Summary
The most recent genome-wide association study (GWAS) of cutaneous melanoma identified 54 risk-associated loci, but functional variants and their target genes for most have not been established. Here, we performed massively parallel reporter assays (MPRAs) by using malignant melanoma and normal melanocyte cells and further integrated multi-layer annotation to systematically prioritize functional variants and susceptibility genes from these GWAS loci. Of 1,992 risk-associated variants tested in MPRAs, we identified 285 from 42 loci (78% of the known loci) displaying significant allelic transcriptional activities in either cell type (FDR < 1%). We further characterized MPRA-significant variants by motif prediction, epigenomic annotation, and statistical/functional fine-mapping to create integrative variant scores, which prioritized one to six plausible candidate variants per locus for the 42 loci and nominated a single variant for 43% of these loci. Overlaying the MPRA-significant variants with genome-wide significant expression or methylation quantitative trait loci (eQTLs or meQTLs, respectively) from melanocytes or melanomas identified candidate susceptibility genes for 60% of variants (172 of 285 variants). CRISPRi of top-scoring variants validated their cis-regulatory effect on the eQTL target genes, MAFF (22q13.1) and GPRC5A (12p13.1). Finally, we identified 36 melanoma-specific and 45 melanocyte-specific MPRA-significant variants, a subset of which are linked to cell-type-specific target genes. Analyses of transcription factor availability in MPRA datasets and variant-transcription-factor interaction in eQTL datasets highlighted the roles of transcription factors in cell-type-specific variant functionality. In conclusion, MPRAs along with variant scoring effectively prioritized plausible candidates for most melanoma GWAS loci and highlighted cellular contexts where the susceptibility variants are functional.
Keywords: melanoma, GWAS, MPRA, cell-type specificity, variant scoring, GWAS follow-up, cell of tumor origin, transcription factors, eQTL, CRISPRi
Long et al. used massively parallel reporter assays combined with variant scoring to identify functional variants from 78% of known melanoma GWAS loci, including those specific to cell of origin versus cancer contexts. Linking prioritized functional variants to eQTLs identified target genes as validated by CRISPRi.
Introduction
Cutaneous melanoma originates from melanocytes and is the deadliest skin cancer,2 with increasing incidence and burden worldwide.3 Melanoma has a substantial heritable germline genetic component explained partly by the 54 genome-wide significant risk loci identified through the most recent genome-wide association study (GWAS)1 including 36,760 cases and 375,188 controls. While a subset of these loci are explained by genetic determinants of pigmentation phenotypes—well-known risk factors of melanoma (e.g., MC1R,4 OCA2,5 SLC45A2,6 and TYR7)—molecular mechanisms of most loci have not been characterized, with a few exceptions (e.g., PARP1,8 MX2,9 TERT,10 and AHR11). Identifying potentially causal variants and their target genes from melanoma GWAS loci is challenging because there are often many co-inherited variants in strong linkage disequilibrium (LD),12 and these variants display statistically indistinguishable associations with disease risk given the current sample size. Further, most of these risk-associated variants are in non-protein-coding regions,13 and therefore it is difficult to pinpoint the target genes.14
Most non-coding GWAS variants most likely function via cis-regulatory mechanisms to regulate target gene expression. Classical reporter assays can test this hypothesis by assessing allelic transcriptional activity of individual variants, and massively parallel reporter assays (MPRAs) allow for scaling the reporter assays to test hundreds to thousands of variants, enabling the identification of functional variants among multiple variants that are indistinguishable as a result of strong LD. Our previous study9 using this approach tested 832 variants from 16 melanoma loci based on a previous GWAS15 and prioritized 39 candidate functional variants from 14 loci in the context of a melanoma cell line. While MPRAs can functionally test individual variants in a reporter system, this approach does not identify candidate susceptibility genes. Quantitative trait locus (QTL) analysis is a powerful tool to link GWAS variants to candidate susceptibility genes.16 Our previous studies established multi-QTL datasets through the use of cultured melanocytes as well as skin cutaneous melanomas from The Cancer Genome Atlas (TCGA)17,18 and demonstrated that a multi-QTL approach of disease-relevant cell/tissue datasets could nominate candidate susceptibility genes for most melanoma GWAS loci. The strategy of combining both MPRAs and cell-type-specific eQTLs identified the most prominent locus to follow up and led to a discovery of MX2 as a pleiotropic gene promoting melanoma in a zebrafish model.9
Despite this progress, a comprehensive understanding of the role of GWAS-identified loci in melanoma susceptibility is still lacking. A more recent melanoma GWAS meta-analysis identified a total of 54 loci reaching genome-wide significance,1 increasing the total number of melanoma risk-associated loci by more than 3-fold, most of which have not been functionally tested. Moreover, beyond our work focusing on a handful of loci with one prominent candidate variant, most loci tend to have multiple functional variants displaying allelic transcriptional activity, and systematic prioritization schemes are needed to guide further time-consuming functional studies on these more challenging loci. Furthermore, there is growing evidence that the cis-regulation of gene expression underlying complex trait susceptibility is cell-type and context specific.19,20 Indeed, our previous studies using LD score regression1 and colocalization/TWAS approaches17 demonstrated that using data from primary human melanocytes, the cell of melanoma origin,21 is more useful for annotating melanoma GWAS data than any tissue type from the GTEx dataset including skin. Still, it is often not clear in studying individual cancer susceptibility variants and genes whether their tumor-promoting potential is more pronounced in the context of early stages (i.e., normal cells) or later stages (i.e., cancer cells) along the evolutionary trajectory of tumorigenesis. Critically, substantial heterogeneity of QTLs between melanocytes and melanomas has been observed in our previous studies,18 highlighting the importance of studying the gene expression regulation in the contexts of both normal and cancer cells. While there have been many approaches and datasets that prioritized functional variants from GWAS loci and linked them to target genes,22,23,24 the relative roles of different trait-relevant cell types in variant functionality have not been systematically compared and incorporated to prioritizing variants, especially for melanoma and other cancer GWASs.
To address these issues and functionally characterize all 54 reported melanoma GWAS loci, we performed MPRAs in both malignant melanoma and normal melanocyte cell lines. Multilayered variant functional features, including motif prediction, epigenomic annotation, and statistical/functional fine-mapping were integrated with MPRA data to further prioritize the plausible candidate causal variants by locus. To link functional variants to potential susceptibility genes, expression QTLs (eQTLs) and DNA methylation QTLs (meQTLs) from melanocytes and melanoma were incorporated. Leveraging these approaches, we prioritized plausible candidates from GWAS loci and highlighted significant cell-type specificity of melanoma susceptibility in relevant tumor and normal cell types.
Material and methods
MPRA variant selection
For MPRA, we selected candidate variants from each of the 54 genome-wide significant loci (Table S1) from the melanoma GWAS meta-analyses by Landi and colleagues1 that meet one of the following three criteria:
-
(1)
variants with log likelihood ratio (LLR) < 1:1,000 relative to the primary lead SNP on the basis of the GWAS p values (fixed-effect model) from the main meta-analysis; for the locus tagged by rs4731207, LLR < 1:150 was applied to test only the strongest candidate variants within an extended/large LD block (∼600 variants with LLR < 1:1,000);
-
(2)
LD R2 > 0.8 (1000 Genomes, phase 3, EUR populations) with the primary lead SNP for any variant not genotyped or successfully imputed in the GWAS (p values not available);
-
3.
LD R2 > 0.8 (1000 Genomes, phase 3, EUR populations) with an additional independent lead SNP(s) identified through a conditional analysis1 within 1 Mb of a primary lead SNP (regardless of LLR); two additional lead SNPs (rs3212371 and rs73069846) reported in the melanoma GWAS1 were not included in the design.
After considering these criteria, 214 variants were dropped because of technical reasons including those that have enzyme digestion sites for either Kpnl, Xbal, or Sfil within the 145 bp encompassing the variant. A total of 1,992 melanoma GWAS variants were tested by MPRAs. A complete list of variants tested are shown in Table S2.
MPRA oligo library design
The oligo library was designed in a similar way to our previous work9 with some modifications. For each variant, 145-base sequences encompassing the variant (+/− 72 bases) with reference and alternative alleles in both forward and reverse directions were extracted from human genome build GRCh37. Strand (forward/reverse) was tested in assessing enhancer function of a sequence element, which models the relative position of enhancer element to gene promoter. Each test sequence was randomly associated with 20 different randomly generated 12-base sequence tags separated by recognition sequences for restriction enzymes, KpnI (GGTACC) and XbaI (TCTAGA), and flanked by binding sequences for PCR primers and a two-base spacer (204 bases oligo sequences: 5′-ACTGGCCGCTTCACTG-145 bases-GGTACCTCTAGA-12 bases tag-AC (spacer)-AGATCGGAAGAGCGTCG-3′; close to maximum test sequence allowed by the oligo synthesis platform). For each variant, a single scrambled sequence of 145-base test sequence was also included and associated with 16 tag sequences (using forward direction and the reference allele) as a background level control for activator/repressor inference (see transcriptional activator/repressor inference). The number of tags is based on down-sampling analysis from a previous study.25 When there are additional SNPs other than the test SNP that fall in the 145 bp region, the major allele in the 1000 Genomes EUR populations was used for both sequences of reference/alternative alleles, ensuring all the sequences are fixed except the tested variant. For indel variants, a 145 total base length was set on the basis of insertion allele, and additional bases were added to each side of the test sequence of the deletion allele to fit 145 bases. For the 12-base tag sequence and scrambled sequences, only homopolymers of <4 bases were used and the enzyme recognition sites for KpnI, XbaI, and SfiI were avoided. A pooled library of 191,232 oligos in a randomized order was synthesized by Agilent Technologies (Santa Clara, CA). A complete list of oligo sequences can be found in Table S3.
MPRA library construction, transfection, and sequencing
MPRA library construction, transfection, and sequencing was performed following published procedures9,26 with some modifications. For library cloning, ten femtomoles of gel-purified (10% TBE-Urea polyacrylamide gel) oligo library was amplified by emulsion PCR with 1.5 μL of Herculase II fusion polymerase (Agilent, Santa Clara, CA), 0.5 mg/mL BSA acetylated, 375 μM dNTP, and 3 μM of primers providing SfiI enzyme sites and 25 cycles amplification per 50 μL reaction, then 3 × 50 μL reactions were combined and cleaned up in column purification step, following the instructions of the Micellula DNA Emulsion and Purification Kit (EURx/CHIMERx, Milwaukee, WI). To verify the oligo sequences, we prepared amplicon libraries by using 100 ng of oligos from emulsion PCR with KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA) following the instructions of the manufacturer and sequenced them with the MiSeq reagent kit v3 (150-cycles). Twelve-base tag sequences plus spacer sequences were used to map each oligo from FASTQ files and count the total read depth. 98% of the designed tag sequences were detected at one or more reads. The sequence-verified oligo library was first cloned into pMPRA1 vector (Addgene, Watertown, MA) with SfiI site followed by electroporation into a 10 times higher number of bacterial cells than the number of unique sequences in the oligo library. Cloned pMPRA1 was further digested on KpnI and XbaI sites between the 145 bp test sequence and the 12 bp barcode sequence, where a luc2 open reading frame (ORF) with a minimal promoter from pMPRAdonor2 (Addgene, Watertown, MA) was inserted. The ligation product was transformed by electroporation into a 10 times higher number of bacterial cells in the same manner. The cloned final library for transfection was verified on the gel as a single band after KpnI digestion.
