Summary
As genomic and personalized medicine becomes mainstream, assessing and understanding the public’s genetic literacy is paramount. Because genetic research drives innovation and involves much of the public, it is equally important to assess its impact on genetic literacy. We designed a survey to assess genetic literacy in three ways (familiarity, knowledge, and skills) and distributed it to two distinct samples: 2,050 members of the general population and 2,023 individuals currently enrolled in a large-scale genetic research study. We compared these data to a similar survey implemented in 2013. The results indicate that familiarity with basic genetic terms in 2021 (M = 5.36 [range 1–7], p < 0.001) and knowledge of genetic concepts in 2021 (M = 9.06 [56.6% correct], p = 0.002) are significantly higher compared to 2013 (familiarity: M = 5.08 [range 1–7]; knowledge: M = 8.72 [54.5% correct]). Those currently enrolled in a genetic study were also significantly more familiar with genetic terms (M = 5.79 [range 1–7], p < 0.001) and more knowledgeable of genetic concepts (M = 10.57 [66.1% correct], p < 0.001), and they scored higher in skills (M = 3.57 [59.5% correct], p < 0.001) than the general population (M = 5.36 [range 1–7]; M = 9.06 [56.6% correct]; M = 2.65 [44.2% correct]). The results suggest that genetic literacy is improving over time, with room for improvement. We conclude that educational interventions are needed to ensure familiarity with and comprehension of basic genetic concepts and suggest further exploration of the impact of genetic research participation on genetic literacy to determine mechanisms for potential interventions.
Keywords: genetic literacy, genomics education, autism, genetic research, ASD, science communication, genetic testing
We compare data from a survey to assess genetic literacy from general population samples in 2013 and 2021 and from individuals enrolled in an autism genetic research study in 2021. The results suggest that familiarity with genetic concepts is higher in 2021 and in research participants, with room for improvement.
Introduction
Genomic research advancements in the last decade and the rapid reduction in genomic sequencing costs have exponentially increased the use of genetic information in everyday life. The cost of whole-genome sequencing has declined from just under $1 million in 2008 to as low as $99 in 2022 (with limited interpretation provided), expanding opportunities for clinical genetic research and direct-to-consumer genetic testing.1,2,3 Increasing numbers of individuals are pursuing ancestry testing, clinical genetic testing, or more personalized testing options on their own.4 A concurrent rise in research involving genome or exome sequencing also exposes individuals to their own genetic data and the implications of those data.5 This increase in the accessibility of genetic information, both within and beyond the healthcare setting, requires the public to interpret genetic testing results and apply them to their own health. To make educated decisions based on genetic information, individuals must have a basic understanding of genetic concepts and be able to effectively communicate with their providers about testing options and results. As the use of genetic information in healthcare increases, there is a need to measure and assess how individuals learn and communicate this information. Genetic literacy is one construct that can measure this ability.
Genetic literacy is defined as the sufficient knowledge and understanding of genetic principles for individuals to make decisions that sustain personal well-being and effective participation in social decisions on genetic issues.6 Distinct from genetic knowledge, genetic literacy evaluates both an individual’s understanding of basic genetic concepts and their ability to apply this understanding to health decisions. Genetic literacy can improve communication between patients and providers as well as help patients make informed decisions for their health.7,8,9 Higher genetic literacy can also improve attitudes toward genetic tests, such as the belief that genetic testing can provide important information to family members.10 Genetics education can also improve understanding of the limitations of genetic testing and its predictive ability, helping individuals make informed decisions about testing options and participation in genomic research.5 In addition, individuals with low genetic literacy benefit the least from advancements in genomic and personalized medicine and are less likely to participate in genetic research.11 Improving literacy in overlooked populations is crucial to limiting gaps in clinical opportunity and quality of care.7,12 Low genetic literacy acts as a potential threat to effective translation of genetic tests and is also associated with higher rates of heath complications, furthering health disparities.10,12
Despite the increase in genetic information in healthcare, medical training in genetics is often lacking, directly impacting genetic literacy. Low genetic literacy can be detrimental to quality of care and impair providers’ ability to effectively discuss genetic contributions to health with patients. A genetics knowledge survey distributed to first-year pathology residents indicated significantly reduced understanding of genetics concepts compared to non-genetics concepts. Further, only 53% of the sample reported interacting with a medical geneticist during medical school.13 In a sample of 10,303 physicians, only 29% reported they received education in pharmacogenetic testing;14 many medical students also self-report insufficient genetics knowledge.15 The consequences of low genetic literacy in providers include misdiagnosis, treatment failure, and unnecessary genetic testing and represent a significant barrier to the implementation of precision health medicine.16,17
Similarly, the general population (GP) in the U.S. is often not adequately trained in basic genetic science. A survey distributed to 5,404 participants with secondary education indicated that only 1.2% of the sample answered all 18 basic genetic knowledge questions correctly.11 Similar trends are seen in educators, with 25% of high school teachers reporting teaching contemporary issues in genetics.5 Another national survey conducted in 2017 indicated that only half of individuals are aware of genetic testing and approximately one-third are aware that genetic testing can contribute to disease treatment.18 Over 30% of a 2,093-person national sample indicated that they were unfamiliar with genetic concepts, whereas only 20% indicated they had personal experience with genetic health issues.19 Despite generally low genetic literacy in both patient and provider populations, interventions that involve and engage the public in genomic science can help assuage the disparity.9,20
Participating in genetic research is one way for individuals to gain exposure to genetic contributions to health, potentially despite insufficient education. Genome-wide association studies (GWASs) are a popular tool in genetic research, used to analyze associations between quantitative traits and known genetic variants.1 Funding within genomic research is also increasing. Since the start of the Human Genome Project, research funding provided by the National Human Genome Research Institute (NHGRI) has increased tenfold.21 Although the practice of involving participants in genetic research is growing, its impact on participants’ genetic literacy is not fully understood. In Japan, promotion of genetic research is associated with higher genetic literacy.22 In addition, the informed consent process can improve participants’ understanding of the limitations and benefits of genome sequencing.23 Despite increased interest and participation in genetic research, we do not fully understand genetic literacy rates in the GP compared to those currently enrolled in genetic research.
The tools used to measure genetic literacy vary, including pronunciation of health jargon, awareness of common genetic terms, and accuracy in factual knowledge.10,24,25 The Rapid Estimate of Adult Literacy in Genetics (REAL-G) assesses individuals’ ability to recognize and recite genetic terms within the clinical context.24 Similarly, the Genetic Literacy And Comprehension (GLAC) measure presents eight terms and asks participants to rate their level of familiarity.10 In contrast, the Genetic Literacy Assessment Instrument (GLAI) and the UNC Genomics Knowledge Scale (UNC-GKS) assess factual genetic knowledge.25,26 The Genomics Knowledge Scale (GKnowM) is a recently developed tool of 26 multiple-choice questions meant to assess knowledge of recent genomic developments and content needed to make informed health decisions.27 While all established measures assess an individual’s knowledge and awareness of genetic concepts, there remains a need for a standardized measure that can be used to measure improvements in genetic literacy.
