Abstract
Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignancies worldwide, because its discovery time is in the late stage of the disease, so it is important to develop HNSCC biomarkers to achieve the purpose of early detection and treatment. Fatty acid desaturase 3 (FADS3), the third member of the FADS family, is involved in sphingolipid biosynthesis. Here, we for the first time investigated FADS3 expression in HNSCC, as well as its potential biological function, prognostic value and its impact on the immune system. In this study, we used bioinformatics for gene expression analysis, clinicopathological analysis, enrichment analysis, and immune infiltration analysis of The Cancer Genome Atlas (TCGA) datasets. Statistical analysis was done using R. Tumor IMmune Estimation Resource (TIMER) and CIBERSORT were used to analyze the effect of FADS3 on immune responses in HNSCC. Gene Expression Profiling Interactive Analysis (GEPIA), Kaplan–Meier (KM) survival analysis, and the Human Protein Atlas (HPA) data were used to validate the results from bioinformatics analysis. Our findings indicate that FADS3 influences HNSCC prognosis. High expression of FADS3 is related to higher lymphatic metastasis, histologic grade, and lymphovascular invasion. Gene set enrichment analysis (GSEA) revealed that FADS3 is related to inhibition of amino acid metabolism. CIBERSORT analysis showed high FADS3 expression correlates with reduced levels of B cells. FADS3 is a marker of HNSCC, and high expression of FADS3 is associated with poor prognosis of HNSCC.
Keywords: biomarker, FADS3, head and neck squamous cell carcinoma, HNSCC, TCGA
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is the 6th most common malignancy, worldwide and is characterized by high recurrence and metastasis rates.[1] HNSCC has an indolent clinical course and is often diagnosed at an advanced stage.[2] The treatment of head and neck malignancies is challenging, with locally advanced disease requiring a multidisciplinary approach involving surgery, radiotherapy, and systemic therapy.[3] However, the 5-year survival rate of patients with locally advanced HNSCC is only 50% using standard chemoradiotherapy.[4] The higher recurrence rate of HNSCC further reduces 5-year survival.[5] Thus, effective biomarkers for predicting HNSCC progression are urgently needed for better treatment outcomes.
Human fatty acid desaturase 3 (FADS3), the 3rd member of the FADS family,[6] is located within a 100 kb region on the long arm of chromosome 11 (11q12–13.1), which is a cancer hot spot.[7] Recent findings have identified FADS3 as a key factor in sphingolipid biosynthesis.[8,9]
Although HNSCC is a common cancer type, the role of FADS3 in HNSCC remains unclear. Here, we analyzed microarray datasets from The Cancer Genome Atlas (TCGA) to determine FADS3 expression in HNSCC samples. We used R (version 3.5.3) to evaluate the correlation between FADS3 expression and clinical parameters and HNSCC prognosis. We used gene set enrichment analysis (GSEA), Kyoto Encyclopedia of Genes and Genomes (KEGG), and enhanced gene ontology (GO) analyses to determine the mechanisms underlying FADS3 activity in HNSCC. CIBERSORT and Tumor IMmune Estimation Resource (TIMER) were used to assess the relationship between FADS3 and tumor infiltrating immune cells (TIIC). Finally, Kaplan–Meier (KM), Gene Expression Profiling Interactive Analysis (GEPIA) and the Human Protein Atlas (HPA) analyses were used to assess the relationship between FADS3 and HNSCC prognosis. Our data show that elevated FADS3 expression correlates with shorter survival in HNSCC patients, highlighting its potential as a prognostic biomarker for HNSCC. Our findings uncover the relationship between FADS3 and HNSCC tumorigenesis.
2. Materials and methods
2.1. TCGA database analysis
In this study, the TCGA (https://www.cancer.gov/about-cancer) database of HNSCC was used to obtain immune system invasion, clinical information (Data Type: Clinical Supplement) and gene expression (workflow type: HTSeq-FPKM).[10] Samples with missing or insufficient data on age, local invasion, overall survival (OS) time, lymph node metastasis, distant metastasis, and tumor-node-metastasis (TNM) stage, were excluded from the analysis. Ribonucleic acid sequencing (RNAseq) and clinical data are reserved for further study.
2.2. GSEA
Normalized ribonucleic acid sequencing (RNAseq) datasets from TCGA were used for GSEA.[11] The number of permutations was set to 1000. GSEA was used for GO term and KEGG pathway analysis to determine the potential functions of FADS3. P < .05 and FDR < 0.05 indicated significantly enriched factors.
