Abstract
Background
Routine use of chlorhexidine or octenidine for antiseptic bathing may have unintended consequences. Our analysis aimed to assess the phenotypic susceptibility of bacterial isolates from clinical samples to chlorhexidine and octenidine collected from intensive care units (ICU) that routinely used 2% chlorhexidine-impregnated wash cloths or 0.08% octenidine wash mitts (intervention) or water and soap (control) for daily patient care.
Methods
This study was conducted within the context of a three armed cluster-randomised controlled decolonisation trial (Registration number DRKS00010475, registration date August 18, 2016). Bacterial isolates were collected prior to and at the end of a 12-month-intervention period from patients with ≥ 3 days length of stay at an ICU assigned to one of two intervention groups or the control group. Phenotypic susceptibility to chlorhexidine and octenidine was assessed by an accredited contract research laboratory determining minimal inhibitory concentrations (MIC) as percentage of extraction solutions used. MIC were reported as estimated concentrations in μg/ml derived from the chlorhexidine and octenidine extraction solutions. Statistical analyses including generalized estimating equation models were applied.
Results
In total, 790 ICU-attributable bacterial isolates from clinical samples (e.g. blood, urine, tracheal aspirate) were eligible for all analyses. Pathogens included were Staphylococcus aureus (n = 155), coagulase-negative staphylococci (CoNS, n = 122), Escherichia coli (n = 227), Klebsiella spp. (n = 150) and Pseudomonas aeruginosa (n = 136). For all species, chlorhexidine and octenidine MIC did not increase from baseline to intervention period in the antiseptic bathing groups. For proportions of bacterial isolates with elevated chlorhexidine / octenidine MIC (≥ species-specific chlorhexidine / octenidine MIC50), adjusted incidence rate ratios (aIRR) showed no differences between the intervention groups and the control group (intervention period).
Conclusion
We found no evidence for reduced phenotypic susceptibilities of bacterial isolates from clinical samples to chlorhexidine or octenidine in ICUs 12 months after implementation of routine antiseptic bathing with the respective substances.
Introduction
Chlorhexidine gluconate and octenidine dihydrochloride are two cationic biocides that can bind covalently to the bacterial cell membrane, subsequently cause depolarization and can lead to bacterial cell death if used at high concentrations [1, 2]. Meta-analyses and clinical trials are available that demonstrate the effectiveness of chlorhexidine bathing to reduce healthcare-associated infections (HAI) including bloodstream infections (BSI) in intensive care units (ICUs) [3–9]. Thus, chlorhexidine is most commonly used for antiseptic bathing of intensive care patients. At the same time, only few clinical trials on the effect of antiseptic bathing with octenidine for BSI prevention are available [10–12]. In Germany, antiseptic bathing of ICU patients is no routine infection control practice. If applied, however, octenidine is the most frequently used substance [13].
We conducted a three-armed cluster-randomised controlled trial (cRCT) in 72 adult ICUs to investigate the effect of daily patient bathing with chlorhexidine, octenidine or routine care with water and soap (control) on central-line associated BSI (CLABSI) rates [14]. This trial found no preventive effect of antiseptic bathing neither with chlorhexidine nor octenidine when comparing CLABSI rates of the intervention groups with the control group [14]. The post hoc before-after analysis of our cluster-randomised decolonisation trial, however, suggests that ICUs with CLABSI rates ≥0.8 CLABSI per 1000 central-line (CL) days might benefit from bathing with 2% chlorhexidine-impregnated cloths [15]. At the same time, octenidine showed no preventive effect on ICU-attributable CLABSI [15].
The use of chlorhexidine or octenidine may have unintended consequences. High usage of these antiseptic substances is associated with increases in chlorhexidine and octenidine minimal inhibitory concentrations (MIC) among clinical isolates [16]. Further, long-term use of chlorhexidine is suspected not only to enhance the development of non-susceptibility, but also to increase antibiotic cross-resistance, decolonization failure or potentially disadvantageous alterations of the skin microbiome [17–20].
Our analysis aimed to answer the question whether daily patient bathing with 2% impregnated chlorhexidine cloths or 0.08% octenidine wash mitts might lead to reduced chlorhexidine or octenidine susceptibilities of ICU-attributable bacterial isolates from clinical samples in the CLIP-ID trial.
Methods
Trial design and participants
This investigation was done within a cluster-randomised controlled trial (cRCT) as part of the project Climate and pathogens–impact of decolonisation (CLIP-ID, registration number DRKS00010475, registration date August 18, 2016). Details of the CLIP-ID trial including the study protocol were published elsewhere [14]. Briefly, our cRCT was conducted on 72 ICUs in 68 hospitals in Germany and Austria. Each ICU represented one cluster and was randomly assigned to one of three decolonisation regimes that had to be applied for 12 consecutive months. The random allocation sequence was computer-generated and applied on the cluster (ICU) level. We stratified ICUs by type of ICU (medical, surgical, interdisciplinary wards in hospitals with < 400 beds and interdisciplinary wards in hospitals with ≥ 400 beds) and size of hospital before randomization. In the chlorhexidine group, all ICUs performed daily patient bathing with 2% chlorhexidine-impregnated cloths (Sage 2% Chlorhexidine Gluconate (CHG) by Stryker) below the jaw line and chlorhexidine-free cloths above the jaw line (Stryker). In the octenidine group, ICUs used 0.08% octenidine disposable wash mitts (Octenisan® by Schülke). In the routine care (control) group, all ICUs applied non-antiseptic soap and water (routine care) for daily patient care.
Collection of the ICU-attributable bacterial isolates from clinical samples
All 72 ICUs participating were requested to collect 10 bacterial isolates from clinical samples prior to and 10 bacterial isolates at the end of the intervention period to analyze phenotypic susceptibility to the antiseptic substances (chlorhexidine and octenidine) applied in this trial. Bacterial isolates collected must met the following inclusion criteria: i) specimen were collected for medical or diagnostic purposes only; ii) bacteria isolated from clinical material (e.g. tracheal aspirate, urine, blood, liquor) not colonization sites (no screening isolates from rectal swabs, stool, nasal-pharyngeal swabs); iii) clinical material from patients with length of stay of at least 3 days on the ICU participating and iv) identification of Staphylococcus (S.) aureus, Enterobacterales (e.g. Escherichia (E.) coli, Klebsiella (K.) subspecies (spp.) or Pseudomonas (P.) aeruginosa from all clinical material or coagulase-negative staphylococci (CoNS) from blood samples only. Bacterial isolates (with Amies transport medium) were sent by the diagnostic laboratories of ICUs participating in the trial to the coordinating study center. The study was not blinded. However, collection of bacterial isolates was done by laboratory personnel who were not informed about assignments of ICUs to one of three study groups. Information on isolates (e.g. ICU name, ICU day of sample collection, anonymous sequential patient number, date, material, and species) were provided by the ICU and their diagnostic laboratories in anonymous form. No patient based data were collected or distributed.
Phenotypic susceptibility to chlorhexidine and octenidine
Phenotypic susceptibility of bacterial isolates from clinical samples to the antiseptic substances contained in the chlorhexidine-impregnated cloths and octenidine disposable wash mitts were analysed by determining minimal inhibitory concentrations (MIC). Phenotypic susceptibility testings were assigned as contract work to an accredited laboratory and conducted according to VAH method 7 (A00040) [21].