In 2013, a comprehensive survey assessing genetic literacy and its relationship to breast cancer risk was administered to a nationally representative sample.6 The survey measured genetic literacy in three facets: familiarity with genetic terms (e.g., heredity, chromosome), accuracy in genetic knowledge (e.g., is a gene bigger than a chromosome?), and ability to synthesize information that applies genetics to human health (e.g., what is the purpose of genetic testing?). These facets are based on previously used measures and, in conjunction, evaluate an individual’s awareness and understanding of basic genetic concepts and their ability to apply this information to a clinical example. Because of genetic research advances in the last decade and the drastic increase in genetic testing, there is a need to update genetic literacy rates in the GP, with consideration of how genetic literacy may relate to common, complex conditions such as autism spectrum disorder (ASD).
Here, we take advantage of elements of the Abrams et al. (2015)6 survey to longitudinally assess genetic literacy as well as its relationship to a different common, complex genetic condition. We assess genetic literacy in the same three facets by replicating the familiarity and knowledge segments, while adapting the skills segment to address genetic susceptibility for ASD. We have two primary aims. The first is to update genetic literacy rates for the GP and compare results to those in Abrams et al. (2015).6 The second is to assess genetic literacy in a population currently enrolled in a genetic research study specifically focused on ASD, called the Simons Powering Autism Research or SPARK study.28 Our hypotheses are that genetic literacy is measurably higher compared to the first administration of the survey and that individuals participating in a genetic research project will have higher genetic literacy.
Material and methods
We implemented the Genetic Literacy Survey (GLS) to assess individuals’ genetic knowledge and ability to apply genetic information to human health decisions. It was developed from the survey used in Abrams et al. (2015),6 including the same three assessments of genetic literacy: familiarity, knowledge, and skills. The survey was completed entirely online. It was designed to be completed in approximately 25 min. The research protocol was deemed exempt by the NIH Office of Research Protections. This study was approved by the institutional review board at the National Human Genome Research Institute (protocol 000399).
Language choices
We note that terms used around autism are fluid and personal and, whenever possible, should depend on what those involved in the research prefer when describing themselves.29,30 For this study, we will use “autism,” “autism spectrum disorder,” or “ASD” when describing the condition and “autistic” when describing individuals.
Participants and survey development
2013 GP sample
A previous version of the GLS described in Abrams et al. (2015)6 was distributed to a sample (N = 1,016) recruited through a third-party contractor. See Abrams et al. (2015)6 for further details.
2021 GP sample
The GLS was distributed to a sample (N = 2,050) recruited from a respondent panel in the U.S. compiled by a third-party contractor. The respondent panel is a group of people who have indicated their interest in completing surveys and have been pre-qualified, both by indicating their willingness to participate and by providing their geographic, demographic, sociographic, and psychographic data. The final sample was designed to represent the GP in age, gender, and education level. The sample over-represents Black participants (27.5%) to replicate the sample distribution in Abrams et al. (2015).6 No participants were excluded, resulting in a final sample of 2,050. Participants in the GP sample completed the electronic survey between April 12, 2021, and April 27, 2021.
SPARK (genetic study) sample
The GLS was distributed to a sample (N = 2,264) identified through the SPARK study, a large-scale database of autistic individuals and families with autism, also in the U.S. Individuals in this database freely elect to share their medical, genetic, and demographic data with SPARK and voluntarily completed the GLS survey through the Research Match platform. Two hundred and forty-one participants were excluded from analysis because they did not complete the survey in its entirety, resulting in a final sample of 2,023. Sixteen participants completed the survey in its entirety yet did not provide demographic data. Because their data contribute to overall genetic literacy scores, they were not excluded. They are marked as “missing” in Table 3 and are included in aggregate scores. Again, the sample over-represents Black participants (20.5%) to replicate previous sample distributions. Participants in the genetic study sample completed the electronic survey between September 20, 2021, and November 21, 2021.
Table 3.
Pearson’s correlations of the three genetic literacy measures in each sample
| Familiarity | Skills | Knowledge | |
|---|---|---|---|
| 2013 general population | |||
| Familiarity | – | 0.343 (p < 0.001) | 0.451 (p < 0.001) |
| Skills | 0.405 (p < 0.001) | – | 0.447 (p < 0.001) |
| Knowledge | 0.511 (p < 0.001) | 0.507 (p < 0.001) | – |
| Education | 0.307 (p < 0.001) | 0.308 (p < 0.001) | 0.357 (p < 0.001) |
| 2021 general population | |||
| Familiarity | – | 0.268 (p < 0.001) | 0.407 (p < 0.001) |
| Skills | 0.299 (p < 0.001) | – | 0.416 (p < 0.001) |
| Knowledge | 0.438 (p < 0.001) | 0.443 (p < 0.001) | – |
| Education | 0.222 (p < 0.001) | 0.199 (p < 0.001) | 0.234 (p < 0.001) |
| SPARK | |||
| Familiarity | – | 0.27 (p < 0.001) | 0.46 (p < 0.001) |
| Skills | 0.29 (p < 0.001) | – | 0.26 (p < 0.001) |
| Knowledge | 0.48 (p < 0.001) | 0.29 (p < 0.001) | – |
| Education | 0.19 (p < 0.001) | 0.15 (p < 0.001) | 0.24 (p < 0.001) |
Simple Pearson’s correlations shown below the diagonal; partial correlations adjusting for education shown above the diagonal.
Survey measures
Term familiarity
Respondents were asked to rate their familiarity with eight terms—genetic, chromosome, susceptibility, mutation, variation, abnormality, heredity, and sporadic—on a scale of 1 (not at all familiar) to 7 (completely familiar). Scores were calculated as an average familiarity across the eight items, with an average Cronbach’s alpha of 0.923. A variation of this scale has been used in previous measures of genetic literacy. We replicated the GLAC measure, which presented the eight common genetic terms and asked respondents to rate their level of familiarity.10 The short version of the REAL-G also uses the same eight terms with established face and predictive validity.24 We added a ninth term, “genome,” to assess awareness surrounding recent advances in genomic medicine, including the Human Genome Project.31 Familiarity with this ninth term is not included in the final average familiarity score.
