Abstract
Prior to implantation, the cells in the mammalian epiblast constitute a naïve pluripotent state, which is distinguished by absence of lineage priming, freedom from epigenetic restriction, and expression of a unique set of transcription factors. However, human embryonic stem cells (hESCs) derived under conventional conditions have exited this naïve state and acquired a more advanced “primed” pluripotent state that corresponds to the post-implantation epiblast. We have developed a cocktail comprising five kinase inhibitors and two growth factors (5i/L/A) that enables induction of defining features of naïve pluripotency in primed hESCs. These conditions can also be applied to induce naïve pluripotency in patient-specific induced pluripotent stem cells (iPSCs). Here we provide a detailed protocol for inducing naïve pluripotency in primed hESCs and iPSCs and methods for the routine validation of naïve identity. We also outline the use of two fluorescent reporter systems to track acquisition of naïve identity in live cells: (i) a GFP reporter linked to an endogenous OCT4 allele in which the primed-specific proximal enhancer has been deleted (OCT4-ΔPE-GFP); and (ii) a dual color reporter system targeted to both alleles of an X-linked gene that reports on the status of the X chromosome in female cells (MECP2-GFP/tdTomato). The conditions described herein have given insight into various aspects of naïve human pluripotent stem cells (hPSCs), including their unique transposon transcription profile, X chromosome status, and extraembryonic potential.
Keywords: Naïve pluripotency, primed pluripotency, embryonic stem cells, induced pluripotent stem cells, 5i/L/A, X chromosome reactivation
1. Introduction
Studies in the mouse system have demonstrated that pluripotency comprises at least two discrete states: a naïve state that resembles the pre-implantation epiblast and a primed state that corresponds to the post-implantation epiblast [1]. While the former can be captured in vitro in the form of mouse embryonic stem cells (mESCs) [2,3], the latter can be isolated in the form of mouse epiblast stem cells (EpiSCs) [4,5]. Biological and molecular similarities between hESCs and mouse EpiSCs suggested that hESCs acquire a primed identity during derivation under conventional culture conditions. These observations prompted widespread interest in methods to isolate naïve hPSCs from pre-existing primed hPSCs, human blastocysts, or during reprogramming of somatic cells. Various transgenic and chemical approaches have been reported that capture facets of naïve pluripotency in human cells [6-13]. In this chapter, we summarize how we developed the 5i/L/A cocktail for induction of naïve hPSCs and characterized the unique molecular properties of these cells. We then provide a detailed protocol for inducing and validating naïve identity in hESC and iPSC lines.
Naïve and primed pluripotent states in the mouse can be distinguished by their reliance on the distal and proximal enhancers of OCT4, respectively [14,5]. We took advantage of this epigenetic difference to develop a specific reporter system for naïve human pluripotency. TALEN-mediated deletion of the proximal OCT4 enhancer in primed hESCs resulted in pronounced downregulation of an endogenous OCT4-GFP reporter allele [9]. The inducible overexpression of KLF2 and NANOG transgenes enabled the activation of OCT4-Δ-PE-GFP activity in combination with inhibitors of MEK and GSK3 (2i) and the cytokine LIF (2i/L), a cocktail that supports efficient derivation and maintenance of naïve mESCs [15]. However, naïve-specific reporter activity in hESCs could not be maintained in 2i/L alone, prompting us to screen for additional compounds. By performing a small molecule screen, we identified a combination of five kinase inhibitors targeting MEK, GSK3, BRAF, SRC, and ROCK together with LIF (5i/L) that supported the expansion of OCT4-ΔPE-GFP+ and transgene-free naïve hESCs. These conditions could also induce naïve reporter activity directly in primed hESCs when combined with Activin A and FGF2 (5i/L/AF). Transcriptome profiling revealed upregulation of a broad cohort of naïve-specific transcription factors under these conditions. The withdrawal of FGF2 had little impact on expression of naïve-specific transcription factors and was subsequently omitted from the 5i/L/A cocktail [9].