Extraction solutions of antiseptic substances (chlorhexidine and octenidine) were harvested by wringing out chlorhexidine-impregnated cloths (Stryker / Sage Products) and octenidine wash mitts (Schülke) provided by the manufacturer. Concentrations of these extraction solutions were estimated to be similar to the concentrations of the ready-to-use products reported by the manufacturers: 2% (20,000 μg/ml) for chlorhexidine and 0.08% (800 μg/ml) for octenidine. Dilutions of antiseptic substances were made in water of standardized water hardness (WSH) according to VAH method 5 [21]. Briefly, bacteria were inoculated in soybean casein digest agar (CSA) at 37°C for 18 – 24h. Concentrations of these bacterial solutions were adjusted to 1.5–5.0 x 108 colony forming units (CFU)/ml. Subsequently, 150 μl of the respective antiseptic dilutions and 150 μl doubled concentrated soybean casein solutions (CSL) were mixed. Microtiter plates were inoculated with 3 μl bacteria (1:10-dilution in CSL) yielding in a final bacterial concentration of 1.5–5.0 x 105 colony forming units (CFU)/ml. Bacteria were inoculated at 37°C for 48 h. The lowest concentration of extraction solutions without bacterial growth (detectable turbidity) was interpreted as minimal inhibitory concentration (MIC) reported in percentage of extraction solution. Percentages were converted to estimated (est.) concentrations of extraction solutions in μg / ml. Controls were inoculated with WSH instead of antiseptic dilutions. Determination of bacteriostatic efficacy and suitable neutralizing agents was done according to VAH method 7 [21]. The following reference strains were included to the analysis: Staphylococcus aureus ATCC6538 (DSM 799), Proteus mirabilis ATCC 14153 (DSM 788), Klebsiella pneumoniae ESBL (DSM 16609), Pseudomonas aeruginosa ATCC 15442 and Escherichia coli NCTC 10538.
Statistical analyses
In the descriptive analysis, numbers with percentages, medians (MIC50) with interquartile ranges (IQR) and / or percentiles (MIC90) were calculated. The variables “sample site” and “study group and study period” were dummy-coded. P-values were calculated by Chi-square test or Fisher’s exact test for categorical, and by Mann-Whitney U Test for continuous variables.
We generated a binary variable to represent phenotypic chlorhexidine and octenidine susceptibilities of bacterial isolates from clinical samples. All bacterial isolates were categorized according to their chlorhexidine and octenidine MIC being ≥ or < the species-specific chlorhexidine and octenidine MIC50 (identified in this study). Bacterial isolates were categorized as “yes” (bacterial isolates with chlorhexidine / octenidine susceptibility ≥ species-specific MIC50) or “no” (chlorhexidine /octenidine susceptibility of bacterial isolates < species-specific MIC50).
In the multivariable analysis, we applied generalised estimating equation (GEE) models to identify effects of chlorhexidine / octenidine interventions on the susceptibility of bacterial isolates from clinical samples to these substances. We used one GEE model for all species to investigate the susceptibility to chlorhexidine, and another GEE model to investigate the susceptibility to octenidine. The outcomes “phenotypic susceptibilities to chlorhexidine / octenidine” were represented by proportions of bacterial isolates with chlorhexidine or octenidine MIC being ≥ the species-specific chlorhexidine / octenidine MIC50. Interaction of study groups (chlorhexidine / octenidine / control) and study periods (baseline / intervention) as well as potential cluster effects were considered in these models [22]. Control group (intervention period) served as reference. Further, ICU day of sample collection and clinical site (blood, tracheal aspirate, urine, wound, or other) were determined as possible confounders and considered in all models. The category “other” as clinical site included e.g. intra-operative samples, intra-abdominal swabs and / or tissue. All parameters added one degree of freedom to the model.
P-values less than 0.05 were considered significant. All analyses were performed using SPSS 27 (IBM SPSS statistics, Somer, NY, USA).
Sample size calculation
Initial sample size calculation of this cRCT was done for the primary outcome CLABSI [14]. However, we performed a power calculation for the outcomes “phenotypic susceptibilities to chlorhexidine / octenidine” represented by proportions of bacterial isolates with chlorhexidine or octenidine MIC being ≥ the species-specific chlorhexidine / octenidine MIC50. The latter were identified in this study (Table 2). We assumed that at the end of the intervention period, 35% of isolates were ≥ MIC50 in the control group (routine care) compared to 65% in the intervention group (antiseptic bathing with chlorhexidine or octenidine). Further, we assumed a ratio of 1:1 between the groups. With individual randomization, a sample size of 70 bacterial isolates in each group would have been required to show the difference with a power of 80%, and a two-sided type 1 error of 0.025. Adjusting for cluster effects with 13 clusters per arm and an intracluster correlation coefficient of 0.1 (adapted from literature [23]), we must have included 101 bacterial isolates per arm. Multiple comparisons to the routine care group were considered by a two-sided type 1 error of 0.025.
Table 2. Chlorhexidine and octenidine minimal inhibitory concentrations (MIC) of ICU-attributable bacterial isolates from clinical samples reported by study group and period.
| Total | Chlorhexidine group | Octenidine group | Routine care group = control | ||||
|---|---|---|---|---|---|---|---|
| Baseline | Intervention | Baseline | Intervention | Baseline | Intervention | ||
| Staphylococcus aureus | |||||||
| n (%) | 155 (100.0%) | 38 (25.0%) | 22 (14.1%) | 24 (15.4%) | 18 (11.5%) | 23 (14.7%) | 30 (19.2%) |
| Chlorhexidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 5.0 (2.0–5.0) | 5.0 (5.0–5.0) | 2.0 (2.0–4.3) | 5.0 (2.0–5.0) | 2.0 (2.0–5.0) | 5.0 (2.0–5.0) | 5.0 (2.8–10.0) |
