan reproducibility of 18F-sodium fluoride coronary microcalcification activity. Journal of Nuclear Cardiology 2020:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kwiecinski J, Cadet S, Daghem M, Lassen ML, Dey D, Dweck MR et al. Whole-vessel coronary 18F-sodium fluoride PET for assessment of the global coronary microcalcification burden. European journal of nuclear medicine and molecular imaging 2020;47:1736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S et al. Generative adversarial nets. Advances in neural information processing systems 2014;27. [Google Scholar]
- (9).Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 1125–34. [Google Scholar]
- (10).Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage 2007;38:95–113. [DOI] [PubMed] [Google Scholar]
- (11).Hernandez M, Bossa MN, Olmos S. Registration of anatomical images using paths of diffeomorphisms parameterized with stationary vector field flows. International Journal of Computer Vision 2009;85:291–306. [Google Scholar]
- (12).Chen Z, Zeng Z, Shen H, Zheng X, Dai P, Ouyang P. DN-GAN: Denoising generative adversarial networks for speckle noise reduction in optical coherence tomography images. Biomedical Signal Processing and Control 2020;55:101632. [Google Scholar]
- (13).Yang Q, Yan P, Zhang Y, Yu H, Shi Y, Mou X et al. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE transactions on medical imaging 2018;37:1348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A et al. Gan augmentation: Augmenting training data using generative adversarial networks. arXiv preprint arXiv:181010863 2018. [Google Scholar]
- (15).Moss AJ, Dweck MR, Doris MK, Andrews JP, Bing R, Forsythe RO et al. Ticagrelor to reduce myocardial injury in patients with high-risk coronary artery plaque. Cardiovascular Imaging 2020;13:1549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Massera D, Doris MK, Cadet S, Kwiecinski J, Pawade TA, Peeters FE et al. Analytical quantification of aortic valve 18F-sodium fluoride PET uptake. Journal of Nuclear Cardiology 2020;27:962–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; 2015. p. 234–41. [Google Scholar]
- (18).Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K et al. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:180403999 2018. [Google Scholar]
- (19).Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. International conference on machine learning; 2015. p. 448–56. [Google Scholar]
- (20).Agarap AF. Deep learning using rectified linear units (relu). arXiv preprint arXiv:180308375 2018. [Google Scholar]
- (21).Shi L, Onofrey JA, Liu H, Liu Y-H, Liu C. Deep learning-based attenuation map generation for myocardial perfusion SPECT. European Journal of Nuclear Medicine and Molecular Imaging 2020;47:2383–95. [DOI] [PubMed] [Google Scholar]
- (22).Cartlidge TR, Doris MK, Sellers SL, Pawade TA, White AC, Pessotto R et al. Detection and prediction of bioprosthetic aortic valve degeneration. Journal of the American College of Cardiology 2019;73:1107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kwiecinski J, Tzolos E, Cartlidge TR, Fletcher A, Doris MK, Bing R et al. Native aortic valve disease progression and bioprosthetic valve degeneration in patients with transcatheter aortic valve implantation. Circulation 2021;144:1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fletcher AJ, Lembo M, Kwiecinski J, Syed MB, Nash J, Tzolos E et al. Quantifying microcalcification activity in the thoracic aorta. Journal of Nuclear Cardiology 2021:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Andrews JP, MacNaught G, Moss AJ, Doris MK, Pawade T, Adamson PD et al. Cardiovascular 18F-fluoride positron emission tomography-magnetic resonance imaging: A comparison study. Journal of Nuclear Cardiology 2021;28:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kwiecinski J, Tzolos E, Fletcher AJ, Nash J, Meah MN, Cadet S et al. Bypass Grafting and Native Coronary Artery Disease Activity. JACC: Cardiovascular Imaging 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Fletcher AJ, Tew YY, Tzolos E, Joshi SS, Kaczynski J, Nash J et al. Thoracic Aortic 18F-Sodium Fluoride Activity and Ischemic Stroke in Patients With Established Cardiovascular Disease. JACC: Cardiovascular Imaging 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tzolos E, Lassen ML, Pan T, Kwiecinski J, Cadet S, Dey D et al. Respiration-averaged CT versus standard CT attenuation map for correction of 18F-sodium fluoride uptake in coronary atherosclerotic lesions on hybrid PET/CT. Journal of Nuclear Cardiology 2020:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bellinge JW, Schultz CJ. Optimizing arterial 18F-sodium fluoride positron emission tomography analysis; 2021. p. 1887–90. [DOI] [PubMed]
