Abstract
Neurons, especially when coupled with muscles, allow animals to interact with and navigate through their environment in ways unique to life on earth. Found in all major animal lineages except sponges and placozoans, nervous systems range widely in organization and complexity, with neurons possibly representing the most diverse cell-type. This diversity has led to much debate over the evolutionary origin of neurons as well as synapses, which allow for the directed transmission of information. The broad phylogenetic distribution of neurons and presence of many of the defining components outside of animals suggests an early origin of this cell type, potentially in the time between the first animal and the last common ancestor of extant animals. Here, we highlight the occurrence and function of key aspects of neurons outside of animals as well as recent findings from non-bilaterian animals in order to make predictions about when and how the first neuron(s) arose during animal evolution and their relationship to those found in extant lineages. With advancing technologies in single cell transcriptomics and proteomics as well as expanding functional techniques in non-bilaterian animals and the close relatives of animals, it is an exciting time to begin unraveling the complex evolutionary history of this fascinating animal cell type.
Keywords: choanoflagellates, cnidarians, ctenophores, placozoans, sponges, synapses
Introduction
Consistent with an early evolutionary origin, many of the core characteristics and molecular protein complexes found in neurons predate animal multicellularity. Neurons are highly polarized cells and rely on polarized secretion during differentiation and neurite development [1–4]. Asymmetric distribution of proteins and cellular structures is a pan-eukaryotic characteristic and the core components of polarized secretion show deep conservation [5–13]. One of the defining characteristics of bilaterian neurons is the localized, rapid, and highly synchronous release of vesicle contents. As the core components and major trafficking of vesicles is conserved, this regulated secretion appears to be an extreme example of constitutive polarized secretion [14]. Important factors in this form of exocytosis are the accumulation of vesicles in close proximity to the membrane and the tight coupling of calcium influx to fusion events [15–18]. This probably represents a key step in the evolution of neuronal signaling. The evolutionary connection between neurons and secretory cells has been nicely reviewed in [19], and we postulate that the assembly of pre-animal sensory, excretory, and signaling complexes paved the way for the first true neurons to arise from sensory-secretory cells following the occurrence of constitutive multicellularity.
Deep conservation of the components of neurons in unicellular eukaryotes
Unlike constitutive exocytosis, regulated secretion relies on maintaining vesicle pools near the plasma membrane while actively preventing fusion from occurring. This is achieved through the activity of various proteins including RIM, Unc13, complexin, and synaptotagmin 1, some of which can be found in the close unicellular relatives to animals [18,20–23] (Figure 1). The clustering of voltage-gated calcium channels (VGCC) at synapses allows for rapid and localized increases in calcium concentration, triggering synchronize vesicle release [15,24]. VGCC predate animal multicellularity, including orthologs of Cav1/2 and Cav3 found in the choanoflagellate (the sister group to animals) Salpingoeca rosetta [25]. The activity of these channels and their function in these organisms are yet to be studied. In animals, clustering of these channels is achieved by Cast/ELKS proteins [15], which are animal specific [23]. However, loss-of-function studies show a decrease, but not complete ablation of calcium-dependent fusion events [26]. The major Ca2+ sensor in neurons, synaptotagmin 1, has not been identified in unicellular eukaryotes [27,28], though, closely related Ca2+ sensors, extended-synaptotagmins, have been identified in animals, yeasts, and plants [29,30]. These proteins have a similar domain organization, but with an additional Synaptotagmin-like, Mitochondrial and Lipid-Binding protein (SMP) domain, as well as different subcellular localization and function in studied organisms [30–32]. With the presence of so many active zone proteins and of polarized and enriched vesicle pools in choanoflagellates [33], it is tempting to say these are sites of regulated exocytosis. However, this remains to be functionally tested.
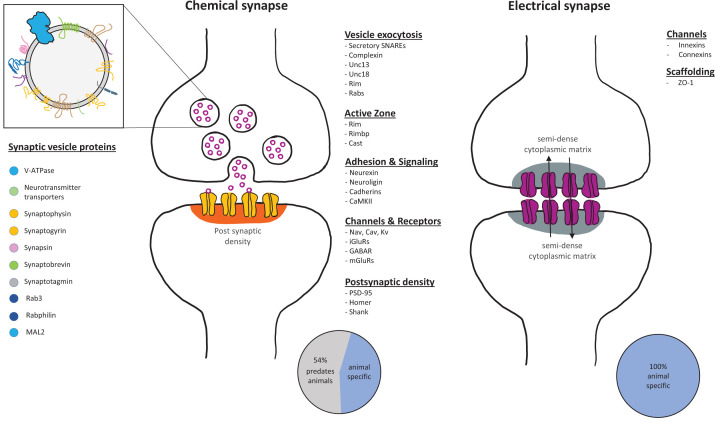
Figure 1. Major components of chemical and electrical synapses.
Chemical synapses are composed of a rich repertoire of conserved proteins involved in bringing the pre- and post-synapse into close proximity, tightly regulating vesicle exocytosis, and clustering of receptors on the post-synapse. Many of these proteins originated before animal multicellularity [33–35,33–35]. Electrical synapses are primarily established through the interaction of the innexins or connexins, neither of which have been identified outside of animals.
Communication through a chemical synapse requires tight regulation of the release of the signaling molecule as well as tightly ordered receptors to detect it at the post synapse. In early animals, these post synaptic structures likely arose from receptor clusters on sensory cells. As with active zone proteins, many post-synaptic proteins predate animals, such as Homer, Shank, PSD-95, and CamKII [34, 36–38] (Figure 1), as well as a wide repertoire of ion channels, transient receptor potential channels (TRP), and G-protein coupled receptors (GPCRs) [39–41], which could function in detection of signal. However, the function of these proteins in the close relatives to animals remains unresolved.
Flagella/cilia are found throughout eukaryotes and can serve as a major guide in polarity and polarized secretion. In animals, cilia (both motile and non-motile) have specialized sensory function in a variety of cell types, both neuronal and non-neuronal. Examples include flow sensation [42,43], hedgehog signaling [44], photoreception [44,45], and a wide variety of sensory neurons [46,47]. Though not nearly as well conserved as the core set of structural proteins, signaling modules associated with cilia also appear to be ancient, with the function as a sensory organelle arising alongside motility in early eukaryotic evolution [48–51]. Apart from a flagellum, choanoflagellates also have a microvilli collar which functions in prey capture. Though structurally distinct from cilia, actin-based microvilli and stereovilli represent cellular projections with high surface area to volume, making ideal domains for receptor presentation. In animals, these structures can serve to increase nutrient absorption as in the brush border epithelial cells [52] but are also found as sensory structures in cell types such as vertebrate lateral line and cochlear hair cells [53–55], cnidarian hair cells and cnidocytes [56–58], and ctenophore sensory neurons [59]. Though specialized feeding cells, sponge choanocytes express many markers of sensory microvilli in bilaterians [60] and may have a sensory function [61]. The similarity of these structures with those of choanoflagellates has long suggested homology [62–65], though it remains unclear whether subtle differences are the result of large evolutionary distances or convergence [66–68]. The presence of both of these cellular structures in non-animal eukaryotes raises the possibility that the sensory modules associated with them are also ancient. Consistent with this, TRP channels, often associated with ciliary signaling [69–71], localized to the cilia of Chlamydomonas have been shown to function in mating behavior [72] as well as mechanosensation [73]. Technical limitations on functional studies of these channels and signaling molecules in non-animal holozoans (ichthyosporea, pluriformea, filasterea, and choanoflagellatea) are lessening [74–77], making it an exciting time to better understand how organized the sensory components of these structures may have been in the first animals.
At the root of neuronal signaling is cell–cell communication. Cell–cell communication exists in all domains of life and is facilitated by a variety of means (reviewed in [78]). In neurons, small molecules like glutamate and GABA, neuropeptides, and diffusible gases, like nitric oxide, are released at chemical synapses [79,80]. In the mammalian central nervous system glutamate is the most highly utilized chemical messenger (reviewed in [81]). Glutamatergic signaling is likely very ancient and arose out of a balance of osmotic regulation, metabolism, and damage response (reviewed in [41]). Peptidergic signaling involves the release of small peptides, which are processed from large prepropeptide proteins by a variety of well conserved enzymes [82]. One common modification of neuropeptides is the amidization of glycine by peptidylglycine α-amidating monooxygenase (PAM) [82,83]. A functional PAM enzyme has been identified in Chlamydomonas, where it was shown to function in ciliogenesis [84,85]. Amidated peptides have also been identified in ciliary exosomes, which function in mating behavior, suggesting the presence of pepidergic signaling between cells [86]. Orthologs for animal neuropeptide-like molecules have also been identified in choanoflagellates, though functional studies have not yet been performed [87]. Nitric oxide is a small diffusible signal that plays an important role in both plant and animal multicellularity [88–92]. It has been shown to serve a variety of functions in eukaryotes [93] and was recently shown to control collective contraction in a colonial choanoflagellates [94].
Direct intercellular communication also occurs through gap junctions at electrical synapses. Gap junctions are established through the extracellular interaction of innexins/connexins (see below) of two cells forming intercellular channels [95] (Figure 1). So far, orthologs for these proteins have not been identified outside of animals, suggesting gap junctions are a metazoan innovation [96]. However, intercellular bridges are seen between cells of choanoflagellate colonies [68,97,98], which could serve a signaling function. These bridges contain electron dense structures near the body of each cell, though the molecular composition of these is unknown.
