Abstract
Cross-talk between Rho- and Arf-family guanosine triphosphatases (GTPases) plays an important role in linking the actin cytoskeleton to membrane protrusions, organelle morphology, and vesicle trafficking. The central actin regulator, WAVE regulatory complex (WRC), integrates Rac1 (a Rho-family GTPase) and Arf signaling to promote Arp2/3-mediated actin polymerization in many processes, but how WRC senses Arf signaling is unknown. Here, we have reconstituted a direct interaction between Arf and WRC. This interaction is greatly enhanced by Rac1 binding to the D site of WRC. Arf1 binds to a previously unidentified, conserved surface on the Sra1 subunit of WRC, which, in turn, drives WRC activation using a mechanism distinct from that of Rac1. Mutating the Arf binding site abolishes Arf1-WRC interaction, disrupts Arf1-mediated WRC activation, and impairs lamellipodia formation and cell migration. This work uncovers a new mechanism underlying WRC activation and provides a mechanistic foundation for studying how WRC-mediated actin polymerization links Arf and Rac signaling in cells.
The Arf-family GTPases can switch on a central actin regulator named the WAVE Complex to promote actin polymerization.
INTRODUCTION
Small guanosine triphosphatases (GTPases) of the Ras superfamily control diverse processes throughout eukaryotic cells (1). Among them, the distantly related Arf-family and Rho-family GTPases play distinct roles and yet have extensive cross-talk in many different processes. Arf GTPases are key players in various steps of membrane trafficking and organelle morphogenesis, where they are best known to promote the assembly of coat proteins to initiate vesicle formation (2–5). Rho GTPases, such as Rac1, are central regulators of the actin cytoskeleton in the formation of various cell membrane protrusions, such as lamellipodia and filopodia, where they are best known to promote cell migration, adhesion, and endocytosis (6, 7). Since it was discovered more than two decades ago (8–11), the cross-talk between Arf- and Rac1-mediated signaling pathways has been recognized as a crucial component for the regulation of actin cytoskeletal dynamics during cell migration, spreading, adhesion, fusion, phagocytosis, and endocytosis (8–16). Nevertheless, our knowledge of the underlying molecular mechanism has remained fragmental.
In addition to the role of Arf in regulating phospholipid compositions (17, 18), endosomal recycling of Rac1 (8, 9, 19), and the localization and activity of various regulators of Rac1 [including guanine nucleotide exchange factors (GEFs) (15, 20–22), GTPase activating protein (GAPs) (23), and adaptor proteins (11, 24)], a plethora of studies have observed that, in many processes, Arf and Rac1 often converge on a central actin nucleation promotion factor known as the WAVE regulatory complex (WRC) (25–32). The WRC is a 400-kDa protein assembly containing five conserved proteins: Sra1 (or Cyfip2), Nap1 (or Hem1), Abi2 (or Abi1 and Abi3), HSPC300, and WAVE1 (or WAVE2 and WAVE3, members of the Wiskott-Aldrich Syndrome protein family). In the basal state, the WRC keeps WAVE autoinhibited in the cytosol by sequestering the WCA (WH2-central-acidic) sequence at the C terminus of WAVE through a collection of interactions with the Sra1 subunit and the “meander” sequence of WAVE (Fig. 1, cartoon) (33–37). Various membrane ligands can directly interact with and recruit the WRC to the plasma membrane and simultaneously activate it to release the WCA, which, in turn, can bind the Arp2/3 complex to nucleate branched actin filaments (25, 38–45). Among these ligands, Rac1 is the canonical activator of the WRC (43). It acts by directly binding to two distinct locations on the opposite ends of the Sra1 subunit, which are named A and D sites, respectively. The two sites have an ~40-fold difference in the affinity for Rac1 (36, 41). Recent cryo–electron microscopy (cryo-EM) structures revealed that the binding of Rac1 to the low-affinity site (A site), but not the high-affinity site (D site), drives a conformational change to allosterically destabilize the WCA leading to WRC activation (46).
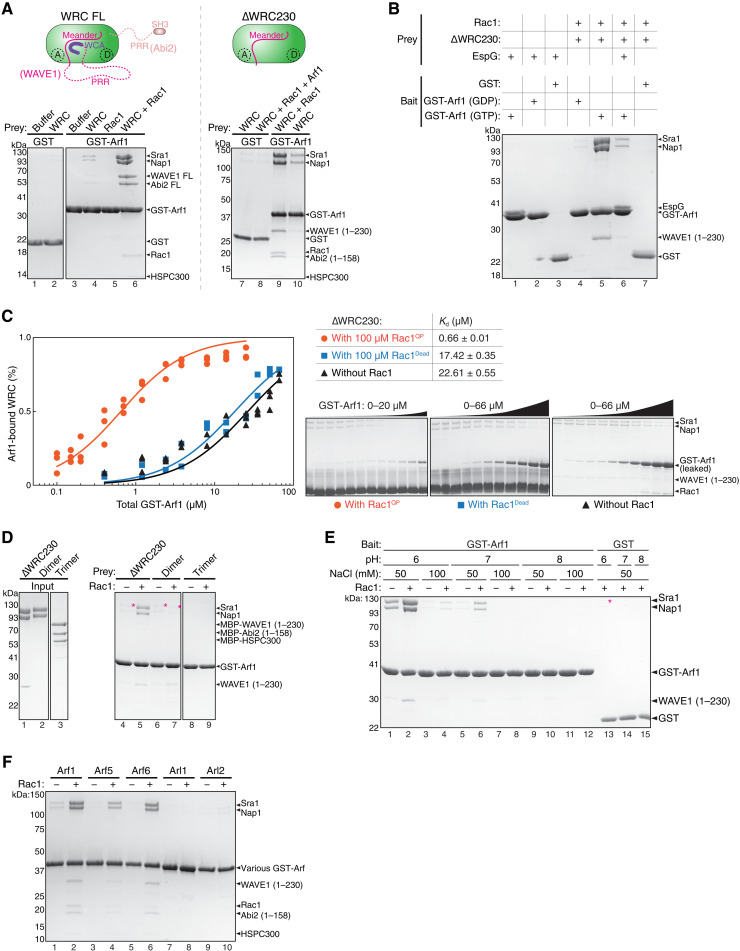
Fig. 1. Arf-WRC interaction is direct and can be greatly enhanced by Rac1.
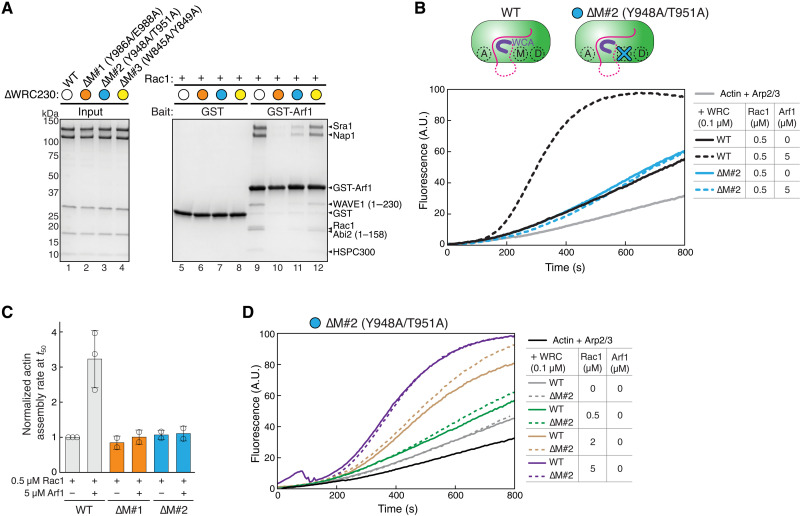
(A) Coomassie blue–stained SDS-PAGE gels showing GST-Arf1 pull down of WRC FL (left) and ∆WRC230 (right) in the presence or absence of untagged Rac1QP. In the schematic of WRCs, dotted lines indicate unstructured sequences. Both the A and D sites for Rac1 binding are indicated. (B) Coomassie blue–stained SDS-PAGE gels showing pull down of ∆WRC230 by GST-Arf1 in indicated nucleotide states or in the presence of the Arf1-binding protein EspG. (C) EPD assay to measure the binding affinity of the Arf1-WRC interaction in the presence of indicated Rac1 variants. On the left is the quantification of the data pooled from two to three independent experiments for each condition and globally fitted to obtain the binding isotherms. The derived Kd and fitting errors are shown in the table. On the right are representative Coomassie blue–stained SDS-PAGE gels of the supernatant samples used for quantification. (D) Coomassie blue–stained SDS-PAGE gels showing GST-Arf1 pull down of WRC subcomplexes in the presence or absence of Rac1QP. The dimer is the Sra1/Nap1 subcomplex. The trimer is the WAVE1(1-230)/Abi2(1-158)/HSPC300 subcomplex. Asterisks indicate weak binding signals. (E) Arf1 binding to the WRC is sensitive to pH and salt concentration. Shown is Coomassie blue–stained SDS PAGE from GST-Arf1 pull down of ∆WRC230 in indicated buffer conditions, in the presence or absence of Rac1QP. Red asterisk indicates increased background binding to GST beads at pH 6, to avoid which we use pH 7 and 50 mM NaCl throughout this study. (F) Coomassie blue–stained SDS-PAGE gel showing pull down of ∆WRC230 by different GST-tagged Arf-family members with or without Rac1QP.
The connection between Arf1, Rac1, and the WRC was initially found by proteomic and cellular studies to identify proteins important for clathrin-adaptor protein 1 (AP-1)–coated carrier biogenesis at the trans-Golgi network (47, 48). A more direct connection was established in a seminal study by Koronakis et al. in 2011 (25) in which they reconstituted WRC activation using lipid-coated beads and mammalian brain lysates. They found lipid-coated beads containing individual Rac1 or Arf1 only bound and activated WRC weakly, but the beads containing both GTPases markedly enhanced WRC membrane recruitment and activation (25). After that, a series of studies further corroborated the connection of Arf with the WRC. For example, Arf79 (the Arf1 homolog in Drosophila) was found to be critical for Sra1 localization and concomitant formation of lamellipodia (26). This function could not be complemented by Rac overexpression but could be restored by expressing human Arf1, underlining the importance of Arf1 for WRC activation and the conserved role of the Arf-WRC interaction across species (26). Furthermore, two different types of bacterial pathogens, Salmonella enterica and enteropathogenic or enterohemorrhagic Escherichia coli (EPEC or EHEC), could both hijack the Arf1-Rac1-WRC signaling axis to facilitate infection, albeit with opposite objectives (from the bacteria point of view) and via distinct mechanisms (27–29). In addition, the cooperative actions of Arf1 (or Arf6) and Rac1 on the WRC were found to be critical for the migration of invasive breast cancer cells (30, 31). Moreover, a missense mutation in Hem1 from patients with an inherited immunologic syndrome named immunodeficiency-72 with an autoinflammation phenotype was found to disrupt Arf1- but not Rac1-mediated WRC activation (49).
Despite the importance of Arf1-Rac1-WRC signaling in various normal and disease-related processes, the mechanism by which Arf1 achieves this function is unknown. Sharing less than 30% sequence identity with Rac1, Arf1 may use a distinct mechanism to regulate the WRC. However, does Arf1 directly interact with the WRC or Rac1 at all? If yes, what is the interaction mechanism, and what is the biochemical and structural basis of the cooperativity between Arf1 and Rac1? To answer these questions, here we have reconstituted a direct interaction between Arf and the WRC in solution by using purified proteins. We find that the interaction is greatly enhanced by Rac1 binding to the WRC mainly on the D site. Once bound to WRC, Arf1 can directly activate it independent of Rac1 binding to the A site. We further identified the Arf1 binding site, which is located at a conserved surface on Sra1 between the D site and the W helix of the WCA domain of WAVE. Mutating the Arf1 binding site abolished Arf1 binding, disrupted Arf1-mediated WRC activation, and impaired lamellipodia formation and cell migration. Together, our work reveals a new mechanism underlying WRC activation and paves the way for understanding how WRC-mediated actin polymerization integrates signals from Arf and Rac in various processes.
