Abstract
Human gingival epithelial cells (HGE) express two antimicrobial peptides of the β-defensin family, human β-defensin 1 (hBD-1) and hBD-2, as well as cytokines and chemokines that contribute to innate immunity. In the present study, the expression and transcriptional regulation of hBD-2 was examined. HBD-2 mRNA was induced by cell wall extract of Fusobacterium nucleatum, an oral commensal microorganism, but not by that of Porphyromonas gingivalis, a periodontal pathogen. HBD-2 mRNA was also induced by the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) and phorbol myristate acetate (PMA), an epithelial cell activator. HBD-2 mRNA was also expressed in 14 of 15 noninflamed gingival tissue samples. HBD-2 peptide was detected by immunofluorescence in HGE stimulated with F. nucleatum cell wall, consistent with induction of the mRNA by this stimulant. Kinetic analysis indicates involvement of multiple distinct signaling pathways in the regulation of hBD-2 mRNA; TNF-α and F. nucleatum cell wall induced hBD-2 mRNA rapidly (2 to 4 h), while PMA stimulation was slower (∼10 h). In contrast, each stimulant induced interleukin 8 (IL-8) within 1 h. The role of TNF-α as an intermediary in F. nucleatum signaling was ruled out by addition of anti-TNF-α that did not inhibit hBD-2 induction. However, inhibitor studies show that F. nucleatum stimulation of hBD-2 mRNA requires both new gene transcription and new protein synthesis. Bacterial lipopolysaccharides isolated from Escherichia coli and F. nucleatum were poor stimulants of hBD-2, although they up-regulated IL-8 mRNA. Collectively, our findings show inducible expression of hBD-2 mRNA via multiple pathways in HGE in a pattern that is distinct from that of IL-8 expression. We suggest that different aspects of innate immune responses are differentially regulated and that commensal organisms have a role in stimulating mucosal epithelial cells in maintaining the barrier that contributes to homeostasis and host defense.
Mucosal epithelial cells play an integral role in innate immune defense by sensing signals from the external environment, generating various molecules to affect growth, development, and function of other cells, and maintaining the balance between health and disease (23). Mucosal epithelial cells express antimicrobial peptides, including the β-defensins human β-defensin 1 (hBD-1) and hBD-2, as well as chemokines that attract monocytes and neutrophils and cytokines that activate the adaptive immune system (23). It is now recognized that the antimicrobial peptide hBD-2 also stimulates antigen-presenting dendritic cells that signal the adaptive immune system (51), in addition to its antimicrobial activity. Therefore, characterization of β-defensin regulation is essential for understanding the role of these peptides in protecting the host by activating both innate and adaptive immune systems and in contributing to the epithelial barrier to inflammatory disease processes.
It is now widely recognized that epithelial cells participate in innate immune responses, yet the bacterial pattern recognition molecules and signaling pathways are not clear and do not necessarily correspond to those of monocytes, macrophages, and endothelial cells (31). Gingival epithelium is a stratified squamous epithelium surrounding the tooth and forming an attachment to the tooth surface. It functions as a protective barrier against pathogenic microorganisms in dental plaque. The oral mucosa of the gingiva is a useful model for studies of innate immune defenses because it is constantly exposed to microorganisms yet generally maintains a homeostasis and balance that is associated with oral health.
The defensin family of antimicrobial peptides is an evolutionarily conserved group (reviewed in reference 47). In mammals, epithelial defensins include α-defensins of the intestinal epithelium and β-defensins of skin and mucosal epithelia. The β-defensins are small cationic peptides, 36 to 42 amino acids in length (4, 14, 40; J. Harder, J. Bartels, E. Christophers, and J. M. Schröder, Letter, Nature 387:861, 1997), with a structure that is stabilized by three disulfide bonds (reviewed in references 12 and 22). Tracheal antimicrobial peptide (TAP) was the first member of the epithelial β-defensin family characterized (14). Up-regulation of TAP mRNA was shown to occur via the CD14-mediated signal transduction pathway in bovine airway epithelial cells challenged with bacterial lipopolysaccharide (LPS) (13) and with tumor necrosis factor alpha (TNF-α) and interleukin 1β (IL-1β) (5, 38). The related lingual antimicrobial peptide (LAP) was shown to be up-regulated in vivo under conditions of infection and inflammation (39).
In humans, two β-defensins, hBD-1 and hBD-2, have been identified exclusively in epithelial tissues. hBD-1 is constitutively expressed in the kidney, pancreas, urinary and respiratory tracts, and oral epithelia (4, 17, 26, 29, 30, 41, 45, 54). hBD-2 was originally isolated from psoriatic-scale keratinocytes (Harder et al., 1997). hBD-2 is poorly expressed in normal epidermal keratinocytes but is induced when keratinocytes are stimulated with gram-negative or gram-positive bacteria, Candida albicans, or TNF-α (Harder et al., 1997), and is up-regulated in inflamed epithelial tissues (27). hBD-2 demonstrates in vitro antimicrobial activities against yeast and both gram-negative and gram-positive bacteria (3, 45; Harder et al., 1997).
We previously reported the constitutive expression of hBD-1 mRNA in gingival epithelial cells (26) and the inducible expression of hBD-2 mRNA in cultured gingival epithelial cells (47). In other studies, the expression of hBD-2 mRNA has also been reported to be induced by IL-1β, TNF-α, and specific microorganisms (29; Harder et al., 1997). In this study, we have established that hBD-2 mRNA is expressed in gingival epithelial cells and tissue and that several natural stimuli induce its expression, but expression is regulated differently from that of IL-8, another aspect of innate host defense. We show evidence for the involvement of multiple pathways of regulation and find differences in the abilities of cell wall extracts of two gram-negative periodontal bacteria, Fusobacterium nucleatum, present in both healthy and diseased sites (18), and Porphyromonas gingivalis, a periodontal pathogen (19, 42), to induce hBD-2 mRNA. Finally, we have detected hBD-2 peptide in stimulated gingival epithelial cells. Our findings suggest that hBD-2 mRNA is regulated at the transcriptional level via several signaling pathways and that its regulation differs from that of IL-8.
MATERIALS AND METHODS
Reagents.
Human serum, Escherichia coli (strain 055:B5) LPS, phorbol myristate acetate (PMA), actinomycin D, and cycloheximide were obtained from Sigma Co. (St. Louis, Mo.). E. coli LPS was diluted in keratinocyte growth medium (KGM); PMA, actinomycin D, and cycloheximide were dissolved in dimethyl sulfoxide (DMSO) (Sigma). In all studies, the concentration of DMSO was always less than 0.1% (vol/vol). TNF-α, anti-human TNF-α monoclonal antibody, and normal mouse immunoglobulin G1 (IgG1) were purchased from R&D Systems Inc. (Minneapolis, Minn.). Lyophilized powder of TNF-α was reconstituted to the stock concentration of 10 μg/ml with sterile phosphate-buffered saline (GIBCO BRL) containing at least 0.1% bovine serum albumin.
Cell culture.
Normal gingival biopsy specimens were surgically removed from young patients who underwent third-molar extraction at the Department of Oral Surgery, School of Dentistry, University of Washington (UW). Primary human gingival epithelial cells (HGE) were isolated from these biopsy specimens and grown in a serum-free KGM (Clonetics, Walkersville, Md.) as previously described (26). Primary human gingival fibroblasts (HGF) were isolated from the biopsy specimens after the epithelium was removed and grown in Dulbecco's modified Eagle's medium (GIBCO BRL) supplemented with 10% fetal calf serum (Gemini, Calabasas, Calif.) and 1% penicillin-streptomycin (GIBCO BRL). Second-passage cultures of both cell types were used for experimental studies after they reached approximately 80% confluence. Cultures of HGF or HGE were stimulated for 24 h unless otherwise indicated.