To examine the correspondence of naïve hPSCs to pluripotent cells in vivo, we compared their transcriptome, DNA methylome, and X chromosome status to that of the human pre-implantation embryo [16]. First, we used RNA sequencing to map the expression of transposable elements, which are expressed in a sequential manner during embryonic development [17]. We identified SVA-D and HERVK integrants as being enriched in naïve hPSCs, while HERVH integrants were more abundantly expressed in primed hPSCs. Intriguingly, SVA-D and HERVK integrants are also activated in human morula embryos, suggesting that naïve hPSCs may have an earlier-than-anticipated developmental identity [16]. Second, we profiled the DNA methylation landscape using whole genome bisulfite sequencing and observed a global reduction in 5-methylcytosine methylation from ~75 % in primed hPSCs to ~30 % in naïve hPSCs. Reexposure of naïve hPSCs to primed media, a process known as “re-priming”, resulted in restoration of global DNA methylation levels, recapitulating the gain in DNA methylation seen upon implantation of the human blastocyst [18]. An important exception to this pattern were imprinted gene loci, which displayed erasure of DNA methylation [16,19]. Third, we examined the status of the X chromosome using female hESC lines containing different color fluorescent reporters targeted to the two alleles of MECP2, an X-linked locus. Naïve induction in 5i/L/A resulted in reactivation of the previously silent allele, indicating a switch from monoallelic to biallelic expression [16]. These observations were corroborated by allele-specific gene expression analysis and RNA fluorescence in situ hybridization for X-linked transcripts [20]. Thus, the 5i/L/A cocktail induces transcriptional and epigenetic hallmarks of naïve human pluripotency. Similar molecular attributes of the pre-implantation epiblast are observed in naïve hPSCs maintained in the alternative t2i/L/Gö formulation [10,21-23].
The isolation of bona fide naïve hPSCs presents new opportunities to model the lineage decisions and epigenetic regulation of the human pre-implantation epiblast. Recent work from our lab and others indicates that naïve hPSCs in 5i/L/A or t2i/L/Gö have an enhanced potential for differentiation into human trophoblast stem cells (hTSCs), which can further differentiate into specialized trophoblast cells [24,25] (see also Chapter 7). The molecular basis for this extraembryonic potential remains unclear, but may reflect the fact that segregation between embryonic and extraembryonic lineages is delayed in the human compared to the mouse blastocyst [26]. Naïve hPSCs may also provide a superior cellular platform for modeling X-linked diseases: according to a recent study naïve hPSCs can complete random X chromosome inactivation upon differentiation when the 5i/L/A cocktail is supplemented with FGF receptor inhibitors [27]. However, it is important to bear in mind that current naïve culture conditions require further modifications to improve their stability during extended passaging. In addition to the aforementioned imprint erasure, naïve hPSC cultures are susceptible to genomic instability, particularly in 5i/L/A [19,9]. It remains to be determined whether conditions can be devised that support all defining features of naïve human pluripotency while safeguarding long-term genetic and epigenetic stability.
2. Materials
2.1. Preparation of iMEF plates
Mitotically inactivated mouse embryonic fibroblasts (iMEFs) (see Note 1).
0.1 % Gelatin solution: dissolve 1 g gelatin in 1 L of 1X Dulbecco’s phosphate-buffered saline (DPBS) on a magnetic stirrer. Sterilize the solution by autoclaving and store at 4 °C for up to 2-3 months.
Fibroblast medium (FM): DMEM supplemented with 10 % fetal bovine serum (FBS), 2 mM GlutaMAX, and 1 % penicillin-streptomycin. Filter-sterilize the solution using a 0.22 μm filter and store at 4 °C for up to 2 weeks.
2.2. Primed hPSCs
Primed hPSC medium: DMEM/F12 supplemented with 15 % FBS, 5 % KSR, 1 mM GlutaMAX, 1 % nonessential amino acids, 1 % penicillin-streptomycin, 0.1 mM 2-mercaptoethanol, and 4-8 ng/mL FGF2 (see Note 2). Store at 4 °C for up to 1-2 weeks.
Collagenase IV solution: dissolve 75 mg Collagenase IV in 50 mL DMEM/F12. Filter-sterilize the solution using a 0.22 μm filter.
mTeSR Plus supplemented with 1 % penicillin-streptomycin.
Matrigel hESC-Qualified Matrix, LDEV-free.
Dispase.
STEMPRO EZPassage tool.
CryoStor CS10.
2.3. Induction and expansion of naïve hPSCs
N2B27 medium: A 1:1 mixture of Neurobasal and DMEM/F12 supplemented with 1X N-2 supplement, 1X B-27 supplement, 2 mM GlutaMAX, 1X MEM nonessential amino acids, 1 % penicillin-streptomycin, 50 mg/mL bovine serum albumin fraction V (BSA), and 0.1 mM 2-mercaptoethanol. Media is filter sterilized using a 0.22 um filter and stored at 4 °C for up to 1-2 weeks.
5i/L/A medium: N2B27 supplemented with 1 μM PD0325901, 1 μM IM-12, 0.5 μM SB590885, 1 μM WH4-023, 10 μM Y-27632 (see Note 3), 10 ng/mL Activin A, and 20 ng/mL recombinant human LIF (see Note 4). Media is stored at 4 °C for up to 1-2 weeks.
TrypLE Express.
2.4. Characterization of naïve hPSCs
FACS buffer: 1X DPBS supplemented with 5 % FBS.
E.Z.N.A Total RNA Kit I.
High Capacity cDNA Reverse Transcription Kit.
PowerUp SYBR Green Master Mix.
anti-SUSD2-PE antibody.
anti-CD75-eFluor-660 antibody.