Non-bilaterian animals provide key insights into the evolutionary origin of synapses and neurons
Neurons are very ancient cell types, with complex nervous systems present across bilaterians. However, the precise origin is difficult to resolve, partly due to the contentious placement of sponges or ctenophores as the sister group to all other animals [99–105]. Of the non-bilaterian lineages, neurons are present in ctenophores and cnidarians, while they appear to be absent from sponges and placozoans. Despite the absence of bona fide neurons in these two lineages, the molecular toolkits for developing a nervous system are largely present [106–109]. Depending on the placement of sponges or ctenophores, it is assumed that either ctenophores independently gained their nervous system [110], the nervous system was lost in the placozoan (and possibly sponge) lineage(s) [111], or a combination of these two [101,112].
Traditionally thought of as ‘simple’ nerve-nets, cnidarian nervous systems are now considered highly complex (reviewed in [113]). There is evidence that both classical small molecule neurotransmitters, such as glutamate, GABA, and acetylcholine, and neuropeptides [70,114–117] play a role in neurotransmission. However, diverse studies have found that small molecule transmitters or genes involved in their processing do not always localize to neurons and may play a role in developmental patterning [118–120]. Furthermore, many of the genes involved in pre- and post-synaptic guidance and organization in bilaterians are not well conserved and the synaptic toolkit of cnidarians remains ambiguous [113]. On the other hand, cnidarians have a rich repertoire of neuropeptides [121] and the presence of dense core vesicles [122] suggests an important role of peptidergic signaling at chemical synapses.
Ctenophores, fierce predators in marine environments all over the world, use long cilia to propel their body through the water column [67,123,124]. The ctenophore nervous system consists of a polygonal subepithelial nerve net, mesogleal neurons, an aboral-sensory organ, and a variety of putative sensory cells with cellular protrusions (cilia or filopodia) covering the epidermis, the pharynx, and the tentacles [110,125–133]. Although there is no direct experimental evidence yet, the interplay of these neuronal cell types is thought to allow the animal to quickly react to different physical and environmental stimuli. In agreement with this, the aboral-sensory organ acts as a gravity sensor and also can detect changes in light intensity and pressure, as putative pressure sensors and photoreceptor cells have been described in this sensory organ [125,134–137]. Their polygonal nerve net displays a unique structure, as neurites from the same cell occur through a continuous membrane and thus are anastomosed [132]. There is evidence of only very few classical small molecule neurotransmitters in ctenophores [110,138]. Glutamate and glycine are candidates for the main neurotransmitters, as some ionotropic glutamate receptors tested are sensitive to glutamate and glycine [139,140]. In addition to small molecule neurotransmitters, many neuropeptides have recently been identified by de novo predictions in the ctenophore Mnemiposis leidyi, some of which showed a functional effect on swimming speed and muscle contraction [132,141]. Furthermore, there is evidence that neurons and colloblast (specialized secretory cells) share common progenitor cells, strengthening the link between these two cell types [142].
Sponges and placozoans both lack nervous systems and muscles, though both display coordinated body wide behaviors. Placozoans are small, generally flat marine animals with a limited number of identifiable cell types [143]. There is currently strong support for their placement as sister group to cnidarians and bilaterians suggesting their morphological simplicity arises from a secondary reduction [108,144]. The genome of Trichoplax adhaerens contains genes for major components of both small molecule and peptidergic neurosecretion and transmission [108]. Functional work has also shown specific responses to treatment with different neuropeptide-like molecules as well as non-overlapping expression of the endogenous peptides [145,146]. Single cell sequencing reveals overlapping expression of neuropeptide-like precursors, with peptide processing genes, and pre-synaptic regulatory proteins in putative peptidergic cells, which are distinct from digestive gland cell [143]. The occurrence of these two populations of cell types presents a powerful system for understanding subtle differences in secretory cell types.
Sponges are sessile filter feeders, which use ciliary action to draw water through their aquiferous system where bacterial prey and nutrients are filtered out primarily by choanocytes [147]. Like placozoans, sponge genomes and transcriptomes have revealed the presence of proteins involved in generation and secretion of classic neurotransmitters and peptides [107,148–151]. Even with the absence of a nervous system and muscles, sponges have been shown to undergo full body contractions following changes in internal flow rate [152,153]. This behavior is complex, but in general terms it appears the changes in flow are detected by non-motile cilia on cells lining the osculum, major excurrent canal, which results in production of the diffusible molecule NO [154]. Glutamate and GABA treatments have also been shown to induce or inhibit contractions in sponges [155,156]. Apart from small molecule transmitters, recent bioinformatic study has shown the same neuropeptide-like molecules identified in choanoflagellates appear to be conserved in some sponges [87]. Single-cell RNA sequencing has also identified a unique neuroid-like cell in the freshwater sponge Spongillia lacustris. This cell type moves through the choanocyte feeding chambers, expresses many proteins involved in regulated secretion, contains putative secretory vesicles and seems to form neurite-like extensions to microvilli of the choanocytes, though no clear synapse-like structure is present [106].
Action potentials have been recorded in glass sponges (hexactinellid), which triggers the arrest of the feeding system [157]. This electrical signaling is achieved through the syncytial nature of glass sponge tissue, and is dependent on Ca2+ and K+, but not Na+ [158]. At the sites of incomplete abscission there are cytoplasmic bridges, which contain electron dense plugged junctions [159]. These structures appear morphologically different from the densities seen in choanoflagellate bridges, though like them, they remain to be characterized at the protein/molecular level [160,161].
Did neurons evolve multiple times?
There are two evolutionary points to consider in unraveling the origin of fundamental animal cell types: (1) the transition to constitutive multicellularity in the lineage that gave rise to animals and (2) the last common ancestor of all extant animal lineages (LCAA). In the late 19th century, Ernst Haeckel proposed a sequence of transition to animal multicellularity in which the first step was a hollow ball of flagellated cells (Blastea), followed by the invagination, which created evolutionary space for differentiation of endo- and ectoderm as well as specific cell types [162]. In a general sense, this suggests that cell types arose after the onset of stable multicellularity. Many recent studies of closely related unicellular or transiently multicellular relatives of modern animals have revealed complex life cycles, with morphologically distinct cell states [68,97,163–166]. In modern choanoflagellates, for example, physical confinement leads to the retraction of collar and flagellum and a switch to amoeboid-like motility [165]. Reconstructions of colonies have also revealed striking differences in cell morphologies, suggesting different cell states/types within the colony [68]. These findings suggest that the ability to dramatically shift cell state existed prior to the onset of stable animal multicellularity. While differing in the basis from which they arose, these two ideas are not mutually exclusive and the transition to animal multicellularity may have involved a combination of the two.
Within a multicellular organism, division of labor provides a strong selective advantage. Within the choanoblastea model, much like extant choanoflagellate colonies, all cells are directly engaged in feeding. In this scenario, a simple division could be represented as one cell type retaining a microvilli collar for feeding and another a motile cilium for generating movement. Coupling of these cells, either through direct cytoplasmic links or secretion, could facilitate the delivery of primary metabolites, while opening evolutionary space for specialization within each cell types (Figure 2A).
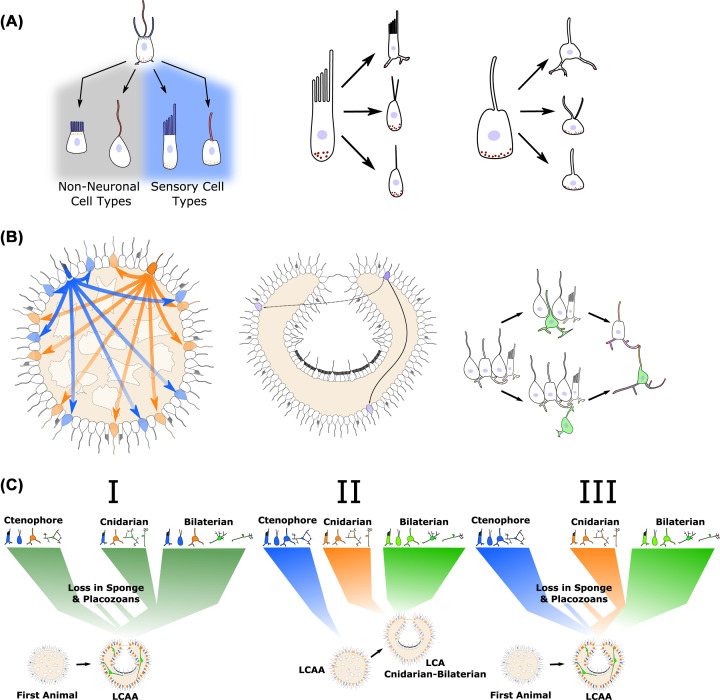
Figure 2. Origin of neurons and neuronal circuits.
(A) Simplified diagram for division of labor of apical structures of a choanoflagellate-like ancestral cell type into villi- (blue) and cilia-based (red) structures on non-neuronal cells and sensory cells. Following the initial division, further specialization can occur under different pressures. (B) Establishment of early neuronal circuits based on diffusion based signaling from peptidergic sensory cells. Distinct sensory cells (blue and orange) release different peptides, which act on cells expressing specific receptors, based on [167]. As bodyplans increase in size and complexity, volumetric signaling is inefficient for full integration and rapid response. Right panel shows hypothetical transitions to directed signaling through interneurons. Synapses are indicated in red. (C) Hypothetical scenarios giving rise to the diversity of neurons and nervous systems seen in extant lineages. First showing a single origin, which diversified in each lineage and was lost in placozoans and possibly sponges. Next, independent origins in major lineages with neurons from a LCAA that did not have true neurons. Third, independent specialization of systems in extant lineages from different aspects of a LCAA with neurons. Better resolving the relationships between the cell types present in extant lineages will help understand which of these occurred.