| MIC90 in est. [μg/ml] | 5.0 | 5.0 | 4.3 | 5.0 | 5.0 | 5.0 | 10.0 |
| Octenidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 2.0 (2.0–2.0) | 2.0 (2.0–4.0) | 2.0 (0.8–2.0) | 2.0 (2.0–2.0) | 2.0 (2.0–2.0) | 2.0 (2.0–4.0) | 2.0 (2.0–2.0) |
| MIC90 in est. [μg/ml] | 2.0 | 4.0 | 2.0 | 2.0 | 2.0 | 4.0 | 2.0 |
| Coagulase-negative staphylococci | |||||||
| n (%) | 122 (100.0%) | 31 (25.4%) | 31 (25.4%) | 17 (13.9%) | 11 (9.0%) | 15 (12.3%) | 17 (13.9%) |
| Chlorhexidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 5.0 (2.0–5.0) | 5.0 (2.0–5.0) | 2.0 (1.0–2.0) | 5.0 (5.0–5.0) | 2.0 (1.0–3.5) | 5.0 (2.0–5.0) | 5.0 (5.0–5.0) |
| MIC90 in est. [μg/ml] | 5.0 | 5.0 | 2.0 | 5.0 | 3.5 | 5.0 | 5.0 |
| Octenidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 2.0 (2.0–4.0) | 2.0 (2.0–4.0) | 2.0 (0.8–2.0) | 2.0 (2.0–4.0) | 0.8(0.8–2.0) | 4.0 (2.0–4.0) | 2.0 (2.0–2.0) |
| MIC90 in est. [μg/ml] | 4.0 | 4.0 | 2.0 | 4.0 | 2.0 | 4.0 | 2.0 |
| Escherichia coli | |||||||
| n (%) | 227 (100.0%) | 64 (28.2%) | 37 (16.3%) | 41 (18.1%) | 18 (7.9%) | 39 (17.2%) | 28 (11.9%) |
| Chlorhexidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 5.0 (2.0–5.0) | 5.0 (2.0–5.0) | 2.0 (2.0–5.0) | 5.0 (5.0–5.0) | 2.0 (2.0–5.0) | 5.0 (5.0–5.0) | 5.0 (2.0–10.0) |
| MIC90 in est. [μg/ml] | 5.0 | 5.0 | 5.0 | 5.0 | 5.0 | 5.0 | 10.0 |
| Octenidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 4.0 (2.0–4.0) | 4.0 (4.0–4.0) | 2.0 (2.0–4.0) | 4.0 (4.0–4.0) | 2.0 (2.0–4.0) | 4.0 (4.0–4.0) | 4.0 (2.0–4.0) |
| MIC90 in est. [μg/ml] | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 |
| Klebsiella spp. | |||||||
| n (%) | 150 (100.0%) | 28 (18.7%) | 28 (18.7%) | 23 (15.3%) | 29 (19.3%) | 20 (13.3%) | 22 (14.7%) |
| Chlorhexidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 50.0 (20.0–50.0) | 50.0 (20.0–50.0) | 20.0 (20.0–50.0) | 50.0 (50.0–50.0) | 50.0 (20.0–50.0) | 50.0 (42.5–50.0) | 50.0 (20.0–50.0) |
| MIC90 in est. [μg/ml] | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 |
| Octenidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 4.0 (4.0–8.0) | 4.0 (4.0–12.0) | 4.0 (4.0–4.0) | 8.0 (4.0–10.0) | 4.0 (4.0–4.0) | 4.0 (4.0–13.0) | 4.0 (4.0–4.0) |
| MIC90 in est. [μg/ml] | 8.0 | 12.0 | 4.0 | 10.0 | 4.0 | 13.0 | 4.0 |
| Pseudomonas aeruginosa | |||||||
| n (%) | 136 (100.0%) | 19 (14.0%) | 18 (13.2%) | 17 (12.5%) | 40 (29.4%) | 18 (13.2%) | 24 (17.6%) |
| Chlorhexidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 50.0 (50.0–50.0) | 50.0 (35.0–50.0) | 50.0 (50.0–50.0) | 50.0 (50.0–50.0) | 50.0 (50.0–50.0) | 50.0 (50.0–50.0) | 50.0 (50.0–50.0) |
| MIC90 in est. [μg/ml] | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 |
| Octenidine | |||||||
| MIC50 in est. [μg/ml] (IQR) | 12.0 (8.0–24.0) | 8.0 (8.0–20.0) | 8.0 (8.0–16.0) | 16.0 (8.0–16.0) | 12.0 (8.0–16.0) | 24.0 (8.0–32.0) | 16.0 (8.0–24.0) |
| MIC90 in est. [μg/ml] | 24.0 | 20.0 | 16.0 | 16.0 | 16.0 | 32.0 | 24.0 |
Median MIC (MIC50) with inter quartile ranges (IQR) and MIC90 are given in estimated concentrations (est. μg/ml) derived from extraction solutions that were harvested from the ready-to-use products. MIC are reported for Staphylococcus aureus, coagulase-negative staphylococci, Escherichia coli, Klebsiella spp. and Pseudomonas aeruginosa in total and by study group (chlorhexidine, octenidine, routine care) and period (baseline, prior to the intervention period, intervention, at the end of the intervention period). Percentages of bacterial isolates per study group and period refer to the total number per species.
Ethics approval
The institutional ethical review board (IRB) of the Charité Universitätsmedizin Berlin granted their approval including a waiver of informed consent for this trial (processing number EA1/093/16). The present study did not include any individual patient data, but analysed aggregated and anonymous data only.
Results
Intervention periods lasted from February 1, 2017 to January 31, 2018 in the octenidine and control group, and from June 1, 2017 to May 31, 2018 in the chlorhexidine group. Baseline periods included 12 months before the interventions started in each study group.
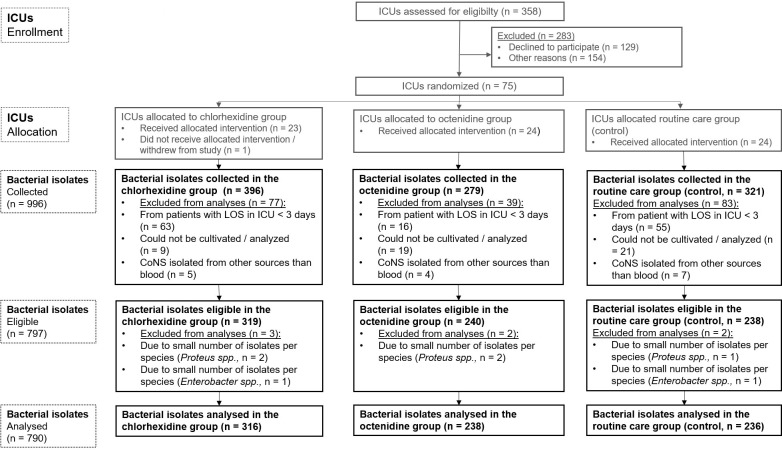
In total, 996 bacterial isolates from clinical samples were collected from participating wards. Among them, 797 bacterial isolates were eligible for analyses of susceptibility to chlorhexidine and octenidine (Fig 1).
Fig 1. Flow chart of ICU-attributable bacterial isolates from clinical samples eligible for analyses of susceptibilities to chlorhexidine and octenidine and included to descriptive analyses.
CoNS, coagulase-negative staphylococci, ICU, intensive care unit. spp., subspecies.
The selected pathogens detected from ICU-attributable clinical isolates were E. coli (n = 227), S. aureus (n = 155), Klebsiella spp. (n = 150), P. aeruginosa (n = 136), CoNS (n = 122), Proteus spp. (n = 5) and Enterobacter spp. (n = 2). Due to the small number of isolates per species, Proteus spp. and Enterobacter spp. were excluded from all further analyses (Fig 1). In consequence, 790 clinical isolates from 60 ICUs were included in the descriptive analyses. Among them, 417 isolates were collected during baseline when all wards used water and soap for daily bathing, and 373 isolates were collected at the end of the intervention period. Clinical isolates from baseline and intervention period were available from 42 of the 60 ICUs (70.0%) responding.
Characteristics of bacterial isolates reported by study group and period are shown in Table 1. No differences in sample site or ICU day of sample collection were detected between study groups neither during baseline nor intervention period (Table 1). The same was true for baseline versus intervention period comparisons in each study group except for the chlorhexidine group. In this group, the samples sites differed between baseline and intervention periods for E. coli and Klebsiella spp. (Table 1).
Table 1. Characteristics of ICU-attributable bacterial isolates from clinical samples included in the analyses (n = 790) in total and reported by study group and period.