2.3. TIMER analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a web resource for the evaluation of the effects of immune cells on various cancers.[12] FADS3 was selected using the “Gene” module input to generate a scatter map of the correlation between FADS3 expression and immune infiltration level in HNSCC. Next, the “Survival” module input was used to select HNSCC life cycle KM curve, which visualized the correlation between 6 immune cell types and HNSCC survival.
2.4. GEPIA
GEPIA (http://gepia.cancer-pku.cn/index.html) is an analytical tool containing data on thousands of tumors and normal control tissue.[13] Patient survival analysis was done using KM analysis for further verify the above research conclusions.
2.5. Human Protein Atlas (HPA) analysis.
The Human Protein Atlas (HPA) (www.proteinatlas.org) was used to examine FADS3 protein levels in normal oral mucosa cells versus HNSCC tissues.[14]
3. Results
3.1. Differential FADS3 expression in HNSCC
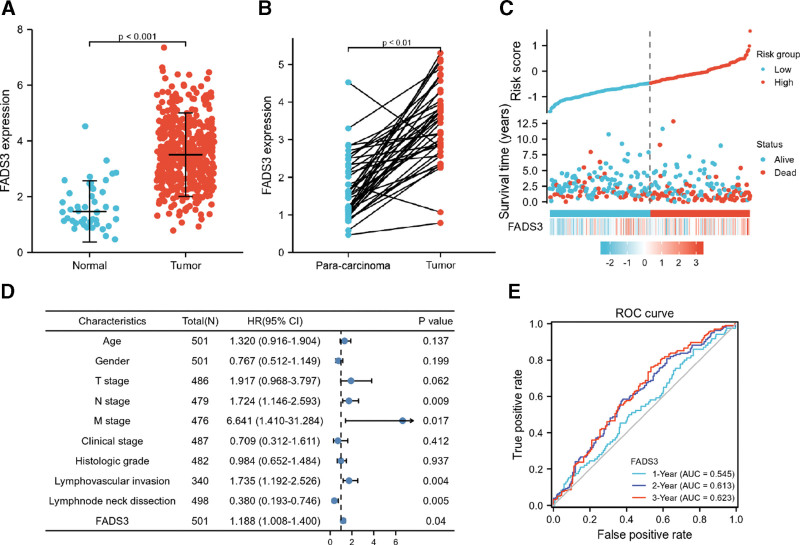
We examined TCGA datasets to determine if FADS3 was differentially expressed in HNSCC tissues versus normal controls. First, 502 tumor information and 44 normal information were converted to the count data closer to the values obtained by the microarray. Comparison of FADS3 expression in normal versus HNSCC samples showed that its expression was significantly higher in HNSCC tissues (Fig. 1A; unpaired Mann–Whitney U test: P < .001). A comparison of FADS3 expression in 43 HNSCC tumor samples and paired, adjacent noncancer tissues showed that in 41 samples, FADS3 levels were significantly higher than in control tissue (Fig. 1B, P < .001). Next, we used Cox analysis to assess the correlation between FADS3 expression and multivariable characteristics and OS of HNSCC patients (Table 1). Univariate correlation analysis showed that N stage (HR = 1.384, P = .026), M stage (HR = 4.745, P = .002), lymphovascular invasion (HR = 1.699, P = .002) and FADS3 expression (HR = 1.205, P = .002) significantly correlated with OS. Multivariate analysis revealed that FADS3 expression (P = .040) was an independent HNSCC prognosis factor (Fig. 1D). Data on HNSCC survival and FADS3 expression profile are shown in Figure 1C. Receiver operator characteristic (ROC) analysis of 3-year survival revealed an area under the curve (AUC) of 0.622, indicating that FADS3 expression can effectively predict HNSCC prognosis (Fig. 1E).
Figure 1.
(A) The expression of FADS3 mRNA between normal and tumor tissues. (B) FADS3 mRNA expression between para-carcinoma tissues and tumor tissues. (C) FADS3 mRNA expression distribution and survival status. (D) Multivariate Cox analysis of FADS3 mRNA expression and clinicopathological variables. (E) ROC curves of FADS3. FADS3 = fatty acid desaturase 3, mRNA = messenger ribonucleic acid, ROC = receiver operator characteristic.
Table 1.
Correlation between overall survival and multivariable characteristics in TCGA patients via Cox regression and multivariate survival model.
