Abstract
Protein interiors contain void space that can bind small gas molecules. Determination of gas pathways and kinetics in proteins has been an intriguing and challenging task. Here, we combined computational methods and the hyperpolarized xenon-129 chemical exchange saturation transfer (hyper-CEST) NMR technique to investigate xenon (Xe) exchange kinetics in maltose-binding protein (MBP). A salt bridge ∼9 Å from the Xe-binding site formed upon maltose binding and slowed the Xe exchange rate, leading to a hyper-CEST 129Xe signal from maltose-bound MBP. Xe dissociation occurred faster than dissociation of the salt bridge, as shown by 13C NMR spectroscopy and variable-B1 hyper-CEST experiments. “Xe flooding” molecular dynamics simulations identified a surface hydrophobic site, V23, that has good Xe binding affinity. Mutations at this site confirmed its role as a secondary exchange pathway in modulating Xe diffusion. This shows the possibility for site-specifically controlling xenon protein-solvent exchange. Analysis of the available MBP structures suggests a biological role of MBP’s large hydrophobic cavity to accommodate structural changes associated with ligand binding and protein-protein interactions.
Significance
It is possible to program the 129Xe NMR signal in maltose-binding protein by engineering two sites—a salt bridge (normally stabilized by maltose binding) and a surface hydrophobic site, V23—which are each separated by 1 nm from a central xenon-binding cavity. The ability to control the linear flow of xenon in a protein has applications for magnetic resonance sensors employing hyperpolarized Xe-129 and is relevant to understanding naturally occurring gas-binding proteins.
Introduction
Proteins are porous materials with internal cavities of various size and duration. Calculation of high-resolution protein crystal structures found that the packing fraction of protein cores is only φ ≈ 0.56, significantly lower than the ideal packing fraction of solid spheres (φ ≈ 0.74 for face-centered cubic crystals) (1). This reveals that the interiors of most natural proteins contain considerable empty space between amino acids. These “packing defects” play an important role in controlling protein dynamics and functions, especially in the case of gas-binding proteins. Internal hydrophobic cavities and pores define pathways for small gas molecules and allow efficient diffusion to the protein active site. For example, X-ray crystallography and photodissociation experiments confirmed that myoglobin possesses multiple preexisting internal cavities that are involved in oxygen diffusion and reactivity (2). Computational methods also provide insights into gas pathways that are difficult to probe by experiments (3). Hydrogenases are another class of proteins where kinetics of gas diffusion is of great interest owing to their bioenergetic metabolism involving H2 conversion (4). One challenge in studying gas diffusion in hydrogenase is that H2 is invisible in X-ray crystallography and difficult to model accurately by computational methods. Heavy noble gases such as xenon and krypton are therefore frequently used as a surrogate for small gaseous molecules such as H2 (5,6) and CO (7,8) to probe protein interiors and gas dynamics given their comparable molecular size (4.3 Å diameter for xenon [Xe], 4.0 Å for krypton [Kr], and 3.4–3.8 Å for H2) (9,10). Analysis on the basis of krypton-pressurized crystal structures and molecular dynamics (MD) simulations predicted possible tunnels for both H2 and the inhibitor O2 (5,11,12,13). However, gas diffusion rates were still unknown without input from experimental approaches, such as protein film voltammetry and isotope exchange assays (14). Moreover, gas delivery in proteins is important in various biological processes that are fundamental to all life, including carbon and nitrogen fixation and noble gas narcosis (15,16,17,18,19,20). Investigation of gas migration pathways largely relied on computational methods, but kinetic information and the mechanisms of gas exchange remain poorly studied. Knowing the kinetics of gas exchange processes in proteins is useful for deciphering catalytic or inhibitory mechanisms and advancing biotechnology applications.
Interestingly, large internal cavities and channels are found not only in gas-reactive proteins and gas vesicle nanostructures (21,22) but also in many other monomeric proteins whose functions are completely unrelated to gas permeability and interactions, such as periplasmic binding proteins, T4 lysozyme, and TEM-1 β-lactamase (23,24,25,26,27,28,29). These proteins can host Xe using a large hydrophobic cavity, thus allowing the spin-1/2 nucleus, 129Xe, to serve as a biocompatible probe for the study of protein-protein interactions (30) and small-molecule sensing (23,24). For example, maltose-binding protein (MBP) in its wild-type form has a cavity volume of 75–95 Å3 (26), which is surprising because an intramolecular cavity of this large size is rarely found in naturally occurring proteins (31) and void space is usually associated with protein instability despite the high stability of MBP. The use of 129Xe NMR spectroscopy enabled detection of the protein cavity and conformational changes in MBP (32). In combination with X-ray crystallography and heteronuclear single-quantum coherence spectroscopy, an Xe-binding cavity was identified in the N domain, and Xe binding affinity was estimated (Ka ∼ 20 ± 10 M−1) (26). Using hyperpolarized 129Xe chemical exchange saturation transfer (hyper-CEST) (33), our laboratory observed a distinctive chemical shift for 129Xe bound to maltose-saturated MBP, shifted ∼90 ppm downfield relative to free aqueous 129Xe (23). However, it remains unexplained how the maltose-bound state, instead of the maltose-free state, produces the unique 129Xe signal. This prompted us to investigate the structural basis for Xe-MBP interactions, which could guide future design of novel Xe-based biosensors.
In this paper, we employ MD simulations to find Xe exchange pathways in MBP and the hyper-CEST technique to characterize Xe dissociation rate. Maltose exchange kinetics and mutations made to a surface hydrophobic site provide insights into additional Xe exchange mechanisms. Cavity volume analysis reveals the possible biological role of the large hydrophobic pocket.
Materials and methods
MD simulations
MD simulations were performed with NAMD 2.14 software (34) using the Bridges-2 Regular Memory system at the Pittsburgh Supercomputing Center (35,36). Initial structures were obtained by adding a Xe atom to MBP crystal structures (PDB: 1LLS for maltose-free form (26) and PDB: 1ANF for maltose-bound form (37)). Each protein was solvated in a TIP3P water box (38), and 150 mM NaCl was used to neutralize total charge. Each system was minimized with 1,000 steps, after which the system was equilibrated at a temperature of 300 K and a pressure of 1 atm using the Langevin thermostat and barostat methods. The CHARMM36 force field was used to perform all MD simulations (39). The Xe atom was treated with only nonbonded interactions (40). Periodic boundary conditions were employed, and the SHAKE algorithm was applied to constrain the lengths of all bonds that involve a hydrogen (41). At least a 43 ns MD trajectory was obtained from each simulation. Salt-bridge distances and root-mean-square deviation data were calculated using VMD 1.9.3. (42).
