Abstract
Summary
The effectiveness of CRISPR/Cas9-mediated genome editing experiments largely depends on the guide RNA (gRNA) used by the CRISPR/Cas9 system for target recognition and cleavage activation. Careful design is necessary to select a gRNA with high editing efficiency at the on-target site and with minimum off-target potential. Here, we present our webserver for gRNA design with a user-friendly graphical interface, which provides interoperability between our on- and off-target prediction tools, CRISPRon and CRISPRoff, for a complete and streamlined gRNA selection.
Availability and implementation
The graphical interface uses the Integrative Genomic Viewer (IGV) JavaScript plugin. The backend tools are implemented in Python and C. The CRISPRon and CRISPRoff webservers and command-line tools are freely available at https://rth.dk/resources/crispr.
1 Introduction
CRISPR/Cas9 is an RNA-guided DNA endonuclease broadly employed as a genome editing tool. The role of the Cas9 complex in editing is recognizing and cleaving on-target DNA sites, which are subsequently repaired to obtain an edit of interest (Haeussler and Concordet, 2016). To recognize a target, Cas9 binds to a short DNA motif called ‘protospacer adjacent motif’ (PAM) and probes flanking DNA for complementarity with its guide RNA (gRNA) (Anders et al., 2014). Because mismatches and bulges in the gRNA–DNA hybrid and in the PAM are tolerated, cleavage by Cas9 can also happen at sites other than the on-target, resulting in off-target edits (Fu et al., 2013). The cleavage efficiency of Cas9 varies at different on- and off-targets, mostly depending on the properties of the gRNA and the target site (Doench et al., 2014, 2016; Peng et al., 2018; Wang et al., 2014; Xiang et al., 2021; Xu et al., 2015).
The goal of gRNA design is to select the gRNA with maximum efficiency and minimal off-target potential among the gRNAs that are suitable to cleave a target region. A major computational challenge of gRNA design is the identification and scoring of potential off-targets (pOTs). This process requires a time- and resource-consuming genome-wide search of gRNA targets and the subsequent scoring of possibly several thousands of pOTs. Training machine and deep learning models for on-target efficiency prediction is also computationally demanding, but once such models are produced and loaded, relatively negligible time is required to generate results compared to the off-target search and evaluation.
To allow for on- and off-target aware gRNA design, we make use of two in silico tools that we previously co-authored. These are available as webservers and command-line tools: CRISPRon (Xiang et al., 2021) for on-target cleavage efficiency prediction, and CRISPRoff (Alkan et al., 2018) for pOT assessment, which searches for pOTs in the genome using RIsearch2 (Alkan et al., 2017). CRISPRon is a deep-learning model that predicts Cas9-mediated indel frequencies at gRNA on-target sites with top prediction performance in the field (Xiang et al., 2021). Compared to other notable on-target cleavage prediction tools available as webservers, such as the Azimuth model (Doench et al., 2016) used in CRISPOR (Concordet and Haeussler, 2018), CRISPick (https://portals.broadinstitute.org/gppx/crispick/public), and CHOPCHOP (Labun et al., 2019), CRISPRon has the advantage to be trained on indel frequencies, which is a more direct measure of cleavage efficiency than the loss-of-protein-function outcomes employed in the training of Azimuth. This aspect makes CRISPRon more suitable to design gRNAs for tasks beyond the functional knockout of protein-coding genes (e.g. knock-in of short genomic variants, knockout of non-coding RNAs, etc.). CRISPRoff is the first computational model for the assessment of pOTs based exclusively on free energy changes, with high prediction accuracy compared to mismatch-based methods and high recall thanks to its consideration for DNA-RNA G-U wobble base pairs (Alkan et al., 2018). On top of this, CRISPRoff is not trained on species-specific data, which makes it suitable to evaluate pOTs in any species.