| Total | Chlorhexidine group | P-valuea | Octenidine group | P-valuea | Routine care group = control | P-valuea | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Intervention | Baseline | Intervention | Baseline | Intervention | |||||
| Staphylococcus aureus | ||||||||||
| n (%) | 155 (100.0) | 38 (25.0) | 22 (14.2) | 24 (15.5) | 18 (11.5) | 23 (14.7) | 30 (19.2) | |||
| ICU day of sample collectionb, median (IQR) | 6 (4–11) | 6 (4–8) | 6 (4–9) | 0.769 | 7 (5–20) | 6 (4–9) | 0.084 | 6 (4–11) | 5 (4–9) | 0.842 |
| Sample site | ||||||||||
| Bloodb, n (%) | 27 (17.3) | 7 (17.9) | 4 (18.2) | 0.633 | 3 (12.5) | 2 (11.1) | 0.639 | 3 (13.0) | 8 (26.7) | 0.193 |
| Tracheal aspirateb, n (%) | 87 (55.8) | 23 (59.0) | 16 (72.2) | 0.340 | 14 (58.3) | 11 (61.1) | 0.856 | 10 (43.5) | 13 (43.3) | 0.992 |
| Urineb, n (%) | 4 (2.6) | 1 (2.6) | 0 (0.0) | 0.633 | 0 (0.0) | 1 (5.6) | 0.420 | 0 (0.0) | 2 (6.7) | 0.316 |
| Woundb, n (%) | 18 (11.6) | 4 (10.3) | 1 (4.5) | 0.389 | 3 (12.5) | 1 (5.6) | 0.420 | 5 (21.7) | 4 (13.3) | 0.328 |
| Othersb, n (%) | 19 (12.3) | 3 (7.7) | 1 (4.5) | 0.532 | 4 (16.7) | 3 (16.7) | 0.665 | 5 (26.3) | 3 (15.8) | 0.213 |
| Coagulase-negative staphylococci | ||||||||||
| n (%) | 122 (100.0) | 31 (25.4) | 31 (25.4) | 17 (13.9) | 11 (9.0) | 15 (12.2) | 17 (13.9) | |||
| ICU day of sample collectionb, median (IQR) | 11 (6–19) | 13 (8–18) | 10 (5–21) | 0.418 | 17 (9–19) | 10 (3–16) | 0.154 | 11 (6–24) | 7 (4–17) | 0.363 |
| Sample site | ||||||||||
| Bloodb, n (%) | 122 (88.4) | 31 (100.0) | 31 (100.0) | n.d. | 17 (100.0) | 11 (100.0) | n.d | 15 (100.0) | 17 (100.0) | n.d. |
| Tracheal aspirateb, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. |
| Urineb, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. |
| Woundb, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. |
| Othersb, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 0 (0.0) | n.d. |
| Escherichia coli | ||||||||||
| n (%) | 227 (100.0) | 64 (28.2) | 37 (16.3%) | 41 (18.1) | 18 (7.9%) | 39 (17.2) | 28 (11.9) | |||
| ICU day of sample collectionb, median (IQR) | 11 (6–19) | 10 (5–16) | 7 (4–20) | 0.584 | 9 (5–20) | 11 (5–15) | 0.947 | 7 (4–14) | 12 (4–22) | 0.154 |
| Sample site | ||||||||||
| Bloodb, n (%) | 16 (7.1) | 2 (3.1) | 5 (13.5) | 0.096 | 3 (7.3) | 1 (5.6) | 1.000 | 2 (5.1) | 3 (10.7) | 0.642 |
| Tracheal aspirateb, n (%) | 87 (38.3) | 30 (46.9) | 8 (21.6) | 0.012* | 16 (39.0) | 8 (44.4) | 0.457 | 13 (33.3) | 12 (42.9) | 0.427 |
| Urineb, n (%) | 50 (22.0) | 8 (12.5) | 13 (35.1) | 0.007* | 11 (26.8) | 4 (22.2) | 1.000 | 10 (25.6) | 4 (14.3) | 0.259 |
| Woundb, n (%) | 33 (14.5) | 8 (12.5) | 6 (16.2) | 0.405 | 3 (7.3) | 4 (22.2) | 0.184 | 8 (20.5) | 4 (14.3) | 0.512 |
| Othersb, n (%) | 41 (18.1) | 16 (25.0) | 5 (13.5) | 0.131 | 8 (19.5) | 1 (5.6) | 0.252 | 6 (15.4) | 5 (17.9) | 1.000 |
| Klebsiella spp. | ||||||||||
| n (%) | 150 (100.0) | 28 (18.7) | 28 (18.7) | 23 (15.3) | 29 (19.3) | 20 (13.3) | 22 (14.7) | |||
| ICU day of sample collectionb, median (IQR) | 10 (5–20) | 11 (6–26) | 8 (5–24) | 0.565 | 12 (6–21) | 11 (6–19) | 0.712 | 9 (4–20) | 9 (5–16) | 0.889 |
| Sample site | ||||||||||
| Bloodb, n (%) | 18 (12.0) | 3 (10.7) | 5 (17.9) | 0.705 | 1 (4.3) | 2 (6.9) | 1.000 | 4 (20.0) | 3 (13.6) | 0.691 |
| Tracheal aspirateb, n (%) | 74 (49.3) | 9 (67.0) | 15 (53.6) | 0.105 | 12 (52.2) | 17 (58.6) | 0.642 | 9 (45.0) | 12 (54.5) | 0.537 |
| Urineb, n (%) | 17 (11.3) | 5 (17.9) | 5 (17.9) | 1.000 | 2 (8.7) | 3 (10.3) | 1.000 | 2 (10.0) | 0 (0.0) | 0.221 |
| Woundb, n (%) | 21 (11.3) | 1 (3.6) | 2 (7.1) | 1.000 | 5 (21.7) | 6 (20.7) | 1.000 | 3 (15.0) | 4 (18.2) | 1.000 |
| Othersb, n (%) | 20 (13.3) | 10 (35.7) | 1 (3.6) | 0.005* | 3 (13.0) | 1 (3.4) | 0.310 | 2 (10.0) | 3 (13.6) | 1.000 |
| Pseudomonas aeruginosa | ||||||||||
| n (%) | 136 (100%) | 19 (14.0%) | 18 (13.2%) | 17 (12.5%) | 40 (29.4%) | 18 (13.2%) | 24 (17.6%) | |||
| ICU day of sample collectionb, median (IQR) | 12 (6–28) | 14 (4–33) | 15 (9–20) | 0.927 | 6 (5–21) | 15 (9–29) | 0.319 | 11 (5–13) | 12 (5–25) | 0.949 |
| Sample site | ||||||||||
| Bloodb, n (%) | 2 (1.5) | 0 (0.0) | 0 (0.0) | n.d. | 0 (0.0) | 1 (2.5) | 1.000 | 1 (5.6) | 0 (0.0) | 0.429 |
| Tracheal aspirateb, n (%) | 82 (60.3) | 7 (36.8) | 12 (66.7) | 0.070 | 12 (70.6) | 26 (65.0) | 0.682 | 11 (61.1) | 14 (58.3) | 0.856 |
| Urineb, n (%) | 16 (7.4) | 2 (10.5) | 2 (11.1) | 1.000 | 2 (11.8) | 4 (10.0) | 1.000 | 2 (11.1) | 4 (16.7) | 0.685 |
| Woundb, n (%) | 15 (11.0) | 4 (21.1) | 2 (11.1) | 0.660 | 1 (5.9) | 4 (10.0) | 1.000 | 3 (16.7) | 1 (4.2) | 0.297 |
| Othersb, n (%) | 21 (15.4) | 6 (31.6) | 2 (11.1) | 0.232 | 22 (11.8) | 5 (12.5) | 1.000 | 1 (5–6) | 5 (20.8) | 0.214 |
P-values were calculated by Chi-quare test or Fisher’s exact test for categorical, and by Mann-Whitney U Test for continuous variables.
aP-values were reported for comparisons between baseline and intervention periods for each study group.
bP-Values were not shown for baseline period comparisons and intervention period comparisons of the three study groups because no significant differences were detected between the study groups in the baseline and the intervention period. baseline, prior to the intervention period, intervention, at the end of the intervention period.
P-values < 0.05 were interpreted as significant (*).
Phenotypic susceptibility of bacterial isolates from clinical samples to chlorhexidine and octenidine
MIC50 and MIC90
For all species, phenotypic susceptibility to chlorhexidine depicted as MIC50 and MIC90 in est. μg/ml did not increase comparing baseline and the intervention periods in all study groups, as well as comparing intervention groups with control group at the end of the intervention (Table 2). The same was true for phenotypic susceptibility to octenidine (Table 2).
Phenotypic susceptibility to chlorhexidine and octenidine of commercially available reference strains (S. aureus, E. coli, P. aeruginosa, K. pneumoniae and Proteus mirabilis) are shown in S1 Table. Chlorhexidine and octenidine MIC50 with IQR and MIC90 in percentage of stock solution are given in S2 Table.
Distribution of chlorhexidine and octenidine MIC for all ICU-attributable bacterial isolates from clinical samples
In the intervention groups, the percentages of bacterial isolates with higher chlorhexidine MIC indicating reduced chlorhexidine susceptibility did not increase comparing baseline and intervention periods for all species (Fig 2A–2E). Further, no increase was observed comparing intervention groups with control group (at the end of the intervention period) for all species. The same observations were made for octenidine MIC (Fig 2A–2E).
Fig 2.
