Abstract
Background
Co-circulation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and other respiratory viruses, such as influenza and respiratory syncytial virus (RSV), can be a severe threat to public health. The accurate detection and differentiation of these viruses are essential for clinical laboratories. Herein, we comparatively evaluated the performance of the Kaira COVID-19/Flu/RSV Detection Kit (Kaira; Optolane, Seongnam, Korea) for detection of SARS-CoV-2, influenza A and B, and RSV in nasopharyngeal swab (NPS) specimens with that of the PowerChek SARS-CoV-2, Influenza A&B, RSV Multiplex Real-time PCR Kit (PowerChek; Kogene Biotech, Seoul, Korea).
Methods
A total of 250 archived NPS specimens collected for routine clinical testing were tested in parallel by the Kaira and PowerChek assays. RNA standards were serially diluted and tested by the Kaira assay to calculate the limit of detection (LOD).
Results
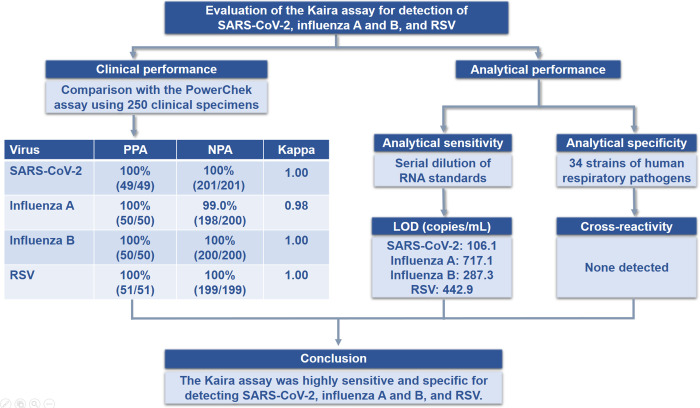
The positive and negative percent agreements between the Kaira and PowerChek assays were as follows: 100% (49/49) and 100% (201/201) for SARS-CoV-2; 100% (50/50) and 99.0% (198/200) for influenza A; 100% (50/50) and 100% (200/200) for influenza B; and 100% (51/51) and 100% (199/199) for RSV, respectively. The LODs of the Kaira assay for SARS-CoV-2, influenza A and B, and RSV were 106.1, 717.1, 287.3, and 442.9 copies/mL, respectively.
Conclusions
The Kaira assay showed comparable performance to the PowerChek assay for detection of SARS-CoV-2, influenza A and B, and RSV in NPS specimens, indicating that the Kaira assay could be a useful diagnostic tool when these viruses are co-circulating.
Introduction
In December 2019, coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in Wuhan, China, and rapidly spread worldwide, achieving pandemic status in March 2020 [1]. As of October 31, 2022, over 627 million people have been infected with SARS-CoV-2 worldwide, resulting in over 6.5 million deaths [2]. To curb the spread of SARS-CoV-2 infection, rapid and accurate laboratory diagnosis is required, and molecular assays are the current gold standard for laboratory diagnosis of SARS-CoV-2 infection [3–5]. More than 200 SARS-CoV-2 molecular assays have been granted emergency use authorization by the US Food and Drug Administration, the majority of which use real-time reverse transcription polymerase chain reaction (rRT-PCR) technology.
During the COVID-19 pandemic, circulation of other respiratory viruses, such as influenza and respiratory syncytial virus (RSV), may pose a tremendous challenge to healthcare systems, as SARS-CoV-2 and these viruses can cause similar symptoms [6–8]. Furthermore, co-infection of SARS-CoV-2 and other respiratory viruses can occur, albeit at a low rate [9, 10]. To address this situation, various molecular assays to simultaneously detect SARS-CoV-2 and other respiratory viruses have been developed and are widely used in clinical settings [11–21].
