Abstract
Endogenous antimicrobial peptides of the cathelicidin family contribute to innate immunity. The emergence of widespread antibiotic resistance in many commonly encountered bacteria requires the search for new bactericidal agents with therapeutic potential. Solid-phase synthesis was employed to prepare linear antimicrobial peptides found in cathelicidins of five mammals: human (FALL39/LL37), rabbit (CAP18), mouse (mCRAMP), rat (rCRAMP), and sheep (SMAP29 and SMAP34). These peptides were tested at ionic strengths of 25 and 175 mM against Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, and methicillin-resistant Staphylococcus aureus. Each peptide manifested activity against P. aeruginosa irrespective of the NaCl concentration. CAP18 and SMAP29 were the most effective peptides of the group against all test organisms under both low- and high-salt conditions. Select peptides of 15 to 21 residues, modeled on CAP18 (37 residues), retained activity against the gram-negative bacteria and methicillin-sensitive S. aureus, although the bactericidal activity was reduced compared to that of the parent peptide. In accordance with the behavior of the parent molecule, the truncated peptides adopted an α-helical structure in the presence of trifluoroethanol or lipopolysaccharide. The relationship between the bactericidal activity and several physiochemical properties of the cathelicidins was examined. The activities of the full-length peptides correlated positively with a predicted gradient of hydrophobicity along the peptide backbone and with net positive charge; they correlated inversely with relative abundance of anionic residues. The salt-resistant, antimicrobial properties of CAP18 and SMAP29 suggest that these peptides or congeneric structures have potential for the treatment of bacterial infections in normal and immunocompromised persons and individuals with cystic fibrosis.
The rapidly expanding prevalence of bacterial strains resistant to conventional antibiotics has prompted a search for new therapeutic agents, including various antimicrobial peptides of animal origin (15). Two broad classes of mammalian antibacterial peptides have been especially well studied: the cysteine-rich α- and β-defensins and various cathelicidins (6, 13, 22, 26, 27, 41, 42). Both classes are produced as precursors that require proteolytic processing to generate the mature antimicrobial peptide. Cathelicidins contain an N-terminal domain called cathelin, for which no function has yet been ascribed, and a C-terminal domain that comprises an antimicrobial peptide (reviewed in references 41 and 42). While the cathelin domains are highly conserved across species, the C-terminal antimicrobial domains are structurally diverse. The first cathelicidin precursor to be described was rabbit CAP18 (20), and its mature peptide was shown to have broad-spectrum bactericidal activity (19). Homologs of CAP18 have since been identified in other species including humans (FALL39/LL37) (1, 19), mice (mCRAMP) (12, 30), rats (rCRAMP), and sheep (SMAP29 and SMAP34) (2, 16, 25, 34). Circular dichroism (CD) measurements indicate that these linear peptides adopt α-helical structure in some solvents (1, 8, 12, 17, 36). These cathelicidin-derived peptides kill bacteria by disrupting the bacterial membrane (28).
Our primary goal in this study was to identify peptides of the cathelicidin family having intrinsically high bactericidal activity toward Pseudomonas aeruginosa and Staphylococcus aureus. These bacteria frequently manifest resistance to conventional antibiotics and pose serious problems for immunocompromised persons and cystic fibrosis patients. We also evaluated the physiochemical properties of each structure that correlated with antimicrobial activity to gain insights that could contribute to the rational design of salt-tolerant peptide antibiotics.
MATERIALS AND METHODS
Peptide synthesis.
All peptides were synthesized on an Applied Biosystems model 433A synthesizer at the 0.1 mM scale, using solid-phase Fastmoc chemistry. Peptides were purified by reverse-phase high-performance liquid chromatography on a Vydac 218TP1022 column. Separations were performed at a flow rate of 10 ml/min employing a linear gradient (0 to 100% solvent B) of aqueous 0.1% trifluoroacetic acid (solvent A) and acetonitrile containing 0.085% trifluoroacetic acid (solvent B). Fractions were collected and subsequently monitored by analytical scale reverse-phase high-performance liquid chromatography on a 4.6 by 250-mm Vydac 218TP54 column employing isocratic elution conditions (40% solvent B) at a flow rate of 1 ml/min. Selected fractions were pooled and lyophilized for further characterization by mass spectrometry and capillary electrophoresis. Mass measurements were performed with a Hewlett-Packard model 1100 MSD equipped with an electrospray ionization source, using flow injection at 0.1 ml/min in 64% acetonitrile containing 0.05% trifluoroacetic acid. Capillary electrophoresis was performed on a Hewlett-Packard 3D instrument equipped with a 75-μm (inner diameter) by 80.5-cm fused-silicate extended-light-path capillary; experiments were conducted at 18°C in 100 mM sodium phosphate (pH 2.9) at 20,000 V. The peptide concentration was determined by quantitative amino acid analysis on a Beckman 6300 amino acid analyzer.
Bacterial strains and antimicrobial assays.
A luminescence assay previously used with Escherichia coli (33, 37) was adapted to examine the potency and killing kinetics of the peptides against P. aeruginosa PAO1. Whereas E. coli DH5α were transformed with Photorhabdus luminescens luminescence genes on plasmid pCGLS1 (11), we transformed P. aeruginosa PAO1 with plasmid pMRP-77, which contains the Vibrio fischeri luminescence genes on plasmid pBBR1MCS-5 (18). The luminescence assays were performed in high- and low-salt media (see below), and antimicrobial activity was measured as the decrease in energy-dependent bacterial luminescence. The conditions for optimal luminescence by P. aeruginosa were somewhat different from those for E. coli. P. aeruginosa were grown to early log phase (2.5 × 107 bacteria/ml) at 30°C in tryptic soy broth containing 25 μg of gentamicin per ml to maintain the pMRP-77 plasmid. Assays were performed at 30°C in 96-well plates (Optiplate; Packard Instruments). Bacteria were appropriately diluted so that each well contained 5 × 104 organisms in 150 μl (final volume) of 3.3% tryptic soy broth and 6.7 mM potassium phosphate (pH 7.4). Antimicrobial peptides and other additions were included as described below. The ionic strength of the standard assay solution for either bacterium was equivalent to 25 mM NaCl (low salt); in some experiments, NaCl was added to increase the ionic strength to 175 mM (high salt). After incubation for the indicated times, relative emission of light (in arbitrary units) was measured with a luminometer (MLX; Packard Instruments). To determine the 50% effective concentration (EC50), we performed assays in duplicate, using several concentrations of each peptide. We fit a logarithmic equation to each curve. The EC50 was defined as the amount of antimicrobial peptide that decreased the luminescence by 50% relative to the peptide-free control.
