Abstract
Reliable point-of-care (POC) rapid tests are crucial to detect infection and contain the spread of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The emergence of several variants of concern (VOC) can reduce binding affinity to diagnostic antibodies, limiting the efficacy of the currently adopted tests, while showing unaltered or increased affinity for the host receptor, angiotensin converting enzyme 2 (ACE2). We present a graphene field-effect transistor (gFET) biosensor design, which exploits the Spike-ACE2 interaction, the crucial step for SARS-CoV-2 infection. Extensive computational analyses show that a chimeric ACE2-Fragment crystallizable (ACE2-Fc) construct mimics the native receptor dimeric conformation. ACE2-Fc functionalized gFET allows in vitro detection of the trimeric Spike protein, outperforming functionalization with a diagnostic antibody or with the soluble ACE2 portion, resulting in a sensitivity of 20 pg/mL. Our miniaturized POC biosensor successfully detects B.1.610 (pre-VOC), Alpha, Beta, Gamma, Delta, Omicron (i.e., BA.1, BA.2, BA.4, BA.5, BA.2.75 and BQ.1) variants in isolated viruses and patient’s clinical nasopharyngeal swabs. The biosensor reached a Limit Of Detection (LOD) of 65 cps/mL in swab specimens of Omicron BA.5. Our approach paves the way for a new and reusable class of highly sensitive, rapid and variant-robust SARS-CoV-2 detection systems.
Keywords: SARS-CoV-2 variants, Biosensor, gFET, Point-of-care, Molecular dynamics, Omicron, Centaurus, Cerberus
Graphical Abstract
We present a graphene field-effect transistor (gFET) biosensor design, which exploits the Spike-ACE2 interaction, essential for infection. Extensive computational analyses show that a chimeric ACE2-Fc construct mimics the native receptor dimer. ACE2-Fc functionalized gFET allows in vitro detection of trimeric Spike, with a limit of detection (LOD) of 20 pg/mL. Our miniaturized POC biosensor successfully detects all prominent virus variants in both isolated viruses and patient’s clinical swabs with a LOD of 65 cps/mL.
Introduction
The pandemic caused by the highly contagious SARS-CoV-2 has greatly impacted human lives and the global economy [1], [2], [3]. To monitor the spread of SARS-CoV-2, several diagnostic tests are being developed [4], with the most widely employed ones being based on viral RNA amplification (i.e., molecular tests) or viral proteins detection via specific antibodies (i.e., antigenic test) [5], [6]. The former represents the gold standard among SARS-CoV-2 tests, but it requires a few hours of turnaround time and specialized machinery. Conversely, most antigen-detecting rapid diagnostic tests (Ag-RDTs) are fast, but show some limitations such as poor sensitivity, time-dependency (accuracy decreases after 3 days of infection), and viral load dependency [7], [8]. Furthermore, the recently emerged SARS-CoV-2 variants bearing mutations in the standard targets of antigenic tests (i.e., viral Spike and Nucleocapsid protein), impact their ability to specifically recognize the virus [9], [10], [11].
Starting from the first recognized variant D614G (i.e., B.1.610) that emerged in March 2020, the virus has rapidly evolved into a series of different variants of concern (VOC) that quickly spread all over the globe. Some of these variants, e.g., the Alpha or the Delta, were shown to be highly transmissible and cause more severe symptomatology [12], [13]. The more recently appeared Omicron, with its sublineages, show many mutations in its genome, seems to be even more contagious than the Delta, inducing a faster circulation of the virus with a higher probability of infection for the population, with an estimated R0 of 3–5 [14], [15], [16], [17].
Virus variants [13], [18] will keep appearing as long as the pandemic is not contained and an incompletely immunized host population exits (i.e., due to slow vaccine roll-out, with delays between the doses, or because of declining protection a few months after the complete vaccination) favouring the selection of antibody-escaping virus variants, as already demonstrated for Omicron [19], [20]. Consequently, Omicron and other variants had an impact on rapid tests, drastically lowering their sensitivity [19], [21]. Thus, alternative variant-robust biosensors, capable of rapidly detecting SARS-CoV-2, have vital importance in monitoring the COVID-19 outbreaks.
Thanks to their sensitivity and rapidity, graphene field-effect transistor (gFET) [22], recently proposed also for virus detection [23], [24], [25], [26], [27], [28], [29], represent a promising biosensing approach. In a gFET, a graphene monolayer connects the source and drain electrodes of a transistor and the graphene is functionalized with a bioreceptor able to specifically bind target molecules. The bioreceptor-target interaction alters graphene’s electronic properties resulting in a readily detectable signal [30]. Thus, gFETs are attractive in POC diagnosis due to their miniaturization, the potential for large-scale manufacture, operability by non-specialized personnel and reusability [31], [32], [33].
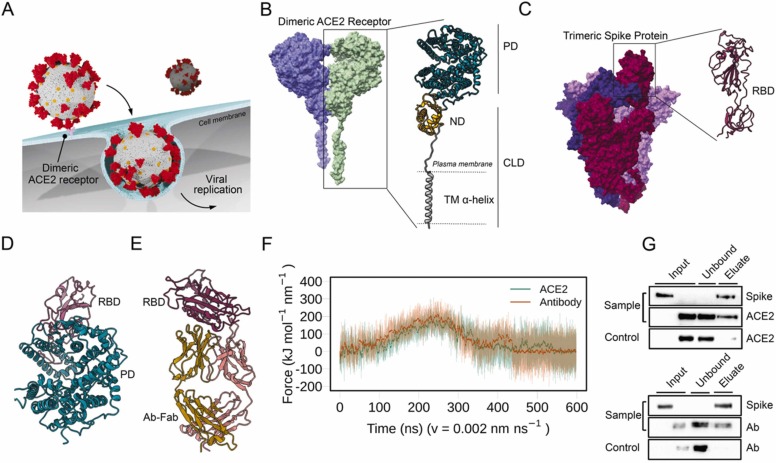
Here, integrating molecular simulations, nanobiotechnology and electronic engineering we developed a POC device that uses ACE2 as bioreceptor (i.e., the same receptor that SARS-CoV-2 uses to enter in cells), aiming to mimic the viral mechanism of host cell access [34] ( Fig. 1A). ACE2 is a widely expressed transmembrane-bound carboxypeptidase dimer composed of a collectrin-like domain (CLD) that ends with a single transmembrane (TM) α-helix and by a peptidase domain (PD) [35] (Fig. 1B). In order to use the dimeric membrane receptor (i.e., ACE2) as a functional bioreceptor on the gFET graphene surface, we used a computer-aided protein design approach to generate a stable dimeric structure of ACE2 in the absence of the cellular membrane. To do that, an ACE2-Fragment crystallizable (ACE2-Fc) chimera was generated by linking the extracellular portion of the ACE2 receptor to the immunoglobulins Fc domain. The ACE2-Fc dimer maintains a high capacity to recognize the SARS-CoV-2 Spike protein. The Spike is a trimeric transmembrane protein whose monomers are composed of two subunits, S1 and S2, with S1 containing the receptor-binding domain, RBD (Fig. 1C) [36]. Contrarily to antibodies (Ab) elicited by a former virus variant, which can be eluded by newly appearing variants (immune escape) [37], the key contact point established by the virus Spike and the host ACE2 conditions the host cell infection, indeed showing increased affinity in some of the most rapidly spreading variants [38], [39], [40], [41].
Fig. 1.
Probing the interactions of ACE2 and Antibodies with SARS-CoV-2 Spike. A) Schematic representation of ACE2-mediated host cell entry mechanism. B) Cryo-EM structure of dimeric ACE2 receptor (the two monomers are colored differently). The two subunits of a monomer are reported: Peptidase domain (PD) and Collectrin-like domain (CLD) that is composed of neck domain (ND) and transmembrane α-helix (TM α-helix). C) Cryo-EM structure of soluble trimeric Spike protein (the three monomers have different colors). A zoomed-in view of the RBD is shown. D) Peptidase Domain of the ACE2 receptor bound to the RBD of the SARS-CoV-2 Spike protein. E) Antibody Anti-Spike CR3022 (Ab-CR3022), bound to the SARS-CoV-2 Spike protein RBD. F) Force profiles from the Steered Molecular Dynamics (SMD) simulations of RBD unbinding from the ACE2 receptor (orange) and Ab-CR3022 (green). G) Pull-down assay of Spike and ACE2 (upper), and Spike and Ab-CR3022 (lower). Control is represented by the same experiment excluding the Spike protein (bait) from the system. The binding of Spike with ACE2 or Ab-CR3022 were monitored by western blot analysis.
We show that our POC device successfully detects B.1.610 (pre-VOC), Alpha, Beta, Gamma, Delta, Omicron (i.e., BA.1, BA.2, BA.4 and BA.5), Omicron BA.2.75 (i.e., Centaurus) and Omicron BQ.1 (i.e., Cerberus) variants in isolated viruses and patient’s clinical samples (i.e., nasopharyngeal swabs), making this ACE2-Fc/Spike pair a very promising approach to variant-robust SARS-CoV-2 sensing.
Materials and methods
Structural clustering of RBD binding modes in experimental complexes
The structural comparison among all available complexes of RBD with Antibody or ACE2 (see Supplementary Materials) revealed 3 different binding modes, shown in Figure S1. We chose a representative structure of each group to simulate (PDB ID: 7BEK, 7MF1 and 6YLA) and compared the steered molecular dynamics (SMD) forces necessary to obtain a complete unbinding.