The Kaira COVID-19/Flu/RSV Detection Kit (Kaira; OPTOLANE, Seongnam, Korea) is a novel rRT-PCR assay that can detect SARS-CoV-2, influenza A and B, and RSV in nasopharyngeal swab (NPS) specimens within 80 min. This assay is a single-tube multiplex assay targeting the open reading frame 1ab (ORF1ab) of SARS-CoV-2, the matrix protein 2 gene of influenza A, the nuclear export protein gene of influenza B, and the matrix protein gene of RSV. Herein, we assessed the performance of the Kaira assay compared with the PowerChek SARS-CoV-2, Influenza A&B, RSV Multiplex Real-time PCR Kit (PowerChek; Kogene Biotech, Seoul, Korea). The graphical abstract of this study is shown in Fig 1.
Fig 1. Graphical abstract of the study.
Material and methods
Clinical specimens
This study included 250 NPS specimens collected for routine clinical testing at Samsung Medical Center, a 1989-bed tertiary care hospital in Seoul, Korea. Positive specimens were selected based on the cycle threshold (Ct) values obtained from routine clinical testing and covered a wide range of Ct values (Table 1). All specimens were stored frozen at −70°C until retrieved for this study. This study was reviewed and approved by the Institutional Review Board (IRB) of Samsung Medical Center (approval number: 2022-08-010). The need for informed consent was waived by the IRB due to the retrospective study design and use of fully anonymized patient data.
Table 1. Distribution of Ct values of positive specimens selected for this study.
| SARS-CoV-2 | Influenza virus | RSV | |||
|---|---|---|---|---|---|
| Ct value* | E | ORF1ab | Influenza A | Influenza B | |
| < 20 | 13 | 12 | 8 | 15 | 3 |
| 20–25 | 6 | 4 | 20 | 13 | 15 |
| 25–30 | 9 | 11 | 18 | 15 | 18 |
| > 30 | 22 | 23 | 4 | 7 | 14 |
| Total no. | 50 | 50 | 50 | 50 | 50 |
* Ct values were obtained by routine clinical testing using the PowerChek SARS-CoV-2 Real-time PCR Kit (Kogene Biotech) for SARS-CoV-2 and the AdvanSure RV-plus real-time RT-PCR (LG Chem, Seoul, Korea) for influenza and RSV.
E, envelope gene; ORF1ab, open reading frame 1ab.
Kaira assay
RNA extraction was performed using QIAamp DSP Viral RNA Mini Kit (Qiagen, Hilden, Germany). The Kaira assay was performed according to the manufacturer’s instructions. In brief, 10 μL of template RNA was added to 12.5 μL of rRT-PCR master mix and 2.5 μL of primer/probe mixture, giving a final reaction volume of 25 μL. The rRT-PCR was performed on the CFX96 system (Bio-Rad, Hercules, CA, USA) using the following cycling conditions: 1 cycle at 50°C for 10 min and 1 cycle at 95°C for 10 min, followed by 45 cycles at 95°C for 10 sec and 57°C for 30 sec. For the SARS-CoV-2 target, a Ct value ≤ 42 was considered a positive result, while for the other three targets, a Ct value ≤ 43 was considered a positive result.
PowerChek assay
The PowerChek assay is a two-tube multiplex rRT-PCR assay and was performed according to the manufacturer’s instructions. In brief, 5 μL of template RNA was added to 10 μL of rRT-PCR master mix and 5 μL of primer/probe mixture, giving a total reaction volume of 20 μL. The rRT-PCR was performed on the CFX96 system using the following cycling conditions: 1 cycle at 50°C for 30 min and 1 cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For all four targets, a Ct value ≤ 38 was considered a positive result. If the results of the Kaira and PowerChek assays were discordant, the BioFire Respiratory Panel 2.1 (RP2.1; bioMérieux, Marcy l’Etoile, France) was conducted.
Analytical performance
The analytical sensitivity of the Kaira assay was assessed using AMPLIRUN TOTAL SARS-CoV-2/FluA/FluB/RSV CONTROL (SWAB) (Vircell, Granada, Spain). This RNA standard was serially diluted in a pool of negative NPS specimens and extracted as described above. Twenty replicates per dilution level were tested using the Kaira assay.
The analytical specificity of the Kaira assay was evaluated using 34 strains of human respiratory pathogens (Table 2). Bacterial and viral strains were tested in duplicate at concentrations of 1 × 106 and 1 × 105 copies/mL, respectively.