Practical skills
Respondents read a one-page information sheet applying genetic information to human health, specifically genetic contributions to autism. This sheet included a cup-and-ball model presented in Hoang et al. (2018)32 that depicts how the combination of environmental and genetic factors can result in an ASD diagnosis. Respondents were then asked six questions about what they read and could refer to the information sheet as they responded to questions. The questions were designed based on those used in Abrams et al. (2015)6 and included both multiple-choice and fill-in-the-blank questions, such as “What is the purpose of genetic testing for autism spectrum disorder?” and “About what percentage of individuals who receive genetic testing are found to have a variant associated with higher risk for autism spectrum disorder?” Correctly answered questions received a score of 1, while all other question answers (e.g., incorrect or unsure responses) received a score of 0. Scores were calculated as a sum of correct answers, creating a final score range of 0–6. Based on Kuder-Richardson 20, the assessment is sufficiently reliable with an alpha of 0.654 based on 2021 GP data.33
Factual knowledge
Respondents were presented with 16 technical genetic statements and asked to respond with “true,” “false,” or “don’t know” for each statement, seven of which were intentionally false. The statements concerned genes, their function and makeup, and potential risk for genetic disease. This measure replicated that used in Abrams et al. (2015).6,25,34,35 Participants received a score of 1 for correct responses and a score of 0 for incorrect or unsure responses, creating a final score range of 0–16. Based on Kuder-Richardson 20, the assessment demonstrates acceptable reliability with an alpha of 0.725.33 We included a 17th, intentionally false, statement, “Environmental factors, such as UV radiation, do not play a role in our genome.” This item was included to reflect the information in the skills module regarding environmental and genetic contributions to autism and was not included in calculating average knowledge scores.
Numeracy
Participants were asked to rate their ability to solve basic math problems—such as “How good are you at figuring out how much a shirt will cost if it is 25% off?”— on a scale from “1—not at all good” to “7—extremely good.” Scores were calculated as an average of all responses.5
ASD in family
Participants were asked if someone in their family (e.g., parent, sibling, or child) has been diagnosed with ASD, responding with “yes,” “no,” or “not sure.”
Experience with ASD
Participants were asked broadly if someone in their life (e.g., friend, coworker, or neighbor) has been diagnosed with ASD, responding with “yes,” “no,” or “not sure.”
ASD diagnosis
Participants were asked if they have received an ASD diagnosis and responded with “yes,” “no,” or “not sure.” This question was only included in the survey distributed to the SPARK sample, which included autistic adults as well as parents of autistic people.
Education
Participants rated their highest level of education completed by selecting one of eight options, ranging from “no schooling completed” to “doctorate degree.” These responses were used to create an ordinal variable dividing participants into four groups: “less than high school,” “high school,” “some college,” and “bachelor’s or graduate degree.”
Age
Participants were asked to select their age from five groups: “18–25,” “26–39,” “40–49,” “50–59,” and “60+.”
Income
Participants were asked to report their income from three options: “less than $49,999,” “$50,000–$99,999,” and “over $100,000.”
Statistical analysis
Analyses were completed using R statistical software. To address our first aim, we conducted analyses of variance (ANOVAs) comparing scores in the three facets of genetic literacy (familiarity, knowledge, and skills) between 2013 and 2021. Subsequent analyses controlled for education level given a two-sample Z-test identified differences in self-reported education across the two samples (p = 0.005). We used a propensity-score-adjusted comparison to control for education, in which we adjusted 2013 GP scores to match baseline variation in education in the 2021 GP. Adjusted mean scores are indicated by “2013 adj.” and can be directly compared to mean scores in the 2021 GP.
To address our second aim, we conducted ANOVAs between scores in the three facets of genetic literacy assessed in the 2021 sample and the SPARK study sample. Given that there was no difference in education level between the 2021 GP and SPARK samples, we did not adjust for education in these analyses.
Exploratory analyses were conducted to determine potential associations between genetic literacy and other demographic and conceptual variables. We conducted simple correlations to determine associations, followed by a fitted regression model to determine significance.
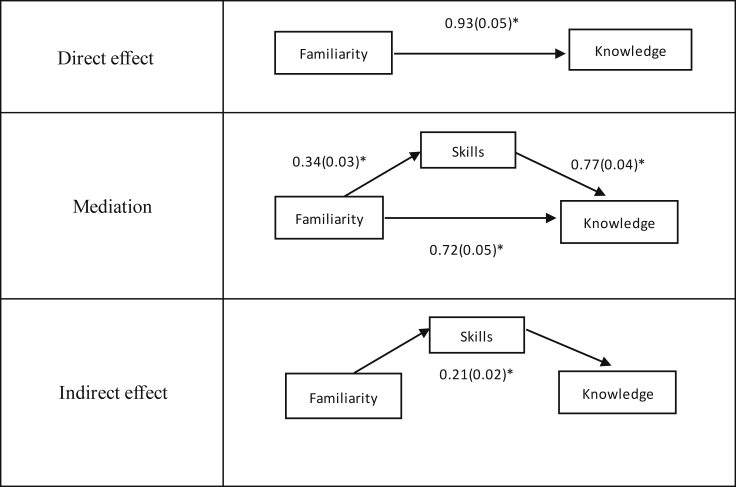
To assess the replacement of the skills module within the genetic literacy measure, we replicated the analyses conducted in Abrams et al. (2015).6 This includes bivariate and partial correlations between the three facets of genetic literacy, controlling for education. We also fitted a regression model to determine if the skills segment mediates the relationship between familiarity and knowledge. Additionally, we conducted a Sobel test to determine the indirect effect between familiarity and knowledge, mediated by skills.
Results
The characteristics of the 2021 GP sample (N = 2,050) and SPARK sample (N = 2,023) as compared to the 2013 GP sample are described in Table 1. The 2021 sample self-reports a higher average education level compared to the 2013 sample; thus, we controlled for education level in subsequent analyses. The SPARK sample, compared to the GP, includes a higher proportion of individuals outside of the gender binary and a slightly younger sample on average, though this difference is not statistically significant.
Table 1.