We used three batches of cloned library to transfect 8 times into UACC903 melanoma cells and 5 times into an immortalized primary melanocyte cell line (C283T),11 aiming for a >100 times higher number of transfected cells than the library complexity in each transfection. The numbers of transfected cells were estimated with transfection efficiency measured by a separate GFP transfection and visualization. For UACC903, cells were transfected with Lipofectamine 3000 and harvested 48 h after transfection for RNA isolation. For C283T, cells were transfected by electroporation with P2 Primary Cell 4D-Nucleofector X Kit L (Lonza, Basel, Switzerland), following manufacturer’s instruction. Nucleofector programs for C283T cell lines were optimized with the P2 Primary Cell 4D-Nucleofector X Kit and GFP visualization. The amount of cloned MPRA library and harvesting time of transfection cells were optimized with qPCR with specific primers (Table S3). Electro-transfected C283T cells were harvested at 24 h for RNA isolation. Total RNA was isolated with Qiagen RNeasy kit (Qiagen, Hilden, Germany), and mRNA was subsequently isolated with PolyA purist MAG kit (Thermo Fisher). cDNA was then synthesized with Superscript III reverse transcriptase, from which short sequences encompassing 12 bp unique tags were amplified with Q5 high-fidelity polymerase (NEB, Ipswich, MA) and primers introducing Illumina TruSeq adapter sequences. Tag sequence libraries were also prepared with input DNA in the same way. Tag sequence libraries were sequenced on NovaSeq 6000 SP flow cells (100 bp dual-indexed single end read) to obtain 125–200 million reads per sample for UACC903 transfections and 218–295 million reads per sample for C283T transfections.
MPRA data analyses
Using FASTQ files from input DNA or RNA transcript (cDNA) sequencing, we counted the number of reads (Illumina read 1) completely matching 12 bp barcode sequences (tag counts) plus spacer sequences and the same downstream sequence context including an XbaI recognition site and the 3′ of luc2. For each transfection, we calculated tag counts per million sequencing reads (TPM) values by dividing each tag count by the total number of sequence-matching tag counts divided by a million. A pseudo count of 1 was added to all TPM values and then TPM ratio was taken as RNA TPM over input DNA TPM and log2 converted: log2 ((RNA TPM + 1)/(DNA TPM + 1)). We defined this log2 transformed TPM ratio as “normalized expression level.”
From each input DNA library, 93.4%–94.9% of designed barcode sequences were detected. From RNA samples 90.2%–92.8% of barcode sequences were detected in melanoma cells and 93.7%–94.3% in melanocytes (Table S4). Median tag counts were 723–810 for DNA input, 412–655 for RNA output from melanoma cells, and 754–1,018 for RNA output from melanocytes (Table S4). In melanoma cells, 96.2%–98.6% of unique tags detected in DNA input were recovered in mRNA output and 98.6%–99.0% were recovered in melanocytes (Table S4). Reproducibility between transfections were assessed by Pearson correlation of normalized expression level of each barcode between replicates of transfection. To avoid low input DNA counts driving variations in RNA/DNA TPM ratios, we removed tags with <2 TPM counts (log2 DNA TPM < 1) from further analyses. The remaining tags account for 82.99% of all the detected tags (Figure S1).
We used the following standard linear regression model to assess the impact of allele (reference or alternative) on the transcriptional activity (normalized expression level defined as log2 ((RNA TPM + 1/DNA TPM + 1)), named “ratio” in following formulas), while adjusting for the effect of strand (forward or reverse) as a binary covariate and the effect of transfection replicate as a categorical
To account for the potential heteroskedasticity in the measurement error, we used the robust sandwich type variance estimate in the Wald test to determine the significance. This analysis was carried out with the R package Sandwich (https://sandwich.r-forge.r-project.org). The Wald test p values were corrected for multiple testing with the procedure of Benjamini and Hochberg.27 We used a corrected p (FDR) < 0.01 to define “MPRA-significant variants” that display significant allelic transcriptional activity in each cell type.
Transcriptional activator/repressor inference
Given that the variants were selected and tested in MPRAs regardless of their functional annotation, we assumed that most of the tested sequences are non-functional, and therefore, the mean normalized expression levels of all the variants were considered as null. Therefore, the putative function of activators (or repressors) was inferred by defining the extreme outliers from the mean expression levels. First, the overall distribution of the normalized expression levels (mean log2((RNA TPM + 1)/(DNA TPM + 1))) of all tags by variants including reference and alternative alleles and scrambled sequences were calculated. The putative function of activators was inferred by defining the extreme outliers from the mean expression levels (upper limit: Q3 + 3 × IQ), where Q3 is 75th percentiles and the interquartile range (IQ) is Q3 – Q1 (25th percentiles). Similarly, putative repressor function was inferred on the basis of extreme lower limits (Q1 − 3 × IQ). For each variant, allele/strand-specific normalized expression levels were then calculated with only the tags for reference-forward, reference-reverse, alternative-forward, or alternative-reverse sub-group. The variants with normalized expression levels of one or more of these sub-groups higher than the upper limits were assigned as activators and vice versa for repressors. These assignments were confirmed by the regression analyses comparing normalized expression levels of scrambled sequences with either reference or alternative allele for each strand separately while still using transfection as a covariate. The Wald test with robust sandwich type variance estimate was used, and false discovery rate (FDR) < 0.01 was applied.
MTSA analyses
MPRA tag sequence analysis (MTSA) is a sequence-based analysis for estimating tag sequence effects on gene expression in an MPRA experiment via the following steps.28 First, tags with low read counts in input DNA (<200 reads) were removed for the purpose of MTSA analysis. Second, the relative expression (tag expression normalized to mean zero across each set of tags associated with a 145 bp sequence) was calculated. Third, a support vector regression (SVR) was trained on the basis of gapped-kmer kernels29 to learn the contribution of each tag sequence to its relative expression level. Fourth, the adjusted expression values (RNA tag counts) were calculated. Finally, the MTSA-corrected FDR and log2FC (log2-transformed fold difference of mean TPM ratio for alternative allele over mean TPM ratio for reference allele) were the outputs. MTSA-corrected FDR is calculated with the approach of linear regression with the robust sandwich type variance estimate in Wald test (see MPRA data analyses). The MTSA-corrected FDRs are compared with original FDRs regarding the significance of allelic transcriptional activity (FDR < 0.01) and allelic direction.
Functional annotations
The melanocyte open chromatin regions were inferred by the human melanocyte DNase I hypersensitive site (DHS) peaks from ENCODE30 (n = 1), Epigenome Roadmap database (n = 2)31, and melanocyte assay for transposase-accessible chromatin using sequencing (ATAC-seq) peaks combined from the cultured melanocytes of six individuals that were generated in our laboratory.11 The melanoma open chromatin regions were inferred by human melanoma short-term culture formaldehyde-assisted isolation of regulatory elements (FAIRE)-seq peaks from one or more individuals of 11 available from Verfaillie et al.31 The enhancer regions were marked if the variant is located within both a human melanocyte H3K27Ac ChIP-seq peak and a H3K4Me1 ChIP-seq peak from at least one individual (n = 2 available through Epigenome Roadmap database). The promoter regions were marked if the variant is located within both a human melanocyte H3K27Ac ChIP-seq peak and a H3K4Me3 ChIP-seq peak from at least one individual (n = 2 available through Epigenome Roadmap database).
Motif analysis
Prediction of variant effects on transcription factor (TF)-binding sites was performed with the motifbreakR package and a comprehensive collection of human TF-binding site models (HOCOMOCO, v11). We selected the information content algorithm and used a threshold of 10−4 as the maximum p value for a transcription-binding site match in motifbreakR. The strong effect is defined by the difference between alternative allele score and reference allele score larger than 0.7.
Melanoma GWAS statistical and functional fine-mapping
For fine-mapping, we used melanoma GWAS summary data derived from both confirmed as well as self-reported melanoma cases from 23andMe and UK Biobank and controls as previously described1; all participants provided informed consent reviewed by institutional review boards (IRBs), including 23andMe participants who gave online informed consent and participation, under a protocol approved by the external AAHRPP-accredited IRB, Ethical and Independent Review Services (E&I Review). Statistical fine-mapping of the 54 genome-wide significant loci from the meta-analysis reported by Landi and colleagues was conducted with FINEMAP v1.4.32 We defined flanking regions as 250 kb on either side of the most significant variant at each locus. Evidence (Z score) for each variant from the GWAS summary statistics and LD matrix (precomputed with n = ∼337,000 unrelated British-ancestry individuals from the UK Biobank33) were the input for the analysis. For loci with one independent signal identified by the conditional analysis in the original GWAS,1 we set the maximum number of causal variants as 2. For loci with multiple conditionally independent signals, we set the maximum number of causal variants equal to the number of independent signals from the GWAS. For an improved fine-mapping efficiency, we also performed a fine-mapping incorporating functional annotation with POLYFUN33 by specifying prior probabilities for FINEMAP analysis. Following the recommended procedure, we incorporated precomputed prior causal probabilities of ∼19 million imputed UK Biobank SNPs with MAF > 0.1%, based on a meta-analysis of 15 UK Biobank traits including hair color. The output includes posterior inclusion probability (PIP) for each variant and the index of the credible set that the variant belongs to. A 95% credible set is comprised of variants that cumulatively reach a probability of 95%. The variants with PIP > 0.1% were considered as being in the 95% credible set.
Integration of MPRA variants with melanoma and melanocyte eQTL and meQTL variants
Significant eQTLs or meQTLs were defined with the empirical genome-wide significance threshold as described in the previous studies.17,18 MPRA-significant variants were linked to target genes if they display significant eQTL or meQTL p values for one of the significant genes (eGenes) or 5'-C-phosphate-G-3' (CpG) sites (meProbes) in melanocytes or melanomas. Gene assignments to each meProbe are presented on the basis of the Illumina HumanMethylation450 BeadChip annotation file, which we define as meGenes. Identification of eQTLs and meQTLs as well as colocalization analyses were previously described.17,18 Briefly, melanocyte eQTLs and meQTLs were obtained from a dataset including 106 individuals mainly of European descent. Melanoma eQTLs and meQTLs were based on our previous analyses of 444 skin cutaneous melanoma (SKCM) samples from TCGA with genotype, expression, and methylation data. The colocalization analysis was performed among melanoma GWAS, eQTL, and meQTL datasets with HyPrColoc and detailed parameters are described in our previous study.18
Variant prioritization scores
We established a system to prioritize variants in each locus by assigning an integrative score to each variant on the basis of multi-layer information. Each variant was first assigned scores in the categories listed below (score 0 for no hit, score 1 for a hit, or score 2 for a strong hit), and scores for all the categories were added up to an integrative score. For each locus, the variant(s) with the highest integrative score were assigned as tier-1 variants. Those with the second-highest scores (no less than 70% of the highest score) were assigned as tier-2 variants and the rest as tier-3 variants.
-
(1)MPRA scores:
-
•variants displaying significant allelic transcriptional activity (FDR < 0.01) in melanoma cells were considered as a hit and those with strong significance FDR < 10−9 as a strong hit;
-
•variants displaying significant allelic transcriptional activity (FDR < 0.01) in melanocytes were considered as a hit and those with strong significance FDR < 10−9 as a strong hit;
-
•an assignment as a transcriptional activator function in either melanoma cells or melanocytes was considered as a hit (see transcriptional activator/repressor inference).
-
•
-
(2)Chromatin annotation scores:
-
•overlap with an accessible chromatin region (genomic regions defined as peaks from ATAC-seq, DHS-seq, or FAIRE-seq data) reported in at least one dataset was considered as a hit and if in more than one dataset (four datasets in total: melanocytes in ENCODE and Epigenome Roadmap datasets, melanocytes from in-house data, and melanoma cultures from Verfaillie et al.31) as a strong hit;
-
•overlap with human melanocyte histone modifications consistent with enhancer (marked by both H3K27Ac ChIP-seq peak and H3K4Me1 ChIP-seq peak) or promoter region (marked by both H3K27Ac ChIP-seq peak and H3K4Me3 ChIP-seq peak) from Epigenome Roadmap database was considered as a hit and overlap with both enhancer and promoter regions as a strong hit.