Primer sequences: RPLP0-F, GCTTCCTGGAGGGTGTCC; RPLP0-R, GGACTCGTTTGTACCCGTTG; KLF17-F, CTGCCTGAGCGTGGTATGAG; KLF17-R, TCATCCGGGAAGGAGTGAGA; DNMT3L-F, TTCTGGATGTTCGTGGACAA; DNMT3L-R, ACATCTGGGATGGTGACTGG; DPPA3-F, GTTACTGGGCGGAGTTCGTA; DPPA3-R, TGAAGTGGCTTGGTGTCTTG; DPPA5-F, TGAAAGATCCAGAGGTGTTC; DPPA5-R, ACTGGTTCACTTCATCCAAG; VIM-F, TGTCCAAATCGATGTGGATGTTTC; VIM-R, TTGTACCATTCTTCTGCCTCCTG; SFRP2-F, ACGGCATCGAATACCAGAACA SFRP2-R, CTCGTCTAGGTCATCGAGGCA ZIC2-F, CCCTTCAAGGCCAAATACAA ZIC2-R, TGCATGTGCTTCTTCCTGTC;
3. Methods
Unless otherwise specified, perform all experiments at room temperature and all cell culture procedures in a sterile laminar flow hood. Warm all cell culture media to room temperature before use. Cells are grown in 6-well tissue culture plates.
3.1. Feeder-free culture of primed hPSCs
Prepare a Matrigel-coated 6-well plate and a 15 mL conical tube containing ~10 mL of FM.
Thaw cryopreserved hPSCs at 37 °C until mostly t hawed. Transfer hPSCs to the 15 mL conical tube containing FM.
Centrifuge cells at 250 x g for 3 min. Aspirate the FM.
Resuspend cells gently in 1 mL mTeSR Plus using a 5 mL serological pipet. Transfer cells to Matrigel-coated plates in mTeSR Plus (see Note 5, 6).
Incubate the cells in 5 % CO2 and 20 % O2.
Replace medium on primed hPSCs with 2 mL mTeSR Plus every 1-2 days. Colonies should show flat, well-defined morphology (Fig. 1B, left panel).
Passage primed hPSCs with dispase (steps 8-12) every 5-6 days or when colonies begin to touch or show signs of differentiation.
Aspirate the media from primed hPSCs and wash twice with 1 mL 1X DPBS.
Add 1 mL dispase and incubate at 37 °C for 7 min .
Aspirate the dispase, wash twice with 1 mL 1X DPBS, and add 1 mL mTeSR Plus.
Cut the primed hPSC colonies into small squares using a sterile STEMPRO EZPassage tool (see Note 7). Scratch the square shaped colonies using a sterile glass Pasteur pipette with a rounded tip under a microscope (see Note 8, 9).
Transfer these square-shaped colonies using 200 μL pipette to a new Matrigel-coated plate containing mTeSR Plus.
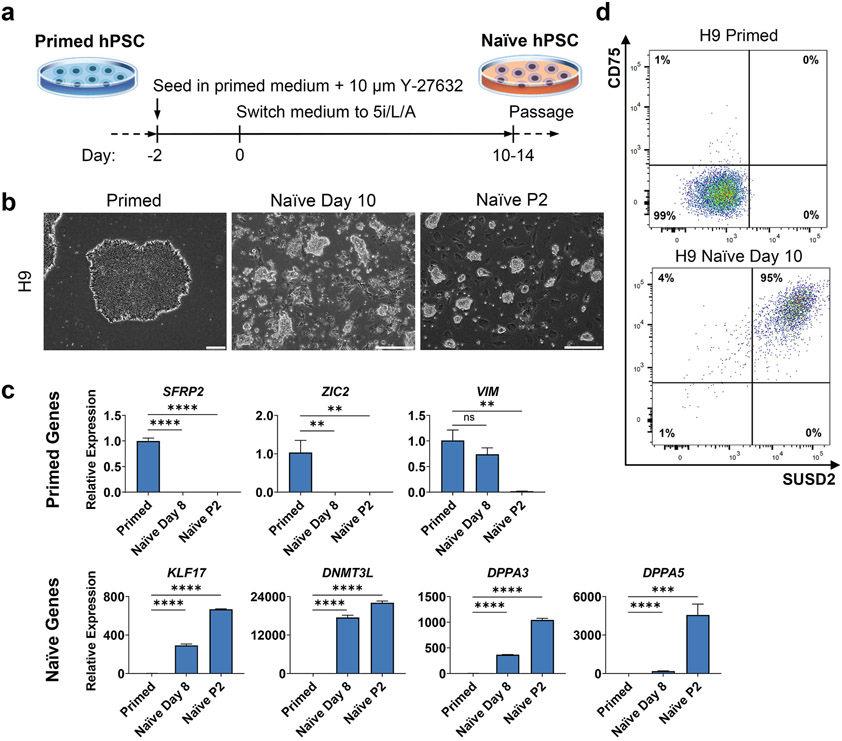
Figure 1. Induction of naïve pluripotency in genetically unmodified H9 primed hESCs using 5i/L/A.