The two-stage radial diffusion assay used in these studies has been described elsewhere (23). Briefly, the purified peptides were serially diluted in acidified water (0.01% acetic acid) that contained 0.1% human serum albumin (essentially globulin free; Sigma A-8763). The bacteria used were E. coli ML-35p (21), E. coli DH5α (GIBCO-BRL, Gaithersburg, Md.), P. aeruginosa MR 3007 (a strain resistant to several aminoglycosides and cephalosporins, obtained from E. A. Wagar, University of California Los Angeles [UCLA]), P. aeruginosa PAO1, S. aureus 930918-3 (from I. A. Holder, Shriners' Hospital, Cincinnati, Ohio), and methicillin-resistant S. aureus ATCC 33591 (MRSA). Bacteria were grown to mid-logarithmic phase in tryptic soy broth and washed with 10 mM phosphate buffer (pH 7.4). Approximately 2 × 105 CFU per ml was incorporated into a thin (1.2-mm) agarose underlay gel that contained 1% (wt/vol) agarose (type I, low electroendoosmosis; Sigma A-6013), 10 mM sodium phosphate buffer (pH 7.4), and 0.3 mg of tryptic soy broth powder per ml, with or without 100 mM NaCl. A regularly spaced, five-by-five array of wells was made in the underlay gel. The wells, 3.2 mm in diameter, had a potential capacity of 10 μl. Six 8-μl aliquots of each peptide (containing 0.79, 2.5, 7.9, 25.0, 79.1, or 250 μg of peptide/ml) were added to the wells. After 3 h, a 10-ml overlay gel consisting of 6% tryptic soy broth powder, 1% agarose, and 10 mM sodium phosphate buffer (pH 7.4) was poured. The plates were incubated overnight to allow surviving organisms to form microcolonies. Zone diameters were measured to the nearest 0.1 mm and expressed in units (1 U = 0.1 mm) after subtracting the diameter of the well. A linear relationship existed between the zone diameter and the base 10 logarithm of the peptide concentration. The MIC was determined by performing least-mean-squares fit and solving for the x intercept.
For colony-counting assays, bacteria were prepared as described for the luminescence assay above, incubated with antimicrobial peptides for 5 h, and then plated on nutrient agar plates. Colonies were counted after 24 to 48 h of incubation at 37°C.
CD spectroscopy.
CD spectroscopy was performed on a 62 DS spectrometer (AVIV Associates, Lakewood, N.J.) equipped with a thermoelectric temperature control at 25°C. Samples contained 0.11 to 0.24 mg of peptide per ml in 50 mM sodium phosphate (pH 7.0); some samples also contained 40% trifluoroethanol or 0.1% (0.22 mM) lipopolysaccharide. Spectra were collected at 0.5-nm intervals with an averaging time of 2 s per data point using a path length of 0.1 cm. The spectra were smoothed once over an interval of five data points prior to plotting, with each spectrum representing the average of two scans. The average of two buffer scans was subtracted prior to data smoothing. The equation fH = Θobs/ΘH was used to calculate the fractional helical content, where Θobs is the observed mean residue ellipticity (MRE) and ΘH is the MRE for a 100% helical peptide of identical length at 25°C calculated by the method of Luo and Baldwin (24). The MRE is expressed in millidegrees × square centimeter per decimole.
Hemolysis assay.
The hemolytic activity of the peptides was assayed with heparinized human red blood cells that had been collected from a normal volunteer and washed three times in phosphate-buffered saline. A 10% suspension of red blood cells was combined with peptide, phosphate-buffered saline (negative control), or 0.2% Triton X-100 (positive control) in a final volume of 200 μl. After a 30-min incubation, cell suspensions were centrifuged for 10 min at 1,300 × g and supernatants were transferred to a flat-bottom 96-well polystyrene microtiter plate and the absorbance (A) was read at 540 nm (Rainbow Spectra; Tecan U.S. Inc., Research Triangle Park, N.C.). The percent hemolysis was calculated using the formula 100 × (Asample − Ablank)/(ATriton − Ablank).
Hydrophobicity calculations.
Peptide hydrophobicity and hydrophobic moments were calculated as described by Eisenberg (10) using the normalized consensus hydrophobicity scales. To determine the gradient of hydrophobicity along the length of each peptide, the mean residue hydrophobicity was calculated using a window size of 11 residues.
RESULTS
Luminescence-based antibacterial assay.
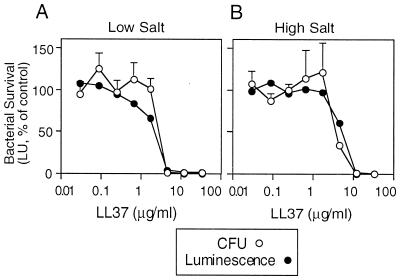
In earlier studies, we developed a luminescence-based assay to measure the peptide-mediated killing of E. coli (33, 37). For this study, a similar light-based assay was developed to monitor the killing of P. aeruginosa PAO1. Figure 1 shows the results of an experiment performed with the synthetic human peptide LL37. The concomitant reduction in light units and CFU for the luminescent P. aeruginosa PAO1 implied that the light-based assay provided a reliable measure of bacterial viability. The antibacterial activity of LL37 was relatively insensitive to salt, consistent with previous reports that LL37 is active in high-salt solutions (1, 3, 17, 38).
FIG. 1.
Antimicrobial activity measured by the luminescence assay. The relationship between viability (CFU) and luminescence of P. aeruginosa PAO1 expressing luminescence genes is shown. Bacteria were incubated with the indicated concentration of LL37 for 5 h, luminescence was measured, and surviving organisms were plated and counted. Studies were performed under low-salt (A) and high-salt (B) conditions. Values are expressed as the percentage of control in the absence of LL37. Symbols indicate mean and standard error of the mean (n = 4).
Relative activity of mammalian peptides.
Antimicrobial peptides encoded by five distinct mammalian cathelicidins genes were synthesized. The peptide sequences are shown in Table 1 and include the CAP18 (20), mCRAMP (12, 30), SMAP34 (16), SMAP29 (2, 25), rCRAMP, and FALL39/LL37 (1, 19) sequences. The rat sequence (rCRAMP) was identified by a homology search with the mouse protein (mCRAMP) sequence and corresponded to GenBank accession no. AA998531.
TABLE 1.