Table 2. Analytical specificity evaluation results of the Kaira assay.
| Organism | Source (code number) | Result |
|---|---|---|
| SARS-CoV-2 B.1.1.7 (Alpha) | Vircell (MBC138-R) | SARS-CoV-2 positive |
| SARS-CoV-2 B.1.351 (Beta) | Vircell (MBC139-R) | SARS-CoV-2 positive |
| SARS-CoV-2 P.1 (Gamma) | Vircell (MBC140-R) | SARS-CoV-2 positive |
| SARS-CoV-2 B.1.617.2 (Delta) | Vircell (MBC141-R) | SARS-CoV-2 positive |
| SARS-CoV-2 B.1.1.529 (Omicron) | Vircell (MBC143-R) | SARS-CoV-2 positive |
| SARS-CoV | Vircell (MBC136-R) | Negative |
| MERS-CoV | Vircell (MBC132) | Negative |
| Human coronavirus 229E | ATCC (VR-740D) | Negative |
| Human coronavirus OC43 | Vircell (MBC135-R) | Negative |
| Human coronavirus NL63 | Vircell (MBC142-R) | Negative |
| Human coronavirus HKU1 | Clinical isolate | Negative |
| Influenza A virus H1N1 | Vircell (MBC028) | Influenza A positive |
| Influenza A virus H3N2 | Vircell (MBC029) | Influenza A positive |
| Influenza A virus H5N1 | Vircell (MBC052) | Influenza A positive |
| Influenza B virus | Vircell (MBC030) | Influenza B positive |
| RSV type A | Vircell (MBC041) | RSV positive |
| RSV type B | Vircell (MBC083) | RSV positive |
| Human parainfluenza virus 1 | Vircell (MBC037) | Negative |
| Human parainfluenza virus 2 | Vircell (MBC038) | Negative |
| Human parainfluenza virus 3 | Vircell (MBC039) | Negative |
| Human parainfluenza virus 4 | Vircell (MBC050) | Negative |
| Enterovirus D68 | Vircell (MBC125) | Negative |
| Enterovirus A71 | Vircell (MBC019) | Negative |
| Rhinovirus B14 | Vircell (MBC091) | Negative |
| Human adenovirus 1 | Vircell (MBC001) | Negative |
| Human bocavirus | ATCC (VR-3251SD) | Negative |
| Human metapneumovirus | Vircell (MBC144-R) | Negative |
| Streptococcus pneumoniae | ATCC (33400D-5) | Negative |
| Haemophilus influenzae | ATCC (51907D-5) | Negative |
| Chlamydophila pneumoniae | ATCC (53592D) | Negative |
| Mycoplasma pneumoniae | ATCC (15531D) | Negative |
| Legionella pneumophila | ATCC (33152D-5) | Negative |
| Bordetella pertussis | ATCC (9797D-5) | Negative |
| Bordetella parapertussis | ATCC (15311D-5) | Negative |
Statistical analysis
Two-by-two tables were used to assess the agreement between the Kaira and PowerChek assays. The positive percent agreement (PPA), negative percent agreement (NPA), Cohen’s kappa values, and two-sided 95% confidence intervals were calculated to evaluate the level of agreement between the two assays. The correlation between Ct values of positive specimens by the two assays was assessed using Pearson correlation coefficient. The limit of detection (LOD) was determined using Probit regression analysis. All statistical analyses were performed using Excel (Microsoft, Redmond, WA, USA) and MedCalc Statistical Software version 19.5 (MedCalc Software Ltd, Ostend, Belgium).