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 501 | 1.252 (0.956–1.639) | .102 | 1.320 (0.916–1.904) | .137 |
| Gender | 501 | 0.764 (0.574–1.018) | .066 | 0.767 (0.512–1.149) | .199 |
| T stage | 486 | 1.245 (0.932–1.661) | .137 | 1.917 (0.968–3.797) | .062 |
| N stage | 479 | 1.384 (1.040–1.842) | .026 | 1.724 (1.146–2.593) | .009 |
| M stage | 476 | 4.745 (1.748–12.883) | .002 | 6.641 (1.410–31.284) | .017 |
| Clinical stage | 487 | 1.217 (0.878–1.688) | .238 | 0.709 (0.312–1.611) | .412 |
| Histologic grade | 482 | 0.939 (0.688–1.282) | .692 | 0.984 (0.652–1.484) | .937 |
| Lymphovascular invasion | 340 | 1.699 (1.211–2.384) | .002 | 1.735 (1.192–2.526) | .004 |
| Lymphnode neck dissection | 498 | 0.731 (0.526–1.016) | .062 | 0.380 (0.193–0.746) | .005 |
| FADS3 | 501 | 1.205 (1.069–1.359) | .002 | 1.188 (1.008–1.400) | .040 |
TCGA = The Cancer Genome Atlas, FADS3 = fatty acid desaturase 3.
3.2. Relationship between FADS3 expression and clinicopathology
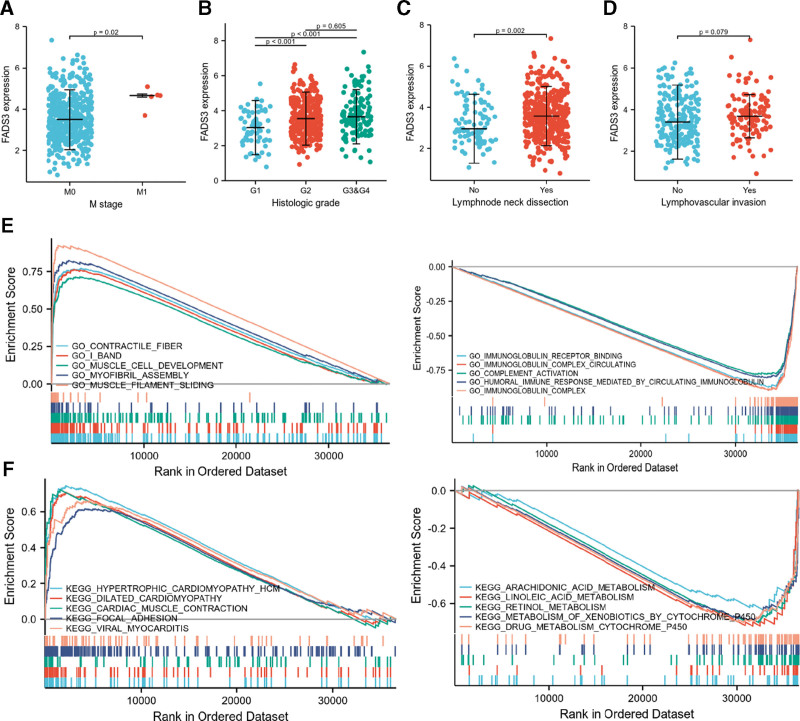
Next, we examined the relationship between FADS3 expression levels and various HNSCC clinicopathological parameters. Analysis of 502 HNSCC samples with FADS3 expression data and accompanying clinical information were obtained from TCGA and revealed that elevated FADS3 levels significantly correlated with tumor M stage (Mann–Whitney U test: P = .020; Fig. 2A), histologic grade (G1 vs G2 & G1 vs G3, Mann–Whitney U test: P < .001; Fig. 2B) and lymph node neck dissection (Mann–Whitney U test: P = .002; Fig. 2C). However, there was no significant correlation between the expression of FADS3 and lymphovascular invasion (Mann–Whitney U test: P = .079, Fig. 2D). Logistic regression (univariate) analysis revealed correlation between FADS3 expression and poor HNSCC prognosis. Increased FADS3 levels in HNSCC significantly correlated with N stage (N1 & N2 & N3 relative to N0, P = .045), histologic grade (G2 & G3 & G4 relative to G1, P = .003), lymphovascular invasion (Yes or No, P = .02), and lymph node neck dissection (Yes or No, P = .002), indicating that relative to HNSCC patients with low FADS3 levels, those with high FADS3 levels are more prone to metastatic disease, lymphovascular invasion, and higher malignant grade (Table 2).
Figure 2.
