Abstract
Although medicinal herbs contain many biologically active ingredients that can act as antibiotic agents, most of them are difficult to dissolve in lipids and absorb through biofilms in the gastrointestinal tract. Besides, silver nanoparticles (AgNPs) have been widely used as a potential antibacterial agent, however, to achieve a bactericidal effect, high concentrations are required. In this work, AgNPs were combined into plant-based antibiotic nanoemulsions using biocompatible alginate/carboxyl methylcellulose scaffolds. The silver nanoparticles were prepared by a green method with an aqueous extract of Allium sativum or Phyllanthus urinaria extract. The botanical antibiotic components in the alcoholic extract of these plants were encapsulated with emulsifier poloxamer 407 to reduce the particle size, and make the active ingredients both water-soluble and lipid-soluble. Field emission scanning electron microscopy (FESEM) and energy-dispersive X-ray (EDX) analysis showed that the prepared nanosystems were spherical with a size of about 20 nm. Fourier transform infrared spectroscopy (FTIR) confirmed the interaction of the extracts and the alginate/carboxyl methylcellulose carrier. In vitro drug release kinetics of allicin and phyllanthin from the nanosystems exhibited a retarded release under different biological pH conditions. The antimicrobial activity of the synthesized nanoformulations were tested against Escherichia coli. The results showed that the nanosystem based on Allium sativum possesses a significantly higher antimicrobial activity against the tested organisms. Therefore, the combination of AgNPs with active compounds from Allium sativum extract is a good candidate for in vivo infection treatment application.
The combination of AgNPs and plant extracts provides synergic antibacterial effect on various E. coli strains.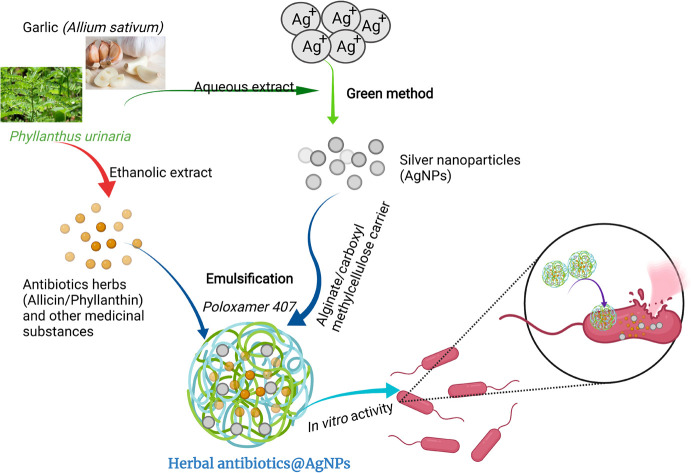
Introduction
Respiratory diseases are common diseases in poultry that significantly affect livestock productivity.1 These diseases often come from bacteria and Escherichia coli is one of the most common types.2 A survey carried out in Ethiopia from 2010 to 2011 indicated that most of the infected yolk sacs in 3- to 5-day-old chicks were caused by E. coli strains, followed by Staphylococcus aureus.3 Currently, antibiotics play an important role in the prevention and treatment of infectious diseases in livestock.4 However, overuse of antibiotics in feed as growth promoters as well as in disease prevention and treatment poses a huge risk of antibiotic resistance and antibiotic residues in food.5
In order to reduce the amount of antibiotics in livestock, great attention has been paid to replacing antibiotics with natural herbs. Recent medical studies have proved that local medicinal plants with antibacterial activities are very rich and effective in treating many different types of infection. Garlic (Allium sativum) and chamber bitter (Phyllanthus urinaria) are known to be the best natural antibiotics.6,7
Allium sativum, or garlic, a popular component in traditional Asian medicine for common cold treatment, has several therapeutic characteristics based on its antibacterial, antiviral, and antifungal activity.8 The three main bioactive ingredients of Allium sativum include allin, diallyl sulfide and ajoene.9 Allin is the strongest and the most important active ingredient of garlic. Allinase enzyme released when garlic cloves are cut or crushed will rapidly turn allin into allicin (C6H10OS2). Allicin has a very strong bactericidal effect against Staphylococcus, typhoid, paratyphoid, dysentery, cholera, and diphtheria bacilli.10,11 However, allicin is unstable and highly reactive to temperature, thus easily decomposed into other stable organosulfur compounds. In addition, garlic also has antioxidant effects by scavenging free radicals and inhibiting the oxidation of low-density lipoproteins.12
Additionally, Phyllanthus urinaria, a frequently used herbal medication, has numerous biological properties including anti-virus, anti-bacteria, and anti-tumor properties.13,14 Lignans, tannins, flavonoids, phenolics, terpenoids, and other secondary metabolites were abundant in P. urinaria. Eldeen et al. reported that P. urinaria inhibited growth of Pseudomonas stutzeri (Gram-negative) with MIC values of 177 μg mL−1. Besides, P. urinaria had a high total phenolic content of 205 mg per GAE per g, which is correlated with DPPH radical scavenging activity.15 The presence of metabolites such as phyllanthin, phyltetralin, rutin, quercetin, trimethyl-3,4-dehydrochebulate, and methyl brevifolincarboxylate is the underlying mechanism of the antibacterial effect of P. urinaria extracts. These chemicals may bind with proteins in the microbial cell membrane, forming persistent water-soluble complexes that cause microbial cell death.16
Although the in vitro and in vivo bactericidal effects of herbs in disease prevention have been identified, the practical uses of herbal antibiotics are limited because of the in vivo instability, aqueous insolubility, and poor absorbability in gut. Herbal antibiotics are also less potent than conventional antibiotics, thus a higher dose and longer time are required, thus affecting livestock productivity. Therefore, a suitable method is necessary to improve the therapeutic effect of herbal antibiotics. One of the effective ways to enhance the antibacterial ability of herbal antibiotics is the application of nanotechnology. Numerous studies have shown that the important active ingredients in plants are enhanced in efficacy and exhibit many advantages in their biological applications using nanotechnology.17 Nanoformulation of herbal antibiotics can enhance solubility, stability and bioavailability, pharmacological activity, as well as improve tissue macrophage distribution.18 Moreover, nanoform of herbal antibiotics can also reduce their toxicity and decomposition by the physiological environment, and the drug release process can be also controlled.19,20
In addition, silver nanoparticles (AgNPs) have been shown to have excellent microbial resistant properties, thus it has long been used as disinfectants in food and water containers and as a disease treatment agent.21,22 The antibacterial properties of AgNPs are mainly attributed to the ability of Ag+ ions to inhibit bacterial cell respiration and prevent bacterial DNA replication.23,24 To synthesize AgNPs, more and more efficient green synthesis methods have been developed in order to overcome the constraints of physical and chemical procedures. The biological synthesis of AgNPs using plants has been proven to be cost-efficient and eco-friendly and is a valuable alternative for large-scale production.25–27 Interestingly, green synthesized AgNPs expressed stronger antibacterial potency than chemically produced ones.28 Yang et al. also demonstrated that the antimicrobial activity of AgNPs and Lonicera japonica Thunb extract was significantly increased when compared with solely AgNPs or herbal extract.29
Herein, the two herbal extracts of Allium sativum and Phyllanthus urinaria were used to synthesize AgNPs. The AgNPs were then combined with each emulsified herbal extract and encapsulated by alginate/carboxymethyl cellulose carrier to form the Allium@AgNPs and Phyllanthus@AgNPs systems. Usually, alginate and carboxymethyl cellulose used in drug delivery are usually formed hydrogel by Ca2+ cross-linking. However, this formulation possessed the notable drawbacks of instability and rapid dissolution.30,31 In the present study, alginate/carboxymethyl cellulose carrier was fabricated by chemical NH–CO bond, thus facilitating the formation of polymeric micelle-based nanoformulation. The physico-chemical characteristics of the nanoformulations were determined to evaluate the effect of different herbal extracts on the Ag+ reduction efficiency. Thereafter, based on the results of antimicrobial activity against E. coli test, a potential antibiotic agent could be recommended for further investigating of its applicability in batling infection at in vivo scale.
Experimental
Materials
The garlic white hybrid variety (Allium sativum L.) and chamber bitter (Phyllanthus urinaria L.) were procured from the Vietnam market. Silver nitrate, ethanol, sodium alginate, poloxamer 407, sodium carboxymethyl cellulose (CMC), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), triethylamine, Luria–Bertani (LB) broth, and phosphate buffer saline (PBS) were purchased from Sigma Aldrich. 3-Aminopropyl triethoxysilane (APTES) was from Merck. The E. coli strains, including V19.11.3; S19.3.6; S19.3.14, S193.3.4, S19.3.11, were provided by the Microbiology – Infectious Laboratory, Faculty of Veterinary Medicine, Vietnam National University of Agriculture.
