Abstract
The gut microbiome has been increasingly suggested as an underlying cause of various human diseases. In this study, we hypothesized that the gut microbiomes of patients with familial adenomatous polyposis (FAP) are different from those of healthy people and attempted to identify the associations between gut microbiome characteristics and FAP.
We collected fecal samples from patients with FAP and healthy volunteers and evaluated the diversity, composition, and distribution of the gut microbiome between the 2 groups via 16S rRNA-based taxonomic profiling of the fecal samples.
Fecal samples were collected from 10 patients with FAP (4 men and 6 women, mean age 39.2 ± 13.8 years) and 10 healthy volunteers (4 men and 6 women, mean age 40.9 ± 9.8 years). The microbial richness in patients with FAP was significantly lower than that in healthy people. Regarding microbial composition, the Firmicutes/Bacteroidetes ratio in patients with FAP was higher than that in healthy people, especially in those with a lower proportion of Bacteroidetes and a higher proportion of Proteobacteria. We also found 7 specific abundant strains in fecal samples of patients with FAP.
Patients with FAP had different Firmicutes/Bacteroidetes ratios and Proteobacteria abundance compared to healthy people and showed the presence of specific bacteria. These findings suggest a promising role of the gut microbiome in patients with FAP, although further studies are needed.
Keywords: familial adenomatous polyposis, gut microbiome, healthy people
1. Introduction
Familial adenomatous polyposis (FAP) is an autosomal dominant genetic disorder where hundreds to tens of thousands of adenomas occur in the colon and rectum, and almost 100% of these colorectal cancers (CRC) occur before the age of 40.[1] A mutation in the adenomatous polyposis coli (APC) gene on chromosome 5q21 is responsible for FAP.[2] Patients with FAP can suffer from several cancers including CRC, duodenal cancer, skin cancer, bone cancer, connective tissue cancer, thyroid cancer, and pancreatic cancer. Therefore, a screening program is essential to prevent life-threatening diseases.[3] Unfortunately, there are few appropriate therapeutic agents that prevent cancer in patients with FAP, and these patients typically develop cancer and then require surgery. A few medications, including metformin and celecoxib, have been suggested for reducing multiple polyps that occur in the gastrointestinal tract of patients with FAP, but these have a nonsignificant effect in clinical practice.[4]
Recently, the gut microbiome has been implicated as an underlying cause of several diseases, including irritable bowel syndrome, inflammatory bowel disease, CRC, rheumatoid arthritis, type 2 diabetes, and obesity.[5] Increasing evidence shows the important role of the gut microbiome in the colorectal carcinogenesis and treatment.[6] Inflammation, immune regulation, metabolism of dietary components, and genotoxin production as main mechanisms in colorectal carcinogenesis are closely linked to the gut microbiome.[6] Several gut microbiome have been expected to play an important role in mediating tumor responses to chemotherapy and immunotherapy in patients with melanoma and lung cancers, affecting the activation of the immune system and tumor responses to treatment.[7–9] A study carried out in APCMin/+ mice presented that the gut microbiome of APC gene-deficient mice differed from that of normal mice and a mutation of the APC gene alters colonic-microbial interactions prior to polyposis.[10] A recent study showed that the Escherichia coli and Bacteroides fragilis strains were characteristically dominant in the colonic mucosa in patients with FAP and genes for colibactin (clbB) and Bacteroides fragilis (bft) were highly expressed in FAP patients’ colonic mucosa compared to healthy individuals.[11] However, there are still insufficient data to clarify the role of the gut microbiome in patients with FAP.
In this study, we hypothesized that the gut microbial diversity and composition of patients with FAP differed from healthy people, and that the gut microbial characteristics of patients with FAP would contribute to the occurrence of numerous colorectal polyps and several cancerous diseases. Herein, we investigated the differences in gut microbiome between patients with FAP and healthy people, and we identified the clinical significance of the gut microbiome in patients with FAP.