Primary sequence and antipseudomonal activity of full-length peptidesa
| Peptide | Peptide sequence | EC50 (μM) against P. aeruginosa PAO1b
|
|
|---|---|---|---|
| Low salt | High salt | ||
| mCRAMP | ISRLAGLLRKGGEKIGEKLKKIGQKIKNFFQKLVPQPEQ | 0.40 ± 0.05 (7) | 1.07 ± 0.16 (6) |
| rCRAMP | ISRLAGLVRKGGEKFGEKLRKIGQKIKEFFQKLALEIEQ | 0.74 ± 0.06 (4) | 2.38 ± 0.61 (3) |
| SMAP34 | GLFGRLRDSLQRGGQKILEKAERIWCKIKDIFRG | 0.37 ± 0.05 (3) | 1.04 ± 0.14 (3) |
| SMAP29 | RGLRRLGRKIAHGVKKYGPTVLRIIRIAG | 0.05 ± 0.01 (8) | 0.06 ± 0.01 (7) |
| CAP18 | GLRKRLRKFRNKIKEKLKKIGQKIQGLLPKLAPRTDY | 0.22 ± 0.05 (6) | 0.11 ± 0.02 (4) |
| FALL39 | FALLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 0.52 ± 0.04 (3) | 0.73 ± 0.08 (3) |
| LL37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 0.81 ± 0.13 (3) | 1.47 ± 0.33 (3) |
Sequences were aligned with the CLUSTAL program (35). The cationic amino acids arginine (R) and lysine (K) are underlined. Activity was determined by the luminescence assay under low-salt (25 mM NaCl) or high-salt (175 mM NaCl) conditions, as described in Materials and Methods. Calculated molecular weights for the peptides are as follows: mCRAMP, 4,419; rCRAMP, 4,473; SMAP34, 3,988; SMAP29, 3,256; FALL39, 4,711; LL37, 4,493; CAP18, 4,434.
Data are mean EC50 ± standard error of the mean, and the number of experiments (n) is indicated.
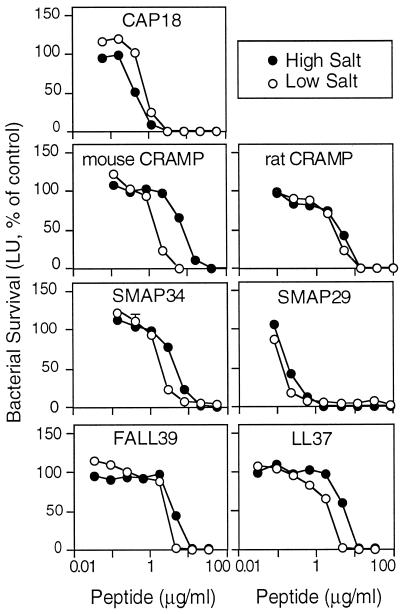
Table 1 and Fig. 2 show the activity of the full-length synthetic peptides against P. aeruginosa, as determined by the luminescence assay performed in low- and high-salt media. Each peptide was effective against P. aeruginosa PAO1 in the presence of 175 mM NaCl, although their activities differed. SMAP29 and CAP18 were clearly the most active peptides against PAO1 since they had the lowest EC50s in low salt of 0.05 and 0.2 μg/ml, respectively. These peptides showed little or no salt sensitivity and killed more rapidly than the other peptides did (see below).
FIG. 2.
Effect of salt on the antimicrobial activity of cathelicidin-derived peptides. P. aeruginosa PAO1 was incubated with the indicated concentrations of antimicrobial peptide in the standard assay buffer at low salt or at high salt. Luminescence was measured after 5 h. Values are percentages of control in the absence of antimicrobial peptide; controls were determined for each salt concentration. Symbols indicate mean and range (n = 2); in most cases, the error bars are covered by the symbol.
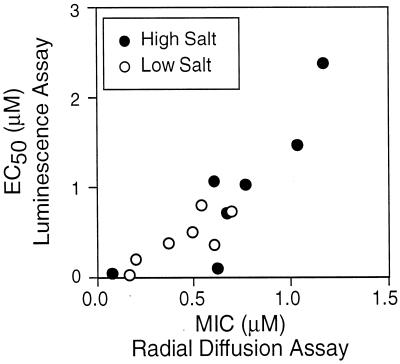
A radial-diffusion method was also employed to examine peptide activity. Table 2 shows radial diffusion data for the same seven peptides tested against two strains of Staphylococcus aureus, one of which was an MRSA strain, E. coli ML-35p, and a second strain of P. aeruginosa, MR3007. There was agreement between the EC50s determined by the luminescence assay and the MICs determined by the radial-diffusion method for P. aeruginosa PAO1 (Fig. 3). As above, SMAP 29 and CAP18 were more effective than the other peptides against P. aeruginosa. In addition, SMAP29 and CAP18 were more active against E. coli, S. aureus, and MRSA than were the other cathelicidins tested.
TABLE 2.
Activity of full-length peptides against gram-negative and gram-positive bacteriaa
| Peptide | MIC (μM) for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
E. coli DH5α
|
E. coli ML-35p
|
P. aeruginosa PAO1
|
P. aeruginosa MR3007
|
MRSA ATCC 33591
|
S. aureus 930918-3
|
|||||||
| Low | High | Low | High | Low | High | Low | High | Low | High | Low | High | |
| mCRAMP | 0.25 | 0.53 | 0.80 | 0.67 | 0.37 | 0.60 | 1.29 | 1.78 | 2.42 | 17.9 | 1.06 | 2.06 |
| rCRAMP | 0.54 | 0.61 | 1.55 | 0.66 | 0.69 | 1.17 | 2.01 | 1.78 | 3.19 | >20.1 | 2.14 | 2.16 |
| SMAP34 | 0.50 | 0.23 | 1.93 | 1.03 | 0.60 | 0.77 | 2.00 | 1.91 | 4.00 | 13.3 | 1.96 | 2.13 |
| SMAP29 | 0.10 | 0.07 | 0.18 | 0.12 | 0.17 | 0.08 | 0.34 | 0.18 | 1.24 | 0.65 | 0.31 | 0.43 |
| CAP 18 | 0.19 | 0.21 | 0.05 | 0.09 | 0.20 | 0.62 | 0.05 | 0.36 | 1.24 | 1.36 | 0.41 | 0.63 |
| FALL39 | 0.16 | 0.20 | 1.68 | 0.93 | 0.49 | 0.69 | 1.78 | 1.29 | 6.90 | >16.8 | 4.25 | 5.48 |
| LL37 | 0.19 | 0.44 | 1.20 | 0.58 | 0.54 | 1.03 | 0.53 | 1.54 | 5.45 | 30.8 | 3.58 | 2.58 |
Data are MICs determined by the radial-diffusion assay with low-salt (no NaCl) and high-salt (100 mM NaCl) underlays.
