Summary
Heterozygous pathogenic variants in DNM1 cause developmental and epileptic encephalopathy (DEE) as a result of a dominant-negative mechanism impeding vesicular fission. Thus far, pathogenic variants in DNM1 have been studied with a canonical transcript that includes the alternatively spliced exon 10b. However, after performing RNA sequencing in 39 pediatric brain samples, we find the primary transcript expressed in the brain includes the downstream exon 10a instead. Using this information, we evaluated genotype-phenotype correlations of variants affecting exon 10a and identified a cohort of eleven previously unreported individuals. Eight individuals harbor a recurrent de novo splice site variant, c.1197−8G>A (GenBank: NM_001288739.1), which affects exon 10a and leads to DEE consistent with the classical DNM1 phenotype. We find this splice site variant leads to disease through an unexpected dominant-negative mechanism. Functional testing reveals an in-frame upstream splice acceptor causing insertion of two amino acids predicted to impair oligomerization-dependent activity. This is supported by neuropathological samples showing accumulation of enlarged synaptic vesicles adherent to the plasma membrane consistent with impaired vesicular fission. Two additional individuals with missense variants affecting exon 10a, p.Arg399Trp and p.Gly401Asp, had a similar DEE phenotype. In contrast, one individual with a missense variant affecting exon 10b, p.Pro405Leu, which is less expressed in the brain, had a correspondingly less severe presentation. Thus, we implicate variants affecting exon 10a as causing the severe DEE typically associated with DNM1-related disorders. We highlight the importance of considering relevant isoforms for disease-causing variants as well as the possibility of splice site variants acting through a dominant-negative mechanism.
Keywords: dynamin-1, DNM1, vesicle fission, synapse, epilepsy, developmental and epileptic encephalopathy, dominant negative, alternative transcripts
Heterozygous pathogenic variants in DNM1 cause developmental and epileptic encephalopathy. Here, we perform RNA sequencing in brain samples and find non-canonical, alternatively spliced transcripts are the most abundant. We highlight the importance of considering relevant isoforms for variant interpretation and the possibility of intronic variants causing disease through dominant-negative mechanisms.
Introduction
Developmental and epileptic encephalopathy (DEE) encompasses a group of disorders characterized by severe epilepsy refractory to medical treatment that affects children and results in delayed development and both neurological and non-neurological comorbidities.1,2,3,4 Genetic factors are increasingly recognized as a major cause of DEEs, and more than 100 monogenic causes for epilepsies and neurodevelopmental disorders have been identified.5,6,7,8 Several common DEE-associated genes, such as SCN1A (MIM: 182389), SCN2A (MIM: 182390), SCN8A (MIM: 600702), and STXBP1 (MIM: 602926), represent prime candidates for precision medicine approaches.9,10,11
Prominent among neurodevelopmental disorders and epilepsies with genetic etiologies are those due to variation in genes related to synaptic transmission. Several genes involved in synaptic vesicle fission and fusion, such as DNM1 (MIM: 602377), AP2M1 (MIM: 601024), STXBP1 (MIM: 602926), STX1B (MIM: 601485), VAMP2 (MIM: 185881), and SNAP25 (MIM: 600322), have been implicated in neurodevelopmental disorders.12,13,14,15,16,17,18,19 In particular, DNM1-related disorders have a distinctive clinical presentation. Individuals with disease-causing variants in DNM1 typically present with profound hypotonia from birth, cortical visual impairment, and refractory infantile spasms with onset at 4–6 months of age that frequently evolve to Lennox-Gastaut syndrome with severe to profound developmental delay and intellectual disability.17,19
DNM1 was first identified as a genetic etiology for epilepsy in 2014.19 DNM1 encodes dynamin-1, a protein essential for clathrin-mediated endocytosis and synaptic vesicle recycling. Dynamin-1 oligomers perform the fission of clathrin-coated vesicles from the plasma membrane, thereby promoting vesicle-mediated neurotransmitter release at the synapse.20,21 Each dynamin-1 unit consists of a GTPase domain, middle domain, pleckstrin homology (PH) domain, GTPase effector domain (GED), and proline rich domain (PRD).20 In mammals, this gene may lead to several isoforms distinguished chiefly by the alternative splicing of the tenth exon—some transcripts contain exon 10b and others contain the further-downstream exon 10a.22
The mechanism of disease in DNM1-related disorders is primarily due to dominant-negative effects of mutant dynamin-1 secondary to missense variants.17,23 The dominant-negative mechanism of disease occurs when the mutant protein blocks the function of the wild-type protein product as well, causing the overall functional gene product to be reduced beyond what is expected from a single loss of function variant. In the case of DNM1, this is predicted to be due to formation of non-functional oligomers including mutant and wild-type dynamin-1. Prior to the current study, it has been estimated that one-third of individuals have a single recurrent missense variant, c.709C>T (GenBank: NM_004408.3) with consequence p.Arg237Trp; this and other recurrent variants, such as c.616A>G (GenBank: NM_004408.3), c.618G>C (GenBank: NM_004408.3), c.1075G>A (GenBank: NM_004408.3), and c.1076G>C (GenBank: NM_004408.3) with consequences p.Lys206Glu, p.Lys206Asn, p.Gly359Arg, and p.Gly359Ala, respectively, are concentrated in the GTPase and middle domains.17
Haploinsufficiency, which occurs when only one wild-type gene copy is insufficient to preserve normal cellular functioning as a result of reduced abundance of the protein product, is not a known mechanism of disease of DNM1; individuals with putative loss-of-function (pLoF) variants in DNM1, such as nonsense, frameshift, or splice site variants, have been identified in the healthy population.24 Nevertheless, there have been two previous reports of a likely disease-causing splicing variant, c.1197−8G>A (GenBank: NM_001288739.1), which affects the alternately spliced exon 10a of this gene.25,26 The discovery of this possibly pathogenic splicing variant underscores the need to better understand the transcriptional profile of DNM1, which includes five known protein-coding transcripts in addition to 24 other predicted transcripts, mostly partial, in the Ensembl database.27
Here, we performed RNA sequencing (RNA-seq) in 39 pediatric brain samples to understand the transcriptional complexity of DNM1. We find that the canonical DNM1 transcript including the alternative exon 10b is not the predominant brain isoform. Instead, we identify transcripts containing exon 10a as the major cortical DNM1 isoform. We extend this finding to understand genotype-phenotype correlations in 11 previously unreported individuals with disease-causing variants in DNM1 affecting either exon 10a or exon 10b and find that disruption of exon 10a leads to the more severe, and more commonly reported, DEE phenotype.
Material and methods
Participant recruitment
Informed consent for participation in this study was obtained from the parents of all subjects in agreement with the Declaration of Helsinki with approval by the institutional review board (IRB) at respective institutions.
Six individuals included in the study were recruited at Children’s Hospital of Philadelphia (CHOP). Four individuals were referred by collaborating institutions: Children’s Hospital of San Antonio, USA (individuals 5 and 6); Universidade Federal de Minas Gerais, Brazil (individual 7); and Assistance Hôpitaux Publique de Paris, France (individual 8). One individual was referred by a US clinical genetic testing laboratory.
Genetic analysis and clinical review
Prior to study inclusion, all individuals underwent diagnostic genetic testing, including gene panel (n = 1) and whole-exome sequencing (WES, n = 9). Gene panel testing in one individual was performed via whole-genome sequencing and targeted analysis of 1,362 genes at Hôpitaux Universitaires de Genève (Geneva, Switzerland). Whole-exome sequencing was performed in seven individuals by GeneDx with the IDT xGen Exome Research Panel v1.0 (Integrated DNA Technologies); in one individual by the Hospital for Sick Children (Toronto, Canada) with the Agilent SureSelect Focused Exome Kit (Agilent); in one individual at Hôpital de la Pitié Salpêtrière (Paris, France) with the SeqCap EZ MedExome Library kit (NimbleGen, Roche Sequencing); and in one individual by CHOP with the Agilent SureSelect Clinical Research Exome Kit (Agilent). Analysis, variant calling, and interpretation by ACMG criteria28 were performed with locally developed pipelines at each institution, and variants were confirmed via additional methods including Sanger sequencing for all probands.