Results
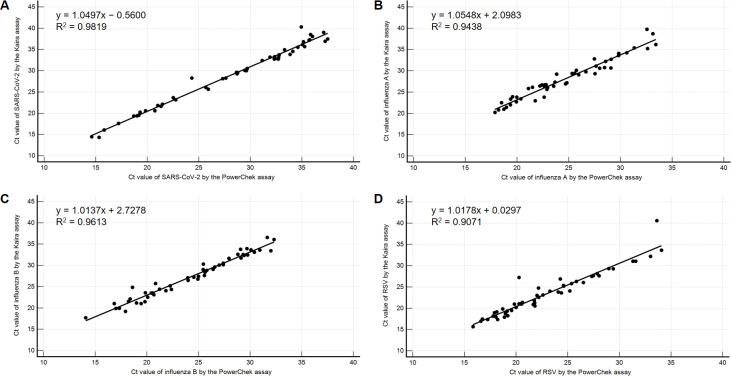
Compared to the PowerChek assay, the PPA and NPA of the Kaira assay for SARS-CoV-2 were 100% (49/49) and 100% (201/201), respectively. The PPA and NPA for influenza A and B were as follows: 100% (50/50) and 99.0% (198/200) for influenza A and 100% (50/50) and 100% (200/200) for influenza B. The PPA and NPA for RSV were 100% (51/51) and 100% (199/199), respectively. Kappa values ranged from 0.98 (influenza A) to 1.00 (SARS-CoV-2, influenza B, and RSV), indicating an almost perfect agreement (Table 3). The Ct values of clinical specimens tested positive by both the Kaira and PowerChek assays were highly correlated, with R2 ranging from 0.9071 to 0.9819 (Fig 2). Only two specimens showed discordant results. These specimens tested positive for influenza A and B by the Kaira assay; however, their Ct values for influenza A (42.6 and 42.3) were near the assay cut-off. They tested positive only for influenza B by the PowerChek and RP2.1 assays (Table 4).
Table 3. Clinical performance of the Kaira assay in comparison with the PowerChek assay.
| Kaira result | PowerChek result | PPA (95% CI) | NPA (95% CI) | Kappa value (95% CI) | ||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| SARS-CoV-2 | Positive | 49 | 0 | 100% (92.7–100%) | 100% (98.2–100%) | 1.00 (1.00–1.00) |
| Negative | 0 | 201 | ||||
| Influenza A | Positive | 50 | 2 | 100% (92.9–100%) | 99.0% (96.4–99.9%) | 0.98 (0.94–1.00) |
| Negative | 0 | 198 | ||||
| Influenza B | Positive | 50 | 0 | 100% (92.9–100%) | 100% (98.2–100%) | 1.00 (1.00–1.00) |
| Negative | 0 | 200 | ||||
| RSV | Positive | 51 | 0 | 100% (93.0–100%) | 100% (98.2–100%) | 1.00 (1.00–1.00) |
| Negative | 0 | 199 | ||||
PPA, positive percent agreement; NPA, negative percent agreement; CI, confidence interval.
Fig 2. Correlation between Ct values of clinical specimens tested positive by both the Kaira and PowerChek assays.
(A) E and ORF1ab Ct values of the PowerChek assay were averaged and plotted against the ORF1ab Ct values of the Kaira assay. (B) Influenza A Ct values of the PowerChek assay were plotted against the influenza A Ct values of the Kaira assay. (C) Influenza B Ct values of the PowerChek assay were plotted against the influenza B Ct values of the Kaira assay. (D) RSV Ct values of the PowerChek assay were plotted against the RSV Ct values of the Kaira assay.
Table 4. Details of two specimens showing discordant results between the Kaira and PowerChek assays.
| Specimen no. | Clinical comparison | Discrepancy resolution | |
|---|---|---|---|
| Kaira result (Ct value) | PowerChek result (Ct value) | RP2.1 result | |
| 124 | Influenza A (42.6)*, Influenza B (22.1) | Influenza B (20.5) | Influenza B |
| 131 | Influenza A (42.3)*, Influenza B (27.2) | Influenza B (25.8) | Influenza B |
* On repeat testing using the Kaira assay, these specimens showed negative results for influenza A.
The LODs of the Kaira assay for SARS-CoV-2, influenza A and B, and RSV were 106.1, 717.1, 287.3, and 442.9 copies/mL, respectively (Table 5), which were comparable to those of the PowerChek assay determined in our previous study (362.7, 1239.8, 90.2, and 634.4 copies/mL, respectively) [14]. In the analytical specificity test, all intended targets of the Kaira assay (SARS-CoV-2, influenza A and B, and RSV) were detected, and no cross-reactivity with other respiratory pathogens was observed (Table 2).