2. Methods
2.1. Subjects and stool collection
Between April 2020 and Mar 2021, patients with FAP and healthy volunteers were enrolled in this study, conducted at Kosin University Gospel Hospital, in Busan, Korea. For healthy volunteers, patients with a history of abdominal surgery, inflammatory bowel disease, or cancer were excluded, and subjects who had taken laxatives, metoclopramide, tegaserod, a proton pump inhibitor, or antibiotics within the month prior to sampling were also excluded. A study investigator explained the study aims and procedures to all participants. Each participant provided written informed consent before enrollment. General patient information, including age, sex, and medical history, was recorded. The subjects were asked to collect their stool using a fecal sample collector kit (Medi4U®, Incheon, Korea) at home and to store it immediately after collection at -20°C in a freezer. Samples were transported to Kosin University Gospel Hospital, enclosed in an insulated foil pack with dry ice, and stored at -80°C in a deep freezer. All samples were transported to Theragen Bio Inc. (Gyeonggi-do, Korea) for analysis. The study protocol was approved by the Kosin University Gospel Hospital (IRB No. KUGH 2020-03-023).
2.2. Sample preparation and data analysis
Fecal DNA was extracted from 200 mg of the stool sample using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quality of the extracted genomic DNA was determined using NanoDrop (NanoDrop 2000c; Thermo Fisher Scientific, Waltham, MA). DNA concentration was measured using an ultraviolet-vis spectrophotometer (Thermo Fisher Scientific) and quantified using the QuantiFluor® ONE dsDNA System (Promega, Madison, WI); thereafter, DNA was stored at -80°C until 16S rDNA library preparation. Library preparation was performed according to the standard instructions of the 16S Metagenomic Sequencing Library Preparation protocol (IlluminaTM, Inc., San Diego, CA). The V3-V4 region of the bacterial 16S rRNA gene was amplified using aliquots of isolated DNA from each sample. The V3 to V4 region was amplified using the following 341F-805R primers.
16s-341F: TCGTCGGCA GCGTCAGAT GTGTATAAGA GACAGCCTAC GGGNGGC WGCAG 16s-805R: GTCTCGTGG GCTCGGAGATG TGTATAAGAGACAGG ACTACHVG GGTATCT AATCC
Polymerase chain reaction products were purified using AMPure XP magnetic beads (Beckman Coulter, Brea, CA) on the DynaMag-96 Side Magnet (Thermo Fisher Scientific). The quality of purified products was controlled using the Agilent Bioanalyzer (Agilent, Santa Clara, CA). A secondary amplification to attach the Illumina Nextera barcodes (Illumina, Inc., San Diego, CA) was then carried out using the i5 forward primer and the i7 reverse primer. The pooled libraries were sequenced using the Illumina MiSeq platform in a 2 × 300 bp paired-end run (Illumina). Reads were sorted using unique barcodes for each polymerase chain reaction product. The barcode, linker, and primer sequences were then removed from the original sequencing reads. Any reads containing 2 or more ambiguous nucleotides, those with a low quality score (average score < 25), or reads shorter than 300 bp were filtered out. Potential chimeric sequences were detected using the Bellerophon method.[12]
2.3. Statistical analysis
Pre-processed reads from each sample were used to calculate the number of operational taxonomic units (OTUs), which was determined by clustering the sequences from each sample using a 97% sequence identity cutoff with QIIME software (v.1.8.0).[13,14] Taxonomic classification was carried out using the RDP (Ribosomal Database Project) database. Non-archaeal/bacterial sequences were removed according to taxonomic classification results. Taxonomic abundance was counted with RDP Classifier v1.1 using a confidence threshold of 0.8 derived from the pre-processed reads for each sample. Microbial composition was normalized using the value calculated from the taxonomy abundance count divided by the number of pre-processed reads for each sample. Alpha- and beta-diversity analyses were used to assess biodiversity based on OTUs and visualized by ggplot in R software package (v4.0.3). Linear discriminant analysis effect size was evaluated to determine the features most likely to explain differences between classes by coupling standard tests for statistical significance using the Galaxy module (https://huttenhower.sph.harvard.edu/galaxy).