Preparation of Allium sativum extracts
Garlic cloves were separated and peeled for the preparation of garlic extracts with different solvents. 0.5 kg of the fresh bulb garlic was peeled off, pounded and pressed to get raw water extract, then extracted in 1 L of ethanol at 60 °C for 5 hours. The extraction was performed 3 times and the ethanolic extract was obtained after filtration through a cellulose membrane (pore diameter 0.22 μm). The residue of ethanolic extraction was further extracted with 1 L of water for 5 hours at 60 °C. The process was, repeated 3 times and the aqueous extract was collected after filtration through a cellulose membrane (pore diameter 0.22 μm). Both aqueous and ethanolic extracts were stored at room temperature until further use.
Preparation of Phyllanthus urinaria extracts
The whole chamber bitter (Phyllanthus urinaria, PU) plant was washed, chopped, and dried at 40–50 °C. 300 grams of pulverized chamber bitter were placed in a cold-soaked flask with 1 L of ethanol and sonicated for 5 hours at 60 °C. The subsequent extraction steps were performed in the same way as for garlic and the final products were obtained with two fractions of ethanolic extract and aqueous extracts.
Synthesis of the herbal antibiotics@AgNPs
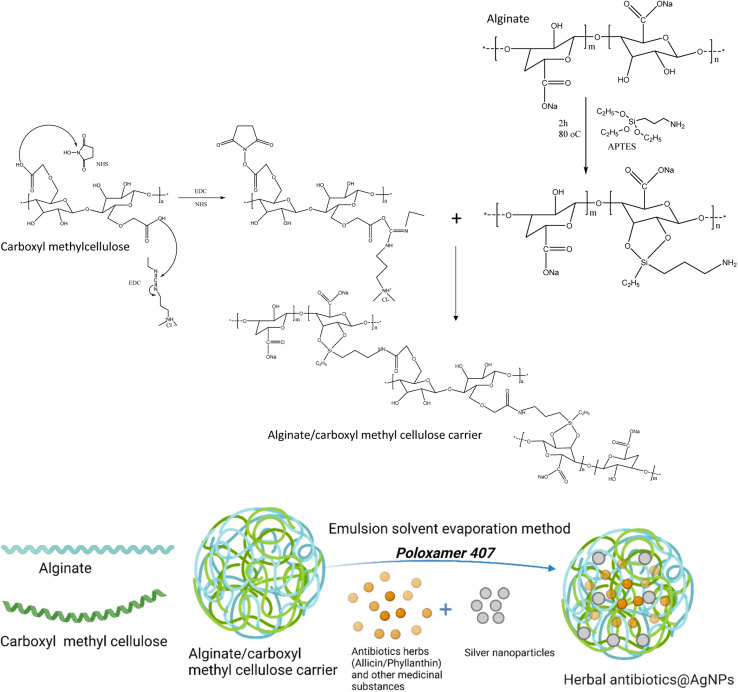
Fig. 1 shows the synthesis process of the herbal antibiotic@AgNPs. First, APTES (10% acid alcohol solution) was added to sodium alginate (1 mg mL−1) and stirred on a magnetic stirrer at 400 rpm for 2 h (80 °C). The APTES reacts with the alginate's hydroxyl groups, promoting silanol group formation with some amine groups in the alginate molecules that are so-called activated alginate. Then, CMC was activated by the presence of EDC and NHS. A premixed solution of NHS (25 mg), EDC (100 mg), and carboxymethyl cellulose (CMC, 150 mg) in 20 mL of deionized water was added. The pH of the solution was then adjusted to 8.5 with triethylamine. The mixture was stirred for 4 h in a closed flask at 55 °C and the activated CMC was obtained. Finally, activated alginate (with –NH2 groups in the molecules) was allowed to react with activated CMC for 24 h at room temperature to obtain the alginate/carboxymethyl cellulose carrier.
Fig. 1. Schematic representation for preparation of herbal antibiotics@AgNPs.
Encapsulation of active compounds in ethanolic plant extracts has been done by an emulsion solvent evaporation method. The ethanolic extract was added dropwise into alginate/CMC solution with a ratio of 1 : 5 (v/v) under vigorously stirring at 350 rpm. Then, 1.0 mL of 0.1 M silver nitrate (AgNO3) was added slowly to the above mixture, followed by the aqueous extract with a 1 : 5 (v/v) ratio of AgNO3: the aqueous extract, and stirred continuously for 1 h. The solution was stirred overnight at room temperature, and then ethanol was evaporated under vacuum pressure. The product was centrifuged at 10 000 rpm for 10 min to remove unloaded compounds. Finally, pre-dispersed overnight at 0–5 °C poloxamer 407 solutions (at various ratios w/v from 0 to 10%) were slowly added to the obtained nanosystem. Thus, we obtained two series of nanosystems based on alginate/CMC carrier, one with Allium sativum extract and AgNPs (referred to as Allium@AgNPs) and the other with Phyllanthus urinaria extract and AgNPs (referred to as Phyllanthus@AgNPs). Similar procedure except for the use of AgNO3 was utilized to prepare non-Ag nanosystems of Allium@NPs, Phyllanthus@NPs for comparison. The control AgNPs were synthesized under the same conditions that used sodium borohydride (NaBH4) as the reducing agent instead of the herbal aqueous extract.
Physico-chemical characterization
UV-vis spectroscopic analysis
The reduction of Ag+ ion and capping of the resulting silver nanoparticles was monitored by UV-vis spectra recorded on a Cary 5000 UV-vis-NIR double beam spectrophotometer (Agilent Technologies, Santa Clara, USA).
X-ray diffraction (XRD)
The crystalline structures of the synthesized systems were analyzed with a Bruker D8-Advance instrument operating at 35 kV and 30 mA, in the reaction mode with Cu Kα line of 1.5406 Å. Data were collected over a 2θ range of 30° to 70°, step size 0.02° and time per step of 4 s at room temperature. The detailed structural characterization was analyzed with the Rietveld method.
Fourier transform infrared (FTIR) spectroscopy
The molecular structure of materials was characterized by Fourier transform infrared spectroscopy (FTIR, SHIMADZU spectrophotometer) using KBr pellets in the wave number region of 400–4000 cm−1.
Field emission scanning electron microscopy
The morphology of the samples was analyzed using a field emission scanning electron microscopy (FESEM) of Hitachi S-4800. Samples were deposited on a Si wafer, dried, and inserted in the instrument without further coating. The measurement was performed at energies between 5 and 10 keV. Energy-dispersive X-ray mapping of the herbal antibiotics@AgNPs was obtained using the FESEM instrument equipped with an energy-dispersive X-ray spectroscopy (EDXS) attachment.
Transmission electron microscopy (TEM)
The morphology and dimensions of the materials were also analyzed in a JEOL JEM-1010 system operating at 120 kV.
Dynamic light scattering (DLS)
The size distribution of the hydrodynamic diameter of the herbal antibiotics@AgNPs and the stability of the suspension were examined using Zetasizer (Zetasizer Nano, Malvern Instruments, UK). The particles were diluted in water at 10 mg L−1 and ultrasonicated (Sonics Vibra Cell, 8 kJ, power 70%, pulse on/off 1 s/1 s). Each sample was measured 3 times.
Thermogravimetric analysis (TGA)
The thermal stability of nanoparticles was determined by thermogravimetric analysis, for which the nanoparticles in powder form were performed on a Discovery TGA (TA Instruments, New Castle, DE, USA). For analyzing TGA, the nanoparticles were placed in an alumina pan and heated from 25 to 800 °C at a ramping time of 10 °C min−1.
Quantification of plant extracts in the nanosystems
Allicin and phyllanthin are standard substances used to quantify the encapsulation of Allium sativum extract and Phyllanthus urinaria extract in the nanosystems, respectively.
The amount of allicin and phyllanthin in the nanosystems was determined by High Performance Liquid Chromatography/Mass Spectrometry (HPLC/MS) method on an HPLC-MS SCIEX-X500R QTOF instrument. Preliminary analysis of allicin and phyllanthin standards using a UV detector (Thermo Scientific, UK) with a spectral range of 200–600 nm was performed under chromatographic conditions. Chromatographic conditions included: Agilent ZORBAX Eclipse Plus 95 Å C18 column, column size of 4.6 × 100 mm, packed particle diameter 5 μm; mobile phase of mix methanol and distilled water with gradient 1–5 minutes (100% H2O), 5–10 minutes (0–50% methanol), 10–12 minutes (70% methanol) and keep 70% until the end of 15 minutes; flow rate of 0.5 mL min−1; sample injection volume of 10 μL; UV detector of DAD wavelength 254 nm and 365 nm. MS conditions were ESI-HRMS positive; voltage 5000 V; temperature 450 °C; collision energy 10 eV; ion source gas: 50 psi.
The encapsulation efficiency of the nanosystem was calculated using eqn (1).
| Encapsulation efficiency (%) = (WExtract on the nanosystem/WTotal extract used) × 100 | 1 |
in which WExtract on the nanosystem is the weight of loaded extract and WTotal extract used is the initial weight of the fed extract. The trials were repeated three times, and average values were obtained.