Sample characteristics
| 2013 GP | 2021 GP | SPARK | |
|---|---|---|---|
| Sample size | N = 1,016 | N = 2,050 | N = 2,023 |
| % (N) | % (N) | % (N) | |
| Gender | |||
| Female | 52.3 (531) | 49.6 (1,017) | 49.09 (993) |
| Male | 47.7 (485) | 49.7 (1,018) | 46.66 (944) |
| Nonbinary | – | 0.56 (12) | – |
| Other | – | 0.15 (3) | 3.46 (70) |
| Missing | – | – | 0.79 (16) |
| Race and ethnicity | |||
| Black or African American | 26.8 (272) | 27.5 (563) | 20.46 (414) |
| Latino or Hispanic American | – | 2.49 (51) | – |
| East Asian or Asian American | – | 4.58 (94) | 5.68(115) |
| South Asian or Indian American or Native Hawaiian | – | 1.07 (22) | 0.94 (19) |
| Middle Eastern or Arab American | – | 0.25 (5) | – |
| White | 59.0 (599) | 57.0 (1,169) | 51.90 (1,050) |
| Other | 2.4 (24) | 2.10 (43) | 5.34(108) |
| Multiple selected | 3.7 (38) | 3.17 (65) | 14.88 (301) |
| Missing | – | – | 0.79 (16) |
| Age | |||
| 18–25 | 15.8 (161) | 15.76 (323) | 8.06 (163) |
| 26–39 | 23.7 (241) | 28.54 (585) | 43.3 (876) |
| 40–49 | 36.2 (368) | 15.76 (323) | 29.16 (590) |
| 50–59 | 24.2 (26) | 11.8 (242) | 13.2 (267) |
| 60+ | 15.8 (161) | 28.15 (577) | 5.49 (111) |
| Missing | – | – | 0.79 (16) |
| Education level | |||
| Less than high school | 9.0 (91) | 5.17 (106) | 4.4(89) |
| High school | 23.2 (236) | 34.0 (697) | 26.5(536) |
| Some college | 29.3 (298) | 17.1 (350) | 20.91 (423) |
| Bachelor’s or graduate degree | 30.2 (307) | 43.8 (897) | 47.4 (959) |
| Missing | – | – | 0.79 (16) |
| Income | |||
| Less than $49,999 | 41.9 (426) | 49.5 (1,015) | 45.8 (926) |
| $50,000–$99,999 | 31.9 (324) | 31.1 (638) | 28.2 (571) |
| Over $100,000 | 26.2 (266) | 19.4 (397) | 25.2 (510) |
| Missing | – | – | 0.79 (16) |
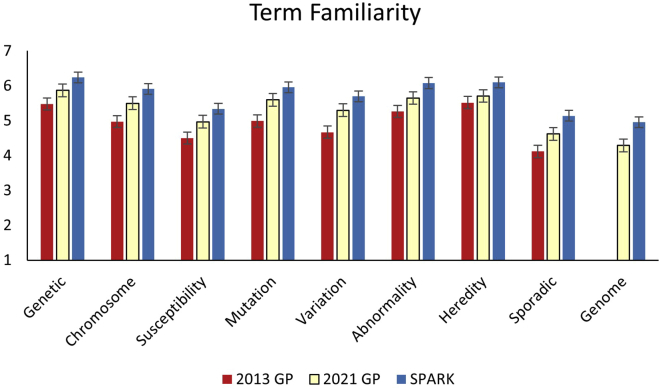
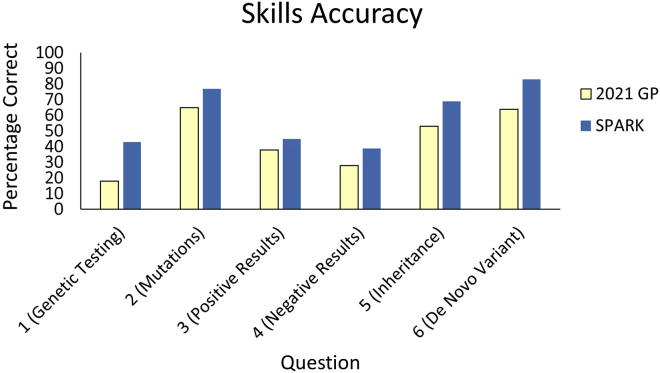
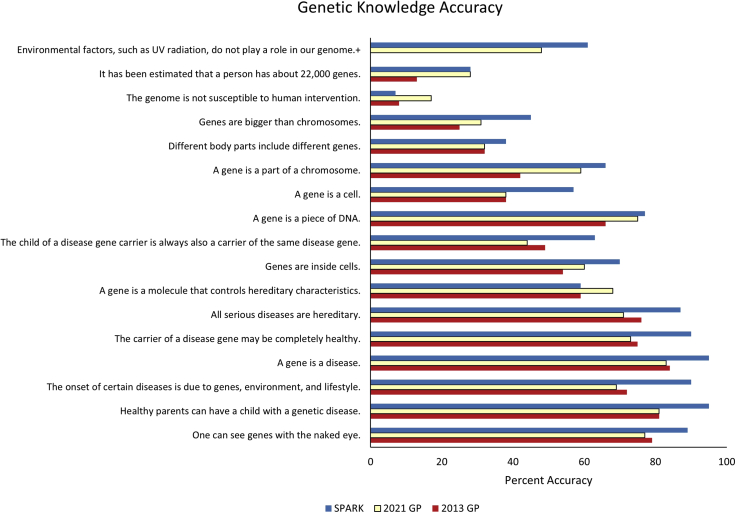
Descriptive statistics of the three facets of genetic literacy within the three samples are presented in Table 2. We present the aggregate results within the three facets of genetic literacy and define “high literacy” as greater than 70% correct, as established in Abrams et al. (2015).6 As such, “high familiarity” indicates an average of at least 5 on the familiarity scale of 1 to 7. In the skills and knowledge segments, “high literacy” is defined as at least 5 out of 6 correct responses and at least 12 out of 6 correct responses, respectively. Average familiarity with the eight common genetic terms, as well as the ninth term, “genome,” in each sample is presented in Figure 1. The 2021 sample on average reported moderate familiarity with the eight genetic terms. Participants were least familiar with the ninth term, “genome” (M = 4.29 [range 1–7], SD = 2.06) which was not included in the average familiarity score. The skills assessment cannot be directly compared to the 2013 data because different assessments and clinical examples were used. Participants in the 2021 GP sample correctly responded to approximately 3 of the 6 skills questions. Average scores for each of the 6 questions in the skills assessment are presented in Figure 2. The 2021 GP sample scored an average of 9 out of 16 on the genetic knowledge assessment compared to approximately 8 in 2013. Average scores for each question in the knowledge assessment, including the additional 17th question, are presented in Figure 3.
Table 2.
Descriptive statistics of the three genetic literacy measures in each sample
| Familiarity | Skills | Knowledge | |
|---|---|---|---|
| Range | 1–7 | 0–6 | 0–16 |
| 2013 General population | |||
| Mean (SD) | 4.98 (1.76) | 3.94 (1.88)a | 8.39 (3.81) |
| % (N) scoring > 70% | 57.6 (585) | 47.5 (483)a | 22.2 (226) |
| 2021 General population | |||
| Mean (SD) | 5.36 (1.36) | 2.65 (1.73) | 9.06 (3.23) |
| % (N) scoring > 70% | 66.9 (1,371) | 18.8 (385) | 23.7 (486) |
| SPARK | |||
| Mean (SD) | 5.79 (1.27) | 3.57 (1.56) | 10.57 (2.96) |
| % (N) scoring > 70% | 79.4 (1,607) | 32.9 (665) | 43.9 (889) |
Different skills module implemented.
Figure 1.
Participants’ self-reported familiarity with common genetic terms
The x axis denotes the scale of familiarity, from 1 (not at all familiar) to 7 (completely familiar). The y axis lists each of the nine terms tested. The error bars represent the standard deviation of responses for each term. Participants in the 2021 general population (GP) reported significantly higher familiarity with each of eight terms (excluding “genome,” only introduced in 2021) compared to the 2013 GP sample (p < 0.004). SPARK participants reported significantly higher familiarity with each of nine terms compared to the 2021 GP (p < 0.001).
Figure 2.
Percentage of each sample that correctly responded to each of six questions in the genetic literacy skills module
The x axis denotes the six items in the skills module with a keyword or phrase describing the content of the question. The y axis denotes the percentage that correctly responded. The full content of the skills module questions is in Table S4.
Figure 3.
Percentage of each sample that correctly identifies each knowledge statement as true or false
The x axis denotes the 17 total statements included in the knowledge module. The y axis indicates the percentage of each sample that correctly responded to the statement with true or false. Eight of the 17 statements are intentionally false. +Not included in 2013 GP assessment.