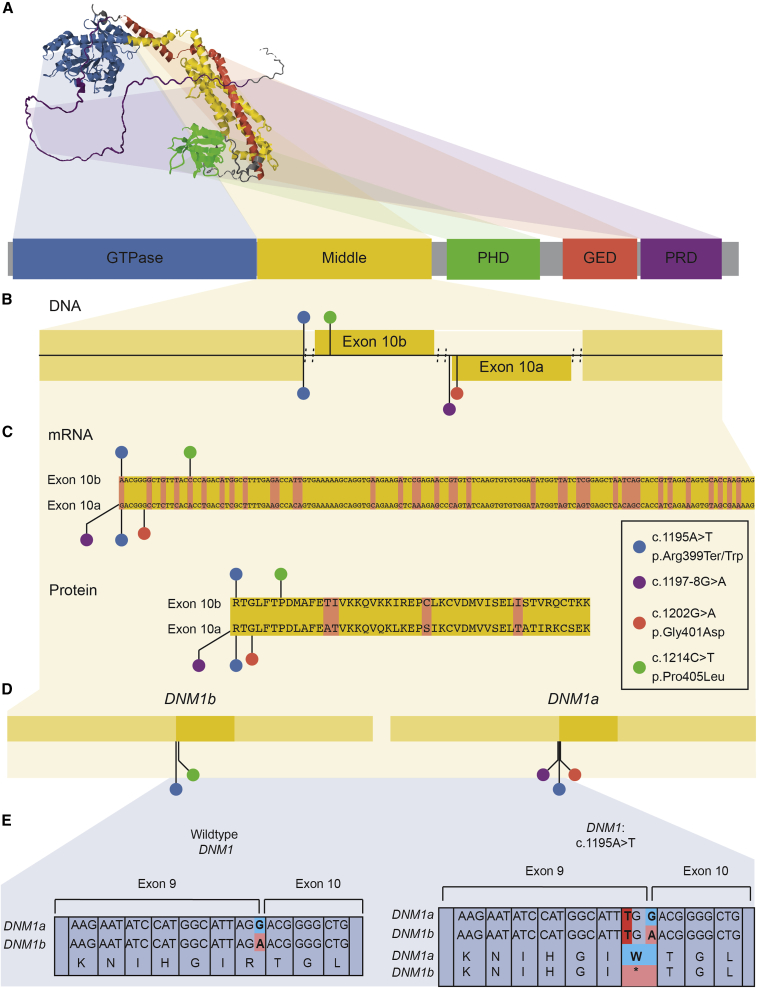
De novo variants in all subjects mapped within or in proximity to exons 10a or 10b within DNM1 (Figures 1A and 1B). Each individual underwent a clinical data review of medical history information obtained from medical records and supplemented in nine individuals by parent interview conducted by a licensed genetic counselor (S.M.R., Table 1). Medical record review included developmental and seizure history, neurological findings, and morphological details. Available electroencephalogram (EEG) and brain imaging data were reviewed for all individuals. Seizure types were classified according to the International League Against Epilepsy (ILAE) classification criteria.4 An overall assessment of developmental status was made via chart review with the Gross Motor Functional Classification Scale (GMFCS) for gross motor function,29 either the Manual Abilities Classification Scale (MACS) for children over 4 years of age30 or the Mini-Manual Ability Classification System (mini-MACS) for children 1–4 years of age for hand use,31 and the Communication Function Classification System (CFCS) for communication skills.32 These ordinal scales were originally developed for children with cerebral palsy and are composed of five levels with scores of “I” indicating higher levels of function and scores of “V” representing severe limitations in function.
Figure 1.
Structure and location of variants in DNM1
(A) Predicted 3D structure of dynamin-1 from AlphaFold and primary sequence of dynamin-1, highlighting distinct domains.
(B and C) Subjects’ variants, including the recurrent variant c.1197−8G>A (GenBank: NM_001288739.1), mapped to the dynamin-1 middle domain primary sequence, including alternative exons. The pathogenic variant-rich middle domain includes the alternatively spliced exon 10 of DNM1, with multiple disease-causing variants identified at positions uniquely affecting exon 10a.
(D) Subjects’ variants have different effects on each isoform. Variants causing severe disease affect exon 10a in the DNM1a isoform.
(E) Exon 10-dependent effects of the c.1195A>T variant. This variant is missense in DNM1a but nonsense in DNM1b; dominant-negative missense, not loss-of-function, variants are the established disease mechanism of DNM1.
Table 1.
Clinical and genetic features in 11 individuals with DNM1-related disorders
| Age at last eval. | Sex | Variant | Exon | Epilepsy/ seizure types | Age of seizure onset | Developmental features | Other notable features | EEG features | MRI features | |
|---|---|---|---|---|---|---|---|---|---|---|
| Individual 1 | 2 years | male | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | CE, ES, MS, SS, refractory | 2 months | profound DD, GMFCS V, CFCS V, MiniMACS V | dystonic movement disorder, profound hypotonia, CVI | Hx of HA, multifocal SW, poorly organized background | markedly decreased cerebral volume, deficiency of WM, small brainstem and CC |
| Individual 2 | 2 years | male | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | CE, ES, refractory | 2 months | profound DD, GMFCS V, CFCS V, MiniMACS V | profound hypotonia, CVI, significant irritability | Hx of HA, multifocal SW, poorly organized background, excessive discontinuity | normal at 1 year |
| Individual 3 | 3 years | female | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | ES, refractory | 2.5 months | profound DD, GMFCS V, CFCS V, MiniMACS V | profound hypotonia, CVI | not available | not available |
| Individual 4 | 7 years | male | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | ES, FE, refractory initially, now SF | 6 months | severe DD, GMFCS V, CFCS IV | profound hypotonia, CVI | Hx of HA | hypoplasia of CC and enlarged ventricles |
| Individual 5 | 2 years | male | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | ES, refractory | 2 months | profound DD, GMFCS V, CFCS V, MiniMACS V | profound hypotonia, CVI, nystagmus, pupils nonreactive | Hx of HA, multifocal discharges | normal at 6 months |
| Individual 6 | 2 years | male | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | ES, refractory | 2 months | profound DD, GMFCS V, CFCS V, MiniMACS V | profound hypotonia, CVI, nystagmus, pupils nonreactive | Hx of HA, multifocal discharges | normal at 6 months |
| Individual 7 | 3 years | female | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | MS, refractory | 2 months | profound DD, GMFCS V, CFCS V | profound hypotonia with peripheral spasticity | not available | normal |
| Individual 8 | deceased at 32 months | male | c.1197−8G>A (GenBank: NM_001288739.1) | 10a | MS, FS, refractory | 3 months | profound DD, GMFCS V, CFCS V | dystonic movement disorder, profound hypotonia, CVI | asynchrony, electroclinical myoclonic seizures | diffuse cerebral atrophy |
| Individual 9 | 3 years | female | c.1202G>A (GenBank: NM_001288739.1) (p.Gly401Asp [GenBank: NP_001275668.1]) | 10a | no clinical seizures | 6 months | profound DD, GMFCS V, CFCS V, MiniMACS V | profound hypotonia, CVI, nystagmus | Hx of HA, mild background slowing, focal spikes and polyspikes | abnormal diffusion restriction attributed to vigabatrin toxicity, delayed myelination, mild enlargement of ventricles, small CC |
| Individual 10 | deceased at 10 months | male | c.1195A>T (GenBank: NM_004408.3) (p.Arg399Ter [GenBank: NP_004399.2]), c.1195A>T (GenBank: NM_001288739.1) (p.Arg399Trp [GenBank: NP_001275668.1]) | 10a | ES, FS, refractory | 3 months | profound DD, GMFCS V, CFCS V | hyperkinetic dystonic movement disorder, profound hypotonia, CVI | HA | not available |
| Individual 11 | 5 years | female | c.1214C>T (GenBank: NM_004408.3) (p.Pro405Leu [GenBank: NP_004399.2]) | 10b | no seizures | N/A | mild DD, GMFCS I, CFCS I, MACS II | mild hypotonia, behavioral problems | moderate increase in beta frequencies | normal at 5 years |
ES, epileptic spasms; MS, myoclonic seizures; SS, subclinical seizures; GE, generalized epilepsy; FE, focal epilepsy; CE, combined epilepsy; LGS, Lennox-Gastaut syndrome; DD, developmental delay; GMFCS, Gross Motor Function Classification System; CFCS, Communication Function Classification System; CVI, cortical visual impairment; HA, hypsarrhythmia; WM, white matter; SF, seizure free; Hx, history; N/A, not applicable.
Assessment of relative transcript expression
RNA-seq was performed on brain tissue samples from 39 individuals from an independent cohort of individuals undergoing resective epilepsy surgery by Novogene Corporation with the NovaSeq 6000 PE150 platform (Illumina). Raw short read files were quantified with Kallisto, version 0.45.0, against the GRCh38 human reference genome.33 Transcript isoform information was downloaded from Ensembl with the R package biomaRt.34 Transcript isoform abundance was estimated with output from Kallisto. Data were subsequently analyzed with R Statistical Framework. We filtered transcript variants to exclude those less than 1,500 bp in length. The remainder were classified as containing exon 10a (“isoform DNM1a” consisting of ENST00000393594 and ENST00000486160) or 10b (“isoform DNM1b” consisting of ENST00000628346, ENST00000341179, and ENST00000372923). We assessed the correlation of proportional expression of each isoform with the age at sample collection by using Pearson’s r, and we used Student’s t test to assess statistical significance of the ratio of DNM1a to DNM1b expression.
Determination of splicing effects of recurrent intronic variant
We used the MaxEnt and NNSPLICE algorithms within the Alamut software (Interactive Biosoftware, France) to predict the effect of the c.1197−8G>A (GenBank: NM_001288739.1) variant on DNM1. Additionally, we used the SpliceAI algorithm to predict any splicing changes caused by this variant as well as splicing or intronic variants in DNM1 that were present in gnomAD by using the default high-precision threshold value of 0.8.35
We then used the pCAS2 splicing reporter minigene assay to experimentally determine the effect of this variant on the splicing pattern in DNM1. The DNM1 amplification was performed with specific primers pair (DNM1_forward 5ʹ-GGGTCTTGTACGGAGCAGGG-3ʹ and DNM1_reverse 5ʹ-GAGTCAGATAGTAAGGGCAAGCAC-3ʹ) with Pfu DNA polymerase (Promega). After the first amplification to select DNM1 fragment of 761 bp, we performed a nested-PCR to construct the 494 bp minigene with the following primers: DNM1_Bam forward 5ʹ-CGGATCCACGCTCACATTGTC-3ʹ and DNM1_Mlu reverse 5ʹ-GACGCGTCATGAGTGCAAGTACC-3ʹ. We inserted the restriction enzyme sites BamHI and MluI into the primer sequences to enable directional cloning. The minigene amplification product was inserted in pGEM T-easy vector (Promega) according to the manufacturer’s instructions. The construction was transformed into E. coli DH5α competent cells. After PCR amplification, we isolated clones with the correct insert size by using the Wizard Plus SV Minipreps DNA Purification System (Promega) and sequenced them to verify their identity. As the variant c.1197−8G>A was found in heterozygosis, a minigene from the wild-type sequence (normal allele) was used as control. pGEM wild-type and variant clones were digested with BamHI and MluI restriction enzymes, respectively. The digested minigenes were purified with Wizard SV Gel and PCR Clean-UP System (Promega) and ligated into a BamHI/MluI opened pCAS2 splicing reporter minigene vector (kindly provided by Dr. Alexandra Martins, Université de Rouen).36 We used the constructions to transform XL1B E. Coli competent cells (Stratagene), and we selected colonies to verify the presence of the DNM1 fragment. Plasmid DNA was isolated with the Midprep kit (MN), and the sequence of the construct was verified by sequencing.