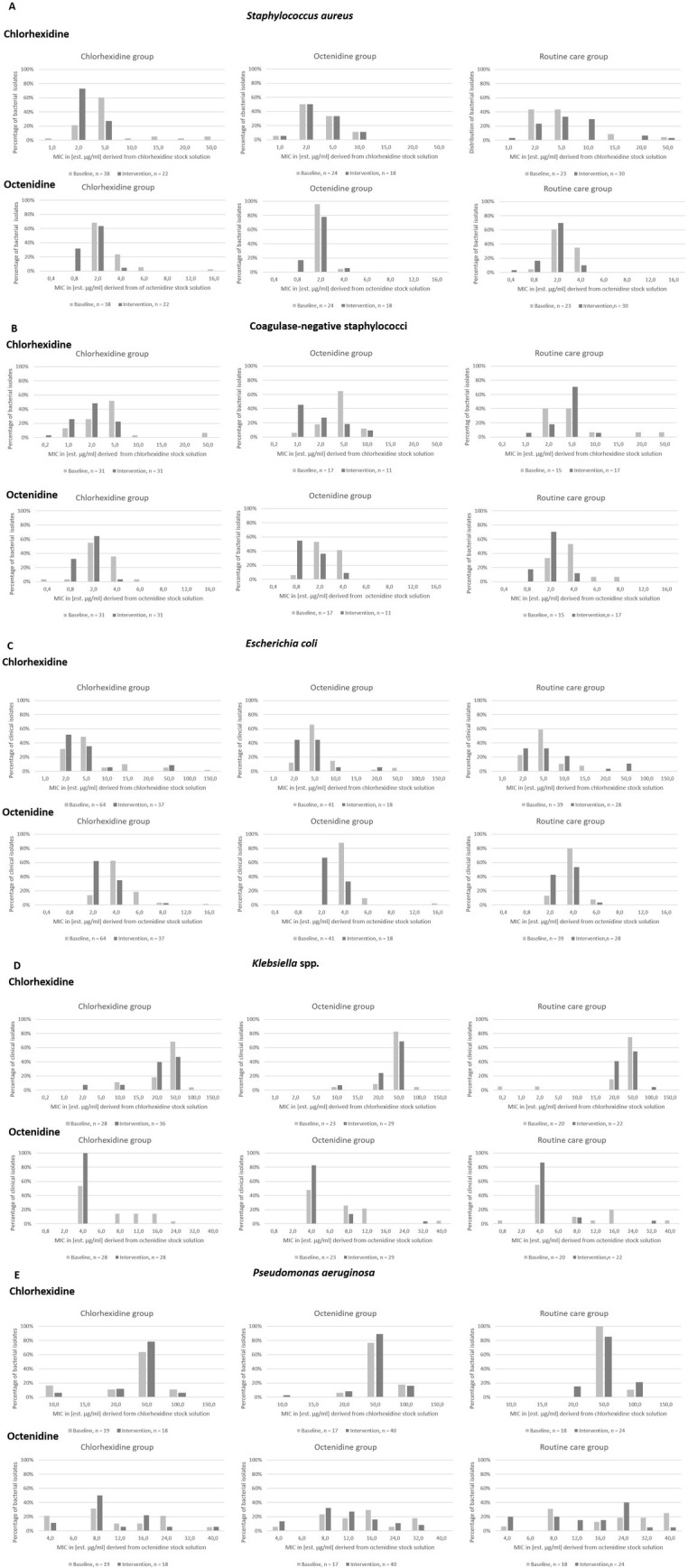
Distribution of chlorhexidine and octenidine MIC for all ICU-attributable bacterial isolates from clinical samples with Staphylococcus aureus (A), coagulase-negative staphylococci (B), Escherichia coli (C), Klebsiella spp. (D) and Pseudomonas aeruginosa (E). MIC were reported as estimated minimal inhibitory concentrations (est. MIC) in [μg/ml] derived from the respective extraction solutions. ICU-attributable bacterial isolates from clinical samples were collected prior to (baseline, light grey bars) and at the end of the intervention period (dark grey bars) applying 2%-chlorhexidine impregnated cloths (chlorhexidine group), 0.08% octenidine wash mitts (octenidine group) or water and soap (routine care = control).
Interestingly, ICUs in the control group that continued bathing with water and soap collected some bacterial isolates with high chlorhexidine and / or octenidine MIC during intervention period even though no antiseptic substances were applied. High chlorhexidine MIC were found in the baseline period and / or in control group for some isolates of S. aureus, E. coli, Klebsiella spp. and P. aeruginosa and high octenidine MIC for P. aeruginosa (Fig 2A and 2C–2E).
Among the 155 S. aureus isolates, 23 (14.8%) had chlorhexidine MIC ≥ 8μg/ml, while this was the case for 10 of 122 CoNS (8.2%). High chlorhexidine MIC (≥ 64 μg/ml) were rare in E. coli and Klebsiella spp. but frequent in P. aeruginosa (≥ 50 μg/ml, Fig 2C–2E).
Characteristics of all 23 S. aureus isolates with chlorhexidine MIC ≥ 8μg/ml can be found in S3 Table.
Multivariable analyses
We used GEE models to investigate the impact of antiseptic bathing with chlorhexidine or octenidine on the chlorhexidine and octenidine susceptibility of bacterial isolates (all species). Characteristics of bacterial isolates (all species) stratified by chlorhexidine or octenidine MIC ≥ species-specific MIC50 (yes / no) are shown in S4 Table. aIRR for the chlorhexidine and the octenidine models are depicted in Table 3. The proportion of bacterial isolates with MIC ≥ species-specific chlorhexidine and octenidine MIC50 did not significantly differ between chlorhexidine or octenidine intervention periods compared with the control group (intervention period, Table 3). We detected significant differences between study groups and study periods with our GEE models (Table 3). However, these differences seemed to be independent from antiseptic bathing. Further, we found evidence that ICU day of sample collection was significantly associated with increased proportions of bacterial isolates with MIC ≥ species-specific MIC50 for chlorhexidine, but not for octenidine. Interestingly, bacterial isolates with chlorhexidine MIC ≥ species-specific chlorhexidine MIC50 were significantly less likely to originate from blood compared with other sample sites. This observation was not found for octenidine.
Table 3. Generalized estimating equation models for the outcomes chlorhexidine and octenidine susceptibility of bacterial isolates (reported by MIC ≥ species-specific chlorhexidine and octenidine MIC50 (yes / no)).
| Chlorhexidine | ||||
|---|---|---|---|---|
| aIRR | 95%CI | p-value | p-value (type III) | |
| Chlorhexidine group in the baseline period | 0.85 | 0.31–2.35 | 0.771 | < 0.001* |
| Chlorhexidine group in the intervention period | 0.29 | 0.12–0.74 | 0.010* | |
| Octenidine group in the baseline period | 1.81 | 0.69–4.78 | 0.229 | |
| Octenidine group in the intervention period | 0.67 | 0.25–1.79 | 0.428 | |
| Control group in the baseline period | 1.05 | 0.49–2.23 | 0.908 | |
| Control group in the intervention period | 1.00 = reference | - | - | |
| ICU day of sample collection | 1.01 | 1.00–1.03 | 0.013 | 0.013* |
| Sample site | 0.015* | |||
| Blood | 0.36 | 0.19–0.70 | 0.002 | |
| Tracheal aspirate | 0.73 | 0.45–1.17 | 0.187 | |
| Urine | 0.82 | 0.43–1.57 | 0.548 | |
| Wound | 0.79 | 0.41–1.51 | 0.470 | |
| Others | 1.00 = reference | |||
| Octenidine | ||||
| Chlorhexidine group in the baseline period | 1.27 | 0.56–2.91 | 0.570 | < 0.001* |
| Chlorhexidine group in the intervention period | 0.52 | 0.25–1.07 | 0.085 | |
| Octenidine group in the baseline period | 2.83 | 1.19–6.76 | 0.019* | |
| Octenidine group in the intervention period | 0.68 | 0.32–1.42 | 0.302 | |
| Control group in the baseline period | 1.63 | 0.83–3.23 | 0.160 | |
| Control group in the intervention period | 1.00 = reference | - | - | |
| ICU day of sample collection | 1.00 | 0.99–1.02 | 0.406 | 0.406 |
| Sample site | 0.627 | |||
| Blood | 1.19 | 0.61–2.36 | 0.616 | |
| Tracheal aspirate | 0.95 | 0.55–1.65 | 0.858 | |
| Urine | 0.69 | 0.36–1.64 | 0.276 | |
| Wound | 0.97 | 0.49–1.95 | 0.945 | |
| Others | 1.00 = reference |
Adjusted incidence rate ratios (aIRR) were estimated for all combinations of study groups (chlorhexidine, octenidine, control) and study periods (baseline, intervention) using control group (intervention period) as a reference. ICU day of sample collection, sample sites, cluster effect and interaction between study groups and periods were considered in all models. P-values < 0.05 were interpreted as significant (*). c P-values were reported for comparisons between intervention groups (chlorhexidine and octenidine) and control group (routine care), adjusted by study period. aIRR, adjusted incidence rate ratio. 95%CI, 95% confidence interval.
