To the Editor: Three sublineages of the B.1.1.529 (omicron) variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have serially transitioned into globally dominant forms — first BA.1, then BA.2, and then BA.5. As of October 2022, most circulating omicron variants belong to BA.5. However, the prevalence of BQ.1.1 (a BA.5 subvariant) and XBB (a BA.2 subvariant) is increasing rapidly in several countries, including the United States and India. BA.2 and BA.5 variants have been shown to have less sensitivity to certain monoclonal antibodies than previously circulating variants of concern.1-5 Notably, as compared with BA.5 and BA.2, BQ.1.1 and XBB carry additional substitutions in the receptor-binding domain of the spike (S) protein, which is the major target for vaccines and therapeutic monoclonal antibodies for coronavirus disease 2019 (Covid-19). These subvariants may, therefore, be more immune-evasive than BA.5 and BA.2.
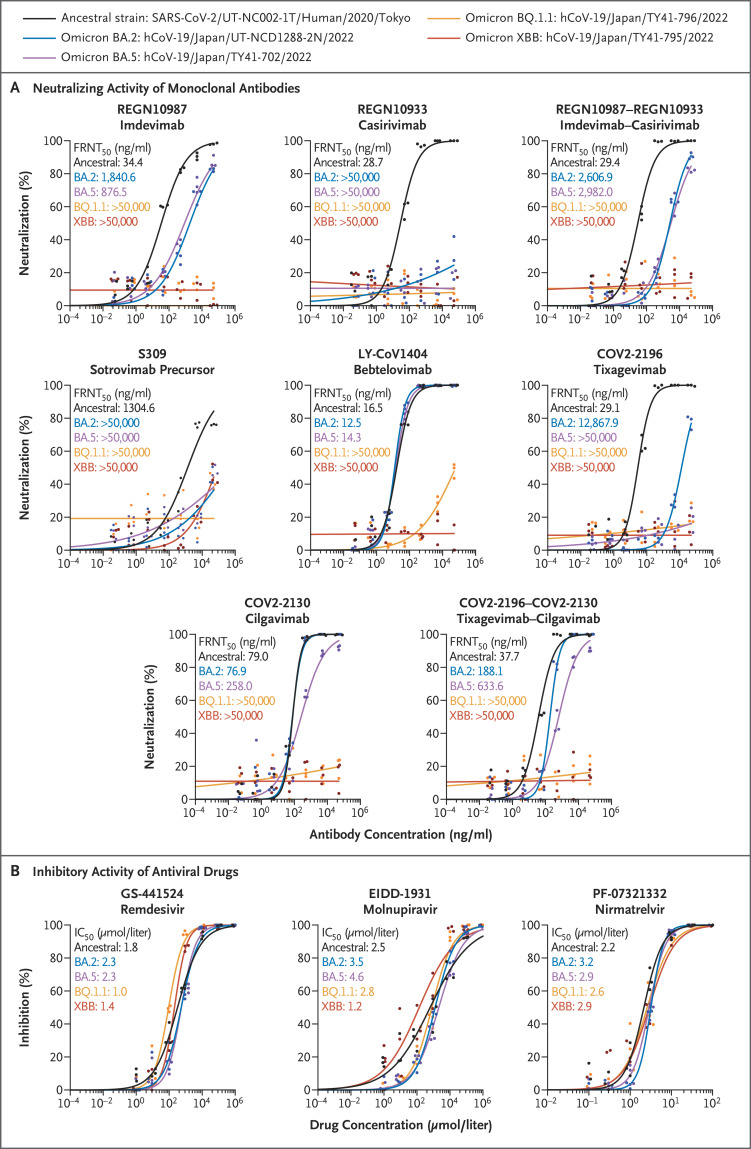
We assessed the efficacy of therapeutic monoclonal antibodies against omicron BQ.1.1 (hCoV-19/Japan/TY41-796/2022; TY41-796) and XBB (hCoV-19/Japan/TY41-795/2022; TY41-795), which were isolated from patients. The BQ.1.1 isolate had three more substitutions (R346T, K444T, and N460K) in its receptor-binding domain than a BA.5 (hCoV-19/Japan/TY41-702/2022) isolate (Fig. S1A in the Supplementary Appendix, available with the full text of this letter at NEJM.org). The XBB isolate had nine more changes (G339H, R346T, L368I, V445P, G446S, N460K, F486S, F490S, and the wild-type amino acid at position 493) in its receptor-binding domain than a BA.2 (hCoV-19/Japan/UT-NCD1288-2N/2022) isolate (Fig. S1B). To examine the reactivity of monoclonal antibodies against these subvariants, we determined the 50% focus reduction neutralization test (FRNT50) titer of the monoclonal antibodies by using a live-virus neutralization assay. REGN10987 (marketed as imdevimab), REGN10933 (marketed as casirivimab), COV2-2196 (marketed as tixagevimab), COV2-2130 (marketed as cilgavimab), and S309 (the precursor of sotrovimab) did not neutralize the BQ.1.1 or XBB isolates even at the highest FRNT50 value (>50,000 ng per milliliter) tested (Figure 1A and Table S1). LY-CoV1404 (marketed as bebtelovimab), which effectively neutralizes1,3-5 omicron BA.1, BA.2, BA.4, and BA.5, had no efficacy against BQ.1.1 or XBB. Both combinations of monoclonal antibodies tested (i.e., imdevimab–casirivimab and tixagevimab–cilgavimab) failed to neutralize either BQ.1.1 or XBB. These results suggest that imdevimab–casirivimab, tixagevimab–cilgavimab, sotrovimab, and bebtelovimab may not be effective against BQ.1.1 or XBB in the clinical setting.