FIG. 3.
Correlation of killing data in low and high salt of P. aeruginosa PAO1 by two assay methods. The EC50s determined by the luminescence assay are plotted against the MICs assessed by the radial-diffusion assay.
Relative activity of truncated CAP18 peptides.
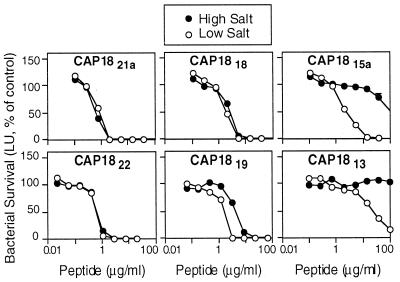
A total of 10 truncated forms of the 37-residue CAP18 molecule were synthesized and tested for antimicrobial activity in the presence of high and low salt. Our intent was to establish the minimal structure conferring bactericidal activity against E. coli, P. aeruginosa, and S. aureus and to assess the relative importance of different regions of the parent molecule to this activity. CAP18 was chosen for this study because (i) it was one of the most active bactericidal peptides we studied, (ii) it was effective in the presence of high salt, and (iii) it was not injurious to eukaryotic cells as assessed by a hemolytic assay (see below) and therefore was a potential candidate for development as a therapeutic agent. Table 3 summarizes the structures synthesized and their respective activities against P. aeruginosa PAO1. Figure 4 presents the antipseudomonal profiles for six of the truncated peptides using luminescence data collected under conditions of 25 and 175 mM NaCl. The first peptide synthesized, CAP1822, comprised the region of CAP18 predicted to be most α-helical. This peptide of 22 residues retained the activity and salt tolerance of the parent structure. Deletion of 16 C-terminal residues of CAP18 (CAP1821a) did not impair antibacterial activity in low and high salt, with the exception of its activity against MRSA. Removal of three N-terminal residues from CAP1821a (CAP1818) had no dramatic effect on activity or salt insensitivity. The 15-mer (CAP1815a) produced by deleting 6 N-terminal residues from CAP21a retained antibacterial activity in low salt but was considerably more salt sensitive, again with the exception of its activity against MRSA. The removal of 9 N-terminal residues of CAP21a (CAP1813) resulted in a peptide with greatly diminished activity in low and high salt. Restoring 2 or 4 C-terminal residues to CAP1813 (CAP1815b or CAP1817, respectively) partially restored the activity but not the salt insensitivity. Thus, we conclude from this assay that the most effective antipseudomonal peptides of the CAP18 truncation series ranged in size from 18 to 22 residues.
TABLE 3.
Primary sequence and antipseudomonal activities of CAP18 truncated peptidesa
| Peptide | Peptide sequence | EC50 (μM) against P. aeruginosa PAO1
|
|
|---|---|---|---|
| Low salt | High salt | ||
| CAP18 | GLRKRLRKFRNKIKEKLKKIGQKIQGLLPKLAPRTDY | 0.22 ± 0.05 (6) | 0.11 ± 0.02 (4) |
| CAP1822 | RKRLRKFRNKIKEKLKKIGQKI | 0.49 ± 0.17 (4) | 0.31 ± 0.04 (4) |
| CAP1821a | GLRKRLRKFRNKIKEKLKKIG | 0.22 ± 0.03 (3) | 0.21 ± 0.04 (3) |
| CAP1818 | KRLRKFRNKIKEKLKKIG | 0.46 ± 0.16 (3) | 0.70 ± 0.13 (3) |
| CAP1815a | RKFRNKIKEKLKKIG | 0.82 ± 0.23 (3) | >10.6 (3) |
| CAP1819 | LRKFRNKIKEKLKKIGQKI | 0.69 ± 0.11 (3) | 1.28 ± 0.15 (3) |
| CAP1821b | LRKFRNKIKEKLKKIGQKIQG | 0.72 ± 0.20 (4) | 1.35 ± 0.01 (2) |
| CAP1813 | KIKEKLKKIGQKI | 14.8 ± 0.90 (3) | >16.1 (3) |
| CAP1815b | KIKEKLKKIGQKIQG | 5.92 ± 0.17 (3) | >46.0 (3) |
| CAP1817 | KIKEKLKKIGQKIQGLL | 1.19 ± 0.11 (3) | >7.9 (3) |
Activities were determined by the luminescence assay under low-salt (25 mM NaCl) or high-salt (175 mM NaCl) conditions. Calculated molecular weights for the peptides are as follows: CAP18, 4,434; CAP1821a, 2,610; CAP1818, 2,284; CAP1815a, 1,886; CAP1822, 2,810; CAP1819, 2,369; CAP1821b, 2,554; CAP1813, 1,554; CAP1815b, 1,739; and CAP1817, 1,966.
Data are mean EC50 ± standard error of the mean, and the number of experiments (n) is indicated.
FIG. 4.
Effect of salt on the antimicrobial activity of truncated CAP18 peptides. P. aeruginosa was incubated with the indicated concentrations of antimicrobial peptide in the standard assay buffer at low salt or at high salt. Luminescence was measured after 5 h. Values are percentages of control in the absence of antimicrobial peptide; controls were determined for each salt concentration. Symbols indicate mean and range (n = 2); in most cases, the error bars are covered by the symbol.
The radial-diffusion assay was also used to assess the activities of the truncated CAP18 peptides (Table 4). CAP1822 was 10-fold less active in high salt against E. coli ML-35p (MIC, 0.94 μM) and P. aeruginosa MR3007 (MIC, 4.2 μM) than was the parent structure. The activity of CAP1822 in high salt toward the gram-positive bacteria (S. aureus 930918-3 and MRSA) was even more adversely affected (MIC, >43.7 μM). Interestingly, the killing efficiency of CAP1821a for E. coli, P. aeruginosa, and methicillin-sensitive S. aureus at high salt compared favorably to that of CAP18 itself. However, this was not observed for MRSA, against which the peptide had greatly diminished activity. CAP1815a, the smallest peptide with significant activity under conditions of low salt in the luminescence assay, was inactive against MRSA (MIC, >130 μM) but otherwise exhibited robust broad-spectrum activity under both low- and high-salt conditions.
TABLE 4.