The pCAS2 wild-type and variant constructs were expressed by transient transfection into HEK293 cells. Approximately 5 × 105 cells grown in each 60 mm diameter tissue culture dish containing 4 mL Dulbecco’s modified Eagle’s medium-high glucose (Sigma-Aldrich) supplemented with 5 mM sodium bicarbonate (Cinética Química Ltda), penicillin (Sigma-Aldrich, 100 units/mL), streptomycin (Sigma-Aldrich, 0.1 mg/mL), amphotericin B (Sigma-Aldrich, 0.25 μg/mL), gentamicin (Schering-Plough, 60 mg/L), and 10% of fetal bovine serum (FBS) (Cripion Biotecnologia). After 24 h, HEK293 cell cultures were transfected with 15 μg of the plasmid DNA via Lipofectamine 2000 (Invitrogen). Transfected cells were harvested 24 h after transfection and total RNA was isolated with RNeasy mini kit (Qiagen) according to the manufacturer’s instructions, followed by DNase Amp Grade (Invitrogen) digestion. First-strand synthesis was performed with Superscript III kit (Invitrogen) according to the manufacturer's recommendations. The cDNA was amplified by PCR with pCAS2-specific primers located in the flanking SERPING1/C1NH exons A and B.36 PCR products were analyzed by electrophoresis on a 1.5% agarose gel and the products were directly sequenced with SERPING1/C1NH exons A and B primers. Transfections were performed five times on different time points, and no difference in the splicing results was observed between any of these independent assays.
Structural modeling of splicing variant
Solved three-dimensional structures of dynamin-1 were obtained by searching the RCSB Protein Data Bank (PDB). Several different X-ray crystal structures of dynamin-1 have been solved (PDB ID: 2X2E, 2X2F, 3ZYC, and 3SNH) in addition to a medium resolution EM structure (PDB ID: 3ZYS) and an X-ray structure showing the tetrameric arrangements (PDB ID: 5A3F). The impact of the splicing variant was modeled on the tertiary structure of dynamin-1 on the basis of these structures.37 Maestro was employed (Schrodinger, New York, NY) and wild-type and mutant structures were minimized with the MMF94s forcefield in Sybyl-X 2.1.1 (Certara L.P., St Louis, MO); the final structure had more than 95% of residues in the allowed region of a Ramachandran plot.
We employed a comprehensive computational platform for analyzing mutation effects on protein structure function and interactions to identify potential molecular mechanisms leading to the observed phenotype. For mutations affecting regions located close to one of the protein-protein interfaces (<10 Å), we employed the mCSM-PPI238 and mmCSM-PPI39 tools to assess effects on oligomerization and binding affinity. Mutation effects on stability and dynamics were also assessed with the mCSM40,41 and DynaMut42,43 platforms. These provide a quantitative assessment of effects of mutations given as differences in Gibbs Free Energy (ΔΔG in Kcal/mol). Potential effects on GTP binding were also assessed with mCSM-lig.44
Neuropathology analysis
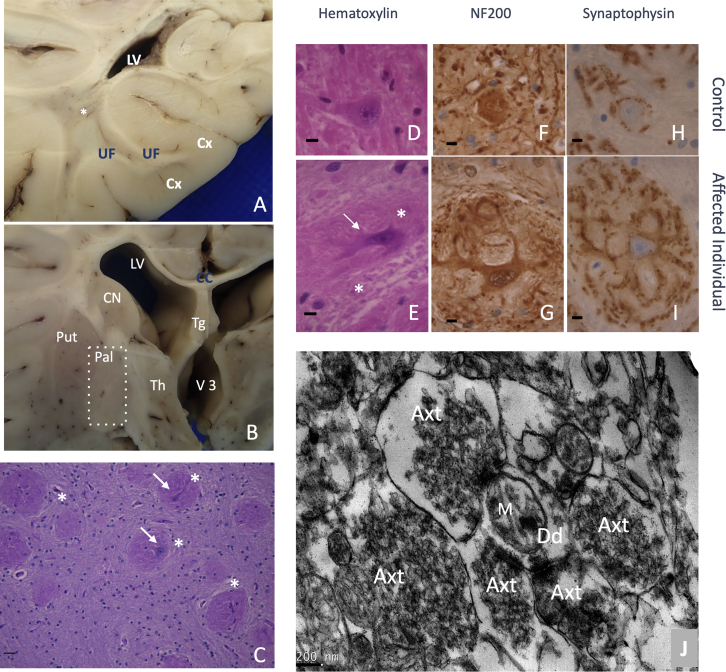
This study included one affected individual and one age-matched control individual obtained from the brain collection “Hôpitaux Universitaires de l’Est Parisien – Neuropathologie du développement” (biobank identification number BB-0033-00082). For both individuals studied, informed consent was obtained for autopsy of the brain and histological examination. Analysis was conducted by a pathologist with expertise in pediatric neuropathology (A.G.) with comparison to controls as well as established standards.45 After removal, brains were fixed with formalin for 5–12 weeks. Macroscopic analysis was performed, allowing the selection and conditioning of samples (paraffin embedding, 7-micron slicing, hematein and periodic acid–Schiff [PAS] staining) of brain tissue for histological analysis. Immunohistochemistry was also conducted with anti-neurofilament 200 (ready to use DAKO clone2F1—reference: IS607) and anti-synaptophysin (ready to use clone DAKO-SYNAPTO—reference: IR660). Brains biopsies (globus pallidus, cervical spinal cord) were performed, and the samples were conditioned for electron microscopy as follows. Tissue was fixed in 3% glutaraldehyde in phosphate buffer (pH 7.4) for 1 h, washed, post-fixed with 1% osmium tetroxide in 0.1 M phosphate buffer, and then gradually dehydrated in 70%, 90%, and 100% ethanol. After 10 min in a 1:2 mixture of epoxy propane and epoxy resin and 10 mn in epon, coverslips were covered by upside down gelatin capsules filled with freshly prepared epoxy resin and polymerized at 60°C for 24 h. After heat separation from the coverslip, ultrathin sections of 90 nm were cut with an ultra-microtome (Reichert ultracut S), stained with uranyl acetate and Reynold’s lead, and observed with a transmission electron microscope (JEOL 1011). Acquisition was performed with a Gatan ES1000W CCD camera.
For this individual and five control specimens obtained from the above-specified biobank, vesicles were measured in samples from the skin and central nervous system. After fixation, samples were washed three times over the course of a few hours. Secondary fixation was performed with 1% osmium tetroxide in phosphate buffer saline for 1 h at room temperature, washed three times in H2O, and further dehydrated with increasing concentrations of ethanol (70%, 90%, and 100% twice). After dehydration, samples were infiltrated with sequential 3:1, 1:1, and 1:3 solutions of resin (Embed 812 kit; Electron Microscopy Sciences, Hatfield, PA) and 100% ethanol for 1 h separately at room temperature. Finally, the samples were incubated with propylene oxide for 2 min and placed in the embedding molds with resin. The embedding resin blocks were polymerized in an oven at 60°C for 24 h. Ultrathin sections (90 nm) were cut from the block surface with an ultramicrotome (Reichert Ultracut S ultramicrotome, Leica Microsystems), stained with uranyl acetate and lead citrate (LFG, France), and observed by TEM (JEOL 1011, JEOL, Japan) at 80 kV of acceleration voltage. Acquisitions were performed by an Orius 1000 CCD Camera (GATAN, USA) with Digital Micrograph software (GATAN, USA) for measurement. Measurements were done on synaptic terminals in the skin, thalamus, cerebellum, globus pallidus, and cervical spinal cord. Skin samples used as controls were from normal biopsies, and brain control samples were from individuals carrying acquired pathologies confirmed by autopsy. In the affected individual and controls, measurements were done in 24 and 35 synaptic terminals, respectively, in skin, and 28 and 16 synaptic terminals, respectively, in brain. For each measured vesicle, when its shape was not spherical, the smallest diameter was considered. We analyzed synaptic vesicle diameter values with Graph Prism 7.03 by using the Shapiro-Wilk normality test and then the Mann-Whitney test to compare values from the affected individuals with those of the controls.
Results
DNM1a is the predominant isoform expressed in the pediatric brain
DNM1 encodes five known protein-coding transcripts in addition to 19 other predicted transcripts, mostly partial, in the Ensembl database.27 We determined relative expression of 24 transcripts, including two full-length transcripts containing exon 10a and three containing exon 10b, from RNA-seq data in brain tissue samples following surgical resection in a pediatric cohort of 39 individuals without disease-causing DNM1 variants (Table S1). Thirteen individuals (33%) were female and 26 male (67%). 30 individuals (77%) had a diagnosis of focal epilepsy, and 9 (23%) had a diagnosis of epileptic encephalopathy. Intellectual disability or developmental delay was documented to be present in 15 individuals (38%) and absent in 24 (62%). Age of seizure onset ranged from 2 months to 13 years with a median of 4 years. The median age of sample collection was 8.6 years with a range of 14 months to 23 years.