There was likely a good amount of time between the onset of multicellularity and the last common ancestor of existing animals [68,168]. During this time fundamental cell types, found across phyla, first arose. However, these were likely multifunctional and highly plastic cell types [65,169]. As single-cell RNA sequencing techniques advance and are applied to a widening variety of animals, the idea of terminally differentiated cell types is being tested, and the ability of morphologically similar cells to deploy distinct transcriptional modalities is being seen [170].
The first ‘neurons’ to arise were likely sensory/secretory cells, which acted at the interface of organism and environment. As stated in the first section, many of the subcellular components necessary for this type of cell can be found in non-animal holozoans. What remains to be fully understood is the capacity for linking sensory activation to tightly regulated secretion. Apart from the polarization of vesicle pools [33], this would also include the tight coupling of Ca2+ influx to the release of exocytic vesicle contents. It has been recently shown that presynaptic active zone organization is established through the linking of vesicle docking proteins to VGCC [171]. At the front end of this would be receptor clustering at cellular projections, either cilia or villi based [172,173].
This hypothesis assumes the first neuronal circuits relied on secretion-based signaling or chemical synapses. The presence of innexins and/or connexins in every phyla that has neurons suggests that gap junctions could have arisen during the time between the first animal and LCAA. However, absence in sponges, placozoans, anthozoan and scyphozoan cnidarians, and the pre-bilaterian bottleneck [174] suggests that losses of the genes has occurred often. Based on this, it remains unclear whether or not the LCAA utilized electrical synapses.
Chemical synapses in extant lineages rely on directed (as in between pre- and post-synaptic regions) or volumetric (peripheral release, generally of neuropeptides) transmission of signals [175,176]. Peptides are well suited for volumetric signaling systems, due to the high potential for variability, tight control of production, and ease of diffusion. This leads to the hypothesis that the first animal signaling systems consisted of peptidergic networks [167]. Well-documented sensory-responsive coupling with NO in choanoflagellates and sponges [154,155,177] suggests, early systems were not strictly peptidergic. However, since information is encoded in a single molecule, this restricts the signaling potential in a diffusion-based system. From a developmental standpoint, regional deployment of cell states and/or receptors on both sensory cells and effector cells allows for a basic wiring system for diffusion-based communication across the body. However, as body plans increase in size and complexity diffusion becomes a limiting factor. Under this hypothesis, neurite-like projections could increase precision and speed of signaling across the body, with development of synapses being an extreme example of precise coupling to an effector cell (Figure 2B).
Effector cells of first neuronal circuits
Movement is a very important aspect of animal biology (even sponges require larval settlement and control over movement of ‘environment’ through their aquiferous system). Two basic modes can be found throughout animals, ciliary-based motility and contractile tissue/muscle-based motility. Coordination of these systems is key to their functioning. At small size scale and large temporal scales, diffusion based signaling nicely allows for this type of coordination (i.e., positional information about signal is transmitted to all cells based on strength of signal, fast enough and with enough precision to allow a response—similar to peptidergic signaling in modern placozoans or diffusion based signaling in sponges). However, as body sizes increase and body plans diversity, targeted release of signaling molecules provides faster and more distant communicative potential (Figure 2B). In modern animals, branching neurites can be found in neuro-secretory cells [178,179], which can extend and regulate the location of signal release. This type of transmission also greatly lowers the amount of transmitter release required to act on distance cells, which could help balance the energetic cost of building neurite structures. The transition to feeding on other multicellular organisms and the ensuing arms race was likely a strong driver for greater speed, integration, and coordination.
Did chemical synapses evolve multiple times?
Chemical synapses bring regulated vesicle release from one cell into extremely close proximity to receptor clusters on another cells. These are highly organized and protein dense, with well conserved molecular components in bilaterians [180–182]. Interestingly, overexpression of some synaptic proteins in non-neuronal cells can induce formation of synapse-like structures [171,183,184], suggesting protein abundance may be an important factor in development. As discussed in the second section, as more detailed information about the molecular components and structure of neurons and synapses in non-bilaterians comes out, the question of a single or multiple origins becomes less clear. Ctenophores possess unique synaptic structures (so-called pre-synaptic triads) and anastomosed neurites in their nerve net, unlike anything reported in other nervous systems [124,202]. Likewise, cnidarians have an incompletely understood synaptic toolkit, many bidirectional synapses, and multifunctional neurons. It has been hypnotized that synapses arose multiple times from extensions of ER [185]. What does this mean for the evolution of neurons? Was there a single origin of neurons and neuronal circuits (or synapses), which has undergone radiations within linages giving rise to the extant phyla, or have neurons and synapses arisen multiple times independently? If the first neurons arose before the LCAA, what did these look like and how did they function (Figure 2C)?
Conclusions and future directions
In order to address these questions and start untangling the events that gave rise to the diversity of neurons we see today it is essential to establish testable hypotheses. An important aspect of both division of labor and temporal to spatiotemporal transitional origins of cell types is the regulated deployment of cellular modules. This creates questions of when the protein–protein interactions essential for neuronal signaling first arise during evolution and their function in the close relatives of animals. The increasing power of prediction software such as AlphaFold [186] coupled with advances using cryo-EM to solve protein structures in vivo [187,188], means addressing these questions in a wide diversity of organisms is more tangible than ever. Another important aspect of this is the co-regulation of key structural genes for building these structures. Within animals, terminal transcription factors appear to be well-conserved and can be a good indicator of cell type homology [170]. Though we have focused our discussion primarily on structural genes, an important aspect of synaptogenesis is the co-regulation and deployment of these genes. This is a multilevel process [189], terminal selector transcription factors appear to play an important role [190–193]. Animals have an extensive transcription factor repertoire, with evidence of the origin of new families and large expansions of existing families occurring within this clade [190], though this probably occurred in a stepwise fashion, from components present in their unicellular ancestors [194]. Though much is known about the gene regulatory networks involved in development of different cell and tissue types [195], less is known about how terminal selectors establish and maintain cell type identities. Recent examples for neurons have been identified and show evidence of conservation [191,196]. Building on this work and expanding into functional studies in the close unicellular relatives of animals will help understand the origin of this level of regulation of synaptic genes. The presence of so many key components of the synaptic toolkit outside of animals suggest that true chemical synapses were assemble in a stepwise manner through co-option of existing genes. As discussed above, the transition to multicellularity opens evolutionary space for deploying existing genes and modules in new cellular contexts. Functional understanding of these genes in the close unicellular relatives of animals as those lacking synapses is essential for understanding the degree in which synaptic machinery was organized prior to the occurrence of neurons.
As functional techniques in these organisms expand [74–77], it is becoming possible to truly understand the role of these factors beyond predictions based on homology. The same applies for the expanded sampling of extant lineages of animals. Single-cell RNA sequencing has proven to be an invaluable tool in understanding cell type evolution as well as the basic biology of a wide variety of animals, and recent mapping of cell atlases across species demonstrates an exciting potential [197,198]. However, at greater evolutionary distances the similarities become less clear and metacell (homogeneous groupings of cells) cutoffs need to be greatly relaxed. Increased sampling within phyla will greatly increase the resolution of this technique, as well as better understand the diversity of neurons within the phyla allowing reconstruction of what was likely present at earlier nodes of these lineages. Also moving these technologies to the close relatives of animals will provide valuable insight into the underlying transcriptional changes that give rise to morphologically different cell states. Traditional RNA sequencing has shown that distinguishable expression changes underlie different life stages in non-animal holozoans [199]. There are also evidence of morphologically distinct cells within choanoflagellate colonies [68,164]. Applying scRNA-sequencing to these organisms will provide valuable insights into how distinct these cell states are as well as the transitional sequences between them. As powerful as scRNA-sequencing appears to be, transcription levels alone do not capture the full picture of different cell types or states. Recent multiplexing has even taken this further to understand the effects of perturbations or genetic manipulations [118], an invaluable contribution to functional studies. As briefly touched on earlier, especially with respect to the synapse, it may not just be presence or absence, but rather abundance of specific proteins that drive development of these structures. Combining transcriptional data with quantitative single cell proteomics [200,201] may be necessary to fully understand the tipping point between a ‘secretory cell’ and a ‘neuron’. Finally, the hypothesis that the first neurons were likely sensory cells relying on peptidergic signaling requires further exploration of peptidergic signaling in non-bilaterian animals as well as their close unicellular relatives. Open questions include the functional understanding of the neuropeptides and their effectors in ctenophores [132]; the function of neuropeptide-like molecules in sponges [87,106]; and is peptidergic signaling present in non-animal holozoans and for what function? Central to addressing these questions is combining thorough study of specific organisms with broad sampling of extant lineages. The recent development of the robust technologies above, makes this more possible than ever, meaning solutions to long standing questions, such as the single or multiple origins of neurons, are finally on the horizon.
Summary
Neurons are found in every major animal lineage except sponges and placozoans and many of the key components of the synaptic toolkit are found outside of animals, especially in their close relatives; ichthyosporeans, pluriformeans, filastereans, and choanoflagellates.
Despite deep conservation of many of the components of neurons, there is a huge amount of diversity within the ‘neuron’ cell type and the traditional view of ctenophores and cnidarians having ‘simple’ nerve-nets underplays the complexity of these nervous systems.