Antimicrobial activities of truncated peptides derived from rabbit CAP18a
| Peptide | MIC (μM) for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
E. coli ML-35p
|
P. aeruginosa MR3007
|
MRSA ATCC 33591
|
S. aureus 930918-3
|
|||||
| Low salt | High salt | Low salt | High salt | Low salt | High salt | Low salt | High salt | |
| CAP18 | 0.05 | 0.09 | 0.04 | 0.36 | 1.90 | 1.40 | 0.41 | 0.63 |
| CAP1822 | 0.05 | 0.94 | 0.37 | 4.20 | 4.50 | >43.7 | 4.40 | >43.7 |
| CAP1821a | 0.88 | 0.15 | 0.27 | 0.34 | 7.40 | >95 | 0.69 | 0.84 |
| CAP1818 | 1.00 | 0.22 | 1.30 | 10.3 | 6.50 | >110 | 4.30 | 12.7 |
| CAP1815a | 0.11 | 0.37 | 0.80 | 2.80 | >130 | >130 | 0.95 | 1.17 |
| CAP1819 | 0.14 | 6.50 | 3.90 | 9.50 | 1.60 | >28.1 | 2.10 | >28.1 |
| CAP1821b | 0.90 | 0.20 | 1.10 | 9.20 | 4.00 | >98.0 | 6.40 | >98 |
| CAP1813 | 14.6 | >160 | >50.9 | >160 | >160 | >160 | >160 | >160 |
| CAP1815b | 0.92 | >143 | >45.4 | >143 | >143 | >143 | >143 | >143 |
| CAP1817 | 4.9 | 0.97 | 2.6 | >127 | >127 | >127 | >127 | >127 |
Data are MICs determined by the radial-diffusion assay performed with low-salt (no NaCl) and high-salt (100 mM NaCl) underlays.
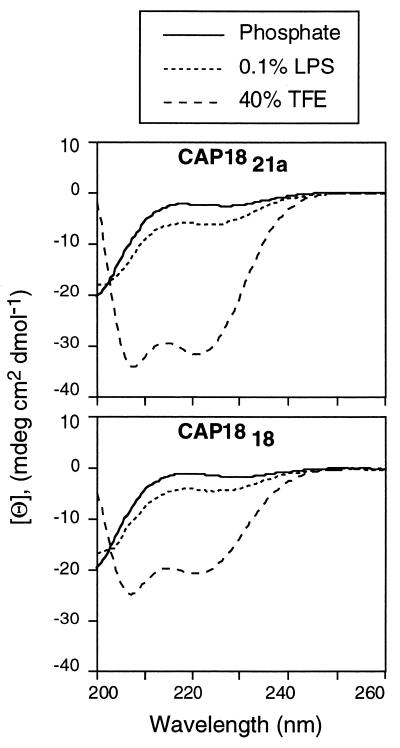
Conformational studies.
The formation of an amphipathic, α-helical structure and its relationship to the bactericidal activity of the cathelicidins has been the subject of previous studies (1, 8, 12, 17, 36). In this study, we analyzed five synthetic peptides (CAP18, CAP1821a, CAP1818, CAP1815a, and CAP1817) by CD spectroscopy. Our intent was to determine if the extent to which the helical structure could be induced in these peptides correlated positively with antimicrobial activity. The combined data in Fig. 5 and Table 5 indicate that each peptide, including the parent molecule of 37 residues, existed primarily in a random-coil conformation in 0.05 M sodium phosphate buffer (pH 7.0). In contrast, each peptide adopted an appreciable α-helical content in the presence of the organic cosolvent 40% trifluoroethanol, a known helix-stabilizing agent (24), and in the presence of 0.1% lipopolysaccharide, the presumed gram-negative bacterial recognition molecule. These data are summarized in Table 5.
FIG. 5.
CD spectroscopy of cathelicidin-derived peptides. The spectra of CAP1821a and CAP1818 were measured in 50 mM sodium phosphate buffer (pH 7.0), 0.1% lipopolysaccharide (LPS), or 40% trifluoroethanol (TFE).
TABLE 5.
Percent helical content of CAP18 peptides analyzed by CD spectroscopya
| Peptide | % helical content in phosphate buffer with:
|
||
|---|---|---|---|
| No addition | 0.1% lipopolysaccharide | 40% trifluoroethanol | |
| CAP18 | 8.1 | 39.2 | 84.6 |
| CAP1821a | 7.7 | 19.2 | 97.0 |
| CAP1818 | 3.8 | 13.4 | 64.8 |
| CAP1817 | 3.1 | 14.6 | 68.8 |
| CAP1815a | 2.7 | 11.5 | 44.7 |
The observed MRE was compared to the MRE for a 100% helical peptide of identical length. CD measurements were performed in 50 mM sodium phosphate buffer (pH 7.0) with additions as indicated.
Rate of killing.
The rate of bacterial killing may be an important factor for assessing the activity of antimicrobial peptides in vivo and for determining their potential use as pharmaceuticals. Therefore, we examined the time course of bacterial killing of several cathelicidin-derived peptides (Table 6). When full-length peptides were tested at 10 times their EC50 concentrations, the time required to achieve half-maximal killing ranged from 2.3 min (SMAP29) to 62 min (LL37). For comparison, the relative time required for killing by the antibiotic tobramycin was 38.9 min. We chose tobramycin because it is frequently administered intravenously and by aerosol to patients with cystic fibrosis (40). The most rapid killers were SMAP29 and CAP18.
TABLE 6.
Relative time for killing of P. aeruginosa PAO1 by several antimicrobial peptides and tobramycina
| Antimicrobial compound | Concn (μg/ml) | Half time of killing (min) |
|---|---|---|
| mCRAMP | 16 | 7.6 ± 3.5 |
| rCRAMP | 30 | 13.9 ± 2.8 |
| SMAP29 | 2 | 2.3 ± 0.8 |
| FALL39 | 24 | 22.7 ± 4.7 |
| LL37 | 36 | 62.0 ± 26.3 |
| CAP18 | 10 | 4.0 ± 1.4 |
| CAP1821a | 6 | 9.2 ± 3.4 |
| CAP1818 | 10 | 19.5 ± 8.2 |
| CAP1815a | 15 | 32.5 ± 13.5 |
| CAP1822 | 14 | 9.8 ± 2.5 |
| CAP1819 | 16 | 8.0 ± 1.6 |
| CAP1817 | 23 | 5.2 ± 2.1 |
| Tobramycin | 0.06 | 38.9 ± 4.5 |
Antimicrobial agents were used at a concentration 10 times their EC50 against P. aeruginosa PAO1 at 25 mM ionic strength (Table 1). Bacteria were incubated with the compound, and luminescence was measured at 1.5-min intervals. The half time of killing is the time at which luminescence was decreased to half its original value. Data are mean ± standard error of the mean (n = 3) for each compound.