In vitro release study of the extracts from the nanocarriers
At four distinct pH levels, the pH-dependent release behavior was examined. The herbal antibiotics@AgNPs were inserted into a dialysis tube having a Molecular Weight Cut Off (MWCO) of 8 kDa. To mimic a neutral pH environment, 2 dialysis tubes containing Allium@AgNPs and Phyllanthus@AgNPs were dipped into 500 mL of 0.1 M phosphate buffer solution (pH 7.4) separately. Similarly, for the acidic environment, 500 mL of 0.1 M acetate buffer solution (pH 5.5) was used and likewise, 500 mL of 0.1 M phosphate buffer solution (pH 6.8) and buffer solution pH 1.2 (reference standard, ∼2.0 g per L sodium chloride and ∼2.917 g per L HCl) was used to achieve simulated gastric fluid. The dialysis tubes were placed into beakers containing solutions to imitate different pH environments. The entire setup was stirred at 37 °C. After a certain time interval, 3 mL of the dialysate was withdrawn from each of the beakers to determine the release profile of the materials. The withdrawn amount of the dialysate was replaced with an equal quantity of the fresh buffer solution to avoid the sink conditions. Measurements were performed in triplicate.
Minimal inhibition concentration (MIC) of nanosystems
The MIC test was carried out based on the method suggested by Netala VR et al. with the minor modification.32 The two folds serial dilution of the four nanosystems (Allium@NPs, Phyllanthus@NPs, Allium@AgNPs, and Phyllanthus@AgNPs) in a sterile flat-bottomed 96-well plate were prepared by the following steps: first, 100 μL of sterile LB broth (Sigma-Aldrich, UK) was added to all wells. Then, 100 μL of each nanosystem was mixed to their corresponded row of the 1st column. Within a row, 100 μL of the well in 1st column was transferred to the well in 2nd column, continue to dilute until the column 10, and then discard 100 μL of each well in last column. Thus, it formed the diluting range from 1/2 to 1/1024. Doxycycline antibiotics and AgNPs solution were used as positive control. The initial concentration of tested nanosystems were presented in Table 1.
Initial concentration of antibiotic agents.
| Sample | Concentration of silver nanoparticles | Initial concentration of antibiotics (μg mL−1) |
|---|---|---|
| Allicin | 200 | |
| Allium@AgNPs | 100 ppm | 200 |
| Phyllanthin | 200 | |
| Phyllanthus@AgNPs | 100 ppm | 200 |
| AgNPs | 100 ppm | |
| Positive control (doxycycline) | 800 |
Cytotoxicity activity of nanoantibiotic system against vero cell
The cytotoxic effects of the herbal antibiotics@AgNPs were evaluated following the previously described methods. The vero cell line (from African Green monkey (Cercopithecus aethiops) kidney, ATCC-CCL-81) was incubated in DMEM (Dulbecco's Modified Eagle Medium) at 37 °C in 5% CO2 for 24 h, and seeded into 96-well microplates. The herbal antibiotics@AgNPs was diluted two-fold, and added to the plate. The concentrations of the tested samples were 16 and 8 μg mL−1. In control group (untreated), 100 μL of DMEM were added in the wells instead of herbal nanoformulations. The plate was subsequently incubated at 37 °C in 5% CO2. The cytotoxicity of the samples against vero cells was estimated based on the different of cell phenotypes between treated and untreated group after 48 h of incubation. The cells were then stained with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) agent for 4 h and solubilized to estimates the cell viability. The cell absorbance was measured at 570 nm (Multiskan SkyHigh Microplate Spectrophotometer, Thermo Fisher Scientific, US). The cell viability was calculated as suggested by Mosmann 1983.33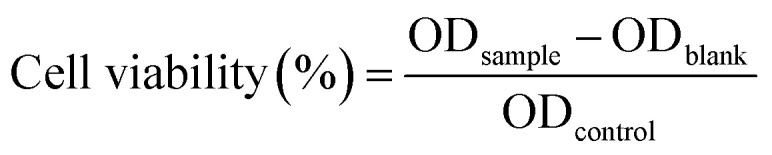
Statistical analysis
Each experiment was performed in triplicate and the data are expressed as the mean ± SD. Statistically significant differences were realized at p < 0.05 via Student's t-test. Statistical analysis was performed using the SigmaPlot 14.0 software.
Results and discussion
Synthesis of herbal antibiotics and AgNPs-encapsulated polymeric nanoparticles (herbal antibiotics@AgNPs)
In this study, in order to bind the activated CMC to the alginate chains, the alginate polymer was treated with APTES (Fig. 1). In the presence of water, APTES hydrolyzes, and the silanol groups condense with the hydroxyl groups of the alginate polymer to create C–O–Si covalent bonds.34 This method provided alginate polymer with the amino group essential to create a CO–NH bond when reacted with the activated carboxylic group of CMC polymers. The final chemical binding system was also confirmed by FTIR spectra (Fig. S1†). Typical oscillations of functional groups in alginate, CMC components and Si–O symmetric stretching vibrations also appeared in FTIR spectrum of the alginate/CMC carrier with some shifts (Table S1†).
The herbal antibiotics@AgNPs were synthesized by emulsion solvent evaporation method. Poloxamer 407 was used in the nanoformulation as a surfactant to optimize the physiochemical properties of the nanosystems. Table 2 summarizes the synthesis parameters and the results obtained from the Allium@AgNPs and Phyllanthus@AgNPs at emulsifier (poloxamer 407) concentrations of 0%, 1%, 3%, 5%, and 10%, respectively. As outlined in Table 2, an increase in poloxamer 407 concentration produces a reduction of both particle size and PDI of the herbal antibiotics@AgNPs. The difference in particle size is comprehensible since, as explained in the literature, more surfactant molecules might cover bigger surfaces of nanoparticles of smaller size.35 The formed nanoparticles had a mean dynamic size of about 160–300 nm. Nanoparticles smaller than 300 nm are preferred for absorption efficacy via oral administration.36,37 The decreased PDI might be owing to an interaction between the encapsulating polymer and the drug. Still, it could also be due to the decreased scattering intensity found at smaller particle sizes.38 Larger particles frequently demonstrate greater scattering intensity. This alters the ratio of the scatter signal of the main particle fraction to the scatter signal of small-particulate impurities, which has an effect on the PDI.39 With the addition of poloxamer 407, the PDI values are below 0.3 indicating the uniformity and narrow size distribution of the herbal antibiotic@AgNPs.40
Characteristics of the herbal antibiotics@AgNPs prepared by the single emulsion method.
| Sample | % poloxamer 407 (w/v) | Size (nm) | Polydispersity index (PDI) | Zeta potential (mV) | Entrapment efficiency (%) |
|---|---|---|---|---|---|
| Allium@AgNPs | 0 | 311.5 ± 8.66 | 0.394 ± 0.19 | −25.2 ± 0.4 | 47.54 ± 1.24 |
| Allium@AgNPs | 1 | 295.82 ± 3.17 | 0.234 ± 0.04 | −33.1 ± 1.1 | 64.12 ± 1.96 |
| Allium@AgNPs | 3 | 262.18 ± 2.05 | 0.188 ± 0.09 | −39.5 ± 0.7 | 72.67 ± 0.63 |
| Allium@AgNPs | 5 | 225.90 ± 6.14 | 0.117 ± 0.01 | −44.3 ± 0.4 | 79.80 ± 1.77 |
| Allium@AgNPs | 10 | 188.36 ± 1.96 | 0.071 ± 0.006 | −38.8 ± 0.6 | 88.24 ± 1.20 |
| Phyllanthus@AgNPs | 0 | 297.9 ± 3.75 | 0.525 ± 0.07 | −31.6 ± 1.5 | 32.17 ± 3.64 |
| Phyllanthus@AgNPs | 1 | 276.37 ± 1.68 | 0.259 ± 0.01 | −35.7 ± 2.1 | 58.33 ± 2.67 |
| Phyllanthus@AgNPs | 3 | 223.38 ± 4.17 | 0.217 ± 0.01 | −38.8 ± 0.5 | 64.25 ± 3.13 |
| Phyllanthus@AgNPs | 5 | 166.11 ± 2.28 | 0.191 ± 0.008 | −39.5 ± 0.5 | 82.68 ± 2.33 |
| Phyllanthus@AgNPs | 10 | 168.25 ± 2.33 | 0.122 ± 0.002 | −42.1 ± 1.1 | 85.17 ± 2.22 |
Poloxamer 407 has an amphiphilic nature with both association and adsorption characteristics, and it can improve chemical solubilization and stability due to the remarkable physiological features and low toxicity. In theory, non-ionic surfactants like poloxamer 407 have no effect on the zeta-potential.41 However, in this study, the absolute value of zeta-potential of the herbal antibiotic@AgNPs increased when the poloxamer concentration changed from 1% to 10% w/v with the least negative zeta potential of −33.1 mV. As the concentration of poloxamer 407 increased, a dense surfactant film was formed at the interface between the nanoparticles and water, increasing the formulation's zeta potential (more negative).42 Furthermore, because poloxamer 407 is an amphoteric surfactant, it can give further emulsion stability via electrostatic repulsion.41 Although there was a slight decrease in the zeta potential of Allium@AgNPs at 10% w/v of poloxamer 407, the zeta potential of −38.8 mV indicates the stability of the nanoparticles. Higher zeta potential reduces aggregation by electrostatic repulsion between similarly charged particles, imparting stability to nanoparticle dispersion.43
The encapsulation efficiency of the herbal antibiotics@AgNPs was calculated based on the content of allicin and phyllanthin by HPLC-MS analysis. Allicin and phyllanthin accounted for 0.025% and 0.2% of the Allium sativum and Phyllanthus urinaria extract, respectively (Fig. S2 and S3†). All herbal@AgNPs formulations prepared with poloxamer 407 showed significantly higher entrapment efficiency than without poloxamer 407. The results also indicate that poloxamer 407 concentration had a significant effect on entrapment efficiency; in which increasing poloxamer concentration also increases entrapment efficiency. With 10% poloxamer 407, the encapsulation efficiency was 88.24% and 85.17% for allicin and phyllanthin, respectively. This might be due to the stronger binding contacts between the drug and the polymeric carrier. The alginate/CMC carrier entrapped hydrophobic allicin and phyllanthin at the interface while poloxamer 407 stabilizes the nanoparticles by diffusing out the water molecules, resulting in the formation of the polymer-rich coacervate during the nanoemulsion process. This is in agreement with other reports.44,45
Overall, nanosystem with 10% poloxamer 407 was found to have the smallest particle size, PDI, and zeta potential with highest entrapment efficiency, which would be the most stable formulation. Therefore, this nanoformulation was chosen for some selected further studies.