(A) Expression of FADS3 mRNA correlated significantly with M stage, (B) histological grade, and (C) lymph node neck dissection. (D) The expression of FADS3 mRNA was not correlated with lymphovascular invasion. (E) GO term analysis revealed 5 positively correlated groups and 5 negatively correlated groups. (F) KEGG pathway showed 5 positively correlated groups and 5 negatively correlated groups. FADS3 = fatty acid desaturase 3, mRNA = messenger ribonucleic acid, GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes.
Table 2.
Association between FAD3 expression and clinicopathologic characteristics using logistic regression.
| Characteristics | Total (N) | Odds ratio (OR) | P value |
|---|---|---|---|
| Age (>60 vs ≤60) | 501 | 0.732 (0.515–1.040) | .082 |
| Gender (Male vs Female) | 502 | 1.130 (0.761–1.681) | .545 |
| T stage (T2 & T3 & T4 vs T1) | 487 | 0.917 (0.448–1.866) | .809 |
| N stage (N1 & N2 & N3 vs N0) | 480 | 1.445 (1.009–2.072) | .045 |
| M stage (M1 vs M0) | 477 | 50497133.569 (0.000–NA) | .992 |
| Clinical stage (Stage II & Stage III & Stage IV vs Stage I) | 488 | 1.358 (0.540–3.564) | .519 |
| Histologic grade (G2 & G3 & G4 vs G1) | 483 | 2.365 (1.360–4.240) | .003 |
| Smoker (Yes vs No) | 492 | 0.850 (0.555–1.297) | .450 |
| Alcohol history (Yes vs No) | 491 | 1.291 (0.884–1.891) | .187 |
| Lymphovascular invasion (Yes vs No) | 341 | 1.705 (1.090–2.683) | .020 |
| Lymphnode neck dissection (Yes vs No) | 499 | 1.952 (1.226–3.151) | .005 |
FADS3 = fatty acid desaturase 3.
3.3. GSEA investigation of FADS3
Next, we used GO and KEGG pathway analyses to investigate the biological functions of FADS3 and selected the most enriched signaling pathways based on normalized enrichment score (NES). GO term analysis revealed that contractile fiber, I band, muscle cell development, myofibril assembly, and muscle filament sliding positively correlated with FADS3 expression (Table 3) and that immunoglobulin receptor binding, immunoglobulin complex circulating, complement activation, immunoglobulin complex, and humoral immune response mediated by circulating immunoglobulin negatively correlated with FADS3 levels. The biological processes and molecular functions that strongly correlated with FADS3 were immune responses and cell movements (Fig. 2E). KEGG pathway analysis revealed that hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy, cardiac muscle contraction, focal adhesion, and viral myocarditis had the strongest positive correlation with FADS3 expression, while arachidonic acid metabolism, linoleic acid metabolism, retinol metabolism, metabolism of xenobiotics by cytochrome P450, and drug metabolism cytochrome P450 had the strongest negative correlation (Fig. 2F). These results indicate that cell metabolism, which is critically important in HNSCC, strongly correlates with FADS3 expression.
Table 3.
Signaling pathways most significantly correlated with FADS3 expression based on their normalized enrichment score (NES) and P-value.
| GO NAME | NES | NOM P-value | FDR q-values | |
|---|---|---|---|---|
| Positive | Contractile fiber | 3.146677819 | .006329114 | 0.075950793 |
| I band | 2.974118691 | .004854369 | 0.075950793 | |
| Muscle cell development | 2.851901095 | .005154639 | 0.075950793 | |
| Myofibril assembly | 2.848879257 | .003496503 | 0.075950793 | |
| Muscle filament sliding | 2.833193806 | .003012048 | 0.075950793 | |
| Negative | Immunoglobulin receptor binding | −2.623954549 | .001381215 | 0.075950793 |
| Immunoglobulin complex circulating | −2.657049121 | .0013947 | 0.075950793 | |
| Complement activation | −2.66081915 | .001234568 | 0.075950793 | |
| Humoral immune response mediated by circulating immunoglobulin | −2.682405634 | .001265823 | 0.075950793 | |
| Immunoglobulin complex | −2.992567084 | .001256281 | 0.075950793 |
| KEGG NAME | NES | NOM P-value | FDR q-values | |
|---|---|---|---|---|
| Positive | Hypertrophic cardiomyopathy (HCM) | 2.661862094 | .003846154 | 0.053072241 |
| Dilated cardiomyopathy | 2.541487035 | .004032258 | 0.053072241 | |
| Cardiac muscle contraction | 2.539996672 | .003703704 | 0.053072241 | |
| Focal adhesion | 2.460728054 | .005494505 | 0.062605588 | |
| Viral myocarditis | 2.285167634 | .003558719 | 0.053072241 | |
| Negative | Arachidonic acid metabolism | −1.879355011 | .002877698 | 0.053072241 |
| Linoleic acid metabolism | −1.893264936 | .001481481 | 0.053072241 | |
| Retinol metabolism | −2.073533352 | .00140647 | 0.053072241 | |
| Metabolism of xenobiotics by cytochrome P450 | −2.080359339 | .001371742 | 0.053072241 | |
| Drug metabolism cytochrome P450 | −2.138592243 | .001336898 | 0.053072241 |
FADS3 = fatty acid desaturase 3, FDR = , GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes, NES = normalized enrichment score, NOM = .