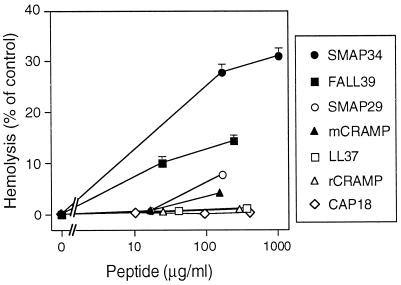
Hemolysis.
A possible limitation to the development of these peptides as antibiotics is their potential to cause injury to mammalian cell membranes. To assess this potential shortcoming, we examined their ability to lyse human erythrocytes (Fig. 6). Of the full-length peptides tested, only SMAP34 and FALL39 were significantly hemolytic. CAP18, rCRAMP, and LL37 (2 residues shorter than FALL39) showed the lowest hemolytic activity. Although these findings do not preclude the occurrence of cytotoxic or secretory responses when other cell types are exposed to these peptides, they provide evidence that the peptides do not have melittin-like, broadly cytolytic properties.
FIG. 6.
Hemolytic activity of cathelicidin-derived peptides. Human erythrocytes were incubated with the indicated concentration of each peptide, and hemolysis was measured as described in Materials and Methods. Values are percentages of control in the absence of antimicrobial peptide. Symbols indicate mean and range (n = 2); in some cases, the error bars are covered by the symbols.
DISCUSSION
We used two methods of assay to examine the relative bactericidal activities of peptides derived from several mammalian cathelicidins. A luminescence procedure was adapted and used to study P. aeruginosa PAO1. A previously described two-stage radial-diffusion method was also used to test strains of P. aeruginosa, E. coli, and S. aureus, one susceptible to methicillin and the other resistant. Both procedures were performed under low- and high-salt conditions. Under low-salt conditions, each of the full-length peptides and several truncated forms of CAP18 killed P. aeruginosa, E. coli, and S. aureus, including MRSA. The full-length peptides retained activity against E. coli, P. aeruginosa, and MRSA in 100 mM NaCl (Table 2), but only SMAP29 and CAP18 showed potent activity against MRSA under high-salt conditions (Table 2).
Retention of antimicrobial activity against gram-negative bacteria in high-salt environments is a feature that distinguishes these cathelicidin-derived peptides (1, 12, 17, 38) from α- and β-defensin peptides (3, 14, 31–33, 39). The β-defensins HBD-1 and HBD-2 were reported to show little activity against E. coli DH5α at NaCl concentrations greater than 50 and 100 mM, respectively (33), although increasing the concentration of either defensin could moderate this inhibitory effect of salt. The activity of our full-length peptides against P. aeruginosa was only marginally diminished at salt concentrations as high as 175 mM, i.e., an ionic strength greater than that of physiological saline (Table 1). CAP1821a, a peptide composed of residues 1 to 21 of CAP18, was as active against E. coli and P. aeruginosa under high-salt conditions as was full-length CAP18. Subsequent deletions indicated that the N-terminal residues of CAP1821a were important for its activity under high-salt conditions (Tables 3 and 4). Peptides which differed from CAP21a by deletion of 3 to 5 N-terminal residues (CAP1818 and especially CAP1819) showed reduced activity against P. aeruginosa MR3007 and P. aeruginosa PAO1 under high- and low-salt conditions (Tables 2 and 4). Removal of additional residues rendered the resulting peptides (CAP1813, CAP1815b, and CAP1817) inactive in high salt. This role for the N terminus is consistent with a previous observation that amino-terminal truncation of mCRAMP resulted in a peptide that was inactive when tested in a high-salt medium (30). Although removal of 16 C-terminal residues of CAP18 (CAP1821a) did not affect the activity of the peptide against P. aeruginosa, it rendered it inactive against MRSA, especially under high-salt conditions. Clearly, the ionic environment exerts complex effects on microbicidal activity, and these are further influenced by the target microorganism. Similar observations were recently reported for LL37 (38).
The most potent full-length peptides, SMAP29 and CAP18, were also the most rapid killers (Table 6). It remains to be determined if the observed differences in the rate of bacterial killing result from differences in binding kinetics or from greater effectiveness in some other step of the killing mechanism, such as peptide assembly or membrane insertion. For pharmaceutical applications, especially those involving topical administration, potency and speed of action are likely to be among the decisive factors. Of the peptides studied here, CAP18 and SMAP29 are clearly without equal in these respects.
What makes a linear peptide devoid of cysteine an effective antimicrobial agent? Among the factors that may influence activity are the ability to form an amphipathic α-helical structure, local or overall charge distribution and density, and some minimal peptide length. The peptides may also vary in their ability to self-associate, which may in turn influence killing activity (38). For the peptides studied here, we found that the degree of helicity was not a simple predictor of antibacterial activity. For example, CAP1821a had a higher helical content and higher killing activity than CAP1818; on the other hand, CAP1817 and CAP1818 had the same helical content, but CAP1817 manifested considerably less killing activity for P. aeruginosa and S. aureus under high-salt conditions. Thus, whereas helical structure may be essential for the antibacterial activity of this group of peptides, it clearly cannot be the sole determinant.
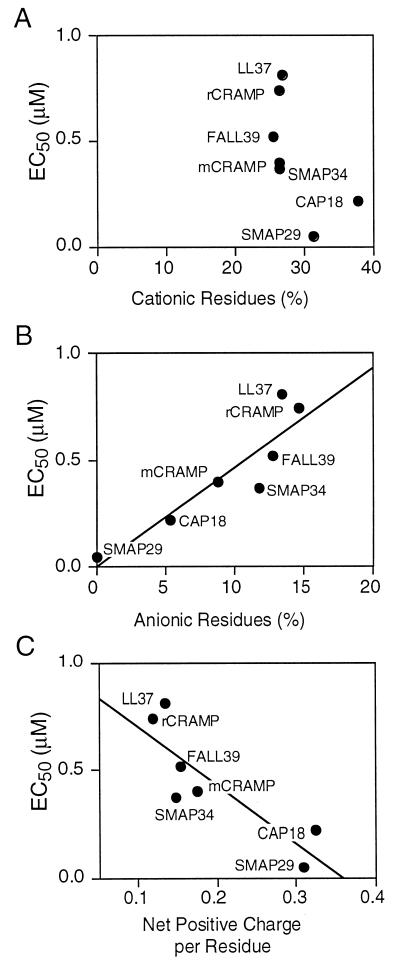
Among the full-length peptides, there was no correlation between the percentage of cationic residues and the level of antimicrobial activity (Fig. 7A). However, there was an inverse correlation between antimicrobial activity and the percentage of anionic residues (Fig. 7B) as well as a positive correlation between antimicrobial activity and the net positive charge of the peptide (Fig. 7C). Similarly, we found no simple correlation between antibacterial activity and the calculated average hydrophobicity or amphipathicity of the peptides or their calculated hydrophobic moment (data not shown). Other investigators have observed that overall hydrophobicity is a contributing factor to hemolytic activity for antimicrobial peptides (5, 9). The reduced hemolytic activity of LL37 compared to FALL39 agrees with this observation.