RESULTS
Arf GTPases directly interact with WRC, and the interaction is greatly enhanced by Rac1
The interaction between Arf1 and WRC was initially found using lipid-coated beads where both Arf1 and Rac1 were anchored on the membrane and incubated with mammalian brain extracts (25). To examine whether the Arf1-WRC interaction is direct and, if yes, to determine the underlying mechanism, we reconstituted this interaction in solution using recombinantly purified proteins. We found that glutathione S-transferase (GST)–tagged Arf1 could directly pull down both full-length (FL) WRC and a truncated WRC named ∆WRC230 (Fig. 1A, lanes 4 and 10) (34, 41). ∆WRC230 represents the minimal, structured core of the WRC, because it lacks the C-terminal, unstructured proline-rich region (PRR) and the WCA sequence of WAVE1, as well as the unstructured PRR and the Src homology-3 (SH3) domain of Abi2 (Fig. 1A, cartoon). Although the binding signals were weak, they were specific in comparison to background signals in GST controls (Fig. 1A, lane 2 versus lane 4 and lane 7 versus lane 10). Thus, Arf1 directly interacts with WRC, and the structured core of WRC is sufficient to bind Arf1.
To test whether and how Rac1 can enhance Arf1 binding to WRC, we used a Rac1 variant that contained two mutations, Q61L and P29S, which greatly enhanced Rac1 binding to the WRC as shown in our previous studies (41, 46). Unless otherwise noted, we refer to this Rac1Q61L/P29S construct as Rac1 or Rac1QP interchangeably in this study. We found that including free Rac1 in the pull-down reactions drastically enhanced GST-Arf1 binding to WRC (Fig. 1A, lanes 6 and 9). Note that Rac1 did not directly interact with Arf1 (Fig. 1A, lane 5) but was co-retained with WRC by GST-Arf1 (Fig. 1A, lanes 6 and 9). These results suggest that Arf1 and Rac1 can simultaneously bind to the same WRC via nonoverlapping binding sites and that Rac1 binding greatly stabilizes Arf1 binding.
As molecular switches, GTPases usually use the GTP state to engage with downstream effector proteins. We found that the Arf1-WRC interaction was also dependent on the nucleotide state of Arf1. Only Arf1 loaded with GTP, but not guanosine diphosphate (GDP), showed robust binding (Fig. 1B, lane 4 versus lane 5). Moreover, the interaction could be specifically blocked by EspG (E. coli secreted protein G) (Fig. 1B, lane 6), a bacterial effector protein secreted into the host cell by EPEC and EHEC during infection (29, 50). EspG directly binds the GTP form of Arf1 and Arf6 (also see Fig. 1B, lanes 1 to 3) (29, 50). This interaction was suggested to disrupt Arf-WRC signaling in host cells, which helped these extracellular pathogens evade WRC-mediated phagocytosis (29). Our data suggest that EspG can achieve this by directly competing off Arf1 (and/or Arf6) binding to WRC. Therefore, Arf1 may use the same surface to interact with the WRC and EspG.
We next used our previously established equilibrium pull-down (EPD) assay to quantitatively measure the enhancement of Arf1 binding by Rac1 (41, 51) (Fig. 1C). We found that in the absence of Rac1, the Arf1-WRC interaction was weak, with a dissociation constant Kd ~ 23 μM (Fig. 1C, black). By contrast, in the presence of 100 μM Rac1, which should saturate both A and D sites of the WRC (41, 46), Arf1 binding affinity was increased nearly 30-fold (Kd ~ 0.66 μM; Fig. 1C, orange). The enhanced binding was not an artifact of high concentration of free Rac1 included in the assay, as a mutant Rac1, in which the entire Switch I motif critical for WRC binding was removed (herein referred to as Rac1Dead; fig. S1A), could not promote Arf1 binding at the same concentration (Fig. 1C, blue). Thus, Rac1 can enhance the weak interaction between Arf1 and WRC by ~30-fold.
We found that Arf1 binding was likely mediated by the Sra1 or Nap1 subunit, but not WAVE1, Abi2, or HSPC300, as only the dimeric subcomplex containing Sra1/Nap1, but not the trimeric subcomplex formed by WAVE1/Abi2/HSPC300, showed weak binding signals comparable to the fully assembled, pentameric WRC (Fig. 1D, lane 4 versus lane 6, asterisks). Unlike binding to the intact WRC, however, the interaction with the Sra1/Nap1 dimer could not be enhanced by Rac1 (Fig. 1D, lane 5 versus lane 7, asterisks), suggesting that although Sra1 or Nap1 may contain the Arf1 binding site, the enhancement of Arf1 binding by Rac1 is dependent on the fully assembled WRC. Moreover, we found that Arf1 binding to WRC was sensitive to both pH and salt concentration, with pH 6 to 7 and 50 mM NaCl, but not pH 8 or 100 mM NaCl being able to sustain the binding (Fig. 1E, lanes 2 and 6). This indicates that the Arf-WRC binding involves polar interactions (see below).
We further tested whether the Arf1-WRC interaction is unique to Arf1 or is general to other Arf-family proteins. In mammals, the Arf family contains six canonical members (Arf1 to Arf6) and various distantly related Arf-like proteins (Arl) (2, 5). On the basis of sequence similarities, Arf1-Arf6 can be further divided into three classes: class I (Arf1 to Arf3), class II (Arf4 and Arf5), and class III (Arf6). We found that besides Arf1, Arf5 and Arf6 also robustly bound WRC in a Rac1-dependent manner, although perhaps with slightly different affinities (Fig. 1F, lanes 1 to 6). By contrast, Arl1 or Arl2 did not show clear binding (Fig. 1F, lanes 7 to 10). These results suggest that the six members of the Arf family, but perhaps not the more divergent Arl proteins, can use the same mechanism to interact with WRC. Together, our biochemical reconstitution established a direct, nucleotide-dependent interaction between Arf-family GTPases and WRC. This interaction is greatly enhanced by Rac1 binding to WRC.
Arf1 binding mainly depends on Rac1 binding to the D site
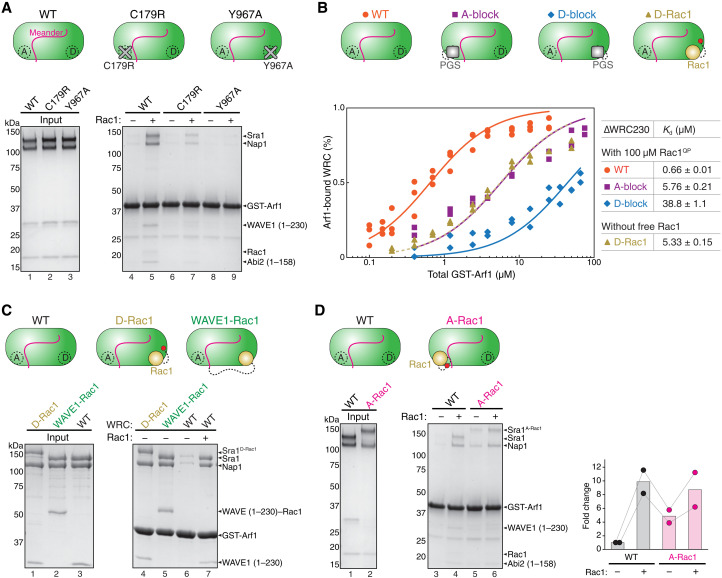
Rac1 can bind to both A and D sites on WRC, albeit with distinct affinities and effects on WRC activation (41, 46). Therefore, we asked which Rac1 binding event was key to promoting Arf1 binding. To answer this question, we first used single–amino acid mutations to specifically disrupt the A or D site from binding to Rac1 (36, 41, 46) (Fig. 2A, cartoon). When Rac1 binding to the A site was disrupted by Sra1C179R, Rac1 binding to the D site still enhanced Arf1 binding, but to a lower extent than WT WRC (Fig. 2A, lanes 6 and 7). By contrast, when Rac1 binding to the D site was disrupted by Sra1Y967A, Rac1 binding to the A site could no longer promote Arf1 binding (Fig. 2A, lanes 8 and 9). These data indicate that Rac1 binding to the D site plays a more important role in promoting Arf1 binding.
Fig. 2. Arf1 binding mainly depends on Rac1 binding to the D site.
(A) Coomassie blue–stained SDS-PAGE gels showing GST-Arf1 pull down of WRC bearing point mutations in Sra1 that specifically disrupt the A or D site. (B) EPD assay measuring the binding affinity of GST-Arf1 for the indicated ∆WRC230 constructs in the presence or absence of 100 μM Rac1QP. Data for each mutant are pooled from two independent experiments. Data for the WT WRC are taken from Fig. 1C and used here as a reference point. See fig. S1 for representative gel images. (C) Coomassie blue–stained SDS-PAGE gels showing GST-Arf1 pull down of WRCs with Rac1 tethered to indicated positions to stabilize Rac1 binding to the D site. (D) Coomassie blue–stained SDS-PAGE gels showing GST-Arf1 pull down of WRCs with Rac1 inserted between Y423/S424 of the surface loop (amino acids 418 to 432) to stabilize Rac1 binding to the A site. Shown on the right is the gel quantification of the Sra1-Nap1 bands normalized to GST-Arf1 bands from two independent repeats, with the data from each repeat connected. In the schematic of WRCs, red dots indicate the tethering points of Rac1 to the A or D site.
To further validate this result, we used EPD assays to directly measure the binding affinities of Arf1 to WRCs with disrupted A versus D site. For this, instead of using the above single–amino acid mutations to disrupt either site, which may retain weak, residual Rac1-binding activity, we inserted an inert protein PGS (glycogen synthase from the extreme thermophile Pyrococcus abysii) into a surface loop at the A or D site to completely block Rac1 binding. We herein name the new variants WRCA-block and WRCD-block, respectively (Fig. 2B, cartoon). Being a small, stable protein and with its N and C termini located in close proximity (6.5 Å), PGS was initially used to insert into the human orexin/hypocretin receptors hOX1R and hOX2R to stabilize an intracellular loop and produce high-resolution diffracting crystals (52, 53). Inserting PGS into the surface loop of the A or D site did not affect WRC assembly or purification (fig. S2, A and B) or the basal level of Arf1-WRC interaction in the absence of free Rac1 (fig. S1C), but further reduced the affinity measurement of Rac1 to WRC (from Kd ~ 2 μM for WRCY967A to ~7.5 μM for WRCD-block; fig. S1B, blue versus orange), likely due to eliminating the residual Rac1 binding to the D site in WRCY967A. When we blocked the A site and subjected the D site to 100 μM Rac1, Arf1 binding was enhanced, although not to the level of WT WRC (Kd ~5.76 μM for WRCA-block versus ~0.66 μM for the WT WRC; Fig. 2B, purple versus orange; and fig. S1D), suggesting that Rac1 binding to the D site was partially sufficient to promote Arf1 binding. By contrast, when we blocked the D site and exposed the A site to 100 μM Rac1, Arf1 binding was not enhanced, but remained similar to that in the absence of Rac1 or in the presence of 100 μM Rac1Dead (Kd ~38.8 μM; Fig. 2B, blue; Fig. 1C, black; and fig. S1C, orange), suggesting that Rac1 binding to the A site alone could not promote Arf1 binding in this specific experimental condition (but see below).
As an alternative strategy to validate the contribution of the D site to Arf1 binding, we stabilized Rac1 binding to the D site by tethering it to the C terminus of Sra1 (which we refer to as ∆WRC230D-Rac1) (46) or the C terminus of WAVE1 that lacked the WCA (which we named ∆WRC230WAVE1-Rac1) (Fig. 2C, cartoon) (41). These constructs stabilize D site Rac1 binding, which had allowed us to solve cryo-EM structures of the WRC with Rac1 bound to the D site (41, 46). We found that, without free Rac1, both ∆WRC230D-Rac1 and ∆WRC230WAVE1-Rac1 were able to enhance Arf1 binding to the level of the WT WRC enhanced by free Rac1 (Fig. 2C, lanes 4, 5, and 7). Furthermore, in the EPD assay, ∆WRC230D-Rac1 without free Rac1 enhanced Arf1 binding to a level nearly identical to that of WRCA-block in the presence of 100 μM Rac1 (Kd ~ 5.33 μM; Fig. 2B, golden dashed versus purple). Therefore, supplying Rac1 to the D site by covalent tethering has the same effect in promoting Arf1 binding as supplying free Rac1 to a WRC with a blocked A site.