- (30).Bellinge JW, Francis RJ, Majeed K, Watts GF, Schultz CJ. In search of the vulnerable patient or the vulnerable plaque: 18F-sodium fluoride positron emission tomography for cardiovascular risk stratification. Journal of Nuclear Cardiology 2018;25:1774–83. [DOI] [PubMed] [Google Scholar]
- (31).Kwiecinski J, Lassen ML, Slomka PJ. Advances in Quantitative Analysis of 18F-Sodium Fluoride Coronary Imaging. Molecular imaging 2021;2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lassen ML, Kwiecinski J, Dey D, Cadet S, Germano G, Berman DS et al. Triple-gated motion and blood pool clearance corrections improve reproducibility of coronary 18F-NaF PET. European journal of nuclear medicine and molecular imaging 2019;46:2610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lassen ML, Kwiecinski J, Cadet S, Dey D, Wang C, Dweck MR et al. Data-driven gross patient motion detection and compensation: Implications for coronary 18F-NaF PET imaging. Journal of Nuclear Medicine 2019;60:830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lassen ML, Beyer T, Berger A, Beitzke D, Rasul S, Büther F et al. Data-driven, projection-based respiratory motion compensation of PET data for cardiac PET/CT and PET/MR imaging. Journal of Nuclear Cardiology 2020;27:2216–30. [DOI] [PubMed] [Google Scholar]
- (35).Doris MK, Otaki Y, Krishnan SK, Kwiecinski J, Rubeaux M, Alessio A et al. Optimization of reconstruction and quantification of motion-corrected coronary PET-CT. Journal of Nuclear Cardiology 2020;27:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Armanious K, Jiang C, Fischer M, Küstner T, Hepp T, Nikolaou K et al. MedGAN: Medical image translation using GANs. Computerized medical imaging and graphics 2020;79:101684. [DOI] [PubMed] [Google Scholar]
- (37).Shan H, Zhang Y, Yang Q, Kruger U, Kalra MK, Sun L et al. 3-D convolutional encoder-decoder network for low-dose CT via transfer learning from a 2-D trained network. IEEE transactions on medical imaging 2018;37:1522–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zhou L, Schaefferkoetter JD, Tham IW, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Medical image analysis 2020;65:101770. [DOI] [PubMed] [Google Scholar]
- (39).Jafari MH, Girgis H, Abdi AH, Liao Z, Pesteie M, Rohling R et al. Semi-supervised learning for cardiac left ventricle segmentation using conditional deep generative models as prior. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019. p. 649–52. [Google Scholar]
- (40).Guo Z, Li X, Huang H, Guo N, Li Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Transactions on Radiation and Plasma Medical Sciences 2019;3:162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage 2018;174:550–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sood RR, Shao W, Kunder C, Teslovich NC, Wang JB, Soerensen SJ et al. 3D registration of pre-surgical prostate MRI and histopathology images via super-resolution volume reconstruction. Medical Image Analysis 2021;69:101957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Tanner C, Ozdemir F, Profanter R, Vishnevsky V, Konukoglu E, Goksel O. Generative adversarial networks for MR-CT deformable image registration. arXiv preprint arXiv:180707349 2018. [Google Scholar]
- (44).Hu Y, Gibson E, Ghavami N, Bonmati E, Moore CM, Emberton M et al. Adversarial deformation regularization for training image registration neural networks. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2018. p. 774–82. [Google Scholar]
- (45).Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Physics in Medicine & Biology 2019;64:215016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Nakazato R, Dey D, Alexánderson E, Meave A, Jiménez M, Romero E et al. Automatic alignment of myocardial perfusion PET and 64-slice coronary CT angiography on hybrid PET/CT. Journal of Nuclear Cardiology 2012;19:482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yu H, Zhou X, Jiang H, Kang H, Wang Z, Hara T et al. Learning 3D non-rigid deformation based on an unsupervised deep learning for PET/CT image registration. Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging; 2019. p. 439–44. [Google Scholar]