3. Results
3.1. Baseline characteristics of patients with FAP and of healthy people
Fecal samples were collected from a total of 10 patients with FAP (4 men and 6 women, aged 39.2 ± 13.8 years) and 10 healthy volunteers (4 men and 6 women, aged 40.9 ± 9.8 years). The baseline characteristics of patients with FAP and healthy volunteers are summarized in Table 1 and Table S1, Supplemental Digital Content, http://links.lww.com/MD/I69, respectively. Among the 10 patients with FAP, 4 had a mutation on exon 15 of APC gene, 1 had a mutation on exon 14 of APC gene, 3 had a mutation on exon 9 of APC gene, and 1 had a mutation on exon 6 of APC gene (1 patient had no data for mutations of APC gene). Among the patients with an APC mutation, F1 and F2 are siblings, F6 is the father of F7, and F8 is the mother of F9. Of the 10 patients, 5 underwent surgery; 2 had rectal cancers, and 3 had numerous colorectal polyps. Extraintestinal manifestations including papillary thyroid cancer, congenial hypertrophy of the retinal pigment epithelium, desmoid tumor, and gallbladder adenoma were present in 4 patients.
Table 1.
Characteristics of patients with FAP.
| No. | Sex/age | BMI (kg/m2) | ABO type | Alcohol history | Current smoker | Co-morbidities | Family history (FAP) | APC mutation | Cancer | Operation | TNM stage | EIM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | M/29 | 18.8 | O | Yes | Yes | None | Father | Exon 15 (codon 951) | None | No | N/A | None |
| F2 | F/25 | 18.3 | AB | No | No | None | Father | Exon 15 (codon 951) | None | No | N/A | PTC |
| F3 | F/27 | 21.1 | A | No | No | None | None | Exon 14 (codon 653) | None | No | N/A | CHRPE |
| F4 | M/46 | 23.1 | AB | No | No | None | Mother | Exon 6 (codon 646) | None | Yes | T0N0M0 | Desmoid tumor |
| F5 | F/42 | 25.9 | A | No | No | None | Mother | N/A | None | Yes | T0N0M0 | None |
| F6 | M/54 | 16.9 | B | Yes | No | None | Mother | Exon 15 (codon 759) | Rectum | Yes | T3N2M0 | None |
| F7 | M/23 | 29.7 | B | No | No | None | Father | Exon 15 (codon 759) | None | Yes | T0N0M0 | None |
| F8 | F/59 | 28.8 | B | Yes | No | None | Father | Exon 9 (codon 358) | None | No | N/A | GB adenoma |
| F9 | F/28 | 18.5 | O | No | No | None | Mother | Exon 9 (codon 358) | None | No | N/A | None |
| F10 | F/59 | 22.9 | O | Yes | No | HTN | Mother | Exon 9 (codon 405) | Rectum | Yes | T2N0M0 | None |
APC = adenomatous polyposis coli, N/A = not available, BMI = body mass index, CHRPE = congenital hypertrophy of the retinal pigment epithelium, EIM = extraintestinal manifestation, FAP = familial adenomatous polyposis, GB = gallbladder, HTN = hypertension, PTC = papillary thyroid cancer.