FIG. 7.
Correlation of physiochemical properties of the cathelicidin-derived peptides with antimicrobial activity. The relationships between antipseudomonal potency and the percentage of cationic residues (A), the percentage of anionic residues (B), and the net positive charge (C) are shown. Each point represents one peptide.
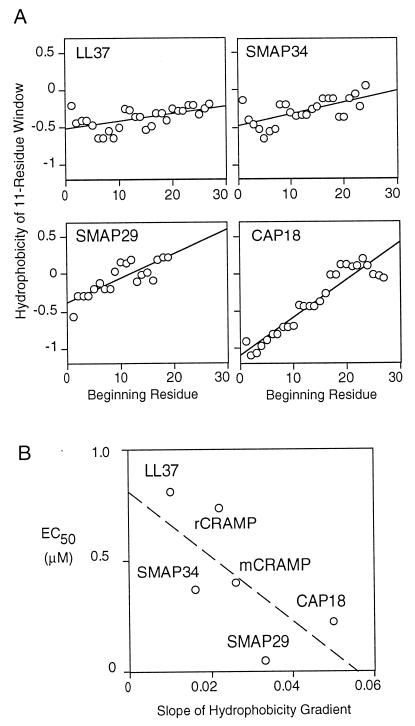
A positive correlation was observed between the bactericidal activity of the full-length cathelicidin-derived peptides and the magnitude of a predicted gradient of hydrophobicity along the helical axis (Fig. 8A). Hydrophobicity gradients are found in signal sequences and viral fusion proteins, where they promote membrane insertion (7). In amphipathic peptides, hydrophobicity gradients appear to promote membrane destabilization by causing the peptide to partially penetrate the bilayer (4, 7, 29). In our series, the peptides with the steepest hydrophobicity gradients, SMAP29 and CAP18, also had the most potent antimicrobial activity (Fig. 8B), implying that such gradients contribute to the killing process.
FIG. 8.
Hydrophobicity gradients of cathelicidin-derived peptides. (A) The hydrophobicity of each 11-residue window was calculated as described by Eisenberg (10), and a line was fit by linear regression. The more hydrophobic regions have higher hydrophobicity values. (B) Relationship between antipseudomonal activity and the magnitude (slope) of the hydrophobicity gradient across the peptide. Each point represents one peptide.
Several properties of the cathelicidin-derived peptides make them attractive candidates for research and drug development. First, they are effective killers of a variety of bacteria, including E. coli, P. aeruginosa, and S. aureus (12, 19, 38). Second, unlike many α- and β-defensins, several of the cathelicidin-derived peptides retain broad-spectrum bactericidal activity at physiologic or elevated salt concentrations—a distinct advantage for potential therapeutic uses. Third, cathelicidin-derived peptides are devoid of disulfide bridges, allowing easier and less costly chemical synthesis. Finally, their high rate of killing should provide an advantage for topical applications, allowing bacterial killing before the peptide is mechanically cleared or inactivated.
Of the peptides tested, CAP18 and SMAP29 were the most active against P. aeruginosa, E. coli, S. aureus, and MRSA. Additional studies are required to evaluate their in vitro activity against clinical isolates, their interactions with constituents of biological fluids, and their in vivo efficacy. It will also be important to learn if these peptides synergize with conventional antibiotics or other endogenous antimicrobial factors. Although much remains to be done, the present work suggests that these peptides are promising candidates for further investigation and development.
ACKNOWLEDGMENTS
We thank Elena Rus and Brian Morrison of the Protein Structure Facility at the University of Iowa for peptide synthesis, Matthew R. Parsek for construction of plasmid pMRP-77, and Michael A. Apicella for the lipopolysaccharide. We are grateful to Linda L. McCarter for her comments on the manuscript.
This work was supported by the National Institutes of Health (grants HL61234, AI29839, and AI43934), the Cystic Fibrosis Foundation, and the Howard Hughes Medical Institute.
ADDENDUM IN PROOF
The peptide sequence of SMAP34 was based on the original description (K. M. Huttner, M. R. Lambeth, H. R. Burkin, D. J. Burkin, and T. E. Broad, Gene 206:85–91, 1998). The sequence has since been corrected (GenBank accession number U60597).