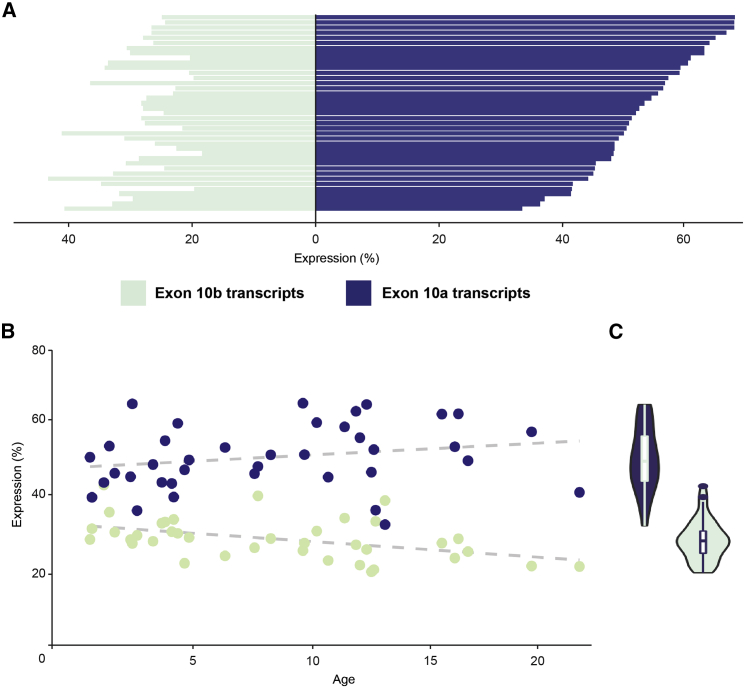
Across this cohort with broad age range and diverse phenotypes, proportional expression of DNM1a, or transcripts containing exon 10a, was consistently higher than that of DNM1b, containing exon 10b, which includes the canonical transcript, with a mean ratio of 2:1 (Student’s t = 10.587, p < 0.001, Figures 2A and S1). This expression pattern for the alternative tenth exons were not strongly correlated with individuals’ age (DNM1a: Pearson’s r = 0.22, p = 0.19; DNM1b: Pearson’s r = −0.46, p = 0.004; Figure 2B), reflecting consistency in the DNM1 transcriptional landscape across early and adolescent development (Figure 2C). As such, DNM1a, which is distinct from the canonical DNM1 sequence, is the primary cortical isoform with double the expression on average compared to DNM1b.
Figure 2.
Expression profiles of DNM1 transcripts from Ensembl in 39 pediatric brain tissue samples
(A) Individual transcript expression, highlighting identifiable transcripts within RefSeq. Bars represent individual samples.
(B) Proportional transcript expression for transcripts containing exon 10a and exon 10b versus age, showing age and expression are not strongly correlated.
(C) Cumulative expression per individual of full-length DNM1 transcripts containing exon 10a (blue) or exon 10b (green). DNM1a has markedly higher expression than DNM1b in the pediatric brain. Proportional transcript expression was computed by quantifying transcripts in TPM, classifying by 10th exon, and normalizing to the total TPM across all DNM1 transcripts. All data are reported as ratios.
Several disease-causing de novo variants in DNM1 affect exon 10a
Given that DNM1 expression patterns strongly favor expression of exon 10a over exon 10b in the brain, we aimed to characterize clinical features of individuals with variants differentially affecting exon 10a or 10b (Figures 1B–1D). Individuals 1–8 had a recurrent splice site variant,25,26 c.1197−8G>A (GenBank: NM_001288739.1). This variant is directly upstream of the splice acceptor preceding exon 10a and as such is expected to affect the splicing pattern of DNM1 transcripts containing exon 10a but not 10b. Individual 9 had a missense variant, c.1202G>A (GenBank: NM_001288739.1) with the protein consequence p.Gly401Asp. This nucleotide substitution is within exon 10a and therefore results in an altered protein product from exon 10a-containing DNM1 transcripts only. Individual 10 had a variant that was identified as nonsense, c.1195A>T (GenBank: NM_004408.3) with the protein consequence p.Arg399Ter. The affected codon includes the final two bases in exon 9 and the first base of exon 10a or 10b depending on the isoform. When assessed with a transcript containing exon 10b, as reported in clinical genetic testing, the nucleotide substitution changed this codon from CGA to TGA, a nonsense mutation inconsistent with known disease-causing variants. However, if exon 10a is considered instead, this substitution results in a change from CGA to TGG with the protein consequence p.Arg399Trp (Figure 1E). This is a missense variant in the middle domain, which is known to be enriched for disease-causing variants with dominant-negative effects.17 Individual 11 had a missense variant, c.1214C>T (GenBank: NM_004408.3) with the protein consequence p.Pro405Leu. This is a nucleotide substitution within exon 10b, leading to a missense variant in isoforms including this exon but not in those including exon 10a.
A recurrent splice variant causes an in-frame insertion disrupting DNM1 oligomerization
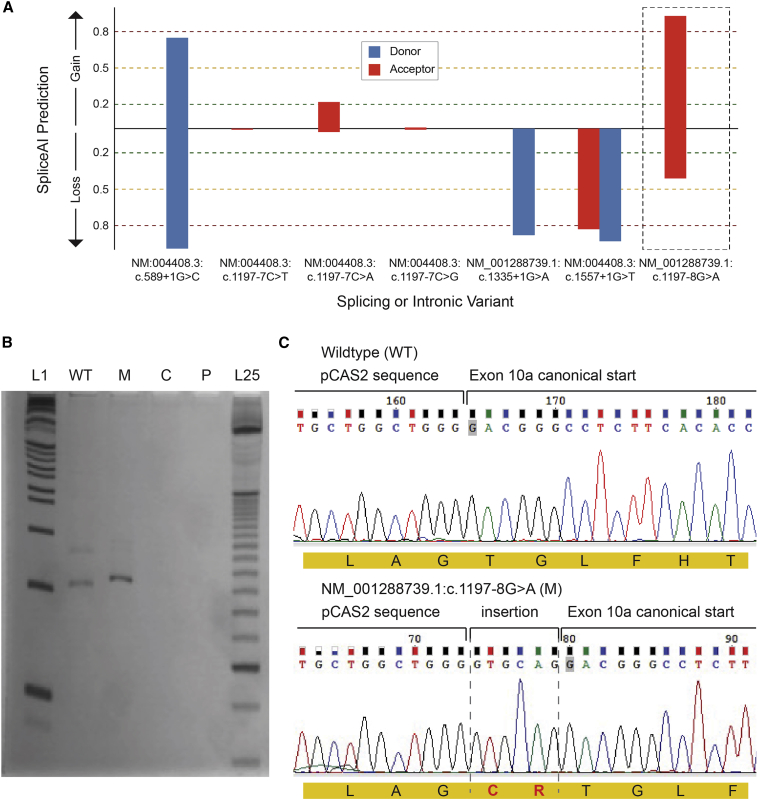
DNM1-related disorders are known to cause DEE through a dominant-negative disease mechanism.17,23 However, although the phenotypes of individuals 1–8 mirrored the previously described DEE phenotype, splice site variants are typically assumed to be putative loss-of-function variants, which do not have dominant-negative effects. Therefore, we investigated the functional consequences of this splice site variant. First, we evaluated the effects of on dynamin-1 monomers. We examined this variant with multiple splice site prediction algorithms and found that the c.1197−8G>A (GenBank: NM_001288739.1) variant is predicted to create a new cryptic acceptor splice site 6 bp upstream of the canonical site of exon 10 in the stalk of the middle domain. The predicted change at the canonical site is −80.9% according to the MaxEnt algorithm and −44.5% according to the NNSPLICE algorithm. Moreover, SpliceAI predictions showed that splicing or intronic variants observed in healthy population controls most likely lead to either donor or acceptor loss, which would not lead to a functional protein product. These population variants include c.589+1G>C (GenBank: NM_004408.3), c.1197−7C>D (GenBank: NM_004408.3), c.1335+1G>A (GenBank: NM_001288739.1), and c.1557+1G>T (GenBank: NM_004408.3). In contrast, the c.1197−8G>A (GenBank: NM_001288739.1) variant had a likelihood of 93% to cause a splice acceptor gain (Figure 3A) with the consequence of a predicted 6 bp insertion.
Figure 3.
Splicing assay assessing consequence of c.1197−8G>A (GenBank: NM_001288739.1) variant
(A) SpliceAI predictions for the consequence of population variants in gnomAD including curated, predicted loss-of-function splicing variants; variants at a position similar to the recurrent disease variant, i.e., c.1197−7 (GenBank: NM:004408.3); and the recurrent disease variant itself. Only the disease variant (c.1197−8G>A [GenBank: NM_001288739]) is strongly predicted to have an effect that may not be loss of function.
(B) cDNAs PCR products. L1, 1 Kb plus DNA ladder (Invitrogen); WT, wild-type amplification; M, mutant amplification; C, amplification from HEK293 cells without minigene transfection; P, PCR control without cDNA; L25, 25 bp ladder (Invitrogen).
(C) Sanger sequencing of wild-type and mutant PCR products. In red, the insertion of “GTGCAG” from 5ʹ of intron 9 to the mutant transcript, which results in a mutant protein with a cysteine and an arginine (CR) in-frame insertion after Arg399 residue.
To verify the validity of these in silico predictions, we also investigated the splicing variant experimentally with the pCAS2 splicing reporter minigene assay. By sequencing amplified cDNA following transfection of wild-type and mutant DNM1 into HEK293 cells, we confirmed that the c.1197−8G>A (GenBank: NM_001288739.1) variant indeed created a new splice site upstream of the canonical splice site, leading to the insertion of two amino acid residues, a cysteine and an arginine (CR), in the beginning of exon 10, between Arg399 and Thr400 in the stalk region of the middle domain (Figures 3B and 3C).