The design of gRNAs requires not only accurate on- and off-target evaluation methods but also an effective user-friendly interface. Here, we present a new interface that allows interoperability between the CRISPRon and CRISPRoff webservers. It is now possible to retrieve genome-wide gRNA off-target information within the CRISPRon platform during gRNA design, as well as obtaining on-target efficiencies for gRNAs tested for off-targets in CRISPRoff. The interoperability of the CRISPRon/off interface enables a complete gRNA design within a single ‘workflow’, speeding up the whole design process and improving usability. In addition to the interoperability, both webservers have been substantially improved in terms of speed and available features compared to the previously published versions. The CRISPRon/off webservers and command line tools are freely available via http://rth.dk/resources/crispr. Via the same link, we also provide a pipeline, CRISPRroots (Corsi et al., 2022), for the post-assessment of on/off-targets in RNA-seq data generated after Cas9-mediated editing experiments.
2 Results and discussion
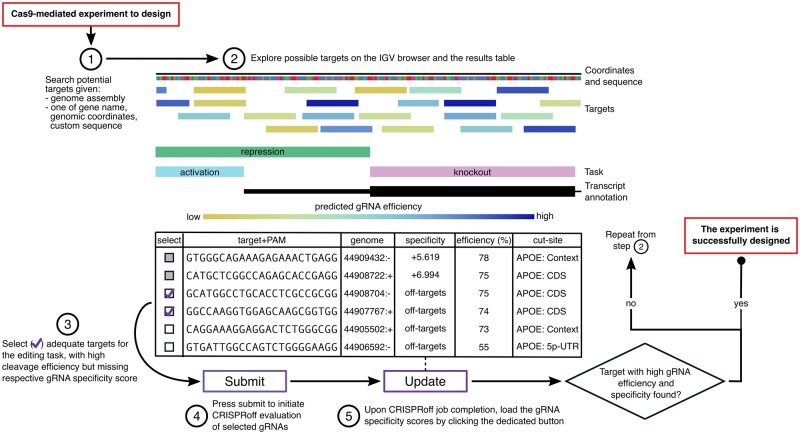
The biggest advance in the CRISPRon/off interface is the interoperability between the CRISPRon and CRISPRoff webservers, which provides huge benefits in the whole gRNA selection process and in terms of user experience. In CRISPRon, users can select multiple gRNA-target candidates based on properties such as promoters and CDS, as well as predicted cleavage efficiency scores (indel frequencies at targets). These can be easily inspected either in the built-in Integrative Genomics Viewer browser (Robinson et al., 2011) or in an interactive table, both provided in the results page. The gRNAs selected in CRISPRon can then be sent directly to CRISPRoff for the evaluation of their off-target potential. CRISPRoff calculates the binding free energy at all the pOTs of each gRNA and summarizes the ability of the Cas9-gRNA complex to bind at the on-target site while accounting for genome-wide pOTs in a single gRNA specificity score [also referred to as CRISPRspec, see Alkan et al. (2018) for details]. The results of the CRISPRoff assessment are then imported in the CRISPRon results page, for a final off-target aware gRNA selection (Fig. 1).
Fig. 1.
Design of gRNAs for Cas9 experiments with CRISPRon/off. Sequence of steps to identify targets that can be edited by Cas9 with high efficiency and with minimum off-target potential. The genomic sequence reported in the table rows is that of the DNA target (5′–3′ strand) which has the same sequence as the gRNA (which binds to the complimentary DNA) and the PAM. The ‘efficiency’ is the predicted indel frequency. The ‘specificity’ is the log10-scaled probability of binding at the on-target site compared to binding anywhere in the genome. The ‘genome’ coordinate is the start position of the target followed by the strand; all targets are in the same region and the same chromosome, which is not shown. IGV, integrative genomic viewer; CDS, coding sequence; 5p-UTR, 5′ untranslated region
The webservers have been updated so that mouse, rat, pig, zebrafish and fruit fly in addition to human are now available for both on- and off-target search. All genomes and annotations are updated to the latest versions as of March 2022 [human: hg38; mouse: mm39; zebrafish: danRer11; fruit fly: dm6; rat: rn6; pig: susScr11; Ensembl annotations version 104 (Cunningham et al., 2022)]. The results of CRISPRon include annotations of a user-selected transcript or, by default, a primary canonical transcript, using either the USCS annotations of the canonical transcript [which only exist for human and mouse, http://genome.ucsc.edu (Lee et al., 2022)] or a simple heuristic for other organisms. The heuristic consists of taking the longest transcript with a biotype matching the gene and, in the case of equally long transcripts, the one with the most exons.