REFERENCES
- 1.Agerberth B, Gunne H, Odeberg J, Kogner P, Boman H G, Gudmundsson G-H. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc Natl Acad Sci USA. 1995;92:195–199. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagella L, Scocchi M, Zanetti M. cDNA sequences of three sheep myeloid cathelicidins. FEBS Lett. 1995;376:225–228. doi: 10.1016/0014-5793(95)01285-3. [DOI] [PubMed] [Google Scholar]
- 3.Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, Wilson J M. Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Investig. 1998;102:874–880. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennik M H J, Vanloo B, Brasseur R, Gorris L G M, Smid E J. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim Biophys Acta. 1998;1373:47–58. doi: 10.1016/s0005-2736(98)00086-8. [DOI] [PubMed] [Google Scholar]
- 5.Bessalle R, Gorea A, Shalit I, Metzger J W, Dass C, Desiderio D M, Fridkin M. Structure-function studies of amphiphilic antibacterial peptides. J Med Chem. 1993;36:1203–1209. doi: 10.1021/jm00061a011. [DOI] [PubMed] [Google Scholar]
- 6.Boman H G. Gene-encoded peptide antibiotics and the concept of innate immunity: an update review. Scand J Immunol. 1998;48:15–25. doi: 10.1046/j.1365-3083.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 7.Brasseur R, Pillot T, Lins L, Vandekerckhove J, Rosseneu M. Peptides in membranes: tipping the balance of membrane stability. Trends Biochem Sci. 1997;22:167–171. doi: 10.1016/s0968-0004(97)01047-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Brock R, Luh F, Chou P J, Larrick J W, Huang R F, Huang T H. The solution structure of the active domain of CAP18—a lipopolysaccharide binding protein from rabbit leukocytes. FEBS Lett. 1995;370:46–52. doi: 10.1016/0014-5793(95)00792-8. [DOI] [PubMed] [Google Scholar]
- 9.Dathe M, Schumann M, Wieprecht T, Winkler A, Beyermann M, Krause E, Matsuzaki K, Murase O, Bienert M. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry. 1996;35:12612–12622. doi: 10.1021/bi960835f. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 11.Frackman S, Anhalt M, Nealson K H. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J Bacteriol. 1990;172:5767–5773. doi: 10.1128/jb.172.10.5767-5773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo R L, Kim K J, Bernfield M, Kozak C A, Zanetti M, Merluzzi L, Gennaro R. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J Biol Chem. 1997;272:13088–13093. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- 13.Ganz T, Lehrer R I. Defensins. Pharmacol Ther. 1995;66:191–205. doi: 10.1016/0163-7258(94)00076-f. [DOI] [PubMed] [Google Scholar]
- 14.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 15.Hancock R E W, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 16.Huttner K M, Lambeth M R, Burkin H R, Burkin D J, Broad T E. Localization and genomic organization of sheep antimicrobial peptide genes. Gene. 1998;206:85–91. doi: 10.1016/s0378-1119(97)00569-6. [DOI] [PubMed] [Google Scholar]
- 17.Johansson J, Gudmundsson G H, Rottenberg M E, Berndt K D, Agerberth B. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem. 1998;273:3718–3724. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- 18.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 19.Larrick J W, Hirata M, Balint R F, Lee J, Zhong J, Wright S C. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect Immun. 1995;63:1291–1297. doi: 10.1128/iai.63.4.1291-1297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larrick J W, Morgan J G, Palings I, Hirata M, Yen M H. Complementary DNA sequence of rabbit CAP18—a unique lipopolysaccharide binding protein. Biochem Biophys Res Commun. 1991;179:170–175. doi: 10.1016/0006-291x(91)91350-l. [DOI] [PubMed] [Google Scholar]
- 21.Lehrer R I, Barton A, Ganz T. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J Immunol Methods. 1988;108:153–158. doi: 10.1016/0022-1759(88)90414-0. [DOI] [PubMed] [Google Scholar]
- 22.Lehrer R I, Ganz T. Antimicrobial peptides in mammalian and insect host defence. Curr Opin Immunol. 1999;11:23–27. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 23.Lehrer R I, Rosenman M, Harwig S S L, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 24.Luo P, Baldwin R L. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 25.Mahoney M M, Lee A Y, Brezinski-Caliguri D J, Huttner K M. Molecular analysis of the sheep cathelin family reveals a novel antimicrobial peptide. FEBS Lett. 1995;377:519–522. doi: 10.1016/0014-5793(95)01390-3. [DOI] [PubMed] [Google Scholar]
- 26.Maloy W L, Kari U P. Structure-activity studies on magainins and other host defense peptides. Biopolymers. 1995;37:105–122. doi: 10.1002/bip.360370206. [DOI] [PubMed] [Google Scholar]
- 27.Martin E, Ganz T, Lehrer R I. Defensins and other endogenous peptide antibiotics of vertebrates. J Leukoc Biol. 1995;58:128–136. doi: 10.1002/jlb.58.2.128. [DOI] [PubMed] [Google Scholar]
- 28.Oren Z, Shai Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Méndez O, Vanloo B, Decout A, Goethals M, Peelman F, Vandekerckhove J, Brasseur R, Rosseneu M. Contribution of the hydrophobicity gradient of an amphipathic peptide to its mode of association with lipids. Eur J Biochem. 1998;256:570–579. doi: 10.1046/j.1432-1327.1998.2560570.x. [DOI] [PubMed] [Google Scholar]
- 30.Popsueva A E, Zinovjeva M V, Visser J W M, Zijlmans J M J M, Fibbe W E, Belyavsky A V. A novel murine cathelin-like protein expressed in bone marrow. FEBS Lett. 1996;391:5–8. doi: 10.1016/0014-5793(96)00692-8. [DOI] [PubMed] [Google Scholar]
- 31.Porter E M, van Dam E, Valore E V, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65:2396–2401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selsted M E, Szklarek D, Lehrer R I. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect Immun. 1984;45:150–154. doi: 10.1128/iai.45.1.150-154.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh P K, Jia H P, Wiles K, Hesselberth J, Liu L, Conway B A, Greenberg E P, Valore E V, Welsh M J, Ganz T, Tack B F, McCray P B., Jr Production of β-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skerlavaj B, Benincasa M, Risso A, Zanetti M, Gennaro R. SMAP-29: a potent antibacterial and antifungal peptide from sheep leukocytes. FEBS Lett. 1999;463:58–62. doi: 10.1016/s0014-5793(99)01600-2. [DOI] [PubMed] [Google Scholar]
- 35.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis service on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 36.Tossi A, Scocchi M, Skerlavaj B, Gennaro R. Identification and characterization of a primary antibacterial domain in CAP18, a lipopolysaccharide binding protein from rabbit leukocytes. FEBS Lett. 1994;339:108–112. doi: 10.1016/0014-5793(94)80395-1. [DOI] [PubMed] [Google Scholar]
- 37.Travis S M, Conway B D, Zabner J, Smith J J, Anderson N N, Singh P K, Greenberg E P, Welsh M J. Activity of abundant antimicrobials of the human airway. Am J Respir Cell Mol Biol. 1999;20:872–879. doi: 10.1165/ajrcmb.20.5.3572. [DOI] [PubMed] [Google Scholar]
- 38.Turner J, Cho Y, Dinh N N, Waring A J, Lehrer R I. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valore E V, Park C H, Quayle A J, Wiles K R, McCray P B, Jr, Ganz T. Human β-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Investig. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsh, M. J., B. W. Ramsey, F. Accurso, and G. R. Cutting. Cystic fibrosis. In C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, and B. Vogelstein (ed.), The metabolic and molecular basis of inherited disease, 8th ed., in press. McGraw-Hill Book Co., New York, N.Y.
- 41.Zanetti M, Gennaro R, Romeo D. The cathelicidin family of antimicrobial peptide precursors: a component of the oxygen-independent defense mechanisms of neutrophils. Ann NY Acad Sci. 1997;832:147–162. doi: 10.1111/j.1749-6632.1997.tb46244.x. [DOI] [PubMed] [Google Scholar]
- 42.Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995;374:1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]