Cavity volume and tunnel analysis
Cavity volume was calculated using the Fpocket algorithm (43), which detects protein cavity based on Voronoi tessellation. Input structures were obtained from PDB. Tunnel analysis was carried out on MD trajectories of MBP using Caver 3.0 software (44).
Site-directed mutagenesis
K15D, D14K/K15D, V23A, and V23L mutations were introduced to the pET His6 MBP TEV LIC cloning vector, a gift from Scott Gradia acquired via Addgene (#29656), using the forward and reverse primers listed in Table S1. The mutated plasmids were amplified in NEB 5α competent E. coli cells (New England Biolabs, Ipswich, MA, USA) and purified using a miniprep kit (Qiagen, Germantown, MD, USA). All mutated sequences were verified at the University of Pennsylvania DNA Sequencing Facility.
Protein expression and purification
Preparation of MBP mutants uses the same procedures as wild-type MBP, which has been described before (23). Briefly, the plasmid encoding the MBP sequence was transformed into E. coli BL21(DE3)-competent cells (New England Biolabs), which were then grown in 1 L lysogeny broth media supplemented with 50 μg/mL kanamycin. Protein expression was induced by addition of 1 mM isopropyl-β-D-thiogalactoside, followed by overnight incubation at 25°C. Cell lysis was carried out using hen egg white lysozyme (Alfa Aesar, Ward Hill, MA, USA) in 20 mM sodium phosphate (pH 7.4) in the presence of benzonase nuclease (Sigma-Aldrich, Burlington, MA, USA). The lysate was subjected to fast protein liquid chromatography. Pure protein was eluted from HisTrap HP Ni column (Cytiva, Marlborough, MA, USA) and HiLoad 16/600 Superdex 75 pg column (GE Life Sciences, Chicago, IL, USA). Protein concentrations were determined from the absorbance at 280 nm using the extinction coefficient 67,840 M−1 cm−1, as calculated by PROTPRAM (45).
129Xe hyper-CEST NMR
HP 129Xe was generated using the spin-exchange optical pumping method (46) with a home-built 129Xe polarizer based on the IGI.Xe.2000 commercial model by GE. A Shark 65 W tunable ultranarrow band diode laser (OptiGrate, Oviedo, FL, USA) set to 794.770 nm was used for optical pumping of Rb vapor. A gas mixture of 88% helium, 10% nitrogen, and 2% natural abundance Xe (Linde Group, Bridgewater, NJ, USA) was used as the hyperpolarizer input. 129Xe hyperpolarization level was roughly 10%–15%. For each data point in the hyper-CEST z-spectra, hyperpolarized (hp) 129Xe was bubbled into the NMR tube through capillaries for 20 s, followed by a 3 s delay to allow the bubbles to collapse. A d-SNOB saturation pulse was used. The pulse length was τpulse = 3.80 ms, number of pulses was npulse = 600, and saturation time was Τsat = 2.28 s. NMR experiments were performed using a Bruker BioDRX 500 MHz NMR spectrometer and 10 mm PABBO probe at 300 K. A 90° hard pulse of this probe has a pulse length of 22 μs. Protein samples were 80 μM in PBS (pH 7.4), with 0.1% (v/v) Pluronic L81 (Aldrich) added to mitigate foaming. All results were averaged over at least three trials.
129Xe exchange rate determination by variable-B1 measurements
Estimation of koff,Xe is based on an approximation to the full hyper-CEST solution (47). As described previously (48), the Xe dissociation rate koff,Xe can be obtained by measuring the CEST effect with various saturation strengths, B1. Exponential Lorentzian fits were applied to model the CEST peaks of protein-bound and free 129Xe. A simplified expression for hyper-CEST is
where and koff represents the 129Xe dissociation rate. Rearrangement of the above equations gives
Therefore, the linear plot of ln−1(1-CEST) versus (γB1)−2 gives koff,Xe = (slope/y-intercept)1/2. Protein samples were prepared at 80 μM concentration in PBS (pH 7.4) with 0.1% (v/v) Pluronic L81. Each z-spectrum was averaged over at least three trials.
13C NMR spectroscopy
0.6 mM protein was mixed with 2 mM D-maltose in PBS (pH 7.4) with 10% D2O. Data collection was performed on a Bruker NEO600 NMR spectrometer. Sample temperature was 300 K. Maltose dissociation rate, koff, maltose, was estimated based on the line shape of 13C signal of the bound maltose. The apparent transverse relaxation of the bound signal results from both intrinsic relaxation and chemical exchange. As a rough estimation, intrinsic relaxation can be neglected given the broad width of the bound signal. Therefore, koff, maltose ≈ 1/T2∗ = π × w1/2.(49).
Fluorescence spectroscopy
For fluorescence quenching of MBP mutants, 1 μM protein was prepared in PBS buffer for measurement. A stock of 100 mM maltose in PBS buffer was added to the protein solution to a final concentration of 1 mM. The excitation wavelength was 280 nm, and fluorescence emission was recorded over a range of 300 to 420 nm. All measurements were obtained at 298 K using a Varian Cary Eclipse fluorescence spectrophotometer.
