Abstract
Chest X-ray (CXR) imaging is a low-cost, easy-to-use imaging alternative that can be used to diagnose/screen pulmonary abnormalities due to infectious diseaseX: Covid-19, Pneumonia and Tuberculosis (TB). Not limited to binary decisions (with respect to healthy cases) that are reported in the state-of-the-art literature, we also consider non–healthy CXR screening using a lightweight deep neural network (DNN) with a reduced number of epochs and parameters. On three diverse publicly accessible and fully categorized datasets, for non–healthy versus healthy CXR screening, the proposed DNN produced the following accuracies: 99.87% on Covid-19 versus healthy, 99.55% on Pneumonia versus healthy, and 99.76% on TB versus healthy datasets. On the other hand, when considering non–healthy CXR screening, we received the following accuracies: 98.89% on Covid-19 versus Pneumonia, 98.99% on Covid-19 versus TB, and 100% on Pneumonia versus TB. To further precisely analyze how well the proposed DNN worked, we considered well-known DNNs such as ResNet50, ResNet152V2, MobileNetV2, and InceptionV3. Our results are comparable with the current state-of-the-art, and as the proposed CNN is light, it could potentially be used for mass screening in resource-constraint regions.
Keywords: Chest X-ray, DNN, Medical imaging, Infectious DiseaseX, Covid-19, Pneumonia, Tuberculosis
Abbreviations: CXR, Chest X-ray; TB, Tuberculosis; DNN, Deep Neural Network; MTB, Mycobacterium Tuberculosis; CT, Computed Tomography; AI, Artificial Intelligence; NN, Neural Network; DL, Deep Learning; ML, Machine Learning; ACC, Accuracy; AUC, Area Under the Curve; SPEC, Specificity; SEN, Sensitivity; CADx, Computer-Aided Diagnosis; CNN, Convolutional Neural Network; WHO, World Health Organization
1. Introduction
Infectious diseaseX is global threat. Covid-19 is caused by the SARS-CoV-2 virus, Pneumonia is caused by viral infections, and Tuberculosis (TB) is caused by Mycobacterium Tuberculosis (MTB) bacteria. All of these affect the lungs. According to the World Health Organization (WHO) [1] 213 million people were sick at the time of this study, and Covid-19 was responsible for 4.45 million deaths. Pneumonia took the lives of around 8,08,000 people in 2017, with children under the age of five accounting for 15% of all deaths. The death rate of the elderly has remained steady since 1990. TB infected 10 million individuals and killed 1.4 million people in 2019. With these data, it is critical to concentrate on understanding pulmonary disorders/abnormalities. Artificial intelligence (AI) has prompted numerous improvements in medical imaging, and X-ray imaging technology is fairly common and less expensive as compared to Computed Tomography (CT) scans. The nature of the screening processes in the Covid-19 period has been enhanced by health tools [2], [3], [4]. Researchers [5] employed deep learning (DL) based algorithms to detect the evidence of Covid-19 infection in lung region, reducing prognosis time and highlighting the need for RT-PCR tests. Custom Neural Networks (NNs) have been suggested for chest CT scans and chest X-rays (CXRs) with the help of Computer-aided Diagnosis (CADx) transfer learning to diagnose pulmonary disease. Other studies also used convolutional neural networks (CNNs) to separately detect Covid-19, Pneumonia, and TB infection in patients [6], [7], [8].
In this paper, we proposed a custom designed Deep Neural Network (DNN) model to detect Covid-19, Pneumonia, and TB in CXRs. We conducted cross validation to analyze and evaluated performance using Covid-19, Pneumonia, and TB CXRs datasets, excluding healthy cases. Our findings/results are comparable with the state-of-the-art results for Covid-19, Pneumonia, and TB positive cases. Overall, let us itemize our research contributions:
-
1.
Lightweight CNN: We aimed at building a lightweight (less number of layers, parameters, and epochs) DNN model to detect pulmonary abnormalities in CXRs due to infectious diseaseX: Covid-19, Pneumonia, and TB.
-
2.
Comprehensive experiments: Unlike state-of-the-art literature, we were not limited to classify non–healthy cases from healthy ones, but also to classify one type of non–healthy cases from another non–healthy ones. To be precise, we produced an accuracy of 99.87% on Covid-19 versus healthy, 99.55% on Pneumonia versus healthy, 99.76% on TB versus healthy datasets. When considering non–healthy X-ray screening, we received an accuracy of 98.89% on Covid-19 versus Pneumonia, 98.99% on Covid-19 versus TB, and 100% on Pneumonia versus TB.
-
3.
Genericity, scalability, and comparison: As experiments were done on three publicly accessible diverse datasets (excluding healthy cases): Covid-19 (1,200 CXRs), Pneumonia (3,875 CXRs), and TB (3,500 CXRs) without changing/modifying parameters, our lightweight CNN is robust enough to be compared with state-the-art results. Our results are also compared with other well-known DNN algorithms.
The rest of the manuscript is organized as follows. Section 2 presents background study of Covid-19, Pneumonia, and TB. Section 3 describes the proposed DNN. Section 4 describes the experimental setup and outcomes in full. Section 5 provides results analysis. We discuss our results (including previous results) in Section 6. Section 7 presents comparison with popular DNNs. The work is concluded in Section 8.
2. Related works
AI contributed a lot to healthcare, and pulmonary screening/diagnosis using CXRs is no exception. DL models made it more possible, reliable, and have a significant impact on biomedical research. In this section, we review up-to-date (recently published, peer-reviewed) articles.
Covid-19:Covid-19’s primary symptoms include headaches, muscular discomfort, cough, frequent colds, periodic fevers, and breathing difficulties in various susceptible instances [9]. Machine learning (ML) and/or DL models are used to prevent potential human life hazards [10], [11]. Researchers continue working on Covid-19 in 2021 on a large scale as compared to 2020. We refer authors few books on Covid-19 screening for further detailed information [12]. Authors, in [13], worked on a new definition of cluster-based effective one-shot learning to detect Covid-19 cases. Das et al.[14] employed pre-trained CNN model to detect Covid-19 positive patients. Mukherjee et al. [15], [16] developed a CNN model to detect Covid-19 and reported an overall accuracies of 96.28% (CT scans + CXRs) and 99.69% (CXRs) respectively. Authors, in [17], [18], used fuzzy color and stacking approaches with DL models. Deep transfer learning has been always common [19], [20], where authors reported an accuracy of more than 98%. DL models namely ‘DarkCovidNet’ and ‘CoroDet’ were proposed [21], [22]. Pre-trained DL models are no exception [23], [24] to detect Covid-19 cases. Ismael and Sengur [25] pre-trained ImageNet, ResNet50, and SVM classifier, and their reported accuracy was 94.74%. Because of the diverse data sets, the results for each study may vary. For further reading, we refer readers to follow up-to-date research article – how big data is big (for medical imaging: Covid-19)? [26].