Table 5. Analytical sensitivity evaluation results of the Kaira assay.
| Target concentration | SARS-CoV-2 | Influenza A | Influenza B | RSV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Copies/mL | Replicates | Detected | Copies/mL | Replicates | Detected | Copies/mL | Replicates | Detected | Copies/mL | Replicates | Detected | |
| #1 | 2500 | 20 | 20 | 3500 | 20 | 20 | 2400 | 20 | 20 | 4000 | 20 | 20 |
| #2 | 1250 | 20 | 20 | 1750 | 20 | 20 | 1200 | 20 | 20 | 2000 | 20 | 20 |
| #3 | 500 | 20 | 20 | 700 | 20 | 18 | 480 | 20 | 20 | 800 | 20 | 20 |
| #4 | 250 | 20 | 20 | 350 | 20 | 0 | 240 | 20 | 17 | 400 | 20 | 18 |
| #5 | 50 | 20 | 12 | 70 | 20 | 0 | 48 | 20 | 3 | 80 | 20 | 11 |
| #6 | 25 | 20 | 7 | 35 | 20 | 0 | 24 | 20 | 0 | 40 | 20 | 8 |
| #7 | 12.5 | 20 | 5 | 17.5 | 20 | 0 | 12 | 20 | 1 | 20 | 20 | 2 |
| Probit LOD (copies/mL) | 106.1 | 717.1 | 287.3 | 442.9 | ||||||||
Discussion
In this study, we compared the performance of the Kaira and PowerChek assays for detection of SARS-CoV-2, influenza A and B, and RSV in NPS specimens. We found that the performance of the Kaira assay was comparable to that of the PowerChek assay.
The COVID-19 pandemic has drastically changed the epidemiology of other respiratory viruses. During the 2020–2021 season, other respiratory viruses circulated at historically low levels due to public health measures to curb the spread of SARS-CoV-2. Notably, the circulation of influenza and RSV was virtually absent during this period [22–25]. However, after relaxation of public health measures, an unexpected out-of-season resurgence of influenza and RSV has recently been observed in many parts of the world [26–30]. Given the changes in the epidemiology of influenza and RSV, molecular assays to simultaneously detect these viruses and SARS-CoV-2 are urgently needed and should be performed throughout the year.
Currently, various molecular assays to simultaneously detect SARS-CoV-2 and other respiratory viruses are commercially available, most of which are sample-to-result rRT-PCR assays [11–21]. Sample-to-result assays such as the RP2.1 and Xpert Xpress SARS-CoV-2/Flu/RSV assays are simple to perform and do not require skilled personnel. Furthermore, these assays enable random-access testing, providing test results to physicians in a timely manner; however, they have relatively low throughput and are suited for small-volume clinical laboratories [14, 19]. By contrast, the Kaira and PowerChek assays are designed for high-throughput batch testing (Kaira assay: up to 96 specimens per batch; PowerChek assay: up to 48 specimens per batch) and suited for high-volume clinical laboratories. The performance of the PowerChek assay has recently been evaluated [14]; however, little is known about the performance of the Kaira assay. To the best of our knowledge, this is the first study to evaluate the performance of the Kaira assay.
In this study, the clinical performance of the Kaira assay was comparable to that of the PowerChek assay, with kappa values ranging from 0.98 (influenza A) to 1.00 (SARS-CoV-2, influenza B, and RSV). Only two specimens gave discordant results (Kaira: positive for influenza A and B; PowerChek: positive for influenza B only), which were resolved by the RP2.1 assay (positive for influenza B only). On repeat testing using the Kaira assay, these specimens showed positive results only for influenza B. As the initial Ct values for influenza A were near the assay cut-off and coinfection of influenza A and B viruses is rare [31, 32], the initial positive results for influenza A are highly likely to be false-positive. In addition, the LODs of the Kaira assay were comparable to those of the PowerChek assay, indicating high sensitivity of the Kaira assay in detecting SARS-CoV-2, influenza A and B, and RSV.