The above assays not only confirm that Rac1 binding to the D site is essential for enhancing Arf1 binding, but also show that mutating or blocking the A site dampens this effect (Fig. 2A, lane 7; and Fig. 2B, purple). This indicates that Rac1 binding to the A site should also play a role, which might have eluded detection in the assays described above due to the low affinity of the A site for Rac1. The potential cooperativity between A and D sites could further reduce A site binding when the D site is disrupted (41, 46). To examine the contribution of the A site more directly, we stabilized Rac1 binding to the A site by inserting a Rac1 between Y423/S424 in a nonconserved surface loop near the A site (termed ∆WRC230A-Rac1; Fig. 2D, cartoon). This strategy had allowed us to determine the cryo-EM structure of the WRC with Rac1 bound to the A site and D site simultaneously (46). We found that, without free Rac1, tethering Rac1 to the A site mildly promoted Arf1 binding (Fig. 2D, lane 3 versus lane 5). Adding free Rac1 to ∆WRC230A-Rac1 to occupy the D site further enhanced Arf1 binding (Fig. 2D, lane 6). These data suggest that Rac1 binding to the A site partially contributes to Arf1 binding. Together, we conclude that Rac1 binding to both A and D sites plays a role in promoting Arf1 binding to the WRC, but the D site has a major contribution as compared to the A site.
Arf1 promotes WRC activation using a novel mechanism distinct from Rac1
Arf1 and Rac1 were shown to cooperatively promote WRC activation on lipid-coated beads (25). Because Rac1 binding to the A site is sufficient to activate the WRC through an allosteric mechanism (46), the question remains: Does Arf1 binding merely increase the membrane recruitment of WRC, contribute to the same allosteric changes driven by Rac1 binding to the A site, or promote WRC activation through an entirely different mechanism?
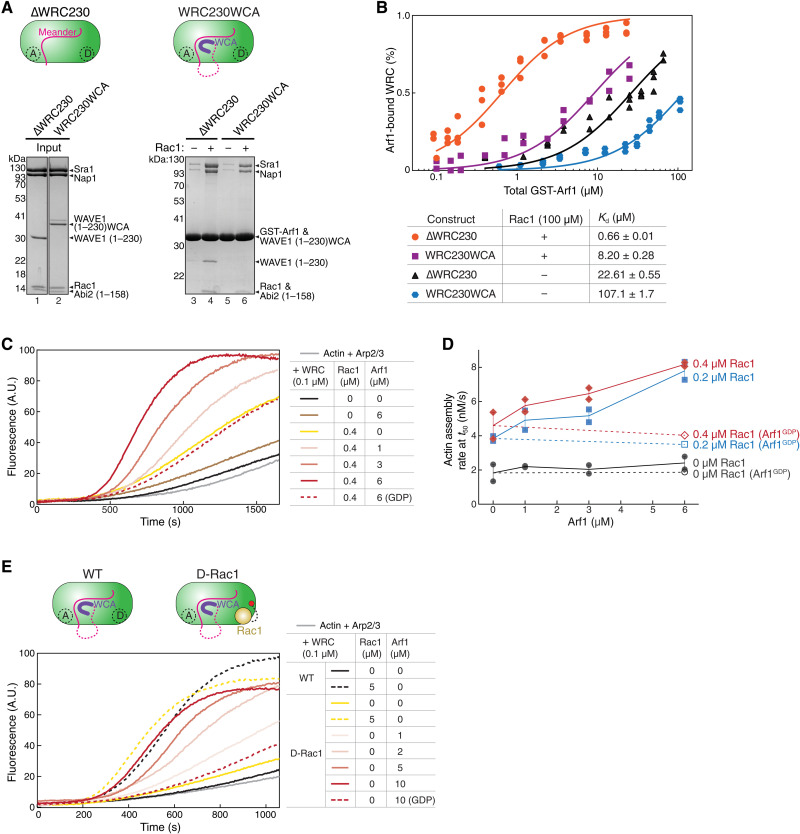
To distinguish between these possibilities, we first tested whether Arf1 differentially binds to the WRC in the autoinhibited (“closed”) or activated (“open”) state. Previous studies showed that, as an activator, Rac1 had higher affinity for the open conformation represented by ∆WRC230 (which lacks the WCA) than for the closed conformation represented by the WRC that contained WCA (WRC230WCA; Fig. 3A, cartoon) (36, 41). If Arf1 is an activator, it should similarly prefer the open conformation. We observed less binding for WRC230WCA than ∆WRC230, both in the presence and in the absence of Rac1 (Fig. 3A). Our EPD assay further confirmed this observation (Fig. 3B and fig. S1E). In the absence of free Rac1, Arf1 had very low binding affinity for WRC230WCA, with a Kd (~107 μM) ~5 times of ∆WRC230 (~22.6 μM) (Fig. 3B, blue versus black). Addition of a saturating concentration of Rac1 (100 μM) enhanced Arf1 binding to both WRC230WCA and ∆WRC230, although not to the same level (Kd ~ 8.2 μM for WRC230WCA versus Kd ~ 0.66 μM for ∆WRC230) (Fig. 3B, purple versus orange). These data indicate that Arf1 distinguishes the closed versus the open conformation and therefore may act as an activator of the WRC.
Fig. 3. Arf1 promotes WRC activation independent of Rac1 binding to the A site.
(A) Coomassie blue–stained SDS-PAGE gels showing GST-Arf1 pull down of WRC with or without the WCA sequence. (B) EPD assay comparing the binding affinity of GST-Arf1 for the WRC with or without the WCA sequence. Data for WRC230WCA are pooled from two independent experiments for each condition. Data for the ∆WRC230 are taken from Fig. 1C and used here as a reference point. See fig. S1 for representative gel images. (C and D) Representative pyrene-actin polymerization assay (C) and quantification of the actin polymerization rate at t50 (D) (70) measuring the activity of WRC230WCA in the presence of indicated concentrations of Rac1QP and Arf1. (E) Pyrene-actin polymerization assay of the WT WRC230WCA versus WRCD-Rac1 in response to the addition of free Rac1QP or Arf1. Reactions in (C) to (E) contain 3.5 μM actin (5% pyrene-labeled), 10 nM Arp2/3 complex, 100 nM WRC, and indicated amounts of Rac1 and/or Arf1. In all actin assays, Arf1 is loaded with the non-hydrolyzable GTP analogue guanosine-5'-[(β,γ)-imido]-triphosphate (GMPPNP), unless it is indicated with GDP. A.U.: arbitrary units.
We next measured whether Arf1 could promote WRC activation in the pyrene-actin polymerization assay in aqueous solution (Fig. 3, C to E) [as opposed to on membranes as in the previous study (25)]. For this, the Arf1 construct used in this study does not contain the N-terminal amphipathic helix (also referred to as Arf1∆N17). This helix is important for Arf1 to bind membranes but is usually dispensable for binding downstream effectors and therefore often removed in biochemical and structural studies (50, 54). In the absence of Rac1, Arf1 had no obvious effect on WRC activity, potentially due to its low binding affinity to WRC230WCA (Fig. 3C, brown, and Fig. 3D, black). In the presence of low concentrations of Rac1, however, Arf1 enhanced WRC activation in a dose-dependent manner (Fig. 3C, red curves, and Fig. 3D, red and blue). The enhanced WRC activation depended on Arf1 GTP binding as Arf1 loaded with GDP did not have such an effect (Fig. 3, C and D, dashed lines).
The above data suggest that Arf1 binding directly contributes to WRC activation. Because of the presence of free Rac1 in the above reactions, however, these data cannot tell whether Arf1 acts by promoting the same conformational changes driven by Rac1 binding to the A site or by directly activating the WRC through a separate mechanism. To distinguish between these two mechanisms, we further tested whether Arf1 could activate the WRC230WCA in which a Rac1 molecule was tethered to the D site (WRCD-Rac1; Fig. 3E, cartoon) (46). In this construct, the tethered Rac1 does not activate the WRC (also see Fig. 3E, yellow solid curve) (46) but can promote Arf1 binding to the WRC (Fig. 2C), allowing us to determine whether Arf1 can activate WRC in the absence of a Rac1 molecule acting through the A site. In the absence of free Rac1, we found that Arf1 activated WRCD-Rac1 in a dose-dependent manner (Fig. 3E, red solid curves), while Arf1 loaded with GDP had no such effect (Fig. 3E, red dashed curve). To rule out the possibility that Arf1 may activate WRC by mimicking Rac1 binding to the A site, we disrupted the A site by the point mutation C179R and found Arf1 still activated WRCD-Rac1 in a dose-dependent manner (fig. S3A), although with reduced potency perhaps because the mutation indirectly weakened Arf1 binding. Together, the above data suggest that Arf1 binding can directly activate WRC, at least in vitro, and the Arf1-mediated activation does not involve an interaction of either Rac1 or Arf1 with the A site. Therefore, Arf1 must use a novel mechanism to drive WRC activation.
It is important to note that the Arf1-mediated WRC activation reached levels similar to those achieved with Rac1 binding to the A site (Fig. 3E, black and yellow dashed curves), suggesting that Arf1 binding activates the WRC by releasing the WCA, instead of by causing protein aggregation (which is believed to cause artificial WRC activation to a much larger extent than the release of WCA) (33, 40, 55). This is consistent with our dynamic light scattering (DLS) measurement showing that Arf1 did not promote WRC aggregation (fig. S2J).
Arf1 binds to a conserved site distinct from Rac1 binding sites
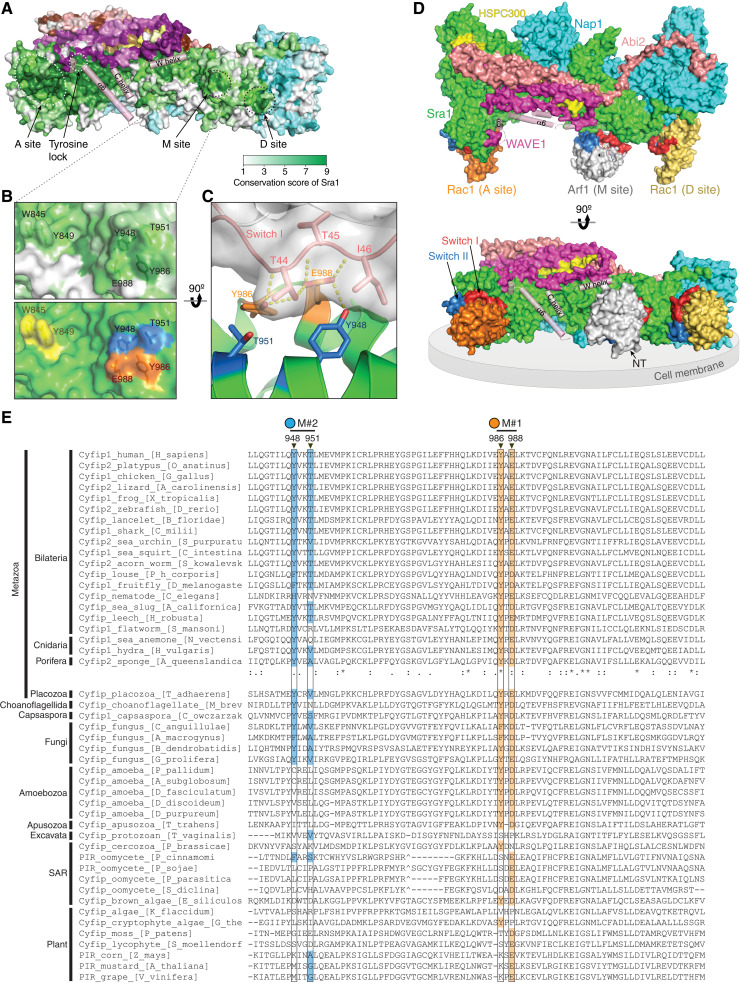
How does Arf1 binding activate WRC? To answer this question, we determined the Arf1 binding site by combining protein docking, surface conservation analysis, mutagenesis, and molecular dynamics (MD) simulation (Figs. 4 and 5 and figs. S4 and S5). We first searched for potential binding sites using several different protein docking programs, including ClusPro (56), HADDOCK (57), InterEvDock (58), FRODOCK (59), and HDOCK (60). During the search, we restrained the Switch I and Switch II motifs of Arf1 in close contact with the WRC, because they usually mediate GTPase-effector interactions. Combining the docking results with the surface conservation analysis of the WRC by Consurf (61), we selected a series of conserved surface patches, mutated the solvent-exposed residues individually or in combination, purified the mutant WRCs, and used pull-down assays to examine whether any mutation could disrupt Arf1 binding (fig. S4).