A. Timeline of primed-to-naïve conversion using 5i/L/A.
B. Phase contrast images of primed and naïve H9 hESCs. Scale bars represent 250 μm.
C. Quantitative gene expression analysis of primed and naïve H9 hESCs for primed markers SFRP2, VIM, and ZIC2, and naïve markers KLF17, DNMT3L, DPPA3, and DPPA5. Error bars indicate ± 1 SD of technical replicates. “ns” indicates p-value of >0.05. “**” indicates p-value of <0.01. “***” indicates p-value of <0.001. “****” indicates p-value of <0.0001.
D. Flow cytometry analysis of primed and naïve H9 hESCs for naïve markers SUSD2 and CD75.
3.2. Preparation of iMEF plates
Coat the 6-well tissue culture plate with gelatin solution (1 mL/well) and leave at room temperature for 10 min (see Note 10).
Thaw cryopreserved iMEFs at 37 °C until mostly t hawed and transfer iMEFs to 15 mL conical tubes with ~10 mL FM (see Note 11).
Centrifuge cells at 250 x g for 3 min. Aspirate medium.
Resuspend cells in 1-3 mL FM and count cells (see Note 12).
Aspirate gelatin solution from the 6-well plate and add 2 mL FM to each well.
Add 250,000 iMEFs to each well. Distribute cells throughout each well by gently moving the plate in an “up-down/left-right” pattern (see Note 13).
Incubate overnight at 37 °C in 5 % CO 2 and 20 % O2 (see Note 14).
3.3. Feeder-based culture of primed hPSCs (for WIBR1-3 hESC lines)
Prepare iMEF-coated plates with 2-4 mL primed hPSC medium (see Note 15) and a 15 mL conical tube containing ~10 mL FM.
Thaw cryopreserved hPSCs at 37 °C until mostly t hawed and transfer hPSCs to the 15 mL conical tube containing FM.
Centrifuge cells at 250 x g for 3 min. Aspirate the FM.
Gently resuspend cells in 1 mL primed hPSC medium using a 5 mL serological pipette and transfer to iMEF-coated plates.
Incubate the cells at 37 °C in 5 % CO 2 and 20 % O2.
Remove primed hPSCs medium and replace with 4 mL fresh primed hPSC medium every 1-2 days. Colonies should show a flat, well-defined morphology. Differentiated areas may require removal using mechanical techniques.
Passage primed hPSCs with Collagenase IV (steps 8-14) every 5-6 days or when colonies begin to touch or show signs of differentiation.
Aspirate the medium from primed hPSCs and wash with 1 mL 1X DPBS.
Add 1 mL Collagenase IV solution to the well and incubate at 37 °C for 20 min or until edges of colonies begin to roll up.
After incubation, aspirate the Collagenase IV solution and transfer cells in 5 mL primed hPSC medium to a 15 mL conical tube. Allow the cells to sediment for 20 min.
Aspirate the supernatant containing single cells. Resuspend the pellet five times to reduce the size of the colonies to smaller clumps via P1000 pipetting in 5 mL primed hPSC medium (avoid generating single cells). Allow the cells to sediment for 10 min.
Aspirate the supernatant and gently resuspend the pellet in the required volume of primed hPSC medium five times with a 5 mL pipette and allow the cells to sediment for another 10 min.
Aspirate supernatant containing single cells, resuspend pellet in required volume of primed hPSC medium, and distribute over new iMEF-coated wells.
3.4. Induction of naïve pluripotency in primed hPSCs using 5i/L/A
Aspirate medium from primed hPSCs.
Add 1 mL TrypLE Express to the cells. Incubate at 37 °C for 5 min.
Prepare a 15 mL conical tube with 5-10 mL FM.
Mechanically dissociate cells into single cells using a P1000 pipette and transfer cells to the 15 mL conical tube containing FM.
Spin at 250 x g for 3 min. Aspirate the medium.
For cells cultured in mTeSR Plus, resuspend pellet in 1-3 mL mTeSR Plus supplemented with 10 μM Y-27632. For cells cultured in primed hPSC medium, resuspend pellet in primed hPSC medium supplemented with 10 μM Y-27632.
Count cells (see Note 12).
Aspirate FM from iMEF plates.
For cells that were maintained in mTeSR Plus, add 2 mL mTeSR Plus supplemented with 10 μM Y-27632 to each well containing iMEFs. For cells that were maintained in primed hPSC medium, add 2 mL primed hPSC medium supplemented with 10 μM Y-27632.
Add 200,000 primed hPSCs to each well. Distribute cells throughout each well by gently moving the plate in an “up-down/left-right” pattern.