An important limitation of the Kaira assay is that it utilizes only one target gene (ORF1ab) for detection of SARS-CoV-2. As mutations in the primer/probe binding sites of the SARS-CoV-2 genome could compromise the rRT-PCR assay’s performance, it is important to use rRT-PCR assays targeting at least two independent regions of the SARS-CoV-2 genome [33–35]. Although the Kaira assay correctly detected all SARS-CoV-2 strains included in this study, clinical laboratories should be aware of this assay’s limitations regarding the use of only one SARS-CoV-2 target.
A major limitation of this study is its retrospective design. A prospective study was not feasible because during the ongoing COVID-19 pandemic, influenza cases, particularly influenza B cases, have rarely been identified in Korea. To obtain a sufficient number of positive specimens, archived NPS specimens previously collected for routine clinical testing were used for this study.
In conclusion, the Kaira assay was found to be highly sensitive and specific for detecting SARS-CoV-2, influenza A and B, and RSV in NPS specimens. During the COVID-19 pandemic, circulation of influenza and RSV may pose a significant challenge to the already overburdened healthcare systems. In this situation, the Kaira assay with a high-throughput capacity (up to 96 specimens per batch) and short turnaround time (80 min) can be useful in clinical settings.
Supporting information
(XLSX)
Acknowledgments
We thank OPTOLANE Technologies Inc. for providing the Kaira assay kits used in this study. OPTOLANE Technologies Inc. had no role in the study design, data collection and analysis, and manuscript writing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Korea Medical Device Development Fund grant (https://www.kmdf.org/) funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry, and Energy, the Ministry of Health & Welfare, and the Ministry of Food and Drug Safety) (project number: 202011A04; PI: HJH) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI; https://www.khidi.or.kr/), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HW20C2130; PI: HJH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. WHO Director-General’s opening remarks at the Mission briefing on COVID-19. 2020. Mar 12 [cited 2022 Oct 4]. Available from: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-mission-briefing-on-covid-19—12-march-2020. [Google Scholar]
- 2.World Health Organization. Coronavirus Disease (COVID-19) Dashboard. 2022. Oct 31 [cited 2022 Nov 1]. Available from: https://covid19.who.int. [Google Scholar]
- 3.Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020;40(5):351–360. doi: 10.3343/alm.2020.40.5.351 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huh HJ, Hong KH, Kim TS, Song SH, Roh KH, Lee H, et al. Surveillance of coronavirus disease 2019 (COVID-19) testing in clinical laboratories in Korea. Ann Lab Med. 2021;41(2):225–229. doi: 10.3343/alm.2021.41.2.225 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong KH, Kim GJ, Roh KH, Sung H, Lee J, Kim SY, et al. Update of guidelines for laboratory diagnosis of COVID-19 in Korea. Ann Lab Med. 2022;42(4):391–397. doi: 10.3343/alm.2022.42.4.391 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Wang J, Wang C, Yang Q, Xu Y, Xu J, et al. Characteristics of respiratory virus infection during the outbreak of 2019 novel coronavirus in Beijing. Int J Infect Dis. 2020;96:266–269. doi: 10.1016/j.ijid.2020.05.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon DA, Sherman AC, Kanjilal S. Influenza in the COVID-19 era. JAMA. 2020;324(13):1342–1343. doi: 10.1001/jama.2020.14661 . [DOI] [PubMed] [Google Scholar]