The SPARK sample also reported moderate familiarity, though higher than the 2021 GP sample. SPARK members correctly responded to 4 out of 6 skills questions, compared to 3 out of 6 in the 2021 GP (Table 2). Participants in the SPARK sample correctly answered 11 of the 16 questions in the knowledge assessment (Table 2).
GP updates in genetic literacy—2013 to 2021
Average familiarity with genetic terms is significantly higher in 2021 (M = 5.36 [range 1–7], SD = 1.36) compared to 2013 (M = 4.98 [range 1–7], SD = 1.76, p < 0.001, d = 0.27). After adjusting for variance due to education, the difference is still significant (2013 adj. M = 5.08 [range 1–7], SD = 1.77, p < 0.001). Participants in the 2021 GP sample also reported significantly higher familiarity with each individual term compared to 2013 (Figure 1).
Scores in the knowledge assessment were significantly higher in the 2021 sample (M = 9.06 [56.6% correct], SD = 3.23) compared to 2013 (M = 8.39 [52.4% correct], SD = 3.81, p < 0.001, d = 0.20). When adjusting for the variance due to education, the difference in average knowledge scores remains statistically significant (2013 adj. M = 8.72 [54.5% correct], SD = 3.76, p = 0.002). At the item level, the 2021 sample performed significantly better on 7 of the 16 items (p < 0.02, Figure 3). Some items showing significant improvement over the 8-year period include, for example, “It has been estimated that a person has about 22,000 genes” (true), “A gene is a part of a chromosome” (true), “A gene is a molecule that controls hereditary characteristics” (true), and “The genome is not susceptible to human intervention” (false). The 2021 sample performed significantly lower on 3 of the 16 items (p < 0.001, Figure 3). These statements are: “The onset of certain diseases is due to genes, environment, and lifestyle” (true), “All serious diseases are hereditary” (false), and “The child of a disease gene carrier is always a carrier of the same disease gene” (false). Forty-eight percent of the GP sample correctly identified the 17th statement, “Environmental factors, such as UV radiation, do not play a role in our genome,” which was not included in average scores as it was new in 2021, as false.
Differences in genetic literacy—GP versus research participants
Participants in the SPARK sample reported significantly higher familiarity with genetic terms overall (M = 5.79 [range 1–7], SD = 1.27) compared to the 2021 GP (M = 5.36 [range 1–7], SD = 1.36, p < 0.001, d = 0.22). The SPARK sample also reported significantly higher familiarity with each of the eight genetic terms, as well as the ninth term, “genome” (Figure 1). Knowledge scores were also significantly higher in the SPARK sample (M = 10.57 [66.1% correct], SD = 2.96) compared to the GP (M = 9.06 [56.6% correct], SD = 3.23, p < 0.001, d = 0.39). A significantly higher proportion of the SPARK sample correctly determined the verity of 12 out of the 16 technical genetics statements (p < 0.001), compared to the 2021 GP (Figure 3). Example items include: “The onset of certain diseases is due to genes, environment, and lifestyle,” “All serious diseases are hereditary,” and “The child of a disease gene carrier is always also a carrier of the same disease gene.”
The SPARK sample also scored significantly higher on the skills module (M = 3.57 [59.5% correct], SD = 1.56) compared to the GP (M = 2.65 [44.2% correct], SD = 1.73, p < 0.001, d = 0.38). Specifically, participants in the SPARK sample scored significantly higher in each of the six questions in the skills module (p < 0.001, Figure 2).
Associations between genetic literacy and other variables—2021 GP and SPARK
In the 2021 GP sample, participants’ self-reported numeracy significantly correlated with scores in familiarity (r = 0.34, p < 0.001), knowledge (r = 0.32, p < 0.001), and skills (r = 0.23, p < 0.001). In the SPARK sample, numeracy significantly correlated with familiarity (r = 0.33, p < 0.001) and knowledge (r = 0.38, p < 0.001) scores. In both samples, participants with a bachelor’s degree or higher scored significantly higher in all three facets compared to those who completed some or all of high school. Genetic literacy scores in both samples were higher in older participants and those with higher income.
Five hundred and ninety-three participants in the 2021 GP sample reported having an autistic person in their life, and these individuals scored higher in familiarity (M = 5.72 [range 1–7], p < 0.001), knowledge (M = 9.77 [61.1% correct], p < 0.001), and skills (M = 2.96 [49.3% correct], p < 0.001) than those without (M = 5.32 [range 1–7]; M = 8.97 [56.1% correct]; M = 2.59 [43.2% correct]).
In the SPARK sample, 838 individuals disclosed an autism diagnosis (this question was only asked in the SPARK sample) and scored marginally higher in all three facets of genetic literacy (M = 5.96 [range 1–7]; M = 10.88 [68% correct]; M = 3.79 [63.2% correct]) compared to non-autistic participants (M = 5.69 [range 1–7]; M = 10.43 [65.2% correct]; M = 3.46 [57.7% correct]). Those who reported knowing an autistic person in their life (N = 1,323) also reported higher familiarity (M = 5.88 [range 1–7]) compared to those without (M = 5.65 [range 1–7]).
Associations between genetic literacy measures and education
Results within each facet of genetic literacy in the 2021 sample positively correlated with each other and with education (Table 3). The same is shown for the SPARK sample. Partial correlations between the three facets of genetic literacy remained significant when adjusting for years of education.
We also fitted a regression model to test whether the skills segment mediates the relationship between familiarity and knowledge, while adjusting for education. Similar to the data in Abrams et al. (2015),6 the results were significant with a Sobel test36 indicating an indirect effect of familiarity on knowledge, mediated by skills (0.21, p < 0.001) (Figure 4).
Figure 4.
Mediation model, indicating effect size (standard deviation) and adjusting for education
∗p < 0.001.
Discussion
Changes in genetic literacy over time
The survey data confirmed our first hypothesis that genetic literacy in 2021 is generally higher than in 2013, reporting significantly higher knowledge of and familiarity with basic genetic concepts compared to Abrams et al. (2015).6 Considering the rapid advances in genomic research and utilization of genetic information in clinical and personal contexts, it follows that the GP’s genetic literacy will improve over time.21 In addition, depictions of genetics in the media, including celebrities undergoing genetic testing, the popularization of direct-to-consumer genetic testing and ancestry testing, and scientific fiction depicting genetic science only increase awareness of the impact of genetics on our lives.6 However, with the rise of genetic information, it is even more important to ensure that genetic education guides interpretation of genetic information in the media, to ensure misconceptions such as genetic determinism and genetic essentialism are not perpetuated.37 It is also necessary to equip the public with the skills and knowledge that will guide personal well-being and decision making. Though our data indicate higher genetic literacy in the current GP, it is important to explore significant changes or gaps in knowledge that have emerged in the past decade.