-
•
-
(3)Fine-mapping scores:
-
•variant included in the 95% credible sets from FINEMAP analyses was considered as a hit and PIP > 0.5 as a strong hit;
-
•variant included in the 95% credible sets from POLYFUN analyses was considered as a hit and PIP > 0.5 as a strong hit.
-
•
-
(4)TF-binding motif scores:
-
•variant displaying a significant match with a TF-binding motif (p < 10−4) predicted by motifbreakR analysis was considered as a hit and those displaying strong effects (allelic differences of binding scores > 0.7) as a strong hit.
-
•
Differentially expressed genes between melanoma and melanocytes
We profiled differentially expressed genes (DE-Gs) from RNA sequencing (RNA-seq) data generated for the same melanoma cells (UACC903, n = 3) and immortalized primary melanocytes (C283T, n = 3) used for MPRAs. Total counts of mappable reads for each annotated gene (hg38) were obtained with featureCounts from the Rsubread package.34 We applied the DESeq2 software35 to perform quality control and determine differential expression on the basis of a negative binomial model by using count data from both melanoma and melanocytes groups. The Wald test p values were corrected for multiple testing with the procedure of Benjamini and Hochberg.27 A total of 4,388 DE-Gs were determined with corrected p (FDR) < 0.01 and |log2-fold change| > 2.
Identification of cell-type-specific variants
To identify whether variants were cell-type specific for either melanocyte or melanoma, we applied the following three criteria and assigned scores for each criterion (score 0 for no hit, score 1 for a hit).
-
(1)MPRA allelic effect is exclusively observed in one cell type
-
•MPRA allelic effect FDR < 10−9 (extreme significance) in one cell type
-
•And MPRA allelic effect FDR > 0.01 (non-significance) in the other cell type
-
•
-
(2)145 bp sequence harboring the variant is an activator in the same cell type where the significant allelic effect is observed
-
•MPRA allelic effect FDR < 0.01 in one cell type
-
•And 145 bp sequence is an activator in the same cell type (see transcriptional activator/repressor inference)
-
•And MPRA allelic effect FDR > 0.01 in the other cell type
-
•
-
(3)Predicted TFs binding to the variant display significantly higher abundance in the same cell type where the significant allelic effect is observed
-
•MPRA allelic effect FDR < 0.01 in one cell type
-
•And the levels of predicted TFs are significantly higher in the same cell type (see “DE-Gs” defined in differentially expressed genes between melanoma and melanocytes)
-
•And MPRA allelic effect FDR > 0.01 in the other cell type
-
•
Cell-type regression analyses
To directly compare the allelic transcriptional activity of variants between melanoma and melanocyte, we applied a standard linear regression to encode the interaction term between the cell_type and allele, after adjusting the effect of strand and transfection:
We used the Wald test with robust sandwich type variance estimate on the interaction term to determine the significance, which was corrected for multiple testing. The cutoff of corrected cell-type FDR < 0.01 was applied.
Variant-TF-gene interaction analyses
Melanoma- or melanocyte-specific candidate variant-TF-gene trios were established separately when variants are (1) significant in the MPRA of the corresponding cell type, (2) predicted to significantly change TF binding by motifbreakR, and (3) linked with genome-wide significant eQTL genes in the corresponding dataset. We identified 38 trios for melanoma and 119 trios for melanocyte datasets. For each trio, a multiple linear regression with interaction model was used for the expression levels of eGene and TF (RNA-seq by expectation maximization [RSEM]36) and variant genotype (alternative allele count) (eGene ∼ SNP + TF + SNP × TF). A Benjamini-Hochberg27 correction was applied to the corrected p value (FDR) across each variant-eGene pair (for testing multiple TFs). The trios with FDR value < 5% in SNP × TF are considered as displaying significant variant-TF-eGene interaction.
CRISPRi experiments
CRISPR interference (CRISPRi) was performed in the UACC903 melanoma cell line. Three different guide RNAs (gRNAs) for each variant were designed to target the genomic regions surrounding three tested variants (rs61935859, rs4384, and rs2111398), and the sequences of gRNAs are listed in Table S5A. Non-targeting gRNA and gRNA targeting the adeno-associated virus site 1 (AAVS1) were used as controls. gRNAs were ligated into the lentiviral vector pRC0608-U6-SpCas9-XPR050-puro-2A-GFP (made by Dr. Raj Chari at Genome Modification Core in the Frederick National Laboratory for Cancer Research). For the generation of lentiviral particles, plasmids encoding gRNA or dCas9-ZIM3 (pRC0528_Lenti-dCas9-ZIM3-Blast from Dr. Raj Chari) were co-transfected into HEK293T cells with psPAX2, pMD2-G, and pCAG4-RTR2 packaging vectors. Virus particles were collected 2 days after transfection, and titer was measured by Lenti-X GoStix Plus (Takara, CA). UACC903 melanoma cells were infected with dCas9-ZIM3 lentivirus and selected by 10 μg/mL blasticidin for generation of UACC903-dCas9-ZIM3 polyclonal stable cell line. UACC903-dCas9-ZIM3 cells were infected with lentivirus harboring gRNA. 24 h after infection, 2 μg/mL of puromycin was applied for selection. Surviving cells were harvested 48 h after puromycin selection for RNA and protein isolation. The experiments were performed in at least three biological replicates in sets of six replicates. Total RNA was isolated with an RNeasy Kit (Qiagen). For optimal synthesis of the relatively large full-length cDNA of MED13L (3.2 kb), SuperScript III First-Strand Synthesis kit (Thermo Fisher) was used. The cDNA of MAFF and GPRC5A/HEBP1/EMP1 was generated with High-Capacity cDNA Reverse Transcription kit (Thermo Fisher). mRNA levels of each gene were measured with a Taqman probe set (Table S5B) and normalized to GAPDH levels. qPCR triplicates (technical replicates) were averaged to be considered as one data point. Proteins were separated on NuPAGE 3%–8% Tris-Acetate Protein Gels (Thermo Fisher) and detected by mouse anti-Cas9 (7A9–3A3, Active Motif) and mouse anti-GAPDH (sc-47724, Santa Cruz) primary antibodies.
Statistical analyses
Cell-based experiments were repeated at least three times with separate cell cultures, and mean values of all the biological replicates are presented. For all plots, individual data points are shown with the median or mean, range (maximum and minimum), and 25th and 75th percentiles (where applicable). The statistical method, number of data points, and number and type of replicates are indicated in each figure legend.
Results
MPRAs identified functional variants in 42 melanoma GWAS loci
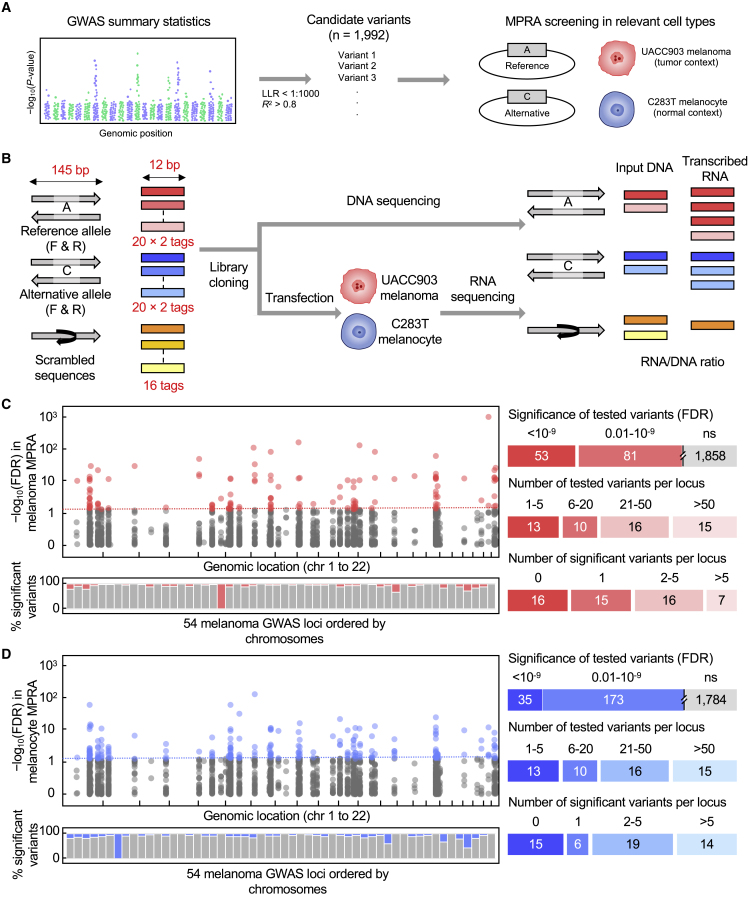
We performed MPRAs to simultaneously identify functional cis-regulatory variants for multiple melanoma GWAS loci. We tested 1,992 variants (median 26.5 variants per locus) from 54 genome-wide significant loci (including 11 additional independent signals) based on the recent melanoma GWAS meta-analysis1 (Table S1). To select these variants, we primarily considered GWAS statistics (log likelihood ratio < 1:1,000 with the primary lead SNPs) and further used LD for the variants that are not present in the imputation reference set or poorly imputed in the GWAS data and for the secondary signals (R2 > 0.8 with the lead SNP) (Figure 1A; Table S2; material and methods). We assessed 145 bp genomic sequences encompassing the reference and alternative alleles of each variant for their potential as a transcriptional enhancer in luciferase constructs in both forward and reverse directions with 20 unique barcodes associated with each tested sequence. A scrambled sequence of the same 145 bp associated with 16 barcodes was also tested as a null for each variant (Figure 1B; material and methods). To test variant function in the cellular contexts representing both tumor and normal states, we transfected the MPRA library into a melanoma cell line (UACC903, n = 8 transfections) and an immortalized primary melanocyte cell line (C283T, n = 5). Each barcode sequence detected in the input DNA or mRNA (cDNA) after transfections was counted by sequencing. Initial quality assessment showed a good correlation of normalized tag counts among transfection replicates by tags (median Pearson R = 0.553 and 0.745 for melanoma and melanocyte, respectively; Figures S2 and S3) and by variants (median Pearson R = 0.938 and 0.947 for melanoma and melanocyte, respectively; Figures S4 and S5). High recovery rates of designed tags were observed in the transcribed output (90.2%–92.8% for melanoma and 93.7%–94.3% for melanocyte; Table S4). Details of quality control measure for downstream analyses are shown in Table S4.
Figure 1.
MPRA analysis of candidate variants and risk loci identified in melanoma GWAS
(A) Overall workflow from melanoma GWAS summary statistics1 to candidate variants for MPRA analysis.
(B) MPRA design. Oligo libraries were synthesized with 145 bp of sequence encompassing each variant with reference or alternative allele in both forward and reverse (F and R) directions, which are associated with 12 bp barcodes (20 tags per unique sequence). For each variant, a scrambled sequence of 145 bp test sequence was also included and associated with 16 tag sequences (using forward direction and reference allele) as a null. Libraries were cloned into luciferase constructs and then transfected into UACC903 melanoma cells or melanocyte cells to generate expressed RNA tag libraries. Both input DNA and RNA libraries were sequenced to assess the tag counts associated with the test sequences.
(C and D) A summary of MPRA results in UACC903 melanoma cells (C) and melanocyte cells (D). FDR values for allelic transcriptional activity of each variant measured by MPRAs are displayed in Manhattan plots (two-sided Wald test with robust sandwich type variance estimate). Horizontal lines represent an FDR cutoff of 0.01 (−log10(FDR) = 2), and variants displaying significant allelic transcriptional activity are shown separately for melanoma (red) and melanocyte (blue) experiments. Bar graphs under the Manhattan plots show the percentage of variants displaying significant allelic transcriptional activity (FDR < 0.01, red for melanoma and blue for melanocyte; ≥ 0.01, gray) by melanoma GWAS loci ordered by chromosomes (defined in Table S1). Bar graphs on the right present the summarized statistics as to the numbers of tested versus MPRA-significant variants in total or by locus for each cell type. Notes: LLR, log likelihood ratio; FDR, false discovery rate; ref, reference allele; alt, alternative alleles.