Pneumonia: In [27], authors examined DL approaches and automated CXR analysis for pneumonia detection so medical practitioners could accurately diagnose Covid-19. Authors, in [28], employed transfer learning technique with a pre-trained ImageNet model for diagnosis of Pneumonia based on lung segmentation (U-Net architecture). Authors employed largely fine-tuned version of MobileNet V2, InceptionResNet V2, and ResNet50 to see how effective single and combined model can be made to diagnose Pneumonia [29]. Using their combined model, they achieved an accuracy of 93.52%. The COVID-DeepNet system [30] is a popular hybrid multimodal deep learning system that helps radiologists in precise and efficient image interpretation with a precision rate of 99.93%. They reported 100 % precision and F1 score 99.93 %. A.K. Jaiswal et al. [31] employed a DL method to diagnose Pneumonia using CXRs. Similarly, authors [32] used deep transfer learning using pre-trained models such as MobileNetV3, InceptionV3, ResNet18, Xception, DenseNet121, and InceptionV3 to detect Pneumonia, and they achieved an accuracy of 98.43%. Pre-trained models are common. In [33], AlexNet was employed to classify between Covid-19 and normal, and viral and bacterial pneumonia. Authors, in [34], [35], employed deep transfer learning using pre-trained models: Inception-V3, VGG16, ResNet18, DenseNet201, Xception, and SqueezeNet to detect Pneumonia. Similarly, pre-trained models such as GoogLeNet, LeNet, and ResNet-50, ResNet152, DenseNet121, and ResNet18 were used [36], [37]. In [38], transfer learning techniques were used to identify treatable diseases like Pneumonia and reported an accuracy of 100.0%. They have not, however, analyzed whether results were biased.
Tuberculosis (TB): TB – a fatal infectious disease – caused by MTB bacteria. In medical imaging, feature selection is crucial, and in [39], authors addressed the importance of correct use of features to achieve optimal performance. Another work [40] evaluates on the impact of image enhancement. In [41], ensemble learning based TB detection in CXRs using hybrid feature descriptors. Another work, in [42], was to eliminate attainable sources of bias in computer assisted CXR analysis for pulmonary TB. In [43], authors suggested CNN models for TB diagnosis based on voting and pre-processing variation ensemble. Their findings show that 97.500% and 97.699% of data accuracy in Montgomery and Shenzhen datasets were achieved with the suggested approach, respectively. Authors of [44] used DL techniques to identify TB using CXRs, and received 94.73% and 98.6% accuracy, respectively. For TB, DCNNs are used [45] for detecting TB using CXR images. Three DNNs, namely CAD4TB, Lunit INSIGHT, and qXR were used [46]. Interestingly, authors used thoracic edge map of CXRs for automatically screening pulmonary abnormalities [47], [48]. Karargyris et al. [49] combined features (local and global), aiming to detect pulmonary abnormalities caused by pneumonia or (TB). Qin et al. [50] explore five different AI algorithms to detect TB from CXRs.
Overall, in all three infectious disease types, more often, we observed that transfer learning techniques with pre-trained models such as VGG16, GoogLeNet, LeNet, ResNet-50, ResNet152, DenseNet121, ResNet18, MobileNetV3, InceptionV3, ResNet18, and Xception were employed. Also, they (most of them) were limited to binary classification: non–healthy versus healthy.
The proposed tools’ primary objective, whether on Covid-19, Pneumonia, and TB, was to train a single DNN architecture and test accordingly. Additional objective is not only to consider non–healthy versus healthy CXR screening but also to check whether non–healthy CXR screening does work using the exact same DNN model. As before, our aim is to develop one DNN architecture so Covid-19, Pneumonia, and TB positive patients (in CXRs) can be detected. On three publicly available datasets, we evaluated the model.
3. DNN architecture
There are various reasons why proposed architecture is a DNN architecture. The first thing we found is that DNNs are quite good at lowering the amount of parameters while maintaining model quality. DNN does not require human feature engineering because it can extract features from an image automatically. We also noted that several of the researchers employed DNN and achieved good image classification and recognition accuracy. Our suggested DNN architecture comprises of three layers: a convolutional layer, a pooling layer, and a fully connected layer to detect Covid-19, Pneumonia, and TB positive patients. To accomplish operations successfully, the layers are fully integrated. We have made our suggested DNN model open to the public1 .
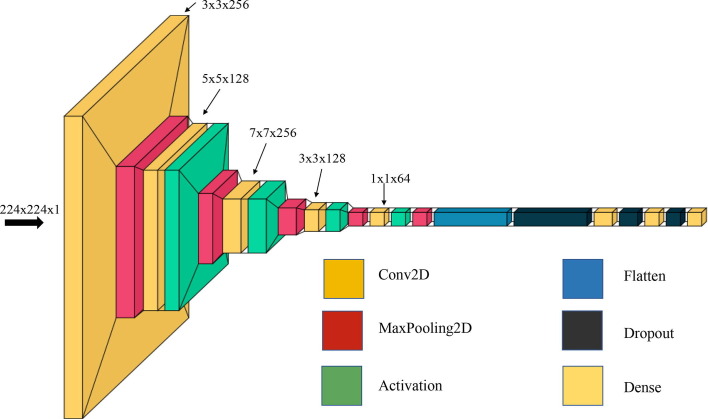
The architecture of proposed model is shown in Fig. 1 . The first layer of the architecture is the input layer, with the input shape (224, 224, 1) with strides 2. The second layer is a convolution layer with 256 filters. The filter size of this layer is followed by activation function ReLU and max pooling layer. The third layer is convolution layer with 128 filters. The filter size of this layer is followed by activation function ReLU and maxpooling layer. Fourth layer is convolution layer with 256 filters. The filter size of this layer is followed by activation function ReLU and maxpooling layer. The fifth layer of the architecture is a convolution layer with 128 filters. The filter size of this layer is followed by activation function ReLU and maxpooling layer. Sixth layer of the architecture is a convolution layer with 64 filters. The filter size of this layer is , followed by activation function ReLU and the maxpooling layer. The seventh layer of the architecture is a flatten or fully connected layer with 0.5 or 50% dropout. The subsequent two layers are dense layers with 256 and 128 neurons with 0.5 or 50% dropout. The eighth and last layer is output layers with a sigmoid activation function. The output shape and number of learning parameters of the proposed DNN architecture are shown in Table 1 .
Fig. 1.
Architecture of the proposed DNN model for abnormality screening (Covid-19, Pneumonia, and TB).
Table 1.
Learning parameters for our proposed architecture (image size: ).
| No | Layer (type) | Output shape | Parameters |
|---|---|---|---|
| 1 | conv2d (Conv2D) | (None, 112, 112, 256) | 256 |
| 2 | max_pooling2d (MaxPooling2) | (None, 56, 56, 256) | 0 |
| 3 | conv2d_1 (Conv2D) | (None, 52, 52, 128) | 819,328 |
| 4 | max_pooling2d_1 (MaxPooling2) | (None, 26, 26, 128) | 0 |
| 5 | conv2d_2 (Conv2D) | (None, 20, 20, 256) | 1,605,888 |
| 6 | max_pooling2d_2 (MaxPooling2) | (None, 10, 10, 256) | 0 |
| 7 | conv2d_3 (Conv2D) | (None, 8, 8, 128) | 2,95,040 |
| 8 | max_pooling2d_3 (MaxPooling2) | (None, 4, 4, 128) | 0 |
| 9 | conv2d_4 (Conv2D) | (None, 4, 4, 64) | 8,256 |
| 10 | max_pooling2d_4 (MaxPooling2) | (None, 4, 4, 64) | 0 |
| 11 | flatten (Flatten) | (None, 1024) | 0 |
| 12 | dropout (Dropout) | (None, 1024) | 0 |
| 13 | dense (Dense) | (None, 256) | 2,62,400 |
| 14 | dropout_1 (Dropout) | (None, 256) | 0 |
| 15 | dense_1 (Dense) | (None, 128) | 32,896 |
| 16 | dropout_2 (Dropout) | (None, 128) | 0 |
| 17 | dense_2 (Dense) | (None, 1) | 129 |
| Total parameters | 3,026,497 | ||
4. Experimental setup
4.1. Datasets
DNN requires a large amount of data. Using multiple resources, we created six different data combinations to train and evaluate the proposed architecture. Table 2 shows all the detailed information about a collection of all images for this study. Few samples from the above-mentioned collections are provided in Fig. 2 . The combinations of six different dataset (D1 to D6) are listed below (also in Table 3 ):
-
1.