Steered molecular dynamics simulations
Structures were first equilibrated with a multistage equilibration protocol adapted from [42] (details in Supplementary Materials). The SMD simulations were performed by harmonically restraining the x component of the distance between the center of mass of the backbone of the two proteins with a force constant of 10 kJ mol-1 Å-2. Two different simulations for each system were performed with a constant velocity of 0.005 nm ns-1 or 0.002 nm ns-1. See Supplementary Materials for details.
Simulation of dimeric ACE2
The three systems (ACE2-Membrane, soluble ACE2 and ACE2-Fc) were built starting from the structure 6M17 [35]. Systems were properly equilibrated with a multistage equilibration protocol [43] (see Supplementary Materials) and then simulated in absence of restraints. The first 30 ns of each simulation were discarded as a further equilibration stage, and the subsequent 500 ns were analyzed.
Correlation matrices were obtained using a modified version of g_covar, available at the GROMACS [44] user contribution page, which computes the matrix of atomic correlation coefficients. The calculation was performed on Cα atoms of PD and CLD domains of ACE2 for all the three dimeric systems, sampling the frames every 100 ps.
Non-reducing SDS-PAGE
The ACE2-Fc dimerization was assessed through SDS-PAGE [45] and carried out under non-reducing and reducing conditions. Briefly, 2 µg of ACE2-Fc or ACE2 samples were placed 10 min at 100 °C under denaturing conditions with Laemmli sample buffer reduced by β-mercaptoethanol or under non-reducing condition using a sample buffer without β-mercaptoethanol. 8% gel was used to correctly separate ACE2-Fc or ACE2 monomers from the dimers.
Purification of Trimeric Spike protein from FreeStyle HEK293-F cells
The plasmid for expression of the SARS-CoV-2 prefusion-stabilized Spike ectodomain in HEK293-F cells (Thermo Fisher) was a generous gift from the McLellan laboratory at the University of Texas at Austin [46]. 350 μg of plasmid and 1.05 mg Polyethylenimine (PEI) (Polysciences Inc.) were used to transfect cells at 1.2 × 106 cells/mL [47]. After five days in suspension culture, the cell supernatant was collected and filtered using a 0.22 µm filter. Protein in the supernatant was bound to Nickel-NTA agarose (Qiagen) while under rotation in buffer composed of 2 mM Tris, 150 mM NaCl and 10 mM imidazole for two hours at 4 °C. The resin was then washed in Tris-NaCl buffer, pH 8, with 20 mM imidazole and protein was eluted in 200 mM imidazole. Following overnight dialysis against PBS, the protein was filtered and stored as 0.2 mg/mL aliquots at − 80 °C.
Pull-down assay
The pull-down assay to validate the interaction between Spike and ACE2 or anti-Spike antibody Ab-CR3022 or ACE2-Fc was performed using a strep-tactin Sepharose resin as previously described [48], with some modifications. Briefly, 80 µL of indicated strep-tactin resin were washed with phosphate-buffered saline (PBS) pH 7.4 (173 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) and incubated with 5 μg of recombinant trimeric Spike protein with a C-terminus strep-tag (Spike-strep) next to a his-tag at room temperature for 1 h. After incubation, Spike-bound beads were washed three times with 500 µL of PBST buffer (PBS and 0.05% Tween-20) and then were aliquoted into different tubes. 5 μg of ACE2 in PBS or 5 μg of anti-Spike or 5 μg of ACE2-Fc in PBS were mixed with spike-bound beads in three different 1.5 mL tubes and incubated at room temperature for 1 h separately. After a 1-hour incubation, beads were washed three times with 500 µL PBST buffer and the bound proteins were eluted using 50 µL of elution buffer (2.5 mM biotin in PBS (Sigma)). The samples were then subjected to SDS–PAGE and analysed by western blotting using an anti-histidine antibody (Thermo Scientific) to detect Spike and ACE2, an anti-rabbit antibody (Santa Cruz Biotechnology) to detect Ab-CR3022 and an anti-ACE2 antibody (EMP Millipore Corp) to detect ACE2-Fc by chemiluminescent revelation [49]. The same protocol, using empty beads (without Spike), was performed as a negative control for each system.
gFET functionalization protocol
Anti-Spike antibody, ACE2, and ACE2-Fc were immobilized over the fabricated gFET chip (Graphenea gFET-S20) through 1-pyrenebutanoic acid succinimidyl ester (PBASE).
PBASE is an heterobifunctional linker whose pyrene group is stably immobilized with graphene by π-π stacking, while the N-hydroxysuccinimide (NHS) ester reacts with primary amines located in several biomolecules such as antibodies, thus establishing covalent bond with them [50].
20 µL of 5 mM PBASE (Thermo Fisher Scientific, Waltham, MA) in dimethylformamide (DMF) was placed on the chip for 2 h at room temperature before being rinsed three times with DMF, deionized water (DI) and dried with N2. Finally, the PBASE-functionalized devices were exposed to 20 µL of 250 μg/mL of anti-Spike (40150-R007; Sino Biological, Inc., China), ACE2 (10108-H08B-100; Sino Biological, Inc., China) or ACE2-Fc (Z03484–1; GenScript Biotech) separately and left overnight in a humidified environment at 4 °C. The sensor was then sequentially rinsed ten times with PBS (pH 7.4, 1X), three times with DI water and dried under N2 flow. The chip was subsequently treated with 20 µL of 100 mM glycine in PBS (pH 8.4, 1X) for 30 min for the termination of excess PBASE NHS groups at room temperature. After glycine treatment, samples were rinsed ten times with PBS (pH 7.4, 1X), three times with DI water and dried with N2. The reusability of GFETs was performed following manufacturer’s protocol (2021_Measurement Protocols GFET-S2X_v2), with some modifications. Briefly, we washed the chips for 30 min with water, then overnight with 100% acetone. After that, the regenerated chip was properly dried with N2 gun. The same chip was used at least 5 times to avoid the doping-reduction.
gFET characterization using Raman and AFM
Atomic Force Microscopy (AFM) measurements were performed with a SOLVER PRO from NT-MDT, RMS was evaluated by using Nova Px software. A Horiba Jobin-Yvon XploRA Raman microspectrometer, equipped with a 532-nm diode laser (∼50 mW laser power at the sample) was used. All measurements were acquired by using a × 100 long working distance objective (LMPLFLN, N.A. 0.8, Olympus). The spectrometer was calibrated to the 520.7 cm-1 line of silicon prior to spectral acquisition. A 2400 lines per mm grating was chosen. The spectra were dispersed onto a 16-bit dynamic range Peltier cooled CCD detector. The spectral range from 1100 to 3000 cm-1 was chosen and spectra were acquired for 3 × 10 s at each measurement spot. Chips were measured before and after PBASE functionalization. For each sample, 10 point/spectra were acquired, and a Raman map was acquired with the same parameters on squared areas (20 µm × 20 µm), with a step size of ∼3 µm, for a total number of 36 spectra. On each Raman map, the following values were calculated: intensity of the band centred at 2690 cm-1 (the 2D band), the intensity of the band at 1592 cm-1 (the G band), and the ratio between these two bands (I2D/IG). False colour images were built by using the I2D/IG ratio.
Electrochemical measurements
Sensing performances of gFETs were evaluated by using a Wentworth probe station equipped with a Bausch & Lomb MicroZoom optical microscope and by using an HP4145B semiconductor parameter analyzer (for recombinant proteins) and POC device (for isolated viruses and nasopharyngeal swabs). Current-voltage curves (Ids-Vds) were acquired (i) by applying a Vds between − 0.1–0.1 V, (ii) by operating in liquid gating condition with PBS solution pH 7.4, and (iii) by using a Vg of 0 V. Transfer curves (Ids-Vg) were obtained (i) by using Vds 0.050 V, (ii) by operating in liquid gating condition with PBS solution at pH 7.4, (iii) by applying Vg swept from 0 to 1.5 V, (iv) by carrying out a relaxation step to obtain a constant equilibrium of ions on the surface of graphene [23]. During this step, Vds and Vg were both applied on the gFET until no variations on the I-V curves were observed; in this way, the same ions screening effect was maintained during the measurements, and the I-V curves, taken on the same gFET at different times, were completely superimposable. After this brief equilibration stage of the ions on the surface of graphene, the blank was recorded by operating in liquid gating condition with PBS solution at pH 7.4. Then, gFET was incubated for 30 min at room temperature with different concentrations of 20 µL of sample (recombinant proteins, viruses or swabs) in PBS pH 7.4 to allow the binding with the attached bioreceptors. To avoid aspecific signals due to adsorption of molecules on the graphene channel, a wash step, using 10 mL of PBS, was performed. Only then, the electrical signal of the sample is recorded, representing the effect of the specific interaction between the bioreceptors and the ligand (Figure S2).
mPRO recombinant protein was kindly provided by Prof. Paolo Mariani from Polytechnic University of Marche [51]. Recombinant MERS-CoV Spike protein was purchased from Sino Biological (40069-V08B).
Statistical analysis
Dirac Point values for Blank and sample are compared using paired t-test implemented in Python. Significance was expressed as reported on the relative figures.