Fig. 4. Arf1 binds to a conserved site distinct from Rac1 binding sites.
(A) Surface conservation of the WRC, with color to white gradients representing the most (ConSurf score = 9) to the least conserved residues (ConSurf score = 1) (61). Important sites on Sra1 are indicated with dotted circles. Semitransparent pink cylinders refer to WAVE1 sequences that are destabilized upon WRC activation by Rac1 (46). (B) Close-up view of the M site showing surface conservation (top) and surface patches to be mutated (bottom, same color scheme as in Fig. 5A). (C) Side view showing the interaction between Arf1 and the M site in the MD-optimized model C8. Contacting residues are shown as sticks. Yellow dashed lines indicate polar interactions. (D) Surface representation of the overall structural model of the WRC bound to two Rac1 molecules (PDB: 7USE) (46) and one Arf1 molecule (PDB: 1J2J). Position of Arf1 shows the MD-optimized docking solution C8. Switch I and II of Rac1 and Arf1 are red and blue, respectively. Gray disc demonstrates the predicted orientation of the WRC at the plasma membrane. “NT” indicates the N terminus of Arf1∆N17 used in this study. (E) Sequence alignments of Sra1 from representative eukaryotic organisms. Surface residues of the M site (black boxes) are highlighted with orange for the M#1 surface patch and blue for M#2, as indicated by black arrowheads on top. Degrees of conservation in metazoans (up to Porifera) are represented with ClustalW symbols (84) (“*” for no change; “:” for conserved; “.” for less conserved changes). “-” for missing amino acids; “^” for amino acid insertions in alignments that are not shown.
Of the more than 12 conserved surfaces that we surveyed, one surface specifically disrupted Arf1 binding (fig. S4A, M1 site, and S4F, lane 6, and Fig. 5A, lane 10). We named this site the M site because it is in the middle of the WRC, sandwiched between the D site and the W helix of the WCA (Fig. 4A). The M site is a small, conserved, and slightly negatively charged surface patch on Sra1 (fig. S4, A and B) (85). Mutating the conserved surface residues at the M site, either Y986A/E988A (∆M#1) or Y948A/T951A (∆M#2), disrupted Arf1 binding, whereas mutating two other conserved residues, W845A/Y849A (∆M#3), near the M site did not disrupt Arf1 binding (Figs. 4B and 5A, lanes 10 to 12, and fig. S4F), suggesting that the effect of ∆M#1 and ∆M#2 was specific to Arf1 binding. Furthermore, the WRC carrying ∆M#1 or ∆M#2 mutations could not be further activated by Arf1 (Fig. 5, B and C, and fig. S3B). It is important to note that the M site mutations only disrupted Arf1-mediated activation but not Rac1-mediated activation (Fig. 5D and fig. S3B). Thus, these surface mutations are specific in disrupting Arf1 binding and Arf1-mediated activation, without affecting WRC folding (fig. S2, C to G) or disturbing Rac1-mediated activation.
Fig. 5. M site mutations disrupt Arf1 binding and Arf1-mediated WRC activation.
(A) Coomassie blue–stained SDS-PAGE gels showing GST pull down of ∆WRC230 bearing the indicated mutations in Sra1 at the M site. (B and C) Representative pyrene-actin polymerization assay (B) and quantification of the actin polymerization rate at t50 normalized to WT WRC230WCA + 0.5 μM Rac1 (C), measuring the effect of M site mutations on WRC activation by Arf1. Reactions contain 3.5 μM actin (5% pyrene labeled), 10 nM Arp2/3 complex, 100 nM WRC230WCA (WT or indicated mutants), and indicated amounts of Rac1QP and/or Arf1 loaded with GMPPNP. Error bars represent SEM. (D) Comparison of the WT WRC to the ∆M#2 (Y948A/T951A) mutant activated by different amounts of Rac1QP. Reactions were performed in the same conditions as in (B).
Note that all four residues at the M site surface are highly conserved in metazoans (Fig. 4E). In particular, Y986 remains strictly tyrosine from human to sponge, while E988 is only exchangeable with aspartate. In non-metazoan species, they are either partially conserved (such as in amoeba) or not conserved (such as in plants) (Fig. 4E). This suggests that the Arf-WRC interaction is important for processes unique to metazoans.
To further define the binding mechanism, we applied MD simulation to optimize binding poses of the top six docking models that placed Arf1 at the M site (fig. S5, A and B). We then evaluated different models by calculating the molecular mechanics/Poisson-Boltzmann surface area/weighted solvent accessible surface area (MM-PBSA-WSAS), free energies of the whole complex and the binding free energy between Arf1 and WRC (fig. S5, C to I). Of the six docking models, model C8 gave the lowest binding free energy (fig. S5, G and I). Introducing ∆M#1 or ∆M#2 mutations onto model C8 increased the binding energy, suggesting that they destabilized Arf1-WRC interaction. By contrast, introducing the control mutation ∆M#3 did not affect the binding energy (fig. S5I). These data are consistent with our pull-down assays showing that only ∆M#1 and ∆M#2, but not ∆M#3, disrupted Arf1 binding (Fig. 5A and fig. S4F).
Note that it was previously shown that the M371V mutation in Hem1 (M373V in Nap1) found in human patients interfered with (but did not abolish) Arf1 binding and WRC activation (49). The above analysis suggests that M371 is not the Arf1 binding site. Rather, the effect of M371V was likely indirect, as this residue is located at the bottom of a deep pocket neighboring the D site, where it was difficult to accommodate an Arf1 molecule (fig. S4A).
The MD-optimized model sheds light on how Arf1 may bind and activate WRC. First, the interaction is mediated by the Switch I motif (Fig. 4C and fig. S5G), the same region that binds to EspG (50), explaining how EspG competes off WRC binding to inhibit phagocytosis during pathogenic E. coli infection (29) (Fig. 1B). Second, the interaction mainly involves hydrogen bonding between Y986 and E988 in Sra1 and T44 and T45 in Arf1, with Y948 or T951 in Sra1 contacting I46 and T44 in Arf1 through van der Waals interactions (Fig. 4C and fig. S5G). This explains why Y986A/E988A (∆M#1) disrupted Arf1 binding more severely than Y948A/T951A (∆M#2) in GST pull-down assays (Fig. 5A and fig. S4F) and is also consistent with our observation that Arf1 binding is sensitive to pH and salt concentration (Fig. 1E). Third, the relative orientation of Arf1 is compatible with the model of how WRC is oriented on the membrane together with two Rac1 molecules (Fig. 4D) (36, 41, 46). In this orientation, the N terminus of Arf1∆17 is near the plasma membrane (Fig. 4D, arrow), which would allow its N-terminal amphipathic helix to associate with membranes. Last, this model explains how Arf1 binding may activate the WRC. Arf1 is located near (but not in direct contact with) the W helix of WCA (Fig. 4D). Therefore, distinct from Rac1-mediated WRC activation, which involves a series of conformational changes propagating from the A site to a conserved region around WAVE1Y151 (referred to as tyrosine lock) to release the WCA (46), Arf1 binding may contribute to WRC activation by directly perturbing the W helix located in its proximity (see models in Discussion).
Arf1 binding acts downstream of Rac1 binding to the D site to promote lamellipodia formation and cell migration
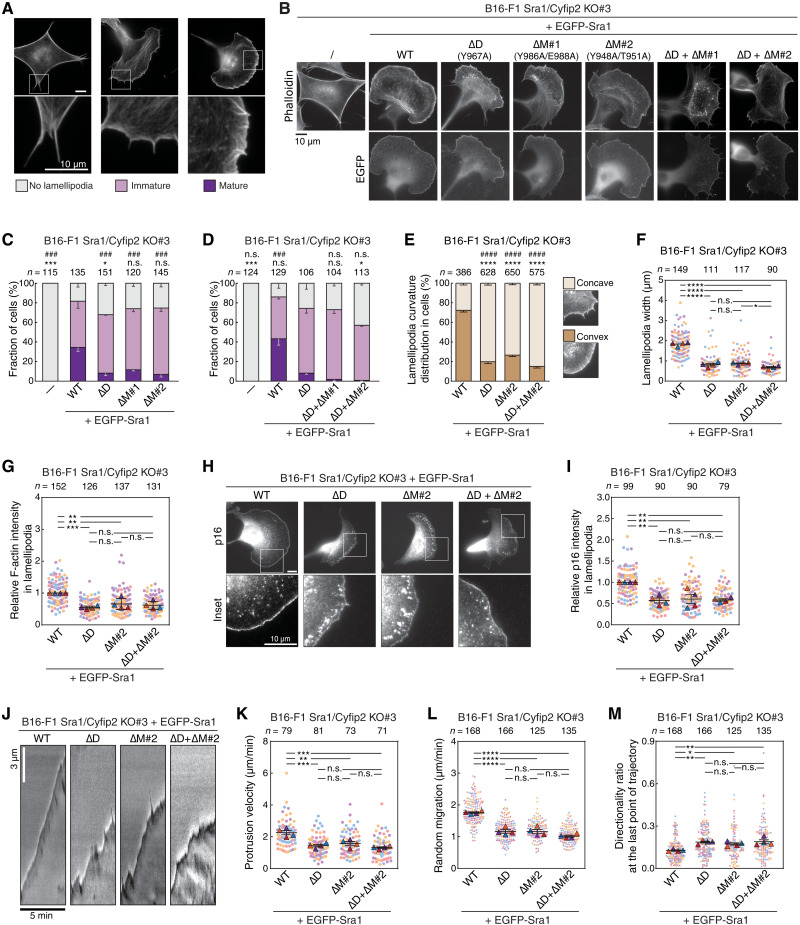
The identification of the M site allowed us to specifically probe the function of the Arf1-WRC interaction in cells. WRC is key to actin polymerization at plasma membranes and formation of sheet-like protrusions known as lamellipodia and membrane ruffles commonly found at the leading edge of migrating or spreading cells (39, 43). We previously established a WRC complementation assay to evaluate the contributions of various Sra1 mutations to lamellipodia formation. In this assay, we could introduce wild-type (WT) or mutant Sra1 variants to rescue loss of lamellipodia formation in B16-F1 cells that were genetically disrupted for both Sra1 and Cyfip2 genes. We previously reported that mutating the A site almost completely abolished WRC-mediated lamellipodia formation, while mutating the D site impaired (but did not eliminate) actin assembly and lamellipodia morphology (62, 63). The morphologies of cells rescued with WT or mutant Sra1 variants fell into three major categories: lacking lamellipodia entirely, displaying partially developed lamellipodia, and displaying fully developed lamellipodia. We termed the latter two categories “immature” and “mature” lamellipodia, respectively (Fig. 6A) (46, 62, 63). Here, using the same approach, we found that mutating the M site produced phenotypes nearly identical to mutating the D site (Fig. 6, B and C). In both cases, mutations produced narrow actin networks and reduced the formation frequency of mature lamellipodia but did not perturb WRC localization or assembly (Fig. 6, B and C, and fig. S6A). When we combined the M site and D site mutations into one construct, they did not aggravate the phenotype, except that the ∆D/∆M#2 dual mutations slightly decreased the total percentage of lamellipodia-containing cells (Fig. 6, B and D, and fig. S6B). Because Arf1 binding mainly depends on Rac1 binding to the D site (Fig. 2), this result indicates that M and D sites may function in the same mechanistic pathway to regulate WRC activity.