3.2. Differences in gut microbial diversity and composition between patients with FAP and healthy people
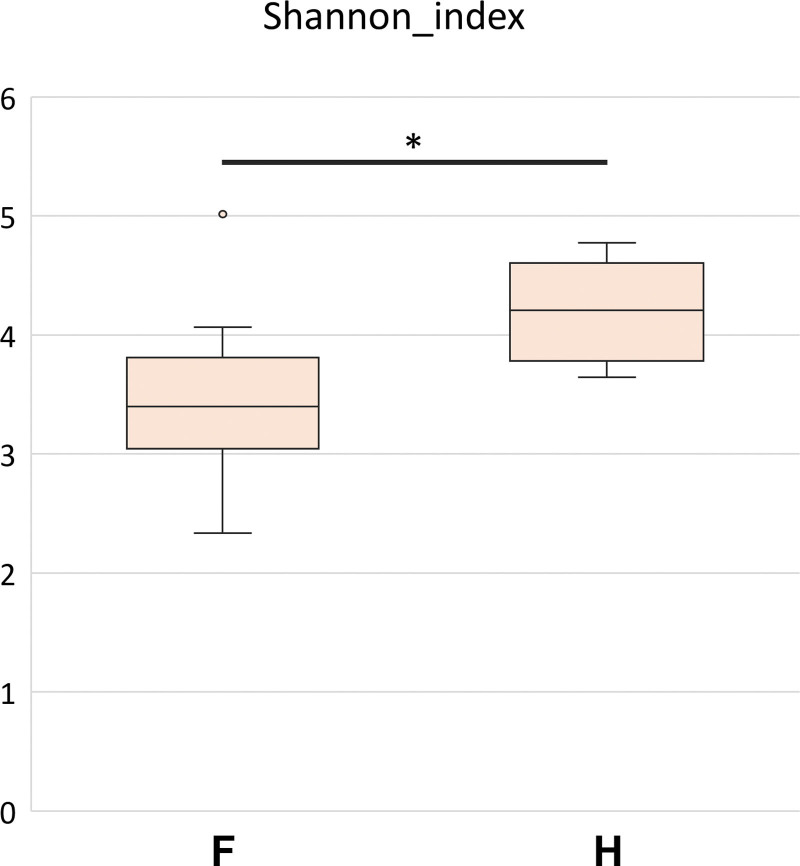
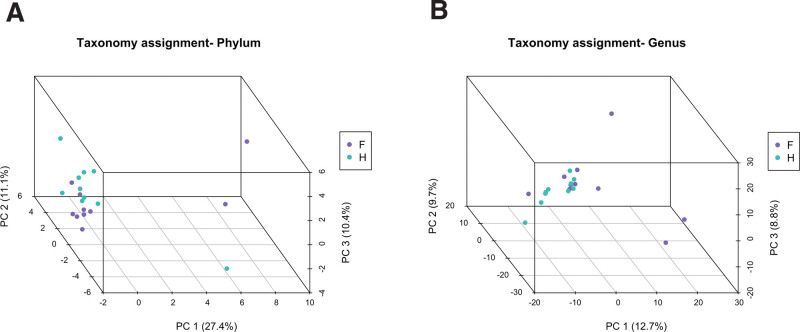
To examine the microbial richness and distribution of fecal samples, we assessed α-diversity and β-diversity. As shown in Figure 1, α-diversity is presented using the Shannon index and was significantly higher in healthy people than in patients with FAP. In the analysis of β-diversity using Bray-Curtis dissimilarity, the samples showed different diversity patterns between patients with FAP and healthy people (Fig. 2a, b).
Figure 1.
Box plot of the alpha-diversity of bacterial communities in the 2 groups – patients with familial adenomatous polyposis (F) and healthy people (H). The box plot presents the full range of values obtained from the source data. The ggplot package for R was used for visualization (*P < .05).
Figure 2.
Beta-diversity visualized using the non-metric multidimensional scaling plot with Bray-Curtis dissimilarity distances of the 2 groups – patients with familial adenomatous polyposis (F, violet color) and healthy people (H, sky-blue color) at the phylum and genus level.