Open questions exist around the evolutionary origin of neurons and synapses. Did neurons and synapses evolve once during animal evolution or has this occurred multiple times?
Advances in single-cell transcriptomics and proteomics as well as developing functional techniques in non-bilaterian animals and the close unicellular relatives to animals are providing new tools to address these long-debated questions.
Abbreviations
- GPCR
G-protein coupled receptor
- LCAA
last common ancestor of all extant animal lineages
- LCA
last common ancestor
- PAM
peptidylglycine α-amidating monooxygenase
- SMP
Synaptotagmin-like, Mitochondrial and Lipid-Binding protein
- TRP
transient receptor potential
- VGCC
voltage-gated calcium channels
Contributor Information
Jeffrey Colgren, Email: jeffrey.colgren@uib.no.
Pawel Burkhardt, Email: pawel.burkhardt@uib.no.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors were supported by the Sars Centre core budget.
Author Contribution
The authors contributed equally to this work.
References
- 1.Martinez-Arca S., Coco S., Mainguy G., Schenk U., Alberts P., Bouillé P.et al. (2001) A common exocytotic mechanism mediates axonal and dendritic outgrowth. J. Neurosci. 21, 3830–3838 10.1523/JNEUROSCI.21-11-03830.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Q., Xiao J. and Liu Y. (2000) Participation of syntaxin 1A in membrane trafficking involving neurite elongation and membrane expansion. J. Neurosci. Res. 61, 321–328 [DOI] [PubMed] [Google Scholar]
- 3.Urbina F.L., Gomez S.M. and Gupton S.L. (2018) Spatiotemporal organization of exocytosis emerges during neuronal shape change. J. Cell Biol. 217, 1113–1128 10.1083/jcb.201709064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega I.E. and Hsu S.C. (2001) The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J. Neurosci. 21, 3839–3848 10.1523/JNEUROSCI.21-11-03839.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu B. and Guo W. (2015) The exocyst at a glance. J. Cell Sci. 128, 2957–2964 10.1242/jcs.156398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu S.-C., TerBush D., Abraham M. and Guo W. (2004) The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 243–265 10.1016/S0074-7696(04)33006-8 [DOI] [PubMed] [Google Scholar]
- 7.He B. and Guo W. (2009) The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 537–542 10.1016/j.ceb.2009.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloch D., Pleskot R., Pejchar P., Potocký M., Trpkošová P., Cwiklik L.et al. (2016) Exocyst SEC3 and phosphoinositides define sites of exocytosis in pollen tube initiation and growth. Plant Physiol. 172, 980–1002 10.1104/pp.16.00690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zajac A., Sun X., Zhang J. and Guo W. (2005) Cyclical regulation of the exocyst and cell polarity determinants for polarized cell growth. Mol. Biol. Cell. 16, 1500–1512 10.1091/mbc.e04-10-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheu Y.J., Barral Y. and Snyder M. (2000) Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235–5247 10.1128/MCB.20.14.5235-5247.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehm C. and Field M.C. (2019) Evolution of late steps in exocytosis: conservation and specialization of the exocyst complex. Wellcome Open Res. 4, 112 10.12688/wellcomeopenres.15142.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cvrčková F., Grunt M., Bezvoda R., Hála M., Kulich I., Rawat A.et al. (2012) Evolution of the land plant exocyst complexes. Front. Plant Sci. 3, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parra-Rivas L.A., Palfreyman M.T., Vu T.N. and Jorgensen E.M. (2022) Interspecies complementation identifies a pathway to assemble SNAREs. iScience 25, 104506 10.1016/j.isci.2022.104506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarto-Jackson I. and Tomaska L. (2016) How to bake a brain: yeast as a model neuron. Curr. Genet. 347–370 10.1007/s00294-015-0554-2 [DOI] [PubMed] [Google Scholar]
- 15.Dong W., Radulovic T., Oliver Goral R., Thomas C., Montesinos M.S., Guerrero-Given D.et al. (2018) CAST/ELKS proteins control voltage-gated Ca2 channel density and synaptic release probability at a mammalian central synapse. Cell Rep. 284.e6–293.e6 10.1016/j.celrep.2018.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai R.G. and Bellen H.J. (2004) The architecture of the active zone in the presynaptic nerve terminal. Physiology 19, 262–270 10.1152/physiol.00014.2004 [DOI] [PubMed] [Google Scholar]
- 17.Jung J.H., Szule J.A., Marshall R.M. and McMahan U.J. (2016) Variable priming of a docked synaptic vesicle. Proc. Natl. Acad. Sci. U.S.A. 113, E1098–E1107 10.1073/pnas.1523054113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahn R. and Fasshauer D. (2012) Molecular machines governing exocytosis of synaptic vesicles. Nature 490, 201–207 10.1038/nature11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moroz L.L. (2021) Multiple origins of neurons from secretory cells. Front. Cell Dev. Biol. 9, 669087 10.3389/fcell.2021.669087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y., Kaeser P., Südhof T. and Schneggenburger R. (2011) RIM determines Ca2+ channel density and vesicle docking at the presynaptic active zone. Neuron 69, 304–316 10.1016/j.neuron.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palfreyman M.T. and Jorgensen E.M. (2017) Unc13 aligns SNAREs and superprimes synaptic vesicles. Neuron 95, 473–475 10.1016/j.neuron.2017.07.017 [DOI] [PubMed] [Google Scholar]
- 22.Zdanowicz R., Kreutzberger A., Liang B., Kiessling V., Tamm L.K. and Cafiso D.S. (2017) Complexin binding to membranes and acceptor t-SNAREs explains its clamping effect on fusion. Biophys. J. 113, 1235–1250 10.1016/j.bpj.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burkhardt P. and Sprecher S.G. (2017) Evolutionary origin of synapses and neurons - bridging the gap. Bioessays 39, 1700024 10.1002/bies.201700024 [DOI] [PubMed] [Google Scholar]
- 24.Miki T., Kaufmann W.A., Malagon G., Gomez L., Tabuchi K., Watanabe M.et al. (2017) Numbers of presynaptic Ca channel clusters match those of functionally defined vesicular docking sites in single central synapses. Proc. Natl. Acad. Sci. U.S.A. 114, E5246–E5255 10.1073/pnas.1704470114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran Y. and Zakon H.H. (2014) The evolution of the four subunits of voltage-gated calcium channels: ancient roots, increasing complexity, and multiple losses. Genome Biol. Evol. 6, 2210–2217 10.1093/gbe/evu177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hida Y. and Ohtsuka T. (2010) CAST and ELKS proteins: structural and functional determinants of the presynaptic active zone. J. Biochem. 148, 131–137 10.1093/jb/mvq065 [DOI] [PubMed] [Google Scholar]
- 27.King N., Jody Westbrook M., Young S.L., Kuo A., Abedin M., Chapman J.et al. (2008) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 783–788 10.1038/nature06617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barber C.F., Jorquera R.A., Melom J.E. and Littleton J.T. (2009) Postsynaptic regulation of synaptic plasticity by synaptotagmin 4 requires both C2 domains. J. Cell Biol. 187, 295–310 10.1083/jcb.200903098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schapire A.L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D.et al. (2008) Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell. 20, 3374–3388 10.1105/tpc.108.063859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saheki Y. and De Camilli P. (2017) The Extended-Synaptotagmins. Biochim. Biophys. Acta Mol. Cell. Res. 1864, 1490–1493 10.1016/j.bbamcr.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Lopez N., Pérez-Sancho J., del Valle A.E., Haslam R.P., Vanneste S., Catalá R.et al. (2021) Synaptotagmins at the endoplasmic reticulum-plasma membrane contact sites maintain diacylglycerol homeostasis during abiotic stress. Plant Cell. 2431–2453 10.1093/plcell/koab122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A. and Emr S.D. (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23, 1129–1140 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- 33.Göhde R., Naumann B., Laundon D., Imig C., McDonald K., Cooper B.H.et al. (2021) Choanoflagellates and the ancestry of neurosecretory vesicles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376, 20190759 10.1098/rstb.2019.0759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkhardt P., Grønborg M., McDonald K., Sulur T., Wang Q. and King N. (2014) Evolutionary insights into premetazoan functions of the neuronal protein homer. Mol. Biol. Evol. 31, 2342–2355 10.1093/molbev/msu178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moroz L.L. and Kohn A.B. (2015) Unbiased view of synaptic and neuronal gene complement in ctenophores: are there pan-neuronal and pan-synaptic genes across metazoa? Integr. Comp. Biol. 55, 1028–1049 10.1093/icb/icv104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alié A. and Manuel M. (2010) The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol. Biol. 10, 34 10.1186/1471-2148-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkhardt P. (2015) The origin and evolution of synaptic proteins - choanoflagellates lead the way. J. Exp. Biol. 218, 506–514 10.1242/jeb.110247 [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharyya M., Stratton M.M., Going C.C., McSpadden E.D., Huang Y., Susa A.C.et al. (2016) Molecular mechanism of activation-triggered subunit exchange in Ca2+/calmodulin-dependent protein kinase II. Elife 5, e13405 10.7554/eLife.13405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himmel N.J. and Cox D.N. (2020) Transient receptor potential channels: current perspectives on evolution, structure, function and nomenclature. Proc. Biol. Sci. 287, 20201309 10.1098/rspb.2020.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schöneberg T., Hofreiter M., Schulz A. and Römpler H. (2007) Learning from the past: evolution of GPCR functions. Trends Pharmacol. Sci. 28, 117–121 10.1016/j.tips.2007.01.001 [DOI] [PubMed] [Google Scholar]