Preparation of bacterial crude cell wall extract and LPS.
Anaerobic cultures of F. nucleatum ATCC 25586 and P. gingivalis ATCC 33277 were grown, and their crude cell wall extract was prepared using a French pressure cell at 15,000 lb/in2 and differential centrifugation as previously described (26). Purified LPS fraction of F. nucleatum ATCC 25586, grown in mycoplasma broth supplemented with hemin and menadione, was prepared by the cold magnesium-ethanol precipitation technique (11) followed by lipid extraction (16) and conversion to sodium salts (34). The LPS preparations were subjected to gas chromatography for analysis of carbohydrates (8) and fatty acids (43) and were found to have compositions consistent with a previous report (33) and to be devoid of phospholipids and procedure-related detergent. The A280 and A260 indicated the LPS preparation was free of detectable levels of protein and nucleic acid contamination.
RNA extraction and analysis.
After stimulation, cells were lysed directly with 300 μl of lysis buffer using an RNAqueous kit (Ambion Inc., Austin, Tex.). Total RNA was extracted according to the manufacturer's protocols and then precipitated with LiCl. The RNA pellet was resuspended in 40 μl of RNA storage buffer (Ambion). One-tenth of the total volume was used to determine optical density values. Reverse transcriptase (RT) PCR was conducted to semiquantitatively analyze mRNA for hBD-2, IL-8, TNF-α, monocyte chemoattractant protein 1 (MCP-1), ribosomal phosphoprotein (RPO) gene (housekeeping gene), and keratin 5 (control for epithelial contribution in tissue) using previously described protocols (26). The specific sequences and annealing temperatures of the oligonucleotide primers are summarized in Table 1. The denaturing and polymerizing temperatures were 95 and 72°C, respectively. Amplification was conducted for 25 cycles (except in the experiment where amplification was conducted for 35 cycles to detect any small amount of hBD-2 expression in HGF) as a means to interpret the differences between the relative amounts of amplified products obtained under different conditions. The identities of purified PCR products of hBD-2 from samples stimulated with F. nucleatum cell wall, PMA, and TNF-α were confirmed by direct sequencing using PCR primers at the DNA Sequencing Facility, Department of Biochemistry, UW. In some experiments where more-quantitative analysis of mRNA targets was required, a ribonuclease protection assay (RPA) was conducted. Total RNA (20 μg) from each sample was probed with biotin-labeled RNA specific for hBD-2, IL-8, and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) overnight at 42°C using an RPA III kit (Ambion). Digestion of nonhybridized RNA with RNase T (1:100) was performed at 37°C for 30 min. Protected RNA fragments were resolved on a 5% denaturing polyacrylamide gel and detected by a nonisotopic detection kit, BrightStar Biodetect, on a BrightStar-Plus positively charged nylon membrane (Ambion). The time of exposure to the X-ray film varied depending on the abundance of the messages, i.e., GAPDH required much less exposure time than the other two messages. The sizes of protected fragments for GAPDH, hBD-2, and IL-8 are 316, 195, and 619 bp, respectively. The intensities of bands were analyzed by densitometry using Kodak 1D analysis software. The relative ratios of the net intensities (after the background intensity was subtracted) of either hBD-2 or IL-8 and GAPDH from each sample were determined and compared for different experimental conditions.
TABLE 1.
Primer sequences and annealing temperatures
| Primer | Sequence | Annealing temp (°C) |
|---|---|---|
| hBD-2 (5′) | CCA GCC ATC AGC CAT GAG GGT | 65 |
| hBD-2 (3′) | GGA GCC CTT TCT GAA TCC GCA | |
| IL-8 (5′) | TTT CTG ATG GAA GAG AGC TCT GTC TGG | 60 |
| IL-8 (3′) | AGT GGA ACA AGG ACT TGT GAA TCC TGG | |
| TNF-α (5′) | TTC TGC CTG CTG CAC TTT GGA CTC AT | 60 |
| TNF-α (3′) | TTG ATG GCA GAG AGG AGG TTG ACC TT | |
| MCP-1 (5′) | CTG CTC ATA GCA GCC ACC TTC ATT | 54 |
| MCP-1 (3′) | GCA CAG ATC TCC TTG GCC ACA ATG | |
| RPO (5′) | AGC AGG TGT TCG ACA ATG GCA | 50 |
| RPO (3′) | ACT CTT CTT TGG CTT CAA CC | |
| Keratin (5′) | GTC CTC TCC ATG GAC AAC AAC | 49 |
| Keratin (3′) | TGT CAA TCT CGG CTC TCA GCC |
Preparation of biotin-labeled RNA probes.
DNA templates for hBD-2 and IL-8 RNA probes were prepared by using a PCR strategy to append a T7 phage promoter sequence at the 5′ end of a downstream primer. The sequence of an upstream primer for hBD-2 was CCT CTT CCA GGT GTT TTT GGT G, and that of an upstream primer for IL-8 was GAG TGA TTG AGA GTG GAC CAC ACT G. The sequence of a downstream primer for hBD-2 was TAA TAC GAC TCA CTA TAG GGA GCC CTT TCT GAA TCC GCA TC, and that of a downstream primer for IL-8 was TAA TAC GAC TCA CTA TAG GCA GAC TAG GGT TGC CAG ATT TAA C. The nucleotides in boldface are a consensus sequence for T7 RNA polymerase. Five separate PCRs (50 μl each) were conducted for 30 cycles with 2.5 U of Pfu DNA polymerase enzyme (Stratagene, La Jolla, Calif.), combined to increase the amount of DNA template, and precipitated with 0.1 M NaCl and 2.5 volumes of cold ethanol at −80°C for 1 h. The DNA pellet was washed with 70% ethanol and resuspended in 20 μl of Tris-EDTA, pH 8.0. The DNA templates were purified by gel electrophoresis and extracted with a QIAquick gel extraction kit (Qiagen Inc., Santa Clarita, Calif.). The sequence of the DNA templates with the appended T7 promoter was confirmed by direct sequencing using an upstream PCR primer. A linearized plasmid of GAPDH template (in pTRIPLEscript vector) was purchased from Ambion. Antisense RNA transcripts of hBD-2, IL-8, and GAPDH were transcribed from 1 μg of the respective DNA templates using an in vitro transcription kit (MAXIscript; Ambion). Full-length RNA transcripts were purified by gel electrophoresis with 5% acrylamide–8 M urea denaturing gel, cut, and eluted in 350 μl of probe elution buffer (Ambion) containing 0.5 M NH4OAC, 1 mM EDTA, and 0.1% sodium dodecyl sulfate overnight at 37°C. The RNA probes were further purified with 1 ml of acid phenol-chloroform (pH 4.5) (Ambion) to remove any contaminating protein. The aqueous phase containing RNA probes was transferred to a new tube and precipitated with 2.5 volumes of cold ethanol. The RNA pellet was resuspended in 20 μl of Tris-EDTA. Five microliters (25%) was used to determine optical density values. The RNA probe (50 ng/μl) was labeled with a nonisotopic labeling kit (BrightStar Psolaren-Biotin; Ambion). The biotin-labeled RNA probes were stored in 5-μl aliquots at −80°C. The amounts of biotinylated probes used in RPA were about 400 pg for GAPDH and less than 100 pg for hBD-2 and IL-8 for 20 μg of total RNA in each sample.
Immunocytochemistry.