[C1:] Tawsifur Rahman’s Covid-19 collection [23], [24] is publicly available. It contains 1,200 Covid-19 positive CXRs (date: October, 2021).
-
2.
[C2:] A publicly available collection [38] (Paul Mooney and his team) is composed of 5,856 CXRs, where 3,875 of them were used in our study.
-
3.
[C3:] We used Kaggle data (Tawsifur Rahman and his team) [44]. It contains 7,000 CXRs (date: October, 2021). This dataset contains 3500 images of TB positive CXRs.
-
4.
[C4:] We collected healthy images from several publicly available sources [23], [24], [38], [44], and 6,182 CXRs were used in our study.
Table 2.
Data collections (open source).
| Data collections | # of CXRs |
|---|---|
| C1: Covid-19 | 1,200 |
| C2: Pneumonia | 3,875 |
| C3: Tuberculosis | 3,500 |
| C4: Healthy | 6,182 |
Fig. 2.
CXR samples (see Table 2): Covid-19 (left-most), Pneumonia (middle-left), TB (middle-right) and healthy (right-most).
Table 3.
Details of dataset used our experiment (Table 1).
| Dataset | Covid-19 |
Pneumonia |
Tuberculosis |
|||
|---|---|---|---|---|---|---|
| +ve | −ve | +ve | −ve | +ve | −ve | |
| D1 | 1,200 | 1,341 | – | – | – | – |
| D2 | – | – | 3,875 | 1,341 | – | – |
| D3 | – | – | – | – | 3,500 | 3,500 |
| D4 | 1,200 | – | 3,875 | – | – | – |
| D5 | 1,200 | – | – | – | 3,500 | – |
| D6 | – | – | 3,875 | – | 3,500 | – |
With these collections, we built six different datasets (D1 to D6) for our experiment:
-
1.
[D1:] It contains a total of 1,200 Covid-19 and 1,341 healthy cases.
-
2.
[D2:] It is composed of 3,875 Pneumonia and 1,341 healthy cases.
-
3.
[D3:] It includes 3,500 TB and 3,500 healthy cases.
-
4.
[D4:] In D4, we have 1,200 Covid-19 and 3,875 Pneumonia cases.
-
5.
[D5:] In D5, 1,200 Covid-19 and 3,500 TB cases were considered.
-
6.
[D6:] In D6, 3,875 Pneumonia and 3,500 TB cases are considered.
The purpose of building the different data organizations (D1 to D3) is to show that our suggested DNN can detect Covid-19, Pneumonia, and TB cases with respect to healthy cases. As Covid-19 could cause Pneumonia, a new dataset (D4) was created to classify Covid-19 positive individuals and those having conventional Pneumonia. TB manifestation were also included in D5 so that we can separate Covid-19 infected patients from TB patients, and vice versa. In addition, D6 was constructed to evaluate whether our model can classify between Pneumonia and TB cases.
CXRs images were scaled down to (grayscale) for this study to match the input dimensions of the proposed DNN model as an input to our architecture. It is also possible to reduce the computational complexity of such a resizing.
4.2. Evaluation protocol and performance metrics
We followed 10-fold cross-validation technique to evaluate our approach on all six different data sets: D1 to D6. To measure the performance, six distinct assessment metrics: accuracy (ACC), sensitivity (SEN), specificity (SEPC), precision (PREC), F1 score, and area under the curve (AUC) were used for all 10 folds, and these are computed as follows:
where ,fn refer to true positive, false positive, true negative, and false negative respectively.
For evaluation, in addition to straightforward classification accuracy, we emphasize other important metrics such as precision, specificity, sensitivity, and F1 score. Sensitivity refers to the probability of a positive test provided that the individual has the disease. And, specificity refers to the probability of a negative test provided that the individual is healthy. A model’s combined performance score, the symphonic mean of its accuracy and sensitivity, is calculated using the F1 score.
4.3. Model validation
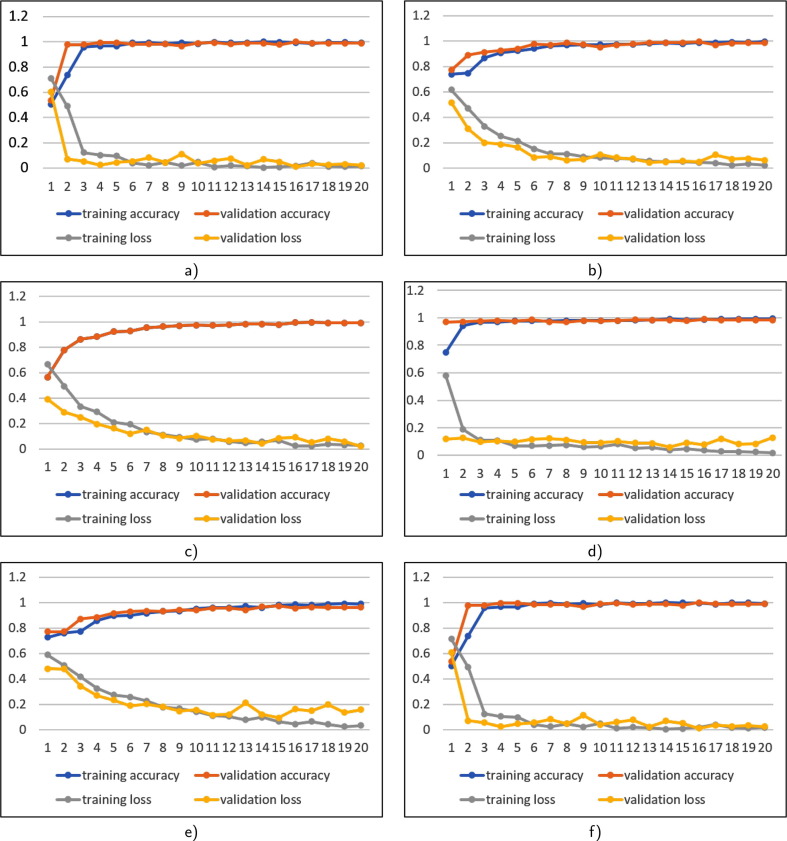
The vision was to build a simple, low-computing, low-epoch model that could identify three different lung abnormalities using the exact same DNN model. We utilized few epochs to train our model, and we achieved an optimal performance (twenty epochs). The entire dataset was partitioned into 70:30 ratios for training and validation purpose. The training and validation accuracy of six data sets are shown in Fig. 3 .
Fig. 3.
Training accuracy versus validation accuracy and training loss versus validation loss on all datasets (see Table 3): a) D1, b) D2, c) D3, d) D4, e.) D5, and f) D6.