Fig. 6. M site mutations impair lamellipodia formation and cell migration.
(A) Categorization of lamellipodia morphology and (B) representative images of B16-F1 Sra1/Cyfip2 double KO#3 cells transfected with indicated EGFP-Sra1 constructs and stained for F-actin. (C and D) Quantification of lamellipodia morphology in indicated Sra1-rescued cells. Statistical significance was assessed for differences between transfected cell groups concerning cell percentages displaying “no lamellipodia” phenotype (*P < 0.05; ***P < 0.001) and with “mature lamellipodia” phenotype (###P < 0.001); n = cell numbers. (E) Quantification of the frequency of convex (****P < 0.0001) versus concave (####P < 0.0001) lamellipodia in Sra1-rescued cells; n = lamellipodia numbers. (F) Quantification of the width of individual lamellipodia; n = cell numbers. (G) Relative F-actin intensities in lamellipodia, normalized to WT Sra1-rescued cells; n = cell numbers. (H) Representative immunostaining images and (I) quantification of Arp2/3 complex subunit ArpC5A (p16) in lamellipodia of Sra1-rescued cells; average intensities normalized to WT Sra1; n = cell numbers. (J) Representative kymographs of phase-contrast movies of Sra1-rescued cells and (K) quantification of derived lamellipodia protrusion rates; n = cell numbers. (L) Quantification of random migration rates and (M) migration directionalities of indicated Sra1-rescued cells. In (M), the directionality ratios at the final time points of all migration trajectories are shown. See Materials and Methods for how directionality ratios are calculated and fig. S6D for the directionality ratios throughout entire migration trajectories. Differentially colored (red, blue, violet, and orange) dots in superplots in (F), (G), (I), and (K) to (M) represent individual data points from separate, independent experiments (n = 4); triangles equal arithmetic means for each experiment, and error bars are SEM. Statistics: one-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not statistically significant.
To further compare the contributions of M versus D sites to WRC function, we quantified an array of parameters for the cells rescued with Sra1 mutants. It is remarkable that, in all assays, we found no difference for Sra1 carrying individual ∆D or ∆M#2 or combined ∆D/∆M#2 mutations (Fig. 2, E to L). We first quantified lamellipodia curvature distribution in rescued cells, as efficiency of protrusions frequently correlates with lamellipodia shape, with mature lamellipodia displaying convex and immature ones adopting more concave shapes (Fig. 6E, inset). This was previously noted qualitatively for the typical, underdeveloped lamellipodia formed upon rescue with the ΔD variant (62) and confirmed here quantitatively for both D site and M site mutations or the combination of both (Fig. 6E). In all other detailed analyses, individual or combined D and M site mutations produced lamellipodia of virtually undistinguishable morphology and dynamics, which is reflected by the width (breadth) of individual lamellipodia (Fig. 6F), total levels of F-actin (Fig. 6G) or Arp2/3 complex (Fig. 6, H and I; p16 intensity) in lamellipodia, lamellipodial protrusion rates (Fig. 6, J and K, and movie S1), and migration efficiency and directionality of these cells (Fig. 6, L and M; fig. S6, C and D; and movie S2). All the above-examined parameters were reduced by mutant WRCs except for migration directionality, which was increased. Similar negative correlations between cell migration rate and directionality were previously observed in B16-F1 cells lacking actin polymerases of the Enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) family, which are another important regulator of lamellipodia formation (64). It is possible that in B16-F1 cells, which mostly exhibit lamellipodia-dependent migration (62), compromised protrusion efficiency and actin assembly at cell leading edges reduce frequent turning activity and hence change of migration direction.
Together, all the aforementioned biochemical and cellular results suggest that the M and D sites act in the same mechanistic pathway—with Arf1 binding to the M site likely acting downstream of Rac1 binding to the D site—to regulate lamellipodia morphology and dynamics. This activity directly contributes to the efficiency of Arp2/3 complex–mediated protrusion and cell migration.
DISCUSSION
By biochemical reconstitution, structural analysis, and cellular assays, our work establishes that Arf1 directly interacts with WRC through a previously unidentified conserved surface located on Sra1. We show that, although intrinsically weak, this interaction can be greatly enhanced by Rac1 binding to the D site. Once bound to the WRC, Arf1 can independently drive WRC activation, at least in vitro, using a mechanism distinct from that mediated by Rac1 binding to the A site. We further demonstrate that disrupting the Arf1-WRC interaction by point mutations specifically abolishes Arf1-mediated (but not Rac1-mediated) WRC activation and impairs WRC-mediated lamellipodia formation and cell migration. Our work has important implications for the regulation of the actin cytoskeleton in many different biological systems.
First, our study established a new mechanism underlying WRC activation. The WRC is a central signaling hub through which a large diversity of membrane ligands can transmit signals to Arp2/3 complex–mediated actin polymerization (38, 40, 41, 43). Despite the long list of WRC ligands, Rac1 has been known as the only activator that is both necessary and sufficient—at least in vitro—to activate WRC (41, 62). While other ligands may act cooperatively with Rac1 to further tune WRC activity, exactly how they do so is completely unknown (25, 38, 40, 65). In particular, how Arf1 facilitates WRC activation has remained enigmatic for many years. It was not known whether Arf1 can directly interact with WRC, and if yes, how Arf works together with Rac1 to promote WRC activity (12). Our work provides firm answers to these questions, revealing that notable Arf1 binding relies on Rac1 binding mainly to the D site, but Arf1 binding can directly promote WRC activity even independently of Rac1 binding the A site. These results establish Arf1 as a second, genuine activator of the WRC and provides a mechanism to explain the cooperativity between Arf1 and Rac1 previously observed both in vitro and in cells (8–10, 25).
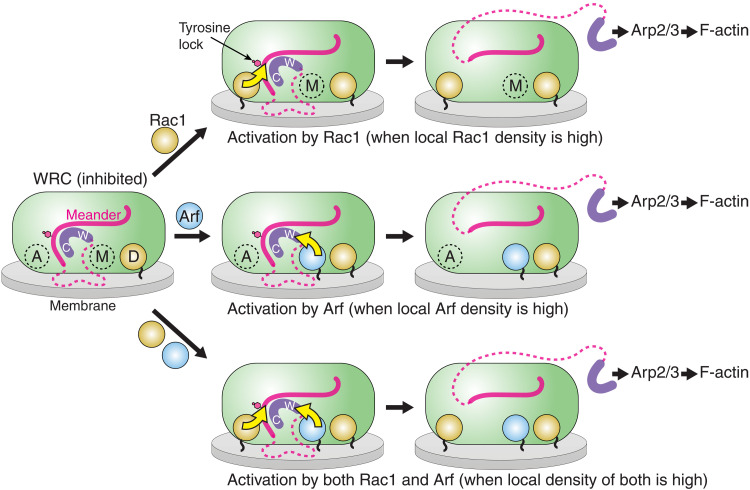
Second, our study lays a foundation for studying how WRC-mediated actin polymerization connects various Arf- and Rac1-mediated processes. Our work identifies point mutations that can specifically disrupt Arf binding and Arf-mediated (but not Rac1-mediated) WRC activation. These mutations will be powerful tools for dissecting the role of the Arf-WRC-Arp2/3-actin signaling axis from the canonical Rac1-WRC-Arp2/3-actin axis. Arf-family GTPases play an important role in various membrane trafficking processes, with some of them tightly connected to actin cytoskeleton regulation (2, 12, 13). On the other hand, new roles for actin, WRC, and Arp2/3 complex are emerging, suggesting their importance in the endomembrane systems beyond their canonical role in driving plasma membrane protrusions (48, 66–68). We thus posit that Arf-mediated WRC activation provides the cell with an additional pathway for promoting WRC activation and actin polymerization, the precise outcome of which will likely depend on relative local membrane densities of Rac1 versus Arf (Fig. 7). Specifically, Rac1 binding to the high-affinity D site may serve as a general recruitment mechanism to prime the WRC on the membrane without causing activation. Then, depending on specific upstream signals in distinct cell types and tissues leading to activation of various Arf- or Rac1-GEFs, the precise tuning of WRC activation in the given condition and system will depend on the local density of activated Rac1 or Arf molecules, which can subsequently trigger WRC activation by distinct structural mechanisms (Fig. 7). Revealing precisely how the established roles of various Arf GTPases in trafficking and the prominent accumulation of these GTPases on endomembranes (e.g., Golgi membranes) (69) relate to their roles in WRC activation and lamellipodial protrusion warrants substantial future investigation.
Fig. 7. Rac1 and Arf may act both cooperatively and separately to promote WRC activation.
Schematic showing how the WRC can be activated by Rac1 (top), Arf (middle), and both (bottom) through specific mechanisms that can arise independently from each other. Structural elements critical to WRC inhibition and activation are shown. Yellow arrows indicate structural pathways leading to WRC activation. Magenta dashed lines represent unstructured sequences in WAVE1. Black wiggly lines attached to Arf and Rac1 represent membrane binding sequences and lipid modifications of the GTPases. Rac1 first engages with the D site because of its relatively high affinity, which primes the WRC on the membrane without causing activation (left). When Rac1 density on the membrane is high (top), further binding of Rac1 to the A site promotes WRC activation by allosterically destabilizing the tyrosine lock region, which subsequently releases Y151 (indicated by pink hexagon) and the WCA (purple) (46). Alternatively, when Arf1 density on the membrane is high (middle), Rac1 at the D site promotes Arf binding to the M site, which, in turn, through its close proximity to the W helix, can perturb WCA binding to promote WRC activation. The remaining part of the schematic displays the functional outcome of both mechanisms operating in cooperation to ensure an optimized output response (bottom).
Third, the Arf binding site is highly conserved in metazoans, from human to sponge, but is only partially conserved in other organisms and is not conserved in plants. This suggests that the function of Arf1-mediated WRC activation is likely important for processes unique to metazoans, such as neuronal outgrowth and synapse formation, immune cell chemotaxis and activation, and cancer cell migration and metastasis, in all of which Arf and WRC play important roles (2, 3, 12, 13, 43). In non-metazoan species, while the sequence analysis of the M site suggests that the direct interaction between Arf and the WRC is perhaps lost (Fig. 4E), considering the conserved importance of Arf and WRC, we cannot rule out the possibility that the M site surface and Arf molecules may still have coevolved to maintain the connection between Arf and WRC. Our work raises the possibility of exploring the role of Arf-related processes in WRC-mediated actin polymerization in both metazoan and non-metazoan organisms. Together, this work uncovers a new, conserved mechanism underlying WRC activation and provides a foundation for exploring the regulation of the actin cytoskeleton in multiple processes in which Rac and the various Arf-family GTPases may intimately cooperate.
MATERIALS AND METHODS
Protein purification
All WRC constructs used in this work were derived from previously published WRC230WCA (also called WRC230VCA or WRCapo) and ΔWRC230 by standard molecular biology procedures and were verified by Sanger sequencing (34, 41, 46). WRC230WCA contains human FL Sra1, FL Nap1, WAVE1(1-230)–(GGS)6–WCA(485-559) [(GGS)6 for six repeats of Gly-Gly-Ser], Abi2(1-158), and FL HSPC300. ∆WRC230 also contains the same subunits except that WAVE1(1-230)–(GGS)6–WCA(485-559) is replaced by WAVE1(1-230). Other WRCs contain modified subunits described in detail in tables S1 and S2.