Gingival epithelial cells were seeded on coverslips (9 by 9 mm) (Bellco Glass Inc., Vineland, N.J.) at 2,000/cm2 in a 24-well tissue culture plate (Corning Costar Corporation, Cambridge, Mass.) with 1 ml of KGM per well. The cells were grown for 4 to 5 days prior to challenge with F. nucleatum cell wall for 24 h. Cells on the coverslips were washed with phosphate-buffered saline twice, fixed in 4% paraformaldehyde in Sorensen's buffer for 5 min, and permeabilized with cold acetone on ice for 5 min. The cells were then blocked with 3% normal serum in 0.05% Tween in Tris-buffered saline (TBS) for 20 min and incubated with polyclonal antibody against hBD-2, kindly provided by Tomas Ganz, University of California—Los Angeles, at 1:500 dilution in Tween-TBS overnight at 4°C. On the following day, the cells were rinsed with Tween-TBS twice and TBS once and reacted with fluorescein isothiocyanate-conjugated secondary antibody (Vector Laboratories Inc.) at 1:200 dilution in TBS for 30 min. The cells were rinsed in TBS twice, reacted with DAPI (4′,6′-diamidino-2-phenylindole) for 5 min, and rinsed in TBS twice and distilled water once. The coverslips were air dried for 10 min and mounted on slides with mounting medium (Molecular Probes, Eugene, Oreg.). Immunofluorescence images were captured by a Photometrics Sensys camera attached to a Nikon microphot-SA epifluorescence microscope. Image capturing was performed with an IP Lab Spectrum program version 3.12. All computer-generated pictures were organized by Adobe Photoshop version 5.0 software using a PowerPC computer.
Analysis of hBD-2 mRNA in gingival-tissue samples.
Clinically normal gingival-tissue samples were obtained from tissue overlying impacted third molars from 15 different patients (age range, 17 to 30 years). Total RNA was immediately harvested by homogenizing fresh tissue samples, and expression of hBD-2, IL-8, and human keratin 5 mRNAs was analyzed as previously described (26).
RESULTS
Expression of hBD-2 mRNA is variable in gingival-tissue samples.
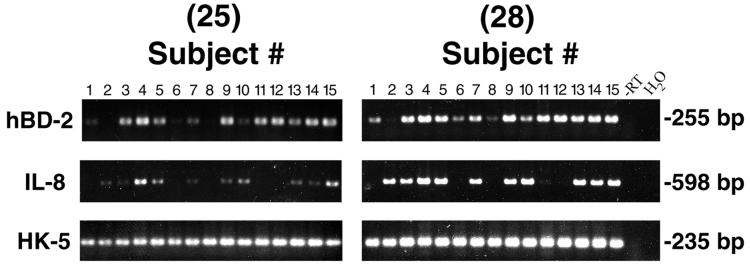
To determine expression of hBD-2 in gingival tissue in vivo, total RNA was extracted from clinically normal tissue freshly obtained from 15 individuals undergoing third-molar extraction. RT-PCR was performed for 25 and 28 cycles to semiquantitatively analyze hBD-2 mRNA. HBD-2 mRNA was expressed in 14 of 15 samples tested, and the level of hBD-2 expression varied between subjects (Fig. 1). For comparisons of tissue samples, an epithelium-specific protein, keratin 5, was used to evaluate the relative contribution of the epithelial compartment in each sample. IL-8 mRNA expression was readily detected in a subset of the normal gingival samples (n = 10) (Fig. 1), suggesting tissue activation even though inflammation was not clinically evident in these tissue samples. No correlation between expression of hBD-2 and IL-8 mRNA was found in the samples. For example, there was no hBD-2 expression but there was IL-8 expression in tissue from subject 2, and there was no IL-8 expression but there was hBD-2 expression in tissue from subjects 1, 6, 8, 11, and 12 (Fig. 1).
FIG. 1.
RT-PCR analysis of hBD-2 mRNA expression in normal gingival-tissue samples. Specific primer pairs (Table 1) were used for an amplification of RNA from multiple noninflamed gingival-tissue samples: hBD-2, 255-bp product; IL-8, 598-bp product; and human keratin 5 (HK-5), 235-bp product. All products were amplified for 25 and 28 cycles. Note that HBD-2 mRNA is expressed in 14 of 15 tissue samples tested, and the level of expression varies between individuals. −RT and H2O samples serve as negative controls.
Stimulated HGE express hBD-2 mRNA and exhibit differential regulation by various stimulants.
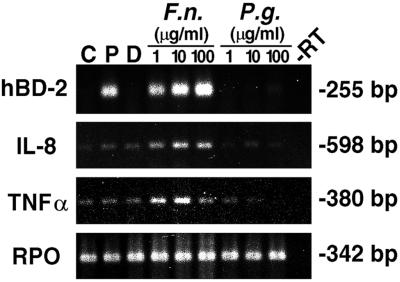
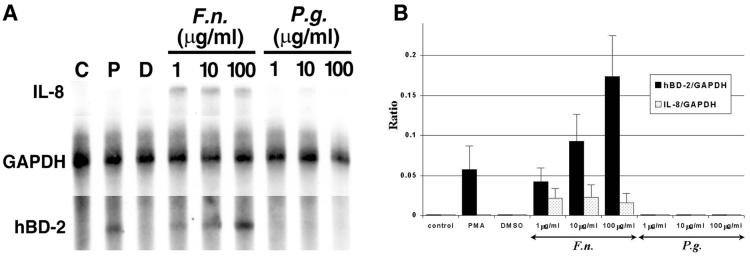
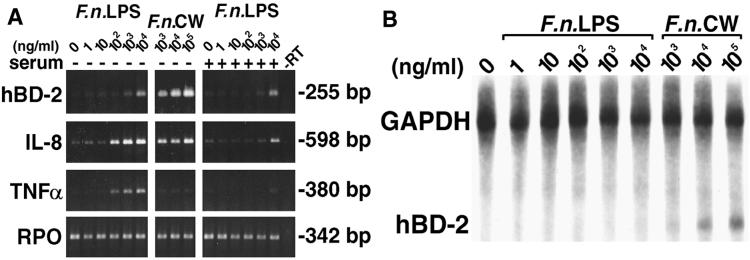
To examine expression of hBD-2 mRNA in vitro, HGE were isolated from gingival biopsy specimens overlying impacted third molars and cultured for a few passages. The HGE were challenged with cell wall extract of two gram-negative periodontal bacteria, F. nucleatum and P. gingivalis, and two potent activators for epithelial cells, TNF-α and PMA, for 24 h. Total RNA was harvested and analyzed by RT-PCR and RPA. HBD-2 mRNA was significantly up-regulated by F. nucleatum cell wall (Fig. 2) and TNF-α (see Fig. 4) in a dose-dependent fashion, as well as by 10 ng of PMA/ml (Fig. 2). The finding of hBD-2 induction by TNF-α is in agreement with a previous study (Harder et al., 1997). In contrast, P. gingivalis cell wall at all doses tested failed to induce hBD-2 mRNA (Fig. 2). Induction of IL-8 and TNF-α mRNAs was also determined as an indication of the state of cell activation. Both cytokines were induced by F. nucleatum cell wall and TNF-α (Fig. 2; see also Fig. 4) but not by P. gingivalis cell wall and PMA (Fig. 2). P. gingivalis cell wall at the maximum dose (100 μg/ml) suppressed the expression of both cytokines (Fig. 2), consistent with a previous study (10). The viability of HGE and the yield of total RNA were checked during each experiment, and no differences were found between untreated and treated cells. The sizes of amplified products were as predicted. The RPO gene is a housekeeping gene control included to show equivalent loading of samples under all conditions. The sequence of amplified products of hBD-2 was confirmed to be identical to the cDNA sequence of hBD-2 (GenBank accession no. Z71389). Control unstimulated gingival epithelial cells (Fig. 2, lane C) did not express hBD-2 mRNA. Consistent with the findings from RT-PCR shown in Fig. 2, we demonstrated the inducible expression of hBD-2 mRNA by PMA and the dose-dependent response by F. nucleatum cell wall with a quantitative RPA analysis (Fig. 3A). The sizes of protected fragments of hBD-2, IL-8, and GAPDH were as predicted based on RNA standards (BrightStar Biotinylated RNA Century-Plus size markers; Ambion). A sample containing yeast RNA hybridized with all three probes served as a control for RNase T treatment and showed no protected fragments (data not shown). GAPDH, a housekeeping gene marker, was approximately equal in all samples (Fig. 3A). The results shown in Fig. 3A as well as those from two separate experiments were analyzed by densitometry relative to GAPDH (Fig. 3B). The expression of hBD-2 relative to that of GAPDH was 0.043 ± 0.017, 0.093 ± 0.034, and 0.174 ± 0.051 for 1, 10, and 100 μg of F. nucleatum cell wall per ml, respectively, and 0.058 ± 0.030 for 10 ng of PMA/ml (n = 3). While the ratio of hBD-2 to GAPDH (Fig. 3B) increased with higher doses of F. nucleatum cell wall, the ratio of IL-8 to GAPDH showed no response (Fig. 3B), suggesting that it was maximally stimulated at the lowest dose used in the study.