The training datasets D1, D2, and D3 contain 807, 27,12, 2,478 positive and 968, 939, 2,422 negative Covid-19, pneumonia, and TB CXRs, respectively. Validation datasets D1, D2 and D3 comprise 393, 1,163, 1,022 positive and 373, 402, 1,078 negative Covid-19, Pneumonia and TB CXRs, respectively. To see how our proposed DNN performs for non–healthy CXR screening, we have trained our DNN with non–healthy CXR screening. The training set of non–healthy CXR screening dataset D4 contains 852 Covid-19 positive and 2,797 Pneumonia positive; D5 contains 8,41 Covid-19 positive and 2,451 TB positive CXRs, and D6 2,711 Pneumonia positive and 2,451 TB positive CXRs. The validation set includes 348 Covid-19 positive and 1,078 Pneumonia positive (D4); 359 Covid-19 positive and 1,049 TB positive CXRs (D5); and 1,164 Pneumonia positive and 1,049 TB positive CXRs (D6). In Table 4 , we provide accuracies and corresponding loss from both training validation of six datasets.
Table 4.
Training and validation performance (after 20 epochs, in %): Training Accuracy (TA), Validation Accuracy (VA), Training Loss (TL), and Validation Loss (VL).
| Dataset | TA | VA | TL | VL | ||
|---|---|---|---|---|---|---|
| D1 | 99.03 | 98.88 | 0.0168 | 0.0229 | ||
| D2 | 99.48 | 98.36 | 0.0223 | 0.0621 | ||
| D3 | 99.12 | 99.18 | 0.0251 | 0.0200 | ||
| D4 | 99.46 | 98.31 | 0.0170 | 0.1280 | ||
| D5 | 98.81 | 96.06 | 0.0314 | 0.1559 | ||
| D6 | 99.68 | 99.61 | 0.0165 | 0.0178 |
5. Results and analysis
In this section, we provide both results and analysis of our experiments. In our experiments, we considered not only healthy versus non–healthy CXR screening but also within non–healthy CXRs.
5.1. Healthy versus non–healthy CXR screening
To provide a quantifiable result, we present the mean findings on each of healthy versus non–healthy CXR screening using the 10-fold cross-validation. In Table 5 , we provide experimental outcomes. Furthermore, detailed performance scores are provided in the form of confusion matrix in Table 6 . Confusion matrix depicts the distribution of correct and inaccurate predictions by class. Note that, in datasets (D1 to D3), we considered non–healthy versus healthy CXR screening.
Table 5.
Performance: 10-fold cross-validation accuracy (in ).
| Dataset | k1 | k2 | k3 | k4 | k5 | k6 | k7 | k8 | k9 | k10 | Avg () |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | 99.61 | 98.82 | 99.22 | 99.61 | 100.0 | 99.61 | 100.0 | 99.22 | 99.61 | 99.61 | 99.53 0.34 |
| D2 | 99.62 | 99.81 | 99.23 | 99.81 | 99.04 | 98.85 | 99.42 | 99.23 | 99.62 | 99.42 | 99.41 0.30 |
| D3 | 99.29 | 99.57 | 99.43 | 99.86 | 99.14 | 99.57 | 99.57 | 98.86 | 99.57 | 99.57 | 99.44 0.27 |
Table 6.
Confusion matrix table for healthy versus non–healthy CXR screening.
| Dataset | ||||||
|---|---|---|---|---|---|---|
| D1 | 393 | 393 | 0 | 373 | 372 | 1 |
| D2 | 1,163 | 1,163 | 0 | 402 | 395 | 7 |
| D3 | 1,022 | 1,021 | 1 | 1,078 | 1,074 | 4 |
Overall, the proposed DNN correctly detected all 393 Covid-19 CXRs from D1 test set, and one of 372 healthy cases was misclassified as Covid-19 (see confusion matrix in Table 6). In D2 dataset, seven (0f 402) normal instances were misclassified as Pneumonia and 1,163 Pneumonia cases were correctly detected. In D3, 1,021 TB cases were successfully detected and 1 TB case was misclassified as healthy, and 4 out of 1,078 healthy cases were misclassified as TB. Following Table 6, we calculated accuracy, sensitivity, specificity, precision, and F1 score for datasets (D1 to D3) to better understand the performance of the proposed model (see Table 7 ).
Table 7.
Performance (in ) on healthy versus non–healthy CXR screening: accuracy, sensitivity, specificity, precision, and F1 score.
| Dataset | ACC | AUC | SEN | SPEC | PREC | F1 score | |
|---|---|---|---|---|---|---|---|
| D1 | 99.87 | 100.00 | 99.75 | 100.00 | 100.00 | 99.87 | |
| D2 | 99.55 | 100.00 | 99.40 | 100.00 | 100.00 | 99.70 | |
| D3 | 99.76 | 100.00 | 99.61 | 99.91 | 99.90 | 99.76 | |
| 99.72 | 100.00 | 99.59 | 99.97 | 99.97 | 99.78 | ||
| 0.133 | 0.00 | 0.181 | 0.051 | 0.058 | 0.086 |
5.2. Non–healthy CXR screening
To provide quantifiable outcome, we present the mean findings on each of the non–healthy CXR datasets (D3 to D6) using 10-fold cross-validation. Table 8 shows experimental results. As before, we provide confusion matrix in Table 9 for better results analysis and/or understanding.
Table 8.
Performance: 10-fold cross-validation accuracy (in ).
| Dataset | k1 | k2 | k3 | k4 | k5 | k6 | k7 | k8 | k9 | k10 | Avg () |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D4 | 99.61 | 98.82 | 99.41 | 99.02 | 98.04 | 99.41 | 99.02 | 99.21 | 98.43 | 99.02 | 99.00 0.45 |
| D5 | 98.73 | 97.88 | 97.45 | 98.51 | 97.88 | 97.66 | 97.66 | 98.30 | 98.73 | 99.36 | 98.22 0.58 |
| D6 | 99.73 | 99.73 | 99.46 | 99.46 | 99.73 | 99.59 | 99.73 | 99.86 | 99.86 | 99.73 | 99.69 0.14 |
Table 9.
Confusion matrix for non–healthy CXR screening.
| Dataset | ||||||
|---|---|---|---|---|---|---|
| D4 | 1,078 | 1,068 | 10 | 348 | 341 | 7 |
| D5 | 1,049 | 1,043 | 6 | 359 | 349 | 10 |
| D6 | 1,049 | 1,049 | 0 | 1,164 | 1,164 | 0 |
For non–healthy CXR screening (D4 to D6 in Table 9), we observe the following. The proposed DNN in D4 misclassified 7 Covid-19 cases as Pneumonia and 10 Pneumonia cases were misidentified as Covid-19. From D5, 10 Covid-19 cases were misidentified as TB and 6 TB cases were misidentified as Covid-19. In D6, both TB and Pneumonia CXRs were correctly classified. With these, Table 10 shows accuracy, sensitivity, specificity, precision, and F1 score. In addition, we provide standard deviation () and mean () that help us check statistical stability of the proposed DNN model. Interestingly, we achieved AUC of 1 across all datasets (D4 to D6).
Table 10.
Performance (in ) on non–healthy CXR screening: accuracy, sensitivity, specificity, precision, and F1 score.
| Dataset | ACC | AUC | SEN | SPEC | PREC | F1 score | |
|---|---|---|---|---|---|---|---|
| D4 | 98.89 | 100.00 | 99.40 | 97.15 | 99.1 | 99.28 | |
| D5 | 98.99 | 100.00 | 99.15 | 98.31 | 99.43 | 99.29 | |
| D6 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| 99.29 | 100.00 | 99.52 | 98.49 | 99.53 | 99.52 | ||
| 0.614 | 0 | 0.437 | 1.433 | 0.433 | 0.413 |
6. Previous studies
In this section, we provide a quick overview of our experiments as well as comparison study with several existing approaches.