The WRCs were expressed and purified essentially as previously described (34, 41). Reconstitution of the recombinant WRC is a multistep process, involving purification of individual proteins from different host cells (prokaryotic cell and insect cell), assembly/purification of subcomplexes (Sra1/Nap1 dimer and WAVE1/Abi2/HSPC300 trimer), and lastly of the WRC pentamer by a series of affinity, ion exchange, and gel filtration chromatographical steps. Mutations introduced into WRC subunits were carefully chosen and typically made to surface-exposed residues, producing complexes that behaved well and identically to the WT WRC during each step of reconstitution (fig. S2). Except Sra1 and Nap1, which were expressed in Tni cells using the ESF 921TM medium (Expression Systems), other proteins were typically expressed in BL21 (DE3)T1R cells (Sigma-Aldrich) at 18°C overnight or ArcticExpress (DE3) RIL cells (Stratagene) at 10°C for 24 hours. GST-Rac1QP and GST-Rac1Dead were purified by Glutathione Sepharose beads (Cytiva), followed by cation-exchange chromatography through a Source SP15 column and gel filtration through a Hiload Superdex 75 column. GST-Arf1 was purified by Glutathione Sepharose beads, followed by anion-exchange chromatography through a Source Q15 column and gel filtration through a Hiload Superdex 75 column. His8 -(GGS)2-Arf1 and His6 -Tev-EspG were purified by Ni-NTA agarose beads (Qiagen), followed by anion-exchange chromatography through a Source Q15 column and gel filtration through a Hiload Superdex 75 column. Untagged Rac1QP and untagged Rac1Dead were purified by SP Sepharose Fast Flow beads, followed by a Source SP15 column and a Hiload Superdex 75 gel filtration column. Proteins including the Arp2/3 complex, actin, WAVE1 WCA, and TEV protease were purified as previously described (34, 35, 41). All ion exchange and gel filtration chromatographical steps were performed using columns from Cytiva on an ÄKTA pure protein purification system.
Nonequilibrium pull-down assay
Nonequilibrium GST pull-down experiments were performed as previously described (41). Typically, 100 to 200 pmol of GST-tagged proteins as baits and 100 to 200 pmol of WRCs as preys were mixed with 20 μl of Glutathione Sepharose beads (Cytiva) in 1 ml of binding buffer [10 mM Hepes (pH 7), 50 mM NaCl, 5% (w/v) glycerol, 0.05% (w/v) Triton X-100, 2 mM MgCl2, and 5 mM β-mercaptoethanol or 1 mM dithiothreitol (DTT)] at 4°C for 30 min, followed by three washes using 1 ml of the binding buffer in each time of wash. Bound proteins were eluted with the GST elution buffer [100 mM Tris-HCl (pH 8.5), 2 mM MgCl2, and 30 mM reduced glutathione] and examined by SDS–polyacrylamide gel electrophoresis (SDS-PAGE).
EPD assay
EPD experiments were performed essentially as previously described (41). Glutathione Sepharose beads (Cytiva) were first equilibrated in EPD buffer [10 mM Hepes (pH 7), 50 mM NaCl, 5% (w/v) glycerol, 2 mM MgCl2, and 1 mM DTT] and stored as a 50% (v/v) slurry. Before use, all protein samples were dialyzed against EPD buffer overnight at 4°C or purified by gel filtration through a column equilibrated with the EPD buffer to maximize buffer match. Each reaction was assembled in 100 μl of total volume of EPD buffer in a 200-μl polymerase chain reaction (PCR) tube (Axygen), which contained 0.1 μM prey (e.g., WRC), varying concentrations of bait (e.g., GST-Arf1), with or without 100 μM untagged Rac1QP or Rac1Dead, 30 μl of the Glutathione Sepharose beads [by aliquoting 60 μl of the 50% (v/v) slurry using a wide-bore pipette tip], and 0.05% (w/v) Triton X-100 to facilitate mixing. The reactions were gently mixed at 4°C on a rotary mixer for 30 min. After a brief centrifugation (~10,000g for 10 s) to pellet the beads, 40 μl of the supernatant was immediately transferred to 8 μl of 6× loading buffer [360 mM Tris-HCl (pH 6.8), 12% (w/v) SDS, 60% (w/v) glycerol, 0.012% (w/v) bromophenol blue, and 140 mM freshly added β-mercaptoethanol] and analyzed by Coomassie blue–stained SDS-PAGE gels. The gels were imaged by a ChemiDoc XRS+ system (Bio-Rad). The total intensity of the Sra1 and Nap1 bands was quantified by ImageJ (Fiji) to determine the unbound WRC. The derived fractional occupancy from two to three independent experiments was pooled to obtain the binding isotherms for global fitting. The program Prism 8 (GraphPad) was used to fit the binding isotherms using the equation below to obtain dissociation constants Kd: , where y is the fractional occupancy, W is the total WRC concentration (typically 0.1 μM), and x is the total GST-Arf1 concentration.
Pyrene-actin polymerization assay
Actin polymerization assays were performed as previously described with some modifications here (41). Each reaction (120 μl) contained 3 to 4 μM actin (5% pyrene-labeled), 10 nM Arp2/3 complex, 100 nM various WRC230WCA constructs or WAVE1 WCA, and desired concentrations of untagged Rac1QP and/or His8-Arf1 in the NMEH20GD buffer [50 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 10 mM Hepes (pH 7.0), 20% (w/v) glycerol, and 1 mM DTT]. We found that compared to the commonly used KMEI20GD buffer [50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole (pH 7.0), 20% (w/v) glycerol, and 1 mM DTT], the NMEH20GD buffer increased the sensitivity of WRC to Rac1 and Arf1, allowing us to use lower protein concentrations and reduce reaction time in actin assembly assays. Pyrene-actin fluorescence was recorded every 5 s at 22 °C, with one reaction per measurement using a single-channel pipette to minimize air bubbles or pipetting errors, using a 96-well flat-bottom black plate (Greiner Bio-One) in a Spark plate reader (Tecan), with an excitation at 365 nm and emission at 407 nm (15-nm bandwidth for both wavelengths). Actin assembly rates at the time where the fluorescence intensity is half of the maximum plateau (t50) were derived from the kinetic curves using previously published Python scripts (70), which is also implemented on a web application of the scripts (https://biochempy.bb.iastate.edu).
DLS measurement
All experiments were performed on a Wyatt DynaPro NanoStar instrument using Dynamics 7.1.7 software. Sample definitions were as follows: Mw-R model: globular proteins; refractive index increment (dn/dC): 0.185 ml/g; RG model: sphere; cuvette: glass cuvette; solvent name: glycerol 5%. Otherwise, default parameters from the instruments were used, including refractive index and viscosity. Proteins and buffers were filtered using 0.22-μm centrifugal filters right before use to ensure that dust was removed from samples. Proteins were mixed directly, and 10 μl was loaded into a quartz microcuvette. Each protein mixture was repeated multiple times, with each repeat undertaking 20 acquisitions (5 s per acquisition). The cuvette was cleaned by washing three times with filtered Milli-Q water and three times with filtered 95% ethanol, then dried using filtered air. Cutoffs for acceptable runs were defined as any run with sum of squares (SOS) less than 10.0 and with a baseline reading between 0.995 and 1.005. Acquisitions exceeding these values were excluded. For each protein mixture, the readings of all acquisitions from multiple repeats were pooled to obtain the average molecule radius and compared for statistical significance of differences using the analysis of variance (ANOVA) test in the software R.
MD simulations
We applied MD simulations and free energies to optimize Arf1 and WRC binding poses. In total, we simulated six binding poses that placed the Arf1 close to the D site. Each MD system consists of one WRC bound to two Rac1 molecules [Protein Data Bank (PDB): 7USE] (46), one Arf1 (PDB: 1J2J), 231,710 water molecules, 400 NaCl (~0.1 M), and 45 extra Na+ to neutralize the system. The proteins and cofactors were described by AMBER FF14SB (71) and GAFF (72) force fields, respectively. MD simulations were performed using a well-established protocol described elsewhere (73–75). Briefly, each MD system was first relaxed by a series of minimizations, followed by four phases of MD simulations, including the relaxation phase (5 ns in total with 1-fs time steps), the system heating-up phase (10 ns in total), the equilibrium phase (10 ns), and the final sampling phase (100 ns). The time step was 2 fs for the last three phases, and the MD simulations of the last two phases were performed at 298 K and 1 bar to produce isothermal-isobaric ensembles. All MD simulations were performed using the pmemd.cuda program in AMBER 18 (76). Besides the root mean square deviation (RSMD)–time curves, a representative MD conformation, which has the smallest RMSD between itself and the average MD structure, was identified for each MD system.
Free-energy calculations
One hundred forty snapshots from the sampling phase (30 to 100 ns) of a trajectory were collected for free-energy calculations. An internal program was applied to calculate the MM-PBSA-WSAS free energies of the complex and the binding free energy between Arf1 and WRC. The polar part of the solvation free energy was calculated using Delphi 95 software (77, 78), and the nonpolar part was estimated by scaling the solvent accessible surface area as described elsewhere (79, 80). The conformational entropy term was predicted using WSAS, a weighted solvent accessible surface area method (81). The interior and exterior dielectric constants of PBSA calculations were set to 1.0 and 80.0, respectively. To study the effect of M-site mutations, we conducted computational mutagenesis using the WT snapshots and calculated the MM-PBSA-WSAS free energies of complex and Arf1 binding.
Cell culture, transfection, and coimmunoprecipitation
B16-F1–derived Sra1/Cyfip2 knockout (KO) cells (clone #3) were previously described (62) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) [glucose (4.5 g/liter); Invitrogen] supplemented with 10% fetal bovine serum (FBS) (Gibco), 2 mM glutamine (Thermo Fisher Scientific), and penicillin (50 U/ml)/streptomycin (50 μg/ml) (Thermo Fisher Scientific). Cells were routinely transfected in six-well plates (Sarstedt) using 1 μg of DNA in total and 2 μl of JetPrime (Polyplus Transfection, Illkirch) per well.
pEGFP-C2-Sra1 (Sra1 also known as Cyfip1, or cytoplasmic FMR1 interacting protein 1) and the derived Y967A mutant construct were described previously (62) and corresponded to the splice variant CYFIP1a, sequence AJ567911, of murine origin. Further point mutations in the M site were introduced by site-directed mutagenesis. The identity of all DNA constructs was verified by sequencing.
For EGFP-immunoprecipitation experiments, B16-F1–derived cell lines ectopically expressing EGFP-tagged variants of Sra1 were lysed with lysis buffer [1% Triton X-100, 140 mM KCl, 50 mM Tris-HCl (pH 7.4) supplemented with 50 mM NaF, 10 mM Na4P2O7, 2 mM MgCl2, and cOmplete Mini, EDTA-free Protease Inhibitor (Roche)]. Lysates were cleared and incubated with GFP-Trap agarose beads (Chromotek) for 60 min. Subsequently, beads were washed three times with lysis buffer lacking protease inhibitor and Triton X-100, mixed with SDS-PAGE loading buffer, boiled for 5 min, and examined by Western blotting using primary antibodies against Sra-1/Cyfip2 (82), Nap1 (82), WAVE (62), and Abi1 (D3G6C, #39444, Cell Signaling Technology), as well as corresponding horseradish peroxidase (HRP)–conjugated secondary antibodies (Invitrogen). Chemiluminescence signals were obtained upon incubation with ECL Prime Western Blotting Detection Reagent (Cytiva) and recorded with an ECL Chemocam imager (Intas, Goettingen, Germany).