- 41.Moroz L.L., Nikitin M.A., Poličar P.G., Kohn A.B. and Romanova D.Y. (2021) Evolution of glutamatergic signaling and synapses. Neuropharmacology 199, 108740 10.1016/j.neuropharm.2021.108740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nauli S.M., Alenghat F.J., Luo Y., Williams E., Vassilev P., Li X.et al. (2003) Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137 10.1038/ng1076 [DOI] [PubMed] [Google Scholar]
- 43.Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y.et al. (1998) Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 10.1016/S0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- 44.Bangs F. and Anderson K.V. (2017) Primary cilia and mammalian hedgehog signaling. Cold Spring Harbor Perspect. Biol. 9a028175 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besharse J.C., Forestner D.M. and Defoe D.M. (1985) Membrane assembly in retinal photoreceptors. III. Distinct membrane domains of the connecting cilium of developing rods. J. Neurosci. 5, 1035–1048 10.1523/JNEUROSCI.05-04-01035.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bae Y.-K. (2008) Sensory roles of neuronal cilia: Cilia development, morphogenesis, and function in C. elegans. Front. Biosci. 15959–5974 10.2741/3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenkins P.M., McEwen D.P. and Martens J.R. (2009) Olfactory cilia: linking sensory cilia function and human disease. Chem. Senses. 34, 451–464 10.1093/chemse/bjp020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloodgood R.A. (2010) Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell Sci. 123, 505–509 10.1242/jcs.066308 [DOI] [PubMed] [Google Scholar]
- 49.Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J.B. and Bettencourt-Dias M. (2011) Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell D.R. (2007) The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 607, 130–140 10.1007/978-0-387-74021-8_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell D.R. (2017) Evolution of cilia. Cold Spring Harbor Perspect. Biol. 9a028290 10.1101/cshperspect.a028290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danielsen E.M. and Hansen G.H. (2006) Lipid raft organization and function in brush borders of epithelial cells. Mol. Membr. Biol. 23, 71–79 10.1080/09687860500445604 [DOI] [PubMed] [Google Scholar]
- 53.Goutman J.D., Elgoyhen A.B. and Gómez-Casati M.E. (2015) Cochlear hair cells: the sound-sensing machines. FEBS Lett. 589, 3354–3361 10.1016/j.febslet.2015.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fettiplace R. and Hackney C.M. (2006) The sensory and motor roles of auditory hair cells. Nat. Rev. Neurosci. 7, 19–29 10.1038/nrn1828 [DOI] [PubMed] [Google Scholar]
- 55.Raible D.W. and Kruse G.J. (2000) Organization of the lateral line system in embryonic zebrafish. J. Comp. Neurol. 421, 189–198 [DOI] [PubMed] [Google Scholar]
- 56.Caprara G.A. and Peng A.W. (2022) Mechanotransduction in mammalian sensory hair cells. Mol. Cell. Neurosci. 120, 103706 10.1016/j.mcn.2022.103706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suli A., Watson G.M., Rubel E.W. and Raible D.W. (2012) Rheotaxis in larval zebrafish is mediated by lateral line mechanosensory hair cells. PLoS ONE 7, e29727 10.1371/journal.pone.0029727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson G.M., Mire P. and Kinler K.M. (2009) Mechanosensitivity in the model sea anemone Nematostella vectensis. Mar. Biol. 156, 2129–2137 10.1007/s00227-009-1243-9 [DOI] [Google Scholar]
- 59.Norekian T.P. and Moroz L.L. (2019) Neuromuscular organization of the Ctenophore Pleurobrachia bachei. J. Comp. Neurol. 527, 406–436 10.1002/cne.24546 [DOI] [PubMed] [Google Scholar]
- 60.Peña J.F., Alié A., Richter D.J., Wang L., Funayama N. and Nichols S.A. (2016) Conserved expression of vertebrate microvillar gene homologs in choanocytes of freshwater sponges. Evodevo 7, 13 10.1186/s13227-016-0050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mah J.L. and Leys S.P. (2017) Think like a sponge: the genetic signal of sensory cells in sponges. Dev. Biol. 431, 93–100 10.1016/j.ydbio.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 62.Clark H.J. (1866) Conclusive proofs of the animality of the ciliate sponges, and of their affinities with the Infusoria flagellata. Am. J. Sci. s2-42320–324 10.2475/ajs.s2-42.126.320 [DOI] [Google Scholar]
- 63.Kent W.S. (1880) A manual of the infusoria: including a description of all known flagellate, ciliate, and tentaculiferous protozoa, british and foreign, and an account of the organization and the affinities of the sponges, London [Google Scholar]
- 64.Nichols S.A., Dayel M.J. and King N. (2009) Genomic, phylogenetic, and cell biological insights into metazoan origins. In Animal Evolution: Genes, Genomes, Fossils and Trees (Telford M.J. and Littlewood D.T.J.eds.) Oxford Academic, Oxford, 10.1093/acprof:oso/9780199549429.003.0003 [DOI] [Google Scholar]
- 65.Brunet T. and King N. (2017) The origin of animal multicellularity and cell differentiation. Dev. Cell. 43, 124–140 10.1016/j.devcel.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mah J.L., Christensen-Dalsgaard K.K. and Leys S.P. (2014) Choanoflagellate and choanocyte collar-flagellar systems and the assumption of homology. Evol. Develop. 16, 25–37 10.1111/ede.12060 [DOI] [PubMed] [Google Scholar]
- 67.Dunn C.W., Leys S.P. and Haddock S.H.D. (2015) The hidden biology of sponges and ctenophores. Trends Ecol. Evol. 30, 282–291 10.1016/j.tree.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 68.Laundon D., Larson B.T., McDonald K., King N. and Burkhardt P. (2019) The architecture of cell differentiation in choanoflagellates and sponge choanocytes. PLoS Biol. 17, e3000226 10.1371/journal.pbio.3000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeCaen P.G., Delling M., Vien T.N. and Clapham D.E. (2013) Direct recording and molecular identification of the calcium channel of primary cilia. Nature 504, 315–318 10.1038/nature12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teilmann S.C., Byskov A.G., Pedersen P.A., Wheatley D.N., Pazour G.J. and Christensen S.T. (2005) Localization of transient receptor potential ion channels in primary and motile cilia of the female murine reproductive organs. Mol. Reprod. Dev. 71, 444–452 10.1002/mrd.20312 [DOI] [PubMed] [Google Scholar]
- 71.Andrade Y.N., Fernandes J., Vázquez E., Fernández-Fernández J.M., Arniges M., Sánchez T.M.et al. (2005) TRPV4 channel is involved in the coupling of fluid viscosity changes to epithelial ciliary activity. J. Cell Biol. 168, 869–874 10.1083/jcb.200409070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang K., Diener D.R., Mitchell A., Pazour G.J., Witman G.B. and Rosenbaum J.L. (2007) Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 179, 501–514 10.1083/jcb.200704069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujiu K., Nakayama Y., Iida H., Sokabe M. and Yoshimura K. (2011) Mechanoreception in motile flagella of Chlamydomonas. Nat. Cell Biol. 13, 630–632 10.1038/ncb2214 [DOI] [PubMed] [Google Scholar]
- 74.Phillips J.E., Santos M., Konchwala M., Xing C. and Pan D. (2022) Genome editing in the unicellular holozoan Capsaspora owczarzaki suggests a premetazoan role for the Hippo pathway in multicellular morphogenesis. eLife 11, e77598 10.7554/eLife.77598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Booth D.S., Szmidt-Middleton H. and King N. (2018) Transfection of choanoflagellates illuminates their cell biology and the ancestry of animal septins. Mol. Biol. Cell. 29, 3026–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Booth D.S. and King N. (2020) Genome editing enables reverse genetics of multicellular development in the choanoflagellate Salpingoeca rosetta. eLife 9, e56193 10.7554/eLife.56193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kożyczkowska A., Najle S.R., Ocaña-Pallarès E., Aresté C., Shabardina V., Ara P.S.et al. (2021) Stable transfection in protist Corallochytrium limacisporum identifies novel cellular features among unicellular animals relatives. Curr. Biol. 31, 4104.e5–4110.e5 10.1016/j.cub.2021.06.061 [DOI] [PubMed] [Google Scholar]