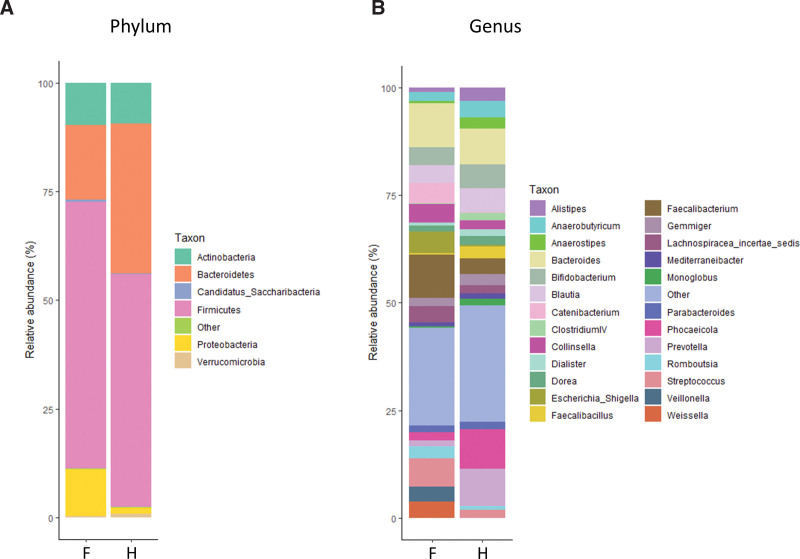
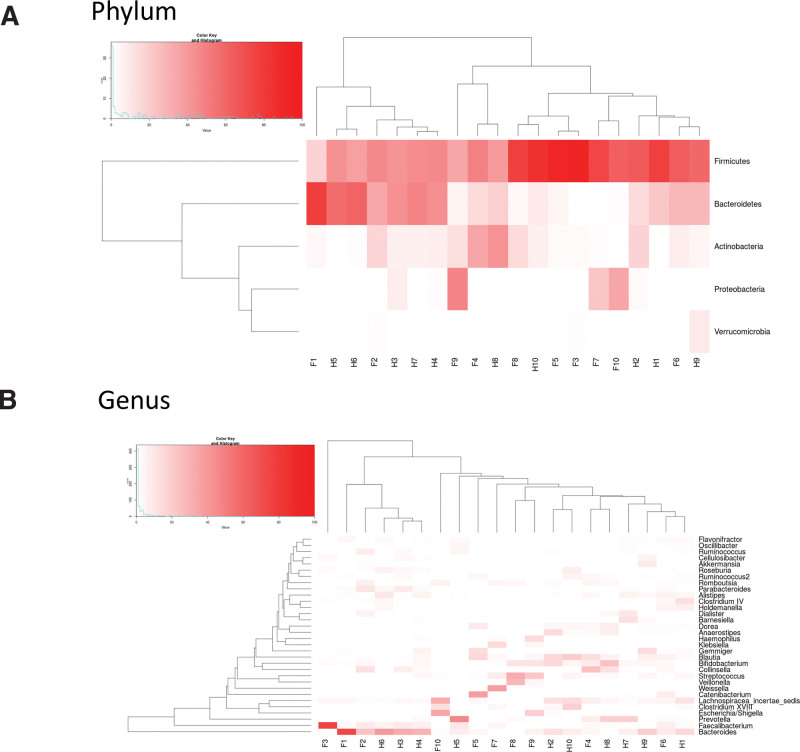
When comparing microbial composition at the phylum level, we found that the Firmicutes/Bacteroidetes ratio in patients with FAP was higher than in healthy people. In addition, Proteobacteria in patients with FAP was relatively more abundant than in healthy people (Fig. 3a). At the genus level, Catenibacterium, Collinsella, Escherichia_Shigella, Faecalibacterium, Streptococcus, Veillonella, and Weissella in patients with FAP were more abundant than in healthy people (Fig. 3b). Microbial composition is also presented using heatmap analysis. Similar to the results presented as a stacked bar chart, the individual fecal samples of patients with FAP tended to have a lower abundance of Bacteroidetes and a higher abundant of Proteobacteria compared with healthy people at the phylum level (Fig. 4a). The results of heatmap analysis at the genus level are presented in Figure 4b and are similar to the results shown in the stacked bar chart.
Figure 3.
Comparison of bacterial composition of the 2 groups presented using stacked bar charts – patients with familial adenomatous polyposis (F) and healthy people (H). (a) Relative abundance at the phylum level. (b) Relative abundance at the Genus level.
Figure 4.
Comparison of bacterial composition of the 2 groups presented in a heat map – patients with familial adenomatous polyposis (F) and healthy people (H). (a) Two-dimensional representation of data at the phylum level. (b) Two-dimensional representation of data at the genus level. Each number indicates an individual participant.