Fluorescence microscopy, phalloidin staining, and immunolabeling
B16-F1–derived cell lines expressing indicated, EGFP-tagged Sra1 constructs or untransfected control cells were seeded onto laminin-coated (25 μg/ml), 15-mm-diameter glass coverslips and allowed to adhere for about 24 hours before fixation. For sole morphological assessments and immunolabeling of the Arp2/3 complex, cells were fixed with prewarmed, 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, and permeabilized with 0.05% Triton X-100 in PBS for 30 s. The actin cytoskeleton was subsequently stained using ATTO 594–conjugated phalloidin (ATTO-TEC GmbH, Germany). Immunolabeling was performed by following standard procedures in which samples were blocked with 5% horse serum in 1% bovine serum albumin (BSA)/PBS for 1 hour, followed by incubation with homemade, mouse monoclonal anti-p16A/ArpC5A 323H3 antibodies (undiluted supernatant) as the primary antibody, and Alexa Fluor 594–conjugated anti-mouse immunoglobulin G (H+L) antibodies (Dianova, Hamburg, Germany; 1:200 dilution in 1% BSA/PBS) as the secondary antibody. Samples were mounted using VectaShield Vibrance antifade reagent and imaged using a 63×/1.4–numerical aperture (NA) Plan Apochromatic oil objective. Phalloidin staining involved the same procedures to fix and permeabilize the cells, except that 0.25% glutaraldehyde was added to the fixation mixture.
Quantification of lamellipodia formation frequency
For assessment of lamellipodia formation, cells were randomly selected and categorized in a blinded manner as follows: “no lamellipodia” if no phalloidin-stained, peripheral lamellipodia–like actin meshwork was detectable; “immature lamellipodia” if the lamellipodia seen were small, narrow, or displayed multiple ruffles; and “lamellipodia” if the protrusive actin meshwork appeared to be fully developed (see representative examples in Fig. 6A).
Determination of lamellipodia curvature distribution and lamellipodium width
Average percentages of convex versus concave lamellipodia were determined by categorizing all lamellipodia formed in phalloidin-stained cells in each experimental condition as displaying one of the two shapes. Only lamellipodia-forming Sra1/Cyfip2-KO#3 cells rescued with respective constructs were categorized. Data in Fig. 6E were derived from 120, 87, 84, and 74 individual cells transfected with WT, ΔD, ΔM#2, and ΔD + ΔM#2 rescued cells, respectively, with multiple lamellipodia assessed for each cell. The width of lamellipodia was quantified with ImageJ using images of phalloidin-stained cells and drawing lines from lamellipodia tips to their distal edges, followed by measuring lengths of drawn lines. For each cell, three randomly chosen lamellipodial areas were measured and averaged.
Assessment of lamellipodial F-actin and Arp2/3 complex intensities
Lamellipodial F-actin intensity was determined by measuring average pixel intensities of lamellipodial regions of phalloidin-stained cells (microspikes excluded), followed by subtraction of background intensities in their immediate extracellular regions. For every independent experiment, the average lamellipodial F-actin intensity of Sra1/Cyfip2-KO#3 cells transfected with EGFP-Sra1 WT was normalized to “1,” to correct for experiment-specific staining variabilities.
Determination of lamellipodial protrusion rate, random cell migration efficiency, and migration directionality
For protrusion rate determination, B16-F1 Sra1/Cyfip2 KO#3 cells were transfected with EGFP-Sra1 WT or mutants (ΔD, ΔM#2, and ΔD + ΔM#2) and, on the second day, transferred into μ-slide microscopy chambers precoated with laminin (25 μg/ml; Sigma-Aldrich). After 1 day of incubation, the growth medium was replaced with microscopy medium, which included Ham’s F12 supplemented with 10% FBS (Gibco), 2 mM glutamine (Thermo Fisher Scientific), and penicillin (50 U/ml)/streptomycin (50 μg/ml) (Thermo Fisher Scientific). The chambers were then mounted on an inverted microscope for live-cell, phase-contrast imaging as previously described (62), except that a 100×/1.4-NA Plan Apochromatic objective, instead of a 63× objective, was used. Movies were taken over 5 min, with 5-s intervals between frames. Recorded movies were analyzed by kymography using ImageJ.
For random cell migration, cells were prepared as described for protrusion rate analysis, but the movies were recorded using a 10×/0.15-NA Plan Neofluar objective over 10 hours, with 5-min intervals between frames. Each movie contained 121 frames in total. Subsequent analysis was done in ImageJ by tracking frame-by-frame the nuclei of cells using the manual tracking plugin. The migration trajectories were further analyzed to obtain migration directionality information using previously published protocols (83). Essentially, at any given time, directionality of a migrating cell is defined as the ratio between dt and Dt (i.e., dt/Dt), where “dt” is the straight distance between migration start and given trajectory points and “Dt” is the cumulative length of the given trajectory. Higher dt/Dt values indicate greater directionality in cell migration.
Image processing and statistical analysis
Where appropriate, brightness and contrast were adjusted uniformly for the entire image using ImageJ. Data analysis and Superplot graphs used ImageJ, Excel 2016, and Graphpad Prism 8 (GraphPad Software, San Diego, CA). Each assay was typically repeated four independent times. Data are shown as arithmetic means ± SEM (error bars). To assess statistical significance, one-way ANOVA with Dunnett’s post hoc test was applied to compare multiple groups with one control group. Statistical analyses were performed using Prism 6 or 8. An error probability below 5% (P < 0.05; * in the figure panels) was considered to imply statistical significance. **, ***, and **** indicated P values ≤0.01, ≤ 0.001, and ≤ 0.0001, respectively.
Acknowledgments
We thank S. Padrick at Drexel University for updating the published Python codes for actin data analysis; A. Sijo-Gonzales, F. Rhodes, L. Loh, G. Prasad, S. Mitra, and the ResearchIT at Iowa State University for migrating the Python codes to the web-based application; S. Nelson at Iowa State for the use of fluorimeters; D. Rosenbaum at UT Southwestern for providing the PGS construct; and N. Alto at UT Southwestern for providing various Arf, Arl, and EspG constructs.
Funding: This work was supported by NIH grant R35-GM128786 to B.C., start-up funds from Iowa State University and Roy J. Carver Charitable Trust to B.C., start-up funds from Stony Brook University to S.C., Deutsche Forschungsgemeinschaft DFG Research Training Group GRK2223 and individual grant RO2414/8-1 to K.R., NIH grant R01-DK107733 to D.D.B., NSF grant 1955260 to J.W., and NIH grant R01-GM147673 to J.W.
Author contributions: B.C. conceived the project and oversaw biochemical work. K.R. oversaw cell biological work. J.W. performed MD simulation and energy calculation. S.Y. purified proteins and performed biochemical experiments. Y.L. and A.J.B. helped with protein purification and biochemical assays. Y.T., M.S., and M.M. performed cellular experiments. B.D. and S.C. helped with structural analysis. D.A.K. performed DLS analysis. L.D., O.A., and D.D.B. helped with cell biological analysis. B.C. wrote the manuscript and prepared the figures with assistance from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
Other Supplementary Material for this : manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.K. Wennerberg, K. L. Rossman, C. J. Der,The Ras superfamily at a glance. J. Cell Sci. 118,843–846 (2005). [DOI] [PubMed] [Google Scholar]
- 2.E. Sztul, P. W. Chen, J. E. Casanova, J. Cherfils, J. B. Dacks, D. G. Lambright, F. J. S. Lee, P. A. Randazzo, L. C. Santy, A. Schürmann, I. Wilhelmi, M. E. Yohe, R. A. Kahn,Arf GTPases and their GEFs and GAPS: Concepts and challenges. Mol. Biol. Cell 30,1249–1271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J. G. Donaldson, C. L. Jackson,ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12,326–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.C. D’Souza-Schorey, P. Chavrier,ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7,347–358 (2006). [DOI] [PubMed] [Google Scholar]
- 5.A. K. Gillingham, S. Munro,The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23,579–611 (2007). [DOI] [PubMed] [Google Scholar]
- 6.N. Mosaddeghzadeh, M. R. Ahmadian,The rho family gtpases: Mechanisms of regulation and signaling. Cell 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Etienne-Manneville, A. Hall,Rho GTPases in cell biology. Nature 420,629–635 (2002). [DOI] [PubMed] [Google Scholar]
- 8.H. Radhakrishna, O. Al-Awar, Z. Khachikian, J. G. Donaldson,ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112,855–866 (1999). [DOI] [PubMed] [Google Scholar]
- 9.R. L. Boshans, S. Szanto, L. van Aelst, C. D’Souza-Schorey,ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20,3685–3694 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L. C. Santy, J. E. Casanova,Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154,599–610 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.C. D’Souza-Schorey, R. L. Boshans, M. McDonough, P. D. Stahl, L. Van Aelst,A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 16,5445–5454 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.V. Singh, A. C. Davidson, P. J. Hume, D. Humphreys, V. Koronakis,Arf GTPase interplay with Rho GTPases in regulation of the actin cytoskeleton. Small GTPases 10,411–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K. R. Myers, J. E. Casanova,Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 18,184–192 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. Phuyal, H. Farhan,Multifaceted Rho GTPase signaling at the endomembranes. Front. Cell Dev. Biol. 7,127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.E. H. Chen, B. A. Pryce, J. A. Tzeng, G. A. Gonzalez, E. N. Olson,Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 114,751–762 (2003). [DOI] [PubMed] [Google Scholar]
- 16.E. L. Hunt, H. Rai, T. J. C. Harris,SCAR/WAVE complex recruitment to a supracellular actomyosin cable by myosin activators and a junctional Arf-GEF during Drosophila dorsal closure. Mol. Biol. Cell 33,br12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Honda, M. Nogami, T. Yokozeki, M. Yamazaki, H. Nakamura, H. Watanabe, K. Kawamoto, K. Nakayama, A. J. Morris, M. A. Frohman, Y. Kanaho,Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99,521–532 (1999). [DOI] [PubMed] [Google Scholar]
- 18.M. Krauss, M. Kinuta, M. R. Wenk, P. De Camilli, K. Takei, V. Haucke,ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Iγ. J. Cell Biol. 162,113–124 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. Balasubramanian, D. W. Scott, J. D. Castle, J. E. Casanova, M. A. Schwartz,Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat. Cell Biol. 9,1381–1391 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L. C. Santy, K. S. Ravichandran, J. E. Casanova,The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 15,1749–1754 (2005). [DOI] [PubMed] [Google Scholar]
- 21.F. Palacios, J. K. Schweitzer, R. L. Boshans, C. D’Souza-Schorey,ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat. Cell Biol. 4,929–936 (2002). [DOI] [PubMed] [Google Scholar]
- 22.T. H. Koo, B. A. Eipper, J. G. Donaldson,Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 8,29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. Hu, B. Shi, M. J. Jarzynka, J. J. Yiin, C. D’Souza-Schorey, S. Y. Cheng,ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res. 69,794–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. Tarricone, B. Xiao, N. Justin, P. A. Walker, K. Rittinger, S. J. Gamblin, S. J. Smerdon,The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411,215–219 (2001). [DOI] [PubMed] [Google Scholar]
- 25.V. Koronakis, P. J. Hume, D. Humphreys, T. Liu, O. Hørning, O. N. Jensen, E. J. McGhie,WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U.S.A. 108,14449–14454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D. Humphreys, T. Liu, A. C. Davidson, P. J. Hume, V. Koronakis,The Drosophila Arf1 homologue Arf79F is essential for lamellipodium formation. J. Cell Sci. 125,5630–5635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D. Humphreys, A. C. Davidson, P. J. Hume, L. E. Makin, V. Koronakis,Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc. Natl. Acad. Sci. U.S.A. 110,16880–16885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D. Humphreys, A. Davidson, P. J. Hume, V. Koronakis,Salmonella virulence effector SopE and host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe 11,129–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D. Humphreys, V. Singh, V. Koronakis,Inhibition of WAVE regulatory complex activation by a bacterial virulence effector counteracts pathogen phagocytosis. Cell Rep. 17,697–707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Lewis-Saravalli, S. Campbell, A. Claing,ARF1 controls Rac1 signaling to regulate migration of MDA-MB-231 invasive breast cancer cells. Cell. Signal. 25,1813–1819 (2013). [DOI] [PubMed] [Google Scholar]