Participants in both the 2013 and 2021 GPs reported moderate familiarity with basic genetic terms, with the 2021 GP sample reporting significantly higher familiarity with each individual term and overall. These data support the notion that as the use of genetic testing and engagement with genomic research increases over time, so does general awareness of basic genetic terms. Like the 2013 sample, participants in 2021 were most familiar with general terms such as “genetic” and “heredity” and less familiar with risk terms such as “susceptibility” and “sporadic.” This suggests a need to expose the public to clinical applications of genetics and community-based genomics education programs in addition to basic technical terms.38,39 Participants in both 2021 samples were least familiar with the term “genome,” indicating a need for increased awareness of recent advances in genomic and personalized medicine. It is important to note, however, that familiarity with a term does not equate to comprehension, and as such it is important to also explore individuals’ factual knowledge. Other large surveys have similarly suggested that the term “genome” is less familiar to a general population and that participants’ “perceived knowledge” or familiarity may be less than their actual knowledge in this area.40
On average, participants in the 2021 GP sample scored higher in the knowledge module, meaning they more accurately identified the 16 factual genetics statements as true or false. They were more accurate in identifying statements related to the definition and function of genes, such as “A gene is a molecule that controls hereditary characteristics” and “A gene is a piece of DNA,” indicating an improved understanding of basic biology. However, participants were also less accurate in identifying one statement as false, “The child of a disease gene carrier is always also a carrier of the same disease gene,” suggesting a need for skills in understanding inheritance patterns. This could be accomplished by applying knowledge of gene function to a clinical example, such as a family with genetic contributions to disease. We included an additional 17th knowledge statement, “Environmental factors, such as UV radiation, do not play a role in our genome,” in order to further assess knowledge of gene/environment interaction. Approximately half of the GP recognized this statement as false. A similar response was seen in a recent national survey assessing the statement “Genes can be altered by the environment,” in which 55.9% of the sample correctly identified it as true.40
The knowledge module offered three response options for each statement: “true,” “false,” or “not sure.” Thus, a 33% correct score would be expected if responses were chosen at random. Here, we found that all three samples scored on average above 50%. While it is possible that responses were chosen at random, given that all three samples scored on average above 50% correct, we conclude that such biases are unlikely.
Somewhat surprisingly, the 2021 GP sample responded correctly to fewer than half of the skills questions, on average. This may be because of the high volume of text used in the infographic compared to that used in Abrams et al. (2015).6 The SPARK sample correctly answered over half, more consistent with the 2013 GP sample. Though both skills modules implemented six fill-in-the-blank or multiple-choice questions, the 2021 survey (for both GP and SPARK samples) included one “select all that apply,” in which the correct response was to select more than one answer choice. We posit that this question added an element of difficulty not included in the Abrams et al. (2015)6 skills module and contributed to the reduction in skills scores. Given the different clinical context (i.e., hereditary breast and ovarian cancer vs. autism) and skills assessment across the two surveys (2013 vs. 2021), we cannot conclude from these data that participants’ performances on this genetic literacy skills assessment have significantly changed between 2013 and 2021.
Genetic literacy in genetic research participants
The results confirmed our second hypothesis that genetic literacy is higher in those enrolled in a large-scale genetic study. Compared to the GP sample recruited in 2021, those enrolled in SPARK reported significantly higher knowledge of and familiarity with basic genetic concepts, as well as improved skills to synthesize information to make health decisions.
The SPARK sample reported moderate familiarity with basic genetic terms, though significantly higher than the 2013 and 2021 GPs for each term and overall. The SPARK sample also scored significantly higher in the knowledge segment on average. Moreover, the SPARK sample was significantly more accurate in determining the verity of more nuanced statements, such as “The onset of certain diseases is due to genes, environment, and lifestyle” (true) and “The child of a disease gene carrier is always also a carrier of the same disease gene” (false). This suggests that the SPARK sample is not only more aware of basic genetic concepts, but also has a better understanding of the interaction between genes and environments and the applications to health. Though the etiology of autism is not fully understood, current data show that it is caused by a complex interaction between genetics and environment, with previous studies indicating 70%–90% heritability.41,42 Most notably, the SPARK sample performed significantly higher in the skills segment, both overall and within each question.
We posit that there are several reasons for the higher observed genetic literacy scores in the SPARK sample: first, participation in the SPARK study involves submission of medical information and genetic samples, along with access to multiple media resources on genetics generally and ASD specifically (examples at https://sparkforautism.org/discover/). Participants also receive regular newsletters with study updates and additional learning resources. Second, this sample’s experience with ASD, a highly heritable condition, may lead to improved recognition and comprehension of basic genetic concepts. This may be through interactions with the healthcare system, self-directed research, participation in advocacy groups, or all of the above. Similarly, a sample of 257 individuals with inflammatory bowel disease (IBD) in the US were asked to complete the same familiarity measure used here and as a group scored a 5.9 in familiarity out of 7 (completely familiar).10
Finally, beyond personal experience with a genetic condition, research experience alone is known to improve scientific literacy and, more specifically, genetic literacy. In the introductory biology classroom, data-based learning, or identifying biological concepts from previously implemented studies, promotes active learning and retention.43 Early introduction to research can also build students’ excitement for and success in science.44 Our data suggest that active participation in a research study, combined with personal experience with a genetic condition, can result in higher genetic literacy. Further research is needed to determine if this trend can be demonstrated in other genetic research participants.
Indeed, we chose to measure genetic literacy around autism in part because of the controversial nature of genetic research in the field.45 Members of the autism community have stated that they prioritize applied research (e.g., for increased daily supports) over basic research such as genetics,46 and some autistic people question whether the use of genetic research in ASD can be justified given past language dehumanizing autistic people and justifying eugenic applications.20 We want to clearly state that any eugenic applications should be abhorrent to biomedical researchers and clinicians. We suggest that understanding of the facts and limitations of genetic research, along with true participatory engagement, is crucial to ensure any partnership with autistic individuals and their families.
Conclusion
Our results suggest that genetic literacy in the GP, specifically familiarity with genetic terms and demonstrable genetic knowledge, has improved in the past decade and that involvement in genetic research can improve one’s genetic literacy. It is important to note, however, that although average changes in genetic literacy measures show significant differences, our effect sizes exhibit a small to moderate effect. This is a limitation to our study and may indicate that although scores in genetic familiarity and knowledge are improving, it is not enough to demonstrate remarkable changes in health outcomes. We acknowledge that there is still much room for improvement in the population’s genetic literacy, particularly as genomic and personalized medicine becomes more mainstream. We recommend that educational interventions to improve genetic literacy are implemented in several domains: the general public, including school-aged children; patients affected by genetic or inherited conditions; and healthcare providers, highlighting both basic facts and the ability to apply information in multiple settings. In addition, educational interventions should focus on the ethical, legal, and social issues of contemporary genetics to combat misconceptions that can perpetuate harmful beliefs. The NHGRI has repeatedly stated that improving genomic literacy is a significant priority, including calling for active engagement between the public and genomic researchers.21,47 By equipping the public with the knowledge and understanding needed for personal health decisions, we are enabling advancements in genomic medicine to create positive change.