3.3. Comparison of a specific gut microbiome of patients with FAP and healthy people
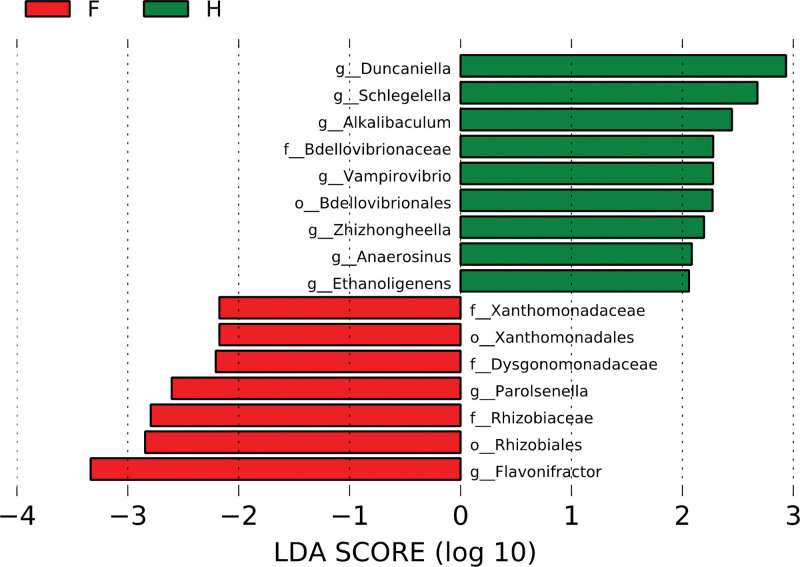
We estimated Linear discriminant analysis effect size to compare the differential abundance of each gut microbiome between FAP patients and healthy people (threshold 2.0). As shown in Figure 5, f_Xanthomonadaceae, o_Xanthomonadales, f_Dysgonomonadaceae, g_Parolsenella, f_Rhizobiaceae, o_Rhizobiales, and g_Flavonifractor gnavus were enriched in patients with FAP. In contrast, g_Duncaniella, g_Schlegelella, g_Alkalibaculum, f_Bdellovibrionaceae, g_Vampirovibrio, o_Bdellovibrionales, g_Zhizhongheella, g_Anaerosinus, and g_Ethanoligenens were enriched in healthy people. Next, we explored each sample’s composition of gut microbiome using Krona plots, which use multilevel and zoomable pie charts to visualize both the most abundant organisms and their most specific classification. As shown in Figure S1, Supplemental Digital Content , http://links.lww.com/MD/I70, the Krona plots of healthy people were generally similar, with a majority of Firmicutes and Bacteroidetes, except in a few instances. Meanwhile, the Krona plots of patients with FAP showed different patterns from healthy people, and some included Proteobacteria and Actinobacteria. Interestingly, the Krona plots for the FAP patients who had the same mutation of APC gene did not show the same patterns (F1/ F2, F6/ F7, and F8/ F9).
Figure 5.
Prominent gut microbiota presented by LDA score of patients with familial adenomatous polyposis (F) and healthy people (H). These data were assessed according to a threshold of 2.0. Taxon level names are abbreviated as p-phylum; c-class; o-order; f-family, and g-genus. LDA = linear discriminant analysis.
4. Discussion
Emerging evidence has shown the crucial role of gut microbiome in the development of colorectal adenomas and CRC. However, data on how gut microbiome contribute to development of colorectal adenoma and CRC in patients with FAP are lacking. In this study, we observed different gut microbial richness and composition between patients with FAP and healthy people, and the Firmicutes/Bacteroidetes ratio in patients with FAP was higher than in healthy people, particularly characterized by a lower proportion of Bacteroidetes and a higher proportion of Proteobacteria. We also observed that several specific gut microbiome entities, including f_Xanthomonadaceae, o_Xanthomonadales, f_Dysgonomonadaceae, g_Parolsenella, f_Rhizobiaceae, o_Rhizobiales, and g_Flavonifractor gnavus, were enriched in patients with FAP compared with healthy people. These findings suggest that the characteristic gut microbial diversity and composition of patients with FAP, which differ from those of healthy people, influence multiple colorectal adenoma and CRC.