To plot comparative response the values were normalized for each gFET as follows: Normalized Vd =.
where VdA is the individual value of the Dirac point for (i) Blank or (ii) Sample: after the addition of: SARS-CoV-2 Spike protein, SARS-CoV-2 mPRO, MERS Spike protein, isolated SARS-CoV-2, HVS-1 and nasopharyngeal swabs. VdB are the Dirac point values of the blanks on each gFET with n = 6 technical replicates (i.e., number of independent graphene pads on each gFET). The limit of detection (LOD) of ACE2-Fc_gFET was calculated by measuring progressively decreasing concentrations of the Spike protein (i.e., Range of Detection: 2 µg/mL-0.002 ng/mL) and indicates the lowest concentration at which the analyte is detected [22], [52]. All experiments, including the LOD calculation, with recombinant proteins (i.e., SARS-CoV-2 Spike protein, SARS-CoV-2 mPRO, MERS Spike protein) were performed in triplicates (i.e., 3 biological samples).
SARS-CoV-2 isolation and virus stocks
Different lineages of SARS-CoV-2 were isolated from RT-PCR positive nasopharyngeal swabs collected at Ospedali Riuniti, Ancona (Italy) using Vero E6 cells (ATCC n° CRL-1586), as described by Alessandrini et al., 2020 [53]. Vero E6 cells, seeded in 75 cm2 flasks, were subsequently infected with 2 mL of the virus from the isolation to a final volume of 12 mL to obtain a larger stock. Supernatants of the infected cells were harvested after 72 h, centrifuged at 3000 rpm for 10 min, filtered using a 0.2 µm filter, aliquoted and stored at − 80 °C. Six virus stocks were sequenced and used for the present study: B.1.610 (EPI_ISL_417491), Alpha (EPI_ISL_778869), Gamma (EPI_ISL_1118260), Beta (EPI_ISL_1118258), Delta (EPI_ISL_2975994) and Omicron BA.1 (EPI_ISL_7897869).
RT-qPCR of patient samples
Clinical Samples used in this study were provided by the U.O.C. of Clinical Pathology from the hospital of Urbino (Italy) “Santa Maria della Misericordia” and from Virology Unit from Ospedali Riuniti, Ancona (Italy). Nasopharyngeal swabs from COVID-19 positive patients and COVID-19 negative were stored in PBS 1X and used. The positivity or negativity of these samples were determined by real-time RT-qPCR following manufacturer's specifications (ALLPLEX SARS-CoV-2 ASSAY and MDS methodologies). From the RT-qPCR a Cycle Threshold (Ct) value is obtained. It indicates the viral load, therefore the number of cycles after which the virus can be detected.
Ethics approval
All subjects provided written consent to be used for research purposes. The Ethics Committee of Region Marche (C.E.R.M.) approved the study on 17/12/20.
Components of C.E.R.M. are listed here:
https://www.ospedaliriuniti.marche.it/portale/index.php?id_sezione= 132&id_doc= 446&sottosezione= 37.
RT-PCR SARS-CoV-2 and Variants Detection
Viral RNA was extracted from nasopharyngeal swabs using the Kit QIAsymphony DSP Virus/Pathogen Midi kit on the QIAsymphony automated platform (QIAGEN, Hilden, Germany) according to manufacturer’s instructions. Multiplex real-time RT-PCR assay was performed using qPCRBIO Probe 1-Step Go No-Rox (PCRBIOSYSTEMS, London, UK) on the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (Thermo Fisher Scientific). The oligonucleotide primers and probes were designed to detect 69/70 deletion and N501Y mutation from virus Spike gene to discriminate alpha and gamma lineage respectively. Variant lineage was confirmed by sequence analysis of Spike gene using ABI Prism 3100 Genetic Analyzer (Applied Biosystems-HITACHI).
Calibration curve was obtained from 10-fold dilutions (105 to 102 cps/rct) of the WHO International Standard for SARS-CoV-2 RNA (cat#20/146), purchased from the National Institute for Biological Standards and Control (NIBSC) to quantify the number of copies of SARS-CoV-2 RNA per milliliter of nasopharyngeal swab ( Table 1, Table 2).
Table 1.
Table showing samples type, and relative Ct and cps/mL of all the patients tested and reported in Fig. 5 C.
| No. Patient | Sample type | Ct Average | cps/mL |
|---|---|---|---|
| 1 | RV/EV | 33,2/31,48 | / |
| 2 | HPIV 3 | 31,2 | / |
| 3 | H3N2 | 28,7 | / |
| 4 | Negative | / | / |
| 5 | Negative | / | / |
| 6 | Negative | / | / |
| 7 | Negative | / | / |
| 8 | B.1.610 | 16.23 | 7.45E+ 07 |
| 9 | Alpha | 18.12 | 9.86E+ 06 |
| 10 | Delta | 23.4 | 1.08E+ 06 |
| 11 | Delta | 14.5 | 2.17E+ 08 |
| 12 | Delta | 18.43 | 6.07E+ 06 |
| 13 | Omicron (BA.1) | 20.24 | 4.46E+ 06 |
| 14 | Omicron (BA.2) | 19.22 | 9.13E+ 06 |
| 15 | Omicron (BA.2) | 15.17 | 1.57E+ 08 |
| 16 | Omicron (BA.2) | 15.39 | 1.34E+ 08 |
| 17 | Omicron (BA.4) | 15.68 | 1.10E+ 08 |
| 18 | Omicron (BA.5) | 15.4 | 1.35E+ 08 |
| 19 | Omicron (BA.5) | 17.54 | 2.97E+ 07 |
| 20 | Omicron (BA.5) | 21.5 | 1.84E+ 06 |
| 21 | Omicron (BA.5) | 20 | 5.28E+ 06 |
| 22 | Omicron (BA.5) | 19 | 1.07E+ 07 |
| 23 | Omicron (BA.5) | 15.26 | 1.47E+ 08 |
| 24 | Omicron (BQ.1) | 23.08 | 6.07E+ 02 |
| 25 | Omicron (BQ.1) | 14.75 | 2.11E+ 05 |
Table 2.
Table showing Ct and cps/mL of all dilutions of the BA.5 swab tested and reported in Fig. 5D.
| Ct Average | cps/mL |
|---|---|
| 15.38 | 1.35E+ 08 |
| 21.84 | 1.45E+ 06 |
| 28.64 | 1.22E+ 04 |
| 32.65 | 7.32E+ 02 |
| 36.1 | 6.5E+ 01 |
| 40.05 | 4 |
Results
Soluble ACE2 and Ab binding with SARS-CoV-2 Spike protein
As a first step in exploring the possibility of using ACE2 as a bioreceptor, we characterized in silico the force profiles of the ACE2-RBD interaction (Fig. 1D), in comparison to an Antibody-RBD (Ab-RBD) complex (Fig. 1E), by performing a series of constant velocity Steered Molecular Dynamics (SMD) simulations. Since it is the interaction domain [35], RBD was used as a proxy for Spike in our simulations. To account for the various known RBD binding sites (Table S1) we used as initial starting points for SMD three representative structures of clusters obtained by structural comparison of over thirty different complexes (Figure S1). Although they show different patterns of interaction, the force needed to dissociate the RBD from ACE2 or from the most stable Ab-RBD complex (group 3) is comparable (Fig. 1F) at both velocities tested (Figure S3). A detailed description of the ACE2-RBD unbinding mechanism is shown in Figure S4. The other two structural groups exhibited lower binding forces and were thus less relevant in the comparison.
The interaction of ACE2, in its soluble truncated form, with the trimeric Spike protein was also probed with pull-down assays and western blot (Fig. 1G). Spike-decorated sepharose beads were used to pull down ACE2, and both were recovered in the eluted fractions, confirming its ability in recognizing the Spike protein. Similar pull-down results were obtained for the anti-Spike Antibody (Ab-CR3022). No Spike leakage was observed in the unbound fractions on each pull-down assay (Fig. 1G, unbound lanes). Taken together, our computational and biochemical results suggest that ACE2 receptor can be used as a bioreceptor for gFETs functionalization.
SARS-CoV-2 Spike protein detection by ACE2_gFET and Ab-CR3022_gFET
Spike detection was performed using a gFET provided by Graphenea (San Sebastian, Spain) consisting of 12 separated single-layer graphene channels connected to source and drain gold electrodes. A non-encapsulated electrode at the center of the chip allows liquid gating through PBS solution ( Fig. 2A). The bifunctional PBASE was used as a linker between graphene and bioreceptor proteins (i.e., ACE2 and Ab-CR3022) (Fig. 2 A). PBASE pyrene group stacks via π-π interactions on the aromatic graphene lattice, while the succinimide covalently binds amino groups of proteins [54]. To obtain efficient coverage, avoiding the formation of multiple layers of pyrene [55], we treated the gate electrode with a 5 mM PBASE in dimethylformamide (DMF) solution. We then characterized the bare and functionalized gFET by AFM (Figure S5) and Raman spectroscopy (Fig. 2B). Electrical characterization of pristine and activated gFET and Raman maps further confirmed the PBASE attachment to graphene (Figure S6).
Fig. 2.
gFET setup and Spike recognition. A) gFET (size 10 mm × 10 mm) is composed of two source electrodes each one connected with six graphene channels and the respective drains. A single gate electrode is used for both sides of gFET. A schematic representation of the PBASE-modified gFET is reported in the inset panel. B) Raman Spectra of gFET (black) and gFET-PBASE (red) (diode laser wavelength 523 nm and laser power 50 mW). C) Schematic representation of gFET modified with ACE2. D-E) Detail of Ids-Vg curves obtained for (D) ACE2_gFET (black) and ACE2_gFET + Spike (2 µg/mL) (red). E) ACE2_gFET (black) and ACE2_gFET + mPRO (2 µg/mL) (red). F) Comparative bar charts of ACE2_gFET before (black bars) and after (red bars) the addition of Spike (2 and 0.2 µg/mL) or mPRO (2 µg/mL). G) Schematic representation of gFET modified with Ab (Ab-CR3022). H-I) Detail of Ids-Vg curves for (H) Ab_gFET (black) and Ab_gFET + Spike (2 µg/mL) (red); (I) Ab_gFET (black) and Ab_gFET + mPRO (2 µg/mL) (red); (J) Comparative bar charts of Ab_gFET before (black bars) and after (red bars) the addition of Spike (2, 0.2 and 0.02 µg/mL) or mPRO (2 µg/mL). Details of Vg from 0 to 1.20 V are shown. In (F) and (J): * ** p < 0.001, * p < 0.05, error bars represent standard deviation (s.d.).