Acknowledgments
This work is funded by a grant from the SPARK Research Match Diversity, Equity, and Inclusion initiative (RM0149) and the Intramural Research Program of the National Human Genome Research Institute (HG200410-01), National Institutes of Health. This study was approved by the institutional review board at the National Human Genome Research Institute (protocol 000399). We would like to thank Daisy Yu of The Emmes Company, LLC for statistical assistance and Madeline Piper for insight into autistic perspectives.
Declaration of interests
The authors declare no competing interests.
Published: November 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.11.005.
Supplemental information
Data and code availability
The published article includes the datasets collected as part of this study as supplemental files.
References
- 1.Knerr S., Ramos E., Nowinski J., Dixon K., Bonham V.L. Human difference in the genomic era: Facilitating a socially responsible dialogue. BMC Med. Genom. 2010;3:20. doi: 10.1186/1755-8794-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Incerti D., Xu X.M., Chou J.W., Gonzaludo N., Belmont J.W., Schroeder B.E. Cost-effectiveness of genome sequencing for diagnosing patients with undiagnosed rare genetic diseases. Genet. Med. 2022;24:109–118. doi: 10.1016/j.gim.2021.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Nebula.org Nebula Genomics - 30x Whole Genome Sequencing - DNA Testing. 2022. https://nebula.org/whole-genome-sequencing-dna-test/
- 4.Roberts M.C., Allen C.G., Andersen B.L. The FDA authorization of direct-to-consumer genetic testing for three BRCA1/2 pathogenic variants: A twitter analysis of the public's response. JAMIA Open. 2019;2:411–415. doi: 10.1093/jamiaopen/ooz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabatello M., Chen Y., Sanderson S.C., Chung W.K., Appelbaum P.S. Increasing genomic literacy among adolescents. Genet. Med. 2019;21:994–1000. doi: 10.1038/s41436-018-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrams L.R., McBride C.M., Hooker G.W., Cappella J.N., Koehly L.M. The many facets of genetic literacy: Assessing the scalability of multiple measures for broad use in survey research. PLoS One. 2015;10:e0141532. doi: 10.1371/journal.pone.0141532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaphingst K.A., Blanchard M., Milam L., Pokharel M., Elrick A., Goodman M.S. Relationships between health Literacy and genomics-related knowledge, self-efficacy, perceived importance, and communication in a medically underserved population. J. Health Commun. 2016;21:58–68. doi: 10.1080/10810730.2016.1144661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milo Rasouly H., Cuneo N., Marasa M., DeMaria N., Chatterjee D., Thompson J.J., Fasel D.A., Wynn J., Chung W.K., Appelbaum P., et al. GeneLiFT: A novel test to facilitate rapid screening of genetic literacy in a diverse population undergoing genetic testing. J. Genet. Counsel. 2021;30:742–754. doi: 10.1002/jgc4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meiser B., Woodward P., Gleeson M., Kentwell M., Fan H.M., Antill Y., Butow P.N., Boyle F., Best M., Taylor N., et al. Pilot study of an online training program to increase genetic literacy and communication skills in oncology healthcare professionals discussing BRCA1/2 genetic testing with breast and ovarian cancer patients. Fam. Cancer. 2022;21:157–166. doi: 10.1007/s10689-021-00261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooker G.W., Peay H., Erby L., Bayless T., Biesecker B.B., Roter D.L. Genetic literacy and patient perceptions of IBD testing utility and disease control: A randomized vignette study of genetic testing. Inflamm. Bowel Dis. 2014;20:901–908. doi: 10.1097/MIB.0000000000000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman R., Likhanov M., Selita F., Zakharov I., Smith-Woolley E., Kovas Y. New literacy challenge for the twenty-first century: Genetic knowledge is poor even among well educated. J. Community Genet. 2019;10:73–84. doi: 10.1007/s12687-018-0363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura S., Narimatsu H., Katayama K., Sho R., Yoshioka T., Fukao A., Kayama T. Effect of genomics-related literacy on non-communicable diseases. J. Hum. Genet. 2017;62:839–846. doi: 10.1038/jhg.2017.50. [DOI] [PubMed] [Google Scholar]
- 13.Haspel R.L., Genzen J.R., Wagner J., Fong K., Undergraduate Training in Genomics UTRIG Working Group. Wilcox R.L. Call for improvement in medical school training in genetics: Results of a national survey. Genet. Med. 2021;23:1151–1157. doi: 10.1038/s41436-021-01100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kampourakis K. In: Molecular Diagnostics. Third Edition. Patrinos G.P., editor. Academic Press; 2017. Chapter 27 - Public Understanding of Genetic Testing and Obstacles to Genetics Literacy; pp. 469–477. [DOI] [Google Scholar]
- 15.Alotaibi A.A., Cordero M.A.W. Assessing medical students' knowledge of genetics: Basis for improving genetics curriculum for future clinical practice. Adv. Med. Educ. Pract. 2021;12:1521–1530. doi: 10.2147/amep.S337756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swandayani Y.M., Cayami F.K., Winarni T.I., Utari A. Familiarity and genetic literacy among medical students in Indonesia. BMC Med. Educ. 2021;21:524. doi: 10.1186/s12909-021-02946-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaibley V.M., Ramos I.N., Woosley R.L., Curry S., Hays S., Ramos K.S. Limited genomics training among physicians remains a barrier to genomics-Based implementation of precision medicine. Front. Med. 2022;9:757212. doi: 10.3389/fmed.2022.757212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krakow M., Ratcliff C.L., Hesse B.W., Greenberg-Worisek A.J. Assessing genetic literacy awareness and knowledge gaps in the US population: Results from the health information national trends survey. Public Health Genomics. 2017;20:343–348. doi: 10.1159/000489117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton A., Milne R., Almarri M.A., Anwer S., Atutornu J., Baranova E.E., Bevan P., Cerezo M., Cong Y., Critchley C., et al. Global public perceptions of genomic data sharing: What shapes the willingness to donate DNA and health data? Am. J. Hum. Genet. 2020;107:743–752. doi: 10.1016/j.ajhg.2020.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botha M., Dibb B., Frost D.M. "Autism is me": an investigation of how autistic individuals make sense of autism and stigma. Disabil. Soc. 2022;37:427–453. doi: 10.1080/09687599.2020.1822782. [DOI] [Google Scholar]
- 21.Green E.D., Gunter C., Biesecker L.G., Di Francesco V., Easter C.L., Feingold E.A., Felsenfeld A.L., Kaufman D.J., Ostrander E.A., Pavan W.J., et al. Strategic vision for improving human health at The Forefront of Genomics. Nature. 2020;586:683–692. doi: 10.1038/s41586-020-2817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishiyama I., Nagai A., Muto K., Tamakoshi A., Kokado M., Mimura K., Tanzawa T., Yamagata Z. Relationship between public attitudes toward genomic studies related to medicine and their level of genomic literacy in Japan. Am. J. Med. Genet. 2008;146a:1696–1706. doi: 10.1002/ajmg.a.32322. [DOI] [PubMed] [Google Scholar]