Figure 1. In Vitro Efficacy of Therapeutic Monoclonal Antibodies and Antiviral Drugs against Omicron Subvariants.
Panel A shows the neutralizing activity of monoclonal antibodies against omicron subvariants. The antibodies used in this analysis were produced in the authors’ laboratories and are not identical to the commercially available products. The 50% focus reduction neutralization test (FRNT50) titer levels were determined by performing a focus reduction neutralization test in Vero E6 cells engineered to express transmembrane serine protease 2 (TMPRSS2) and human angiotensin-converting enzyme 2 (ACE2). The data shown are the mean values for triplicate experiments. Panel B shows the inhibitory activity of antiviral drugs against omicron subvariants. The drugs used were purchased from MedChemExpress. The in vitro 50% inhibitory concentration (IC50) values were determined by performing a focus reduction assay in Vero E6 cells engineered to express TMPRSS2 and ACE2. The data shown are the mean values for triplicate experiments. GS-441524 (the main metabolite of remdesivir) and EIDD-1931 (the active form of molnupiravir) are RNA-dependent RNA polymerase inhibitors. PF-07321332 (nirmatrelvir) is a main protease inhibitor.
The Food and Drug Administration approved remdesivir (an inhibitor of the RNA-dependent RNA polymerase [RdRp] of SARS-CoV-2) for the treatment of Covid-19 and issued an emergency use authorization for molnupiravir (an RdRp inhibitor) and nirmatrelvir (an inhibitor of the main protease of SARS-CoV-2). We therefore tested these antiviral drugs by determining the in vitro 50% inhibitory concentration (IC50) of each compound against BQ.1.1 and XBB. Unlike the amino acid sequence of the reference strain Wuhan/Hu-1/2019, the BQ.1.1 and XBB isolates encode the P3395H substitution in their main protease (Fig. S1C and S1D). The BQ.1.1 and XBB isolates also have two (Y264H and P314L) and three (P314L, M659I, and G662S) substitutions in their RdRp, respectively. The susceptibilities of BQ.1.1 and XBB to the three compounds were similar to those of the ancestral strain (SARS-CoV-2/UT-NC002-1T/Human/2020/Tokyo). For BQ.1.1, the IC50 value was lower by a factor of 0.6 with remdesivir and higher by factors of 1.1 and 1.2 with molnupiravir and nirmatrelvir, respectively. For the XBB subvariant, the IC50 value was lower by a factor of 0.8 with remdesivir, lower by a factor of 0.5 with molnupiravir, and higher by a factor of 1.3 with nirmatrelvir (Figure 1B). These results suggest that remdesivir, molnupiravir, and nirmatrelvir are efficacious against both BQ.1.1 and XBB in vitro.
Our data suggest that the omicron sublineages BQ.1.1 and XBB have immune-evasion capabilities that are greater than those of earlier omicron variants, including BA.5 and BA.2. The continued evolution of omicron variants reinforces the need for new therapeutic monoclonal antibodies for Covid-19.
Supplementary Appendix
Disclosure Forms
This letter was published on December 7, 2022, at NEJM.org.
Footnotes
Supported by grants from the Center for Research on Influenza Pathogenesis and Transmission (75N93021C00014, to Dr. Kawaoka), funded by the National Institute of Allergy and Infectious Diseases; a Research Program on Emerging and Reemerging Infectious Diseases (JP21fk0108552 and JP21fk0108615, to Dr. Kawaoka); a Project Promoting Support for Drug Discovery (JP21nf0101632, to Dr. Kawaoka); the Japan Program for Infectious Diseases Research and Infrastructure (JP22wm0125002, to Dr. Kawaoka); and the Japan Agency for Medical Research and Development (JP223fa627001, to Dr. Kawaoka).
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Iketani S, Liu L, Guo Y, et al. Antibody evasion properties of SARS-CoV-2 omicron sublineages. Nature 2022;604:553-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takashita E, Kinoshita N, Yamayoshi S, et al. Efficacy of antiviral agents against the SARS-CoV-2 omicron subvariant BA.2. N Engl J Med 2022;386:1475-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y, Yisimayi A, Jian F, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by omicron infection. Nature 2022;608:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takashita E, Yamayoshi S, Simon V, et al. Efficacy of antibodies and antiviral drugs against omicron BA.2.12.1, BA.4, and BA.5 subvariants. N Engl J Med 2022;387:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Guo Y, Iketani S, et al. Antibody evasion by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022;608:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.