We first focused on the variants displaying allelic transcriptional activity in each cell type, identifying 134 (7% of tested variants) in UACC903 melanoma (Figure 1C; Table S6) and 208 (10% of tested variants) in C283T melanocyte cell lines (Figure 1D; Table S7) that pass an FDR < 0.01 cutoff (two-sided Wald test with robust sandwich type variance estimate; multiple testing correction by Benjamini and Hochberg27 method; material and methods). We defined these 285 unique variants (FDR < 0.01 in either cell line; 14% of tested variants) as “MPRA-significant variants.” 78% of the melanoma GWAS loci (42 of 54 loci) displayed at least one MPRA-significant variant. For 83% of these loci (35 of 42 loci), MPRA-significant variants were identified from both cell types, while the rest were from only one cell type (three loci in melanoma and four loci in melanocyte). For eight loci, a single MPRA-significant variant was identified, while 2–36 MPRA-significant variants were identified for 34 loci.
We further inferred a putative transcriptional activator/repressor function of the 145 bp around MPRA-significant variants by applying two criteria: (1) the sequence containing either allele displays an extreme outlier expression level (three-time interquartile range above 75th or below 25th percentiles, material and methods) compared to the mean expression level distribution of all the tested tags (assuming that most of the tested variants do not display transcriptional activity) and (2) the same sequence also shows a significantly higher/lower expression level than the matched scrambled sequence (FDR < 0.01) (material and methods). Among these 285 variants, 57 variants were assigned as activators in the melanoma cells (Figure S6A), 28 variants in melanocytes (Figure S6B), and 15 in both cell lines. Only one variant (rs2911405) was identified as a repressor in melanocytes, which displayed significantly lower expression level than the mean value as well as that of scrambled sequence.
Notably, our MPRA design included 206 variants that have been tested in the same UACC903 cell line from our previous study, and 93.3% of them (194 of 208) displayed consistent results between two studies regarding the significance of allelic transcriptional activity (FDR < 0.01) and allelic direction (Table S6). To detect potential bias from tag sequences in measured cis-regulatory activity, we applied a sequence-based correction method, MPRA tag sequence analysis (MTSA)28 (material and methods). The regression using MTSA-corrected expression levels demonstrated that 284 of 285 MPRA-significant variants displayed consistent allelic direction before and after the correction. Moreover, 85% (melanoma) and 78% (melanocyte) of the MPRA-significant variants (FDR < 0.01) still displayed an allelic difference at a relaxed criteria (FDR < 0.1) after correction (Table S8). These results supported that the allelic differences detected in this study are robust and reproducible, and we therefore used the normalized expression values before applying MTSA-correction throughout the study.
Fine-mapping and motif prediction of functional variants
To supplement and compare with the variant prioritization based on MPRAs, we performed a fine-mapping analysis of the melanoma GWAS data. Statistical fine-mapping of 54 melanoma GWAS loci with FINEMAP32 nominated 2 to 101 variants per locus (median = 32.5) in 95% credible sets. We also performed a fine-mapping with POLYFUN,33 incorporating functional annotations (precomputed prior causal probabilities based on a meta-analysis of 15 UK Biobank traits) following the recommended procedure, which further narrowed down the credible set to 2 to 84 variants per locus (median = 19) (Table S9; material and methods). Complementing and refining these prioritizations, MPRAs identified between 1 and 36 candidate functional variants per locus (median 5 variants) that display significant allelic transcriptional activity from 42 melanoma GWAS loci (Figure S7). MPRA-significant variants displayed slightly higher posterior inclusion probability (PIP) and larger proportion of “high” probability score variants (PIP > 0.1) compared to non-significant variants, resulting in a higher percentage being included in the 95% credible sets of FINEMAP and POLYFUN, although the enrichments were not statistically significant (Figure S8; Table S10).
To assess the roles of TFs in variant functionality, we predicted the allelic TF binding affinity of each MPRA tested variant by using motifbreakR37 (material and methods). A substantial proportion of MPRA-significant variants (167/285, 58.6%) were predicted to have effects on at least one TF-binding site (Table S11). These predicted allelic binding scores displayed a significant correlation with allelic transcriptional activities measured from our MPRAs in C283T melanocyte data (Spearman R = 0.249, p = 0.006) and a non-significant but similar pattern in UACC903 melanoma data (R = 0.155, p = 0.172) (Figure S9). MPRA-significant variants more frequently overlapped with the genomic regions annotated as open chromatin (32% versus 28%; Chi-squared p = 0.0026) or promoter/enhancer (15% versus 11%; Chi-squared p = 0.1998) in melanoma or melanocyte datasets compared to non-significant variants (Figure S10; material and methods). These results suggested that some of the observed allelic differences from MPRAs could be attributed to differential binding of TFs and potentially driven by functional cis-regulatory elements in melanocyte or melanoma cells.
Nominating the most plausible candidate variants with an integrative scoring system
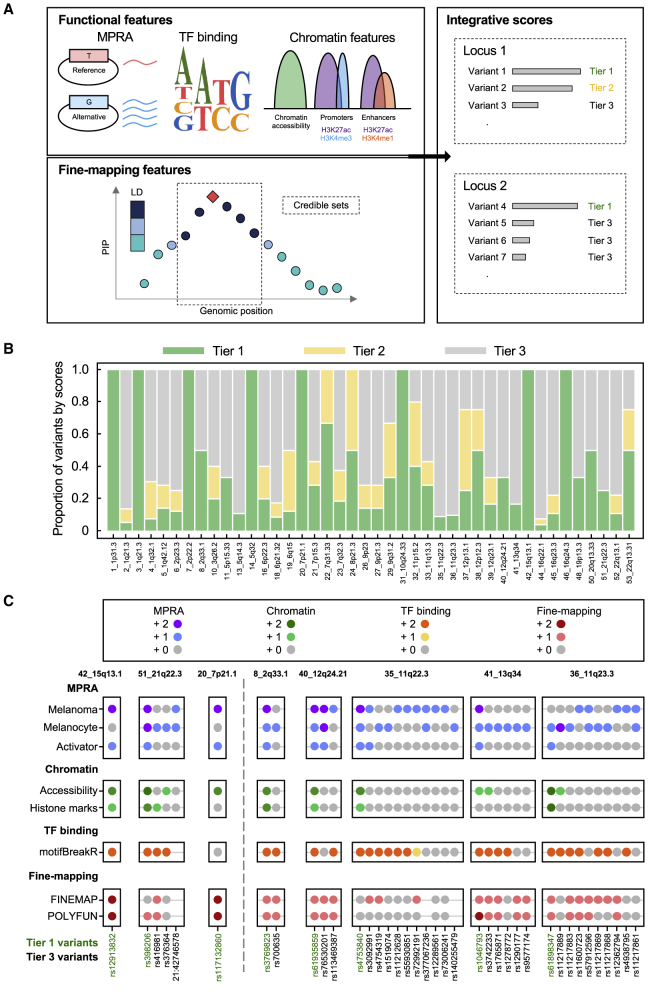
To further nominate the most plausible variants for in-depth follow-up from each locus, we integrated multi-layer functional annotations and fine-mapping data to the 285 variants prioritized by MPRAs. Given that our MPRA system evaluates variants in an episomal setting, we incorporated chromatin features of the genomic regions around these 285 variants in melanocyte and melanoma cells. We previously profiled accessible chromatin regions in primary cultures of melanocytes by using ATAC-seq11 (n = 6 individuals) and compiled other melanocyte and melanoma cell chromatin features (accessible chromatin, promoter, and enhancer histone marks) from public databases and published studies30,31,38 (material and methods). We also incorporated the information from the statistical fine-mapping and motif prediction analyses described earlier. To systematically integrate these multi-layer features, we established a scoring system by assigning three-level scores (0 = no hit, 1 = hit, 2 = strong hit) to each of the eight components under four categories (MPRA, chromatin annotation, TF binding, and fine-mapping) for 285 MPRA significant variants (Figure 2A; material and methods). Within each locus, the variant(s) displaying the highest integrative score on the basis of these four categories were assigned as tier-1 variants. For some loci, there were variants displaying lower but similar scores to the tier-1 variants (>70% of the highest score), which were assigned as tier-2 variants. The rest were assigned as tier-3 variants (material and methods; Table S12).
Figure 2.
Integrative scores for prioritizing plausible candidate variants
(A) The functional (MPRA, motif prediction, and chromatin annotations) and fine-mapping features (credible sets and posterior possibility, PIP) were incorporated to evaluate the candidate variants. For each locus, the variant(s) with the highest combined score were assigned as tier-1 variants (green) and those with the second-highest scores (no less than 70% of the highest score) were assigned as tier-2 variants (yellow).
(B) The overall prioritization from MPRA-significant variants to tier-1 (green) and tier-2 (yellow) variants are shown. Each bar represents a melanoma GWAS locus.
(C) Examples of melanoma GWAS loci with known functional variants (the first three loci on the left side of vertical dashed line) or substantial prioritization performance (five loci on the right side of vertical dashed line). For each variant, hits are given a score of 1 (MPRA, blue dots; chromatin annotation, light green dots; motif, yellow dots; and fine-mapping, light red dots). Strong hits are given a score of 2 (MPRA, purple dots; chromatin annotation, dark green dots; motif, orange dots; and fine-mapping, dark red dots). No hits are shown with gray dots. Definition of hits and strong hits are presented in material and methods. No dots (gray lines) are presented if functional/fine-mapping features are unavailable for the given variant.
Using this system, we nominated 86 top-score variants including 52 tier-1 and 34 tier-2 variants across the 42 loci (Figure 2B; Table S12), with between one and six top-score variants per locus (median = 2) and a single top-score variant for 18 of the 42 loci (43%). Among them were well-characterized functional variants including the top two variants with the highest scores (Figure 2C). For example, rs12913832 in the locus at 15q13.1 (42_15q13.1) is a known functional variant in a melanocyte enhancer element mediating allelic OCA2 expression.5 rs398206 in the locus at 21q22.3 (51_21q22.3) was shown to regulate MX2 expression in melanocytes via allelic binding of YY1, and MX2 accelerated melanoma formation in a zebrafish model.9 A third variant (displaying the fourth highest score), rs117132860 in the locus at 7p21.1 (20_7p21.1), is a functional variant driving ultraviolet B (UVB)-responsive allelic expression of AHR with a prolonged effect in melanocyte growth and cellular response to UVB exposure.11 Re-identification of these known functional variants supported the validity of our scoring system for variant prioritization. For 15 other loci with a single top-score variant (Figure 2C; Table S12), prioritized variants include top candidates from our previous study9 (e.g., rs3769823 at 8_2q33.1) as well as those from ten newly discovered loci by the recent GWAS (e.g., rs61935859 at 40_12q24.21, rs4753840 at 35_11q22.3, rs1046793 at 41_13q34, and rs61898347 at 36_11q23.3).1 These data demonstrated that most of the melanoma GWAS loci (78%) harbor potential functional variants via cis-regulatory mechanisms (i.e., allelic transcriptional activity) either with a single prominent candidate (42% of loci) or multiple (up to six) functional candidate variants (58% of the loci) based on the multi-layer functional features.