Physico-chemical characterization of herbal antibiotics@AgNPs
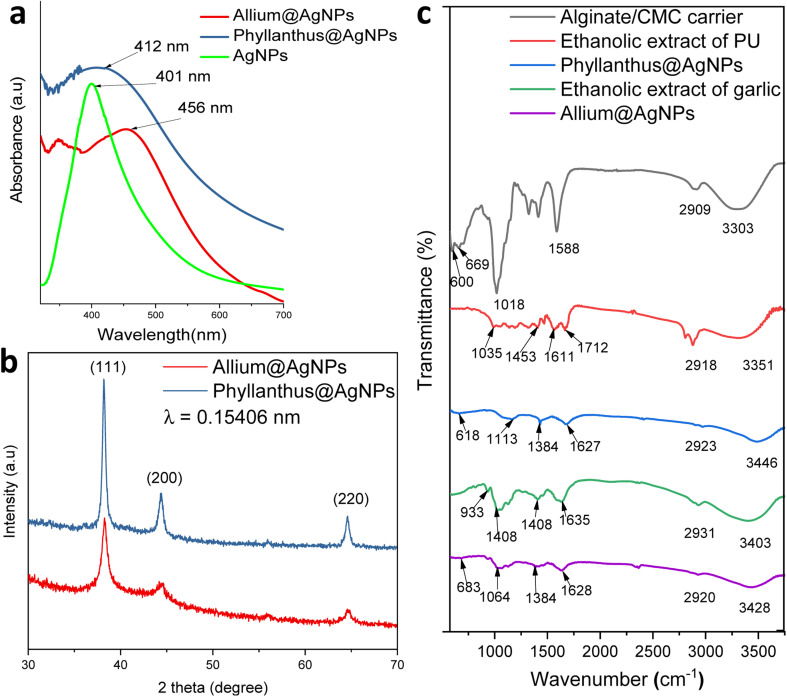
The formation of silver nanoparticles in the herbal antibiotics@AgNPs systems has been elucidated by FESEM and UV-vis spectra. As presented in Fig. 2a, in the wavelength range of 300–800 nm, the AgNPs showed a surface plasmon resonance (SPR) peak at 401 nm. According to previous studies,25,46,47 green synthetic silver nanoparticles have plasmon resonance absorption peak between 400 nm and 460 nm. The SPR peaks of Allium@AgNPs and Phyllanthus@AgNPs were red shifted with different degrees compared to those of AgNPs, at 456 nm and 412 nm, respectively. According to Mie's theory, this shifts were caused by the local dielectric effect.48,49 Furthermore, the existence of an SPR band in the UV-vis spectrum of herbal antibiotics@AgNPs nanostructures was indicative of silver crystal formation on the nanosystems, which was completely consistent with the TEM image data (Fig. 3c & d).
Fig. 2. (a) UV-vis absorption spectra, (b) X-ray diffractogram patterns of herbal antibiotics@AgNPs, and (c) FTIR spectra of alginate/CMC carrier, ethanolic extract of PU, Phyllanthus@AgNPs, ethanolic extract of garlic and Allium@AgNPs.
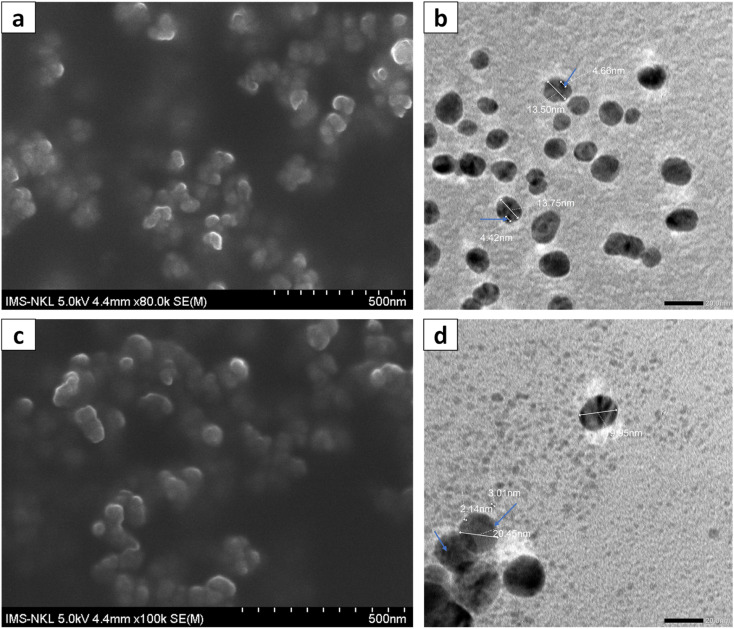
Fig. 3. FESEM (a) and TEM (b) images of Allium@AgNPs; FESEM (c) and TEM (d) images of Phyllanthus@AgNPs (in which silver nanoparticles were marked with blue arrows).
XRD patterns of the synthesized herbal antibiotics@AgNPs are shown in Fig. 2b. Diffraction peaks (Braggs reflections) at 38°, 44° and 64° corresponding to the crystal planes (111), (200) and (220), respectively, were observed in XRD patterns of both Allium@AgNPs and Phyllanthus@AgNPs, revealed the high crystalline nature of pure Ag NPs with a dominant (111) phase.50 The calculated particles size of AgNPs using the Scherrer equation51 was equal to approximately 4.98 nm and 3.25 nm for Allium@AgNPs and Phyllanthus@AgNPs, respectively. On the other hand, there were no sign of Ag2S or Ag2O diffraction peaks. According to the XRD patterns, it seems like only silver metal were formed in both Allium@AgNPs and Phyllanthus@AgNPs. Similar results have been established by Wei and Sun for the biosynthesis of AgNPs using Chinese herbal medicine as well as tea leaf extract, respectively.25,52
Fig. 2c shows FTIR spectra of alginate/CMC carrier, two herbal antibiotics@AgNPs in comparison with those of the two extracts (Allium sativum and Phyllanthus urinaria). The alginate/CMC carrier exhibited a broad band around 3303 cm−1 for the OH stretching vibrations, and a medium peak at 2909 cm−1 belonged to the stretching vibrations of C–H (sp3). Two bands at 1588 cm−1 and 600 cm−1 for N–H bending. Two bands were observed at 1413 cm−1 and 1323 cm−1 for –COO− symmetric stretching vibration and –C–O stretching, respectively, while the antisymmetric C–O–C stretch was observed at 1080 cm−1. Also, the Si–O symmetric stretching mode was observed at 659 cm−1. In the FTIR spectrum of Phyllanthus urinaria's ethanolic extract, a reasonably wide band at wavenumber 3351 cm−1 indicates the presence of strain vibrations of the O–H group, and the peak at 1035 cm−1 was due to C–O–C stretching vibration. The broad absorption peak at 1712 cm−1 and 1611 cm−1 corresponded to C O asymmetric stretching vibration, and the narrow absorption peak at 1453 cm−1 corresponded to –COO− symmetric stretching vibration. The appearance of these characteristic bonds is consistent with the presence of phyllanthin, phenolic, and flavonoid compounds in Phyllanthus urinaria extract.14,16,53 Major peaks of Phyllanthus@AgNPs showed at 3446, 2923, 1627, 1384 and 1113 cm−1 which have been assigned to the O–H stretching, –CH stretching, the asymmetric, the symmetric vibration of –COO stretching and C–O–C stretching, respectively. The absorption peak of Phyllanthus@AgNPs at the wave number of 638 cm−1 was caused by Si–O symmetric stretching vibration. The FT-IR analysis indicated the involvement of amides, carboxyl, amino groups, and polyphenols in the Phyllanthus@AgNPs. From this fact, it was inferred that the organic compounds in the ethanolic extract was encapsulated successfully in Phyllanthus@AgNPs systems by the alginate/CMC carrier.