Microbial richness in the human gut is an important parameter in host-microbe symbiosis.[15] Lower bacterial diversity has been observed in people with inflammatory bowel disease, psoriatic arthritis, type 1 and 2 diabetes, atopic eczema, celiac disease, obesity, and arterial stiffness compared with healthy controls.[16] Recent studies have reported that patients with CRC present lower microbial richness in fecal samples and intestinal mucosa compared with healthy people.[17,18] Gut microbial diversity is likely a generally good indicator of a “healthy gut,”[16,19] although greater richness is not always a sign of healthy gut microbiota and can alternatively be due to overgrowth of a variety of harmful bacteria or archaea.[20] Considering that our data showed lower gut microbial richness in patients with FAP than in healthy people, we posit a link between disease occurrence in patients with FAP and lower abundance of the gut microbiome.
Bacteroidetes and Firmicutes are common dominant phyla of bacteria in the gut, generally comprising half or more of the population.[21,22] Bacteroidetes mostly colonize the colon and participate in harvesting energy from the diet through fermentation of indigestible polysaccharides.[23,24] Several studies reported a higher Firmicutes/Bacteroidetes ratio in obese people compared with lean controls.[25–27] The Firmicutes/Bacteroidetes ratio increases from birth to adulthood and decreases from adulthood to elderly age, and this alteration might be involved in metabolic diseases.[28] Proteobacteria is one of the most abundant phyla in the human gut microbiome and contains several human pathogenic genera including Brucella, Rickettsia, Neisseria, Escherichia, Shigella, Salmonella, Yersinia, and Helicobacter. Proteobacteria are often overexpressed in intestinal and extraintestinal inflammatory diseases, although casualty is not fully understood.[29] Shen et al found a higher abundance of Proteobacteria and a lower abundance Bacteroidetes in colorectal adenoma cases compared with controls.[30] Our finding that patients with FAP had a lower proportion of Bacteroidetes and a higher proportion of Proteobacteria compared with healthy people could help confirm the oncogenic role of Bacteroidetes and Proteobacteria in patients with FAP, although further studies are needed.
We identified specific characteristics of gut microbiota in patients with FAP. As shown in Figure 5, 7 OTUs were more abundant in patients with FAP than healthy people. Conversely, 9 OTUs were more abundant in healthy people than patients with FAP. Of the strains that were more abundant in patients with FAP, f_Xanthomonadaceae, o_Xanthomonadales, f_Rhizobiaceae, and o_Rhizobiales belong to the Proteobacteria phylum; f_Dysgonomonadaceae belongs to Bacteroidetes; g_Parolsenella belongs to Actinobacteri; and g_Flavonifractor gnavus belongs to Firmicutes. Of the strains that were more abundant in healthy people, g_Schlegelella, f_Bdellovibrionaceae, o_Bdellovibrionales, and g_Zhizhongheella belong to the Proteobacteria phylum; g_Duncaniella belongs to Bacteroidetes; g_Vampirovibrio belongs to Cyanobacteria; and g_Alkalibaculum, g_Anaerosinus, and g_Ethanoligenens belong to Firmicutes. Although the function of each bacterium is uncertain, these results suggest the crucial role of a specific microbiome in patients with FAP and healthy people, respectively.
This study has several limitations. First, the number of patients in this study was small, which could reduce the reliability of our results. To overcome this limitation, further prospective studies with larger patient groups are needed. Second, 5 FAP patients who underwent the operation were included in the analysis of this study. Since the patients who underwent operation have no remaining colon, it was unlikely that our data represent the results of the entire gut microbiome of FAP patients. Third, patients included in our study had mutations in various locations of the APC gene including on exon 15, exon 14, exon 9, and exon 6, even 1 had no APC gene test data. We could not evaluate the characteristics of gut microbiome according to the mutation of each exon, because the number of patients with mutations in each exon was small. Fourth, this study was trial included only Korean participants and was conducted in a single institution. Therefore, the results may not be directly generalized to populations of other institutions and countries.