Next, we investigated the sensing performances of the gFET functionalized with the soluble portion of the ACE2 receptor (residues 1–740) (Fig. 2C and F) and with the antibody Ab-CR3022 (Fig. 2G and J). Experiments were conducted fixing the drain-source voltage to Vds= 50 mV and varying the gate voltage Vg from 0 to 1.5 V. In order to detect the binding between the Spike and bioreceptors, we measured the shift in the charge neutrality point (CNP) value, i.e., the gate voltage associated with the minimum of the transfer curve, also called Dirac point, that represents the most widely used electrical metric in gFET sensing [22]. The Dirac point position is altered when the electronic structure of graphene is perturbed (i.e., by target binding with its associated charges) [30]; in principle the negatively-charged molecules induce p-doping of graphene, resulting to a more positive voltage (i.e., shift to the right); oppositely, positively-charged targets prompt to n-doping and the Dirac point shifts to more negative voltages (i.e., shift to the left). However, this basic scenario is valid for simple systems tested like ions, while in the case of bigger molecules like glucose [31], [56], nucleic acids [57], [58], or proteins [59], [60], this model shows some inconsistencies. Indeed, the direction of the CNP change may be affected by several factors, such as Debye length, pH of the solution, environment, and spatial-dynamical complexity of the bioreceptor-target interaction, which involves several characteristics such as the orientation of the bioreceptor and bioreceptor-target binding mode [22], [61]. Nevertheless, the shift of the CNP, before and after the addition of the analyte, is a reporter of binding [22].
Decreasing concentrations of purified Spike protein were tested. For ACE2_gFET (Fig. 2C and F), after the addition of the Spike solution at 2 μg/mL we observed a shift of transfer curves (Fig. 2D) while the use of a Spike concentration of 0.2 μg/mL instead resulted in no significant differences (Figure S7). When we tested the Ab_gFET (Fig. 2G and J), a shift was observed both at 2 (Fig. 2H) and 0.2 μg/mL (Figure S7). Subsequent dilution (0.02 μg/mL) did not elicit a detectable Dirac point shift (Figure S7). The specificity of the bioreceptor-target binding is supported by the absence of significant shifts observed when using mPRO, the main protease of the same virus (Figs. 2E, I and S7).
ACE2-Fc chimera mimics the transmembrane ACE2 dimer
Although Ab and ACE2 bind Spike RBD with similar strength their impact on the performance of gFETs is different. To overcome the lower sensitivity of soluble truncated ACE2_gFET as compared to Ab_gFET, we modeled a chimeric version of ACE2 fused with an Fc-tag at its C-terminus (ACE2-Fc). We expected that the disulfide bridges present in the Fc-tag [62] would enforce the formation and stability of dimeric ACE2 complex in solution, mimicking what occurs in physiological conditions on the cell membrane [35].
To test this hypothesis, we computationally characterized via Molecular Dynamics (MD) simulations three systems: i) a complete ACE2 system with transmembrane (TM) helices embedded in a POPC:CHOL (90:10) bilayer ( Fig. 3A and B); ii) the soluble truncated form of ACE2 (Fig. 3 C); and iii) a chimeric system composed of the soluble portion (PD, CLD, ND) of ACE2 linked to Fc (Fig. 3D, ACE2-Fc). In the ACE2-Fc chimeric system, after a transient period of about 100 ns, we observed that the Fc domain, that has been connected to the soluble portion of the ACE2 receptor through two small unstructured regions (Fig. 3D), bends significantly (Fig. 3E), thus stabilizing the region of the receptor that is directly involved in the recognition of the Spike protein. This fast conformational change causes a more generalized structure stabilization, forcing the ACE2-Fc to maintain a stable dimeric conformation for the rest of the 500 ns sampled (Fig. 3G). This dimeric conformation is very similar to the membrane-embedded one, as shown by the distance between the PD domains center of mass, used to monitor the intra-monomer distance (Fig. 3E, F). In both membrane-embedded ACE2 and chimeric ACE2-Fc systems, the distance fluctuates around a value of 7 nm, rarely sampling more open conformations (up to 8 nm). On the contrary, in soluble ACE2 the monomers tend to separate more from each other (Fig. 3H, grey curve). The rearrangement of monomers is also evident from the time evolution of the number of contacts between them involving the PD and CLD regions, with the soluble ACE2 system losing more contacts over time (Fig. 3G, grey curve), and from the comparison of the RMSD of the two PD domains (Figure S8A). Furthermore, the presence of the Fc domain, despite its significant size (∼26 kDa), allows to considerably increase the compactness of the ACE2-Fc construct, decreasing its length. This is clearly underlined by the analysis of the gyration radius showing how the soluble ACE2, despite being smaller than the ACE2-Fc, tends to reach a more extended shape (Figs. S8C, 3E). The acquisition of a globular conformation does not influence the stabilization of the dimeric conformation, indeed an even more comprehensive reporter on the dimer dynamics is the cross-correlation of the motion of individual residues during the simulation. The corresponding maps (Fig. 3I full length vs. truncated and 3 L full length vs. ACE2-Fc) clearly highlight how the internal dynamics of ACE2-Fc resembles much more the membrane-embedded ACE2, especially in the PD domain involved in the Spike recognition. On the contrary, the soluble truncated ACE2 presents more marked differences both in the profile and intensities of the correlated motions. Overall, these results indicate that the chimeric ACE2-Fc system better preserves the dynamical properties of ACE2 in the membrane (Movie S1). This can be ascribed to an overstabilization of the CLD domains due to the close distance of the CLD-Fc connecting linker (Figure S8B) that are held together by the disulfide bridges in the Fc region (Figure S9).
Fig. 3.
MD simulations of full length, soluble and Fc tagged ACE2 dimers. A) Representative structure of the full-length ACE2 dimer embedded in a membrane. B) Same as (A) but the membrane is omitted to show the TM helices. C) Soluble truncated ACE2 conformation. D) Starting configuration of the ACE2-Fc chimera. For B-D two orthogonal views are shown. E) Representative snapshots of ACE2-Fc structures sampled during the MD trajectory, side view. The PD centers of mass distance is shown by a dashed red line. F) Top view of soluble ACE2, the PD centers of mass distance is shown by a dashed black line. G) Number of contacts between the two PD-CLD regions of monomers for membrane embedded full length ACE2 (in black), ACE2-Fc (red) and soluble ACE2 (grey). H) Time evolution of the intermonomer distance measured between the PD domains, color code as in (G). Comparative Dynamics as reported by the cross-correlation matrices of concerted motions of the residues of our three dimeric systems during the MD simulations. In (I) correlations in the full length ACE2 embedded in the membrane (upper triangle) and soluble truncated ACE2 (lower triangle). L) Same comparison as (I) between full length ACE2 embedded in the membrane (upper triangle) and ACE2-Fc (lower triangle).
The following is the Supplementary material related to this article Video S1
.
The dimerization propensity of the ACE2-Fc chimera inferred from the computational investigation was confirmed by performing SDS-PAGE electrophoresis with (Wβ) and without (W/o β) β-mercaptoethanol. When ACE2-Fc is in reducing conditions ( Fig. 4A, Wβ), the disulfide bonds in the Fc region are reduced, and the protein migrates as a monomer. On the other hand, ACE2-Fc in non-reducing conditions runs as a dimer (Fig. 4A, W/o β), consistently with the computational results. Soluble ACE2 is not affected by reducing conditions, as expected by the absence of disulfide bonds. In addition, the interaction of ACE2-Fc with the trimeric Spike protein was also confirmed with pull-down assays and western blot (Fig. 4B).
Fig. 4.
ACE2-Fc as bioreceptor and LOD calculation. A) SDS-PAGE under reducing (w β lanes) and non-reducing (w/o β lanes) conditions of soluble ACE2 and ACE2-Fc; B) Pull-down assay of Spike and ACE2-Fc. The binding of Spike with ACE2-Fc was monitored by western blot. C) Schematic representation of gFET modified with ACE2-Fc. D) Comparative bar chart showing the ACE2-Fc_gFET response to different concentrations of SARS-CoV-2 Spike. E) Comparative bar chart showing the ACE2-Fc_gFET response to MERS-CoV Spike protein (2 µg/mL) and SARS-CoV-2 mPRO (2 µg/mL). In (D) and (E): * ** p < 0.001, * * p < 0.01 and * p < 0.05, error bars represent s.d.