The FTIR spectrum of garlic's ethanolic extract showed the presence of functional groups like hydroxyl, carbonyl, carboxylic and organosulfur compounds (Fig. 2c). The broad peak at 3403 cm−1 is due to the O–H stretching of a hydroxyl group. The peak at 1635 cm−1 corresponds to C O stretching of peptide linkages or C O stretching of carbonyl and carboxylic groups while the peak at 1408 cm−1 indicates the O–H bend of carboxylic acids. These characteristic peaks showed the existence of polyhydroxy compounds such as flavonoids, tannins, saponins and glycosides in the garlic ethanolic extract.54,55 Another peak at 1028 cm−1 was due to the S O group for the presence of organosulfur compounds including allin, allicin, and diallyl disulfide.56 A peak at 933 cm−1 belongs to C–S single bond. For the spectrum of Allium@AgNPs, additional bands, compared with garlic's ethanolic extract and alginate/CMC carrier, were detected at 3248 cm−1 (–OH), 2920 cm−1 (CH stretching), 1628 cm−1 (C O stretching), 1384 cm−1 (O–H bend of carboxylic acids), 1064 cm−1 (S O group), 930 cm−1 (C–S bond), and 683 cm−1 (Si–O), confirming the successful preparation of Allium@AgNPs.
The morphology and particle size of the herbal antibiotics@AgNPs are shown in FESEM and TEM images. The TEM images (Fig. 3b and d) showed that most of herbal antibiotics@AgNPs were mono-dispersed with spherical shapes, which was similar with the FESEM image (Fig. 3a and c). The Allium@AgNPs were distributed separately, while some Phyllanthus@AgNPs were trapped in the blurred membrane and were observed as clusters. The particle sizes determined from TEM analysis were 13.5 and 20.45 nm for Allium@AgNPs and Phyllanthus@AgNPs, respectively. Meanwhile, the silver nanoparticles (the black particles present in the polymeric nanocarriers) integrated on the Phyllanthus@AgNPs system are 2–3 nm in size, smaller than those in the Allium@AgNPs system (4–5 nm).
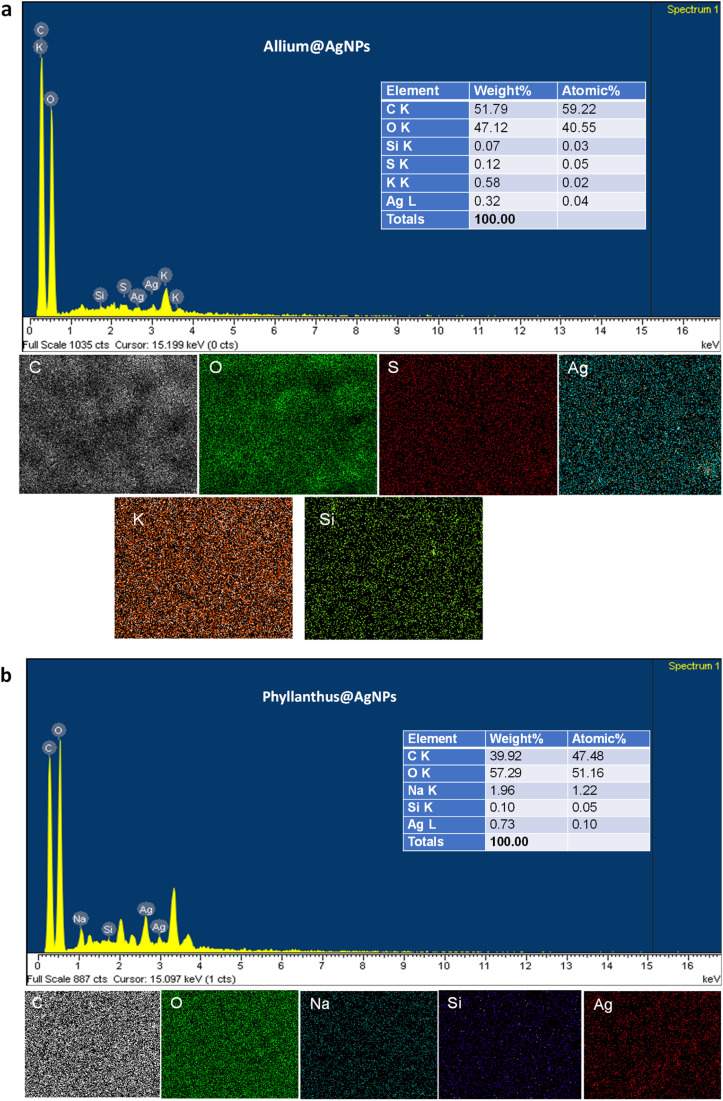
To corroborate the chemical analysis, an EDX analysis of the samples was also carried out. The EDX spectra showed the peaks characterized for silver elements at 3 keV and 2.7 keV, which confirms the formation of AgNPs synthesized with the aqueous extract of Allium sativum and Phyllanthus urinaria, respectively. Next, we performed elemental mapping of the selected region in the SEM scanned image of the nanoparticles. The mapping results showed a clear map of silver elements in Allium@AgNPs and Phyllanthus@AgNPs scanned images. The EDXS Analysis confirms the presence of carbon (C), oxygen (O), sulphur (S), silica (Si), and silver (Ag) in Allium@AgNPs as shown in Fig. 4a. Similarly, Fig. 4b depicts the presence of carbon (C), oxygen (O), sodium (Na), silica (Si), and silver (Ag) in Phyllanthus@AgNPs. The peak for C and O suggests the organic ingredients of herbal antibiotics@AgNPs such as the polymer alginate, chitosan and the bioactive compound in Allium sativum or Phyllanthus urinaria extracts.
Fig. 4. Energy dispersive X-ray (EDX) spectrum and mapping analysis of (a) Allium@AgNPs and (b) Phyllanthus@AgNPs. C: carbon; O: oxygen; Na: sodium; S: sulphur; Si: silica; Ag: silver.
To further confirm the presence of organic compounds in the nanoparticles, the thermal behavior of the obtained herbal antibiotics@AgNPs was investigated (Fig. S4†). The evaporation of moisture adsorbed on the surface of the herbal antibiotics@AgNPs might be attributed to the first weight loss step of the heating process, which occurred between 80 and 120 °C. The next steps in the TGA curve could be assigned to the degradations of organic compounds present in the nanomaterials and the carbonization of the polymeric carrier (Table S2†). The TGA results clearly showed that the bioactive organic compounds from the herbal extract were encapsulated in the produced nanoparticles.57,58
In vitro drug release of herbal antibiotics@AgNPs
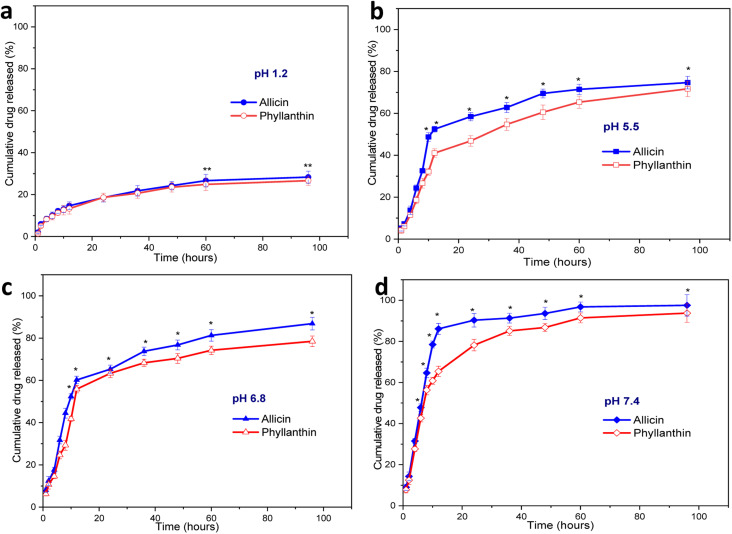
A drug release study of allicin and phyllanthin from the herbal antibiotics@AgNPs was carried out using four different conditions at 37 °C. Different buffer solutions were employed to simulate the gastric medium (pH 1.2), the macrophage environment (pH 5.5), the intestinal condition (pH 6.8), and the physiological condition (pH 7.4) (Fig. 5).