- 31.V. Marchesin, G. Montagnac, P. Chavrier,ARF6 promotes the formation of Rac1 and WAVE-dependent ventral F-actin rosettes in breast cancer cells in response to epidermal growth factor. PLOS ONE 10,e0121747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.V. Singh, A. C. Davidson, P. J. Hume, V. Koronakis,Arf6 can trigger wave regulatory complex-dependent actin assembly independent of arno. Int. J. Mol. Sci. 21,2457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Eden, R. Rohatgi, A. V. Podtelejnikov, M. Mann, M. W. Kirschner,Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418,790–793 (2002). [DOI] [PubMed] [Google Scholar]
- 34.B. Chen, S. B. Padrick, L. Henry, M. K. Rosen,Biochemical reconstitution of the WAVE regulatory complex. Methods Enzymol. 540,55–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. M. Ismail, S. B. Padrick, B. Chen, J. Umetani, M. K. Rosen,The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 16,561–563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Z. Chen, D. Borek, S. B. Padrick, T. S. Gomez, Z. Metlagel, A. M. Ismail, J. Umetani, D. D. Billadeau, Z. Otwinowski, M. K. Rosen,Structure and control of the actin regulatory WAVE complex. Nature 468,533–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.E. Derivery, B. Lombard, D. Loew, A. Gautreau,The wave complex is intrinsically inactive. Cell Motil. Cytoskelet. 66,777–790 (2009). [DOI] [PubMed] [Google Scholar]
- 38.B. Chen, K. Brinkmann, Z. Chen, C. W. Pak, Y. Liao, S. Shi, L. Henry, N. V. Grishin, S. Bogdan, M. K. Rosen,The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156,195–207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.T. Takenawa, S. Suetsugu,The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8,37–48 (2007). [DOI] [PubMed] [Google Scholar]
- 40.A. M. Lebensohn, M. W. Kirschner,Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell 36,512–524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. Chen, H.-T. T. Chou, C. A. Brautigam, W. Xing, S. Yang, L. Henry, L. K. Doolittle, T. Walz, M. K. Rosen,Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife 6,e29795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. B. Padrick, H. C. Cheng, A. M. Ismail, S. C. Panchal, L. K. Doolittle, S. Kim, B. M. Skehan, J. Umetani, C. A. Brautigam, J. M. Leong, M. K. Rosen,Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32,426–438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.K. Rottner, T. E. B. Stradal, B. Chen,WAVE regulatory complex. Curr. Biol. 31,R512–R517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.W. Zou, X. Dong, T. R. Broederdorf, A. Shen, D. A. Kramer, R. Shi, X. Liang, D. M. Miller, Y. K. Xiang, R. Yasuda, B. Chen, K. Shen,A dendritic guidance receptor complex brings together distinct actin regulators to drive efficient F-actin assembly and branching. Dev. Cell 45,362–375.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. A. Kramer, H. K. Piper, B. Chen,WASP family proteins: Molecular mechanisms and implications in human disease. Eur. J. Cell Biol. 101,151244 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.B. Ding, S. Yang, M. Schaks, Y. Liu, A. J. Brown, K. Rottner, S. Chowdhury, B. Chen,Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase. Nat. Commun. 13,5444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.T. Baust, C. Czupalla, E. Krause, L. Bourel-Bonnet, B. Hoflack,Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc. Natl. Acad. Sci. U.S.A. 103,3159–3164 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. Anitei, C. Stange, I. Parshina, T. Baust, A. Schenck, G. Raposo, T. Kirchhausen, B. Hoflack,Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin-AP-1-coated carrier biogenesis at the TGN. Nat. Cell Biol. 12,330–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. A. Cook, W. A. Comrie, M. C. Poli, M. Similuk, A. J. Oler, A. J. Faruqi, D. B. Kuhns, S. Yang, A. Vargas-Hernández, A. F. Carisey, B. Fournier, D. E. Anderson, S. Price, M. Smelkinson, W. A. Chahla, L. R. Forbes, E. M. Mace, T. N. Cao, Z. H. Coban-Akdemir, S. N. Jhangiani, D. M. Muzny, R. A. Gibbs, J. R. Lupski, J. S. Orange, G. D. E. Cuvelier, M. Al Hassani, N. Al Kaabi, Z. Al Yafei, S. Jyonouchi, N. Raje, J. W. Caldwell, Y. Huang, J. K. Burkhardt, S. Latour, B. Chen, G. El Ghazali, V. K. Rao, I. K. Chinn, M. J. Lenardo,HEM1 deficiency disrupts mTORC2 and F-actin control in inherited immunodysregulatory disease. Science 369,202–207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.N. Dong, Y. Zhu, Q. Lu, L. Hu, Y. Zheng, F. Shao,Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150,1029–1041 (2012). [DOI] [PubMed] [Google Scholar]
- 51.T. D. Pollard,A guide to simple and informative binding assays. Mol. Biol. Cell 21,4061–4067 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Yin, K. Babaoglu, C. A. Brautigam, L. Clark, Z. Shao, T. H. Scheuermann, C. M. Harrell, A. L. Gotter, A. J. Roecker, C. J. Winrow, J. J. Renger, P. J. Coleman, D. M. Rosenbaum,Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat. Struct. Mol. Biol. 23,293–299 (2016). [DOI] [PubMed] [Google Scholar]
- 53.J. Yin, J. C. Mobarec, P. Kolb, D. M. Rosenbaum,Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519,247–250 (2015). [DOI] [PubMed] [Google Scholar]
- 54.X. Ren, G. G. Farías, B. J. Canagarajah, J. S. Bonifacino, J. H. Hurley,Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 152,755–767 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. Gautreau, H. Y. H. Ho, J. Li, H. Steen, S. P. Gygi, M. W. Kirschner,Purification and architecture of the ubiquitous WAVE complex. Proc. Natl. Acad. Sci. U.S.A. 101,4379–4383 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.I. T. Desta, K. A. Porter, B. Xia, D. Kozakov, S. Vajda,Performance and its limits in rigid body protein-protein docking. Structure 28,1071–1081.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.G. C. P. Van Zundert, J. P. G. L. M. Rodrigues, M. Trellet, C. Schmitz, P. L. Kastritis, E. Karaca, A. S. J. Melquiond, M. Van Dijk, S. J. De Vries, A. M. J. J. Bonvin,The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428,720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 58.C. Quignot, J. Rey, J. Yu, P. Tufféry, R. Guerois, J. Andreani,InterEvDock2: An expanded server for protein docking using evolutionary and biological information from homology models and multimeric inputs. Nucleic Acids Res. 46,W408–W416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.E. Ramírez-Aportela, J. R. López-Blanco, P. Chacón,FRODOCK 2.0: Fast protein-protein docking server. Bioinformatics 32,2386–2388 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Y. Yan, H. Tao, J. He, S. Y. Huang,The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15,1829–1852 (2020). [DOI] [PubMed] [Google Scholar]
- 61.H. Ashkenazy, S. Abadi, E. Martz, O. Chay, I. Mayrose, T. Pupko, N. Ben-Tal,ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44,W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Schaks, S. P. Singh, F. Kage, P. Thomason, T. Klünemann, A. Steffen, W. Blankenfeldt, T. E. Stradal, R. H. Insall, K. Rottner,Distinct interaction sites of Rac GTPase with WAVE regulatory complex have non-redundant functions in vivo. Curr. Biol. 28,3674–3684.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. Schaks, M. Reinke, W. Witke, K. Rottner,Molecular dissection of neurodevelopmental disorder-causing mutations in CYFIP2. Cell 9,1355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.J. Damiano-Guercio, L. Kurzawa, J. Mueller, G. Dimchev, M. Schaks, M. Nemethova, T. Pokrant, S. Brühmann, J. Linkner, L. Blanchoin, M. Sixt, K. Rottner, J. Faix,Loss of Ena/VASP interferes with lamellipodium architecture, motility and integrin-dependent adhesion. eLife 9,e55351 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.X. J. Chen, A. J. Squarr, R. Stephan, B. Chen, T. E. Higgins, D. J. Barry, M. C. Martin, M. K. Rosen, S. Bogdan, M. Way,Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell 30,569–584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Y. Sung, O. Engmann, M. A. Teylan, A. C. Nairn, P. Greengard, Y. Kim,WAVE1 controls neuronal activity-induced mitochondrial distribution in dendritic spines. Proc. Natl. Acad. Sci. U.S.A. 105,3112–3116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R. Kang, D. Tang, Y. Yu, Z. Wang, T. Hu, H. Wang, L. Cao,WAVE1 regulates Bcl-2 localization and phosphorylation in leukemia cells. Leukemia 24,177–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.A. Cheng, T. V. Arumugam, D. Liu, R. G. Khatri, K. Mustafa, S. Kwak, H. P. Ling, C. Gonzales, O. Xin, D. G. Jo, Z. Guo, R. J. Mark, M. P. Mattson,Pancortin-2 interacts with WAVE1 and Bcl-xL in a mitochondria-associated protein complex that mediates ischemic neuronal death. J. Neurosci. 27,1519–1528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.V. Popoff, J. D. Langer, I. Reckmann, A. Hellwig, R. A. Kahn, B. Brügger, F. T. Wieland,Several ADP-ribosylation factor (Arf) isoforms support COPI vesicle formation. J. Biol. Chem. 286,35634–35642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.L. K. Doolittle, M. K. Rosen, S. B. Padrick,Measurement and analysis of in vitro actin polymerization. Methods Mol. Biol. 1046,273–293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser, C. Simmerling,ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11,3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman, D. A. Case,Development and testing of a general Amber force field. J. Comput. Chem. 25,1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Y. Zhang, X. He, J. Zhai, B. Ji, V. H. Man, J. Wang,In silico binding profile characterization of SARS-CoV-2 spike protein and its mutants bound to human ACE2 receptor. Brief. Bioinform. 22,bbab188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.L. Su, Y. Wang, J. Wang, Y. Mifune, M. D. Morin, B. T. Jones, E. M. Y. Moresco, D. L. Boger, B. Beutler, H. Zhang,Structural basis of TLR2/TLR1 activation by the synthetic agonist diprovocim. J. Med. Chem. 62,2938–2949 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.P. Kim, H. Li, J. Wang, Z. Zhao,Landscape of drug-resistance mutations in kinase regulatory hotspots. Brief. Bioinform. 22,bbaa108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D. A. Case, R. C. Walker, T. E. Cheatham, C. Simmerling, A. Roitberg, K. M. Merz, R. Luo, T. Darden, Amber 2018 (University of California, San Francisco, 2018).
- 77.L. Li, C. Li, S. Sarkar, J. Zhang, S. Witham, Z. Zhang, L. Wang, N. Smith, M. Petukh, E. Alexov,DelPhi: A comprehensive suite for DelPhi software and associated resources. BMC Biophys. 5,9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.W. Rocchia, E. Alexov, B. Honig,Extending the applicability of the nonlinear Poisson-Boltzmann equation: Multiple dielectric constants and multivalent ions. J. Phys. Chem. B 105,6507–6514 (2001). [Google Scholar]
- 79.E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Z. H. Zhang, T. Hou,End-point binding free energy calculation with MM/PBSA and MM/GBSA: Strategies and applications in drug design. Chem. Rev. 119,9478–9508 (2019). [DOI] [PubMed] [Google Scholar]
- 80.J. Wang, T. Hou, X. Xu,Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr. Comput. Aided-Drug Des. 2,95–103 (2006). [Google Scholar]
- 81.J. Wang, T. Hou,Develop and test a solvent accessible surface area-based model in conformational entropy calculations. J. Chem. Inf. Model. 52,1199–1212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.A. Steffen, K. Rottner, J. Ehinger, M. Innocenti, G. Scita, J. Wehland, T. E. B. Stradal,Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23,749–759 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R. Gorelik, A. Gautreau,Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9,1931–1943 (2014). [DOI] [PubMed] [Google Scholar]
- 84.J. D. Thompson, D. G. Higgins, T. J. Gibson,CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22,4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.E. Jurrus, D. Engel, K. Star, K. Monson, J. Brandi, L. E. Felberg, D. H. Brookes, L. Wilson, J. Chen, K. Liles, M. Chun, P. Li, D. W. Gohara, T. Dolinsky, R. Konecny, D. R. Koes, J. E. Nielsen, T. Head-Gordon, W. Geng, R. Krasny, G. W. Wei, M. J. Holst, J. A. McCammon, N. A. Baker,Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27,112–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2
Movies S1 and S2