In the literature, majority of the researchers used deep transfer learning approaches [19], [16], [21] using pre-trained models to detect Covid-19, Pneumonia, and TB using either CXRs. Of all, few authors [14], [21] presented DL models that took into account non–healthy CXR screening for Covid-19 and Pneumonia detection. No significant research works took into account non–healthy CXR screening for multiple disease types: Covid-19, TB, and Pneumonia.
In Table 11, Table 12, Table 13 , we provide comparative studies. Table 11 shows the findings of some of the most recent methodologies reported for Covid-19 detection. We observed that most of the studies at the beginning of the coronavirus pandemic did not have enough Covid-19 positive CXRs, but later in the year 2020, we there exist relatively large data. In this category, the proposed DNN model achieved 99.87% accuracy, which is the third highest accuracy among them. Note that, our comparison may not be fair as we do not have exact same dataset as well as size. As before, Table 12 shows findings of some of the most recent methodologies reported on Pneumonia detection. In contrast, the proposed model reported an accuracy of 99.55%, which outperforms the existing models. However, it is important to note that our comparison may not be fair as results were produced from using different datasets. In a similar fashion, several modern approaches were found for TB detection; and few recent works are provided in Table 13. As before, our results (e.g., 99.76% accuracy) are comparable with the state-of-the-art results.
Table 11.
Performance comparison for Covid-19 detection. Also, Covid-19 positive cases are provided.
| Authors | Dataset size | Performance (in %) |
|||
|---|---|---|---|---|---|
| ACC | AUC | SPEC | SEN | ||
| Das et al. (2020) [14] | 18,524 CXRs: Covid-19 (972) | 98.77 | 99.00 | 99.00 | 95.00 |
| Togaçar et al. (2020) [17] | 458 CXRs: Covid-19 (295) | 97.78 | – | 95.74 | 98.86 |
| Minaee et al. (2020) [19] | 5,420 CXRs: Covid-19 (420) | 98.00 | – | 95.80 | 91.00 |
| Ozturk et al. (2020) [21] | 1,127 CXRs: Covid-19 (127) | 98.08 | – | 95.30 | 95.13 |
| Chowdhury et al. (2020) [23] | 3,487 CXRs: Covid-19 (423) | 99.70 | 100.00 | 99.55 | 99.70 |
| Aradhya et al. (2021) [13] | 306 CXRs: Covid-19 (69) | 79.76 | – | – | – |
| Ismael et al. (2021) [25] | 380 CXRs: Covid-19 (180) | 92.63 | – | – | – |
| Narin et al. (2021) [18] | 14,194 CXRs: Covid-19 (341) | 96.01 | – | 96.60 | 91.80 |
| Mukherjee et al. (2021) [15] | 336 CXRs: Covid-19 (168) | 95.83 | 97.31 | 98.21 | 93.45 |
| Mukherjee et al. (2021) [16] | 260 CXRs: Covid-19 (130) | 96.92 | 99.22 | 100.00 | 94.20 |
| Bassi & Attux (2021) [20] | 2,064 CXRs: Covid-19 (439) | 100.00 | – | 99.98 | 99.99 |
| Hussain et al. (2021) [22] | 7,390 CXRs: Covid-19 (2,843) | 99.12 | – | 97.36 | 95.36 |
| Rahman et al. (2021) [24] | 18,479 CXRs: Covid-19 (3,616) | 99.65 | 100.00 | 95.59 | 94.56 |
| Proposed DNN | 2,541 CXRs: Covid-19 (1,200) | 99.87 | 100.00 | 100.00 | 100.00 |
Table 12.
Performance comparison for Pneumonia detection. Also, Pneumonia positive cases are provided.
| Authors | Dataset size | Performance (in %) |
|||
|---|---|---|---|---|---|
| ACC | AUC | SPEC | SEN | ||
| Kermany, et al. (2018) [38] | 5,232 CXRs: Pneumonia (3,883) | 93.40 | 98.80 | 94.00 | 96.60 |
| Jaiswal et al. (2019) [31] | 2,5684 CXRs: Pneumonia (11,500) | 98.18 | – | – | – |
| Stephen et al. (2019) [36] | 5,856 CXRs: Pneumonia (4,273) | 95.31 | – | – | – |
| Hashmi et al. (2020) [32] | 5,856 CXRs: Pneumonia (4,273) | 98.43 | 99.76 | 98.65 | 99.00 |
| Jain et al. (2020) [34] | 5,856 CXRs: Pneumonia(4,273) | 92.31 | – | – | 98.00 |
| Chouhan et al. (2020) [35] | 5,229 CXRs: Pneumonia (3,883) | 96.40 | 99.34 | – | 99.62 |
| Hammoudi et al. (2021) [27] | 5,232 CXRs: Pneumonia (3,883) | 97.97 | – | – | – |
| Manickam et al. (2021) [28] | 5,229 CXRs: Pneumonia (3,883) | 93.06 | – | – | 96.78 |
| Asnaoui et al. (2021) [29] | 6,087 CXRs: Pneumonia (4,273) | 95.09 | – | 98.31 | 94.43 |
| Waisy et al. (2021) [30] | 800 CXRs: Pneumonia (400) | 99.93 | – | 100.00 | 99.90 |
| Ibrahim et al. (2021) [33] | 11,568 CXRs: Pneumonia(4,450) | 94.43 | – | 100.00 | 97.44 |
| Cha et al. (2021)[37] | 5,856 CXRs: Pneumonia (4,273) | 96.63 | 96.03 | – | 98.46 |
| Proposed DNN | 5,216 CXRs: Pneumonia (3,875) | 99.55 | 100.00 | 100.00 | 99.40 |
Table 13.
Performance comparison for TB detection. Also, TB positive cases are provided.