Results
Salt bridge K15-E111 controls Xe exchange
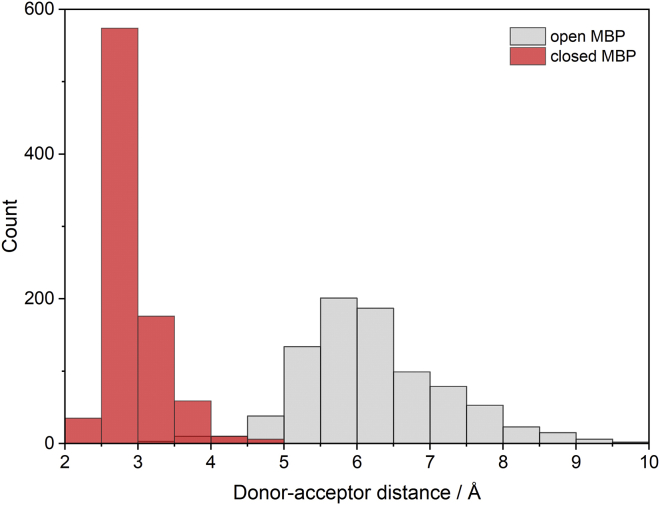
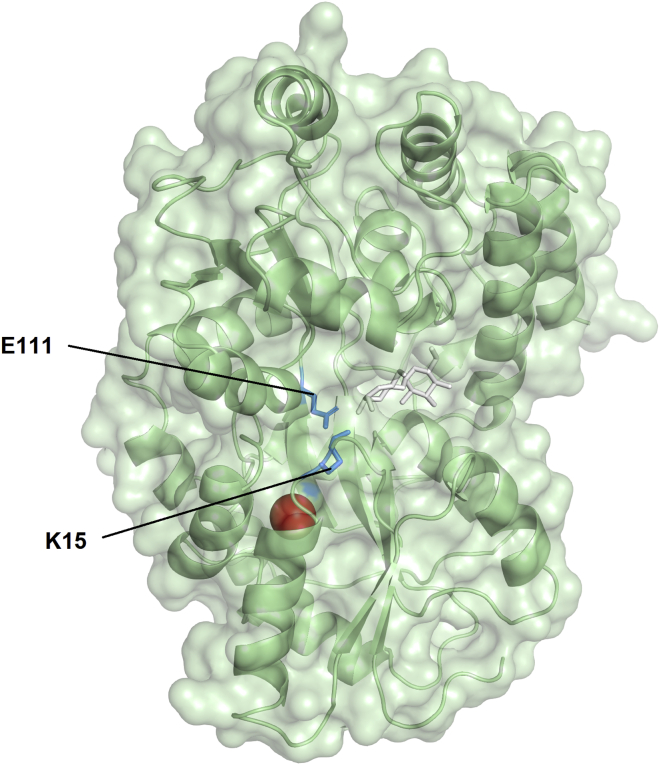
The different 129Xe hyper-CEST response in the presence and absence of maltose indicates a difference in either the Xe binding environment or exchange kinetics. In order to characterize the differences in dynamics, we performed MD simulations on MBP in both the maltose-bound “closed” conformation and maltose-free “open” conformation, initiated with a single Xe atom in the well-defined Xe-binding cavity (23). In the closed conformation, the salt bridge K15-E111 forms an H-bond network with hydroxyl groups of maltose, whereas the same side chains show no interactions in the open conformation (Fig. 1). Interestingly, the Xe-binding pocket resides underneath this salt bridge at a distance of ∼9 Å (Fig. 2), which leads us to postulate that the K15-E111 salt bridge forms a barrier between the bound Xe atom and solvent. Superposition of the cavity-containing N domains of both MBP conformations shows almost identical structure, with an average root-mean-square deviation of only 0.33 Å, indicative of no change in the Xe binding environment in different conformations of MBP. In addition to protein dynamics, we also analyzed Xe trajectories. Notably, in the closed state, Xe is constrained by the protein matrix during the 43 ns simulation with a root-mean-square fluctuation of 1.6 Å, only ∼38% of the van der Waals diameter of Xe. In contrast, during the same simulation length of the open state, Xe can escape from the initial hydrophobic cavity through the crevice between K15 and E111 and finally become completely solvated. Caver tunnel analysis estimates the average width of this crevice to be at least 2.2 ± 0.3 Å in the open state and 1.8 ± 0.4 Å in the closed state during the entire simulation length (50). Despite this narrow bottleneck, Xe can pass the tunnel probably due to transient rearrangement of its surrounding amino acids. Further analysis revealed the most frequently involved bottleneck residues include I11, K15, and L262.
Figure 1.
Donor-to-acceptor distance between K15 and E111 in maltose-free open MBP (gray) and maltose-bound closed MBP (red). To see this figure in color, go online.
Figure 2.
Cartoon representation of closed MBP with a bound Xe atom (red sphere). Maltose is shown as white sticks, and side chains of K15 and E111 are shown as blue sticks. To see this figure in color, go online.
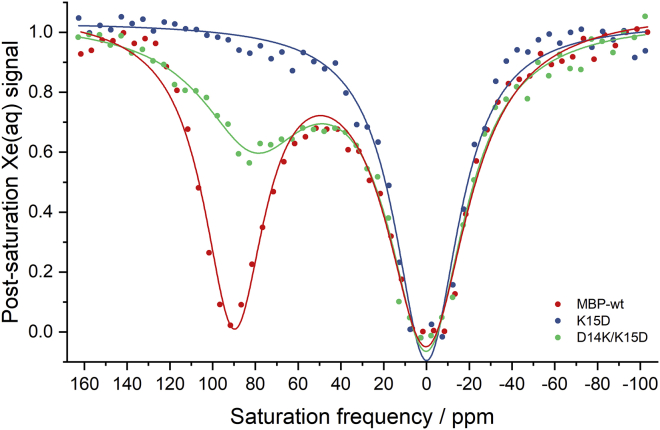
We set out to test experimentally our hypothesis about the effect of the K15-E111 salt bridge on Xe motion. To this end, we attempted to destroy the salt-bridge interaction with a K15D mutation and then restore it with a second mutation, D14K. Salt-bridge analysis using MD trajectories of both mutants showed distinct results. Even though both mutants can bind maltose as observed in MD simulations and fluorescence quenching experiments (Fig. S1), the salt bridge exists only in the D14K/K15D double mutant, with an occupancy of 87% (Fig. S2). As a result, K15D protein exhibits no bound 129Xe NMR signal, similar to the open state, whereas D14K/K15D is able to restore the bound 129Xe NMR signal at +80 ppm (Fig. 3). Our results confirmed the important role of this proximal salt bridge in controlling Xe exchange.
Figure 3.
Hyper-CEST z-spectra of wild-type MBP (red), K15D (blue), and D14K/K15D (green) in presence of 1 mM maltose. 80 μM protein was dissolved in PBS (pH 7.4) at 298 K. Wild-type MBP gives a hyper-CEST signal at +90 ppm, referenced to the free 129Xe in PBS. The K15D mutant completely lost its 129Xe@protein signal due to faster Xe exchange, while D14K/K15D was able to restore the hyper-CEST signal at +80 ppm. To see this figure in color, go online.
