Abstract
Brucella melitensis is a facultative intracellular pathogen which is able to survive and replicate within phagocytic cells. Therefore, it has to adapt to a range of different hostile environments. In order to understand the mechanisms of intracellular survival employed by virulent B. melitensis 16M, an initial approach consisting of analysis of the differences in patterns of protein synthesis in response to heat, oxidative, and acid pH stresses by two-dimensional (2-D) polyacrylamide gel electrophoresis was used. Depending on the stress, this involved about 6.4 to 12% of the 676 protein spots detected in 2-D gel electrophoresis. On the basis of N-terminal sequence analysis and database searching, 19 proteins whose level of synthesis was up- or down-regulated by stress conditions were identified. Some of them were previously reported for Brucella, such as BvrR, DnaK, GroEL, and Cu-Zn superoxide dismutase (SOD). Eight other proteins closely matched proteins found in other bacteria: AapJ, alpha-ETF, ClpP, Fe and/or Mn SOD, malate dehydrogenase, IalB, 30S ribosomal protein S1, and pyruvate dehydrogenase E1 component beta subunit. Results indicated that B. melitensis could bring specific regulatory mechanisms into play in response to stress conditions. For example, the ribosome releasing factor in B. melitensis appeared to be a heat shock protein, whereas the ClpP protein, described as a heat shock protein for Escherichia coli, was strongly down-regulated in B. melitensis in response to heat stress. Some of the identified proteins and their potential specific regulation could be required for the adaptation of B. melitensis to environmental stresses encountered in phagocytic cells and possibly for bacterial virulence.
Brucellae are facultative intracellular pathogenic bacteria that cause diseases in animals and humans (1). Six species are recognized within the genus Brucella: B. abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae (9). This classification is mainly based on differences in pathogenicity and host preference (9). The main pathogenic species worldwide are B. abortus and B. melitensis, which are involved in bovine and ovine brucellosis, respectively. Brucellae belong to a tight phylogenetic family, the family Rhizobiaceae of the α-2 subgroup of the class Proteobacteria, which includes Ochrobactrum, Bartonella, Rhizobium, and Agrobacterium (7, 10, 15, 25, 36, 37, 64, 67). In this family, bacteria live associated pericellularly or intracellularly with plant or animal cells.
The capacity of brucellae to induce disease is dependent on their ability to survive and to multiply within both host professional and host nonprofessional phagocytes (17, 55). In trophoblasts and in Vero cells, B. abortus replicates within cisternae of the rough endoplasmic reticulum (2, 3, 13, 14). The intracellular environment of phagocytic cells is, however, potentially hostile for microorganisms, threatening their viability by oxidative (myeloperoxidase-H2O2-halide) or nonoxidative (cationic proteins, proteases, lactoferritin, lysozyme, and acidification characterized by a pH of 5.4 ± 0.4) bactericidal mechanisms (22, 34). The molecular properties used by Brucella to withstand them have not yet been elucidated. But evidence suggests, for B. abortus, that this phenomenon is likely mediated by both inhibition of fusion of phagocytic vacuoles (phagosomes) to lysosomes and resistance to oxidative killing (21, 43, 47, 48).
Attempts to identify Brucella virulence factors have been performed. The first studies reported that intracellular multiplication of brucellae was attributable to erythritol (40). However, recently Sangari et al. (51) demonstrated that the genetic region implicated in erythritol catabolism was not related to virulence. Other gene products potentially implicated in virulence have been reported, such as DnaK (28), HtrA (16, 42, 49, 59), Cu-Zn superoxide dismutase (SOD) (4, 57), and RecA (58). These studies showed that Brucella deletion mutants for these proteins survived less well than the parent strains in the initial stages of host colonization. However, this attenuation was not sufficient to prevent the stage of chronic infection. Based on these observations, Sangari and Agüero (50) suggested that the virulence of Brucella could be the result of a multifactorial phenomenon. Recently, Sola-Landa et al. (56) characterized the first Brucella two-component regulatory proteic system, named BvrS-BvrR, shown to be essential for intracellular replication of B. abortus in macrophages or HeLa cells. They suggested that this system could control multiple genes not yet identified. In 1996, Rafie-Kolpin et al. (46) showed that several environmental conditions induced variations in B. abortus at the level of expression of a set of proteins. Pathogen-host interactions during bacterial infection expose bacteria to multiple physiological and biological stresses, and intracellular pathogens are known to adapt to changes in their environment, avoiding degradation by host cell defense systems by coordinated regulation of gene expression (38).