Incubate overnight at 37 °C in 5 % CO 2 and 20 % O2. This constitutes Day-2 of the naïve conversion protocol (Fig. 1A).
Two days after seeding primed hPSCs, aspirate the medium (see Note 16). Add 4-6 mL 5i/L/A to each well. This constitutes Day 0 of the naïve conversion protocol.
Continue to replace media with 4-6 mL 5i/L/A every 1-2 days (see Note 17, 18). Small dome-shaped naïve colonies should emerge after an initial wave of widespread cell death (Fig. 1B).
Passage cells to a fresh plate of iMEFs between days 10-14 of 5i/L/A treatment. Passage cells 1:1 or 1:2 using TrypLE Express (see Note 19-21).
The GSK3 inhibitor IM-12 can be titrated down to 0.5 μM or removed altogether to promote the proliferation of naïve hPSCs, buts its omission results in a flatter colony morphology [16].
3.5. Cryopreserving naïve and primed hPSCs
Harvest primed hPSCs as colonies (as described in section 3.1 and 3.3). Harvest naïve hPSCs as single cells (as described in section 3.4).
Transfer the harvested cells into 15 mL conical tubes containing 10 mL FM.
Centrifuge the cells at 250 x g for 3 min. Aspirate the FM.
Gently resuspend the cells in the required amount of CryoStor CS10 (see Note 22).
Transfer 1 mL of this resuspension into a cryovial. Place the cryovial in an isopropanol-filled freezing chamber and store at −80 °C overnight. Move the cryovial to liquid nitrogen for long term storage.
3.6. Validating naïve pluripotent identity by qRT-PCR
Aspirate cell culture media from confluent wells of naïve and primed hPSCs.
Add 1 mL TrypLE Express and incubate at 37 °C fo r 5 min.
Thoroughly dissociate the cells into single cells by pipetting up and down, then transfer the cells to a 15 mL conical tube containing 5-10 mL FM.
Centrifuge the cells at 250 x g for 3 min. Aspirate the media.
Immediately isolate total RNA from the cells using the E.Z.N.A. total RNA kit I (see Note 23) or flash freeze the cell pellets in liquid nitrogen and store at −80 °C to isolate the RNA at a later time. Store the isolated total RNA at −80 °C.
Measure RNA concentration (see Note 24).
Perform cDNA synthesis from the isolated total RNA using the High Capacity cDNA Reverse Transcription Kit (see Note 25). Store the cDNA at −20 °C.
Perform qRT-PCR using the PowerUp SYBR Green Master Mix on the StepOnePlus Real-Time PCR System on cDNA samples from naïve and primed hPSCs. RPLP0 is used as the housekeeping control. Primed marker genes include SFRP2, VIM, and ZIC2. Naïve marker genes include KLF17, DNMT3L, DPPA3, and DPPA5.
Marker genes for primed pluripotency should begin to decrease by day 8 of naïve conversion, with little to no expression at passage 2. Marker genes for naïve pluripotency should begin to increase by day 8 of naïve conversion, with high expression at passage 2 (Fig. 1C). KLF17, DPPA3, and DPPA5 should show a 600-4000-fold increase in the naïve state compared to primed, while DNMT3L should show a 20,000-fold increase.
3.7. Cell surface marker analysis by flow cytometry
Aspirate cell culture media from confluent wells of naïve and primed hPSCs.
Add 1 mL TrypLE Express and incubate at 37 °C fo r 5 min.
Thoroughly dissociate the cells into single cells by pipetting up and down, then transfer the cells to a 15 mL conical tube containing 5-10 mL FM.
Centrifuge the cells at 250 x g for 3 min at 4 °C. Aspirate the medium.
Wash the cell pellet by resuspending in 1 mL cold FACS buffer and pipetting up and down (see Note 26).
Centrifuge the cells at 250 x g for 3 min at 4 °C. Aspirate the FACS buffer.
Resuspend each cell pellet with approximately 1 million cells in 98 μL cold FACS buffer.
Add 1 μL of each antibody to each sample (see Note 27). Naïve cell surface markers include SUSD2 [28] and CD75 [29]. Mix well by flicking the tubes.
Incubate on ice and in the dark for 45 min.
Following incubation, wash the cells by adding 1 mL cold FACS buffer.
Centrifuge the cells at 250 x g for 3 min at 4 °C. Aspirate the FACS buffer.
Resuspend each cell pellet in 300-500 μL cold FACS buffer and pass through a 40 μm cell strainer into a polystyrene round-bottom tube.
Analyze the cells for surface markers expression on a BD LSRFortessa X-20 flow cytometer using a yellow/green laser (561 nm) to detect SUSD2-PE and a red laser (640 nm) to detect CD75-eFlour660. Analyze the results with FlowJo software.