Alternative Xe exchange pathways
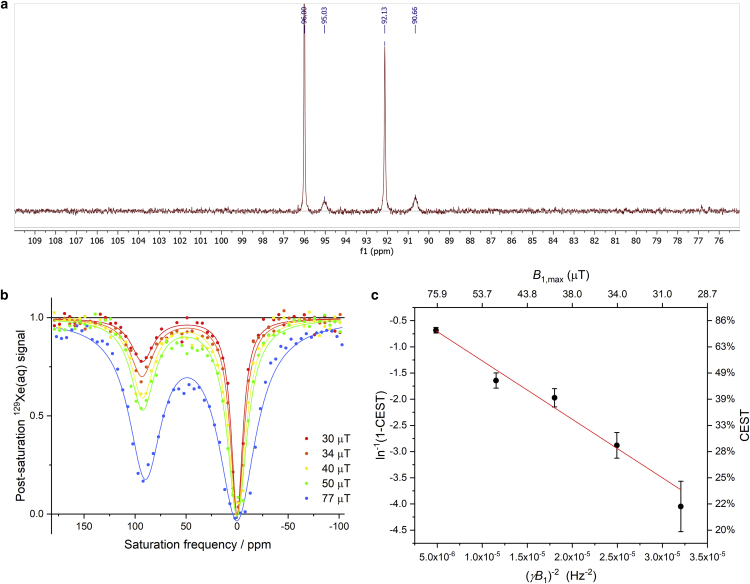
It remains unresolved whether the Xe egress pathway in the closed MBP structure is the same salt-bridge site because it is possible that other pathways that are minor in the open MBP structure could become significant upon closure of the K15-E111 salt bridge. If there are no alternative pathways, the rate of Xe exchange should be comparable to or slower than the dynamics of salt-bridge formation, which in turn depends on maltose dissociation rate. Therefore, we characterized the maltose dissociation rate using [1-13C]-labeled maltose and the Xe dissociation rate via variable-B1 hyper-CEST method (47,48). From the line fitting and peak-width analysis (koff ∼ π × w1/2) of the 13C NMR signal of bound maltose, we can estimate the maltose off rate, koff, maltose, to be 1.2 × 102 s−1 for β-maltose and 1.3 × 102 s−1 for α-maltose (Fig. 4). This value agrees with the result of the stopped-flow fluorescence measurement (51) and is larger than other reported values by one order of magnitude (52,53,54). This is probably because 13C NMR signal reflects the H-bonding of the hydroxyl group directly attached to 1-13C and intrinsic fluorescence is affected by the local environment of nearby tryptophan, whereas other techniques measured the rate of overall protein conformational change. In this aspect, our result better represents the local dynamics of the salt bridge as the hydroxyl group of 1-13C forms a direct H-bond with the K15 side chain. Next, we estimated the 129Xe dissociation rate via the variable-B1 hyper-CEST method (47,48). A series of z-spectra using varying saturation pulse strengths was acquired, and the bound 129Xe signal was analyzed to obtain a Xe dissociation rate, koff,Xe = (8.6 ± 2.1) × 102 s−1 (Fig. 4, b and c). The fact that koff,Xe is roughly four times greater than koff, maltose suggests that alternative Xe pathways contribute significantly to gas diffusion in the closed MBP structure.
Figure 4.
(a) 13C NMR spectrum of 2 mM [1-13C]-maltose and 0.6 mM MBP in PBS (pH 7.4). From high frequency to low frequency are free β-maltose (δ = 96.00 ppm), bound β-maltose (δ = 95.03 ppm), free α-maltose (δ = 92.13 ppm), and bound α-maltose (δ = 90.66 ppm). The peak width (full width at half maximum) is 37.7 Hz for bound β-maltose and 40.4 Hz for bound α-maltose. (b) Hyper-CEST z-spectra of wild-type MBP taken in a series of saturation pulse strengths from 30 to 77 μT at 300 K. (c) Linear fitting of ln−1(1-CEST) versus (γB1)−2 yields the slope = (−1.1 ± 0.1) × 105 s−2, y-intercept = −0.15 ± 0.09, and koff,Xe = (slope/y-intercept)1/2 = (8.6 ± 2.1) × 102 s−1. To see this figure in color, go online.
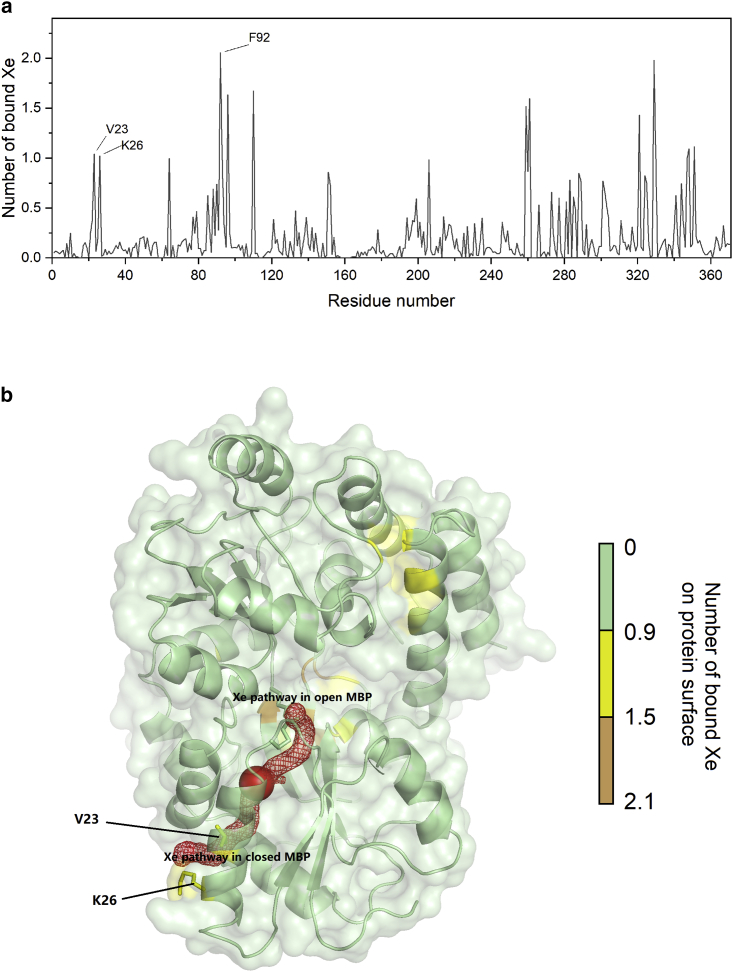
Tunnel prediction using the hard sphere model may not correctly identify all Xe exchange pathways since tunnel hydrophobicity is not considered. Also, evidence for alternative pathways might be hidden in the single-Xe simulations due to insufficient sampling. Therefore, the “Xe flooding” approach, in which Xe atoms were initially placed in the surrounding solvent, was applied, and the concentration of Xe was artificially increased to ∼150 mM to enhance Xe diffusion through the protein. (This is equivalent to nearly 30 atm Xe at room temperature (55). Using this computational method, we identified several protein surface sites that associate with Xe exchange due to transient Xe affinity, defined by the average number of bound Xe atoms, n, at each residue of the protein (Fig. 5 a). These sites include V23, F92, and the hinge region of MBP, all of which consist of nonpolar residues. Meanwhile, Xe has relatively low affinity (n < 0.05) toward the K15-E111 region, while maltose is stably bound during the simulation. F92 and the hinge region residues were not taken into further consideration because of their long distance (>18 Å) from the Xe cavity. Importantly, the solvent-exposed residue V23 is only ∼10 Å from the Xe binding pocket with a relatively high Xe affinity (n = 1.0) (Fig. 5 b). Xe accumulation at this site and its close proximity to the cavity may lead to a considerable rate of Xe exchange through V23. In order to examine the contribution of the V23 pathway, we mutated V23 to the smaller alanine and the bulkier leucine and performed variable-B1 measurements. Both mutations caused less than 5 ppm difference in chemical shift of the bound 129Xe NMR signal, meaning that the Xe binding pocket was minimally perturbed. Moreover, V23A and V23L mutations do not affect maltose-MBP interactions or salt-bridge dynamics, as confirmed by 13C NMR spectroscopy (Figs. S3 and S4). Consequently, the Xe dissociation rate, koff,Xe, increased to (1.2 ± 0.5) × 103 s−1 with V23A mutation (Fig. S5) and decreased to (5.4 ± 0.3) × 102 s−1 with the introduction of V23L (Fig. S6). We then inspected the “Xe flooding” MD trajectories of V23A and V23L proteins. Surprisingly, V23L no longer exhibits high Xe affinity (n = 0.4) at this site, although other affinity sites are mostly unchanged, whereas V23A still has a similar affinity (n = 0.8) compared with wild-type (Fig. S7). These results confirm that V23 is one of the exchange pathways, which can be explained by a hydrophobic cryptic site between the V23 side chain and the aliphatic part of K26.