| Authors | Dataset size | Performance (in %) |
|||
|---|---|---|---|---|---|
| ACC | AUC | SPEC | SEN | ||
| Karargyris et al. (2016) [49] | 615 CXRs: TB (275) | 89.60 | 93.00 | 79.10 | 89.60 |
| Santosh et al. (2016) [47] | 682 CXRs: TB (400) | 86.36 | 94.00 | – | – |
| Santosh et al. (2017) [48] | 1,160 CXRs:: TB (478) | 91.00 | 96.00 | – | – |
| Lakhani et al. (2017) [45] | 1,007 CXRs: TB (492) | 99.00 | 99.00 | 100.00 | 97.30 |
| Vajda et al. (2018) [39] | 814 CXRs: TB (392) | 97.03 | 99.00 | – | – |
| Qin et al. (2019) [46] | 1,196 CXRs: TB (218) | 96.00 | 94.00 | 95.00 | 95.00 |
| Munadi et al. (2020) [40] | 662 CXRs: TB (336) | 67.55 | – | – | 94.08 |
| Rahman et al. (2020) [44] | 7,000 CXRs: TB (3,500) | 96.47 | – | 96.40 | |
| Khan et al. (2020) [42] | 2,198 CXRs: TB (272) | – | – | 75.00 | 93.00 |
| Ayaz et al. (2021) [41] | 662 CXRs: TB (336) | 97.59 | 99.00 | – | – |
| Qin et al. (2021) [50] | 23,954 CXRs: TB (10,837) | 91.29 | – | 95.00 | 95.00 |
| Proposed DNN | 7,000 CXRs: TB (3,500) | 99.76 | 100.00 | 99.91 | 99.61 |
7. Popular DNNs
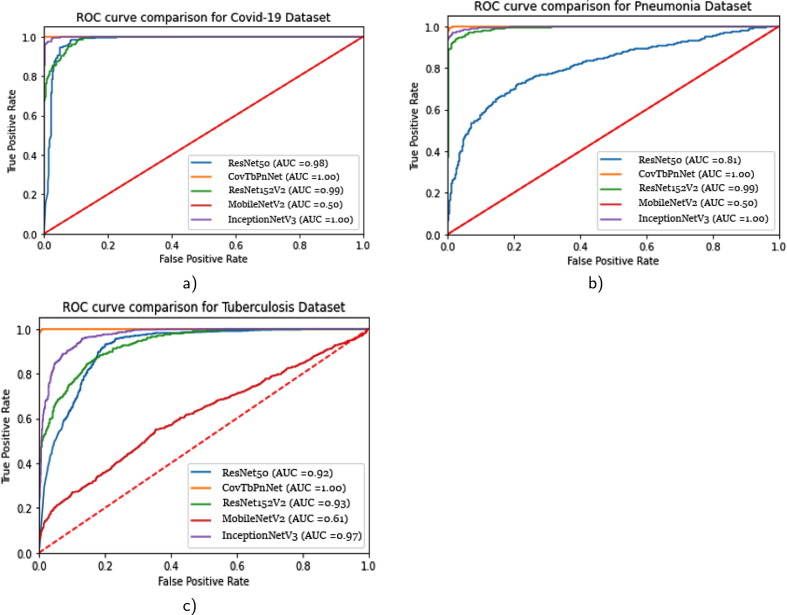
To provide a fair comparison with the popular DNNs, we used exact same datasets (D1 to D3) and evaluation protocol. In our study, we were limited to the following popular DNNs: ResNet50, ResNet152V2, MobileNetV2, and InceptionNetV3. With these, we computed ROC curves and provided in Fig. 4 . ROC curve (the area beneath the receiver operating characteristics) is an important statistical assessment. As compared to ResNet50, ResNet152V2, MobileNetV2, and InceptionV3, our proposed DNN (Covtben) performed better for all three different infectious diseases types: Covid-19 (D1 dataset), Pneumonia (D2 dataset), and TB (D3 dataset).
Fig. 4.
AUC comparison: the proposed DNN (CovTbPnNet), ResNet50, ResNet152V2, MobileNetV2, and InceptionNetV3 a) Covid-19, b) Pneumonia, and c) Tuberculosis.
For a statistical significance test, we employed the Friedman statistics on three different datasets that follows ROC curve in Fig. 4. With this, we have DNN models (InceptionNetV3, CovTbPnNet (proposed model), ResNet152V2, ResNet52, and MobileNetV2) applied on different datasets. For this test, we used AUC scores. To know what models perform the best, let us consider be the rank of model on dataset. We then computed mean of the ranks of all models on all datasets as,
In Table 14 , we provide detalied ranking information of all models we employed. Our proposed model (CovTbPnNet) ranked 1.333 as opposed to 1.667 (InceptionNetV3). Using null hypothesis, we observed that models do not show significant difference even though our model ranked first. We then computed the Friedman statistics using a chi-squared score,
Table 14.
Statistical significance test using AUC scores of five different DNN models on three different datasets
| Dataset/Model | InceptionNetV3 | CovTbPnNet | ResNet152V2 | ResNet52 | MobileNetV2 | |
|---|---|---|---|---|---|---|
| D1 | 1.00 (1.5) | 1.00 (1.5) | 0.99 (3) | 0.98 (4) | 0.50 (5) | |
| D2 | 1.00 (1.5) | 1.00 (1.5) | 0.99 (3) | 0.81 (4) | 0.50 (5) | |
| D3 | 0.97 (2) | 1.00 (1) | 0.93 (3) | 0.92 (4) | 0.61 (5) | |
| Mean rank | 1.667 | 1.333 | 3 | 4 | 5 |
With degrees of freedom in our test, was 4.83. For = 0.05, upper-tail critical value of chi-square distribution was 5.435. This means that we observed no significant difference.
8. Conclusion
In this paper, we have presented a lightweight (9-layered) deep neural network (DNN) to detect pulmonary abnormalities in chest x-rays (CXRs) due to infectious diseaseX: Covid-19, Pneumonia, and Tuberculosis (TB). In our experiments, we were not just limited to healthy versus non–healthy CXR screening, we also extended to non–healthy CXR screening. The latter part of the experiments helped us analyze how well multiple disease types can be used for classification. In all scenarios, performance scores can be compared with existing models (for Covid-19, Pneumonia, and TB). Further, popular DNNs were compared as previous studies used different dataset sizes.
As we have received the highest possible accuracy of more than 99.50%, we could see possible screening tool for infectious diseaseX detection. Note that such a tool could help in assisting radiologists to make clinical decisions. Further, we are encouraged to work on cross-population train/test models under the scope of activities as well as federated learning.
Ethics declarations
Funding: NA.
Ethical approval: This article does not contain any studies with human participants performed by any of the authors.
CRediT authorship contribution statement
Md. Kawsher Mahbub: Methodology, Writing – original draft. Milon Biswas: Methodology, Writing – original draft, Writing – review & editing. Loveleen Gaur: Dicsussion. Fayadh Alenezi: Writing – review & editing. KC Santosh: Methodology, Conceptualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
NA
Biographies

Md. Kawsher Mahbub is currently working as a Lecturer in the department of Computer Science & Engineering, BUBT. He had received his Bachelor of Engineering degree from Bangladesh University of Business and Technology (BUBT). He enjoy learning about the new advancement in technology, especially concerning IT, Artificial Intelligence, Machine learning, and robotics. His aim is to work innovatively for the enhancement and betterment of education. He enjoys learning about new advancements in technology. He has experienced working in Python, Keras, TensorFlow, Sklearn, and Scipy. His aim is to work innovatively for the enhancement and betterment of education. His particular research interests include health informatics, deep learning, computer vision, and robotics.

Assistant Professor Milon Biswas, is Coordinator of Diploma holders of the Department of Computer Science and Engineering at the Bangladesh University of BUsiness and Technology (BUBT). He also serves Bangladesh University of Professionals as an Adjunct Assistant Professor. Artificial intelligence, machine learning, pattern recognition, computer vision, medical image processing, and data mining with applications are some of his research interests. He was awarded the National Science and Technology Fellowship by ICT Ministry of People’s Republic of Bangladesh for his research work during graduate studies.

Professor Loveleen Gaur is the Professor and Program Director (Artificial Intelligence Data Analytics of the Amity international Business School, Amity University, Noida, India. She is a senior IEEE member and Series Editor with CRC and Wiley Her expertise in Artificial intelligence and its applications in the business and healthcare domain. Prof Gaur has significantly contributed to enhancing scientific understanding of Artificial Intelligence by participating in over three hundred scientific conferences, symposia, and seminars, by chairing technical sessions and delivering plenary and invited talks. She has chaired various positions in international Conferences of repute and is a reviewer with top rated journals of IEEE, SCI and ABDX Journals. She has been honored with prestigious National and international awards.