FIG. 2.
HGE express hBD-2 mRNA and exhibit differential regulation by various stimulants. HGE were stimulated with various doses of different stimuli for 24 h. Total RNA was extracted and analyzed by RT-PCR using 3 μg of total RNA as described in Materials and Methods. The sizes of the amplified products for hBD-2, IL-8, TNF-α, and RPO are indicated and were as predicted. The hBD-2 products were sequenced and confirmed to be identical to the predicted sequence. C, unstimulated control cells; P, PMA stimulation (10 ng/ml); D, DMSO, the solvent control for PMA. Other stimulants include crude cell wall extracts of F. nucleatum (F.n.) and P. gingivalis (P.g.) at 1, 10, and 100 μg/ml; minus-RT control (−RT) is shown. RPO, a housekeeping gene control, was uniformly expressed. The data shown are representative of three independent experiments.
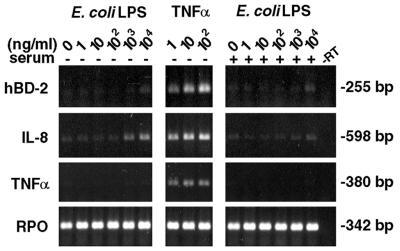
FIG. 4.
Comparison of hBD-2 induction by E. coli LPS and TNF-α. HGE were stimulated with various doses of E. coli LPS or TNF-α in the absence (−) or presence (+) of 1% human serum for 24 h. Total RNA (3 μg) of each sample was used for RT-PCR analysis for hBD-2, IL-8, TNF-α and RPO. The results shown are representative of three independent experiments. Note that there was no additional effect on hBD-2 induction with the presence of human serum. −RT, minus-RT control.
FIG. 3.
HBD-2 up-regulation assessed by RPA. (A) RPA analysis. Total RNA (20 μg) was probed with three different biotin-labeled RNA probes: IL-8, hBD-2, and GAPDH, a housekeeping gene control. RPA was conducted as described in Materials and Methods. The abbreviations are the same as in Fig. 2. The data shown are representative of three separate experiments. (B) Densitometric analysis of RPA. The relative ratios of hBD-2 and IL-8 to GAPDH were determined as described in Materials and Methods. The y axis represents the ratios; the x axis represents a control sample and samples treated with various doses of different stimulants as shown in panel A. The results are represented as means plus standard deviations of three separate experiments. F.n., F. nucleatum; P.g., P. gingivalis.
Bacterial LPS is a poor stimulant for the induction of hBD-2 mRNA in HGE.
E. coli LPS is a known stimulant for bovine tracheal antimicrobial peptide (13). To determine if LPS is the active component in F. nucleatum cell wall extract, purified LPS fraction of F. nucleatum and E. coli LPS were tested for the ability to up-regulate hBD-2 mRNA. HGE were stimulated with various doses of E. coli LPS in comparison with TNF-α (Fig. 4) and various doses of F. nucleatum LPS in comparison with F. nucleatum cell wall (Fig. 5). Total RNA was harvested and analyzed by RT-PCR (Fig. 4 and 5A) and RPA (Fig. 5B). In contrast to significant hBD-2 induction by F. nucleatum cell wall (Fig. 2 and 5A) and TNF-α (Fig. 4), both E. coli and F. nucleatum LPSs poorly induced hBD-2 mRNA (Fig. 4 and 5A). A slight hBD-2 induction was seen in HGE stimulated with the maximum dose (104 ng/ml) of E. coli and F. nucleatum LPSs used in this study (Fig. 4 and 5A). Interestingly, the slight induction observed by RT-PCR (Fig. 5A) was not detected by RPA (Fig. 5B), indicating differences in the sensitivities of the assays. When the mRNA expression for IL-8 was examined, we found doses of 103 ng of E. coli LPS/ml and 102 ng of F. nucleatum LPS/ml or greater induced IL-8 mRNA (Fig. 4 and 5A). Furthermore, doses of 102 ng of F. nucleatum LPS/ml or greater induced TNF-α mRNA (Fig. 5A). Surprisingly, addition of human serum appeared to inhibit up-regulation of IL-8 and TNF-α mRNAs by both bacterial LPSs compared with HGE stimulated with the same dose of LPS in the absence of serum (Fig. 4 and 5A). However, serum did not affect expression of hBD-2 mRNA in any dose of either bacterial LPS (Fig. 4 and 5A) or F. nucleatum cell wall (data not shown).
FIG. 5.
Comparison of hBD-2 induction by F. nucleatum LPS and cell wall. (A) RT-PCR analysis. HGE were stimulated with various doses of F. nucleatum (F.n.) LPS or cell wall in the absence (−) or presence (+) of human serum for 24 h. Total RNA (3 μg) of each sample was used for RT-PCR analysis for hBD-2, IL-8, TNF-α, and RPO. The results shown are representative of three independent experiments. Note that there was slight hBD-2 induction by F. nucleatum LPS at 104 ng/ml, but the degree of hBD-2 induction by F. nucleatum LPS was still less than that by 104 ng of F. nucleatum cell wall/ml. Note the dose-dependent response for IL-8 and TNF-α by F. nucleatum LPS and an inhibitory effect of serum on up-regulation of these two cytokines. −RT, minus-RT control. (B) RPA analysis of some RNA samples in panel A. Note the dose-dependent induction of hBD-2 by F. nucleatum cell wall; however, the slight increase in hBD-2 expression by F. nucleatum LPS (104 ng/ml) observed by RT-PCR was not detected by RPA. The data shown are representative of three independent experiments.
HGF do not express hBD-2 mRNA.
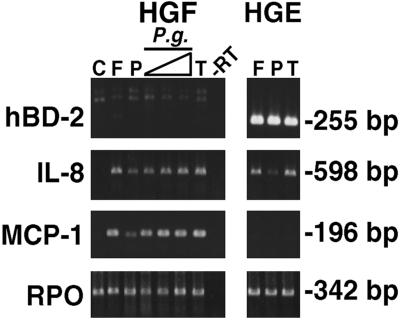
HGF were stimulated for 24 h with different stimulants known to up-regulate hBD-2 in HGE. All stimulants failed to induce hBD-2 expression in HGF either in the absence (Fig. 6) or presence (data not shown) of human serum compared with hBD-2 expression induced in HGE by three activators identified in this study, F. nucleatum cell wall (10 μg/ml), PMA (10 ng/ml), and TNF-α (10 ng/ml) (Fig. 6). This indicates that hBD-2 is derived from epithelial tissue, consistent with other reports showing the expression of hBD-2 in the epithelial lining of several organs as well as epidermis. The mRNAs for IL-8, a marker for cell activation, and for MCP-1, a chemokine known to be expressed in fibroblasts, mononuclear phagocytes, and endothelial cells (20, 53), were induced in HGF stimulated with all of the stimulants used (Fig. 6). In contrast to HGF, HGE did not express MCP-1 under any conditions known to induce hBD-2 and IL-8 (Fig. 6). Interestingly, the P. gingivalis cell wall was effective in up-regulating IL-8 and MCP-1 mRNAs in HGF (Fig. 6) but not hBD-2 and IL-8 mRNAs in HGE (Fig. 2 and 3A), indicating the efficacy of this extract.