Figure 5.
(a) Average number of Xe atoms bound to each residue of MBP during the “Xe flooding” simulation. Xe affinity for the protein surface is represented by peaks, which indicate the number of bound Xe atoms. (b) Visualization of Xe affinity sites on MBP surface. Interior Xe binding site is depicted as the red sphere. Xe exchange pathway switches from the site of K15-E111 salt bridge to surface hydrophobic site V23 upon formation of the K15-E111 salt bridge. To see this figure in color, go online.
Analysis of cavity size in different states of MBP
In addition to gas exchange rate and pathways in MBP, there is also curiosity regarding the origin of gas binding. Xe binding to MBP’s unusually large internal cavity serves no obvious function, and it is unclear about its biological role in different states of MBP. It has been well characterized by crystallography that MBP interacts with a variety of binding substrates, including maltodextrins of various lengths and other oligosaccharides, in different conformations (37,56,57), as well as the maltose transporter complex, MalFGK2 (58). Cavity volume was then calculated using Fpocket (43) based on the crystal structures of MBP in complex with various biomolecules. The largest cavity found in each structure corresponds to the same site as the Xe pocket. As listed in Table 1, the unliganded MBP harbors a cavity of 97 Å3, very similar to the cavity volume found in Xe-occupied cryptophane-A and its derivatives (85–89 Å3) (59). Binding of maltooligosaccharides causes only minor volume change (10%–20%). However, the cavity shrinks by 40% when maltodextrin-bound MBP interacts with MalFGK2, which cannot be explained merely by crystal packing effects. Instead, the extensive protein-protein interaction probably requires MBP to arrange itself into a more compact structure.
Table 1.
Cavity analysis of MBP in different structures.
Discussion
Our previous work has discussed the possibility of different Xe exchange rates in open and closed MBP conformations, but the structural basis was elusive, and no kinetic information was obtained (23). The current study reveals that the salt bridge K15-E111 is pivotal to Xe exchange rate by combining computational analysis and hyper-CEST experiments. In the maltose-bound closed conformation, this salt bridge helps to constrain Xe in the hydrophobic cavity, resulting in a koff,Xe of (8.6 ± 2.1) × 102 s−1. However, in the absence of the K15-E111 salt bridge, MBP in the open conformation can release Xe at a much faster rate. Xe escape from the protein cavity adopts a strategy similar to the “breathing motion” of gas tunnels in myoglobin. Although the bottleneck of the Xe tunnel is only about half of its van der Waals diameter, the dynamics of the bottleneck are highly dependent on the salt bridge, as K15 participates in the formation of a bottleneck. Transient rearrangement of the local environment promoted by the lack of the salt-bridge interaction allows faster Xe diffusion. Notably, the bound maltose is barely involved in the bottleneck, suggesting that the salt bridge itself, instead of the bound maltose, determines Xe exchange. This conclusion may be applied to study the rate-determining step of general gas-protein interactions. According to the quantitative hyper-CEST model (47,60), the CEST signal is optimal when Xe dissociation rate koff,Xe is close to the saturation pulse strength. Considering the broad peak width of protein hyper-CEST signal and the potential heating effect from strong pulses, the saturation pulse strength γB1 used in hyper-CEST experiments is usually limited to ∼5 × 102 Hz (B1,max ≈ 77 μT using dSNOB pulses). Therefore, the koff,Xe in the closed conformation is nearly optimal to allow efficient depolarization of 129Xe, giving a strong hyper-CEST signal of the maltose-MBP-bound 129Xe. The fast off rate, koff,Xe, in the open conformation reaches the “weak saturation” condition, leading to inadequate depolarization of the fast exchanging 129Xe by the same pulse strength. Consequently, the bound 129Xe signal can only be observed in the presence of maltose even though the Xe-binding cavity remains the same without maltose. We also showed that the 129Xe CEST signal could be manipulated by programming the salt bridge. Switching off the salt bridge by K15D mutation can completely turn off the bound 129Xe signal, whereas a reconstructed salt bridge D14K-K15D was able to restore the bound 129Xe NMR signal.