Dr. Fayadh Alenezi is an Assistant Professor in the Department of Electrical Engineering at Jouf University, Sakaka, Saudi Arabia. He received the B.Sc. degree (Hons.) in Electrical Engineering Electronics and Communications Track from Jouf University, Saudi Arabia in 2012, and the M.S. degree Electrical Engineering from Southern Illinois University Carbondale, Illinois, the USA in 2015, and the Ph.D. degree in Electrical Engineering from the University of Toledo, Ohio, the USA in 2019. Alenezi has authored many journal and conference papers. His research interests include artificial intelligence, image processing, signal processing, image enchantment, machine learning, neural networks, and facial recognition

Professor KC Santosh is Chair of the Department of Computer Science at the University of South Dakota (USD). He also serves International Medical University as an Adjunct Professor (Full). Before joining USD, he worked as Research Fellow at the US National Library of Medicine (NLM), National Institutes of Health (NIH). He was Postdoctoral Research Scientist at the Loria Research Centre (with industrial partner, ITESOFT (France)). He has demonstrated expertise in artificial intelligence, machine learning, pattern recognition, computer vision, image processing, and data mining with applications such as medical imaging informatics, document imaging, biometrics, forensics, and speech analysis. His research projects are funded (of more than $2 m) by multiple agencies, such as SDCRGP, Department of Education, National Science Foundation, and Asian Office of Aerospace Research and Development. He is the proud recipient of the Cutler Award for Teaching and Research Excellence (USD, 2021), the President’s Research Excellence Award (USD, 2019), and the Ignite from the U.S. Department of Health & Human Services (2014).
Footnotes
GitHub: https://github.com/Kawsher/A-unified-deep-learning-model.git
References
- 1.Fan Wu., Zhao Su., Bin Yu., Chen Yan-Mei, Wang Wen, Song Zhi-Gang, Yi Hu., Tao Zhao-Wu, Tian Jun-Hua, Pei Yuan-Yuan, et al. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Feng, Wang Jun, Shi Jun, Ziyan Wu., Wang Qian, Tang Zhenyu, He Kelei, Shi Yinghuan, Shen Dinggang. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for covid-19. IEEE Rev. Biomed. Eng. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 3.McCall Becky. Covid-19 and artificial intelligence: protecting health-care workers and curbing the spread. Lancet Digital Health. 2020;2(4):e166–e167. doi: 10.1016/S2589-7500(20)30054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishya Raju, Javaid Mohd, Khan Ibrahim Haleem, Haleem Abid. Artificial intelligence (ai) applications for covid-19 pandemic. Diabetes Metabolic Syndrome: Clin. Res. Rev. 2020;14(4):337–339. doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albahri Ahmed Shihab, Hamid Rula A, Alwan Jwan K, Al-Qays Z.T., Zaidan A.A., Zaidan B.B., Albahri A.O.S., AlAmoodi A.H., Khlaf Jamal Mawlood, Almahdi E.M., et al. Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (covid-19): a systematic review. J. Med. Syst. 2020;44:1–11. doi: 10.1007/s10916-020-01582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xueyan Mei, Hao-Chih Lee, Kai-yue Diao, Mingqian Huang, Bin Lin, Chenyu Liu, Zongyu Xie, Yixuan Ma, Philip M Robson, Michael Chung, et al., Artificial intelligence–enabled rapid diagnosis of patients with covid-19, Nat. Med. 26(8) (2020) 1224–1228. [DOI] [PMC free article] [PubMed]
- 7.Yujin Oh., Park Sangjoon, Ye Jong Chul. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 8.Fan Deng-Ping, Zhou Tao, Ji Ge-Peng, Zhou Yi, Chen Geng, Fu Huazhu, Shen Jianbing, Shao Ling. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 9.Huang Chaolin, Wang Yeming, Li Xingwang, Ren Lili, Zhao Jianping, Hu Yi, Zhang Li, Fan Guohui, Xu Jiuyang, Gu Xiaoying, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santosh K.C. Ai-driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on multitudinal/multimodal data. J. Med. Syst. 2020;44(5):1–5. doi: 10.1007/s10916-020-01562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santosh K.C. Covid-19 prediction models and unexploited data. J. Med. Syst. 2020;44(9):1–4. doi: 10.1007/s10916-020-01645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santosh K.C., Joshi Amit. Springer; 2021. COVID-19: prediction, decision-making, and its impacts. [Google Scholar]
- 13.V.N. Manjunath Aradhya, Mufti Mahmud, D.S. Guru, Basant Agarwal, M. Shamim Kaiser, One-shot cluster-based approach for the detection of covid–19 from chest x–ray images, Cogn. Comput. (2021) 1–9. [DOI] [PMC free article] [PubMed]
- 14.Das Dipayan, Santosh K.C., Pal Umapada. Truncated inception net: Covid-19 outbreak screening using chest x-rays. Phys. Eng. Sci. Med. 2020;43(3):915–925. doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee Himadri, Ghosh Subhankar, Dhar Ankita, Obaidullah Sk Md, Santosh K.C., Roy Kaushik. Deep neural network to detect covid-19: one architecture for both ct scans and chest x-rays. Appl. Intell. 2021;51(5):2777–2789. doi: 10.1007/s10489-020-01943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee Himadri, Ghosh Subhankar, Dhar Ankita, Obaidullah Sk Md, Santosh K.C., Roy Kaushik. Shallow convolutional neural network for covid-19 outbreak screening using chest x-rays. Cogn. Comput. 2021:1–14. doi: 10.1007/s12559-020-09775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toğaçar Mesut, Ergen Burhan, Cömert Zafer. Covid-19 detection using deep learning models to exploit social mimic optimization and structured chest x-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narin Ali, Kaya Ceren, Pamuk Ziynet. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minaee Shervin, Kafieh Rahele, Sonka Milan, Yazdani Shakib, Soufi Ghazaleh Jamalipour. Deep-covid: Predicting covid-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassi Pedro R.A.S., Attux Romis. A deep convolutional neural network for covid-19 detection using chest x-rays. Research on. Biomed. Eng. 2021:1–10. [Google Scholar]
- 21.Ozturk Tulin, Talo Muhammed, Yildirim Eylul Azra, Baloglu Ulas Baran, Yildirim Ozal, Acharya U Rajendra. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emtiaz Hussain, Mahmudul Hasan, Md Anisur Rahman, Ickjai Lee, Tasmi Tamanna, and Mohammad Zavid Parvez. Corodet: A deep learning based classification for covid-19 detection using chest x-ray images. Chaos, Solitons & Fractals, 142:110495, 2021. [DOI] [PMC free article] [PubMed]
- 23.Muhammad EH Chowdhury, Tawsifur Rahman, Amith Khandakar, Rashid Mazhar, Muhammad Abdul Kadir, Zaid Bin Mahbub, Khandakar Reajul Islam, Muhammad Salman Khan, Atif Iqbal, Nasser Al Emadi, et al. Can ai help in screening viral and covid-19 pneumonia? IEEE Access, 8:132665–132676, 2020.
- 24.Tawsifur Rahman, Amith Khandakar, Yazan Qiblawey, Anas Tahir, Serkan Kiranyaz, Saad Bin Abul Kashem, Mohammad Tariqul Islam, Somaya Al Maadeed, Susu M Zughaier, Muhammad Salman Khan, et al. Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images. Computers in biology and medicine, 132:104319, 2021. [DOI] [PMC free article] [PubMed]
- 25.Ismael Aras M, Şengür Abdulkadir. Deep learning approaches for covid-19 detection based on chest x-ray images. Expert Syst. Appl. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.KC Santosh and Sourodip Ghosh Covid-19 imaging tools: How big data is big? J. Med. Syst. 2021;45(7):1–8. doi: 10.1007/s10916-021-01747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammoudi Karim, Benhabiles Halim, Melkemi Mahmoud, Dornaika Fadi, Arganda-Carreras Ignacio, Collard Dominique, Scherpereel Arnaud. Deep learning on chest x-ray images to detect and evaluate pneumonia cases at the era of covid-19. J. Med. Syst. 2021;45(7):1–10. doi: 10.1007/s10916-021-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adhiyaman Manickam, Jianmin Jiang, Yu Zhou, Abhinav Sagar, Rajkumar Soundrapandiyan, and R Dinesh Jackson Samuel. Automated pneumonia detection on chest x-ray images: A deep learning approach with different optimizers and transfer learning architectures. Measurement, page 109953, 2021.