FIG. 6.
HGF do not express hBD-2 mRNA. Primary HGF were stimulated with different stimulants for 24 h. RT-PCR analysis was performed using total RNA (3 μg) from each sample. Amplification was conducted for 25 cycles for IL-8, MCP-1, and RPO primers and 35 cycles for the hBD-2 primer pair to detect any small amount of expression. Stimulants included 10 μg of F. nucleatum cell wall/ml (F); 10 ng of PMA/ml (P); 10 ng of TNF-α/ml (T); and 1, 10, and 100 μg of P. gingivalis cell wall/ml (P.g.). C, unstimulated HGF. RPO was uniformly expressed. As a positive control, HGE were stimulated with 10 μg of F. nucleatum cell wall/ml (F), 10 ng of PMA/ml (P), and 10 ng of TNF-α/ml (T). The data shown are representative of two independent experiments. −RT, minus-RT control.
Kinetics of hBD-2 mRNA up-regulation.
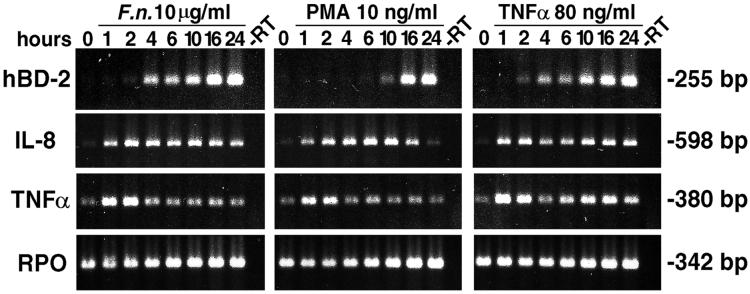
To study the kinetics of hBD-2 up-regulation in HGE, mRNAs for hBD-2 as well as for IL-8 and TNF-α were analyzed by RT-PCR at different times after HGE were challenged with either 10 μg of F. nucleatum cell wall/ml, 10 ng of PMA/ml, or 80 ng of TNF-α/ml. The results (Fig. 7) show that hBD-2 was up-regulated by at least two different signaling pathways. While hBD-2 induction by F. nucleatum cell wall and TNF-α was rapid (2 to 4 h), that by PMA was not seen until 10 h after stimulation. The induction by these three stimulants continuously increased up to 24 h, the maximum period of incubation in the study (Fig. 7). Interestingly, the kinetics of IL-8 and TNF-α mRNA expression were different from each other and from that of hBD-2. IL-8 was rapidly up-regulated at 1 h and peaked a short time later with stimulation by all three stimulants (Fig. 7). IL-8 induction by PMA declined to the baseline level at 24 h (Fig. 7). This was consistent with the absence of IL-8 induction in HGE by PMA at 24 h in other experiments (Fig. 2, 3A, and 6). In contrast, TNF-α mRNA was transiently induced during the first few hours with all three stimulants (Fig. 7). The strong and transient induction of TNF-α prior to hBD-2 induction suggests a possible functional role of TNF-α as an intermediary molecule in hBD-2 regulation.
FIG. 7.
Kinetics of hBD-2 mRNA up-regulation. HGE were stimulated with three activators identified in this study, 10 μg of F. nucleatum (F.n.) cell wall/ml, 10 ng of PMA/ml, and 80 ng of TNF-α/ml for hBD-2 induction for the indicated times. RT-PCR analysis for 25 cycles was performed using total RNA (3 μg). The results show an early hBD-2 induction by F. nucleatum cell wall and TNF-α but a much later induction by PMA. Note that TNF-α mRNA was transiently induced before hBD-2 induction by all three stimulants. The results shown are representative of two independent experiments. −RT, minus-RT control.
Anti-TNF-α does not inhibit hBD-2 induction by F. nucleatum cell wall.
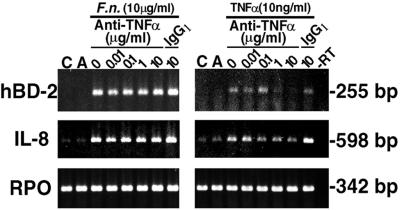
To determine whether TNF-α functions as an intermediary molecule in hBD-2 regulation, various doses of antibody directed to TNF-α were preincubated with either 10 μg of F. nucleatum cell wall/ml or 10 ng of TNF-α (a positive control for inhibition)/ml for 30 min before HGE were challenged for 10 h. Total RNA was harvested and analyzed by RT-PCR (Fig. 8). Anti-TNF-α at 10 μg/ml completely neutralized the effect of TNF-α on hBD-2 induction compared with no inhibitory effect on hBD-2 induction by IgG1, an isotype antibody control. However, anti-TNF-α had no effect at all on hBD-2 induction by F. nucleatum cell wall. Similarly, anti-TNF-α had an inhibitory effect on IL-8 induction by TNF-α but not by F. nucleatum cell wall. Anti-TNF-α alone at 10 μg/ml (Fig. 8, lanes A) had no effect on either hBD-2 or IL-8 expression.
FIG. 8.
Anti-TNF-α does not inhibit hBD-2 induction by F. nucleatum cell wall. Various doses of antibody directed to TNF-α (0.01, 0.1, 1, or 10 μg/ml) were preincubated with either 10 μg of F. nucleatum (F.n.) cell wall/ml or 10 ng of TNF-α/ml for 30 min before they were added to stimulate HGE for 10 h. IgG1 was used as an isotype control antibody for anti-TNF-α. C, unstimulated HGE; A, HGE incubated with anti-TNF-α alone for the same period of time. Total RNA (3 μg) of each sample was analyzed by RT-PCR for 25 cycles. Note that increasing concentrations of the anti-TNF-α antibody result in decreased induction of hBD-2 and IL-8 mRNA by TNF-α; however, the anti-TNF-α antibody has no effect on hBD-2 and IL-8 induction by F. nucleatum cell wall. The results shown are representative of two independent experiments. −RT, minus-RT control.
hBD-2 induction requires new protein synthesis and new gene transcription.
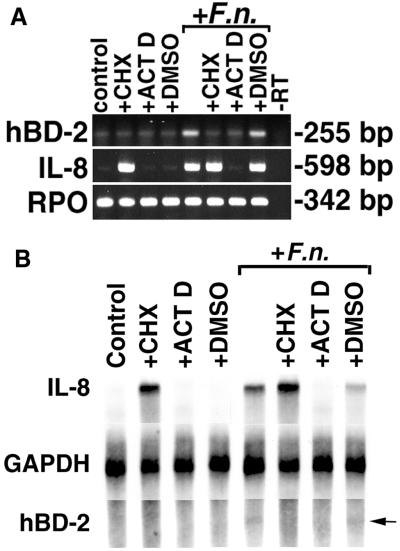
To examine the mechanism(s) for inducible expression of hBD-2 in response to F. nucleatum cell wall, HGE were pretreated with either 10 μg of cycloheximide (an inhibitor of protein synthesis) per ml, 1 μg of actinomycin D (an inhibitor of RNA transcription) per ml, or DMSO (vehicle control) 1 h before exposure to F. nucleatum cell wall for an additional 6 h. The viability of HGE and total RNA yield were checked after each treatment, and no differences were found between experimental and control untreated cells. Because stimulation was short (only 6 h), the level of hBD-2 mRNA expression was low. Nevertheless, both cycloheximide and actinomycin D completely blocked the up-regulation of hBD-2 mRNA in response to F. nucleatum cell wall stimulation, suggesting that both new protein synthesis and new gene transcription are required for F. nucleatum-induced hBD-2 mRNA expression (Fig. 9). Pretreatment with vehicle alone showed no difference in hBD-2 mRNA induction. Similar to induction of hBD-2 mRNA, induction of IL-8 mRNA also required new gene transcription because pretreatment with actinomycin D could completely inhibit F. nucleatum-induced IL-8 expression (Fig. 9). Interestingly, in contrast to the inhibition of hBD-2 induction by pretreatment with cycloheximide, IL-8 mRNA was induced in the presence of cycloheximide regardless of stimulation with F. nucleatum cell wall.