The integrated genome browser is enriched with genomic variants for human from dbSNP (Sherry et al., 2001). The presence of a SNP at the target site is likely to affect the editing efficiency of a gRNA designed to target the reference sequence. Thus, unless the exact sequence of the target is known, gRNAs for all variants of the target should be tested independently. Moreover, we added indications for target regions suitable for specific editing tasks in protein-coding genes:
Knockout: 90% of the protein-coding sequence translated from the target’s primary canonical transcript starting from the N-terminus, optimal to obtain loss of function of the target protein (Doench et al., 2016).
Activation: from 300 nt upstream from the start of the transcription start site (TSS) of the primary canonical transcript, until the start of the TSS.
Repression: from 200 nt upstream from the start of the TSS of the primary canonical transcript, until 200 nt after the TSS start or until the start of the first CDS for genes with 5′ untranslated regions (UTRs) shorter than 200 nt.
The off-target assessment by CRISPRoff is speeded up significantly for human by keeping the indexed genome in the memory and for all organisms by filtering gRNAs for repeat-like sequences prior the extensive off-target search. A gRNA is marked as repeat-like if it maps more than 100 times with up to two mismatches in the genome in a bowtie1 search (Langmead et al., 2009). In the off-target assessment process for gRNAs extraneous to the reference, it is now possible for users to mask out the input and exclude from the search genomic regions that could potentially interfere with the on/off-target lists and scores. This is useful when the user supplies a sequence which differs from the reference genome, to avoid calling potential off-targets in both the target sequence and the reference genome, leading to incorrect off-targets and a skewed specificity score.
3 Conclusion
The integration of the CRISPR/Cas9 on- and off-target webservers, the additional features and the speed-ups of the platform will facilitate user navigation and enhance gRNA selection, allowing to better design genome editing experiments maximizing on-target efficiency and simultaneously minimizing potential off-target effects. Future improvements include the pre-calculation of all the on-targets and, for key gRNAs, of the off-targets and the specificity score.
Funding
This work was supported by the Independent Research Fund Denmark, FTP [9041-00317B]; and by the Novo Nordisk Foundation [NNF21OC0068988].
Conflict of Interest: No conflict of interest to declare.
Contributor Information
Christian Anthon, Center for Non-Coding RNA in Technology and Health, Department of Veterinary and Animal Sciences, University of Copenhagen, Frederiksberg 1871, Denmark.
Giulia Ilaria Corsi, Center for Non-Coding RNA in Technology and Health, Department of Veterinary and Animal Sciences, University of Copenhagen, Frederiksberg 1871, Denmark.
Jan Gorodkin, Center for Non-Coding RNA in Technology and Health, Department of Veterinary and Animal Sciences, University of Copenhagen, Frederiksberg 1871, Denmark.
References
- Alkan F. et al. (2018) CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol., 19, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan F. et al. (2017) RIsearch2: suffix array-based large-scale prediction of RNA–RNA interactions and siRNA off-targets. Nucleic Acids Res., 45, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C. et al. (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature, 513, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet J.-P., Haeussler M. (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res., 46, W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi G.I. et al. (2022) CRISPRroots: on- and off-target assessment of RNA-seq data in CRISPR–Cas9 edited cells. Nucleic Acids Res., 50, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F. et al. (2022) Ensembl 2022. Nucleic Acids Res., 50, D988–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G. et al. (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol., 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G. et al. (2014) Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol., 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol., 31, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Concordet J.-P. (2016) Genome editing with CRISPR-Cas9: can it get any better? J. Genet. Genomics, 43, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K. et al. (2019) CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Research, 47, W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. et al. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.T. et al. (2022) The UCSC genome browser database: 2022 update. Nucleic Acids Research, 50, D1115–D1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. et al. (2018) CRISPR/Cas9 cleavage efficiency regression through boosting algorithms and markov sequence profiling. Bioinformatics, 34, 3069–3077. [DOI] [PubMed] [Google Scholar]
- Robinson J.T. et al. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T. et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science, 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X. et al. (2021) Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun., 12, 3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res., 25, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]