SARS-CoV-2 Spike protein detection by ACE2-Fc_gFET: sensitivity and specificity evaluation
gFETs were functionalized with ACE2-Fc (Fig. 4C) and the sensitivity of the system was tested using different concentrations of trimeric Spike protein (Fig. 4D and Figure S10). ACE2-Fc_gFET is able to detect the Spike protein at lower concentrations, achieving a LOD of 0,02 ng/mL (20 pg/mL), in a range of detection that spans from 2 μg/mL to 2 pg/mL of Spike (Fig. 4D and Figure S10). No significant differences are observed for mPRO (Fig. 4E). Thus, the dimeric ACE2-Fc bioreceptor is specific for Spike and provides the highest sensitivity, far outperforming both the soluble truncated ACE2 (no signal for 0.2 μg/mL, Fig. 2F) and the Ab-based system (no signal for 0.02 μg/mL, Fig. 2J). Notably, our sensitivity is comparable to that of lateral-flow based devices [63] and to other SARS-CoV-2 biosensors (Table S4). Since ACE2 is the receptor for only three coronaviruses, one of which is contained and the other, HcoV-NL63, has low binding affinity, specificity is reasonably assured [64]. To further confirm the specificity of the interaction, we also tested the closely related MERS-CoV Spike protein. We did not observe any significant signal using the MERS-CoV Spike protein in solution at 2 μg/mL (Fig. 4E and Figure S10).
Detection of SARS-CoV-2 variants from both cultured viruses and nasopharyngeal swabs by POC device
Mutations along the genome characterizing SARS-CoV-2 variants have impacted virus transmissibility and antigenicity, while influencing the sensitivity of rapid diagnostic tests [10], [11], [65]. All Spike residues of B.1.610, Alpha, Delta and Omicron subvariants for which substitutions affected recognition by antibodies are summarized in Fig. 5A. On the other hand, ACE2 is still recognized by all SARS-CoV-2 variants. Therefore, the development of a portable POC device that exploits ACE2-Fc as bioreceptor can allow to detect all circulating variants of the virus.
Fig. 5.
POC device detects SARS-CoV-2 variants in clinical samples up to low viral concentrations. A) Detailed amino acid mutations of SARS-CoV-2 Spike proteins in B.1.610, Alpha, Delta, Omicron (BA.1, BA.2, BA.4, BA.5 and BQ.1) variants, compared to the wild-type Whuan-Hu1 of SARS-CoV-2. Positions of mutations are shown both on a schematic domain representation of the protein and on the 3D model (PDB ID: 7DWZ). B) Photograph of the gFET Cartridge Unit and the Signal acquisition modules connected to form the entire POC. A reference dimension bar is reported. Also, a schematic representation of gFET modified with ACE2-Fc tested with different SARS-CoV-2 samples is shown. C) Bar graph reporting ACE2-Fc_gFET signal before (black) and after the addition of nasopharyngeal swab samples from patients (red). D) Comparative bar chart showing the ACE2-Fc_gFET response to different dilutions of Omicron BA.5 swab. Ct and cps/mL of each patient or dilution are shown in Tables 1 or 2, respectively. In (C) and (D): ***p < 0.001, **p < 0.01 and * p < 0.05, error bars represent s.d.
To improve portability, we designed and tested a miniaturized, reusable POC device accommodating the gFET chip (Fig. 5B and Figure S11), that yields the same readouts of the lab-scale probe station used during sensor implementation and testing (Figure S12). The manufacturing process is described in the Supplementary Materials, as well as the solutions adopted to realize an energy-efficient, battery-powered, Bluetooth device able to reliably detect currents around 10 μA and Dirac voltage shifts of a few millivolts.
The ability of the prototype to detect different SARS-CoV-2 variants was assessed with ACE2-Fc_gFET loaded on our POC device, testing both isolated viruses and clinical nasopharyngeal swabs. To assess the performance of the device, different isolated SARS-CoV-2 variants were tested (Figures S13 and S14). From the analysis of pre-VOC B.1.610 and four VOC, including the recently emerged Omicron BA.1, a shift of the Dirac point is always observed (Figures S13 and S14).
We report also that, for a limited number of gFETs, the direction of the CNP shift is toward lower voltages. This is not so surprising since the sensing response of a gFET is influenced by different parameters, such as the total number of adsorbed bioreceptors, their orientation on the graphene surface [22], [61], [66], the net charges and subsequent surrounding presence of anions and/or cations [22], [67] and the Debye Length [22], [68]. All these factors could lead in some cases to have the CNP shift in a different direction than expected (Figure S15A, B and C). In particular, through an accurate analysis of the MD trajectory, we observe for the ACE2-Fc construct that despite the protein remains in a stable dimeric conformation in solution increasing the degree of compactness (Figs. S8C, 3E and 3 F), it continues to sample various conformational sub-states thanks to its intrinsic structural plasticity (Figure S15D). The conformational sub-states are characterized by the fluctuation in the length of the protein and mainly by the changing of the PD domains orientation with respect to the Fc domain (Figure S15D, E and S16). These conformational sub-states can be essentially divided in two subgroups in which one conformational sub-state is sampled more often than the other by shifting the conformational balance (Figure S15D). In this scenario, it is therefore conceivable that these few differences in the direction of the Dirac point shift could be explained by the presence of different conformations of ACE2-Fc, as seen in the MD simulations, which can reorient the bioreceptor charge closer to or farther away from the FET channel, changing the surface potential thus altering the doping state of graphene in its initial value (Figure S15A, B and C). There are several examples in which the bioreceptor structural plasticity and therefore conformational changes play a crucial role in the detection of biomolecules and signals analysis in biosensing experiments [61], [69], [70], [71], [72], [73], however, this mechanism will need to be further investigated.
Nevertheless, the specificity of the target-receptor interaction allowed us to always have a significant signal in presence of the sample, while yielding no changes with our various controls (see Fig. 2, Fig. 4, Fig. 5 and Figures S7, S10, S13, S17).
Specificity was also probed using isolated Herpes Simplex virus 1 (HSV-1) that recognizes a different receptor. Even with such more complex systems, the negative samples do not show any change in the Dirac point, confirming that the specificity of the interaction and the thorough washing steps in our protocol (see Materials and Methods and Figure S2) yield significant differences only in presence of the correct viral target.
Furthermore, we tested the detection performance of the ACE2-Fc_gFET using clinical samples (Fig. 5 C, S17 and Table 1). Nasopharyngeal swabs with minimal processing, only resuspended in PBS, were used. The shift of the Dirac point (Fig. 5 C and S17) demonstrates that our POC device clearly discriminates between SARS-CoV-2 positive and negative samples. RT-qPCR results indicate that eighteen patients were positive for SARS-CoV-2, carrying one B.1.610, one Alpha, three Delta, thirteen Omicron (one BA.1, three BA.2, one BA.4, six BA.5 and two of the recently appeared BQ.1), whereas the other seven tested negative (Fig. 5C and Table 1). Among negatives, one resulted positive to RhinoVirus (RV)/EnteroVirus (EV), one to Human Parainfluenza Virus Type 3 (HPIV3) and one to Influenza A Virus (H3N2). These results confirm once again the specificity of our biosensor toward SARS-CoV-2, showing no cross-reaction with other common human viruses (Figs. 5C, S17 and Table 1). The average Ct values for both isolated viruses and patient swabs are reported in Figure S14 and Table 1 respectively, together with the corresponding cps/mL.
In addition, we have evaluated the detection limit of the real samples measuring a nasopharyngeal swab specimen (Omicron BA.5 variant) at different dilutions (Fig. 5D and Table 2). The ACE2-Fc_gFET sensor responded to patient samples diluted as much as Ct 36 (determined by RT-qPCR), which corresponds to 65 cps/mL, that we consider as our empirical LOD. This result indicates that ACE2-Fc_gFET biosensor has the potential to be used for COVID-19 diagnosis for all known variants. For the sake of completeness, we also tested samples in Universal Transport Medium (UTM) but due to the complexity of this medium (i.e., amino acids, salts and BSA), we did not achieve clear results (data not shown).
Finally, to ensure that different ACE2-Fc_gFET performed similarly, we carried out reproducibility assays using three different POC devices and three functionalized gFET (Figure S18). The tests were run on different days and yielded reproducible results.
Taken together, our findings show that our ACE2-Fc_gFET biosensor successfully detected SARS-CoV-2 from isolated virus and clinical samples without any pre-processing steps.
Discussion
Rapid, sensitive and variant-robust detection systems have been immediately acknowledged as crucial in the COVID-19 global containment strategy. Since generalized social restrictions (i.e., lockdowns) are not applicable anymore for their high social and economic costs, and vaccines are unevenly rolling out in the world, with efficacies that seem to be temporary, the virus is far from being contained. Thus, accurate detection tests able to provide quick response are even more necessary. Highly sensitive PCR-based molecular tests still require a few hours for the result, lab scale facilities and specialized personnel, whereas antigen-detecting rapid diagnostic tests (Ag-RDTs) are characterized by lower accuracy [74], which is further affected by newly emerging virus variants [21], [75]. Most Ag-RDTs were evaluated for their performance (i.e., specificity and sensitivity) far before the emergence and spread in the world population of SARS-CoV-2 VOC [75]. For example, Omicron, and its subvariants Centaurus and Cerberus, accumulated several mutations in both Spike and Nucleocapsid protein and this results in a lowered reliability of Ag-RDTs [9], [10], [11], [21], [76], [77], [78], [79]. Moreover, both testing approaches (i.e., PCR-based and Ag-RDTs) employ disposable plastic supplies in their procedures raising concerns about the environmental impacts associated with the global scale testing campaigns [80].