3.4. Relationship between FADS3 expression and tumor immune cells
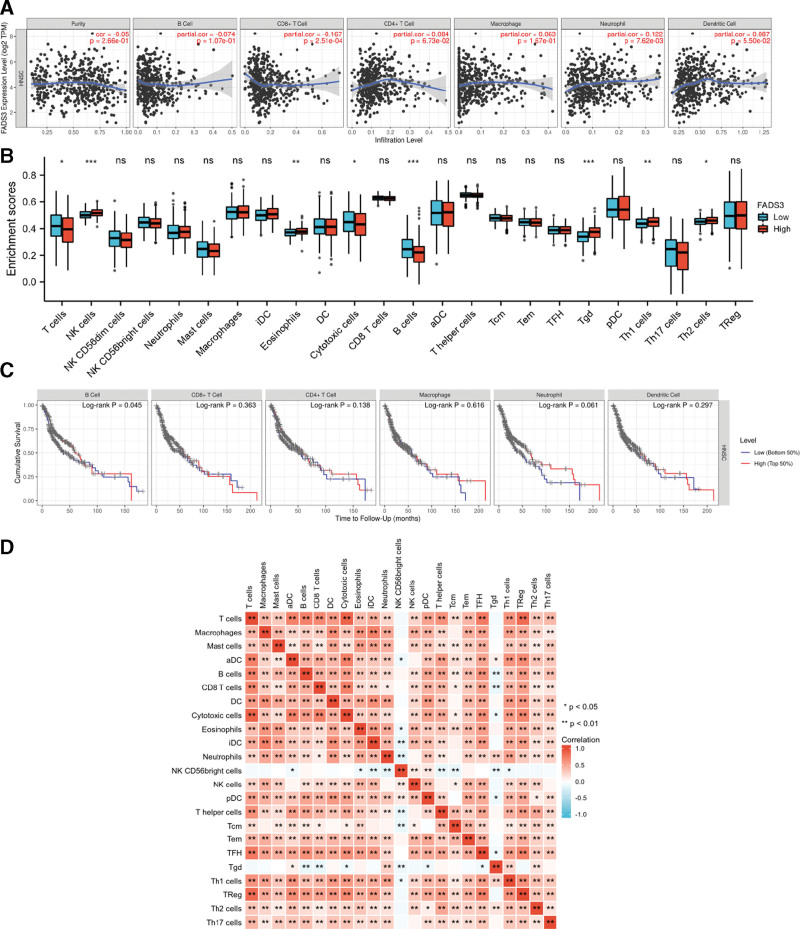
TIMER analysis of the correlation between FADS3 expression and immune infiltration in HNSCC revealed that FADS3 expression negatively correlated with the level of CD8 + T-cells infiltration (P = 2.51 × 10−4) and positively correlated with the level of neutrophil infiltration (P = 7.62 × 10−4) (Fig. 3A). These results indicate that in HNSCC, FADS3 plays an important role in immune response. Next, we investigated differences in the tumor immune microenvironment in HNSCC patients with high FADS3 levels relative to those with low FADS3 levels. To this end, 502 HNSCC tumors were divided into the high FADS3 expression and low FADS3 expression group (251 samples per group). This study used the established computational resource TCGA to explore gene expression profiles of the downloaded samples to determine the levels of 24 immune cell types. The gene set variation analysis (GSVA) package on R was used to investigate the levels of 24 immune cell subtypes in the high FADS3 expression group relative to the low expression groups (Fig. 3B). This analysis revealed that T-cells, NK cells, eosinophils, cytotoxic cells, B cells, Tgd, Th1 cells, and Th2 cells were affected by FADS3 expression. Marked differences were observed between the levels of B cells, NK cells, and Tgd in the high FADS3 expression group relative to the low expression group. NK cells (P < .001), eosinophils (P < .01), Tgd (P < .001), Th1 cells (P < .01), and Th2 cells (P < .05) were higher in the high expression group than in the low expression group. However, the levels of activated T-cells (P < .05), cytotoxic cells (P < .05) and B cells (P < .001) were lower in the high FADS3 expression group. Analysis of the relationship between immune cells and OS in HNSCC patients revealed that only reduced B cell levels correlated with HNSCC survival (Fig. 3C, P = .045). The expression levels of CD8 + T cells, CD4 + T cells, macrophages, neutrophils and dendritic cells showed no significant difference in the survival time of HNSCC patients. Analysis of the correlations between the 24 immune cell types revealed weak to moderate correlation between tumor-infiltrating immune cell subsets (Fig. 3D).