- 78.Combarnous Y. and Nguyen T.M.D. (2020) Cell communications among microorganisms, plants, and animals: origin, evolution, and interplays. Int. J. Mol. Sci. 21, 8052 10.3390/ijms21218052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bloom F.E. (1985) Neurotransmitter diversity and its functional significance. J. R. Soc. Med. 78, 189–192 10.1177/014107688507800303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toda N. (2000) Nitric oxide and the peripheral nervous system, Portland Press, London [Google Scholar]
- 81.Hampe C.S., Mitoma H. and Manto M. (2018) GABA and glutamate: their transmitter role in the CNS and pancreatic islets In GABA and Glutamate (Samardzic J.ed.) IntechOpen; 10.5772/intechopen.70958 [DOI] [Google Scholar]
- 82.Russo A.F. (2017) Overview of neuropeptides: awakening the senses? Headache 57, 37–46 10.1111/head.13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chufán E.E., De M., Eipper B.A., Mains R.E. and Amzel L.M. (2009) Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure 17, 965–973 10.1016/j.str.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar D., Strenkert D., Patel-King R.S., Leonard M.T., Merchant S.S., Mains R.E.et al. (2017) A bioactive peptide amidating enzyme is required for ciliogenesis. Elife 6, e25728 10.7554/eLife.25728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar D., Mains R.E., Eipper B.A. and King S.M. (2019) Ciliary and cytoskeletal functions of an ancient monooxygenase essential for bioactive amidated peptide synthesis. Cell. Mol. Life Sci. 76, 2329–2348 10.1007/s00018-019-03065-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luxmi R., Kumar D., Mains R.E., King S.M. and Eipper B.A. (2019) Cilia-based peptidergic signaling. PLoS Biol. 17, e3000566 10.1371/journal.pbio.3000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yañez-Guerra L.A., Thiel D. and Jékely G. (2022) Premetazoan origin of neuropeptide signaling. Mol. Biol. Evol. 39, msac051, 10.1093/molbev/msac051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Correa-Aragunde N., Graziano M. and Lamattina L. (2004) Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905 10.1007/s00425-003-1172-7 [DOI] [PubMed] [Google Scholar]
- 89.Huang X., Stettmaier K., Michel C., Hutzler P., Mueller M.J. and Durner J. (2004) Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218, 938–946 10.1007/s00425-003-1178-1 [DOI] [PubMed] [Google Scholar]
- 90.Nussler A.K. and Billiar T.R. (1993) Inflammation, immunoregulation, and inducible nitric oxide synthase. J. Leukoc. Biol. 54, 171–178 10.1002/jlb.54.2.171 [DOI] [PubMed] [Google Scholar]
- 91.Gong L., Pitari G.M., Schulz S. and Waldman S.A. (2004) Nitric oxide signaling: systems integration of oxygen balance in defense of cell integrity. Curr. Opin. Hematol. 11, 7–14 10.1097/00062752-200401000-00003 [DOI] [PubMed] [Google Scholar]
- 92.Moncada S., Radomski M.W. and Palmer R.M. (1988) Endothelium-derived relaxing factor. Identification as nitric oxide and role in the control of vascular tone and platelet function. Biochem. Pharmacol. 37, 2495–2501 10.1016/0006-2952(88)90236-5 [DOI] [PubMed] [Google Scholar]
- 93.Astuti R.I., Nasuno R. and Takagi H. (2018) Nitric oxide signalling in yeast. Adv. Microb. Physiol. 72, 29–63 10.1016/bs.ampbs.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 94.Reyes-Rivera J., Wu Y., Guthrie B.G.H., Marletta M.A., King N. and Brunet T.. Nitric oxide signaling controls collective contractions in a colonial choanoflagellate, 32, 2539–2547.e5, . 10.1016/j.cub.2022.04.017 [DOI] [PubMed] [Google Scholar]
- 95.Martin E.A., Lasseigne A.M. and Miller A.C. (2020) Understanding the molecular and cell biological mechanisms of electrical synapse formation. Front Neuroanat. 14, 12 10.3389/fnana.2020.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai X. (2008) Unicellular Ca2 Signaling “Toolkit” at the Origin of Metazoa. Mol. Biol. Evol. 1357–1361 10.1093/molbev/msn077 [DOI] [PubMed] [Google Scholar]
- 97.Dayel M.J., Alegado R.A., Fairclough S.R., Levin T.C., Nichols S.A., McDonald K.et al. (2011) Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev. Biol. 357, 73–82 10.1016/j.ydbio.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hake K.H., West P.T., McDonald K., Laundon D., De Las Bayonas A.G., Feng C.et al.. Colonial choanoflagellate isolated from Mono Lake harbors a microbiome, bioRxiv 10.1101/2021.03.30.437421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dunn C.W., Hejnol A., Matus D.Q., Pang K., Browne W.E., Smith S.A.et al. (2008) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 [DOI] [PubMed] [Google Scholar]
- 100.Whelan N.V., Kocot K.M. and Halanych K.M. (2015) Employing phylogenomics to resolve the relationships among cnidarians, ctenophores, sponges, placozoans, and bilaterians. Integr. Comp. Biol. 55, 1084–1095 [DOI] [PubMed] [Google Scholar]
- 101.Jékely G., Paps J. and Nielsen C. (2015) The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whelan N.V., Kocot K.M., Moroz T.P., Mukherjee K., Williams P., Paulay G.et al. (2017) Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746 10.1038/s41559-017-0331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Philippe H., Derelle R., Lopez P., Pick K., Borchiellini C., Boury-Esnault N.et al. (2009) Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712 10.1016/j.cub.2009.02.052 [DOI] [PubMed] [Google Scholar]
- 104.Nosenko T., Schreiber F., Adamska M., Adamski M., Eitel M., Hammel J.et al. (2013) Deep metazoan phylogeny: when different genes tell different stories. Mol. Phylogenet. Evol. 67, 223–233 10.1016/j.ympev.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 105.Redmond A.K. and McLysaght A. (2021) Evidence for sponges as sister to all other animals from partitioned phylogenomics with mixture models and recoding. Nat. Commun. 12, 1783 10.1038/s41467-021-22074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Musser J.M., Schippers K.J., Nickel M., Mizzon G., Kohn A.B., Pape C.et al. (2021) Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science 374, 717–723 10.1126/science.abj2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Riesgo A., Farrar N., Windsor P.J., Giribet G. and Leys S.P. (2014) The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31, 1102–1120 10.1093/molbev/msu057 [DOI] [PubMed] [Google Scholar]
- 108.Srivastava M., Begovic E., Chapman J., Putnam N.H., Hellsten U., Kawashima T.et al. (2008) The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 10.1038/nature07191 [DOI] [PubMed] [Google Scholar]
- 109.Nikitin M. (2015) Bioinformatic prediction of Trichoplax adhaerens regulatory peptides. Gen. Comp. Endocrinol. 212, 145–155 10.1016/j.ygcen.2014.03.049 [DOI] [PubMed] [Google Scholar]
- 110.Moroz L.L., Kocot K.M., Citarella M.R., Dosung S., Norekian T.P., Povolotskaya I.S.et al. (2014) The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114 10.1038/nature13400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ryan J.F. and Chiodin M. (2015) Where is my mind? How sponges and placozoans may have lost neural cell types Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150059 10.1098/rstb.2015.0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moroz L.L. and Kohn A.B. (2016) Independent origins of neurons and synapses: insights from ctenophores. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150041 10.1098/rstb.2015.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sprecher S.G. (2022) Neural cell type diversity in cnidaria. Front. Neurosci. 16, 909400 10.3389/fnins.2022.909400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kass-Simon G. and Pierobon P. (2007) Cnidarian chemical neurotransmission, an updated overview. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 146, 9–25 10.1016/j.cbpa.2006.09.008 [DOI] [PubMed] [Google Scholar]
- 115.Faltine-Gonzalez D.Z. and Layden M.J. (2019) Characterization of nAChRs in Nematostella vectensis supports neuronal and non-neuronal roles in the cnidarian-bilaterian common ancestor. EvoDevo 10, 27 10.1186/s13227-019-0136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Attenborough R.M.F., Hayward D.C., Wiedemann U., Forêt S., Miller D.J. and Ball E.E. (2019) Expression of the neuropeptides RFamide and LWamide during development of the coral Acropora millepora in relation to settlement and metamorphosis. Dev. Biol. 446, 56–67 10.1016/j.ydbio.2018.11.022 [DOI] [PubMed] [Google Scholar]
- 117.Koch T.L. and Grimmelikhuijzen C.J.P. (2020) A comparative genomics study of neuropeptide genes in the cnidarian subclasses Hexacorallia and Ceriantharia. BMC Genomics 21, 666 10.1186/s12864-020-06945-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chari T., Weissbourd B., Gehring J., Ferraioli A., Leclère L., Herl M.et al. (2021) Whole-animal multiplexed single-cell RNA-seq reveals transcriptional shifts across medusa cell types. Sci Adv. 7, eabh1683 10.1126/sciadv.abh1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oren M., Brickner I., Appelbaum L. and Levy O. (2014) Fast neurotransmission related genes are expressed in non nervous endoderm in the sea anemone Nematostella vectensis. PLoS ONE 9, e93832 10.1371/journal.pone.0093832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Levy S., Brekhman V., Bakhman A., Malik A., Sebé-Pedrós A., Kosloff M.et al. (2021) Ectopic activation of GABAB receptors inhibits neurogenesis and metamorphosis in the cnidarian Nematostella vectensis. Nat. Ecol. Evol. 5, 111–121 10.1038/s41559-020-01338-3 [DOI] [PubMed] [Google Scholar]