Linking functional variants to target genes with eQTLs/meQTLs
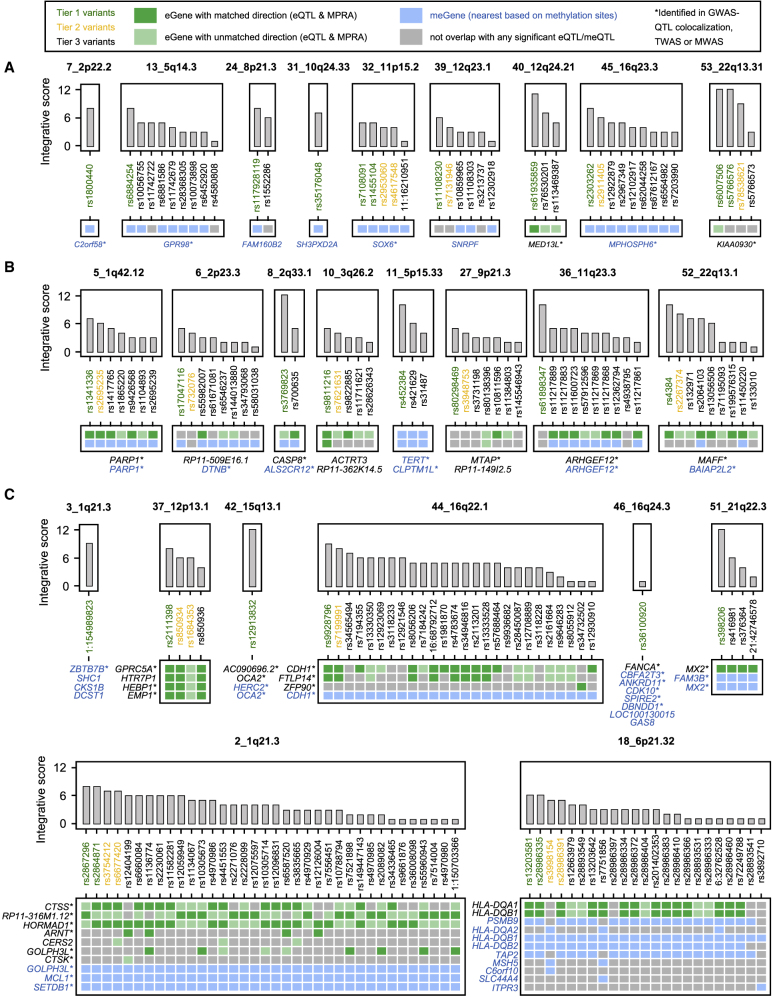
To link the candidate functional variants to target susceptibility genes, we used eQTLs and meQTLs of melanocytes from 106 individuals and of melanoma tissues from 444 individuals with skin cutaneous melanomas from TCGA. We previously identified 597,335 significant cis-eQTLs and 1,497,502 cis-meQTLs (+/−1 Mb of transcription start site or CpG sites, FDR < 0.05, not LD-pruned) in melanocytes, and 209,393 significant cis-eQTLs and 3,794,446 cis-meQTLs in melanomas.17,18 60% of the MPRA-significant variants (172/285) overlapped genome-wide significant eQTLs or meQTLs in melanocytes or melanomas, nominating 31 candidate eGenes (Table S13) and 42 assigned genes for meProbes (which we define as meGenes) in 26 loci (Table S14). Among these loci, nine loci were mapped to a single eGene or meGene (Figure 3A), eight loci to two eGenes/meGenes (Figure 3B, including those at 5_1q42.12 and 36_11q23.3 to the same gene by both eQTL and meQTL), while eight loci were mapped to three or more eGenes/meGenes (Figure 3C). A total of 23 eGenes (23/31, 74.2%) and 25 meGenes (25/42, 59.5%) were further supported by GWAS-QTL colocalization or TWAS/MWAS.13 Furthermore, a total of 93 MPRA-significant variants from 14 loci displayed a consistent direction between MPRAs and eQTL, in which the direction of allelic expression of local genes matches those of MPRA allelic transcriptional levels (Table S13). We limited the allelic direction matching analysis to eQTL genes because of the intrinsic complexity of association between DNA methylation levels and target gene expression levels.
Figure 3.
Linking the candidate variants to the target genes via QTLs
MPRA-significant variants in melanoma or melanocyte datasets are presented by locus if they display genome-wide significant eQTL or meQTL p values for one of the significant genes (eGenes with gene names in black, dark green square for matched direction in eQTL and MPRAs, and light green square for not matched direction) or the nearest gene based on methylation sites (meGenes with gene names in blue, blue square) in melanocyte or melanoma datasets. The loci were presented on the basis of the total number of eGenes and meProbes, with 1 in (A), 2 (including those with the same assigned gene name for eGene and meGene) in (B), and 3 or more in (C). Variants are ordered by integrative scores for each locus with tier-1 variants shown in green, tier-2 variants in yellow, and tier-3 variants in black. Asterisks next to the gene names indicate the genes identified in GWAS-QTL colocalization, TWAS, or MWAS.
For example, rs61935859, a single tier-1 top-score variant in the locus at 12q24.21 (40_12q24.21), is linked to a single eGene, MED13L (Figures 2C and 3A), with a matched direction of allelic expression. Namely, the melanoma-risk-associated G allele displayed 1.6- and 1.05-fold higher transcriptional activity in MPRAs (FDR = 5.48 × 10−92 and 4.93 × 10−4 in UACC903 and C283T, respectively) and is correlated with higher MED13L levels in the melanocyte dataset (slope 0.48 relative to G allele and eQTL p = 5.49 × 10−9). In the locus at 22q13.1 (52_22q13.1), the tier-1 variant, rs4384 (Figure 3B), was the only tier-1 top-score variant and also with a matched direction of allelic expression with eGene MAFF, where the melanoma-risk-associated G allele increased transcription by 1.3-fold in MPRAs (FDR = 5.05 × 10−41 in UACC903) and is also correlated with higher MAFF levels in the melanocyte dataset (slope 0.89 relative to G allele and eQTL p = 7.11 × 10−25). MAFF encodes a basic leucine zipper (bZIP) TF and has been reported to be involved in multiple cancers. In the locus at 16q22.1 (44_16q22.1), two top-score variants, rs9928796 and rs7199991 (Figure 3C), are linked to CDH1 (increased with risk) and FTLP14 (increased with risk) with matched directions of allelic expression in the melanocyte eQTL dataset and MPRAs in UACC903. While FTLP14 is a pseudogene, CDH1 encodes E-cadherin. E-cadherin is a cell adhesion molecule responsible for the adhesion of melanocytes to keratinocytes,39 and loss of E-cadherin was observed in melanoma progression,40 in line with its roles in epithelial-to-mesenchymal transitions.41 In the locus at 12p13.1 (37_12p13.1), rs2111398 and rs850934 (Figure 3C) are the top-score variants linked to four eGenes (GPRC5A, HTR7P1, HEBP1, and EMP1; decreased gene expression associated with risk for all four genes) with matched directions of allelic expression in the melanocyte eQTL dataset and MPRAs in UACC903. We note that some of these variants displaying strong allelic function in UACC903, including those at 22q13.1, 16q22.1, and 12p13.1, were only significant eQTLs/meQTLs in melanocyte dataset, potentially because of relatively lesser statistical power in the heterogeneous TCGA tumor tissue dataset. Significantly enriched pathways in these 31 eGenes and 42 meGenes (67 unique genes) consistently highlighted those relevant to cellular immune response and apoptosis signaling (Table S15). Thus, by combining MPRAs and molecular QTLs in melanomas and melanocytes, we nominated candidate susceptibility genes linked to one or more plausible functional variants from 48% of the known melanoma GWAS loci.
Validation of functional variants and target genes by CRISPRi
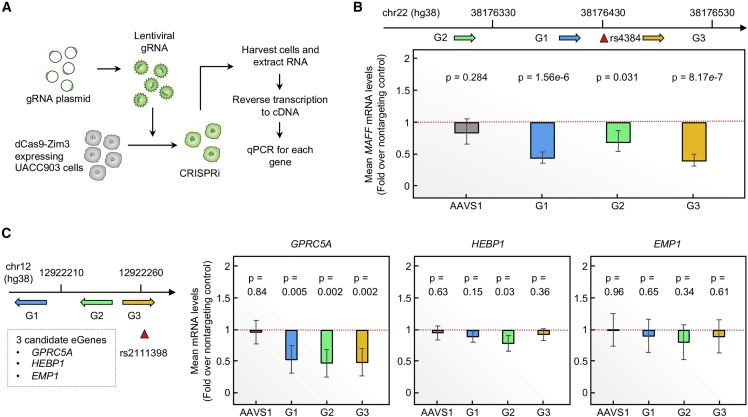
To further determine whether the genomic regions encompassing the prioritized functional variants regulate expression levels of target genes, we performed CRISPRi of three representative top-tier variants by using the dCas9-ZIM3 system in the UACC903 melanoma cell line (Figure 4A, material and methods). We focused on loci (1) that have not been previously characterized, (2) with eGenes identified in GWAS-eQTL colocalization or TWAS, (3) with eGenes and tier-1 variants displaying a matching allelic direction between eQTL and MPRAs, and (4) with the variants located in annotated enhancers/promoters in melanomas or melanocytes. Using these criteria, we selected five variant-eGene pairs from three loci (rs61935859-MED13L at 12q24.21, rs4384-MAFF at 22q13.1, and rs2111398-GPRC5A/HEBP1/EMP1 at 12p13.1) and targeted each SNP by using three different gRNAs. CRISPRi followed by qPCR demonstrated a 31%–60% reduction of MAFF levels upon targeting the region encompassing rs4384 for all three gRNAs (p = 1.56 × 10−6, 0.031, and 8.17 × 10−7, two-tailed t test, n = 24, combined from four biological replicates; Figure 4B). We also observed a 27%–30% reduction of GPRC5A levels for all three gRNAs targeting rs2111398 (p = 0.005, p = 0.002, and p = 0.002, two-tailed t test, n = 24, combined from four biological replicates; Figure 4C). No significant changes of HEBP1 or EMP1 levels were observed for all three gRNAs in this locus (at p < 0.017 cutoff for testing three genes). For MED13L, we did not observe significant changes in three biological replicates (Figure S11). These data identified MAFF and GPRC5A as plausible melanoma susceptibility genes regulated by functional cis-regulatory variants and demonstrated that our scoring strategy could nominate the most plausible loci, functional variants, and candidate susceptibility genes for further in-depth characterization.
Figure 4.
CRISPRi with gRNAs targeting prioritized variants in UACC903 cells
(A) gRNA plasmids were packed into lentiviral particles in HEK293T cells and then transduced into dCas9-Zim3 expressing UACC903 cells. 24 h after infection, transduced UACC903-dCas9-ZIM3 cells were selected with 2 μg/mL of puromycin. Survived cells were harvested 48 h after puromycin selection for RNA isolation.
(B) CRISPRi with three gRNAs (G1, G2, and G3) targeting the region (genomic coordinates in hg38) surrounding rs4384. The levels of MAFF transcript (GAPDH-normalized) are shown as fold change over those from non-targeting gRNA. Four biological replicates of n = 6 were combined (total n = 24). Error bars refer to the standard error. p values are calculated by two-sample t test (two-sided) with unequal variance from non-targeting controls (dotted red lines).
(C) CRISPRi with three gRNAs targeting the region surrounding rs2111398. The levels of GPR5CA/HEBP1/EMP1 transcripts (GAPDH-normalized) are shown as fold change over those from non-targeting gRNA. Replicates, error bars, and p values are the same as described in (B).