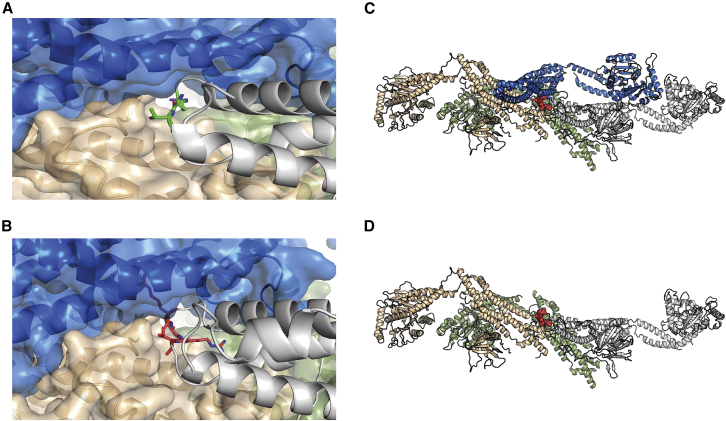
To discern the impact of this two amino acid insertion on the tertiary and quaternary structures of dynamin-1, we modeled this variant on the basis of established structures of the protein. Using several previously described computational tools,37,38,39,40,41,42,43,44 we found that in the protomeric state the insertions were on a solvent-exposed loop with minimal effect to monomer stability (ΔΔG = +0.2 Kcal/mol). However, the insertion was located at one of the interfaces within the dynamin-1 tetramer, and two extra residues would sterically hinder the formation of the active tetramer. The tetramer is formed by a pair of parallel protomers. When a mutant protomer containing the CR insertion formed part of the complex, the corresponding parallel protomer (Figures 4A and 4B; blue) could no longer bind because of steric hindrance (Figures 4A and 4B). Consequently, the insertion results in a mutant protein with normal interactions through the GTPase domain and with one anti-parallel protomer but disrupted interactions with one parallel protomer and the remaining anti-parallel protomer (Figures 4C and 4D), forming an oligomeric complex without the capacity for oligomerization-induced GTPase activation. By thus sequestering wild-type proteins needed to form the active complexes, the c.1197−8G>A (GenBank: NM_001288739.1) variant inactivates the enzyme without affecting monomeric structure. As such, a subset of wild-type dynamin-1 would be prevented from exhibiting normal function, reflecting a dominant-negative mechanism that is known to cause severe disease.
Figure 4.
Structural modeling of the effects of the c.1197−8G>A (GenBank: NM_001288739.1) variant
(A) Mutation site in dynamin-1 corresponding to c.1197−8G>A (GenBank: NM_001288739.1). Green, two residues neighboring the CR insertion.
(B) Model of the CR insertion (pink). Given the lack of space to accommodate, the new residues steric clashes would be created, disrupting the interactions to the neighboring protomers.
(C and D) Tetrameric organization of DNM1 (PDB ID: 5A3F). The tetramer is formed by two sets of parallel protomers facing in the same direction. Gray and green and blue and orange. When a mutant protomer containing the CR insertion (bottom: gray chain) forms part of the complex, the corresponding parallel protomer (blue) can no longer bind as a result of steric hindrance.
Neuropathology reveals vesicular accumulation in an individual with c.1197−8G>A (GenBank: NM_001288739.1)
Having established that the recurrent variant c.1197−8G>A (GenBank: NM_001288739.1) is predicted to cause dominant-negative disease, we next sought to assess its effects on native vesicular function. In the presynaptic terminal, neurotransmitter release relies on vesicles fusing with the membrane and subsequently being recycled via endocytosis. Dynamin-1 is known to perform vesicle fission, or the process of pinching synaptic vesicles off the plasma membrane. Thus, we hypothesized that this variant would lead to evidence of impaired synaptic vesicle fission. In one deceased individual (individual 8), neuropathology analysis permitted detailed exploration of the effects of this recurrent splice variant on the synapse. This individual had significantly reduced brain weight of 902 g, compared to the age-specific normal value (95% CI: 1,083.1–1,215.3 g).45 This individual’s brain also showed diffuse atrophy of cortical gray and white matter as well as central reduction of white matter (Figures 5A and 5B). Infratentorial structures appeared macroscopically preserved.
Figure 5.
Neuropathological hallmarks of the c.1197−8G>A (GenBank: NM_001288739.1) variant
(A and B) Brain macroscopic aspect. (A) White matter displayed severe atrophy with lateral ventriculomegaly, spindly gyri, and extremely thin corpus callosum. A central myelin discoloration was observed consisting of a myelin defect sparing U fiber. (B) Among gray structures, thalamus was severely atrophic associated with a third ventricle distention. Cortical thickness seemed preserved but was difficult to estimate considering the associated severe white matter atrophy. CC, corpus callosum; Cx, cortex; LV, lateral ventricle; Put, putamen; Pal, pallidum; Th, thalamus; Tg, trigone; UF, U fibers; V3, third ventricles, ∗, deep white matter.
(C–J) Pallidal synaptic dysplasia. (C) Pallidal glomeruli (star) appeared as rounded formations containing intermingled eosinophilic neurites (arrow). (D–I) After immunohistochemistry and control matching, these glomeruli (E) (star) consisted of exuberant neurites proliferation (arrow) (NF200(+)) that were covered by diffuse synaptic areas (synaptophysin). (D), (F), and (H) are age-matched controls; (F) and (G) are immunohistochemistry with anti-neurofilament 200; (H) and (I) are with anti-synaptophysin; initial magnification ×100. (J) Brain biopsy at pallidum level after electron microscopy conditioning. Initial magnification: ×7,500. Five hyperplastic synaptic terminations (Axt) surrounding one receptive dendrite (Dd). Synaptic vesicles appeared abnormally numerous and packed. M, mitochondria.
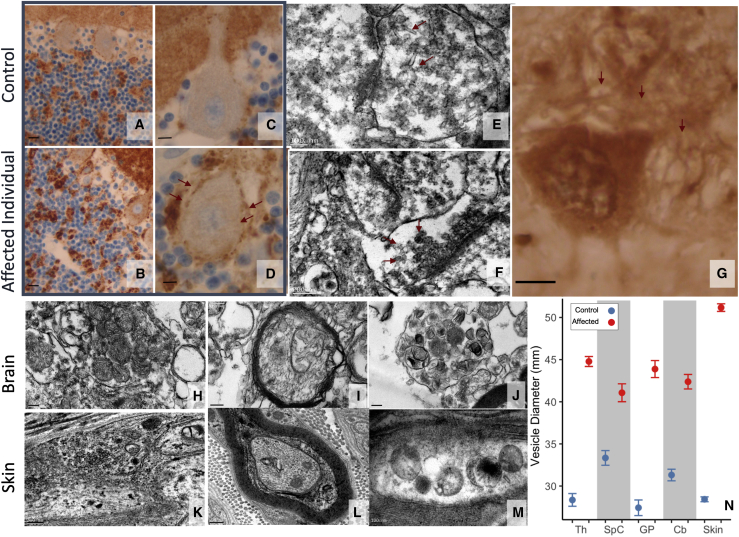
At the histological level, we identified three types of lesions, which were clearly absent from the controls and are not observed in normal brain tissue and are therefore reflective of pathology in the affected individual. (1) In gray structures, we observed decreased neuronal density, which was severe in the thalamus and pyramidal layers of the cortex and less marked in the striatum and globus pallidus. (2) In white matter, we observed that the leukodystrophy was related to a decrease in the density of axons whose myelin was preserved, i.e., no naked axons or hypercellularity. (3) We found a diffuse synaptic dystrophy, particularly marked at the histological level in the internal globus pallidus, where the soma of the pyramidal neurons were embedded by glomerules consisting of a lace of exuberant dendrites displaying hyperplastic synaptic zones (Figures 5C–5I and 6G). Similarly, compared to control samples, in the cerebellar cortex, synaptic areas were hypertrophic in the internal granular layer and increased in size and number around the soma of Purkinje cells (Figures 6A–6D). At the ultrastructural level, hypertrophic synaptic terminals (Figure 5J) were filled by vesicles that were irregular in size and shape, sometimes displaying a tubular shape (Figure 6E), and densely packed against the cell membrane (Figure 6F); these features were consistent with failed vesicular fission. The observed vesicles were significantly larger in diameter than those measured in controls in multiple central nervous system areas (p < 0.001, Figure 6N). In addition, we observed signs of intra-axonal trafficking defect, with accumulation of organelles such as vesicular bodies or mitochondria (Figures 6H–6J), and the latter displayed abnormalities in size, shape, and crest organization. Comparable features, i.e., excessive visibility of synaptic zones, axonal trafficking defect, and significantly enlarged vesicles, were also observed in the peripheral nervous system by ultrastructural analysis of the skin biopsy samples (Figures 6K–N). Taken together, these findings reveal marked synaptic structural abnormalities with an accumulation of vesicles at the plasma membrane, consistent with disruption of vesicular fission due to mutant dynamin-1.
Figure 6.
Synaptic dysplasia expression in brain and skin biopsy
(A–D) Cerebellar cortex revealed an excess of synapses as disorganization of granular glomeruli (arrows); (A) and (B) are with initial magnification ×40 and (C) and (D) are with ×100; (A) and (C) are age-matched controls.
(E–H) Synaptic dysplasia consisted of (G) dendrite proliferation (arrows) evident in pallidal glomeruli; (E) and (F) show excess synaptic vesicles in thalamus with abnormal shape, irregular size, densely packed and adherent to synaptic membrane (arrows); (H)–(J) show excess mitochondria in cervical spinal cord axon with abnormal appearance, with crests irregular in size, rounded and disorganized. (E) with initial magnification ×15,000, (F) with ×12,000, (G) with ×100, (H) with ×7,500, (I) with ×12,000, (J) with ×7,500.
(K–M) In skin biopsy, most of the observed axons contained heterogenous membranous vesicles that are not pathognomonic features but suggest altered trafficking. However, synaptic areas (axonal zone filled by synaptic vesicles), which are typically difficult to find in skin biopsy, were easily observed in this individual. (K) with initial magnification ×12,000, (L) with ×10,000, (M) with ×15,000.
(N) Vesicle diameters in control and affected neuropathological samples from thalamus (Th), cervical spinal cord (SpC), globus pallidus (GP), cerebellum (Cb), and skin. Points and bars are mean ± SEM. Vesicles were significantly enlarged (Mann-Whitney p < 0.001) in the affected individual in all tissue types measured.