- 121.Hayakawa E., Watanabe H., Menschaert G., Holstein T.W., Baggerman G. and Schoofs L. (2019) A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella vectensis. PLoS ONE 14, e0215185 10.1371/journal.pone.0215185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Koizumi O., Wilson J.D., Grimmelikhuijzen C.J. and Westfall J.A. (1989) Ultrastructural localization of RFamide-like peptides in neuronal dense-cored vesicles in the peduncle of Hydra. J. Exp. Zool. 249, 17–22 10.1002/jez.1402490105 [DOI] [PubMed] [Google Scholar]
- 123.Tamm S.L. (2014) Cilia and the life of ctenophores. Invertebr Biol. 133, 1–46 10.1111/ivb.12042 [DOI] [Google Scholar]
- 124.Jokura K., Shibata D., Yamaguchi K., Shiba K., Makino Y., Shigenobu S.et al. (2019) CTENO64 is required for coordinated paddling of ciliary comb plate in ctenophores. Curr. Biol. 29, 3510.e4–3516.e4 10.1016/j.cub.2019.08.059 [DOI] [PubMed] [Google Scholar]
- 125.Hertwig R. (1880) Über den Bau der Ctenophoren, Fischer [Google Scholar]
- 126.Hernandez-Nicaise M.-L. (1973) Le systeme nerveux des cténaires: structure et ultrastructure des réseaux épithéliaux, Springer-Verlag; [PubMed] [Google Scholar]
- 127.Hernandez-Nicaise M.-L. (1991) Ctenophora In Microscopic Anatomy of Invertebrates: Placozoa, Porifera, Cnidaria, and Ctenophora, vol. 2, (Harrison F.W., Ruppert E.E. and Kohn A. J.) Wiley-Liss, New York [Google Scholar]
- 128.Jager M., Chiori R., Alié A., Dayraud C., Quéinnec E. and Manuel M. (2011) New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Müller, 1776). J. Exp. Zool. B. Mol. Dev. Evol. 316B, 171–187 10.1002/jez.b.21386 [DOI] [PubMed] [Google Scholar]
- 129.Norekian T.P. and Moroz L.L. (2019) Neural system and receptor diversity in the ctenophore Beroe abyssicola. J. Comp. Neurol. 527, 1986–2008 10.1002/cne.24633 [DOI] [PubMed] [Google Scholar]
- 130.Norekian T.P. and Moroz L.L. (2020) Comparative neuroanatomy of ctenophores: Neural and muscular systems in Euplokamis dunlapae and related species. J. Comp. Neurol. 528, 481–501 10.1002/cne.24770 [DOI] [PubMed] [Google Scholar]
- 131.Norekian T.P. and Moroz L.L. (2021) Development of the nervous system in the early hatching larvae of the ctenophore Mnemiopsis leidyi. J. Morphol. 282, 1466–1477 10.1002/jmor.21398 [DOI] [PubMed] [Google Scholar]
- 132.Sachkova M.Y., Nordmann E.-L., Soto-Àngel J.J., Meeda Y., Górski B., Naumann B.et al. (2021) Neuropeptide repertoire and 3D anatomy of the ctenophore nervous system. Curr. Biol. 31, 5274.e6–5285.e6 10.1016/j.cub.2021.09.005 [DOI] [PubMed] [Google Scholar]
- 133.Courtney A., Liegey J., Burke N., Hassett A.R., Lowery M. and Pickering M. (2022) Characterization of geometric variance in the epithelial nerve net of the ctenophore Pleurobrachia pileus. J. Comp. Neurol. 530, 1438–1458 10.1002/cne.25290 [DOI] [PubMed] [Google Scholar]
- 134.Horridge G.A. (1964) Presumed photoreceptive cilia in a ctenophore. J. Cell Sci. s3-105, 311–317 10.1242/jcs.s3-105.71.311 [DOI] [Google Scholar]
- 135.Aronova M. (1974) Electron microscopic observation of the aboral organ of Ctenophora. I. The gravity receptor. Z. Mikrosk. Anat. Forsch. 88, 401–412 [PubMed] [Google Scholar]
- 136.Horridge G.A. (1974) Recent studies on the Ctenophora In Coelenterate Biology (Muscatine L.ed.) Academic Press, New York: 10.1016/B978-0-12-512150-7.50016-5 [DOI] [Google Scholar]
- 137.Jokura K. and Inaba K. (2020) Structural diversity and distribution of cilia in the apical sense organ of the ctenophore Bolinopsis mikado. Cytoskeleton 77, 4422940–2956455 10.1002/cm.21640 [DOI] [PubMed] [Google Scholar]
- 138.Burkhardt P. and Jékely G. (2021) Evolution of synapses and neurotransmitter systems: The divide-and-conquer model for early neural cell-type evolution. Curr. Opin. Neurobiol. 71, 127–138 10.1016/j.conb.2021.11.002 [DOI] [PubMed] [Google Scholar]
- 139.Alberstein R., Grey R., Zimmet A., Simmons D.K. and Mayer M.L. (2015) Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes. Proc. Natl. Acad. Sci. U. S. A. 112, E6048–E6057 10.1073/pnas.1513771112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu A., Alberstein R., Thomas A., Zimmet A., Grey R., Mayer M.L.et al. (2016) Molecular lock regulates binding of glycine to a primitive NMDA receptor. Proc. Natl. Acad. Sci. U. S. A. 113, E6786–E6795 10.1073/pnas.1607010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hayakawa E., Guzman C., Horiguchi O., Kawano C., Shiraishi A., Mohri K.et al. (2022) Mass spectrometry of short peptides reveals common features of metazoan peptidergic neurons. Nat. Ecol. Evol. 2022, 10.1038/s41559-022-01835-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Babonis L.S., DeBiasse M.B., Francis W.R., Christianson L.M., Moss A.G., Haddock S.H.D.et al. (2018) Integrating embryonic development and evolutionary history to characterize tentacle-specific cell types in a ctenophore. Mol. Biol. Evol. 35, 2940–2956 10.1093/molbev/msy171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sebé-Pedrós A., Chomsky E., Pang K., Lara-Astiaso D., Gaiti F., Mukamel Z.et al. (2018) Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2, 1176–1188 10.1038/s41559-018-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ryan J.F., Pang K., Schnitzler C.E., Nguyen A.-D., Moreland R.T., Simmons D.K.et al. (2013) The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 10.1126/science.1242592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Senatore A., Reese T.S. and Smith C.L. (2017) Neuropeptidergic integration of behavior in, an animal without synapses. J. Exp. Biol. 220, 3381–3390 10.1242/jeb.162396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mayorova T.D., Hammar K., Winters C.A., Reese T.S. and Smith C.L. (2019) The ventral epithelium of Trichoplax adhaerens deploys in distinct patterns cells that secrete digestive enzymes, mucus or diverse neuropeptides. Biol. Open 8, bio045674 10.1242/bio.045674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Simpson T.L. (1984) The cell biology of sponges, Springer, Berlin Heidelberg, New York [Google Scholar]
- 148.Wong E., Mölter J., Anggono V., Degnan S.M. and Degnan B.M. (2019) Co-expression of synaptic genes in the sponge Amphimedon queenslandica uncovers ancient neural submodules. Sci. Rep. 9, 15781 10.1038/s41598-019-51282-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sakarya O., Armstrong K.A., Adamska M., Adamski M., Wang I.-F., Tidor B.et al. (2007) A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 2, e506 10.1371/journal.pone.0000506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kenny N.J., Francis W.R., Rivera-Vicéns R.E., Juravel K., de Mendoza A., Díez-Vives C.et al. (2020) The genomic basis of animal origins: a chromosomal perspective from the sponge Ephydatia muelleri, bioRxiv 10.1101/2020.02.18.954784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M.E.A., Mitros T.et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Elliott G.R.D. and Leys S.P. (2007) Coordinated contractions effectively expel water from the aquiferous system of a freshwater sponge. J. Exp. Biol. 210, 3736–3748 10.1242/jeb.003392 [DOI] [PubMed] [Google Scholar]
- 153.Nickel M. (2004) Kinetics and rhythm of body contractions in the sponge Tethya wilhelma (Porifera: Demospongiae). J. Exp. Biol. 207, 4515–4524 10.1242/jeb.01289 [DOI] [PubMed] [Google Scholar]
- 154.Ludeman D.A., Farrar N., Riesgo A., Paps J. and Leys S.P. (2014) Evolutionary origins of sensation in metazoans: functional evidence for a new sensory organ in sponges. BMC Evol. Biol. 3 10.1186/1471-2148-14-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Elliott G.R.D. and Leys S.P. (2010) Evidence for glutamate, GABA and NO in coordinating behaviour in the sponge, Ephydatia muelleri (Demospongiae, Spongillidae). J. Exp. Biol. 213, 2310–2321 10.1242/jeb.039859 [DOI] [PubMed] [Google Scholar]
- 156.Ellwanger K., Eich A. and Nickel M. (2007) GABA and glutamate specifically induce contractions in the sponge Tethya wilhelma. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 193, 1–11 10.1007/s00359-006-0165-y [DOI] [PubMed] [Google Scholar]
- 157.Leys S.P. and Mackie G.O. (1997) Electrical recording from a glass sponge. Nature 387, 29–30 10.1038/387029b0 [DOI] [Google Scholar]
- 158.Leys S.P., Mackie G.O. and Meech R.W. (1999) Impulse conduction in a sponge. J. Exp. Biol. 202, 1139–1150 10.1242/jeb.202.9.1139 [DOI] [PubMed] [Google Scholar]
- 159.Mackie G.O. and Singla C.L. (1983) Studies on Hexactinellid Sponges: 1. Histol. Rhabdocalyptus Dawsoni (Lambe, 1873) [Google Scholar]
- 160.Leys S.P., Mackie G.O. and Reiswig H.M. (2007) The biology of glass sponges. Adv. Mar. Biol. 52, 1–145 10.1016/S0065-2881(06)52001-2 [DOI] [PubMed] [Google Scholar]
- 161.Leys S.P. (2003) The significance of syncytial tissues for the position of the hexactinellida in the metazoa. Integr. Comp. Biol. 43, 19–27 10.1093/icb/43.1.19 [DOI] [PubMed] [Google Scholar]
- 162.Haeckel E. (1875) Die Gastraea-Theorie, die phylogenetische Classification des Thierreichs und die Homologie der Keimblätter [Google Scholar]