Cell-type specificity of melanoma-associated functional variants
Given that MPRA-significant variants displayed cell-type-dependent allelic activity, we further inspected the cell-type-specific functionality of these variants. Namely, 57 variants displayed significant allelic transcriptional activity in both melanoma and melanocyte cell lines, while 77 variants were only significant for melanoma and 151 variants only for melanocytes (Figure 5A). Notably, 1.6 times more variants were identified in melanocytes, even though the total number of transfected cells was greater for UACC903 (transfection events = 8 in UACC903 and 5 in C283T), potentially because of higher transfection efficiency of C283T cells. On the other hand, allelic differences in transcriptional activities were significantly larger for 134 variants significant in melanoma (median 1.14-fold, range 1.07- to 2.88-fold, Table S6) than for 208 variants significant in melanocyte (median 1.06-fold; range 1.03- to 2.47-fold, Table S7) (p = 5 × 10−15, two-tailed unpaired t test), which is consistent with elevated global transcription levels observed in cancer cells.42,43 For the 57 variants that are significant in both cell types, allelic differences displayed a similar pattern with a larger effect size in melanoma (median 1.19-fold in melanoma versus 1.08-fold in melanocyte) and a significant difference between two cell types in a paired test (p = 0.00037, two-tailed paired t test). For example, the variant rs398206 in the locus at 21q22.3 (51_21q22.3) showed significant allelic transcriptional activity both in melanoma (MPRA FDR = 0) and melanocyte (MPRA FDR = 3.83 × 10−18), but its allelic effect was stronger in melanoma (allelic difference of 2.88-fold in melanoma versus 1.16-fold in melanocyte; Figure S12A). We further inspected cell-type-dependent “activator” (material and methods) function of the DNA sequences harboring MPRA-significant variants. Among 285 MPRA-significant variants, ∼2-fold more variants also displayed activator function in melanoma (57 variants) compared to melanocytes (28 variants). Moreover, 32% of 77 melanoma-only allelic variants were also located in melanoma-only activators, while 5% of 151 melanocyte-only variants were in melanocyte-only activators. These observations suggested substantial cell-type specificity of melanoma-associated functional variants between melanoma and melanocyte and their potentially larger allelic effect sizes accompanied by stronger transcriptional activity in melanoma cells in our system.
Figure 5.
Cell-type specificity at the level of variants and genes
(A) Overall analysis from 285 MPRA-significant variants to 77 variants only significant in melanoma (red dots represent melanoma MPRA FDR < 0.01) and 151 variants only significant in melanocyte (blue dots represent melanocyte MPRA FDR < 0.01). Three criteria were applied to further prioritize variants with cell-type specificity, including MPRA FDR < 10−9, putative role of activator, and TF identified as the high-expressed DE-Gs in the specific cell type. Variants meeting at least one criterion were further prioritized.
(B–D) Representative loci with variants showing cell-type specificity in melanoma (B), both (C), or melanocyte (D) are shown. Asterisks next to variant IDs represent the number of criteria that are met for that variant.
(E) Variants meeting at least one criterion in the MPRAs of melanoma cells are presented if they are also a genome-wide significant eQTL (eGenes, green) or meQTL (meGenes, blue) in the TCGA melanoma QTL dataset. The variants are grouped and ordered by GWAS loci with locus IDs shown at the top of each group of variants.
(F) Variants meeting at least one criterion in the MPRAs of melanocyte cells are presented if they are also a genome-wide significant eQTL (eGenes, green) or meQTL (meGenes, blue) in the melanocyte QTL dataset. The variants are grouped and ordered by GWAS loci with locus IDs shown at the top of each group of variants.
To formally nominate the cell-type-specific variants, we further assessed 77 melanoma-only and 151 melanocyte-only variants in MPRAs (FDR < 1%, Figure 5A). We applied three criteria for these variants as follows: (1) a variant shows strong allelic transcriptional activity (MPRA FDR < 10−9) in the same cell type, (2) 145 bp encompassing the variant is a cis-activator from the MPRA of the same cell type, or (3) the level of TF predicted to show allelic binding is significantly higher in the same cell type based on the differentially expressed gene analysis between UACC903 melanoma and C283T melanocyte cells (Figure 5A). We reasoned that an extreme allelic significance cutoff (MPRA FDR < 10−9) could help reduce potential false positives coming from technical differences (e.g., transfection efficiency, potential tag sequence effect). Further, we hypothesized that potential drivers of cell-type dependency in allelic transcriptional activity could be enhancer strength and/or differential availability of allele-preferential binding TFs between two cell types used in MPRAs. To test this hypothesis, we performed a transcriptome analysis of UACC903 and C283T cells by sequencing the same mRNA samples from MPRA transfections (n = 3 from each cell type). A total of 4,388 differentially expressed genes (DE-Gs; p < 0.01 and |log2-fold change| > 2) were identified with DESeq2. After applying the three criteria, a total of 36 of 77 variants met at least one criterion in melanoma (Table S16) and 45 of 151 variants in melanocytes (Table S17), which we define as melanoma-specific and melanocyte-specific variants, respectively (Figure 5A). One example is rs4384 in the locus at 22q13.1 (52_22q13.1), which only showed significant allelic transcriptional activity in melanoma (MPRA FDR = 5.05 × 10−41, Figure S12B) and was nominated by all three criteria. To confirm the cell-type specificity, we applied a linear regression to encode the interaction between cell type and allelic effect (material and methods). Notably, all five variants nominated by all three criteria displayed a significant interaction between allelic effect and cell type (FDR < 0.01). Moreover, 82% (melanoma specific) and 75% (melanocyte specific) of variants nominated by at least two criteria displayed a significant interaction between allelic effect and cell type (FDR < 0.01) (Tables S16 and S17). These results further validated the cell-type-specific variants nominated with our three criteria.
Notably, we observed ten loci with only melanoma-specific variants (examples in Figure 5B), 12 loci with both melanoma- and melanocyte-specific variants (an example in Figure 5C), and 11 loci with only melanocyte-specific variants (examples in Figure 5D). We further looked into the QTL-based target genes assigned to these cell-type-specific variants in the matching cell type. As shown in Figure 5E, a total of five eGenes and 12 meGenes from melanomas are linked with melanoma-specific variants, while 12 eGenes and eight meGenes from melanocytes are linked with melanocyte-specific variants (Figure 5F). In the locus at 5p15.33 (11_5p15.33), two of the three MPRA-significant variants are melanoma-specific variants and also meQTLs for TERT (only in melanomas) and CLPTM1L (in both melanomas and melanocytes). Notably, TERT expression is re-activated in transformed melanoma cells but not in differentiated melanocytes. In the locus at 1q42.12 (5_1q42.12), a single variant rs1865220 is melanocyte specific and an eQTL for PARP1, consistent with its role of mediating melanocyte growth.8 Many other loci displayed both melanoma- and melanocyte-specific variants that are linked with target genes. Two variants in the locus at 6p21.32 (18_6p21.32) are melanoma specific and eQTLs for two HLA genes, HLA-DQA1 and HLA-DQB1, in melanomas, while two variants in the same locus are melanocyte specific and meQTLs for an immunoproteasome gene, PSMB9, in melanocytes. The locus at 1q21.3 (2_1q21.3) presents two each of melanocyte-specific and melanoma-specific variants, where CTSS in melanocytes and HORMAD1 in melanomas are representative target genes. The locus at 16q22.1 (44_16q22.1) presents five melanocyte-specific and seven melanoma-specific variants with the common target gene, CDH1.
For ten of 36 melanoma-specific variants, the expression levels of TFs predicted to show allelic binding were higher in UACC903 melanoma cells compared to C283T melanocytes (DE-Gs with FDR < 0.01 and |log2-fold change| > 2, n = 3, material and methods; Table S16). Notably, HES1 and HEY2, which are known targets of NOTCH signaling pathway44 and induced in cancers, were linked to four variants from distinct loci (52_22q13.1, 11_5p15.33, 16_6p22.3, and 44_16q22.1), and three of these variants are in melanoma-specific activators. For 31 of 45 melanocyte-specific variants, the levels of predicted allelic TFs were higher in C283T melanocytes compared to UACC903 melanomas (Table S17). For 22 variants among them, differentially expressed TFs (EGR4, HIC1, TBX5, TCF4, THRB, ARID3A, FOSL2, JUN, JUNB, FOXF2, KLF8, MEIS1, IRF1, IRF7, and IRF9) were linked to melanocyte-specific variants from more than one locus. These data suggested that melanoma risk-associated variants within and across multiple GWAS loci could be functional in different cellular contexts representing normal/primary melanocytes and transformed/melanoma cells, and TF levels could potentially contribute to the context dependency.
Effect of transcription factors on allelic expression of susceptibility genes
Given the suggested roles of TFs in the allelic transcriptional activity of melanoma-associated variants including cell-type-specific ones, we further investigated the interaction of MPRA-significant variants and the levels of allelic binding TFs on target eGene expression in large-scale eQTL datasets. For this, we included 38 variant-TF-eGene trios from melanoma data by selecting MPRA-significant variants in UACC903 (FDR < 0.01), significant allelic binding of a TF to the variant predicted by motifBreakR, and genome-wide significant eQTL target gene for the variant in TCGA melanomas. We included 119 trios from melanocyte data, similarly selecting MPRA-significant variants in C283T, their predicted TFs, and target genes in melanocyte eQTL dataset.
Using a multiple linear regression interaction model45 (material and methods), we identified significant variant-TF-eGene interactions for seven melanoma trios and seven melanocyte trios at FDR 5% (Table S18). In the melanoma analysis, four variants from the locus at 1q21.3 (2_1q21.3) significantly interacted with seven different TFs (ATF6, E4F1, REST, ESRRG, ZNF143, ATF5, and FOXJ3) and all were linked to an eGene, HORMAD1 (Table S18). Notably, this locus has a large LD block with multiple functional variants including two tier-1, two tier-2, two melanoma-specific, and two melanocyte-specific variants with nine potential target genes (Figures 3C, 5E, and 5F). HORMAD1 is a melanoma-specific eQTL gene for multiple MPRA-significant variants, and rs10305673 (melanoma-specific variant), among them, showed a significant interaction with a TF, REST, in the TCGA melanoma dataset (FDR = 0.000488; Table S18). Further, one of the tier-1 variants at this locus, rs2864871, showed a significant interaction with three TFs in the TCGA melanoma dataset (ATF6, E4F1, and ESRRG; FDR = 0.000423, 0.000423, and 0.00493, respectively; Table S18). These data suggested that these TF-interacting MPRA functional variants potentially mediate HORMAD1 expression regulation that might contribute to melanoma susceptibility at this locus. In melanocyte analysis, three variants from three loci (including two melanocyte-specific variants) significantly interacted with six different TFs (FLI1, THRB, ETV4, ELF1, ETS1, and POU3F1) and four eGenes (GPRC5A, CDH1, HEBP1, and CASP8) (Table S18). Notably, the variant rs850936 (melanocyte cell type score = 1) showed an interaction with four ETS-domain TFs (FLI1, ETV4, ELF1, and ETS1) on the expression of GPRC5A and/or HEBP1. Among them, FLI1 was a DE-G displaying higher levels in C283T compared to UACC903, suggesting that FLI1 might mediate cell-type-specific allelic function of this variant in melanocytes (Figure S13A). Moreover, the variant rs4783674 (melanocyte cell type score = 2) showed an interaction with a TF, THRB, on CDH1 levels in melanocytes. Notably, the level of THRB was significantly higher in C283T melanocytes compared to UACC903 melanoma cells, which further supported the hypothesis that THRB mediates melanocyte-specific variant functionality altering CDH1 expression to contribute to melanoma risk in this locus (Figure S13B). Together these data suggested that a subset of MPRA-significant variants including cell-type-specific variants also interact with TFs to affect the target eQTL gene levels, and TF availability might play an important role in variant functionality including their cell-type specificity.