Primed hPSCs should be double-negative for SUSD2 and CD75 using the gating strategy shown in Fig. 1D. Naïve hPSCs should be >85 % double-positive for SUSD2 and CD75. See also Chapter 16 for further information on flow cytometry with naïve and primed hPSCs.
3.8. Monitoring naïve-specific reporter activity by flow cytometry
Grow naïve and primed WIBR3 OCT4-ΔPE-GFP reporter cells (Fig. 2A-C) or WIBR2 MECP2-GFP/tdTomato cells (Fig. 2D-E) to confluence.
Dissociate and wash cells as described in Section 3.7 steps 1-6.
Resuspend the cell pellet in media or FACS buffer and pass through a 40 μm cell strainer into a polystyrene round-bottom tube.
Analyze the cells on a BD LSRFortessa X-20 flow cytometer using a yellow/green laser (561 nm) to detect tdTomato and a blue laser (488 nm) to detect GFP. Analyze the results with FlowJo software.
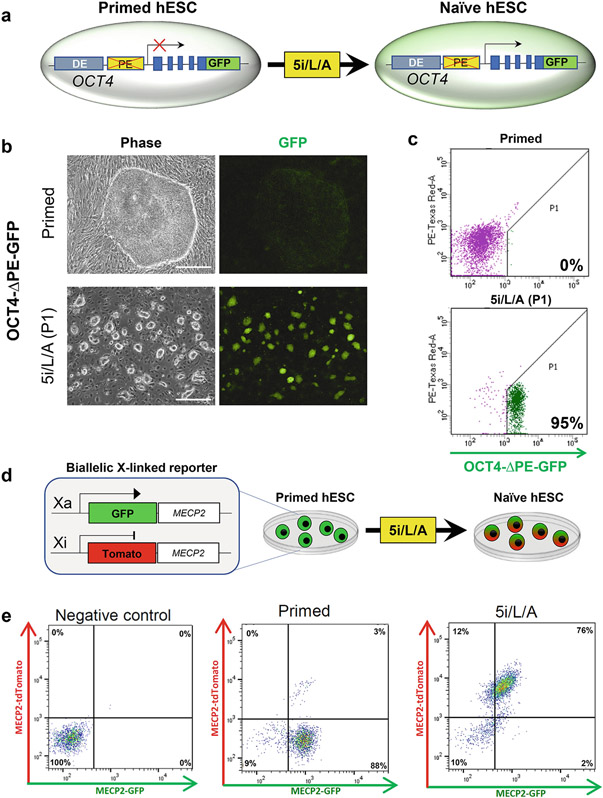
Primed hESCs should be negative for OCT4-ΔPE-GFP reporter activity using the gating strategy shown in Fig. 2C. Naïve hPSCs should be >85 % positive for OCT4-ΔPE-GFP reporter activity. Primed hPSCs should only express a single MECP2-tdTomato/GFP reporter allele (Fig. 2E), while naïve hPSCs should contain a substantial fraction (>75%) of double-positive cells within two passages (see Note 28).
Figure 2. Fluorescent reporter systems to monitor the kinetics of naïve induction in live hESCs.
A. Schematic of the OCT4-ΔPE-GFP reporter allele [9]: TALENs were used to disrupt the primed-specific proximal enhancer from an OCT4-2A-GFP reporter allele in WIBR3 hESCs. GFP activity is reduced in primed conditions, but stimulated upon naïve induction in 5i/L/A.
B. Phase contrast and fluorescent images of primed WIBR3 OCT4-ΔPE-GFP hESCs and naïve cells at P1 post-induction.
C. Flow cytometry analysis of OCT4-ΔPE-GFP activity in primed and naïve hESCs.
D. Schematic of a dual color reporter system targeted to both alleles of an X-linked gene (MECP2) that reports on the status of the X chromosome in female cells (MECP2-GFP/tdTomato). In the example shown, only the GFP allele is active in the primed state, but both fluorophores become expressed upon naïve induction.
E. Flow cytometry analysis of WIBR2 MECP2GFP-ON/Tom-OFF primed cells in primed hPSC medium and after conversion to the naïve state in 5i/L/A. This figure is adapted from our prior study [16].
4. Notes
We use fibroblasts isolated from E13.5 Swiss-Webster mice and passaged a maximum of 4 times before inactivation by Mitomycin C. Once inactivated, cells are cryopreserved and stored under liquid nitrogen conditions.
FGF2 is reconstituted as a 25 μg/mL solution in filter-sterilized 0.5 % BSA in phosphate-buffered saline.
PD0325901, IM-12, SB590885, WH4-023, and Y-27632 are reconstituted as 10 mM solutions in dimethyl sulfoxide.
Activin A and human LIF are reconstituted as 50 μg/mL solutions in filter-sterilized 0.1 % BSA in water.
Primed hPSCs may also be cultured on Matrigel in mTeSR1.