FIG. 9.
Complete inhibition of hBD-2 induction by pretreatment of HGE with cycloheximide and actinomycin D. (A) RT-PCR analysis. HGE were pretreated with 10 μg of cycloheximide/ml (+CHX), 1 μg of actinomycin D/ml (+ACT D), or DMSO solvent (+DMSO) for these two reagents for 1 h, and some samples were then challenged with 10 μg of F. nucleatum cell wall extract/ml (+F.n.) for 6 h. The 6-h time point was chosen because overnight incubation of HGE with cycloheximide and actinomycin D was toxic. At 6 h, there was no significant difference in viability between control and treated cells as assessed by staining with trypan blue. Furthermore, the yields of total RNA from the samples were compared and showed no difference. Total RNA was extracted and analyzed by RT-PCR. Note that in this experiment, PCR for hBD-2 was performed for 28 cycles, which resulted in an increased background of hBD-2 expression in the unstimulated (control) sample and the samples treated with CHX or ACT D in the presence or absence of F. nucleatum cell wall. The data shown are representative of three independent experiments. −RT, minus-RT control. (B) RPA analysis. Total RNA (20 μg) from the samples in panel A was analyzed by RPA as described in Materials and Methods. GAPDH hybridization is shown as a normalization control. Note that hBD-2 mRNA induction is inhibited with both CHX and ACT D, while IL-8 mRNA expression is inhibited only with ACT D. Also note the superinduction of IL-8 mRNA in the sample treated with both CHX and F. nucleatum cell wall and that the degree of hBD-2 expression was lower than that in Fig. 3A due to a shorter time of stimulation (6 h). The arrow indicates a protected fragment of hBD-2 in the samples treated with F. nucleatum and F. nucleatum cell wall plus DMSO.
hBD-2 peptide is detected in HGE by immunofluorescence.
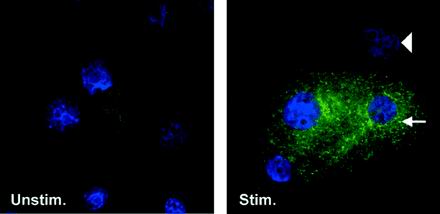
To investigate expression of hBD-2 peptide in HGE, cells were cultured on coverslips, exposed to F. nucleatum cell wall for 24 h, and then reacted with polyclonal antibody against hBD-2. Using immunofluorescence, hBD-2 peptide was detected in the cytoplasm of HGE stimulated with F. nucleatum cell wall but not in unstimulated HGE (Fig. 10), consistent with inducible expression of hBD-2 mRNA. The staining revealed a punctate distribution with concentration of the reaction adjacent to the nucleus (Fig. 10). hBD-2 peptide is not detected in every cell, possibly because of variation in staining of individual cells or because hBD-2 peptide is synthesized in a subpopulation of the epithelial cells.
FIG. 10.
Localization of hBD-2 peptide in F. nucleatum cell wall-stimulated cultured epithelial cells. HGE grown on coverslips were stimulated with 10 μg of F. nucleatum cell wall/ml for 24 h (Stim.) or left unstimulated (Unstim.) for the same time. The cells were fixed and reacted with polyclonal antibody against hBD-2, fluorescein isothiocyanate-conjugated secondary antibody (green), and, briefly, DAPI (blue) as described in Materials and Methods. Note the punctate localization of the signal, which is concentrated adjacent to the nucleus (arrow). The arrowhead indicates no immunoreactivity with polyclonal antibody against hBD-2 in another cell. Magnification, ×75. The data shown are representative of three independent experiments. F. nucleatum-stimulated HGE incubated with normal rabbit serum as a negative control showed no reactivity (data not shown).
DISCUSSION
Our studies of β-defensin expression in oral gingival epithelia show several new characteristics of the innate immune response. First, hBD-2 expression is seen in clinically noninflamed gingival tissue. Second, inducible expression of hBD-2 mRNA occurs in cells via multiple pathways. Third, hBD-2 mRNA is induced by TNF-α and cell wall extract of a representative commensal (F. nucleatum), but not a pathogenic (P. gingivalis) oral microorganism. Fourth, hBD-2 peptide is detected in cultured epithelial cells challenged with F. nucleatum cell wall extract. Taken together, these findings suggest that oral mucosal cells are in an activated state with respect to expression of hBD-2 and that this state is part of the normal barrier function of oral epithelium. We also show that only minimal up-regulation of hBD-2 occurs in response to F. nucleatum or E. coli LPS. Our analyses show consistent differences between hBD-2 and IL-8 regulation (another marker of innate immune response) in response to LPS, in the kinetic analysis, in the requirement for protein synthesis, and in the in vivo tissue analysis. These differences offer evidence that innate immune responses of HGE are differentially regulated and suggest that multiple complex interactions occur between microbial stimulants and host cells.
Gingival epithelium is a useful model for these studies because we can investigate expression of hBD-2 mRNA in gingival tissue as well as its regulation in cultured gingival epithelial cells challenged with natural stimuli found in the oral cavity. We found a dramatic difference in the regulation of hBD-2 by cell wall extracts of F. nucleatum and P. gingivalis. F. nucleatum is a gram-negative anaerobic bacterium which is commonly found in healthy and diseased sites of periodontal tissue (6, 18). Although F. nucleatum is commonly associated with clinical infections of other body sites, this microorganism is not considered causative in periodontal disease. Rather, it is viewed as a “bridge” between early colonizers of the tooth pellicle and the subsequent adherence of pathogenic microorganisms, such as P. gingivalis. F. nucleatum cell wall was found to up-regulate hBD-2 mRNA, as well as IL-8 and TNF-α mRNAs, while P. gingivalis cell wall did not. It has been shown previously that P. gingivalis inhibits the activation of IL-8 by commensal bacteria, including F. nucleatum (10, 28). The absence of hBD-2 mRNA induction by the P. gingivalis cell wall is therefore consistent with the ability of this organism to evade stimulation of host defense mechanisms, while F. nucleatum may help keep gingival epithelial cells in a stimulated state for effective and continuous host defense. Consistent with these in vitro findings, hBD-2 was detected in clinically noninflamed gingival tissue of 14 of 15 subjects in our study, as well as in recent work by Mathews and coworkers (29). Thus, in contrast to the epidermis, in which hBD-2 mRNA is seen primarily in association with inflammation or disease (27; Harder et al., 1997), our results show that clinically noninflamed oral epithelium is in a partially stimulated state and suggest that this may be due to its exposure to oral commensal microorganisms. It will be important to determine if the difference between commensal and pathogenic microorganisms in regulation of β-defensins is seen in other mucosal epithelia that have region-specific ecologies with response to the balance of commensal and pathogenic microorganisms.