It is conceivable that the salt bridge can be functionalized by rational protein engineering. Metal coordination and organic-covalent interactions can form a switchable and controllable bridge at this site to regulate 129Xe exchange and generate different 129Xe NMR responses, which holds great promise for 129Xe NMR/magnetic resonance imaging applications, such as genetically encoded metal biosensors and light/electro-controlled contrast agents. Design and characterization of novel MBP-based 129Xe NMR biosensors will be elaborated in future publications.
Formation of the K15-E111 salt bridge blocks the fastest Xe exchange pathway with an opening rate of ∼102 s−1, as estimated by NMR line shape analysis. This method measures ligand exchange at a millisecond-to-second scale with reasonable approximations, and 1-13C-labeled maltose represents the local dynamics of the K15-E111 salt bridge. Xe residence time (1/koff,Xe = 1.2 ± 0.3 × 10−3 s) is shorter than the salt-bridge lifetime (∼10−2 s), and prior studies showed that the conformational transition rate of maltose-bound MBP is even slower (∼1 s), suggesting that Xe can dissociate without opening the K15-E111 salt bridge or switching protein conformation. Our “Xe flooding” simulation found that Xe has relatively high affinity for several hydrophobic sites on the protein surface but almost no affinity toward the polar environment of the K15-E111 salt bridge and maltose-binding site. In addition, solvent water and salt ions may form a shell at the maltose-binding site via H-bonding and electrostatic interactions, preventing Xe access and egress through this site. On the other hand, surface hydrophobic residues such as V23 can increase local Xe concentration by providing a transient binding site and help to desolvate Xe atoms. Similar surface aggregation of inert gas molecules near hydrophobic regions was also observed in pepsin (61). As a result, surface hydrophobic residues reduce the energy barrier for Xe migration, thus enhancing exchange between solvent and a proximal Xe-binding cavity. This action may be analogous to the “breathing motion” as well (62). Our variable-B1 experiments confirmed that V23 impacts the Xe dissociation rate without affecting the Xe-binding cavity and salt bridge. Importantly, the V23L mutation can inhibit Xe exchange, probably by decreasing local Xe concentration at the surface hydrophobic site. If we hypothetically inhibited all hydrophobic Xe pathways, the low limit of Xe exchange rate can approximate the dynamics of salt-bridge opening. Because the optimal saturation power is found to be √2 times the Xe dissociation rate (60), a narrower and stronger hyper-CEST signal will be observed with a slower Xe exchange. It is meaningful to constrain Xe in protein to a greater extent because protein-bound 129Xe is usually in rapid exchange and its chemical shift is not directly observable in a simple 129Xe NMR measurement. It has been reported that koff,Xe values in myoglobin and hemoglobin are on the order of 104–105 s−1 (63). MBP in its closed conformation has a significantly slower koff,Xe by more than one order of magnitude, which is still not slow enough to give a directly detectable 129Xe NMR signal. Identification and reshaping gas pathways in these proteins could potentially afford protein-based biosensors that are useful in direct-detection 129Xe NMR/magnetic resonance imaging applications.
Investigation of Xe-MBP interactions not only improves our understanding of gas exchange but also raises interesting questions as to the origin of the large packing defect in MBP, which fundamentally determines gas-protein interactions. An internal cavity is often considered to destabilize protein (64,65), but our prior study found that the cavity-filling mutation V293L in unliganded MBP does not increase its thermal stability (melting temperature [Tm] = 53.9°C ± 0.7°C for wild-type and 53.8°C ± 0.8°C for V293L) (23). Hence, the internal cavity volume may not correlate with protein stability. More interestingly, although ligand binding to MBP does not change the cavity as evidenced by MD simulations and volume analysis (Table 1), filling the cavity in maltose-bound MBP has a destabilizing effect (Tm = 61.0°C ± 0.8°C for wild-type and 59.5°C ± 0.9°C for V293L), suggesting that the cavity may facilitate ligand binding. Presumably, void space strengthens salt-bridge and H-bond interactions, e.g., K15-E111, due to its low dielectric constant and separation from solvent. Furthermore, the N domain of MBP can undergo considerable compression when interacting with the transmembrane complex MalFGK2 as the cavity becomes 40% smaller. This supports the functions of the cavity in providing structural flexibility to engage with MalFGK2 and prepare for the release of maltodextrins. Also, compression of the cavity may be detectable using hyper-CEST and push 129Xe chemical shift further downfield because it has been noted that smaller cavities in proteins are correlated with larger downfield 129Xe NMR shifts (26).
In summary, we have demonstrated the structural details behind the turn on 129Xe hyper-CEST signal in MBP and verified the role of the K15-E111 salt bridge in regulating Xe exchange. Dissociation rate of Xe in maltose-bound MBP exceeds the opening rate of the salt bridge, which led to the discovery that Xe exchange can also occur near a surface hydrophobic site, V23. Finally, we discuss the role of the hydrophobic cavity in strengthening ligand binding and protein-protein interactions and its flexibility in different states of the protein. These findings will foster future development and applications of 129Xe-based bioreporters and contrast agents and provide general insights into mechanisms of gas-protein interactions.
Author contributions
Z.Z. and I.J.D. designed the study. Z.Z., N.A.R., and J.H. performed all experiments and analyzed the data. Z.Z. and I.J.D. wrote the manuscript.
Acknowledgments
MD simulations in this work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges-2 system, which is supported by NSF award number ACI-1928147, at the Pittsburgh Supercomputing Center (PSC). This work was supported by NIH grant R35-GM-131907 to I.J.D.
Declaration of interests
The authors declare no competing interests.
Editor: Wendy Shaw.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.10.025.