10 μM Y-27632 may be added for one day upon thawing cryopreserved primed hPSCs to increase viability.
The STEMPRO EZPassage tool may be washed thoroughly and flame-sterilized for reuse.
To create this glass tool, use a flame to create a 90° angle in a glass Pasteur pipette 3 cm from the tip. Then, place the 3 cm segment horizontally over the flame and use forceps to slowly pull off the tip of the glass pipette. Use the flame to shape the resulting pointed tip into a rounded surface.
Use a stereo microscope (we use a Leica M80) housed inside a sterile laminar flow hood.
Gelatin-coated plates may be stored at 4 °C for up to 1-2 weeks.
Freshly inactivated iMEFs may also be used.
We use a Countess II cell counting system, but other cell counters/hemocytometers are acceptable. Use Trypan blue to exclude dead cells.
It is important to ensure even coverage of iMEFs throughout the well. If desired, iMEFs may be resuspended in a larger volume of FM so long as the total volume in each well does not exceed 3 mL.
iMEF plates are ideally used the day after seeding but have been used up to five days post-seeding with no adverse effects. Ensure the iMEFs look healthy and provide 85-95 % plate coverage before using.
The WIBR1, WIBR2, and WIBR3 hESC lines were derived in primed hPSC medium containing 15 % FBS and 5 % KSR on iMEFs [30]. We continue to use these primed conditions for these specific hESC lines, although the cells can also be adapted to Matrigel in mTeSR Plus.
Primed hPSC colonies should be growing among the iMEFs. If primed colonies are sparse or show little growth, wait one additional day before starting 5i/L/A treatment. In this case, replace media with 2 mL mTeSR Plus supplemented with 10 μM Y-27632.
Do not wash cells as they can easily detach from the plate.
You should expect to see a significant amount of cell death around days 2-8 of 5i/L/A treatment. Colonies of naïve cells should become apparent starting on day 7-10. These cells are mostly SUSD2/CD75 double-positive by flow cytometry (Fig. 1C) and show significant upregulation of DNMT3L and KLF17 transcripts by qRT-PCR (Fig. 1E).
StemPro Accutase may also be used for dissociating naïve hPSCs.
Cells typically become confluent around day 10-14. Ideally, wait until you see prominent dome-shaped colonies before passaging. Non-naïve cells will still be present at this time, and passaging will result in a purer culture. Naïve cells grow best at medium-to-high density, so we typically split 1:1 the first passage. After the first full passage, naïve cells can be split 1:1 - 1:3 as necessary. A single passage may require 5-7 days before splitting, depending on the density and growth rate.
We advise using naïve hPSCs < 10 passages in 5i/L/A media to reduce the risk of genomic abnormalities [19,9].
1 mL of CryoStor CS10 is often used per well of a 6-well plate.
Other RNA isolation kits/protocols may be used.
We use a NanoDrop spectrophotometer.
Other reverse transcription kits/protocols may be used.
Resuspend large pellets in > 1 mL FACS buffer.
Prepare unstained, single-stained, and fluorescence-minus-one samples as appropriate.
Note that the maximum intensity of MECP2-GFP is significantly weaker than MECP2-tdTomato.
Acknowledgements
Work in our laboratory is supported by the NIH Director's New Innovator Award (DP2 GM137418) and grants from the Shipley Foundation Program for Innovation in Stem Cell Science, the Edward Mallinckrodt, Jr. Foundation, and the Washington University Children's Discovery Institute. No federal NIH/NIGMS funds are used to develop 3D models of early human development. We thank Kyoung-mi Park for comments on the manuscript.