Fig. 5. Cumulative release of allicin and phyllanthin from Allium@AgNPs and Phyllanthus@AgNPs, respectively at preselected time intervals in several condition (a) pH 1.2; (b) pH 5.5; (c) pH 6.8; (d) pH 7.4 PBS. Results were reported as mean ± SD, n = 3, **p < 0.001; and *p < 0.05.
At pH 1.2, the amount of allicin released was very low, only 12.2% and 26.67% of allicin were released after 8 hours and 60 hours, respectively (Fig. 5a). In the same case, the amount of phyllanthin released were 11.33% after 8 hours and 24.8% after 96 hours of testing. The release of allicin and phyllanthin were very low due to the poor aqueous solubility of the substances at a strong acidic pH.59,60 Therefore, the drug encapsulated nanoformulations were quite stable and a very limited drug were released in this acidic environment.
When the pH was raised to 5.5, there was a significant increase in drug release at each time point. From Allium@AgNPs, 58.5% of allicin was released after 24 hours, and after 60 hours, the amount of allicin released reached 71.45% (Fig. 5b). Besides, after 24 hours, 46.8% of phyllanthin was released from the Phyllanthus@AgNPs nanoparticles, and after 60 hours, 60.5% the amount of phyllanthin were released. While the extracellular pH in the blood remains stable at 7.35–7.45, inflammatory situations are associated with acidification, with pH levels ranging from 5.5 to 7.0.61 Acidic microenvironments are also described in the pathological environment of inflammatory exudates as well as the intracellular environment of infected macrophages.62 With the characteristic of gradual release at pH 5.5, both Allium@AgNPs and Phyllanthus@AgNPs have the potential to maintain the effect of the drug at the pathological site.
With the further increase of the pH to 6.8 (characterized pH of the colon environment, the first part of the small intestine of the digestive system, where most of the absorption of the drug into the blood occurs), the nanoparticles showed an even higher rate of drug release. The amount of allicin released in the first 8 hours and after 6 hours was 44.5% and 81.35%, respectively (Fig. 5c). Under same condition, the released amount of phyllanthin, was 29.3% in the first 8 hours and 74.27% after 60 hours.
Under physiological conditions (pH 7.4), both drugs were almost completely released from the nanosystems after 96 hours, 93.76% and 97.55% after 95 hours for Allium@AgNPs and Phyllanthus@AgNPs, respectively. The release profiles of Allium@AgNPs and Phyllanthus@AgNPs nanosystems showed two-stage release with an initial burst release within the first 12 hours followed by a slower and continuous release over the next 84 hours (Fig. 5d). Moreover, in all the four pH conditions, the released amount of allicin was higher than that of phyllanthin (p < 0.001). This can be due to the fact that allicin has better water solubility than phyllanthin, so it easily diffuses through the alginate/carboxyl methyl cellulose carrier.60,63
It is noted that both allicin and phyllanthin release profile of the herbal antibiotics@AgNPs is pH dependent due to the carboxyl methyl cellulose was complexed with sodium alginate to form a polyelectrolyte that provided a pH-sensitive shell layer.64,65 The substances were released faster at a higher pH than around neutral and acidic pH (pH 7.4 > pH 6.8 > pH 5.5 > pH 1.2). In low pH environment, both alginate and CMC are in acid form which are slightly hydrophobic thus preventing the interaction between water and the active ingredient. But in neutral environment, they were both in base form, and the drugs were released more easily due to ionization.64,66 As expected, drug release was prolonged in pH 5.5, whereas in pH 7.4, drug release was rapid in the first 24 h. Efficient allicin release was achieved at pH 7.4 (90.3%), while only 52% release was observed at pH 5.5 within 24 h. The release result was similar for phyllanthin. Thus, both types of nanosystems are suitable candidates to be used as nano drug delivery systems for oral administration.67,68
Estimating the MIC of nanoantibiotic systems against E. coli
Table 3 showed that all tested E. coli strains resisted to conventional doxycycline and Phyllanthus@NPs at all tested concentrations. However, the integration of AgNPs improved the antibacterial activity of Phyllanthus@AgNPs. The Phyllanthus@AgNPs had the MIC value of 4 μg mL−1 against E. coli V19.11.3. On the other hand, Allium@NPs alone could inhibit the growth of V19.11.3, V19.3.6, and V19.3.14 at a concentration of 2 μg mL−1. Compared to the Allium@AgNPs, the antibacterial activity of AgNPs solution was lower with the MIC value of 8 μg mL−1 for S19.3.6 and 4 μg mL−1 for S19.3.14. The reduction of ion Ag+ to AgNPs by Allium sativum extract to form the Allium@AgNPs further enhanced Allium@AgNPs bacteriostatic against other E. coli strains such as V19.3.4 and V19.3.11, which previously resisted to Allium@NPs. The results were in agreement with previous studies, in which the synergic effect of AgNPs and herbal extracts against various pathogenic bacteria was proved.69–73 In term of particle size, the small size of AgNPs (4–5 nm) as well as the small size of Allium@AgNPs (13.5 nm) might contribute to high antibacterial property of the nanosystem.74–76 Patra and Baek et al. suggested that the AgNPs could strongly bind to the bacterial cell wall, then disrupt its structure.77 Thus, other antibacterial agents such as herbal extracts could easily penetrate bacteria cells and exert their effects. Besides, Cheow et al. suggested that the lower initial antibiotic exposure, the higher the survival chance of biofilm cells were.78 In this study, obtained results proved the faster release rate of allicin from the nanosystem, compared to phyllanthin at all test pH conditions. Thus, apart from the AgNPs corporation, the burst release of allicin might also play an important role in increasing the overall antibacterial property of the Allium@AgNPs compared to the Phyllanthus@AgNPs.
The MIC values of prepared nano antibiotic systems against E. coli strains (μg mL−1) (n = 3, mean ± SD)a.
| Antibiotic agents | V19.11.3 | S19.3.6 | S19.3.14 | S19.3.4 | S19.3.11 |
|---|---|---|---|---|---|
| Phyllanthus@NPs | R | R | R | R | R |
| Allium@NPs | 2 | 2 | 2 | R | R |
| Phyllanthus@AgNPs | 4 | R | R | R | R |
| Allium@AgNPs | 8 | 4 | 2 | 2 | 4 |
| AgNPs | 8 | 8 | 4 | 2 | 2 |
| Doxycycline | R | R | R | R | R |
R: resistance; (—): not detected.
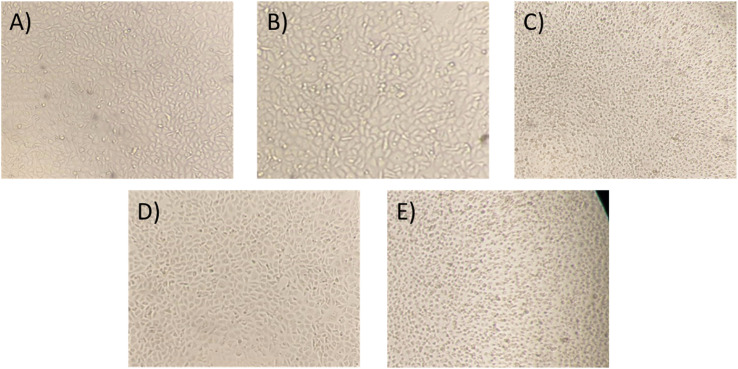
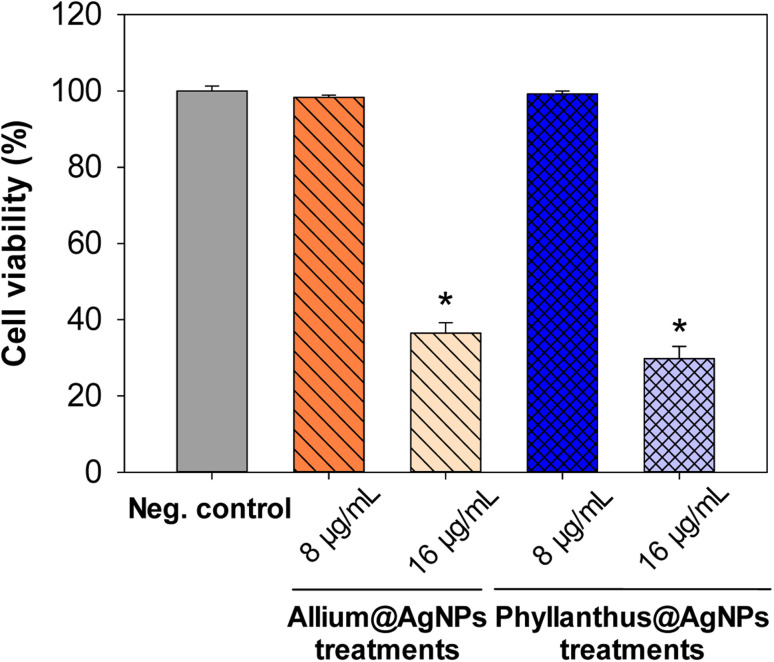
Moreover, to confirm the safety of the antibiotics@AgNPs, the cytotoxicity test using vero cell lines was conducted. Fig. 6 showed the different in cell appearance upon the exposure to high antibiotics@AgNPs concentration of 16 μg mL−1 after 48 h of incubation. These cells tended to look rounded and started to detach from the microplate surface. However, at the antibiotics@AgNPs of 8 μg mL−1, the antibiotics treated cells had almost similar phenotype compared to the negative control. The cell viability in 8 μg mL−1 antibiotic treatments was in a range of 98.3 ± 0.6 to 99.2 ± 0.8%, that was not significantly different from the value of 97.5 ± 1.3% in negative control (Fig. 7). Therefore, the obtained results indicated that both of the Allium@AgNPs and Phyllanthus@AgNPs antibiotics could effectively inhibit the growth of pathogenic bacteria, but did not cause side effects to the normal cell at the required treatment dose in in vitro experiment.