Supporting material
References
- 1.Gaines J.C., Smith W.W., et al. O'Hern C.S. Random close packing in protein cores. Phys. Rev. E. 2016;93:032415. doi: 10.1103/PhysRevE.93.032415. [DOI] [PubMed] [Google Scholar]
- 2.Brunori M., Gibson Q.H. Cavities and packing defects in the structural dynamics of myoglobin. EMBO Rep. 2001;2:674–679. doi: 10.1093/embo-reports/kve159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen J., Arkhipov A., et al. Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys. J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubitz W., Ogata H., et al. Reijerse E. Hydrogenases. Chem Rev. 2014;114:4081–4148. doi: 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- 5.Montet Y., Amara P., et al. Fontecilla-Camps J.C. Gas access to the active site of Ni-Fe hydrogenases probed by X-ray crystallography and molecular dynamics. Nat. Struct. Biol. 1997;4:523–526. doi: 10.1038/nsb0797-523. [DOI] [PubMed] [Google Scholar]
- 6.Ilina Y., Lorent C., et al. Dobbek H. X-Ray crystallography and vibrational spectroscopy reveal the key determinants of biocatalytic dihydrogen cycling by [NiFe] hydrogenases. Angew. Chem., Int. Ed. Engl. 2019;131:18883–18887. doi: 10.1002/anie.201908258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doukov T.I., Blasiak L.C., et al. Drennan C.L. Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry. 2008;47:3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biester A., Dementin S., Drennan C.L. Visualizing the gas channel of a monofunctional carbon monoxide dehydrogenase. J. Inorg. Biochem. 2022;230:111774. doi: 10.1016/j.jinorgbio.2022.111774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batsanov S.S. Van der Waals radii of elements. Inorg. Mater. 2001;37:871–885. [Google Scholar]
- 10.Bondi A. Van der Waals volumes and radii. J. Phys. Chem. A. 1964;68:441–451. [Google Scholar]
- 11.Cohen J., Kim K., et al. Schulten K. Finding gas diffusion pathways in proteins: application to O2 and H2 transport in CpI [FeFe]-hydrogenase and the role of packing defects. Structure. 2005;13:1321–1329. doi: 10.1016/j.str.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J., Kim K., et al. King P. Molecular dynamics and experimental investigation of H2 and O2 diffusion in [Fe]-hydrogenase. Biochem. Soc. Trans. 2005;33:80–82. doi: 10.1042/BST0330080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felbek C., Arrigoni F., et al. Léger C. Mechanism of hydrogen sulfide-dependent inhibition of Fe-Fe hydrogenase. ACS Catal. 2021;11:15162–15176. [Google Scholar]
- 14.Leroux F., Dementin S., et al. Léger C. Experimental approaches to kinetics of gas diffusion in hydrogenase. Proc. Natl. Acad. Sci. USA. 2008;105:11188–11193. doi: 10.1073/pnas.0803689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond M.L., Wilson A.K., Cundari T.R. Carbon dioxide migration pathways in proteins. J. Phys. Chem. Lett. 2012;3:830–833. doi: 10.1021/jz3001085. [DOI] [PubMed] [Google Scholar]
- 16.Gee L.B., Leontyev I., et al. Cramer S.P. Docking and migration of carbon monoxide in nitrogenase: the case for gated pockets from infrared spectroscopy and molecular dynamics. Biochemistry. 2015;54:3314–3319. doi: 10.1021/acs.biochem.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collazo L., Klinman J.P. Control of the position of oxygen delivery in soybean lipoxygenase-1 by amino acid side chains within a gas migration channel. J. Biol. Chem. 2016;291:9052–9059. doi: 10.1074/jbc.M115.709154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahinthichaichan P., Gennis R.B., Tajkhorshid E. All the O2 consumed by Thermus thermophilus cytochrome ba3 is delivered to the active site through a long, open hydrophobic tunnel with entrances within the lipid bilayer. Biochemistry. 2016;55:1265–1278. doi: 10.1021/acs.biochem.5b01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahinthichaichan P., Gennis R.B., Tajkhorshid E. Characterizations of substrate delivery pathways in the nitric oxide reductase. Biophys. J. 2016;110:314a. [Google Scholar]
- 20.Trudell J.R., Koblin D.D., Eger E.I., 2nd A molecular description of how noble gases and nitrogen bind to a model site of anesthetic action. Anesth. Analg. 1998;87:411–418. doi: 10.1097/00000539-199808000-00034. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro M.G., Ramirez R.M., et al. Bajaj V.S. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat. Chem. 2014;6:629–634. doi: 10.1038/nchem.1934. [DOI] [PubMed] [Google Scholar]
- 22.Kunth M., Lu G.J., et al. Schröder L. Protein nanostructures produce self-adjusting hyperpolarized magnetic resonance imaging contrast through physical gas partitioning. ACS Nano. 2018;12:10939–10948. doi: 10.1021/acsnano.8b04222. [DOI] [PubMed] [Google Scholar]
- 23.Roose B.W., Zemerov S.D., Dmochowski I.J. Nanomolar small-molecule detection using a genetically encoded 129Xe NMR contrast agent. Chem. Sci. 2017;8:7631–7636. doi: 10.1039/c7sc03601a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zemerov S.D., Roose B.W., et al. Dmochowski I.J. 129Xe NMR-protein sensor reveals cellular ribose concentration. Anal. Chem. 2020;92:12817–12824. doi: 10.1021/acs.analchem.0c00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowery T.J., Rubin S.M., et al. Wemmer D.E. Design of a conformation-sensitive xenon-binding cavity in the ribose-binding protein. Angew. Chem. Int. Ed. Engl. 2004;43:6320–6322. doi: 10.1002/anie.200460629. [DOI] [PubMed] [Google Scholar]
- 26.Rubin S.M., Lee S.Y., et al. Wemmer D.E. Detection and characterization of xenon-binding sites in proteins by 129Xe NMR spectroscopy. J. Mol. Biol. 2002;322:425–440. doi: 10.1016/s0022-2836(02)00739-8. [DOI] [PubMed] [Google Scholar]
- 27.Desvaux H., Dubois L., et al. Matthews B.W. Dynamics of xenon binding inside the hydrophobic cavity of pseudo-wild-type bacteriophage T4 lysozyme explored through xenon-based NMR spectroscopy. J. Am. Chem. Soc. 2005;127:11676–11683. doi: 10.1021/ja053074p. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Roose B.W., et al. Dmochowski I.J. A genetically encoded beta-lactamase reporter for ultrasensitive 129Xe NMR in mammalian cells. Angew. Chem., Int. Ed. Engl. 2016;128:9130–9133. doi: 10.1002/anie.201604055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roose B.W., Zemerov S.D., et al. Dmochowski I.J. A structural basis for 129Xe hyper-CEST signal in TEM-1 beta-lactamase. ChemPhysChem. 2019;20:260–267. doi: 10.1002/cphc.201800624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z., Roose B.W., et al. Dmochowski I.J. Detecting protein-protein interactions by Xe-129 NMR. Chem. Commun. 2020;56:11122–11125. doi: 10.1039/d0cc02988b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard S.J., Gross K.H., Argos P. Intramolecular cavities in globular proteins. Protein Eng. 1994;7:613–626. doi: 10.1093/protein/7.5.613. [DOI] [PubMed] [Google Scholar]