- 23.Kaphingst K.A., Facio F.M., Cheng M.R., Brooks S., Eidem H., Linn A., Biesecker B.B., Biesecker L.G. Effects of informed consent for individual genome sequencing on relevant knowledge. Clin. Genet. 2012;82:408–415. doi: 10.1111/j.1399-0004.2012.01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erby L.H., Roter D., Larson S., Cho J. The rapid estimate of adult literacy in genetics (REAL-G): a means to assess literacy deficits in the context of genetics. Am. J. Med. Genet. 2008;146a:174–181. doi: 10.1002/ajmg.a.32068. [DOI] [PubMed] [Google Scholar]
- 25.Bowling B.V., Acra E.E., Wang L., Myers M.F., Dean G.E., Markle G.C., Moskalik C.L., Huether C.A. Development and evaluation of a genetics literacy assessment instrument for undergraduates. Genetics. 2008;178:15–22. doi: 10.1534/genetics.107.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langer M.M., Roche M.I., Brewer N.T., Berg J.S., Khan C.M., Leos C., Moore E., Brown M., Rini C. Development and validation of a genomic knowledge scale to advance informed decision making research in genomic sequencing. MDM Policy Pract. 2017;2 doi: 10.1177/2381468317692582. 238146831769258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linderman M.D., Suckiel S.A., Thompson N., Weiss D.J., Roberts J.S., Green R.C. Development and validation of a comprehensive genomics knowledge scale. Public Health Genomics. 2021;24:291–303. doi: 10.1159/000515006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feliciano P., Daniels A.M., Snyder L.G., Beaumont A., Camba A., Esler A., Gulsrud A.G., Mason A., Gutierrez A., Nicholson A., Paolicelli A.M. SPARK: a US cohort of 50, 000 families to accelerate autism research. Neuron. 2018;97:488–493. doi: 10.1016/j.neuron.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny L., Hattersley C., Molins B., Buckley C., Povey C., Pellicano E. Which terms should be used to describe autism? Perspectives from the UK autism community. Autism. 2016;20:442–462. doi: 10.1177/1362361315588200. [DOI] [PubMed] [Google Scholar]
- 30.Bottema-Beutel K., Kapp S.K., Lester J.N., Sasson N.J., Hand B.N. Avoiding ableist language: Suggestions for autism researchers. Autism in Adulthood. 2021;3:18–29. doi: 10.1089/aut.2020.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gates A.J., Gysi D.M., Kellis M., Barabási A.L. A wealth of discovery built on the Human Genome Project - by the numbers. Nature. 2021;590:212–215. doi: 10.1038/d41586-021-00314-6. [DOI] [PubMed] [Google Scholar]
- 32.Hoang N., Cytrynbaum C., Scherer S.W. Communicating complex genomic information: A counselling approach derived from research experience with Autism Spectrum Disorder. Patient Educ. Counsel. 2018;101:352–361. doi: 10.1016/j.pec.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 33.Kuder G.F., Richardson M.W. The theory of estimation of test reliability. Psychometrika. 1937;2:151–160. doi: 10.1007/BF02288391. [DOI] [Google Scholar]
- 34.Jallinoja P., Aro A.R. Knowledge about genes and heredity among Finns. New Genet. Soc. 1999;18:101–110. doi: 10.1080/14636779908656892. [DOI] [Google Scholar]
- 35.Haga S.B., Barry W.T., Mills R., Ginsburg G.S., Svetkey L., Sullivan J., Willard H.F. Public knowledge of and attitudes toward genetics and genetic testing. Genet. Test. Mol. Biomarkers. 2013;17:327–335. doi: 10.1089/gtmb.2012.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobel M.E. Asymptotic confidence intervals for indirect effects in structural equation models. Socio. Methodol. 1982;13:290–321. doi: 10.2307/270723. [DOI] [Google Scholar]
- 37.Donovan B.M., Weindling M., Salazar B., Duncan A., Stuhlsatz M., Keck P. Genomics literacy matters: Supporting the development of genomics literacy through genetics education could reduce the prevalence of genetic essentialism. J. Res. Sci. Teach. 2020;58:520–550. doi: 10.1002/tea.21670. [DOI] [Google Scholar]
- 38.Koehly L.M., Morris B.A., Skapinsky K., Goergen A., Ludden A. Evaluation of the families SHARE workbook: An educational tool outlining disease risk and healthy guidelines to reduce risk of heart disease, diabetes, breast cancer and colorectal cancer. BMC Publ. Health. 2015;15:1120. doi: 10.1186/s12889-015-2483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Haye K., Whitted C., Koehly L.M. Formative evaluation of the families SHARE disease risk tool among low-income African Americans. Public Health Genomics. 2021;24:280–290. doi: 10.1159/000517309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaeger J., Hellwig A., Schiavoni E., Brace-Macdonald B., Lamb N.A., Tumiel-Berhalter L., Halfon M.S., Vishwanath A., Surtees J.A. Challenges in improving genomic literacy: Results from national and regional surveys of genomic knowledge, attitudes, concerns, and behaviors. bioRxiv. 2022 doi: 10.1101/2022.08.26.505444. [DOI] [Google Scholar]
- 41.Johannessen J., Nærland T., Bloss C., Rietschel M., Strohmaier J., Gjevik E., Heiberg A., Djurovic S., Andreassen O.A. Parents’ attitudes toward genetic research in autism spectrum disorder. Psychiatr. Genet. 2016;26(2):74–80. doi: 10.1097/YPG.0000000000000121. [DOI] [PubMed] [Google Scholar]
- 42.Genovese A., Butler M.G. Clinical assessment, genetics, and treatment approaches in autism spectrum disorder (ASD) Int. J. Mol. Sci. 2020;21:47266–E4818. doi: 10.3390/ijms21134726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finby B., Heyer L.J., Malcolm Campbell A. Data-rich textbook figures promote core competencies: Comparison of two textbooks. Biochem. Mol. Biol. Educ. 2021;49:392–406. doi: 10.1002/bmb.21488. Epub 2021 Jan 9. PMID: 33421340; PMCID: PMC8248048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jordan T.C., Burnett S.H., Carson S., Caruso S.M., Clase K., DeJong R.J., Dennehy J.J., Denver D.R., Dunbar D., Elgin S.C.R., et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5 doi: 10.1128/mBio.01051-13. e01051–e01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanderson K. High-profile autism genetics project paused amid backlash. Nature. 2021;598:17–18. doi: 10.1038/d41586-021-02602-7. [DOI] [PubMed] [Google Scholar]
- 46.Roche L., Adams D., Clark M. Research priorities of the autism community: A systematic review of key stakeholder perspectives. Autism. 2021 Feb;25:336–348. doi: 10.1177/1362361320967790. PMID: 33143455. [DOI] [PubMed] [Google Scholar]
- 47.Hurle B., Citrin T., Jenkins J.F., Kaphingst K.A., Lamb N., Roseman J.E., Bonham V.L. What does it mean to be genomically literate?: National Human Genome Research Institute Meeting Report. Genet. Med. 2013;15:658–663. doi: 10.1038/gim.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes the datasets collected as part of this study as supplemental files.