Figure 3.
(A) Correlations between FADS3 mRNA expression and immune infiltration levels. (B) The varied proportions of 24 subtypes of immune cells in high and low FADS3 mRNA expression groups in tumor samples. (C) Relationship between the expression levels of 6 kinds of immune cells and the survival time of HNSCC patients. (D) Heatmap of 23 immune infiltration cells in tumor samples. FADS3 = fatty acid desaturase 3, mRNA = messenger ribonucleic acid, HNSCC = head and neck squamous cell carcinoma.
3.5. Data validation
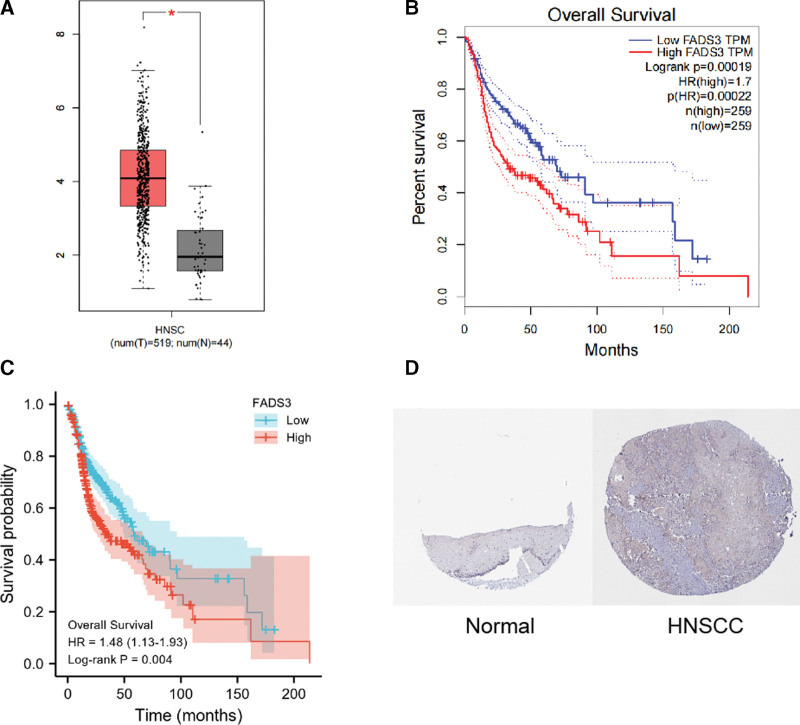
It was found in the GEPIA database that FADS3 messenger ribonucleic acid (mRNA) expression in HNSCC was significantly higher than in the normal group (P < .05, |log2FC| > 1) (Fig. 4A), and there were differences between FADS3 messenger ribonucleic acid (mRNA) level and a shorter OS period significant correlation (P-value < .001, Fig. 4B). This was confirmed using KM analysis, which showed that high FADS3 expression correlated with poor OS (P = .004, Fig. 4C). Moreover, HPA (immunohistochemical) analysis showed that FADS3 levels in tumor tissue were higher than in non-tumor tissue (Fig. 4D).
Figure 4.
(A) FADS3 mRNA expression levels in normal and HNSCC tissues, as obtained from GEPIA. (B) Levels of FADS3 mRNA expression and overall survival based on data obtained from GEPIA. (C) Further validation of the correlation between FADS3 expression and overall survival, as shown in K–M survival plot. (D) Oral mucosa cells expression of FADS3 protein was visualized using immunohistochemistry via HPA. FADS3 = fatty acid desaturase 3, K–M = Kaplan–Meier, mRNA = messenger ribonucleic acid, HNSCC = head and neck squamous cell carcinoma, GEPIA = gene expression profiling interactive analysis, HPA = Human Protein Atlas.