Clinical features in individuals with exon 10a variants reflect the classical DNM1 phenotype
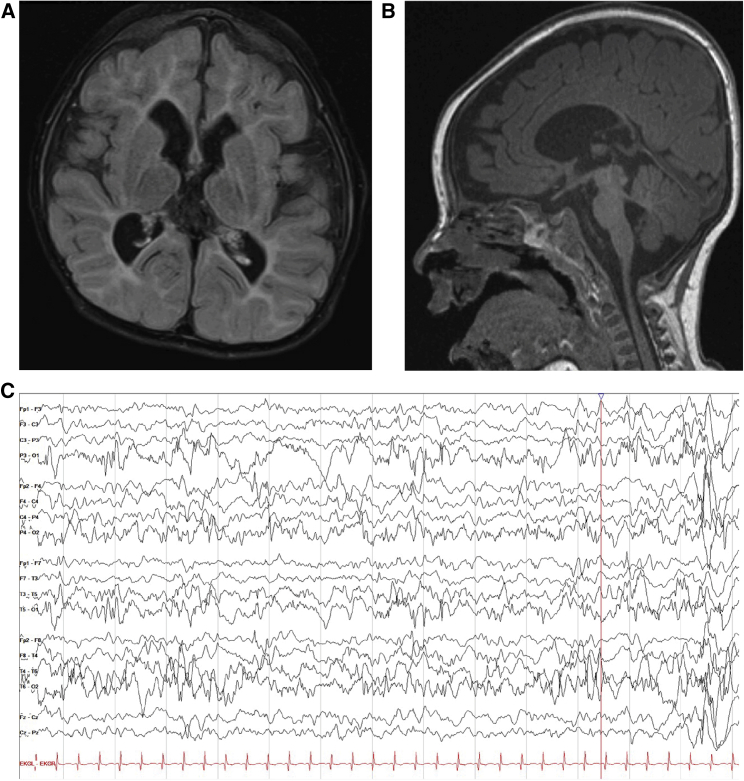
Ten individuals in this cohort had de novo variants in DNM1 affecting exon 10a. All ten of these individuals presented with a DNM1-related disorder consistent with the previously described DEE phenotype.17 Individuals 1–10 presented with early-infantile onset epileptic encephalopathies, including highly abnormal EEGs with or without clinical seizures (n = 10). Two individuals passed away, one in infancy and one in young childhood, as a result of complex feeding difficulties and severe failure to thrive. Individuals presented with either profound (n = 9) or severe (n = 1) global developmental delay. All children were classified as level V on the GMFCS, MACS/mini-MACS, and CFCS, with the exception of individual 4, indicating severe functional limitations in gross motor, fine motor, and communication skills (Table 1). The oldest child included in the cohort, individual 4, a 7 year old, was able to communicate verbally with a few individual words and by using a picture board (CFCS level IV), and he was able to stand, but not walk, with assistance (GMFCS level V). Developmental and seizure outcomes for the remaining (n = 9) individuals are limited by their young age, but the remaining individuals had achieved neither head control nor the ability to communicate with spoken language by the time of the last clinical evaluation. Eye contact and visual tracking were reduced because of cortical visual impairment (n = 9). Epilepsy presentations in all individuals were characterized by early-infantile onset seizures and abnormal EEG findings in 8/8 individuals (Figure 7C) or onset of infantile spasms with hypsarrhythmia at 6 months of age (n = 2). One individual had highly abnormal EEGs significant for polymorphic slow waves seen diffusely as well as multifocal epileptiform discharges, at times concerning for hypsarrhythmia. She had not experienced clinical or subclinical seizures. Seizures remained refractory for most individuals (n = 9); individual 4, however, had initially very refractory epilepsy but achieved excellent seizure control with combination therapy of medicinal marijuana extract, valproic acid, and lamotrigine. Notable MRI features (Figures 7A and 7B) included decreased cerebral volume over time (3/7 individuals) and decreased myelination (2/7).
Figure 7.
Imaging and electroencephalography from individual 1 with the recurrent splice variant c.1197−8G>A (GenBank: NM_001288739.1)
(A and B) MRI showing markedly decreased cerebral volume, with deficiency of cerebral white matter.
(A) Coronal FLAIR. Slightly dysmorphic lateral ventricles are enlarged, with prominent frontal horns and mild prominence of the temporal horn.
(B) Sagittal T1. Small brainstem and small corpus callosum.
(C) Abnormal EEG from the same individual with poorly organized, diffusely slow background; nearly continuous focal slowing in the left occipital region; and multi-focal epileptiform discharges.
One individual with a variant exclusively affecting exon 10b has atypically mild features
A 5-year-old girl presented with a de novo variant in DNM1, c.1214C>T (GenBank: NM_004408.3), which is located within exon 10b. This child presented to care when she was first noted to have global developmental delays in infancy and diffuse hypotonia was diagnosed at this time. She achieved walking independently at 18 months of age, and at age 4 she was able to verbally communicate with multiple-word short sentences. When she was evaluated at age 4, was classified as level I on the GMFCS and CMFCS, which indicates that she able to walk and climb stairs independently, as well communicate easily with both familiar and unfamiliar people. However, because of her hypotonia and cognitive delays, she had decreased standing balance during gross motor play activities and used supramalleolar orthotics to increase ankle stability. She had difficulty following one-step verbal instructions and had behavioral problems with decreased safety awareness. She had distinct difficulties with visual-motor skills resulting in a MACS classification of level II, indicating she can manipulate objects with decreased speed and quality of movement (Table 1). An MRI obtained following DNM1 diagnosis was normal, and her EEG was significant only for increased beta frequencies. At the time of the last clinical evaluation at age 4, she never had clinical seizures.
Discussion
Here, we report the genetic and clinical features of eleven individuals with DNM1-related disorders, including eight individuals with a recurrent de novo splicing variant in DNM1 and three individuals with previously unreported de novo variants. We provide several lines of evidence suggesting the identified recurrent de novo splicing variant acts through a dominant-negative mechanism, including (1) the syndrome phenocopies the DEE previously ascribed to the established DNM1-related disorder typically due to dominant-negative missense variants; (2) functional studies demonstrate the mutant transcript creates a new splice site upstream of the canonical splice site, leading to the insertion of two amino acid residues; (3) structural modeling shows that mutant dynamin-1 forms an oligomeric complex without the capacity for oligomerization-induced GTPase activation; and (4) neuropathological samples reveal an accumulation of synaptic vesicles at the plasma membrane, suggestive of impaired vesicular fission. We further demonstrate that variants involving exon 10a lead to a more severe disease than variants affecting exon 10b, most likely because of relatively decreased expression of transcripts including exon 10b in the brain.
Among the individuals with variants affecting exon 10a, eight had a recurrent intronic variant, yet their clinical presentations were highly similar to individuals with missense variants expected to result in dominant-negative effects.17 This observation is unexpected given that intronic variants near splice sites are typically expected to result in loss of function, causing haploinsufficiency as approximately 50% of the protein product is nonfunctional. Instead, we provide evidence here that this splicing variant instead leads to dominant-negative effects, which would recapitulate typical DNM1-related disorders at the genotype level.26 We predict that this effect is unique to this particular splice site variant, as other splice site variants, most likely leading to loss of function of DNM1, are present in the population. For example, c.589+1G>C (GenBank: NM_004408.3), c.1557+1G>T (GenBank: NM_004408.3), and c.1335+1G>A (GenBank: NM_001288739.1) are variants located directly within splice donors within the GTPase and middle domain portions of the sequence. All are present in gnomAD in apparently healthy individuals and are expected to disrupt splicing, leading to loss of function and haploinsufficiency. Furthermore, this splice variant differs from a similar, frequent intronic site of population variation, c.1197−7C>D (GenBank: NM_004408.3), with a total allele count of 674 in gnomAD. As depicted in Figure 3A, no variant at this position is predicted to interfere with splicing as assessed with SpliceAI.35 Additionally, this site precedes exon 10b, not exon 10a, and would therefore only affect the minor isoform, leaving most DNM1 products intact.
While it is unusual for intronic or splice site variants to lead to dominant-negative disease, it is not unprecedented. A dominant-negative intronic variant has been reported in GABRG2 (MIM: 137164), as the truncated protein product forms aberrant multimers with other GABAA receptor subunits that fail to traffic to the plasma membrane.46 In contrast to the previously identified GABRG2 variant, we found that this recurrent DNM1 variant is not protein truncating but instead forms an alternate splice acceptor upstream of the normal exon 10a acceptor. The resulting insertion of six bases to the exon and two amino acids to the dynamin-1 protein would be expected to disrupt normal oligomerization, leading to stable multimers expected to lack oligomerization-dependent protein function.
We traced the pathogenic mechanism of this splice variant further with neuropathological specimens from one deceased individual. The dense packing of vesicles directly abutting the plasma membrane (Figure 6) demonstrates that vesicles can form from the plasma membrane but are unable to be subsequently removed from the membrane, which is thought to be secondary to impaired vesicular fission. It is possible that the observed synaptic hyperplasia with increased density of synaptic boutons is due to increased axonal sprouting in an effort to circumvent dysfunctional synapses. Similar observations have been made for known dominant-negative missense variants in DNM1, with a reduced quantity of cytoplasmic vesicles in mouse models and an excess of membrane invaginations and membrane-adjacent vesicles in HeLa cells.23 In the future, further functional analysis of this variant’s effects in a model system may reveal effects on single-vesicle release, as previously demonstrated.47 Thus, our findings affirm that the splicing variant c.1197−8G>A (GenBank: NM_001288739.1) has dominant-negative effects identical to those of known disease-causing missense variants in DNM1, consistent with the disease presentation of the eight individuals with this variant, which is indistinguishable from typical DNM1-related disorders and inconsistent with individuals’ harboring a single loss-of-function allele.