- 8.Zayet S, Kadiane-Oussou NJ, Lepiller Q, Zahra H, Royer PY, Toko L, et al. Clinical features of COVID-19 and influenza: a comparative study on Nord Franche-Comte cluster. Microbes Infect. 2020;22(9):481–488. doi: 10.1016/j.micinf.2020.05.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020;323(20):2085–2086. doi: 10.1001/jama.2020.6266 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YG, Park H, Kim SY, Hong KH, Kim MJ, Lee JS, et al. Rates of coinfection between SARS-CoV-2 and other respiratory viruses in Korea. Ann Lab Med. 2022;42(1):110–112. doi: 10.3343/alm.2022.42.1.110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creager HM, Cabrera B, Schnaubelt A, Cox JL, Cushman-Vokoun AM, Shakir SM, et al. Clinical evaluation of the BioFire® Respiratory Panel 2.1 and detection of SARS-CoV-2. J Clin Virol. 2020;129:104538. doi: 10.1016/j.jcv.2020.104538 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung HY, Jian MJ, Chang CK, Lin JC, Yeh KM, Chen CW, et al. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg Microbes Infect. 2021;10(1):161–166. doi: 10.1080/22221751.2021.1873073 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarrett J, Uhteg K, Forman MS, Hanlon A, Vargas C, Carroll KC, et al. Clinical performance of the GenMark Dx ePlex respiratory pathogen panels for upper and lower respiratory tract infections. J Clin Virol. 2021;135:104737. doi: 10.1016/j.jcv.2021.104737 . [DOI] [PubMed] [Google Scholar]
- 14.Kim TY, Kim JY, Shim HJ, Yun SA, Jang JH, Huh HJ, et al. Comparison of the PowerChek SARS-CoV-2, Influenza A&B, RSV Multiplex Real-time PCR Kit and BioFire Respiratory Panel 2.1 for simultaneous detection of SARS-CoV-2, influenza A and B, and respiratory syncytial virus. J Virol Methods. 2021;298:114304. doi: 10.1016/j.jviromet.2021.114304 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mostafa HH, Carroll KC, Hicken R, Berry GJ, Manji R, Smith E, et al. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV test. J Clin Microbiol. 2021;59(3):e02955–20. doi: 10.1128/JCM.02955-20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun J, Park JH, Kim N, Roh EY, Shin S, Yoon JH, et al. Evaluation of three multiplex real-time reverse transcription PCR assays for simultaneous detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus in nasopharyngeal swabs. J Korean Med Sci. 2021;36(48):e328. doi: 10.3346/jkms.2021.36.e328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akashi Y, Horie M, Kiyotaki J, Takeuchi Y, Togashi K, Adachi Y, et al. Clinical performance of the cobas Liat SARS-CoV-2 & Influenza A/B assay in nasal samples. Mol Diagn Ther. 2022;26(3):323–331. doi: 10.1007/s40291-022-00580-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cassidy H, van Genne M, Lizarazo-Forero E, Niesters HGM, Gard L. Evaluation of the QIAstat-Dx RP2.0 and the BioFire FilmArray RP2.1 for the rapid detection of respiratory pathogens including SARS-CoV-2. Front Microbiol. 2022;13:854209. doi: 10.3389/fmicb.2022.854209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TY, Kim JY, Shim HJ, Yun SA, Jang JH, Huh HJ, et al. Performance evaluation of the PowerChek SARS-CoV-2, Influenza A & B Multiplex Real-Time PCR Kit in comparison with the BioFire Respiratory Panel. Ann Lab Med. 2022;42(4):473–477. doi: 10.3343/alm.2022.42.4.473 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mboumba Bouassa RS, Tonen-Wolyec S, Veyer D, Péré H, Bélec L. Analytical performances of the AMPLIQUICK® Respiratory Triplex assay for simultaneous detection and differentiation of SARS-CoV-2, influenza A/B and respiratory syncytial viruses in respiratory specimens. PLoS One. 2022;17(1):e0262258. doi: 10.1371/journal.pone.0262258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhen W, Manji R, Smith E, Wuitschick J, Lucic D, Berry GJ. Evaluation of the Alinity m Resp-4-Plex Assay for the detection of severe acute respiratory syndrome coronavirus 2, influenza A virus, influenza B virus, and respiratory syncytial virus. Microbiol Spectr. 2022;10(1):e0109021. doi: 10.1128/spectrum.01090-21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole S, Brendish NJ, Clark TW. SARS-CoV-2 has displaced other seasonal respiratory viruses: Results from a prospective cohort study. J Infect. 2020;81(6):966–972. doi: 10.1016/j.jinf.2020.11.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Roh YH, Ahn JG, Kim MY, Huh K, Jung J, et al. Respiratory syncytial virus and influenza epidemics disappearance in Korea during the 2020–2021 season of COVID-19. Int J Infect Dis. 2021;110:29–35. doi: 10.1016/j.ijid.2021.07.005 . [DOI] [PubMed] [Google Scholar]