4. Discussion
HNSCC has a high incidence and is associated with poor outcomes.[15] Hence, effective biomarkers for identifying HNSCC patients with poor prognosis are urgently needed. FADS3 is known to be involved in sphingase biosynthesis and has also been implicated in cancer development.[6,8,16] High sphingolipid levels have been associated with HNSCC development and poor prognosis,[17–19] suggesting that FADS3 influences HNSCC pathogenesis. Here, analysis of the relationship between FADS3 and HNSCC pathogenesis and immune cell infiltration found that FADS3 has independent prognostic value for HNSCC. Our data show that HNSCC patients with high FADS3 levels are more likely to have higher lymph node metastasis, histological stage, and lymphovascular invasion when compared to those with low FADS3 levels. Elevated FADS3 levels may affect HNSCC tumorigenesis and immune responses to HNSCC.
GO and KEGG pathway analyses showed that FADS3 upregulation mainly correlates with fiber movement, immune response, and amino acid metabolism. Our findings show that FADS3 overexpression in HNSCC patients may alter the immune microenvironment, migration and invasion of cancer cells, and amino acid metabolism. In the HNSCC tumor microenvironment, anti-tumor immune responses are highly immunosuppressed.[20,21] Our data indicate that FADS3 influences the immune microenvironment of HNSCC. Arachidonic acid metabolism is also implicated in tumorigenesis and its expression in HNSCC is markedly lower than in normal tissues.[22,23] Thus, we speculated that FADS3 changes inhibit amino acid metabolism. Further studies should determine the mechanisms by which FADS3 influences in HNSCC in vivo. Using TIMER analysis, we revealed the relationship between FADS3 expression and the level of immune invasion in HNSCC. Additionally, using CIBERSORT analysis, we show that FADS3 levels moderately or strongly correlated with immune cells infiltration levels, especially NK cells, eosinophils, cytotoxic cells, B cells, Tgd, Th1 cells, and Th2 cells. Our data show that high FADS3 levels correlated with higher levels of NK cells and Tgd and lower B cells levels. Studies show that B cell levels positively correlate with HNSCC prognosis,[24,25] which is consistent with our findings (Fig. 3C). These results may suggest the cytochrome P450 by which FADS3 regulates B cell function in HNSCC. These findings indicate that in HNSCC, FADS3 plays a key role in the regulation and recruitment of infiltrating immune cells. However, the relationship between FADS3 and B cells needs further investigation.
5. Conclusion
We for the first time show that FADS3 is a potential prognostic factor and diagnostic value in HNSCC. Our findings improve our understanding of the role of FADS3 in HNSCC immune cell infiltration.
Author contributions
Guihong Xuan designed the study and prepared the protocol.
Conceptualization: Guihong Xuan.
Data curation: Kuiwei Su.
Formal analysis: Ying Wang.
Investigation: Hefeng Gu, Lan Ma.
Methodology: Kuiwei Su.
Project administration: Ying Wang, Guihong Xuan.
Resources: Ying Wang, Hefeng Gu.
Software: Kuiwei Su.
Supervision: Lan Ma, Guihong Xuan.
Validation: Lan Ma.
Writing – original draft: Kuiwei Su.
Writing – review & editing: Guihong Xuan.
All the authors scrutinized and confirmed the final protocol.
Abbreviations:
- FADS3 =
- fatty acid desaturase 3
- GEPIA =
- gene expression profiling interactive analysis
- GO =
- gene ontology
- GSEA =
- gene set enrichment analysis
- HNSCC =
- head and neck squamous cell carcinoma
- HPA =
- Human Protein Atlas
- KEGG =
- Kyoto encyclopedia of genes and genomes
- KM =
- Kaplan–Meier
- OS =
- overall survival
- TCGA =
- The Cancer Genome Atlas
- TIMER =
- Tumor IMmune Estimation Resource
This work was supported by Health technology Plan of Zhejiang Province (grant no. 2022KY403 to Guihong Xuan).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
All data in this study can be obtained from The Cancer Genome Atlas (TCGA; https://www.cancer.gov/about-cancer), Tumor IMmune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer/), The Human Protein Atlas (www.proteinatlas.org) and Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html).
How to cite this article: Su K, Wang Y, Gu H, Ma L, Xuan G. Overexpression of fatty acid desaturase 3 predicts poor prognosis in head and neck squamous cell carcinoma. Medicine 2022;101:49(e32119).