Cellular innate immune responses are postulated to be initiated by microbial components via pattern recognition receptors (31). Recent discoveries have shown expression of Toll-like receptors (TLR) on human cells (9, 32, 37) that serve this function. TLR4 is implicated in LPS responses in mice (35, 36, 44). TLR2 mediates the response to bacterial LPS-CD14 signaling in transfected kidney epithelial cell lines (25, 52) and to bacterial lipoprotein (2, 7). It is speculated that there may be one or more TLR expressed in other cell types that mediate recognition of microbial components to signal the human innate immune response. Thus, a logical active bacterial component for hBD-2 regulation in the F. nucleatum cell wall extract is LPS, a major fraction of the crude cell wall extract. In addition, mRNA expression for the bovine β-defensins, TAP and LAP, was previously shown to be mediated via bacterial LPS and a CD14-dependent signaling pathway (13, 38). In contrast to the bovine system, our findings in human gingival epithelial cells show induction of IL-8 mRNA by both E. coli LPS and F. nucleatum LPS and induction of TNF-α mRNA by F. nucleatum LPS but a poor response for hBD-2 mRNA to LPSs of both bacteria compared to the F. nucleatum cell wall. This suggests that another cell wall component(s) stimulates hBD-2 mRNA induction. Up-regulation of hBD-2 mRNA in gingival epithelial cells in response to E. coli LPS was previously shown by Mathews and coworkers (29); however, the stimulation in their study was also low compared to the effect of the proinflammatory cytokine IL-1β. Addition of serum as a source of soluble CD14 and LPS-binding protein (LBP) (24) does not activate or inhibit hBD-2 induction in gingival epithelial cells stimulated by bacterial LPS. The addition of serum inhibited induction of IL-8 and TNF-α, possibly because serum proteins, such as LBP and others, enable binding and neutralization of LPS (48, 49, 50). Finally, hBD-2 induction by F. nucleatum cell wall was not inhibited by pretreatment of the cell wall extract with polymyxin B sulfate, an inhibitor of LPS signaling (data not shown). These findings indicate that, unlike induction of cytokines, hBD-2 up-regulation in gingival epithelium is poorly responsive to bacterial LPS; they also support a CD14-independent pathway and suggest multiple cellular responses for eliciting various aspects of innate immune defenses in epithelial cells. Our results are in agreement with findings for human uroepithelial cells that respond poorly to E. coli LPS but effectively to P-fimbriated E. coli cell wall extracts (21) and with other examples of human epithelial cell signaling of innate immune responses that differ from those of monocytes, macrophages, etc. (15, 24).
Kinetic analysis of hBD-2 mRNA induction indicates involvement of multiple signaling pathways and further supports the distinction between hBD-2 and IL-8 regulation. HBD-2 mRNA is rapidly induced by F. nucleatum cell wall and TNF-α and accumulates during the 24-h period of study, as would be expected for an innate immune defense response to microbial and inflammatory stimuli. In contrast to the profile of hBD-2 induction, induction of TNF-α is rapid and transient, while that of IL-8 is rapid but with longer duration. We thought that the transient induction of TNF-α might indicate its role as an intermediary molecule involved in hBD-2 up-regulation by all three stimulants. This hypothesis was disproved (Fig. 8) by using a specific antibody to neutralize both exogenously added and endogenously synthesized TNF-α.
The accumulation of hBD-2 mRNA may be due to new gene transcription, altered mRNA stability, or both, since inhibitors of both transcription and translation block hBD-2 mRNA expression. The inhibitory effect of actinomycin D on hBD-2 and IL-8 mRNA induction implies that both genes are regulated at the transcriptional level. The inhibition of hBD-2 induction by cycloheximide implicates the requirement for new protein synthesis for hBD-2 induction by F. nucleatum cell wall; these proteins may include cell receptors, intermediary proteins in signaling pathways, transcription factors, or proteins that alter mRNA stability. Elevated levels of IL-8 mRNA following inhibition of protein synthesis can be most readily explained by loss of rapidly turned-over proteins necessary for RNA degradation. This is very likely the case for cytokine genes such as IL-8, whose transcripts contain an AU-rich element, a specific binding site for labile proteins, in the 3′ untranslated region (1, 46).
Immunofluorescence detection of hBD-2 peptide within the cytoplasm of stimulated gingival epithelial cells is consistent with the inducible expression of hBD-2 mRNA. The punctate perinuclear immunostaining is suggestive of endoplasmic reticulum and Golgi apparatus localization, typical of a secreted product with a signal peptide, such as is found in both hBD-1 and hBD-2 mRNAs (17, 41, 45; Harder et al., 1997).
In conclusion, our results indicate that gingival epithelial cells and tissue express messages for hBD-2 and produce hBD-2 peptide in response to inflammatory mediators and to continuous challenges from the cell walls of commensal bacteria which are naturally present in the oral cavity. Interestingly, the cell wall of a periopathogenic bacterium, P. gingivalis, does not induce hBD-2 mRNA and therefore appears to have a strategy of evading this aspect of the host innate immune response, in addition to others shown previously (10, 28). The difference in stimulation by commensal and pathogenic bacteria may be important in understanding the molecular aspects of host-bacterial interaction as well as for potential new preventive therapies for mucosal infection. The production of hBD-2 as part of the epithelial barrier may be especially important in innate host defense at confrontational mucosal sites in the gingival sulcus and therefore may contribute to overall oral health and disease susceptibility.
ACKNOWLEDGMENTS
We are grateful to Tomas Ganz, Department of Medicine and Will Rogers Institute for Pulmonary Research, School of Medicine, University of California, Los Angeles, for a generous gift of polyclonal antibody to hBD-2. We thank the faculty and staff from the Oral Surgery Department, School of Dentistry, UW, for providing gingival biopsy specimens. We also thank Philip Fleckman and Martine Michel for their assistance with cell culture and use of the Dermatology Core Lab facilities, Frank Roberts, who provided useful advice and help in setting up an RPA, and Robert Underwood for his assistance with the immunofluorescence microscope.
This work was supported by the University of Washington Royalty Research Fund (to B.A.D.), and NIH-NIDCR grants 2P50 DE08229 (Research Center in Oral Biology), P20DE12380, and P60DE97002 (Comprehensive Center for Oral Health Research).