With this affirmation that the recurrent variant operates by a mechanism similar to established pathogenic variants, we were able to classify it as a variant that uniquely affects exon 10a of DNM1. Accordingly, taking into account this variant along with the three previously unreported disease-causing variants, we find that in spite of the inclusion of exon 10b in the canonical transcript of DNM1, exon 10a represents the overwhelmingly dominant cortical isoform in developing brains, accounting for double the proportion of dynamin-1 in cortical samples compared to exon 10b. Given our brain samples were obtained from individuals with epilepsy and not unaffected control individuals, we also analyzed adult expression data from the GTEx Portal. These data confirm our findings and demonstrate that DNM1a expression is 5.7-fold higher than DNM1b in the brain cortex (Figure S2). As a result, dominant-negative missense variants affecting both types or DNM1a alone are expected to result in similar, severe clinical features, as the majority of dynamin-1 is altered in either case. In contrast, variants affecting DNM1b alone would leave the majority of dynamin-1 intact, resulting in a milder expected presentation. The clinical patterns we report confirm these predictions, as all ten individuals with DNM1a-only variants replicated the severe DNM1 phenotype, while the individual with a DNM1b-only variant had significantly milder symptoms.
Although a single individual with variant in DNM1b and a mild phenotype may not fully establish a clear genotype-phenotype correlation, the striking phenotypic difference suggests a pattern that could be confirmed by more individuals with such variants. However, we expect that identification of these individuals may be limited, as genetic testing is more likely to be considered for individuals with more severe presentations. If DNM1b variants do correlate with a milder presentation, these variants may be underreported because of ascertainment bias in clinical genetic testing. Additionally, even though the transcripts of DNM1 exhibit nearly identical constraint for missense variation, there is a higher missense variant burden in the population in exon 10b than exon 10a, demonstrating that missense variants are observed in DNM1b but not found in a disease context (Figure S3). Accordingly, we expect that with time, as genetic testing becomes more widely available, more individuals will be identifiable with DNM1b variants, highlighting transcript-specific phenotypic differences.
Exon-dependent variability in DNM1-related disease is further supported by mouse models of DNM1. The fitful mouse, a line which was found to have the spontaneous mutation c.1222G>A (GenBank: NM_001301737.1) in exon 10a of the mouse ortholog Dnm1 with consequence p.Ala408Thr, presents with severe seizure disorder. The fitful phenotype is present in mice with variation in Dnm1a but not Dnm1b, and the heterozygous condition in humans is phenotypically equivalent to the homozygous fitful mouse.22 However, the biological difference between isoforms, if any, remains indeterminate. Mouse models with knockout of either tenth exon show that the isoforms have some differences in intracellular localization, yet mice expressing only DNM1a or only DNM1b are phenotypically normal. Previous functional studies of disease-causing variants have only assessed variants that affect both isoforms. Hence, the alternative tenth exons have unclear structural and functional consequences. It was previously thought that these splicing patterns were relevant to mouse but not human disease, yet our findings suggest that the need for homozygosity in mouse models reflects the significant skew towards exon 10a in human cortex. In fact, two human families with homozygous nonsense variants in DNM1 have disease similar to typical DNM1-related disorders.48 Thus, a vast majority of DNM1 product in humans and mice must be affected in order to replicate the effects of a single missense variant in human DNM1a, emphasizing the significance of this isoform alone in causing severe disease.
Disease-causing variations in alternative, non-canonical transcripts are also known to cause disease in a wide range of conditions beyond DNM1. Pathogenic variants have been identified in alternative coding regions of well-known epilepsy genes such as CACNA1A (MIM: 601011) and STXBP1; these variants would go undetected if testing were limited to a single, canonical transcript. CACNA1A and other voltage-gated calcium channels are particularly prone to alternative patterns of expression that may affect disease phenotypes. For example, a second cistron of CACNA1A, which encodes a transcription factor with wider expression than the full-length channel isoform, has been implicated in some symptoms of spinocerebellar ataxia type 6.49 Moreover, we have previously described an individual with a variant in an alternatively spliced exon of STXBP1 leading to a relatively mild phenotype, including the ability to walk independently, communicate with several words, and no history of seizures.18 As such, it is imperative to account for transcript- and isoform-specific clinical patterns in diagnosis and care of individuals with genetic conditions, including DNM1-related disorders.
By elucidating transcript-dependent disease-causing variation in DNM1, our findings also can inform clinical variant interpretation. For instance, a large testing company in the US stated that a transcript containing exon 10a was only added to their epilepsy gene panel analysis in late 2019 (S.M. Ruggiero, personal communication). Individuals 2 and 3 had thus undergone gene panel testing in which the DNM1 variant was not identified; the variant was only revealed after subsequent WES included this transcript. Moreover, individual 10’s variant was initially considered a predicted null variant, and therefore unlikely to be disease-causing, as sequence analysis was initially performed with only DNM1b. As such, inclusion of exon 10a into emerging standards for clinical genetic testing is warranted. Currently, exon 10a is absent from the Matched Annotation from NCBI and EMBL-EBI (MANE) Select and MANE Plus Clinical transcript sets, which are standardized for use in clinical genetics and variant interpretation.50 Future iterations of transcript standardization may benefit from accounting for the DNM1a isoform to facilitate molecular diagnosis in individuals with exon 10a-specific disease-causing variants in DNM1. Ultimately, given the evidence that isoforms expressing exon 10a account for the most severe features of DNM1-related disorders, our findings illuminate the necessity of assessing the relevant transcript in clinical genetic testing in order to make accurate genetic diagnoses in individuals with possible DNM1-related neurodevelopmental disorders.
In summary, we identify DNM1a, characterized by alternative splicing of exon 10a, as the predominant isoform responsible for the classical DNM1 phenotype in contrast to the milder presentation caused by a variant in DNM1b. Our findings underscore the importance of including the DNM1a sequence in clinical genetic testing as well as the need more broadly to include relevant transcripts in diagnosis of genetic neurodevelopmental disorders. Finally, we highlight the continued need to consider splice site variants as causative of dominant-negative disease.
Acknowledgments
The authors thank the patients and families who selflessly contributed their time and medical information to this study. The authors acknowledge and thank Priya Vaidiswaran, Azzedine Yacia, Jean Marc Masse, Eve Brochot, Stephanie May, and Vincent Grammont. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (NIH) and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Some data used for the analyses described in this manuscript were obtained from the GTEx Portal on 07/07/22. S.D.J.P. and F.C.S. were supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico of Brazil (CNPq) and Rede Mineira de Genômica Populacional e Medicina de Precisão (RED00314-16) of Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG). D.B.A. and D.E.V.P. were supported by the National Health and Medical Research Council (NHMRC) of Australia (GNT1174405) and the Victorian Government's Operational Infrastructure Support Program. I.H. was supported by The Hartwell Foundation (Individual Biomedical Research Award), NINDS (K02NS112600, U24NS120854-01, U54NS108874-04), the Eunice Kennedy Shriver National Institute of Child Health and Human Development through the Children's Hospital of Philadelphia and the University of Pennsylvania (U54HD086984), the German Research Foundation (HE5415/3-1, HE5415/5-1, HE5415/6-1, HE5415/7-1), the National Center for Advancing Translational Sciences of the NIH (UL1TR001878), the Institute for Translational Medicine and Therapeutics’ (ITMAT) at the Perelman School of Medicine of the University of Pennsylvania, and by Children's Hospital of Philadelphia through the Epilepsy NeuroGenetics Initiative (ENGIN). V.A.C. was supported by NINDS (T32NS007413). The content is the sole responsibility of the authors and does not necessarily represent the official views of the NINDS of the NIH.
Declaration of interests
The authors declare no competing interests.
Published: November 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.11.002.
Web resources
Online Mendelian Inheritance in Man, http://www.omim.org
Supplemental information
Data and code availability
The published article includes all datasets generated during this study for transcript quantification. The raw RNA-seq dataset supporting the current study has not been deposited in a public repository because consent was not obtained from subjects for deposition of sequence data into an unprotected database, but data are available from the corresponding author on request. The code generated during this study is publicly available at https://github.com/helbig-lab/DNM1. The splicing analysis did not generate or analyze datasets or code. Original/source data for neuropathology analysis in the paper are available from Hôpitaux Universitaires de l’Est Parisien – Neuropathologie du développement (biobank identification number BB-0033-00082).