Contributor Information
Kuiwei Su, Email: sukuiwei@126.com.
Ying Wang, Email: 04wangying@sina.com.
Hefeng Gu, Email: 740085164@qq.com.
Lan Ma, Email: malan1008@163.com.
Reference
- [1].Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- [2].Uppaluri R, Campbell KM, Egloff AM, et al. Neoadjuvant and adjuvant pembrolizumab in resectable locally advanced, human papillomavirus-unrelated head and neck cancer: a multicenter, phase II trial. Clin Cancer Res. 2020;26:5140–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mody M, Rocco J, Yom S, et al. Head and neck cancer. Lancet (London, England). 2021. [DOI] [PubMed] [Google Scholar]
- [4].Powell SF, Gold KA, Gitau MM, et al. Safety and efficacy of pembrolizumab with chemoradiotherapy in locally advanced head and neck squamous cell carcinoma: a phase IB study. J Clin Oncol. 2020;38:2427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Muzaffar J, Bari S, Kirtane K, et al. Recent advances and future directions in clinical management of head and neck squamous cell carcinoma. Cancers (Basel). 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marquardt A, Stöhr H, White K, et al. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–83. [DOI] [PubMed] [Google Scholar]
- [7].Park WJ, Kothapalli KS, Lawrence P, et al. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS One. 2011;6:e28186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Karsai G, Lone M, Kutalik Z, et al. FADS3 is a Δ14Z sphingoid base desaturase that contributes to gender differences in the human plasma sphingolipidome. J Biol Chem. 2020;295:1889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Henriquez-Henriquez M, Acosta MT, Martinez AF, et al. Mutations in sphingolipid metabolism genes are associated with ADHD. Transl Psychiatry. 2020;10:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA). Methods Mol Biol. 2016;1418:111–41. [DOI] [PubMed] [Google Scholar]
- [11].Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lánczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–46. [DOI] [PubMed] [Google Scholar]
- [15].Johnson D, Burtness B, Leemans C, et al. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Blanchard H, Legrand P, Pédrono F. Fatty acid desaturase 3 (Fads3) is a singular member of the Fads cluster. Biochimie. 2011;93:87–90. [DOI] [PubMed] [Google Scholar]
- [17].Li L, Yang Q, Jiang Y, et al. Interplay and cooperation between SREBF1 and master transcription factors regulate lipid metabolism and tumor-promoting pathways in squamous cancer. Nat Commun. 2021;12:4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].da Silva G, de Matos LL, Kowalski LP, et al. Profile of sphingolipid-related genes and its association with prognosis highlights sphingolipid metabolism in oral cancer. Cancer Biomark. 2021;32:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Faedo RR, da Silva G, da Silva RM, et al. Sphingolipids signature in plasma and tissue as diagnostic and prognostic tools in oral squamous cell carcinoma. Biochim Biophys Acta Mol Cell Biol Lipids. 2022;1867:159057. [DOI] [PubMed] [Google Scholar]
- [20].Theodoraki MN, Yerneni SS, Hoffmann TK, et al. Clinical significance of PD-L1(+) exosomes in plasma of head and neck cancer patients. Clin Cancer Res. 2018;24:896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mahata B, Pramanik J, van der Weyden L, et al. Tumors induce de novo steroid biosynthesis in T cells to evade immunity. Nat Commun. 2020;11:3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Park SW, Heo DS, Sung MW. The shunting of arachidonic acid metabolism to 5-lipoxygenase and cytochrome p450 epoxygenase antagonizes the anti-cancer effect of cyclooxygenase-2 inhibition in head and neck cancer cells. Cell Oncol (Dordr). 2012;35:1–8. [DOI] [PubMed] [Google Scholar]
- [23].Wang J, Li X, Xu O, et al. [The role and clinical significance of 12-LOX passway in arachidonic acid metabolism induced by phospholipase Cgamma-2 in laryngeal squamous cell carcinoma]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2013;27:1355–9. [PubMed] [Google Scholar]
- [24].Ruffin AT, Cillo AR, Tabib T, et al. B cell signatures and tertiary lymphoid structures contribute to outcome in head and neck squamous cell carcinoma. Nat Commun. 2021;12:3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim SS, Shen S, Miyauchi S, et al. B cells improve overall survival in HPV-associated squamous cell carcinomas and are activated by radiation and PD-1 blockade. Clin Cancer Res. 2020;26:3345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]