Discussion
In this study, we performed MPRAs of 1,992 variants selected from 54 melanoma GWAS loci to narrow down to a small set (285, 14%) of functional variants displaying allelic transcriptional activity. To further reduce this set, we constructed a score that leveraged multi-layer genetic and functional features including epigenomic annotation from relevant cell types, GWAS fine-mapping scores, and motif prediction, in addition to allelic functionality measured by MPRAs. This score nominated a small number of top-score variants for 42 of 54 known melanoma GWAS loci, most of which had not been functionally tested before. The validity of the MPRA-significant variants and the scoring system was demonstrated by re-identification of the well-characterized variants from three melanoma loci as the top two variants5,9 and another high-ranking variant11 among all 285 variants. By integrating this variant scoring system with expression and methylation QTLs from disease-relevant cell types (melanoma and melanocyte), we linked the functional variants to their potential target eGenes or meGenes. Some of these variant-gene connections were validated with a CRISPRi system in a relevant cell type. Given that in vitro and in vivo characterization of candidate susceptibility genes is laborious and time consuming, a tiered nomination of loci, variants, and genes for 48% of melanoma GWAS loci by our study will inform future functional follow-up studies. Compared to our previous study,9 the current study presents significant advances regarding the number of tested loci (>3-fold more loci), cellular context (primary melanocytes melanoma cells were formally compared), further variant prioritization via scoring system, and variant-to-gene linkage via both eQTLs and meQTLs.
Our systematic profiling of melanoma GWAS loci provided a few general observations regarding genetic susceptibility to melanoma. Unbiased testing of all the known melanoma GWAS loci identified at least one functional variant for 78% of these loci, adding support to the body of knowledge that transcriptional regulation is a main mechanism that GWAS variants exert their function. As expected, the loci that are mainly explained by coding variants of pigmentation genes (e.g., 5p13.2, 11q14.3) did not present strong functional variants based on MPRAs. Our integrative variant scoring system indicated that in 42% of the cases melanoma GWAS loci presented a single prominent variant based on the overlap of variant transcriptional activity and multiple functional annotation features. On the other hand, a larger proportion of the loci (58%) exhibited more than one equally plausible functional variants, suggesting that multiple functional variants could potentially contribute to one or more target genes in each locus. This observation is somewhat consistent with the recent study that identified multiple causal regulatory variants that are in high-LD for a subset of lymphoblastoid cell eQTLs with MPRAs.46
We provided further support to a few melanoma susceptibility genes that have not been studied before by validating the connections between the top-score variants and their target eGenes by using CRISPRi system. For the locus at 22q13.1, MAFF was identified as a target of the top-score variant, rs4384. Higher levels of MAFF are correlated with the melanoma-risk-associated allele in melanocytes, matching the allelic activity of rs4384 in MPRAs. MAFF encodes a bZIP TF that lacks a transactivation domain that forms heterodimers with several regulators of antioxidant responses (e.g., NRF247 and BACH148), regulating genes in stress response and detoxification pathways.49 MAFF has been shown to act as an oncogene that plays a vital role in tumor invasion and metastasis.48 The variant rs4384 is also a melanoma-specific variant predicted to bind HES1 in melanoma context. Although the interaction of rs4384 and HES1 on MAFF expression could not be tested because MAFF was not a significant eGene in the TCGA melanoma dataset, HES1-mediated MAFF regulation in melanoma can be investigated as a potential mechanism of melanoma susceptibility in this locus. For the locus at 12p13.1, GPRC5A was validated as a target of the region harboring rs2111398, the top-score variant of the locus, with CRISPRi to target this region and assessing multiple eQTL target. GPRC5A is an orphan G protein-coupled receptor that has an important role in growth and survival of cancer cells50 and sustaining cell adhesion.51 The melanoma-risk-associated allele is correlated with lower expression of GPRC5A in melanocytes, which is consistent with the allelic activity of rs2111398 in MPRAs. We did not observe significant effect of CRISPRi on MED13L levels in our system. Given that MED13L plays an essential role in general transcription regulation as well as embryonic development,52 it is possible that multiple layers of redundant regulatory mechanism53 hindered the detection of relatively small effects of a single enhancer. It is also possible that there are other target gene(s) that were not detected in our QTL datasets.
Our study highlighted the cell-type-specific functionality of cancer-associated variants in the contexts of tumor and cell of tumor origin. We identified a subset of MPRA-significant variants as melanoma- (13%) or melanocyte-specific (16%) variants, while most of the variants are functional in both. Notably, these cell-type-/context-specific variants were distributed evenly across melanoma GWAS loci, suggesting that both tumor and cell-of-origin contexts may play a role across melanoma loci. For example, two top melanoma-specific variants (rs452384 and rs31487) in the locus at 5p15.33 were identified on the basis of their strong allelic transcription and enhancer activity restricted to melanoma cell line. Notably, these variants are also significant meQTLs for CpG probes within TERT in the TCGA melanoma dataset but not in the melanocyte dataset. Given that TERT expression is re-activated in most cancers including melanoma,54 two of three MPRA-significant variants at this locus being melanoma-specific is consistent with their contributing to target gene expression in tumor context rather than normal melanocyte context. Moreover, rs452384 in the locus at 5p15.33 and another top melanoma-specific variant, rs4384 in the locus at 22q13.1, are both predicted to modulate the binding of a NOTCH1 target, HES1, which displayed elevated expression in the UACC903 melanoma cell line compared to C283T melanocytes and has previously been shown to promote tumorigenesis.55 This observation and identification of two other melanoma-specific variants (rs6914598 at 6p22.3 and rs57688464 at 16q22.1) potentially recruiting another NOTCH1 target, HEY2, suggested that tumor-specific activation of TFs could mediate the activity of melanoma-specific variants across multiple loci. Consistent with this observation, our previous TWAS analysis demonstrated that increased NOTCH2 levels (located in 1p12) in melanocytes are associated with melanoma risk.1 NOTCH signaling is involved in maintaining melanocyte stem cells and melanoblasts,56 and Notch1 was shown to reprogram mature melanocytes into stem-like cells.57 NOTCH1 was also shown to be elevated in melanomas and promote growth and survival of melanoma cells.58 Interaction analysis of the functional variants and their TF partners further validated a melanoma-specific variant (REST-rs10305673-HORMAD1) and two melanocyte-specific variants (FLI1-rs850936-GPRC5A and THRB-rs4783674-CDH1) identified through MPRAs in the large-scale expression datasets. These data further supported the roles of TFs in mediating cell-type-specific variant function contributing to melanoma susceptibility. Future studies exploring the effects of these TFs on target gene expression in relevant cell types using CRISPR knockout/knockin of TF motifs or direct modulation of TF levels will be informative. Although we identified more melanocyte-specific functional variants than melanoma-specific ones through MPRAs, we observed larger allelic effect sizes and stronger enhancer activities of MPRA-significant variants in the UACC903 melanoma cell line in general. This could be due to increased global transcription levels in cancer cells by oncogene-induced activation and amplification of general transcription that have been observed before.42,43
We acknowledge several limitations of the current study. First, MPRA-significant variants were not identified for 22% (12/54) of the melanoma GWAS loci. While these loci might have alternative mechanisms that could not be tested by MPRAs, incorporating additional cell types (e.g., immune cells) and relevant exposures or contexts (e.g., exposure to UV radiation59) as well as adopting a lentiviral system to reflect genomic context in MPRA approaches could potentially identify additional functional variants. Second, 38% (16/42) of the loci with MPRA-significant variants are not supported by any genome-wide significant QTLs in melanoma or melanocyte datasets. This could be attributed to limited statistical power for lower-frequency variants and heterogeneity in melanoma tumor samples further limiting the eQTL detection17 as well as cellular contexts of eQTL detection that were not incorporated in these datasets. The power issue in the tumor eQTL dataset as well as potential differences between episomal enhancer activity tested in MPRA and endogenous expression measured in QTL datasets also limited our variant-TF interaction analyses as many melanoma-specific variants (e.g., rs4384 and rs2111398) are showing stronger allelic activities in UACC903 cell line but are linked to melanocyte eQTLs. To complement eQTL-based approaches, adopting chromatin interaction methods (e.g., capture-Hi-C60) will be beneficial for better sensitivity in variant-gene linkage. For example, the activity-by-contact (ABC) model utilizes epigenomic features and Hi-C data to predict the enhancer-gene connections.61 An initial query of the ABC model based on skin fibroblasts data (foreskin_fibroblast-Roadmap, ABC scores no less than 0.015) nominated candidate genes for nine variants among 285 MPRA-significant variants, which includes two variants that are not linked to any gene based on eQTL/meQTL (Table S19).
In conclusion, we provide a strategy to profile multiple cancer GWAS loci by using high-throughput variant screening and prioritization while incorporating the contexts of tumor and cell of tumor origin, which could be applied to other cancer GWAS follow-up studies.
Acknowledgments
This work has been supported by the Intramural Research Program (IRP) of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, US National Institutes of Health. The Leeds Melanoma Cohort was funded by the Cancer Research UK (under project grant C8216/A6129 and program award C588/A19167) and by the National Institutes of Health (NIH) (R01 CA83115) and EU FP6 Network of Excellence award to GenoMEL. This work utilized the Biowulf cluster computing system at the NIH. The results appearing here are in part based on data generated by the TCGA Research Network. We would like to thank members at the National Cancer Institute Cancer Genomics Research Laboratory (CGR) for help with sequencing efforts. We also thank all the cohorts, funders, and investigators who contributed to the melanoma GWAS, as originally acknowledged by Landi, 23andMe, and colleagues1; data from this GWAS were used toward MPRA variant selection and fine-mapping. We would like to thank the research participants and employees of 23andMe for making this work possible. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Declaration of interests
The authors declare no competing interests.
Published: November 23, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.11.006.
Web resources
FINEMAP, http://www.christianbenner.com/
POLYFUN, https://github.com/omerwe/polyfun
motifbreakR, https://github.com/Simon-Coetzee/motifBreakR
NIH Biowulf Cluster, http://hpc.nih.gov
Rsubread, https://bioconductor.org/packages/release/bioc/html/Rsubread.html
The Cancer Genome Atlas (TCGA) Research Network, http://cancergenome.nih.gov/
Supplemental information
Data and code availability
The sequencing data generated during this study (MPRA sequencing and RNA-seq data) are accessible through Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession GEO: GSE210356. A complete list of MPRA oligo sequences can be found in Table S3. The raw Illumina HumanMethylation450 BeadChips data are accessible through GEO under the accession GEO: GSE166069; melanocyte genotype data, RNA-seq expression data, and all eQTL/meQTL association results are accessible through Genotypes and Phenotypes (dbGaP) under accession dbGaP: phs001500.v2.p1. Data from the 2020 melanoma GWAS meta-analysis performed by Landi and colleagues were obtained from dbGaP (dbGaP: phs001868.v1.p1), with the exclusion of self-reported data from 23andMe and UK Biobank. The full GWAS summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data. Summary data from the remaining self-reported cases are available from the corresponding authors of that manuscript1 (Matthew Law, matthew.law@qimrberghofer.edu.au; Mark Iles, m.m.iles@leeds.ac.uk; and Maria Teresa Landi, landim@mail.nih.gov).