Here, we present a gFET sensor that exploits as bioreceptor a chimeric version of the human receptor ACE2 which outperforms an anti-Spike antibody in detecting specifically the SARS-CoV-2 Spike protein. Recently, other ACE2-based sensors have been developed [81] (Table S4), but none of them are optimized to resemble its physiological counterpart and none of them tested all the known variants. We believe that using a chimeric version of ACE2 composed of the soluble part of the protein and the Fc region of the antibodies is a better bioreceptor choice to detect SARS-CoV-2. In fact, the fusion of the Fc-tag on ACE2's C-terminus promotes stable protein dimerization, making it more similar to its native state on the cell membrane [62], as shown by our computational and in vitro biochemical analyses (Fig. 3, Fig. 4). By mimicking the actual virus-host interaction for cell infection, whose affinity has been shown to have improved in some of the late variants [38], [39], [82], our sensor is robust to current and future virus mutations. The LOD achieved by our ACE2-Fc_gFET were 20 pg/mL and 65 cps/mL for recombinant Spike and swab specimens respectively. Notably, electrochemical measurements using nasopharyngeal swab specimens did not show an optimal linear concentration-dependent response (Fig. 5 C and 5D), therefore reliable virus quantification cannot be acquired. Thus, qualitative (yes/no) response can be obtained from our gFET biosensor. Anyhow, the empirical LOD achieved using both recombinant Spike and swabs from patients confirms the high sensitivity of our gFET device, especially the one obtained with nasopharyngeal swabs which is comparable with most of the existing tests (some of them are reported for comparison in Table S4), including the gold standard RT-qPCR [83]. Nevertheless, we believe that RT-qPCR remains the most solid method to confirm a COVID-19 diagnosis. Considering this, our intent is not to substitute this test but to provide a faster screening approach.
Our biosensor, aptly miniaturized into a reusable, low-waste user-friendly POC device, was successfully tested on complex samples, such as isolated viruses and clinical samples from patients infected by the most prominent virus variants: B.1.610, Alpha, Beta, Gamma, Delta, Omicron BA.1, BA.2, BA.4, BA.5, BA.2.75 and BQ.1. The number of different types of assays also proves the robustness of the ACE2-Fc_gFET (Table S5). Our approach can detect all the circulating variants within ∼40 min using 20 µL of the sample without any pre-processing steps. The ready-to-use gFET needs only a few preparation phases, widely described in the literature and well-characterized for other gFET technologies, therefore this procedure can be easily applicable in any laboratory (Figure S2). Anyhow, drawbacks are also present, such as the cost and the commercial availability of manufactured bare-gFET chips (See Supplementary Materials text).
This is the first study to reliably detect almost all SARS-CoV-2 variants, among others the latest appeared Omicron in all its sub-lineages and the currently circulating Centaurus and Cerberus variants in different specimens (Fig. 5, S14 and S15), dissimilarly from others comparable biosensor developed (Table S4). We strongly believe that our technology could be considered as a new tool for COVID-19 management, especially for future variants, taking into account that growing evidence demonstrated that antigen tests are less sensitive for Omicron detection [77].
Furthermore, using our graphene-based POC device, with its digitized electrical recordings, ensures higher performance in the collection, handling and screening of the data as compared to most rapid tests.
Conclusions
Through a multidisciplinary effort we developed a novel point-of-care graphene-based device able to detect all known SARS-CoV-2 variants. Driven by computational and biochemical approaches that were useful to characterize the structural and dynamical properties of the chimeric ACE2-Fc construct, which mimics the functional dimeric conformation of the receptor, we attained an optimal sensitivity and specificity for the detection of SARS-CoV-2 in nasopharyngeal swabs. Our technology can complement the gold standard PCR methods and can be considered as an additional instrument in the fight against the COVID-19 pandemic, leveraging on its robustness to the virus variants, a possible pitfall of Ag-RDTs. In addition, modifications of the ACE2 amino acid sequence, which are expected to increase binding affinity with the viral Spike [62], [84], could also further improve the sensitivity of our ACE2-Fc gFET-based biosensor. In general, our novel biosensor sets the basis for a class of highly sensitive, fast, reusable and variant-robust SARS-CoV-2 detection systems that, in a close future can be extended for the detection of other diagnostic relevant biomarkers, i.e., other viruses or extracellular vesicles.
CRediT authorship contribution statement
Alice Romagnoli, Mattia D’Agostino and Eleonora Pavoni contributed equally to this work. Daniele Di Marino conceived and designed the study. Mattia D’Agostino, Eleonora Pavoni, Alice Romagnoli and Anna La Teana designed the protocols and performed experiments on gFET. Chiara Ardiccioni, Davide Mencarelli and Paolo Crippa contributed to electrical measurements. Giorgio Biagetti and Claudio Turchetti designed the electronics of the device. Valentina Notarstefano performed Raman experiments. Stefano Motta, Daniele Di Marino and Ilda D’Annessa performed and analyzed the MD simulations. Eleonora Pavoni performed AFM experiments. Simone Barocci and Sara Caucci contributed to patient samples collection and analysis. Jesmina Rexha and Stefano Menzo carried out virus isolation and culture. Brianna K. Costabile and Filippo Mancia expressed the Spike protein. Daniele Di Marino, Marco Farina, Luca Pierantoni, Francesco Cicconardi, Mauro Chinappi, Nunzio Perta, Blasco Morozzo della Rocca, and Emiliano Trucchi contributed to data interpretation. Gabriele Colasurdo designed the case and carrier units of POC. Richard Al Hadi contributed to the design of the POC. Stefano Motta, Daniele Di Marino, Mauro Chinappi, Eleonora Pavoni, Mattia D’Agostino and Blasco Morozzo della Rocca prepared the figures. Daniele Di Marino, Emiliano Trucchi, Francesco Cicconardi, Filippo Mancia, Ilda D’Annessa, Mauro Chinappi, Mattia D’Agostino, Alice Romagnoli, Stefano Motta and Blasco Morozzo della Rocca wrote the original draft. All authors reviewed the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The ACE2-Fc functionalization of gFET and the design of the POC device are under patent pending, applied by Polytechnic University of Marche. D.DM., M.DA., C.A., A.R., I.DA., D.M., E.P., P.C., G.B., L.P., M.F. are the inventors of the patent application N. 102021000000533 filed in 01/13/2021. All other authors declare they have no competing interests.
Acknowledgements
DDM acknowledges funding from the Italian Ministry of Research, Project N. FISR2020IP_03475. We acknowledge the CINECA as part of the agreement with the University of Milano-Bicocca for the availability of high-performance computing resources.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nantod.2022.101729.
Appendix A. Supplementary material
Supplementary material
Data availability
Data will be made available on request.
References
- 1.Grubaugh N.D., Hodcroft E.B., Fauver J.R., Phelan A.L., Cevik M. Cell. 2021;184:1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(2021). https://outbreak.info/ (accessed September 17, 2021).
- 3.Barlow P., van Schalkwyk M.C., McKee M., Labonté R., Stuckler D. Lancet planet. Heal. 2021;5:e102–e107. doi: 10.1016/S2542-5196(20)30291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Huang Z., Zhu J., Li C., Fang Z., Chen K., Zhang Y. Bioeng. Transl. Med. 2022 doi: 10.1002/btm2.10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toropov N., Osborne E., Joshi L.T., Davidson J., Morgan C., Page J., Pepperell J., Vollmer F. ACS Sens. 2021;6:2815–2837. doi: 10.1021/acssensors.1c00612. [DOI] [PubMed] [Google Scholar]
- 6.Yüce M., Filiztekin E., Özkaya K.G. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C., Lam E.T., Chan R.C., Tsang D.N. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krüger L.J., Tanuri A., Lindner A.K., Gaeddert M., Köppel L., Tobian F., Brümmer L.E., Klein J.A.F., Lainati F., Schnitzler P., Nikolai O., Mockenhaupt F.P., Seybold J., Corman V.M., Jones T.C., Drosten C., Gottschalk C., Weber S.F., Weber S., Ferreira O.C., Mariani D., dos Santos Nascimento E.R., Pereira Pinto Castineiras T.M., Galliez R.M., Faffe D.S., de I., Leitão C., dos Santos Rodrigues C., Frauches T.S., Nocchi K.J.C.V., Feitosa N.M., Ribeiro S.S., Pollock N.R., Knorr B., Welker A., de Vos M., Sacks J., Ongarello S., Denkinger C.M. EBioMedicine. 2022;75 doi: 10.1016/j.ebiom.2021.103774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention, 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (2021) (accessed September 17, 2021).