- 24.Olsen SJ, Winn AK, Budd AP, Prill MM, Steel J, Midgley CM, et al. Changes in influenza and other respiratory virus activity during the COVID-19 pandemic—United States, 2020–2021. MMWR Morb Mortal Wkly Rep. 2021;70(29):1013–1019. doi: 10.15585/mmwr.mm7029a1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knudsen PK, Lind A, Klundby I, Dudman S. The incidence of infectious diseases and viruses other than SARS-CoV-2 amongst hospitalised children in Oslo, Norway during the Covid-19 pandemic 2020–2021. J Clin Virol Plus. 2022;2(1):100060. doi: 10.1016/j.jcvp.2021.100060 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi T, Kawano Y. Resurgence of respiratory syncytial virus infection during an atypical season in Japan. J Pediatric Infect Dis Soc. 2021;10(10):982–983. doi: 10.1093/jpids/piab065 . [DOI] [PubMed] [Google Scholar]
- 27.Eden JS, Sikazwe C, Xie R, Deng YM, Sullivan SG, Michie A, et al. Off-season RSV epidemics in Australia after easing of COVID-19 restrictions. Nat Commun. 2022;13(1):2884. doi: 10.1038/s41467-022-30485-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faico-Filho KS, Barbosa GR, Bellei N. Peculiar H3N2 outbreak in São Paulo during summer and emergence of the Omicron variant. J Infect. 2022;85(1):90–122. doi: 10.1016/j.jinf.2022.04.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J-H, Kim HY, Lee M, Ahn JG, Baek JY, Kim MY, et al. Respiratory syncytial virus outbreak without influenza in the second year of the coronavirus disease 2019 pandemic: a national sentinel surveillance in Korea, 2021–2022 season. J Korean Med Sci. 2022;37(34):e258. doi: 10.3346/jkms.2022.37.e258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SS, Viboud C, Petersen E. Understanding the rebound of influenza in the post COVID19 pandemic period holds important clues for epidemiology and control. Int J Infect Dis. 2022;122:1002–1004. doi: 10.1016/j.ijid.2022.08.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falchi A, Arena C, Andreoletti L, Jacques J, Leveque N, Blanchon T, et al. Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. J Clin Virol. 2008;41(2):148–151. doi: 10.1016/j.jcv.2007.11.003 . [DOI] [PubMed] [Google Scholar]
- 32.Cunha BA, Connolly JJ, Abruzzo E. Clinical implications of dual-positive rapid influenza diagnostic tests during influenza season: Co-colonization, coinfection, or false positive test? Am J Infect Control. 2014;42(10):1139–40. doi: 10.1016/j.ajic.2014.06.016 . [DOI] [PubMed] [Google Scholar]
- 33.Tahan S, Parikh BA, Droit L, Wallace MA, Burnham CD, Wang D. SARS-CoV-2 E gene variant alters analytical sensitivity characteristics of viral detection using a commercial reverse transcription-PCR assay. J Clin Microbiol. 2021;59(7):e0007521. doi: 10.1128/JCM.00075-21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi H, Hwang M, Lukey J, Jinadatha C, Navarathna DH. Presumptive positive with the Cepheid Xpert Xpress SARS-CoV-2 assay due to N mutations in the Delta variant. Diagn Microbiol Infect Dis. 2022;103(3):115699. doi: 10.1016/j.diagmicrobio.2022.115699 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong KH, In JW, Lee J, Kim SY, Lee KA, Kim S, et al. Prevalence of a single-nucleotide variant of SARS-CoV-2 in Korea and its impact on the diagnostic sensitivity of the Xpert Xpress SARS-CoV-2 assay. Ann Lab Med. 2022;42(1):96–99. doi: 10.3343/alm.2022.42.1.96 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.