References
- 1.Landi M.T., Bishop D.T., MacGregor S., Machiela M.J., Stratigos A.J., Ghiorzo P., Brossard M., Calista D., Choi J., Fargnoli M.C., et al. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat. Genet. 2020;52:494–504. doi: 10.1038/s41588-020-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shain A.H., Bastian B.C. From melanocytes to melanomas. Nat. Rev. Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 3.Karimkhani C., Green A.C., Nijsten T., Weinstock M.A., Dellavalle R.P., Naghavi M., Fitzmaurice C. The global burden of melanoma: results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017;177:134–140. doi: 10.1111/bjd.15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaumont K.A., Shekar S.N., Shekar S.L., Newton R.A., James M.R., Stow J.L., Duffy D.L., Sturm R.A. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum. Mol. Genet. 2007;16:2249–2260. doi: 10.1093/hmg/ddm177. [DOI] [PubMed] [Google Scholar]
- 5.Visser M., Kayser M., Palstra R.-J. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsetskhladze Z.R., Canfield V.A., Ang K.C., Wentzel S.M., Reid K.P., Berg A.S., Johnson S.L., Kawakami K., Cheng K.C. Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS One. 2012;7:e47398. doi: 10.1371/journal.pone.0047398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halaban R., Cheng E., Zhang Y., Moellmann G., Hanlon D., Michalak M., Setaluri V., Hebert D.N. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc. Natl. Acad. Sci. USA. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J., Xu M., Makowski M.M., Zhang T., Law M.H., Kovacs M.A., Granzhan A., Kim W.J., Parikh H., Gartside M., et al. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat. Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. [DOI] [PubMed] [Google Scholar]
- 9.Choi J., Zhang T., Vu A., Ablain J., Makowski M.M., Colli L.M., Xu M., Hennessey R.C., Yin J., Rothschild H., et al. Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma. Nat. Commun. 2020;11:2718. doi: 10.1038/s41467-020-16590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang J., Jia J., Makowski M., Xu M., Wang Z., Zhang T., Hoskins J.W., Choi J., Han Y., Zhang M., et al. Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148. Nat. Commun. 2018;9:16159. doi: 10.1038/ncomms16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu M., Mehl L., Zhang T., Thakur R., Sowards H., Myers T., Jessop L., Chesi A., Johnson M.E., Wells A.D., et al. A UVB-responsive common variant at chromosome band 7p21.1 confers tanning response and melanoma risk via regulation of the aryl hydrocarbon receptor. Am. J. Hum. Genet. 2021;108:1611–1630. doi: 10.1016/j.ajhg.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 13.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher M.D., Chen-Plotkin A.S. The post-GWAS era: From association to function. Am. J. Hum. Genet. 2018;102:717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law M.H., Bishop D.T., Lee J.E., Brossard M., Martin N.G., Moses E.K., Song F., Barrett J.H., Kumar R., Easton D.F., et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Ye Y., Gong J., Han L. Expression quantitative trait loci (eQTL) analysis in cancer. Methods Mol. Biol. 2020;2082:189–199. doi: 10.1007/978-1-0716-0026-9_13. 2082. [DOI] [PubMed] [Google Scholar]
- 17.Zhang T., Choi J., Kovacs M.A., Shi J., Xu M., NISC Comparative Sequencing Program, Melanoma Meta-Analysis Consortium. Goldstein A.M., Trower A.J., Bishop D.T., et al. Cell-type-specific eQTL of primary melanocytes facilitates identification of melanoma susceptibility genes. Genome Res. 2018;28:1621–1635. doi: 10.1101/gr.233304.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang T., Choi J., Dilshat R., Einarsdóttir B.Ó., Kovacs M.A., Xu M., Malasky M., Chowdhury S., Jones K., Bishop D.T., et al. Cell-type-specific meQTLs extend melanoma GWAS annotation beyond eQTLs and inform melanocyte gene-regulatory mechanisms. Am. J. Hum. Genet. 2021;108:1631–1646. doi: 10.1016/j.ajhg.2021.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim-Hellmuth S., Aguet F., Oliva M., Muñoz-Aguirre M., Kasela S., Wucher V., Castel S.E., Hamel A.R., Viñuela A., Roberts A.L., et al. Cell type-specific genetic regulation of gene expression across human tissues. Science. 2020;369:eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Fukunaga-Kalabis M., Li L., Herlyn M. Developmental pathways activated in melanocytes and melanoma. Arch. Biochem. Biophys. 2014;563:13–21. doi: 10.1016/j.abb.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mountjoy E., Schmidt E.M., Carmona M., Schwartzentruber J., Peat G., Miranda A., Fumis L., Hayhurst J., Buniello A., Karim M.A., et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet. 2021;53:1527–1533. doi: 10.1038/s41588-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boer C.G., Hatzikotoulas K., Southam L., Stefánsdóttir L., Zhang Y., Coutinho de Almeida R., Wu T.T., Zheng J., Hartley A., Teder-Laving M., et al. Deciphering osteoarthritis genetics across 826, 690 individuals from 9 populations. Cell. 2021;184:6003–6005. doi: 10.1016/j.cell.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Arensbergen J., Pagie L., FitzPatrick V.D., de Haas M., Baltissen M.P., Comoglio F., van der Weide R.H., Teunissen H., Võsa U., Franke L., et al. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet. 2019;51:1160–1169. doi: 10.1038/s41588-019-0455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulirsch J.C., Nandakumar S.K., Wang L., Giani F.C., Zhang X., Rogov P., Melnikov A., McDonel P., Do R., Mikkelsen T.S., Sankaran V.G. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G., Jr., Kinney J.B., et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 28.Lee D., Kapoor A., Lee C., Mudgett M., Beer M.A., Chakravarti A. Sequence-based correction of barcode bias in massively parallel reporter assays. Genome Res. 2021;31:1638–1645. doi: 10.1101/gr.268599.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D. LS-GKM: a new gkm-SVM for large-scale datasets. Bioinformatics. 2016;32:2196–2198. doi: 10.1093/bioinformatics/btw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verfaillie A., Imrichova H., Atak Z.K., Dewaele M., Rambow F., Hulselmans G., Christiaens V., Svetlichnyy D., Luciani F., Van den Mooter L., et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 2015;6:6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benner C., Spencer C.C.A., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissbrod O., Hormozdiari F., Benner C., Cui R., Ulirsch J., Gazal S., Schoech A.P., van de Geijn B., Reshef Y., Márquez-Luna C., et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet. 2020;52:1355–1363. doi: 10.1038/s41588-020-00735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coetzee S.G., Coetzee G.A., Hazelett D.J. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–3849. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R., et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang A., Eller M.S., Hara M., Yaar M., Hirohashi S., Gilchrest B.A. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. J. Cell Sci. 1994;107:983–992. doi: 10.1242/jcs.107.4.983. ( Pt 4) ( Pt 4) [DOI] [PubMed] [Google Scholar]
- 40.Hsu M.Y., Wheelock M.J., Johnson K.R., Herlyn M. Shifts in cadherin profiles between human normal melanocytes and melanomas. J. Investig. Dermatol. Symp. Proc. 1996;1:188–194. [PubMed] [Google Scholar]
- 41.Onder T.T., Gupta P.B., Mani S.A., Yang J., Lander E.S., Weinberg R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 42.Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotsantis P., Silva L.M., Irmscher S., Jones R.M., Folkes L., Gromak N., Petermann E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016;7:13087. doi: 10.1038/ncomms13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aster J.C., Pear W.S., Blacklow S.C. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flynn E.D., Tsu A.L., Kasela S., Kim-Hellmuth S., Aguet F., Ardlie K.G., Bussemaker H.J., Mohammadi P., Lappalainen T. Transcription factor regulation of eQTL activity across individuals and tissues. PLoS Genet. 2022;18:e1009719. doi: 10.1371/journal.pgen.1009719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abell N.S., DeGorter M.K., Gloudemans M.J., Greenwald E., Smith K.S., He Z., Montgomery S.B. Multiple causal variants underlie genetic associations in humans. Science. 2022;375:1247–1254. doi: 10.1126/science.abj5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon E.J., Giaccia A. Dual roles of NRF2 in tumor prevention and progression: possible implications in cancer treatment. Free Radic. Biol. Med. 2015;79:292–299. doi: 10.1016/j.freeradbiomed.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon E.J., Mello S.S., Li C.G., Chi J.-T., Thakkar K., Kirkland J.G., Lagory E.L., Lee I.J., Diep A.N., Miao Y., et al. The HIF target MAFF promotes tumor invasion and metastasis through IL11 and STAT3 signaling. Nat. Commun. 2021;12:4308. doi: 10.1038/s41467-021-24631-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kannan M.B., Solovieva V., Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim. Biophys. Acta. 2012;1823:1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Hirano M., Zang L., Oka T., Ito Y., Shimada Y., Nishimura Y., Tanaka T. Novel reciprocal regulation of cAMP signaling and apoptosis by orphan G-protein-coupled receptor GPRC5A gene expression. Biochem. Biophys. Res. Commun. 2006;351:185–191. doi: 10.1016/j.bbrc.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 51.Bulanova D.R., Akimov Y.A., Rokka A., Laajala T.D., Aittokallio T., Kouvonen P., Pellinen T., Kuznetsov S.G. Orphan G protein-coupled receptor GPRC5A modulates integrin β1-mediated epithelial cell adhesion. Cell Adh. Migr. 2017;11:434–446. doi: 10.1080/19336918.2016.1245264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Utami K.H., Winata C.L., Hillmer A.M., Aksoy I., Long H.T., Liany H., Chew E.G.Y., Mathavan S., Tay S.K.H., Korzh V., et al. Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum. Mutat. 2014;35:1311–1320. doi: 10.1002/humu.22636. [DOI] [PubMed] [Google Scholar]
- 53.Payne J.L., Wagner A. Mechanisms of mutational robustness in transcriptional regulation. Front. Genet. 2015;6:322. doi: 10.3389/fgene.2015.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akincilar S.C., Unal B., Tergaonkar V. Reactivation of telomerase in cancer. Cell. Mol. Life Sci. 2016;73:1659–1670. doi: 10.1007/s00018-016-2146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao F., Zhang Y., Wang S., Liu Y., Zheng L., Yang J., Huang W., Ye Y., Luo W., Xiao D. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci. Rep. 2014;4:3963. doi: 10.1038/srep03963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moriyama M., Osawa M., Mak S.-S., Ohtsuka T., Yamamoto N., Han H., Delmas V., Kageyama R., Beermann F., Larue L., Nishikawa S.I. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J. Cell Biol. 2006;173:333–339. doi: 10.1083/jcb.200509084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zabierowski S.E., Baubet V., Himes B., Li L., Fukunaga-Kalabis M., Patel S., McDaid R., Guerra M., Gimotty P., Dahmane N., et al. Direct reprogramming of melanocytes to neural crest stem-like cells by one defined factor. Stem Cell. 2011;29:1752–1762. doi: 10.1002/stem.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bedogni B. Notch signaling in melanoma: interacting pathways and stromal influences that enhance Notch targeting. Pigment Cell Melanoma Res. 2014;27:162–168. doi: 10.1111/pcmr.12194. [DOI] [PubMed] [Google Scholar]
- 59.Watson M., Holman D.M., Maguire-Eisen M. Ultraviolet radiation exposure and its impact on skin cancer risk. Semin. Oncol. Nurs. 2016;32:241–254. doi: 10.1016/j.soncn.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baxter J.S., Leavy O.C., Dryden N.H., Maguire S., Johnson N., Fedele V., Simigdala N., Martin L.-A., Andrews S., Wingett S.W., et al. Capture Hi-C identifies putative target genes at 33 breast cancer risk loci. Nat. Commun. 2018;9:1028. doi: 10.1038/s41467-018-03411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fulco C.P., Nasser J., Jones T.R., Munson G., Bergman D.T., Subramanian V., Grossman S.R., Anyoha R., Doughty B.R., Patwardhan T.A., et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 2019;51:1664–1669. doi: 10.1038/s41588-019-0538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated during this study (MPRA sequencing and RNA-seq data) are accessible through Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession GEO: GSE210356. A complete list of MPRA oligo sequences can be found in Table S3. The raw Illumina HumanMethylation450 BeadChips data are accessible through GEO under the accession GEO: GSE166069; melanocyte genotype data, RNA-seq expression data, and all eQTL/meQTL association results are accessible through Genotypes and Phenotypes (dbGaP) under accession dbGaP: phs001500.v2.p1. Data from the 2020 melanoma GWAS meta-analysis performed by Landi and colleagues were obtained from dbGaP (dbGaP: phs001868.v1.p1), with the exclusion of self-reported data from 23andMe and UK Biobank. The full GWAS summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data. Summary data from the remaining self-reported cases are available from the corresponding authors of that manuscript1 (Matthew Law, matthew.law@qimrberghofer.edu.au; Mark Iles, m.m.iles@leeds.ac.uk; and Maria Teresa Landi, landim@mail.nih.gov).