- 10.Adamson B., Sikka R., Wyllie A.L., Premsrirut P. MedRxiv. 2022 doi: 10.1101/2022.01.04.22268770. [DOI] [Google Scholar]
- 11.Barrera-Avalos C., Luraschi R., Vallejos-Vidal E., Mella-Torres A., Hernández F., Figueroa M., Rioseco C., Valdés D., Imarai M., Acuña-Castillo C., Reyes-López F.E., Sandino A.M. Front. Public Heal. 2022;9 doi: 10.3389/fpubh.2021.780801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., Pandey R., Brown J., Zhou J., Goonawardane N., Mishra S., Whittaker C., Mellan T., Marwal R., Datta M., Sengupta S., Ponnusamy K., Radhakrishnan V.S., Abdullahi A., Charles O., Chattopadhyay P., Devi P., Caputo D., Peacock T., Wattal C., Goel N., Satwik A., Vaishya R., Agarwal M., Chauhan H., Dikid T., Gogia H., Lall H., Verma K., Dhar M.S., Singh M.K., Soni N., Meena N., Madan P., Singh P., Sharma R., Sharma R., Kabra S., Kumar S., Kumari S., Sharma U., Chaudhary U., Sivasubbu S., Scaria V., Oberoi J.K., Raveendran R., Datta S., Das S., Maitra A., Chinnaswamy S., Biswas N.K., Parida A., Raghav S.K., Prasad P., Sarin A., Mayor S., Ramakrishnan U., Palakodeti D., Seshasayee A.S.N., Thangaraj K., Bashyam M.D., Dalal A., Bhat M., Shouche Y., Pillai A., Abraham P., Potdar V.A., Cherian S.S., Desai A.S., Pattabiraman C., Manjunatha M.V., Mani R.S., Udupi G.A., Nandicoori V., Tallapaka K.B., Sowpati D.T., Kawabata R., Morizako N., Sadamasu K., Asakura H., Nagashima M., Yoshimura K., Ito J., Kimura I., Uriu K., Kosugi Y., Suganami M., Oide A., Yokoyama M., Chiba M., Saito A., Butlertanaka E.P., Tanaka Y.L., Ikeda T., Motozono C., Nasser H., Shimizu R., Yuan Y., Kitazato K., Hasebe H., Nakagawa S., Wu J., Takahashi M., Fukuhara T., Shimizu K., Tsushima K., Kubo H., Shirakawa K., Kazuma Y., Nomura R., Horisawa Y., Takaori-Kondo A., Tokunaga K., Ozono S., Baker S., Dougan G., Hess C., Kingston N., Lehner P.J., Lyons P.A., Matheson N.J., Owehand W.H., Saunders C., Summers C., Thaventhiran J.E.D., Toshner M., Weekes M.P., Maxwell P., Shaw A., Bucke A., Calder J., Canna L., Domingo J., Elmer A., Fuller S., Harris J., Hewitt S., Kennet J., Jose S., Kourampa J., Meadows A., O’Brien C., Price J., Publico C., Rastall R., Ribeiro C., Rowlands J., Ruffolo V., Tordesillas H., Bullman B., Dunmore B.J., Fawke S., Gräf S., Hodgson J., Huang C., Hunter K., Jones E., Legchenko E., Matara C., Martin J., Mescia F., O’Donnell C., Pointon L., Pond N., Shih J., Sutcliffe R., Tilly T., Treacy C., Tong Z., Wood J., Wylot M., Bergamaschi L., Betancourt A., Bower G., Cossetti C., De Sa A., Epping M., Fawke S., Gleadall N., Grenfell R., Hinch A., Huhn O., Jackson S., Jarvis I., Krishna B., Lewis D., Marsden J., Nice F., Okecha G., Omarjee O., Perera M., Potts M., Richoz N., Romashova V., Yarkoni N.S., Sharma R., Stefanucci L., Stephens J., Strezlecki M., Turner L., De Bie E.M.D.D., Bunclark K., Josipovic M., Mackay M., Rossi S., Selvan M., Spencer S., Yong C., Allison J., Butcher H., Caputo D., Clapham-Riley D., Dewhurst E., Furlong A., Graves B., Gray J., Ivers T., Kasanicki M., Le Gresley E., Linger R., Meloy S., Muldoon F., Ovington N., Papadia S., Phelan I., Stark H., Stirrups K.E., Townsend P., Walker N., Webster J., Scholtes I., Hein S., King R., Mavousian A., Lee J.H., Bassi J., Silacci-Fegni C., Saliba C., Pinto D., Irie T., Yoshida I., Hamilton W.L., Sato K., Bhatt S., Flaxman S., James L.C., Corti D., Piccoli L., Barclay W.S., Rakshit P., Agrawal A., Gupta R.K. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. Aleem, A.B. Akbar Samad, A.K. Slenker, Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19), 2022. http://www.ncbi.nlm.nih.gov/pubmed/34033342 (accessed January 5, 2022). [PubMed]
- 14.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K.R., Dillen J.R., Powell A.E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N.M., Logue J., Iqbal N.T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P.E., Franzetti-Pellanda A., Garzoni C., Halfmann P.J., Kawaoka Y., Hebner C., Purcell L.A., Piccoli L., Pizzuto M.S., Walls A.C., Diamond M.S., Telenti A., Virgin H.W., Lanzavecchia A., Snell G., Veesler D., Corti D. Nature. 2021 doi: 10.1038/s41586-021-04386-2. [DOI] [Google Scholar]
- 15.Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., Liu X., Lambe T., Crook D., Stuart D.I., Mongkolsapaya J., Nguyen-Van-Tam J.S., Snape M.D., Screaton G.R. Lancet. 2022;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., Karim F., Bernstein M., Lustig G., Archary D., Smith M., Ganga Y., Jule Z., Reedoy K., Hwa S.-H., Giandhari J., Blackburn J.M., Gosnell B.I., Abdool Karim S.S., Hanekom W., Davies M.-A., Hsiao M., Martin D., Mlisana K., Wibmer C.K., Williamson C., York D., Harrichandparsad R., Herbst K., Jeena P., Khoza T., Kløverpris H., Leslie A., Madansein R., Magula N., Manickchund N., Marakalala M., Mazibuko M., Moshabela M., Mthabela N., Naidoo K., Ndhlovu Z., Ndung’u T., Ngcobo N., Nyamande K., Patel V., Smit T., Steyn A., Wong E., von Gottberg A., Bhiman J.N., Lessells R.J., Moosa M.-Y.S., Davenport M.P., de Oliveira T., Moore P.L., Sigal A. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trucchi E., Gratton P., Mafessoni F., Motta S., Cicconardi F., Mancia F., Bertorelle G., D’Annessa I., Di Marino D. Mol. Biol. Evol. 2021;38:1966–1979. doi: 10.1093/molbev/msaa337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. Science. 2022 doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bekliz M., Perez-Rodriguez F., Puhach O., Adea K., Melancia S.M., Baggio S., Corvaglia A.-R., Jacquerioz-Bausch F., Alvarez C., Essaidi-Laziosi M., Escadafal C., Kaiser L., Eckerle I. Clinical Microbiology. 2022 doi: 10.1128/spectrum.00853-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Béraud A., Sauvage M., Bazán C.M., Tie M., Bencherif A., Bouilly D. Analyst. 2021;146:403–428. doi: 10.1039/D0AN01661F. [DOI] [PubMed] [Google Scholar]
- 23.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.I.S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I.S.-J. ACS Nano. 2020;14:12257–12258. doi: 10.1021/acsnano.0c06726. [DOI] [PubMed] [Google Scholar]
- 24.Reina G., Iglesias D., Samorì P., Bianco A. Adv. Mater. 2021;33:1–10. doi: 10.1002/adma.202007847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong D., Wang X., Gu C., Guo M., Wang Y., Ai Z., Zhang S., Chen Y., Liu W., Wu Y., Dai C., Guo Q., Qu D., Zhu Z., Xie Y., Liu Y., Wei D. J. Am. Chem. Soc. 2021;143:17004–17014. doi: 10.1021/jacs.1c06325. [DOI] [PubMed] [Google Scholar]
- 26.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava A.K., Dwivedi N., Dhand C., Khan R., Sathish N., Gupta M.K., Kumar R., Kumar S. Mater. Today Chem. 2020;18 doi: 10.1016/j.mtchem.2020.100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmieri V., Papi M. Nano Today. 2020;33 doi: 10.1016/j.nantod.2020.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macchia E., Kovács-Vajna Z.M., Loconsole D., Sarcina L., Redolfi M., Chironna M., Torricelli F., Torsi L. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao S., Chang J., Pu H., Lu G., He Q., Zhang H., Chen J. Chem. Soc. Rev. 2017;46:6872–6904. doi: 10.1039/C6CS00827E. [DOI] [PubMed] [Google Scholar]
- 31.Huang C., Hao Z., Qi T., Pan Y., Zhao X., Mater J. 2020;6:308–314. doi: 10.1016/j.jmat.2020.02.002. [DOI] [Google Scholar]
- 32.Yu Y., Li Y.-T., Jin D., Yang F., Wu D., Xiao M.-M., Zhang H., Zhang Z.-Y., Zhang G.-J. Anal. Chem. 2019;91:10679–10686. doi: 10.1021/acs.analchem.9b01950. [DOI] [PubMed] [Google Scholar]
- 33.Sharma P.K., Kim E.-S., Mishra S., Ganbold E., Seong R.-S., Kaushik A.K., Kim N.-Y. ACS Sens. 2021;6:3468–3476. doi: 10.1021/acssensors.1c01437. [DOI] [PubMed] [Google Scholar]
- 34.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. Circ. Res. 2000;87 doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 35.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 37.Hie B., Zhong E.D., Berger B., Bryson B. Science. 2021;371:284–288. doi: 10.1126/science.abd7331. [DOI] [PubMed] [Google Scholar]