References
- 1.Nichols J, Smith A (2009) Naive and primed pluripotent states. Cell Stem Cell 4 (6):487–492. doi: 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- 2.Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78 (12):7634–7638. doi: 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292 (5819):154–156 [DOI] [PubMed] [Google Scholar]
- 4.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448 (7150):191–195. doi: 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- 5.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD (2007) New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448 (7150):196–199. doi: 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- 6.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, Amit I, Tanay A, Massarwa R, Novershtern N, Hanna JH (2013) Derivation of novel human ground state naive pluripotent stem cells. Nature 504 (7479):282–286. doi: 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- 7.Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH (2013) Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13 (6):663–675. doi: 10.1016/j.stem.2013.11.015 [DOI] [PubMed] [Google Scholar]
- 8.Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, Tesar PJ, Okada J, Margaretha L, Sperber H, Choi M, Blau CA, Treuting PM, Hawkins RD, Cirulli V, Ruohola-Baker H (2014) Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A 111 (12):4484–4489. doi: 10.1073/pnas.1319738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D'Alessio A, Zhang J, Gao Q, Dawlaty MM, Young RA, Gray NS, Jaenisch R (2014) Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15 (4):471–487. doi: 10.1016/j.stem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A (2014) Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158 (6):1254–1269. doi: 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmerlin L, Park TS, Huo JS, Verma K, Pather SR, Talbot CC Jr., Agarwal J, Steppan D, Zhang YW, Considine M, Guo H, Zhong X, Gutierrez C, Cope L, Canto-Soler MV, Friedman AD, Baylin SB, Zambidis ET (2016) Tankyrase inhibition promotes a stable human naive pluripotent state with improved functionality. Development 143 (23):4368–4380. doi: 10.1242/dev.138982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin H, Hejna M, Liu Y, Percharde M, Wossidlo M, Blouin L, Durruthy-Durruthy J, Wong P, Qi Z, Yu J, Qi LS, Sebastiano V, Song JS, Ramalho-Santos M (2016) YAP Induces Human Naive Pluripotency. Cell Rep 14 (10):2301–2312. doi: 10.1016/j.celrep.2016.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R (2010) Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A 107 (20):9222–9227. doi: 10.1073/pnas.1004584107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR (1996) Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122 (3):881–894 [DOI] [PubMed] [Google Scholar]
- 15.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A (2008) The ground state of embryonic stem cell self-renewal. Nature 453 (7194):519–523. doi: 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theunissen TW, Friedli M, He Y, Planet E, O'Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, Duc J, Cohen MA, Wert KJ, Castanon R, Zhang Z, Huang Y, Nery JR, Drotar J, Lungjangwa T, Trono D, Ecker JR, Jaenisch R (2016) Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 19 (4):502–515. doi: 10.1016/j.stem.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedli M, Trono D (2015) The Developmental Control of Transposable Elements and the Evolution of Higher Species. Annual Review of Cell and Developmental Biology 31 (1):429–451. doi: 10.1146/annurev-cellbio-100814-125514 [DOI] [PubMed] [Google Scholar]
- 18.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, Jin X, Shi X, Liu P, Wang X, Wang W, Wei Y, Li X, Guo F, Wu X, Fan X, Yong J, Wen L, Xie SX, Tang F, Qiao J (2014) The DNA methylation landscape of human early embryos. Nature 511:606. doi: 10.1038/nature13544 https://www.nature.Com/articles/nature13544#supplementary-information [DOI] [PubMed] [Google Scholar]
- 19.Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT (2016) Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 18 (3):323–329. doi: 10.1016/j.stem.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R, Plath K (2017) Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 20 (1):87–101. doi: 10.1016/j.stem.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J (2016) Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Reports 6 (4):437–446. doi: 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stirparo GG, Boroviak T, Guo G, Nichols J, Smith A, Bertone P (2018) Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development 145 (3). doi: 10.1242/dev.158501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M (2016) A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537 (7618):57–62. doi: 10.1038/nature19096 [DOI] [PubMed] [Google Scholar]
- 24.Dong C, Beltcheva M, Gontarz P, Zhang B, Popli P, Fischer LA, Khan SA, Park KM, Yoon EJ, Xing X, Kommagani R, Wang T, Solnica-Krezel L, Theunissen TW (2020) Derivation of trophoblast stem cells from naive human pluripotent stem cells. Elife 9. doi: 10.7554/eLife.52504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cinkornpumin JK, Kwon SY, Guo Y, Hossain I, Sirois J, Russett CS, Tseng HW, Okae H, Arima T, Duchaine TF, Liu W, Pastor WA (2020) Naive Human Embryonic Stem Cells Can Give Rise to Cells with a Trophoblast-like Transcriptome and Methylome. Stem Cell Reports 15 (1):198–213. doi: 10.1016/j.stemcr.2020.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F (2016) Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 165 (4):1012–1026. doi: 10.1016/j.cell.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An C, Feng G, Zhang J, Cao S, Wang Y, Wang N, Lu F, Zhou Q, Wang H (2020) Overcoming Autocrine FGF Signaling-Induced Heterogeneity in Naive Human ESCs Enables Modeling of Random X Chromosome Inactivation. Cell Stem Cell 27 (3):482–497 e484. doi: 10.1016/j.stem.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 28.Bredenkamp N, Stirparo GG, Nichols J, Smith A, Guo G (2019) The Cell-Surface Marker Sushi Containing Domain 2 Facilitates Establishment of Human Naive Pluripotent Stem Cells. Stem Cell Reports. doi: 10.1016/j.stemcr.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, Corcoran AE, Walker R, Douagi I, Lanner F, Rugg-Gunn PJ (2017) Comprehensive Cell Surface Protein Profiling Identifies Specific Markers of Human Naive and Primed Pluripotent States. Cell Stem Cell 20 (6):874–890 e877. doi: 10.1016/j.stem.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, Santagata S, Powers D, Barrett CB, Young RA, Lee JT, Jaenisch R, Mitalipova M (2010) Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141 (5):872–883. doi: 10.1016/j.cell.2010.04.010 [DOI] [PubMed] [Google Scholar]