- 163.Tikhonenkov D.V., Hehenberger E., Esaulov A.S., Belyakova O.I., Mazei Y.A., Mylnikov A.P.et al. (2020) Insights into the origin of metazoan multicellularity from predatory unicellular relatives of animals. BMC Biol. 18, 39 10.1186/s12915-020-0762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Naumann B. and Burkhardt P. (2019) Spatial Cell Disparity in the Colonial Choanoflagellate Salpingoeca rosetta. Front. Cell Developmental Biol. 10.3389/fcell.2019.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brunet T., Albert M., Roman W., Coyle M.C., Spitzer D.C. and King N. (2021) A flagellate-to-amoeboid switch in the closest living relatives of animals. Elife 10, 10.7554/eLife.61037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sebé-Pedrós A., Peña M.I., Capella-Gutiérrez S., Antó M., Gabaldón T., Ruiz-Trillo I.et al. (2016) High-Throughput Proteomics Reveals the Unicellular Roots of Animal Phosphosignaling and Cell Differentiation. Dev. Cell. 39, 186–197 10.1016/j.devcel.2016.09.019 [DOI] [PubMed] [Google Scholar]
- 167.Jékely G. (2021) The chemical brain hypothesis for the origin of nervous systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376, 20190761 10.1098/rstb.2019.0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ros-Rocher N., Pérez-Posada A., Leger M.M. and Ruiz-Trillo I. (2021) The origin of animals: an ancestral reconstruction of the unicellular-to-multicellular transition. Open Biol. 11, 200359 10.1098/rsob.200359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sogabe S., Hatleberg W.L., Kocot K.M., Say T.E., Stoupin D., Roper K.E.et al. (2019) Pluripotency and the origin of animal multicellularity. Nature 570, 519–522 10.1038/s41586-019-1290-4 [DOI] [PubMed] [Google Scholar]
- 170.Arendt D., Musser J.M., Baker C.V.H., Bergman A., Cepko C., Erwin D.H.et al. (2016) The origin and evolution of cell types. Nat. Rev. Genet. 744–757 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- 171.Tan C., Wang S.S.H., de Nola G. and Kaeser P.S. (2022) Rebuilding essential active zone functions within a synapse. Neuron 110, 1498.e8–1515.e8 10.1016/j.neuron.2022.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nachury M.V. (2018) The molecular machines that traffic signaling receptors into and out of cilia. Curr. Opin. Cell Biol. 51, 124–131 10.1016/j.ceb.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ghosh S., Di Bartolo V., Tubul L., Shimoni E., Kartvelishvily E., Dadosh T.et al. (2020) ERM-Dependent Assembly of T Cell Receptor Signaling and Co-stimulatory Molecules on Microvilli prior to Activation. Cell Rep. 30, 3434.e6–3447.e6 10.1016/j.celrep.2020.02.069 [DOI] [PubMed] [Google Scholar]
- 174.Welzel G. and Schuster S. (2022) Connexins evolved after early chordates lost innexin diversity. Elife 11, 10.7554/eLife.74422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Agnati L.F., Zoli M., Strömberg I. and Fuxe K. (1995) Intercellular communication in the brain: Wiring versus volume transmission. Neuroscience 711–726 10.1016/0306-4522(95)00308-6 [DOI] [PubMed] [Google Scholar]
- 176.Fuxe K., Borroto-Escuela D.O., Romero-Fernandez W., Zhang W.-B. and Agnati L.F. (2013) Volume transmission and its different forms in the central nervous system. Chin. J. Integr. Med. 19, 323–329 10.1007/s11655-013-1455-1 [DOI] [PubMed] [Google Scholar]
- 177.Reyes-Rivera J., Wu Y., Guthrie B.G.H., Marletta M.A., King N. and Brunet T. (2022) Nitric oxide signaling controls collective contractions in a colonial choanoflagellate. Curr. Biol. 32, 2539.e5–2547.e5 10.1016/j.cub.2022.04.017 [DOI] [PubMed] [Google Scholar]
- 178.Kim B. (2019) Evolutionarily conserved and divergent functions for cell adhesion molecules in neural circuit assembly. J. Comp. Neurol. 527, 2061–2068 10.1002/cne.24666 [DOI] [PubMed] [Google Scholar]
- 179.Jin H. and Kim B. (2020) Neurite branching regulated by neuronal cell surface molecules in Caenorhabditis elegans. Front. Neuroanatomy 14, 10.3389/fnana.2020.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Collins M.O., Husi H., Yu L., Brandon J.M., Anderson C.N.G., Blackstock W.P.et al. (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 10.1111/j.1471-4159.2005.03507.x [DOI] [PubMed] [Google Scholar]
- 181.Schoch S. and Gundelfinger E.D. (2006) Molecular organization of the presynaptic active zone. Cell Tissue Res. 326, 379–391 10.1007/s00441-006-0244-y [DOI] [PubMed] [Google Scholar]
- 182.Fernández E., Collins M.O., Uren R.T., Kopanitsa M.V., Komiyama N.H., Croning M.D.R.et al. (2009) Targeted tandem affinity purification of PSD‐95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Systems Biol. 2009, 269 10.1038/msb.2009.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Park D., Wu Y., Lee S.-E., Kim G., Jeong S., Milovanovic D.et al. (2021) Cooperative function of synaptophysin and synapsin in the generation of synaptic vesicle-like clusters in non-neuronal cells. Nat. Commun. 12, 263 10.1038/s41467-020-20462-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nam C.I. and Chen L. (2005) Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc. Natl. Acad. Sci. U.S.A. 102, 6137–6142 10.1073/pnas.0502038102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Moroz L.L. and Romanova D.Y. (2021) Selective advantages of synapses in evolution. Front. Cell Dev. Biol. 9, 726563 10.3389/fcell.2021.726563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O.et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu Y.-T. and Tao C.-L. (2022) Digitalizing neuronal synapses with cryo-electron tomography and correlative microscopy. Curr. Opin. Neurobiol. 76, 102595 10.1016/j.conb.2022.102595 [DOI] [PubMed] [Google Scholar]
- 188.Benjin X. and Ling L. (2020) Developments, applications, and prospects of cryo-electron microscopy. Protein Sci. 29, 872–882 10.1002/pro.3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hobert O. (2016) A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip. Rev. Dev. Biol. 5, 474–498 10.1002/wdev.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hobert O. (2016) Terminal selectors of neuronal identity. Curr. Top. Dev. Biol. 116, 455–475 10.1016/bs.ctdb.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 191.Kratsios P., Pinan-Lucarré B., Kerk S.Y., Weinreb A., Bessereau J.-L. and Hobert O. (2015) Transcriptional coordination of synaptogenesis and neurotransmitter signaling. Curr. Biol. 25, 1282–1295 10.1016/j.cub.2015.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dubois L. and Vincent A. (2001) The COE - Collier/Olf1/EBF - transcription factors: structural conservation and diversity of developmental functions. Mech. Dev. 108, 3–12 10.1016/S0925-4773(01)00486-5 [DOI] [PubMed] [Google Scholar]
- 193.Li Y., Osuma A., Correa E., Okebalama M.A., Dao P., Gaylord O.et al. (2020) Establishment and maintenance of motor neuron identity via temporal modularity in terminal selector function. Elife 9, 10.7554/eLife.59464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.de Mendoza A., Sebé-Pedrós A., Šestak M.S., Matejcic M., Torruella G., Domazet-Loso T.et al. (2013) Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. U.S.A. 110, E4858–E4866 10.1073/pnas.1311818110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fiers M.W.E.J., Minnoye L., Aibar S., Bravo González-Blas C., Kalender Atak Z. and Aerts S. (2018) Mapping gene regulatory networks from single-cell omics data. Brief. Funct. Genomics 17, 246–254 10.1093/bfgp/elx046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Catela C. and Kratsios P. (2021) Transcriptional mechanisms of motor neuron development in vertebrates and invertebrates. Dev. Biol. 475, 193–204 10.1016/j.ydbio.2019.08.022 [DOI] [PubMed] [Google Scholar]
- 197.Tarashansky A.J., Musser J.M., Khariton M., Li P., Arendt D., Quake S.R.et al. (2021) Mapping single-cell atlases throughout Metazoa unravels cell type evolution. Elife 10, e66747 10.7554/eLife.66747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang J., Sun H., Jiang M., Li J., Zhang P., Chen H.et al. (2021) Tracing cell-type evolution by cross-species comparison of cell atlases. Cell Rep. 34, 108803 10.1016/j.celrep.2021.108803 [DOI] [PubMed] [Google Scholar]
- 199.Fairclough S.R., Chen Z., Kramer E., Zeng Q., Young S., Robertson H.M.et al. (2013) Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 14, R15 10.1186/gb-2013-14-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Schoof E.M., Furtwängler B., Üresin N., Rapin N., Savickas S., Gentil C.et al. (2021) Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat. Commun. 12, 3341 10.1038/s41467-021-23667-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Derks J., Leduc A., Wallmann G., Huffman R.G., Willetts M., Khan S.et al. (2022) Increasing the throughput of sensitive proteomics by plexDIA. Nat. Biotechnol. 10.1038/s41587-022-01389-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Burkhardt P., Medhus A., Digel L., Naumann-Joan B., Soto-Àngel J., Norman E., Sachkova M., Kittelmann M. (2022) Syncytial nerve net in a ctenophore sheds new light on the early evolution of nervous systems. bioRxiv 10.1101/2022.08.14.503905 [DOI] [PMC free article] [PubMed] [Google Scholar]