Discussion
In a large sample of ICU-attributable bacterial isolates collected during the cluster-randomized decolonization trial CLIP-ID, susceptibility to chlorhexidine and octenidine was not reduced 12 months after implementation of daily antiseptic bathing with the respective substances. Some bacterial isolates had enhanced chlorhexidine and octenidine MIC. Most of them were observed in the control group and / or in the baseline period when ICUs did not apply any antiseptic substances. Thus, in our setting, daily bathing with chlorhexidine or octenidine did not enhance the development of non-susceptibilities among bacterial isolates of clinical samples to these substances.
Several in vitro and clinical studies suggest an association of high chlorhexidine use and the development of non-susceptibilities of bacterial isolates to chlorhexidine [17–19, 24, 25]. However, our findings confirmed data from the REDUCE MRSA trial suggesting that chlorhexidine susceptibility of S. aureus isolates was not reduced after implementation of routine chlorhexidine bathing in ICUs [1]. At the same time, data on octenidine susceptibility of bacterial isolates are scarce. Most studies did not report development of non-susceptibility or resistance to octenidine [17, 26–29]. Interestingly, for P. aeruginosa, development of tolerances to both antiseptics, chlorhexidine and octenidine, has been shown in vitro [30]. We observed high chlorhexidine and octenidine MIC for P. aeruginosa. In 90% of P. aeruginosa isolates, chlorhexidine MIC was ≥ 50 μg/ml that represents the species-specific epidemiological cut-off value (Ecoff) as reported by Kampf et al. [18]. However, no P. aeruginosa clinical isolate showed higher MIC than the reference strain.
In our study, the proportions of S. aureus and CoNS isolates with elevated chlorhexidine MIC was high (14.8%; 23 of 155 and 8.2%, 10 of 122) when using MIC ≥ 8μg/ml as Ecoff [1, 18]. This observation, however, was independent from the implementation of antiseptic bathing. For S. aureus isolates, elevated chlorhexidine MIC (32 μg/ml) and octenidine MIC (3 μg/ml) were reported from hospital trusts in the United Kingdom (UK) after high usage of chlorhexidine and implementation of octenidine usage, respectively [16].
Other studies reported lower proportion of S. aureus and CoNS isolates with MIC ≥ 8μg/ml but did not include clinical and / or ICU-attributable isolates [1, 31–33]. In our trial, bacterial isolates were collected from patients who stayed at least 3 days on the ICU. Approximately 70% of ICU patients receive antibiotic therapy [34]. Thus, it is most likely that bacterial isolates from clinical samples in our study were collected from patients previously or currently exposed to antibiotics that might lead to elevated chlorhexidine MIC of their bacterial isolates. However, comparisons to other studies and / or epidemiological cut-off values are only possible to a limited extend. MIC reported here could only be estimated by the concentration of our extraction solutions. Please see limitations for more details.
Our data confirms previous findings that chlorhexidine is highly active against gram-positive and shows lower activity against gram-negative bacteria [35]. Further, estimated MIC required to inactivate bacterial isolates (especially Klebsiella spp. and P. aeruginosa) was higher for chlorhexidine compared with octenidine. These observations confirm in vitro analyses suggesting superiority of octenidine compared with chlorhexidine in the laboratory [36].
The highest estimated chlorhexidine MIC in our study was observed for an E.coli isolate (150 μg/ml) collected during the baseline period. The concentration of chlorhexidine in 2% chlorhexidine-impregnated cloths (= 20.000 μg/ml) is more than 130 times higher. In consequence, the concentration applied to the patient’s skin during antiseptic bathing is expected to be sufficient (20). For octenidine, the highest estimated MIC in this analysis was 40 μg/ml for P. aeruginosa and Klebsiella spp. The octenidine concentration in the 0.08% octenidine (= 800 μg/ml) wash mitts is only 20 times higher, but might still be adequate to reduce the bacterial load on the patient’s skin. Unfortunately, no studies investigating the biocide concentrations that remain on the patient’s skin after bathing with these antiseptic ready-to-use products are available.
Our GEE model found evidence that length of ICU stay (represented by ICU day of sample collection) was associated with higher proportions of bacteria with increased chlorhexidine MIC. Patients with high length of stay on the ICU most likely received more interventions (e.g. invasive procedures, medication, antibiotic therapy) over longer durations of time. As antibiotic resistance is a known indicator of chlorhexidine susceptibility, longer ICU stays (and in consequence presumably more antibiotic treatment) might have an impact on the chlorhexidine susceptibility of bacterial isolates from clinical samples (37). Further, bacterial isolates with high chlorhexidine MIC were significantly less likely to originate from blood cultures compared with other sources. We can only speculate on the reasons for this finding. One possible explanation might be that bacteria isolated from blood might be less affected by ICU interventions including antiseptic bathings compared with other sample sites, e.g. wounds. However, it is unclear, why we did not see these observations with octenidine.
Strengths and limitations
This is one of the largest multicenter studies investigating the susceptibility of bacterial isolates from clinical samples to the antiseptic substances chlorhexidine and octenidine. Bacterial isolates from 60 different ICUs were analyzed, the majority (70.0%) of them sent isolates before and at the end of the intervention period. Further, we included clinical isolates from patients with length of stay on ICU of at least 3 days. Thereby, clinical isolates were collected from patients receiving all treatments applied in the ICU including the respective bathing procedures (chlorhexidine-impregnated cloths, octenidine wash mitts or water and soap) for at least 3 days. Median ICU day of sample collection was 9 days for all species. In consequence, clinical isolates included in this analyses can be designated as ICU-attributable.