REFERENCES
- 1.Abruzzo L V, Thornton A J, Liebert M, Grossman H B, Evanoff H, Westwick J, Strieter R M, Kunkel S L. Cytokine-induced gene expression of interleukin-8 in human transitional cell carcinomas and renal cell carcinomas. Am J Pathol. 1992;140:365–373. [PMC free article] [PubMed] [Google Scholar]
- 2.Aliprantis A O, Yang R-B, Mark M R, Suggett S, Devaux B, Radolf J D, Klimpel G R, Godowski P J, Zychlinsky A. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 3.Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, Wilson J M. Human β-defensin-2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Investig. 1998;102:874–880. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensch K W, Raida M, Magert H J, Schulz K P, Forssmann W G. hBD 1: a novel beta defensin from human plasma. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 5.Bevins C L, Russell J P, Diamond G. Inducible expression of a mammalian antibiotic peptide gene in tracheal cells. Pediatr Pulmonol Suppl. 1996;13:324. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolstad A I, Jensen H B, Bakken V. Taxonomy, biology and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brightbill H D, Libraty D H, Krutzik S R, Yang R-B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, Brennan P J, Bloom B R, Godowski P J, Modlin R L. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 8.Bryn K, Jantzen E. Analysis of lipopolysaccharides by methanolysis, trifluoroacetylation, and gas chromatography on a fused-silica capillary column. J Chromatogr. 1982;240:405–413. [Google Scholar]
- 9.Chaudhary P M, Ferguson C, Nguyen V, Nguyen O, Massa H F, Eby M, Jasmin A, Trask B J, Hood L, Nelson P S. Cloning and characterization of two Toll/Interleukin-1 receptor-like genes TIL3 and TIL4: evidence for a multi-gene receptor family in humans. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- 10.Darveau R P, Belton C M, Reife R A, Lamont R J. Local chemokine paralysis: a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darveau R P, Hancock R E W. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. 1983;155:831–838. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond G, Bevins C L. Molecule of the month, β-defensins: endogenous antibiotics of the innate host defense response. Clin Immunol Immunopathol. 1998;88:221–225. doi: 10.1006/clin.1998.4587. [DOI] [PubMed] [Google Scholar]
- 13.Diamond G, Russell J P, Bevins C L. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond G, Zasloff M, Eck H, Brasseur M, Maloy W L, Bevins C L. Tracheal antimicrobial peptide, a cysteine rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckmann L, Kagnoff M F, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folch J, Lees M, Stanley G. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 17.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human beta defensin 1 is a salt sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 18.Haffajee A D, Cugini M A, Tanner A, Pollack R P, Smith C, Kent R L., Jr Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 19.Haffajee A D, Socransky S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 20.Hanazawa S, Kawata Y, Takeshita A, Kumada H, Okithu M, Tanaka S, Yamamoto Y, Masuda T, Umemoto T, Kitano S. Expression of monocyte chemoattractant protein 1 (MCP-1) in adult periodontal disease: increased monocyte chemotactic activity in crevicular fluids and induction of MCP-1 expression in gingival tissues. Infect Immun. 1993;61:5219–5224. doi: 10.1128/iai.61.12.5219-5224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedlund M, Wachtler C, Johansson E, Hang L, Somerville J E, Darveau R P, Svanborg C. P fimbriae-dependent, lipopolysaccharide-independent activation of epithelial cytokine responses. Mol Microbiol. 1999;33:693–703. doi: 10.1046/j.1365-2958.1999.01513.x. [DOI] [PubMed] [Google Scholar]
- 22.Kagan B L, Ganz T, Lehrer R I. Defensins: a family of antimicrobial and cytotoxic peptides. Toxicology. 1994;87:131–149. doi: 10.1016/0300-483x(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 23.Kagnoff M F, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Investig. 1997;100:S51–S55. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielian T L, Blecha F. CD14 and other recognition molecules for lipopolysaccharide: a review. Immunopharmacology. 1995;29:187–205. doi: 10.1016/0162-3109(95)00003-c. [DOI] [PubMed] [Google Scholar]
- 25.Kirschning C J, Wesche H, Merrill Ayres T, Rothe M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krisanaprakornkit S, Weinberg A, Perez C N, Dale B A. Expression of the peptide antibiotic human β-defensin-1 in cultured gingival epithelial cells and gingival tissue. Infect Immun. 1998;66:4422–4428. doi: 10.1128/iai.66.9.4222-4228.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Wang L, Jia H P, Zhao C, Heng H H Q, Schutte B C, McCray P B, Jr, Ganz T. Structure and mapping of the human beta-defensin HBD-2 gene and its expression at sites of inflammation. Gene. 1998;222:237–244. doi: 10.1016/s0378-1119(98)00480-6. [DOI] [PubMed] [Google Scholar]
- 28.Madianos P N, Papapanou P N, Sandros J. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect Immun. 1997;65:3983–3990. doi: 10.1128/iai.65.10.3983-3990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews M, Jia H P, Guthmiller J M, Losh G, Graham S, Johnson G K, Tack B F, McCray P B., Jr Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCray P B, Jr, Bentley L. Human airway epithelia express a β-defensin. Am J Respir Cell Mol Biol. 1997;16:343–349. doi: 10.1165/ajrcmb.16.3.9070620. [DOI] [PubMed] [Google Scholar]
- 31.Medzhitov R, Janeway C A., Jr Innate immunity: the virtues of a monoclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 32.Medzhitov R, Preston-Hurlburt P, Janeway C A., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 33.Onoue S, Niwa M, Isshiki Y, Kawahara K. Extraction and characterization of the smooth-type lipopolysaccharide from Fusobacterium nucleatum JCM 8532 and its biological activities. Microbiol Immunol. 1996;40:323–331. doi: 10.1111/j.1348-0421.1996.tb01075.x. [DOI] [PubMed] [Google Scholar]
- 34.Peterson A A, Haug A, Haug E, McGroarty E J. Physical properties of short- and long-O-antigen-containing fractions of lipopolysaccharide from Escherichia coli O111:B4. J Bacteriol. 1986;165:116–122. doi: 10.1128/jb.165.1.116-122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poltorak A, He X, Smirnova I, Liu M-Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 36.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell J P, Diamond G, Tarver A P, Scanlin T F, Bevins C L. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators and lipopolysaccharide. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 40.Selsted M E, Tang Y Q, Morris W L, McGuire P A, Novotny M J, Smith W, Henschen A H, Cullor J S. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J Biol Chem. 1993;268:6641–6648. [PubMed] [Google Scholar]
- 41.Singh P K, Jia H P, Wiles K, Hesselberth J, Liu L, Conway B A, Greenberg E P, Valore E V, Welsh M J, Ganz T, Tack B F, McCray P B., Jr Production of beta-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slots J, Rams T E. Microbiology of periodontal disease. In: Slots J, Taubman M A, editors. Contemporary oral microbiology and immunology. St. Louis, Mo: Mosby Year Book; 1992. pp. 425–443. [Google Scholar]
- 43.Somerville J E, Cassiano L, Bainbridge B, Cunningham M D, Darveau R P. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J Clin Investig. 1996;97:359–365. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 45.Valore E V, Park C H, Quayle A, Wiles K R, McCray P B, Jr, Ganz T. Human β-defensin-1, an antimicrobial peptide of urogenital tissues. J Clin Investig. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang E, Ma W J, Aghajanian C, Spriggs D R. Posttranscriptional regulation of protein expression in human epithelial carcinoma cells by adenine-uridine-rich elements in the 3′-untranslated region of tumor necrosis factor-alpha messenger RNA. Cancer Res. 1997;57:5426–5433. [PubMed] [Google Scholar]
- 47.Weinberg A, Krisanaprakornkit S, Dale B A. Epithelial antimicrobial peptides: review and significance for oral applications. Crit Rev Oral Biol Med. 1998;9:399–414. doi: 10.1177/10454411980090040201. [DOI] [PubMed] [Google Scholar]
- 48.Wurfel M M, Kunitake S T, Lichenstein H, Kane J P, Wright S D. Lipopolysaccharide (LPS)-binding protein is carried on lipoproteins and acts as a cofactor in the neutralization of LPS. J Exp Med. 1994;180:1025–1035. doi: 10.1084/jem.180.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wurfel M M, Wright S D. Lipopolysaccharide (LPS) binding protein catalyzes binding of LPS to lipoproteins. Prog Clin Biol Res. 1995;392:287–295. [PubMed] [Google Scholar]
- 50.Wurfel M M, Wright S D. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayers: preferential interaction with particular classes of lipid. J Immunol. 1997;158:3925–3934. [PubMed] [Google Scholar]
- 51.Yang D, Chertov O, Bykovskaia S N, Chen Q, Buffo M J, Shogan J, Anderson M, Schröder J M, Wang J M, Howard O M Z, Oppenheim J J. β-Defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 52.Yang R B, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signaling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 53.Yu X, Graves D T. Fibroblasts, mononuclear phagocytes, and endothelial cells express monocyte chemoattractant protein-1 (MCP-1) in inflamed human gingiva. J Periodontol. 1995;66:80–88. doi: 10.1902/jop.1995.66.1.80. [DOI] [PubMed] [Google Scholar]
- 54.Zhao C, Wang I, Lehrer R I. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 1996;396:319–322. doi: 10.1016/0014-5793(96)01123-4. [DOI] [PubMed] [Google Scholar]