In summary, this study found that patients with FAP have different gut microbial richness, distribution, and composition compared with healthy people. We also identified specific bacteria existing in the fecal samples of patients with FAP. These results suggest a promising role of gut microbiome evaluation for screening and treating patients with FAP, although more investigations are needed. Our findings illustrate another potential means by which the gut microbiome can influence the development of colorectal adenoma and CRC in patients with FAP.
Author contributions
Conceptualization: Jae Hyun Kim, Yeon Ji Kim.
Data curation: Yeon Ji Kim.
Funding acquisition: Jae Hyun Kim.
Investigation: Yeon Ji Kim, Gyu Man Oh, Woohyuk Jung.
Methodology: Gyu Man Oh, Woohyuk Jung.
Supervision: Seun Ja Park.
Visualization: Jae Hyun Kim.
Writing – original draft: Jae Hyun Kim, Yeon Ji Kim.
Writing – review & editing: Jae Hyun Kim, Gyu Man Oh, Woohyuk Jung, Seun Ja Park.
Supplementary Material
Abbreviations:
- APC =
- adenomatous polyposis coli
- CRC =
- colorectal cancers
- FAP =
- familial adenomatous polyposis,
- OTUs =
- operational taxonomic units
This study was supported by National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT), No. 2020R1C1C1012694.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental Digital Content is available for this article.
The authors have no conflicts of interest to disclose.
How to cite this article: Kim JH, Kim YJ, Oh GM, Jung W, Park SJ. How is gut microbiome of patients with familial adenomatous polyposis different from healthy people?. Medicine 2022;101:49(e32194).
Contributor Information
Yeon Ji Kim, Email: lovelover9@naver.com.
Gyu Man Oh, Email: kyumangeoje6669@gmail.com.
Woohyuk Jung, Email: jungwh85@gmail.com.
Seun Ja Park, Email: parksj6406@daum.net.
References
- [1].Powell SM, Petersen GM, Krush AJ, et al. Molecular diagnosis of familial adenomatous polyposis. N Engl J Med. 1993;329:1982–7. [DOI] [PubMed] [Google Scholar]
- [2].Leoz ML, Carballal S, Moreira L, et al. The genetic basis of familial adenomatous polyposis and its implications for clinical practice and risk management. Appl Clin Genet. 2015;8:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vasen HF, Möslein G, Alonso A, et al. Guidelines for the clinical management of familial adenomatous polyposis (FAP). Gut. 2008;57:704–13. [DOI] [PubMed] [Google Scholar]
- [4].Walcott FL, Patel J, Lubet R, et al. Hereditary cancer syndromes as model systems for chemopreventive agent development. Semin Oncol. 2016;43:134–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. [DOI] [PubMed] [Google Scholar]
- [6].Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690–704. [DOI] [PubMed] [Google Scholar]
- [7].Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- [10].Son JS, Khair S, Pettet DW, 3rd, et al. Altered interactions between the gut microbiome and colonic mucosa precede polyposis in APCMin/+ mice. PLoS One. 2015;10:e0127985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–9. [DOI] [PubMed] [Google Scholar]
- [13].Chiu CM, Huang WC, Weng SL, et al. Systematic analysis of the association between gut flora and obesity through high-throughput sequencing and bioinformatics approaches. Biomed Res Int. 2014;2014:906168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zaura E, Keijser BJ, Huse SM, et al. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79. [DOI] [PubMed] [Google Scholar]
- [16].Valdes AM, Walter J, Segal E, et al. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Saffarian A, Mulet C, Regnault B, et al. Crypt-and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio. 2019;10:e01315–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sommer F, Anderson JM, Bharti R, et al. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–8. [DOI] [PubMed] [Google Scholar]
- [20].Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- [21].Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22:763–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–42. [DOI] [PubMed] [Google Scholar]
- [25].Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- [27].Million M, Lagier JC, Yahav D, et al. Gut bacterial microbiota and obesity. Clin Microbiol Infect. 2013;19:305–13. [DOI] [PubMed] [Google Scholar]
- [28].Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rizzatti G, Lopetuso L, Gibiino G, et al. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017:9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.