Our study has some limitations. First, the number of bacterial isolates tested per species and study group was low. Second, the observation period of 12 months might be too short to draw any final conclusions. In consequence, results must be interpreted with caution. Further, epidemiological cut-off values for chlorhexidine were not available for all species (e.g. CoNS), and not available at all for octenidine (18). Thus, we used species-specific MIC50 as threshold to generate binary variables of phenotypic susceptibilities to chlorhexidine and octenidine for our GEE models. This approach was chosen to apply the same methods for all species and substances (chlorhexidine and octenidine). However, we did not collect all bacterial isolates from clinical samples or a representative sample set from all ICUs participating. Thus, statistical analyses that could be performed to control for potential confounders including cluster-effects were not fully applicable. Fourth, we did not collect any patient-specific data in our analyses (e.g. age, antimicrobial therapy, medication). Potential differences between patient populations of different ICUs could not be considered. Fifth, we did not perform any molecular analyses on the presence of qac genes, efflux pumps, antiseptic resistance genes or any analyses on antimicrobial susceptibility of bacterial isolates. However, this was not subject of our analyses. Several studies are available that investigate the correlation of biocide susceptibility with the absence or presence of efflux pump genes such as qac genes [1, 16, 25, 31, 32]. The correlation of antimicrobial and antiseptic susceptibility is known and has been shown for bacteria such as methicillin-resistant S. aureus (MRSA) and Extended spectrum Beta-Lactamase (ESBL) producing Enterobacterales [31, 37, 38]. Sixth, MIC comparisons to other studies are only possible to a limited extent as no standardized methods for testing phenotypic susceptibility to the antiseptic substances chlorhexidine and octenidine are available [16, 20]. Even though, MIC and minimal bactericidal concentrations (MBC) are most frequently used in this context, no consensus breakpoints for determining susceptibility to chlorhexidine or octenidine exist [16, 20, 25]. Further, MIC tests were not performed with pure chlorhexidine and octenidine substances but with extraction solutions harvested by wringing out the antiseptic products. Pure stock solutions were not provided by the manufacturers. In consequence, MIC reported here could only be estimated from the concentrations reported for the antiseptic products (2μg / ml for chlorhexidine and 0.08 μg/ml of octenidine) and the dilutions (MIC in percentages) derived from of our extraction solutions. This might also limit comparisons to other studies. However, the main focus of our study was whether susceptibilities of bacterial isolates from clinical samples to chlorhexidine and octenidine might decrease from baseline to intervention periods as well as between antiseptic bathing and control group(s). The method of harvesting antiseptic solutions for MIC tests does not have an impact on these results. In fact, using extraction solutions harvested from the antiseptic products is even more close to reality as it included all additional detergents and ingredients from the ready-to-use product that might reduce the properties of pure chlorhexidine and octenidine substances. Further, this approach might consider fractions of chlorhexidine and octenidine remaining in the cloths / mitts during the bathing process. This would not be factored in when using pure stock solutions. Thus, we consider our approach being appropriate and even more conservative. However, our estimated MIC might be overestimated and true MICs of bacterial isolates from clinical samples to chlorhexidine and octenidine might be lower. Finally, MIC of bacterial isolates from clinical samples were determined on ICU- not individual patient level. The longitudinal analyses of bacterial isolates from individual patients before, during and at the end of their ICU stay would allow to directly investigate the potential development of any non-susceptibilities of bacterial isolates from clinical samples to chlorhexidine or octenidine during routine antiseptic bathing. Such elaborate sample collections were conducted for analyses of the skin microbiome including antibiotic resistance genes in this project. Results are currently in preparation. Our study has a high likelihood of being underpowered to show smaller, but also clinically relevant increases in the proportions of non- or less susceptible bacterial isolates from clinical samples (all species) to chlorhexidine or octenidine. However, we discussed limitations in detail and add important data on chlorhexidine and octenidine susceptibilities of bacterial isolates from clinical samples to the scarce literature available on this topic.
Outlook
More analyses on susceptibility of bacterial isolates from clinical samples to antiseptic substances frequently used such as chlorhexidine or octenidine are needed. A standardized protocol would be necessary to compare MIC between different settings and to interpret own findings.
Conclusions
We found no evidence for reduced chlorhexidine or octenidine susceptibilities of bacterial isolates from clinical samples in ICUs after implementation of daily patient bathing with these antiseptics. However, the observation period of 12 months might be too short and the number of bacteria per species and study group too small to draw any final conclusions. In consequence, results must be interpreted with caution.
Supporting information
(XLSX)
(DOCX)
Median MIC (MIC50) with inter quartile ranges (IQR) and MIC90 are given in in [%] of extraction solution and are reported for Staphylococcus aureus, coagulase-negative staphylococci, Escherichia coli, Klebsiella spp. and Pseudomonas aeruginosa stratified by study group (chlorhexidine, octenidine, routine care) and period (baseline = prior to the intervention period, intervention = at the end of the intervention period).
(DOCX)
(DOCX)
Chlorhexidine and octenidine susceptibility of bacterial isolates were reported as binary variable (chlorhexidine / octenidine MIC of bacterial isolates ≥ species-specific chlorhexidine/ octenidine MIC50 (yes / no). aP-values were reported for comparisons between chlorhexidine/ octenidine MIC50 ≥ species-specific chlorhexidine/ octenidine MIC50 = “yes” or “no”. b percentage of columns. c percentage of rows. P-values < 0.05 were interpreted as significant (*). n, number. (%), percent.
(DOCX)
(DOCX)
Acknowledgments
We thank all infection control practitioners, nurses, physicians, healthcare staff and technicians from all ICUs and laboratories participating in and supporting the CLIP-ID trial.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was funded by the German Federal Ministry of Education and Research within the scope of the InfectControl consortium (Grant No. 03ZZ0807A) awarded to PG. Sage Products / Stryker and Schülke, funded the (antiseptic) products for the intervention and supported the investigation of tolerances to chlorhexidine and octenidine by an independent accredited contract laboratory. Dr. Brill + Partner GmbH was the independent contract-laboratory assigned by Schülke and Stryker /Sage to conduct the investigations of tolerances to chlorhexidine and octenidine (by minimum inhibitory concentration testing). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hayden MK, Lolans K, Haffenreffer K, Avery TR, Kleinman K, Li H, et al. Chlorhexidine and Mupirocin Susceptibility of Methicillin-Resistant Staphylococcus aureus Isolates in the REDUCE-MRSA Trial. J Clin Microbiol. 2016;54(11):2735–42. doi: 10.1128/JCM.01444-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hübner NO, Siebert J, Kramer A. Octenidine dihydrochloride, a modern antiseptic for skin, mucous membranes and wounds. Skin pharmacology and physiology. 2010;23(5):244–58. doi: 10.1159/000314699 [DOI] [PubMed] [Google Scholar]
- 3.Afonso E, Blot K, Blot S. Prevention of hospital-acquired bloodstream infections through chlorhexidine gluconate-impregnated washcloth bathing in intensive care units: a systematic review and meta-analysis of randomised crossover trials. Euro Surveill. 2016;21(46). doi: 10.2807/1560-7917.ES.2016.21.46.30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, et al. Effect of daily chlorhexidine bathing on hospital-acquired infection. The New England journal of medicine. 2013;368(6):533–42. doi: 10.1056/NEJMoa1113849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frost SA, Alogso MC, Metcalfe L, Lynch JM, Hunt L, Sanghavi R, et al. Chlorhexidine bathing and health care-associated infections among adult intensive care patients: a systematic review and meta-analysis. Critical care (London, England). 2016;20(1):379. doi: 10.1186/s13054-016-1553-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frost SA, Hou YC, Lombardo L, Metcalfe L, Lynch JM, Hunt L, et al. Evidence for the effectiveness of chlorhexidine bathing and health care-associated infections among adult intensive care patients: a trial sequential meta-analysis. BMC Infect Dis. 2018;18(1):679. doi: 10.1186/s12879-018-3521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, et al. Targeted versus universal decolonization to prevent ICU infection. The New England journal of medicine. 2013;368(24):2255–65. doi: 10.1056/NEJMoa1207290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musuuza JS, Guru PK, O’Horo JC, Bongiorno CM, Korobkin MA, Gangnon RE, et al. The impact of chlorhexidine bathing on hospital-acquired bloodstream infections: a systematic review and meta-analysis. BMC Infect Dis. 2019;19(1):416. doi: 10.1186/s12879-019-4002-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Horo JC, Silva GL, Munoz-Price LS, Safdar N. The efficacy of daily bathing with chlorhexidine for reducing healthcare-associated bloodstream infections: a meta-analysis. Infect Control Hosp Epidemiol. 2012;33(3):257–67. doi: 10.1086/664496 [DOI] [PubMed] [Google Scholar]
- 10.Gastmeier P, Kämpf KP, Behnke M, Geffers C, Schwab F. An observational study of the universal use of octenidine to decrease nosocomial bloodstream infections and MDR organisms. The Journal of antimicrobial chemotherapy. 2016;71(9):2569–76. doi: 10.1093/jac/dkw170 [DOI] [PubMed] [Google Scholar]
- 11.Meißner A, Hasenclever D, Brosteanu O, Chaberny IF. EFFECT of daily antiseptic body wash with octenidine on nosocomial primary bacteraemia and nosocomial multidrug-resistant organisms in intensive care units: design of a multicentre, cluster-randomised, double-blind, cross-over study. BMJ Open. 2017;7(11):e016251. doi: 10.1136/bmjopen-2017-016251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messler S, Klare I, Wappler F, Werner G, Ligges U, Sakka SG, et al. Reduction of nosocomial bloodstream infections and nosocomial vancomycin-resistant Enterococcus faecium on an intensive care unit after introduction of antiseptic octenidine-based bathing. The Journal of hospital infection. 2019;101(3):264–71. doi: 10.1016/j.jhin.2018.10.023 [DOI] [PubMed] [Google Scholar]
- 13.Denkel LA GP, Leistner R, Geffers C. Antiseptic bathing of intensive care patients–What is the current routine practice in Germany? Jahrestagung der Deutschen Gesellschaft für Hygiene und Mikrobiologie (DGHM); Würzburg, Germany: DGHM; 2017.