- 38.Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. Lancet Infect. Dis. 2021;21:1070. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han P., Su C., Zhang Y., Bai C., Zheng A., Qiao C., Wang Q., Niu S., Chen Q., Zhang Y., Li W., Liao H., Li J., Zhang Z., Cho H., Yang M., Rong X., Hu Y., Huang N., Yan J., Wang Q., Zhao X., Gao G.F., Qi J. Nat. Commun. 2021;12:6103. doi: 10.1038/s41467-021-26401-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J., Peng P., Cao X., Wu K., Chen J., Wang K., Tang N., Huang A. Cell. Mol. Immunol. 2022 doi: 10.1038/s41423-021-00836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., Yu J., Chik K.K.-H., Yuen T.T.-T., Yoon C., To K.K.-W., Chen H., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Yuen K.-Y., Ho D.D. Nature. 2021 doi: 10.1038/s41586-021-04388-0. [DOI] [Google Scholar]
- 42.Motta S., Minici C., Corrada D., Bonati L., Pandini A. PLOS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Marino D., Achsel T., Lacoux C., Falconi M., Bagni C. J. Biomol. Struct. Dyn. 2014;32:337–350. doi: 10.1080/07391102.2013.768552. [DOI] [PubMed] [Google Scholar]
- 44.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. Softw. 1–. 2015;2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 45.Laemmli U.K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., Chou C.-W., Byrne P.O., Hjorth C.K., Johnson N.V., Ludes-Meyers J., Nguyen A.W., Park J., Wang N., Amengor D., Lavinder J.J., Ippolito G.C., Maynard J.A., Finkelstein I.J., McLellan J.S. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longo P.A., Kavran J.M., Kim M.-S., Leahy D.J. Transient Mamm. Cell Transfection Polyethylenimine (PEI) 2013:227–240. doi: 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai X., Wang R., Filloux A., Waksman G., Meng G. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D’Agostino M., Motta S., Romagnoli A., Orlando P., Tiano L., La Teana A., Di Marino D. Front. Chem. 2020;8:1170. doi: 10.3389/fchem.2020.609942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Georgakilas V., Otyepka M., Bourlinos A.B., Chandra V., Kim N., Kemp K.C., Hobza P., Zboril R., Kim K.S. Chem. Rev. 2012;112:6156–6214. doi: 10.1021/cr3000412. [DOI] [PubMed] [Google Scholar]
- 51.Silvestrini L., Belhaj N., Comez L., Gerelli Y., Lauria A., Libera V., Mariani P., Marzullo P., Ortore M.G., Palumbo Piccionello A., Petrillo C., Savini L., Paciaroni A., Spinozzi F. Sci. Rep. 2021;11:9283. doi: 10.1038/s41598-021-88630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai C., Liu Y., Wei D. Chem. Rev. 2022 doi: 10.1021/acs.chemrev.1c00924. [DOI] [PubMed] [Google Scholar]
- 53.Alessandrini F., Caucci S., Onofri V., Melchionda F., Tagliabracci A., Bagnarelli P., Di Sante L., Turchi C., Menzo S. Genes (Basel) 2020;11:929. doi: 10.3390/genes11080929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kodali V.K., Scrimgeour J., Kim S., Hankinson J.H., Carroll K.M., de Heer W.A., Berger C., Curtis J.E. Langmuir. 2011;27:863–865. doi: 10.1021/la1033178. [DOI] [PubMed] [Google Scholar]
- 55.Thodkar K., Cazade P.-A., Bergmann F., Lopez-Calle E., Thompson D., Heindl D. ACS Appl. Mater. Interfaces. 2021;13:9134–9142. doi: 10.1021/acsami.0c18485. [DOI] [PubMed] [Google Scholar]
- 56.You X., Pak J.J. Sens. Actuators B Chem. 2014;202:1357–1365. doi: 10.1016/j.snb.2014.04.079. [DOI] [Google Scholar]
- 57.Cai B., Wang S., Huang L., Ning Y., Zhang Z., Zhang G.-J. ACS Nano. 2014;8:2632–2638. doi: 10.1021/nn4063424. [DOI] [PubMed] [Google Scholar]
- 58.Chen T.-Y., Loan P.T.K., Hsu C.-L., Lee Y.-H., Tse-Wei Wang J., Wei K.-H., Lin C.-T., Li L.-J. Biosens. Bioelectron. 2013;41:103–109. doi: 10.1016/j.bios.2012.07.059. [DOI] [PubMed] [Google Scholar]
- 59.Kim D., Oh H., Park W., Jeon D., Lim K., Kim H., Jang B., Song K. Sensors. 2018;18:4032. doi: 10.3390/s18114032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park D., Kim J.H., Kim H.J., Lee D., Lee D.S., Yoon D.S., Hwang K.S. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112505. [DOI] [PubMed] [Google Scholar]
- 61.Abrantes M., Rodrigues D., Domingues T., Nemala S.S., Monteiro P., Borme J., Alpuim P., Jacinto L. J Nanobiotechnol. 2022;20:495. doi: 10.1186/s12951-022-01695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beacon T.H., Delcuve G.P., Davie J.R. Genome. 2021;64:386–399. doi: 10.1139/gen-2020-0124. [DOI] [PubMed] [Google Scholar]
- 65.Jungnick S., Hobmaier B., Mautner L., Hoyos M., Haase M., Baiker A., Lahne H., Eberle U., Wimmer C., Hepner S., Sprenger A., Berger C., Dangel A., Wildner M., Liebl B., Ackermann N., Sing A., Fingerle V. Eurosurveillance. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.16.2100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trilling A.K., Beekwilder J., Zuilhof H. Analyst. 2013;138:1619. doi: 10.1039/c2an36787d. [DOI] [PubMed] [Google Scholar]
- 67.Fu W., Jiang L., van Geest E.P., Lima L.M.C., Schneider G.F. Adv. Mater. 2017;29:1603610. doi: 10.1002/adma.201603610. [DOI] [PubMed] [Google Scholar]
- 68.Kase H., Negishi R., Arifuku M., Kiyoyanagi N., Kobayashi Y. J. Appl. Phys. 2018;124 doi: 10.1063/1.5036538. [DOI] [Google Scholar]
- 69.Sudibya H.G., He Q., Zhang H., Chen P. ACS Nano. 2011;5:1990–1994. doi: 10.1021/nn103043v. [DOI] [PubMed] [Google Scholar]
- 70.He Q., Wu S., Yin Z., Zhang H. Chem. Sci. 2012;3:1764. doi: 10.1039/c2sc20205k. [DOI] [Google Scholar]
- 71.Wang L., Wang X., Wu Y., Guo M., Gu C., Dai C., Kong D., Wang Y., Zhang C., Qu D., Fan C., Xie Y., Zhu Z., Liu Y., Wei D. Nat. Biomed. Eng. 2022;6:276–285. doi: 10.1038/s41551-021-00833-7. [DOI] [PubMed] [Google Scholar]
- 72.Nakatsuka N., Yang K.-A., Abendroth J.M., Cheung K.M., Xu X., Yang H., Zhao C., Zhu B., Rim Y.S., Yang Y., Weiss P.S., Stojanović M.N., Andrews A.M. Science. 2018;362:319–324. doi: 10.1126/science.aao6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon J., Lee Y., Lee T., Ahn J.-H. Anal. Chem. 2020;92:5524–5531. doi: 10.1021/acs.analchem.0c00348. [DOI] [PubMed] [Google Scholar]
- 74.Jegerlehner S., Suter-Riniker F., Jent P., Bittel P., Nagler M. Int. J. Infect. Dis. 2021;109:118–122. doi: 10.1016/j.ijid.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bekliz M., Adea K., Essaidi-Laziosi M., Sacks J.A., Escadafal C., Kaiser L., Eckerle I. Lancet Microbe. 2021;2 doi: 10.1016/S2666-5247(21)00147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jian M.-J., Chung H.-Y., Chang C.-K., Lin J.-C., Yeh K.-M., Chen C.-W., Lin D.-Y., Chang F.-Y., Hung K.-S., Perng C.-L., Shang H.-S. Int. J. Infect. Dis. 2022;114:112–114. doi: 10.1016/j.ijid.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osterman A., Badell I., Basara E., Stern M., Kriesel F., Eletreby M., Öztan G.N., Huber M., Autenrieth H., Knabe R., Späth P.M., Muenchhoff M., Graf A., Krebs S., Blum H., Durner J., Czibere L., Dächert C., Kaderali L., Baldauf H.-M., Keppler O.T. Med. Microbiol. Immunol. 2022 doi: 10.1007/s00430-022-00730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., Wang Y., Wang P., Zhao L., Sun H., Yu L., Yang S., Niu X., Xiao T., Gu Q., Shao F., Hao X., Xu Y., Jin R., Shen Z., Wang Y., Xie X.S. BioRxiv. 2022 doi: 10.1101/2022.09.15.507787. [DOI] [Google Scholar]
- 79.Qu P., Evans J.P., Faraone J., Zheng Y.-M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., Panchal A., Saif L.J., Oltz E.M., Xu K., Gumina R.J., Liu S.-L. BioRxiv. 2022 doi: 10.1101/2022.10.19.512891. 2022.10.19.512891. [DOI] [Google Scholar]
- 80.Celis J.E., Espejo W., Paredes-Osses E., Contreras S.A., Chiang G., Bahamonde P. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei H., Zhang C., Du X., Zhang Z. Talanta. 2023;251 doi: 10.1016/j.talanta.2022.123813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffmann M., Zhang L., Pöhlmann S. Signal Transduct. Target. Ther. 2022;7:118. doi: 10.1038/s41392-022-00965-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fung B., Gopez A., Servellita V., Arevalo S., Ho C., Deucher A., Thornborrow E., Chiu C., Miller S. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glasgow A., Glasgow J., Limonta D., Solomon P., Lui I., Zhang Y., Nix M.A., Rettko N.J., Zha S., Yamin R., Kao K., Rosenberg O.S., Ravetch J.V., Wiita A.P., Leung K.K., Lim S.A., Zhou X.X., Hobman T.C., Kortemme T., Wells J.A. Proc. Natl. Acad. Sci. 2020;117:28046–28055. doi: 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.