Fig. 6. Phenotype of vero cell in (A) antibiotic-free; (B) Allium@AgNPs at 8 μg mL−1; (C) Allium@AgNPs at 16 μg mL−1; (D) Phyllanthus@AgNPs at 8 μg mL−1; (E): Phyllanthus@AgNPs at 16 μg mL−1 media after 48 h.
Fig. 7. Effect of different herbal nanoformulation treatment on vero cell viability. Results were reported as mean ± SD, n = 3. *p < 0.05.
Conclusions
In the present study, the herbal antibiotics@AgNPs from Allium sativum and Phyllanthus urinaria were successfully synthesized and their properties were assessed by different characterization techniques including UV-VIS, XRD, FESEM, TEM, EDX and HPLC-MS. The optimum conditions were 10% (w/v) poloxamer 407 for both Allium@AgNPs and Phyllanthus@AgNPs. The drug-nanocarrier formulations exhibited much sustained drug release for a prolonged period of time, especially in inflammatory environment. The combination of herbal antibiotics and silver nanoparticles has shown an in vitro antibacterial effect on E. coli. In comparision, Allium@AgNPs nanosystem has better drug release ability and higher inhibitory activity against E. coli than the Phyllanthus@AgNPs nanosystem. As a result, we conclude that the synthesized Allium@AgNPs have the potential to be employed as an eco-friendly agent with antibacterial properties in biomedical applications. In vivo studies of the Allium@AgNPs formulation mentioned in this study will be conducted in the future.
Author contributions
Phan Ke Son, Nguyen Thi Minh, To Xuan Thang, Dong Thi Nham, Le Thi Thu Huong, Nguyen Thanh Trung: investigation; formal analysis; writing – original draft. Pham Kim Dang, Hoang Phuong Ha, Dang Dinh Kim, Phan Thi Hong Tuyet: methodology; writing – review & editing. Mai Thi Thu Trang: conceptualization; methodology, writing – review & editing. Ha Phuong Thu: conceptualization; writing – review & editing; funding acquisition; supervision.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the National Foundation for Science and Technology Development of Vietnam-NAFOSTED under Grant no. 06/2020/TN.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra06847h
Notes and references
- Swelum A. A. Elbestawy A. R. El-Saadony M. T. Hussein E. O. S. Alhotan R. Suliman G. M. Taha A. E. Ba-Awadh H. El-Tarabily K. A. Abd El-Hack M. E. Poult. Sci. 2021;100:101039. doi: 10.1016/j.psj.2021.101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez H. M. Attia Y. A. Front. Vet. Sci. 2020;7:516. doi: 10.3389/fvets.2020.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reda A. Amin A. Shiferaw A. Nazir S. Am.-Eurasian J. Sci. Res. 2013;8(1):10–14. [Google Scholar]
- Kirchhelle C. Palgrave Communications. 2018;4:96. [Google Scholar]
- Van T. T. H. Yidana Z. Smooker P. M. Coloe P. J. J. Global Antimicrob. Resist. 2020;20:170–177. doi: 10.1016/j.jgar.2019.07.031. [DOI] [PubMed] [Google Scholar]
- Darmawan K. JPSCR: Journal of Pharmaceutical Science and Clinical Research. 2017;2:110. [Google Scholar]
- Soni N. Dinda A. Kumar V. J. Herb. Med. 2022;33:100559. [Google Scholar]
- Mozaffari Nejad A. S. Shabani S. Bayat M. Ebrahim Hosseini S. Jundishapur J. Microbiol. 2014;7:e13134. doi: 10.5812/jjm.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang A. Cao S.-Y. Xu X.-Y. Gan R.-Y. Tang G.-Y. Corke H. Mavumengwana V. Li H.-B. Foods. 2019;8:246. doi: 10.3390/foods8070246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P. Moore P. K. Whiteman M. Zhu Y.-Z. Molecules. 2019;24:4006. doi: 10.3390/molecules24214006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratti C. Araya-Farias M. Mendez L. Makhlouf J. Drying Technol. 2007;25:349–356. [Google Scholar]
- Capasso A. Molecules. 2013;18:690–700. doi: 10.3390/molecules18010690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S.-G. Song J. Hong E.-H. Lee B.-R. Kwon Y. Chang S.-Y. Kim S. Lee S. Park J.-H. Ko H.-J. Arch. Pharmacal Res. 2015;38:193–202. doi: 10.1007/s12272-014-0390-9. [DOI] [PubMed] [Google Scholar]
- Nisar M. F. He J. Ahmed A. Yang Y. Li M. Wan C. Molecules. 2018;23:2567. doi: 10.3390/molecules23102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldeen I. M. S. Seow E.-M. Abdullah R. Sulaiman S. F. S. Afr. J. Bot. 2011;77:75–79. [Google Scholar]
- Geethangili M. Ding S.-T. Front. Pharmacol. 2018;9:1109. doi: 10.3389/fphar.2018.01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L. Yang H. Gao S. Li L. Fu X. Wei Q. J. Mater. Chem. B. 2022;10:1908–1922. doi: 10.1039/d1tb02470a. [DOI] [PubMed] [Google Scholar]
- Patra J. K. Das G. Fraceto L. F. Campos E. V. R. Rodriguez-Torres M. del P. Acosta-Torres L. S. Diaz-Torres L. A. Grillo R. Swamy M. K. Sharma S. Habtemariam S. Shin H.-S. J. Nanobiotechnol. 2018;16:71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-Fuentes J. C. Chavez-Santoscoy R. A. Foods. 2021;10:2701. doi: 10.3390/foods10112701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahcheraghi N. Golchin H. Sadri Z. Tabari Y. Borhanifar F. Makani S. 3 Biotech. 2022;12:65. doi: 10.1007/s13205-021-03108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Li X. Zheng Y. Wang Z. Ma Z. Yang Q. Yao B. Zhao Y. Zhang H. New J. Chem. 2018;42:2882–2888. [Google Scholar]
- Gahlawat G. Choudhury A. R. RSC Adv. 2019;9:12944–12967. doi: 10.1039/c8ra10483b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S. K. Jha E. Sahoo B. Panda P. K. Thirumurugan A. Parashar S. K. S. Suar M. RSC Adv. 2017;7:40034–40045. [Google Scholar]
- Yin I. X. Zhang J. Zhao I. S. Mei M. L. Li Q. Chu C. H. Int. J. Nanomed. 2020;15:2555–2562. doi: 10.2147/IJN.S246764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q. Cai X. Li J. Zheng M. Chen Z. Yu C.-P. Colloids Surf., A. 2014;444:226–231. [Google Scholar]
- Roy A. Bulut O. Some S. Mandal A. K. Yilmaz M. D. RSC Adv. 2019;9:2673–2702. doi: 10.1039/c8ra08982e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmy H. M. Mosleh A. M. Elghany A. A. Shams-Eldin E. Abu Serea E. S. Ali S. A. Shalan A. E. RSC Adv. 2019;9:20118–20136. doi: 10.1039/c9ra02907a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibo D. Borbón-Nuñez H. A. de León J. N. D. García Mendoza E. Estrada I. Toledano-Magaña Y. Tiznado H. Ovalle-Marroquin M. Soto-Ramos A. G. Blanco A. Rodríguez J. A. Romo O. A. Chávez-Almazán L. A. Susarrey-Arce A. Sci. Rep. 2020;10:12805. doi: 10.1038/s41598-020-69606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. Aguilar Z. P. Qu F. Xu H. Xu H. Wei H. IET Nanobiotechnol. 2016;10:28–32. doi: 10.1049/iet-nbt.2015.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemiyeh P. Mohammadi S. -Samani. 2019;5:7–24. [Google Scholar]
- Zhang K. Wang Y. Wei Q. Li X. Guo Y. Zhang S. Gels. 2021;7:115. doi: 10.3390/gels7030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netala V. R. Bukke S. Domdi L. Soneya S. G Reddy S. Bethu M. S. Kotakdi V. S. Saritha K. V. Tartte V. Artif. Cells, Nanomed., Biotechnol. 2018;46:1138–1148. doi: 10.1080/21691401.2018.1446967. [DOI] [PubMed] [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Deshmukh K. Kovářík T. Křenek T. Docheva D. Stich T. Pola J. RSC Adv. 2020;10:33782–33835. doi: 10.1039/d0ra04287k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig N. Kammakakam I. Falath W. Mater. Adv. 2021;2:1821–1871. [Google Scholar]