- 14.Denkel LA, Schwab F, Clausmeyer J, Behnke M, Golembus J, Wolke S, et al. Effect of antiseptic bathing with chlorhexidine or octenidine on central-line associated bloodstream infections in intensive care patients: a cluster-randomised controlled trial. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2022. [DOI] [PubMed] [Google Scholar]
- 15.Denkel LA, Schwab F, Clausmeyer J, Behnke M, Golembus J, Wolke S, et al. Central-line associated bloodstream infections in intensive care units before and after implementation of daily antiseptic bathing with chlorhexidine or octenidine—A post-hoc analysis of a cluster-randomised controlled trial Antimicrobial resistance and infection control. 2022;Under Revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy K, Sunnucks K, Gil H, Shabir S, Trampari E, Hawkey P, et al. Increased Usage of Antiseptics Is Associated with Reduced Susceptibility in Clinical Isolates of Staphylococcus aureus. mBio. 2018;9(3). doi: 10.1128/mBio.00894-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Htun HL, Hon PY, Holden MTG, Ang B, Chow A. Chlorhexidine and octenidine use, carriage of qac genes, and reduced antiseptic susceptibility in methicillin-resistant Staphylococcus aureus isolates from a healthcare network. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2019;25(9):1154.e1-.e7. doi: 10.1016/j.cmi.2018.12.036 [DOI] [PubMed] [Google Scholar]
- 18.Kampf G. Acquired resistance to chlorhexidine—is it time to establish an ’antiseptic stewardship’ initiative? The Journal of hospital infection. 2016;94(3):213–27. doi: 10.1016/j.jhin.2016.08.018 [DOI] [PubMed] [Google Scholar]
- 19.Suwantarat N, Carroll KC, Tekle T, Ross T, Maragakis LL, Cosgrove SE, et al. High prevalence of reduced chlorhexidine susceptibility in organisms causing central line-associated bloodstream infections. Infect Control Hosp Epidemiol. 2014;35(9):1183–6. doi: 10.1086/677628 [DOI] [PubMed] [Google Scholar]
- 20.Babiker A, Lutgring JD, Fridkin S, Hayden MK. Assessing the Potential for Unintended Microbial Consequences of Routine Chlorhexidine Bathing for Prevention of Healthcare-associated Infections. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2021;72(5):891–8. doi: 10.1093/cid/ciaa1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbund für Angewandte Hygiene e.V. (VAH). Anforderungen und Methoden zur VAH-Zertifizierung chemischer Desinfektionsverfahren: Desinfektionsmittel-Kommission im VAH; 2019. Available from: https://vah-online.de/files/download/ebooks/eBook_VAH_Methoden_Anforderungen.pdf.
- 22.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44(4):1049–60. [PubMed] [Google Scholar]
- 23.Donner A, Birkett N, Buck C. Randomization by cluster. Sample size requirements and analysis. Am J Epidemiol. 1981;114(6):906–14. doi: 10.1093/oxfordjournals.aje.a113261 [DOI] [PubMed] [Google Scholar]
- 24.Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2010;50(2):210–7. doi: 10.1086/648717 [DOI] [PubMed] [Google Scholar]
- 25.Horner C, Mawer D, Wilcox M. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? The Journal of antimicrobial chemotherapy. 2012;67(11):2547–59. doi: 10.1093/jac/dks284 [DOI] [PubMed] [Google Scholar]
- 26.Sedlock DM, Bailey DM. Microbicidal activity of octenidine hydrochloride, a new alkanediylbis[pyridine] germicidal agent. Antimicrob Agents Chemother. 1985;28(6):786–90. doi: 10.1128/AAC.28.6.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Doori Z, Goroncy-Bermes P, Gemmell CG, Morrison D. Low-level exposure of MRSA to octenidine dihydrochloride does not select for resistance. The Journal of antimicrobial chemotherapy. 2007;59(6):1280–1. doi: 10.1093/jac/dkm092 [DOI] [PubMed] [Google Scholar]
- 28.Spencer C, Orr D, Hallam S, Tillmanns E. Daily bathing with octenidine on an intensive care unit is associated with a lower carriage rate of meticillin-resistant Staphylococcus aureus. The Journal of hospital infection. 2013;83(2):156–9. doi: 10.1016/j.jhin.2012.10.007 [DOI] [PubMed] [Google Scholar]
- 29.Krishna BV, Gibb AP. Use of octenidine dihydrochloride in meticillin-resistant Staphylococcus aureus decolonisation regimens: a literature review. The Journal of hospital infection. 2010;74(3):199–203. doi: 10.1016/j.jhin.2009.08.022 [DOI] [PubMed] [Google Scholar]
- 30.Shepherd MJ, Moore G, Wand ME, Sutton JM, Bock LJ. Pseudomonas aeruginosa adapts to octenidine in the laboratory and a simulated clinical setting, leading to increased tolerance to chlorhexidine and other biocides. The Journal of hospital infection. 2018;100(3):e23–e9. doi: 10.1016/j.jhin.2018.03.037 [DOI] [PubMed] [Google Scholar]
- 31.Azrad M, Shmuel C, Leshem T, Hamo Z, Baum M, Rokney A, et al. Reduced Susceptibility to Chlorhexidine among Staphylococcus aureus Isolates in Israel: Phenotypic and Genotypic Tolerance. Antibiotics (Basel). 2021;10(3). doi: 10.3390/antibiotics10030342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.do Vale BCM, Nogueira AG, Cidral TA, Lopes MCS, de Melo MCN. Decreased susceptibility to chlorhexidine and distribution of qacA/B genes among coagulase-negative Staphylococcus clinical samples. BMC Infectious Diseases. 2019;19(1):199. doi: 10.1186/s12879-019-3823-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soma VL, Qin X, Zhou C, Adler A, Berry JE, Zerr DM. The effects of daily chlorhexidine bathing on cutaneous bacterial isolates: a pilot study. Infect Drug Resist. 2012;5:75–8. doi: 10.2147/IDR.S30662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent JL, Sakr Y, Singer M, Martin-Loeches I, Machado FR, Marshall JC, et al. Prevalence and Outcomes of Infection Among Patients in Intensive Care Units in 2017. Jama. 2020;323(15):1478–87. doi: 10.1001/jama.2020.2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karpiński TM, Szkaradkiewicz AK. Chlorhexidine—pharmaco-biological activity and application. Eur Rev Med Pharmacol Sci. 2015;19(7):1321–6. [PubMed] [Google Scholar]
- 36.Koburger T, Hubner NO, Braun M, Siebert J, Kramer A. Standardized comparison of antiseptic efficacy of triclosan, PVP-iodine, octenidine dihydrochloride, polyhexanide and chlorhexidine digluconate. The Journal of antimicrobial chemotherapy. 2010;65(8):1712–9. doi: 10.1093/jac/dkq212 [DOI] [PubMed] [Google Scholar]
- 37.Kõljalg S, Naaber P, Mikelsaar M. Antibiotic resistance as an indicator of bacterial chlorhexidine susceptibility. The Journal of hospital infection. 2002;51(2):106–13. doi: 10.1053/jhin.2002.1204 [DOI] [PubMed] [Google Scholar]
- 38.Leshem T, Gilron S, Azrad M, Peretz A. Characterization of reduced susceptibility to chlorhexidine among Gram-negative bacteria. Microbes Infect. 2022;24(2):104891. doi: 10.1016/j.micinf.2021.104891 [DOI] [PubMed] [Google Scholar]