- 32.Rubin S.M., Spence M.M., et al. Wemmer D.E. Detection of a conformational change in maltose binding protein by 129Xe NMR spectroscopy. J. Am. Chem. Soc. 2001;123:8616–8617. doi: 10.1021/ja0110325. [DOI] [PubMed] [Google Scholar]
- 33.Schröder L., Lowery T.J., et al. Pines A. Molecular imaging using a targeted magnetic resonance hyperpolarized biosensor. Science. 2006;314:446–449. doi: 10.1126/science.1131847. [DOI] [PubMed] [Google Scholar]
- 34.Phillips J.C., Braun R., et al. Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nystrom N.A., Levine M.J., et al. Scott J.R. Proceedings of the 2015 XSEDE Conference: Scientific Advancements Enabled by Enhanced Cyberinfrastructure. ACM; 2015. Bridges: a uniquely flexible HPC resource for new communities and data analytics; pp. 1–8. [Google Scholar]
- 36.Towns J., Cockerill T., et al. Wilkins-Diehr N. XSEDE: accelerating scientific discovery. Comput. Sci. Eng. 2014;16:62–74. [Google Scholar]
- 37.Quiocho F.A., Spurlino J.C., Rodseth L.E. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen W.L., Chandrasekhar J., et al. Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 39.Best R.B., Zhu X., et al. MacKerell A.D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verlet L., Weis J.-J. Perturbation theory for the thermodynamic properties of simple liquids. Mol. Phys. 1972;24:1013–1024. [Google Scholar]
- 41.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinf. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chovancova E., Pavelka A., et al. Damborsky J. Caver 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasteiger E., Hoogland C., et al. Bairoch A. In: The Proteomics Protocols Handbook. Walker J.M., editor. Humana Press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- 46.Mortuza M.G., Anala S., et al. Meersmann T. Spin-exchange optical pumping of high-density xenon-129. J. Chem. Phys. 2003;118:1581–1584. [Google Scholar]
- 47.Kunth M., Witte C., Schröder L. Quantitative chemical exchange saturation transfer with hyperpolarized nuclei (qHyper-CEST): sensing xenon-host exchange dynamics and binding affinities by NMR. J. Chem. Phys. 2014;141:194202. doi: 10.1063/1.4901429. [DOI] [PubMed] [Google Scholar]
- 48.Du K., Zemerov S.D., et al. Dmochowski I.J. Paramagnetic organocobalt capsule revealing xenon host-guest chemistry. Inorg. Chem. 2020;59:13831–13844. doi: 10.1021/acs.inorgchem.9b03634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Dmochowski I.J. Cucurbit[6]uril is an ultrasensitive 129Xe NMR contrast agent. Chem. Commun. 2015;51:8982–8985. doi: 10.1039/c5cc01826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavelka A., Sebestova E., et al. Damborsky J. CAVER: algorithms for analyzing dynamics of tunnels in macromolecules. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016;13:505–517. doi: 10.1109/TCBB.2015.2459680. [DOI] [PubMed] [Google Scholar]
- 51.Miller D.M., Olson J.S., et al. Quiocho F.A. Rates of ligand-binding to periplasmic proteins involved in bacterial transport and chemotaxis. J. Biol. Chem. 1983;258:13665–13672. [PubMed] [Google Scholar]
- 52.Telmer P.G., Shilton B.H. Insights into the conformational equilibria of maltose-binding protein by analysis of high affinity mutants. J. Biol. Chem. 2003;278:34555–34567. doi: 10.1074/jbc.M301004200. [DOI] [PubMed] [Google Scholar]
- 53.Li X., Lee K.H., et al. Chen M. Different anomeric sugar bound states of maltose binding protein resolved by a cytolysin A nanopore tweezer. ACS Nano. 2020;14:1727–1737. doi: 10.1021/acsnano.9b07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo M.H., Park J., et al. Kim H.S. Protein conformational dynamics dictate the binding affinity for a ligand. Nat. Commun. 2014;5:3724. doi: 10.1038/ncomms4724. [DOI] [PubMed] [Google Scholar]
- 55.Clever H.L. Pergamon; 1979. Krypton, Xenon and Radon. [Google Scholar]
- 56.Boos W., Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharff A.J., Rodseth L.E., Quiocho F.A. Refined 1.8-Å structure reveals the mode of binding of beta-cyclodextrin to the maltodextrin binding protein. Biochemistry. 1993;32:10553–10559. doi: 10.1021/bi00091a004. [DOI] [PubMed] [Google Scholar]
- 58.Oldham M.L., Chen S., Chen J. Structural basis for substrate specificity in the Escherichia coli maltose transport system. Proc. Natl. Acad. Sci. USA. 2013;110:18132–18137. doi: 10.1073/pnas.1311407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taratula O., Hill P.A., et al. Dmochowski I.J. Crystallographic observation of 'induced fit' in a cryptophane host-guest model system. Nat. Commun. 2010;1:148. doi: 10.1038/ncomms1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunth M., Witte C., Schröder L. Continuous-wave saturation considerations for efficient xenon depolarization. NMR Biomed. 2015;28:601–606. doi: 10.1002/nbm.3307. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Zhang Y., et al. Fang H. Inert gas deactivates protein activity by aggregation. Sci. Rep. 2017;7:10176. doi: 10.1038/s41598-017-10678-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomita A., Sato T., et al. Adachi S.I. Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin. Proc. Natl. Acad. Sci. USA. 2009;106:2612–2616. doi: 10.1073/pnas.0807774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tilton R.F., Jr., Kuntz I.D., Jr. Nuclear magnetic resonance studies of xenon-129 with myoglobin and hemoglobin. Biochemistry. 1982;21:6850–6857. doi: 10.1021/bi00269a035. [DOI] [PubMed] [Google Scholar]
- 64.Eriksson A.E., Baase W.A., et al. Matthews B.W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 65.Xu J., Baase W.A., et al. Matthews B.W. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7:158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.