- 29.El Asnaoui Khalid. Design ensemble deep learning model for pneumonia disease classification. International Journal of Multimedia Information Retrieval. 2021;10(1):55–68. doi: 10.1007/s13735-021-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.AS Al-Waisy, Mazin Abed Mohammed, Shumoos Al-Fahdawi, MS Maashi, Begonya Garcia-Zapirain, Karrar Hameed Abdulkareem, SA Mostafa, Nallapaneni Manoj Kumar, and Dac Nhuong Le. Covid-deepnet: hybrid multimodal deep learning system for improving covid-19 pneumonia detection in chest x-ray images. Computers, Materials and Continua, 67(2), 2021.
- 31.Amit Kumar Jaiswal, Prayag Tiwari, Sachin Kumar, Deepak Gupta, Ashish Khanna, and Joel JPC Rodrigues. Identifying pneumonia in chest x-rays: a deep learning approach. Measurement, 145:511–518, 2019.
- 32.Mohammad Farukh Hashmi, Satyarth Katiyar, Avinash G Keskar, Neeraj Dhanraj Bokde, and Zong Woo Geem. Efficient pneumonia detection in chest xray images using deep transfer learning. Diagnostics, 10(6):417, 2020. [DOI] [PMC free article] [PubMed]
- 33.Abdullahi Umar Ibrahim, Mehmet Ozsoz, Sertan Serte, Fadi Al-Turjman, and Polycarp Shizawaliyi Yakoi. Pneumonia classification using deep learning from chest x-ray images during covid-19. Cognitive Computation, pages 1–13, 2021. [DOI] [PMC free article] [PubMed]
- 34.Rachna Jain, Preeti Nagrath, Gaurav Kataria, V Sirish Kaushik, and D Jude Hemanth. Pneumonia detection in chest x-ray images using convolutional neural networks and transfer learning. Measurement, 165:108046, 2020.
- 35.Vikash Chouhan, Sanjay Kumar Singh, Aditya Khamparia, Deepak Gupta, Prayag Tiwari, Catarina Moreira, Robertas Damaševičius, and Victor Hugo C De Albuquerque. A novel transfer learning based approach for pneumonia detection in chest x-ray images. Applied Sciences, 10(2):559, 2020.
- 36.Stephen Okeke, Sain Mangal, Maduh Uchenna Joseph, Jeong Do-Un. An efficient deep learning approach to pneumonia classification in healthcare. Journal of healthcare engineering. 2019, 2019. doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cha So-Mi, Lee Seung-Seok, Ko Bonggyun. Attention-based transfer learning for efficient pneumonia detection in chest x-ray images. Applied Sciences. 2021;11(3):1242. [Google Scholar]
- 38.Daniel S Kermany, Michael Goldbaum, Wenjia Cai, Carolina CS Valentim, Huiying Liang, Sally L Baxter, Alex McKeown, Ge Yang, Xiaokang Wu, Fangbing Yan, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell, 172(5), 1122–1131, 2018. [DOI] [PubMed]
- 39.Szilárd Vajda, Alexandros Karargyris, Stefan Jaeger, KC Santosh, Sema Candemir, Zhiyun Xue, Sameer Antani, and George Thoma. Feature selection for automatic tuberculosis screening in frontal chest radiographs. Journal of medical systems, 42(8):1–11, 2018. [DOI] [PubMed]
- 40.Munadi Khairul, Muchtar Kahlil, Maulina Novi, Pradhan Biswajeet. Image enhancement for tuberculosis detection using deep learning. IEEE Access. 2020;8:217897–217907. [Google Scholar]
- 41.Ayaz Muhammad, Shaukat Furqan, Raja Gulistan. Ensemble learning based automatic detection of tuberculosis in chest x-ray images using hybrid feature descriptors. Physical and Engineering Sciences in Medicine. 2021;44(1):183–194. doi: 10.1007/s13246-020-00966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faiz Ahmad Khan, Arman Majidulla, Gamuchirai Tavaziva, Ahsana Nazish, Syed Kumail Abidi, Andrea Benedetti, Dick Menzies, James C Johnston, Aamir Javed Khan, and Saima Saeed. Chest x-ray analysis with deep learning-based software as a triage test for pulmonary tuberculosis: a prospective study of diagnostic accuracy for culture-confirmed disease. The Lancet Digital Health, 2(11), e573–e581, 2020. [DOI] [PubMed]
- 43.Tasci Erdal, Uluturk Caner, Ugur Aybars. 2021. A voting-based ensemble deep learning method focusing on image augmentation and preprocessing variations for tuberculosis detection. Neural Computing and Applications; pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tawsifur Rahman, Amith Khandakar, Muhammad Abdul Kadir, Khandaker Rejaul Islam, Khandakar F Islam, Rashid Mazhar, Tahir Hamid, Mohammad Tariqul Islam, Saad Kashem, Zaid Bin Mahbub, et al. Reliable tuberculosis detection using chest x-ray with deep learning, segmentation and visualization. IEEE Access, 8:191586–191601, 2020.
- 45.Lakhani Paras, Sundaram Baskaran. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 46.Zhi Zhen Qin, Melissa S Sander, Bishwa Rai, Collins N Titahong, Santat Sudrungrot, Sylvain N Laah, Lal Mani Adhikari, E Jane Carter, Lekha Puri, Andrew J Codlin, et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: A multi-site evaluation of the diagnostic accuracy of three deep learning systems. Scientific reports, 9(1):1–10, 2019. [DOI] [PMC free article] [PubMed]
- 47.KC Santosh, Szilárd Vajda, Sameer Antani, and George R Thoma. Edge map analysis in chest x-rays for automatic pulmonary abnormality screening. International journal of computer assisted radiology and surgery, 11(9):1637–1646, 2016. [DOI] [PubMed]
- 48.KC Santosh and Sameer Antani Automated chest x-ray screening: Can lung region symmetry help detect pulmonary abnormalities? IEEE transactions on medical imaging. 2017;37(5):1168–1177. doi: 10.1109/TMI.2017.2775636. [DOI] [PubMed] [Google Scholar]
- 49.Alexandros Karargyris, Jenifer Siegelman, Dimitris Tzortzis, Stefan Jaeger, Sema Candemir, Zhiyun Xue, KC Santosh, Szilárd Vajda, Sameer Antani, Les Folio, et al. Combination of texture and shape features to detect pulmonary abnormalities in digital chest x-rays. International journal of computer assisted radiology and surgery, 11(1):99–106, 2016. [DOI] [PMC free article] [PubMed]
- 50.Zhi Zhen Qin, Shahriar Ahmed, Mohammad Shahnewaz Sarker, Kishor Paul, Ahammad Shafiq Sikder Adel, Tasneem Naheyan, Rachael Barrett, Sayera Banu, and Jacob Creswell. Tuberculosis detection from chest x-rays for triaging in a high tuberculosis-burden setting: an evaluation of five artificial intelligence algorithms. The Lancet Digital Health, 3(9), e543–e554, 2021. [DOI] [PubMed]