- Wang Y. Pi C. Feng X. Hou Y. Zhao L. Wei Y. Int. J. Nanomed. 2020;15:6295–6310. doi: 10.2147/IJN.S257269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Yin L. Tang C. Yin C. Biomaterials. 2012;33:8569–8578. doi: 10.1016/j.biomaterials.2012.07.063. [DOI] [PubMed] [Google Scholar]
- Malm A. Corbett J. Sci. Rep. 2019;19:13519. doi: 10.1038/s41598-019-50077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajba-Horváth E. Fodor-Kardos A. Shah N. Wacker M. Feczkó T. Int. J. Mol. Sci. 2021;22:13069. doi: 10.3390/ijms222313069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaei M. Dehghankhold M. Ataei S. Hasanzadeh Davarani F. Javanmard R. Dokhani A. Khorasani S. Mozafari M. Pharmaceutics. 2018;10:57. doi: 10.3390/pharmaceutics10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés H. Hernández-Parra H. Bernal-Chávez S. A. Prado-Audelo M. L. D. Caballero-Florán I. H. Borbolla-Jiménez F. V. González-Torres M. Magaña J. J. Leyva-Gómez G. Materials. 2021;14:3197. doi: 10.3390/ma14123197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClements D. J. Jafari S. M. Adv. Colloid Interface Sci. 2018;251:55–79. doi: 10.1016/j.cis.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Lowry G. Hill R. Harper S. Rawle A. Hendren C. Klaessig F. Nobbmann U. Sayre P. Rumble J. Environ. Sci.: Nano. 2016;3:953–965. [Google Scholar]
- Sharma N. Madan P. Lin S. Asian J. Pharm. Sci. 2016;11:404–416. [Google Scholar]
- Pezeshki A. Ghanbarzadeh B. Mohammadi M. Fathollahy I. Hamishehkar H. Adv. Pharm. Bull. 2014;4:563–568. doi: 10.5681/apb.2014.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi K. Venugobal J. Ramasamy V. Appl. Nanosci. 2016;6:399–408. [Google Scholar]
- Balan L. Chandrasekaran S. Gajendiran M. Nanjian R. J. Mol. Struct. 2021;1242:130695. [Google Scholar]
- Peng S. McMahon J. M. Schatz G. C. Gray S. K. Sun Y. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14530–14534. doi: 10.1073/pnas.1007524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. Upadhyay C. Mater. Res. Express. 2017;4:105401. [Google Scholar]
- Parit S. B. Karade V. C. Patil R. B. Pawar N. V. Dhavale R. P. Tawre M. Pardesi K. Jadhav U. U. Dawkar V. V. Tanpure R. S. Kim J. H. Jadhav J. P. Chougale A. D. Mater. Today Chem. 2020;17:100285. [Google Scholar]
- Marinkovic N. Sasaki K. Adzic R. J. Electrochem. Soc. 2018;165:J3222–J3230. [Google Scholar]
- Wei S. Wang Y. Tang Z. Xu H. Wang Z. Yang T. Zou T. RSC Adv. 2021;11:1411–1419. doi: 10.1039/d0ra08287b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartini K. Hardianti D. Hadiyat M. Pharmaciana. 2021;11:251. [Google Scholar]
- Youssef A. M. El-Sayed H. S. EL-Nagar I. El-Sayed S. M. RSC Adv. 2021;11:22571–22584. doi: 10.1039/d1ra03819b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Y. Tsai T.-Y. Chen B.-H. Antioxidants. 2021;10:1611. doi: 10.3390/antiox10101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songsungkan J. Chanthai S. Int. Food Res. J. 2014;21:2377–2385. [Google Scholar]
- Yallappa S. Manjanna J. Dhananjaya B. L. Spectrochim. Acta, Part A. 2015;137:236–243. doi: 10.1016/j.saa.2014.08.030. [DOI] [PubMed] [Google Scholar]
- Elemike E. E. Fayemi O. E. Ekennia A. C. Onwudiwe D. C. Ebenso E. E. Molecules. 2017;22:E701. doi: 10.3390/molecules22050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Li X. Liu X. Sato S. Zhang X. Song J. J. Sci. Food Agric. 2015;95:1838–1844. doi: 10.1002/jsfa.6884. [DOI] [PubMed] [Google Scholar]
- Hanh N. D. Sinchaipanid N. Mitrevej A. Drug Dev. Ind. Pharm. 2014;40:793–802. doi: 10.3109/03639045.2013.788010. [DOI] [PubMed] [Google Scholar]
- Behnen M. Möller S. Brozek A. Klinger M. Laskay T. Front. Immunol. 2017;8:184. doi: 10.3389/fimmu.2017.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I. M. Patankar Y. R. Berwin B. Am. J. Physiol. 2018;314:L225–L235. doi: 10.1152/ajplung.00338.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlinghaus J. Albrecht F. Gruhlke M. Nwachukwu I. Slusarenko A. Molecules. 2014;19:12591–12618. doi: 10.3390/molecules190812591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. Hu S. Zhang S. Dong S. Hu J. Kang L. Yang X. Sci. Rep. 2021;11:9142. doi: 10.1038/s41598-021-88503-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaladhare S. Das D. J. Mater. Chem. B. 2022;10:1923–1945. doi: 10.1039/d1tb02848k. [DOI] [PubMed] [Google Scholar]
- Rizwan M. Yahya R. Hassan A. Yar M. Azzahari A. D. Selvanathan V. Sonsudin F. Abouloula C. N. Polymers. 2017;9:137. doi: 10.3390/polym9040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L. Huang T. Chen X. Li W. Yang X. Zhang W. Li M. Gao R. Oncol. Rep. 2021;46:1–14. doi: 10.3892/or.2021.8179. [DOI] [PubMed] [Google Scholar]
- Rajamäki K. Nordström T. Nurmi K. Åkerman K. E. O. Kovanen P. T. Öörni K. Eklund K. K. J. Biol. Chem. 2013;288:13410–13419. doi: 10.1074/jbc.M112.426254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagashetty A. Ganiger S. K. Shashidhar Heliyon. 2019;5:e02794. doi: 10.1016/j.heliyon.2019.e02794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul Das M. Rebecca Livingstone J. Veluswamy P. Das J. J. Food Drug Anal. 2018;26:917–925. doi: 10.1016/j.jfda.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo R. K. Tamuli K. J. Narzary B. Bordoloi M. Sharma H. K. Gogoi K. Bhattacharyya D. R. Chem. Phys. Lett. 2020;738:136893. [Google Scholar]
- Bautista-Guzman J. Gomez-Morales R. Asmat-Campos D. Checca N. R. Molecules. 2021;26:7633. doi: 10.3390/molecules26247633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen M. W. Sufyan M. Nazir R. Naseem A. Shah R. Sheikh A. A. Iqbal M. Appl. Nanosci. 2021;11:2479–2488. [Google Scholar]
- Dong Y. Zhu H. Shen Y. Zhang W. Zhang L. PLoS One. 2019;14:e0222322. doi: 10.1371/journal.pone.0222322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skomorokhova E. A. Sankova T. P. Orlov I. A. Savelev A. N. Magazenkova D. N. Pliss M. G. Skvortsov A. N. Sosnin I. M. Kirilenko D. A. Grishchuk I. V. Sakhenberg E. I. Polishchuk E. V. Brunkov P. N. Romanov A. E. Puchkova L. V. Ilyechova E. Y. Nanotechnol., Sci. Appl. 2020;13:137–157. doi: 10.2147/NSA.S287658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna T. Maldonado-Bravo F. Jara P. Caro N. Int. J. Mol. Sci. 2021;22:7202. doi: 10.3390/ijms22137202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra J. K. Baek K.-H. Front. Microbiol. 2017;8:167. doi: 10.3389/fmicb.2017.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheow W. S. Chang M. W. Hadinoto K. Pharm. Res. 2010;27:1597–1609. doi: 10.1007/s11095-010-0142-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.