References
- 1.Helbig I., Heinzen E.L., Mefford H.C., International League Against Epilepsy Genetics Commission Genetic literacy series: Primer part 2-Paradigm shifts in epilepsy genetics. Epilepsia. 2018;59:1138–1147. doi: 10.1111/epi.14193. [DOI] [PubMed] [Google Scholar]
- 2.Helbig I., Tayoun A.A.N. Understanding genotypes and phenotypes in epileptic encephalopathies. Mol. Syndromol. 2016;7:172–181. doi: 10.1159/000448530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McTague A., Howell K.B., Cross J.H., Kurian M.A., Scheffer I.E. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15:304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 4.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L., et al. ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helbig K.L., Farwell Hagman K.D., Shinde D.N., Mroske C., Powis Z., Li S., Tang S., Helbig I. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet. Med. 2016;18:898–905. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]
- 6.Heyne H.O., Singh T., Stamberger H., Abou Jamra R., Caglayan H., Craiu D., De Jonghe P., Guerrini R., Helbig K.L., Koeleman B.P.C., et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018;50:1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]
- 7.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindy A.S., Stosser M.B., Butler E., Downtain-Pickersgill C., Shanmugham A., Retterer K., Brandt T., Richard G., McKnight D.A. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia. 2018;59:1062–1071. doi: 10.1111/epi.14074. [DOI] [PubMed] [Google Scholar]
- 9.Stamberger H., Weckhuysen S., De Jonghe P. STXBP1 as a therapeutic target for epileptic encephalopathy. Expert Opin. Ther. Targets. 2017;21:1027–1036. doi: 10.1080/14728222.2017.1386175. [DOI] [PubMed] [Google Scholar]
- 10.Wengert E.R., Wagley P.K., Strohm S.M., Reza N., Wenker I.C., Gaykema R.P., Christiansen A., Liau G., Patel M.K. Targeted Augmentation of Nuclear Gene Output (TANGO) of Scn1a rescues parvalbumin interneuron excitability and reduces seizures in a mouse model of Dravet Syndrome. Brain Res. 2022;1775:147743. doi: 10.1016/j.brainres.2021.147743. [DOI] [PubMed] [Google Scholar]
- 11.Brunklaus A., Feng T., Brünger T., Perez-Palma E., Heyne H., Matthews E., Semsarian C., Symonds J.D., Zuberi S.M., Lal D., Schorge S. Gene variant effects across sodium channelopathies predict function and guide precision therapy. Brain. 2022:awac006. doi: 10.1093/brain/awac006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helbig I., Lopez-Hernandez T., Shor O., Galer P., Ganesan S., Pendziwiat M., Rademacher A., Ellis C.A., Hümpfer N., Schwarz N., et al. A recurrent missense variant in AP2M1 impairs clathrin-mediated endocytosis and causes developmental and epileptic encephalopathy. Am. J. Hum. Genet. 2019;104:1060–1072. doi: 10.1016/j.ajhg.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klöckner C., Sticht H., Zacher P., Popp B., Babcock H.E., Bakker D.P., Barwick K., Bonfert M.V., Bönnemann C.G., Brilstra E.H., et al. De novo variants in SNAP25 cause an early-onset developmental and epileptic encephalopathy. Genet. Med. 2021;23:653–660. doi: 10.1038/s41436-020-01020-w. [DOI] [PubMed] [Google Scholar]
- 14.Salpietro V., Malintan N.T., Llano-Rivas I., Spaeth C.G., Efthymiou S., Striano P., Vandrovcova J., Cutrupi M.C., Chimenz R., David E., et al. Mutations in the neuronal vesicular SNARE VAMP2 affect synaptic membrane fusion and impair human neurodevelopment. Am. J. Hum. Genet. 2019;104:721–730. doi: 10.1016/j.ajhg.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert J., Siekierska A., Langlois M., May P., Huneau C., Becker F., Muhle H., Suls A., Lemke J.R., de Kovel C.G.F., et al. Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat. Genet. 2014;46:1327–1332. doi: 10.1038/ng.3130. [DOI] [PubMed] [Google Scholar]
- 16.Verhage M., Sørensen J.B. SNAREopathies: diversity in mechanisms and symptoms. Neuron. 2020;107:22–37. doi: 10.1016/j.neuron.2020.05.036. [DOI] [PubMed] [Google Scholar]
- 17.von Spiczak S., Helbig K.L., Shinde D.N., Huether R., Pendziwiat M., Lourenço C., Nunes M.E., Sarco D.P., Kaplan R.A., Dlugos D.J., et al. DNM1 encephalopathy: A new disease of vesicle fission. Neurology. 2017;89:385–394. doi: 10.1212/WNL.0000000000004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xian J., Parthasarathy S., Ruggiero S.M., Balagura G., Fitch E., Helbig K., Gan J., Ganesan S., Kaufman M.C., Ellis C.A., et al. Assessing the landscape of STXBP1-related disorders in 534 individuals. Brain. 2021;145:1668–1683. doi: 10.1093/brain/awab327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project, and De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. Epi4K Consortium [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chappie J.S., Dyda F. Building a fission machine--structural insights into dynamin assembly and activation. J. Cell Sci. 2013;126:2773–2784. doi: 10.1242/jcs.108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson S.M., De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asinof S., Mahaffey C., Beyer B., Frankel W.N., Boumil R. Dynamin 1 isoform roles in a mouse model of severe childhood epileptic encephalopathy. Neurobiol. Dis. 2016;95:1–11. doi: 10.1016/j.nbd.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhindsa R.S., Bradrick S.S., Yao X., Heinzen E.L., Petrovski S., Krueger B.J., Johnson M.R., Frankel W.N., Petrou S., Boumil R.M., Goldstein D.B. Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis. Neurol. Genet. 2015;1:e4. doi: 10.1212/01.NXG.0000464295.65736.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devanna P., van de Vorst M., Pfundt R., Gilissen C., Vernes S.C. Genome-wide investigation of an ID cohort reveals de novo 3'UTR variants affecting gene expression. Hum. Genet. 2018;137:717–721. doi: 10.1007/s00439-018-1925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahly A.N., Krochmalnek E., St-Onge J., Srour M., Myers K.A. Severe DNM1 encephalopathy with dysmyelination due to recurrent splice site pathogenic variant. Hum. Genet. 2020;139:1575–1578. doi: 10.1007/s00439-020-02224-5. [DOI] [PubMed] [Google Scholar]
- 27.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palisano R.J., Avery L., Gorter J.W., Galuppi B., McCoy S.W. Stability of the gross motor function classification system, manual ability classification system, and communication function classification system. Dev. Med. Child Neurol. 2018;60:1026–1032. doi: 10.1111/dmcn.13903. [DOI] [PubMed] [Google Scholar]
- 30.Eliasson A.C., Krumlinde-Sundholm L., Rösblad B., Beckung E., Arner M., Ohrvall A.M., Rosenbaum P. The Manual Ability Classification System (MACS) for children with cerebral palsy: scale development and evidence of validity and reliability. Dev. Med. Child Neurol. 2006;48:549–554. doi: 10.1017/S0012162206001162. [DOI] [PubMed] [Google Scholar]
- 31.Eliasson A.C., Ullenhag A., Wahlström U., Krumlinde-Sundholm L. Mini-MACS: development of the Manual Ability Classification System for children younger than 4 years of age with signs of cerebral palsy. Dev. Med. Child Neurol. 2017;59:72–78. doi: 10.1111/dmcn.13162. [DOI] [PubMed] [Google Scholar]
- 32.Hidecker M.J.C., Paneth N., Rosenbaum P.L., Kent R.D., Lillie J., Eulenberg J.B., Chester K., Jr., Johnson B., Michalsen L., Evatt M., Taylor K. Developing and validating the communication function classification system for individuals with cerebral palsy. Dev. Med. Child Neurol. 2011;53:704–710. doi: 10.1111/j.1469-8749.2011.03996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 34.Kinsella R.J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Gaildrat P., Killian A., Martins A., Tournier I., Frébourg T., Tosi M. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol. Biol. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- 37.Pires D.E.V., Chen J., Blundell T.L., Ascher D.B. In silico functional dissection of saturation mutagenesis: Interpreting the relationship between phenotypes and changes in protein stability, interactions and activity. Sci. Rep. 2016;6:19848. doi: 10.1038/srep19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues C.H.M., Myung Y., Pires D.E.V., Ascher D.B. mCSM-PPI2: predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res. 2019;47:W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. mmCSM-PPI: predicting the effects of multiple point mutations on protein-protein interactions. Nucleic Acids Res. 2021;49:W417–W424. doi: 10.1093/nar/gkab273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pires D.E.V., Ascher D.B., Blundell T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires D.E.V., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut2: assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 2021;30:60–69. doi: 10.1002/pro.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pires D.E.V., Blundell T.L., Ascher D.B. mCSM-lig: quantifying the effects of mutations on protein-small molecule affinity in genetic disease and emergence of drug resistance. Sci. Rep. 2016;6:29575. doi: 10.1038/srep29575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Husain A.F., Stocker J.T., Dehner L.P. Wolters Kluwer; 2021. Stocker and Dehner’s Pediatric Pathology. [Google Scholar]
- 46.Tian M., Macdonald R.L. The intronic GABRG2 mutation, IVS6+2T->G, associated with childhood absence epilepsy altered subunit mRNA intron splicing, activated nonsense-mediated decay, and produced a stable truncated gamma2 subunit. J. Neurosci. 2012;32:5937–5952. doi: 10.1523/JNEUROSCI.5332-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keighron J.D., Wigström J., Kurczy M.E., Bergman J., Wang Y., Cans A.S. Amperometric detection of single vesicle acetylcholine release events from an artificial cell. ACS Chem. Neurosci. 2015;6:181–188. doi: 10.1021/cn5002667. [DOI] [PubMed] [Google Scholar]
- 48.Yigit G., Sheffer R., Daana M., Li Y., Kaygusuz E., Mor-Shakad H., Altmüller J., Nürnberg P., Douiev L., Kaulfuss S., et al. Loss-of-function variants in DNM1 cause a specific form of developmental and epileptic encephalopathy only in biallelic state. J. Med. Genet. 2021;59:549–553. doi: 10.1136/jmedgenet-2021-107769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du X., Wang J., Zhu H., Rinaldo L., Lamar K.M., Palmenberg A.C., Hansel C., Gomez C.M. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell. 2013;154:118–133. doi: 10.1016/j.cell.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morales J., Pujar S., Loveland J.E., Astashyn A., Bennett R., Berry A., Cox E., Davidson C., Ermolaeva O., Farrell C.M., et al. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature. 2022;604:310–315. doi: 10.1038/s41586-022-04558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during this study for transcript quantification. The raw RNA-seq dataset supporting the current study has not been deposited in a public repository because consent was not obtained from subjects for deposition of sequence data into an unprotected database, but data are available from the corresponding author on request. The code generated during this study is publicly available at https://github.com/helbig-lab/DNM1. The splicing analysis did not generate or analyze datasets or code. Original/source data for neuropathology analysis in the paper are available from Hôpitaux Universitaires de l’Est Parisien – Neuropathologie du développement (biobank identification number BB-0033-00082).