A description of the pattern of proteic expression in response to different environmental conditions would possibly provide insights into the intracellular behavior of Brucella at the molecular level. The two-dimensional (2-D) electrophoresis method has proved to be an appropriate tool to study protein expression in general and to identify stress response proteins in particular. Indeed, this technology permits the analysis of complex protein mixtures, since several hundred gene products can be visualized in one single gel (39). Previously, we have used this tool to initiate the study of the proteome of Brucella, i.e., the total set of expressed proteins (60, 61). Based on these works, we further studied the synthesis of B. melitensis proteins in response to environmental stress conditions, which are thought to be relevant to the intracellular environment encountered by brucellae during their dissemination. In order to achieve a better understanding of the impact of environmental stresses, we attempted to identify the polypeptides whose synthesis is altered by heat, oxidative, or acid pH stresses.
MATERIALS AND METHODS
Bacterial strain.
The reference and virulent strain B. melitensis 16M was provided by J.-M. Verger and M. Grayon (Brucella Culture Collection, Laboratoire de Pathologie Infectieuse et Immunologie, Institut National de la Recherche Agronomique, Nouzilly, France). Bacteria were grown on Trypticase soy agar (Bio-Mérieux) supplemented with 0.1% (wt/vol) yeast extract (Difco).
Culture conditions and radiolabeling of proteins.
After a preculture in Trypticase soy broth at 37°C, B. melitensis cells were grown to the mid-log growth phase (optical density at 600 nm of 1.0). The protocol described by Köhler et al. (28) was then followed. A 5-ml aliquot of culture was centrifuged, and the pellet was resuspended in 0.25 ml of methionine-free RPMI 1640, which was further subjected to the desired stress conditions. For all experiments, a control culture in the same medium in which the experiment was conducted and in the absence of the stress was grown at 37°C. In the experiments with acid pH, 1.0 N HCl was added to shift the pH from 7.6 to 5.5. To induce a heat shock response, the culture temperature was shifted from 37 to 42°C. For oxidative stress, the medium was supplemented with 50 mM H2O2. After preincubation with shaking for 30 min, a further 0.25 ml of medium supplemented with 150 μCi of 35S protein labeling mix was added. The bacteria were incubated for an additional 3 h and then chased with nonradioactive methionine and cysteine (10 mM final concentration). Finally, they were heat killed at 65°C for 1 h 30 min and washed three times in phosphate-buffered saline prior to analysis.
2-D gel electrophoresis of radiolabeled B. melitensis proteins.
Cell extracts for 2-D gel electrophoresis were prepared by sonic disruption with a Branson Sonifier (model 250). All cultures were disrupted in 0.20 ml of 10 mM Tris buffer (pH 7.4)–5 mM MgCl2 and a cocktail of protease inhibitors [200 μM 4-(2-aminoethyl)-benzenesulfonyl fluoride (Pefabloc-SC), 2 μM leupeptin, 1 μM pepstatin, and 100 μM EDTA]. The sonication was performed at low intensity for two or three 30-s bursts, with cooling in an ice-water bath between bursts. The mixture was then treated with 50 μg of DNase and RNase, at 37°C for 30 min. The solubilization of radiolabeled proteins and their separation by 2-D polyacrylamide gel electrophoresis were achieved according to our standardized procedure (60). Equal numbers of counts per minute (5 × 105 cpm) were loaded on each first-dimensional isoelectric focusing gel. Second-dimensional slab gels were fixed and dried at 80°C under vacuum. Radiolabeled proteins were visualized by exposing dried gels to Kodak BioMax MR-1 films for 14 days, to produce autoradiographs.
Computer-aided analysis of 2-D gels.
Autoradiographs were scanned by a charge-coupled device camera (Kodak Eikonix 1412) and analyzed with LSB Kepler software (version 8.2D). Each stress experiment was performed at least twice, and at least three repetitive 2-D gels were run for each extract. The changes in the protein expression were deemed valid if they were observed in at least n − 1 gels, n being the number of gels run for a given sample, and if they were observed in a minimum of two separate experiments. The results were analyzed using Student's t test (confidence level, 0.05), which ensured that only significant changes in the value of protein spots were taken into consideration.
Identification of proteins by N-terminal microsequencing.
To obtain the N-terminal amino acid sequences of the selected proteins, these were transferred to polyvinylidene difluoride membranes (Hyperbond; Porton Instruments, Inc., Tarzana, Calif.) and microsequenced using an automatic Beckman/Porton LF3000 protein sequencer. Searches for sequence homology were performed with the FASTA program (41).
RESULTS
General description of protein synthesis in B. melitensis 16M.
In order to elucidate patterns of response of protein expression of B. melitensis 16M to a variety of environmental stress conditions, [35S]methionine- and [35S]cysteine-labeled proteins were separated by 2-D gel electrophoresis and analyzed by a computerized analysis system. About 676 distinct protein spots were analyzed by Kepler software, in the experimental window of molecular mass of 14 to 90 kDa and with the pI range of 4.7 to 7.
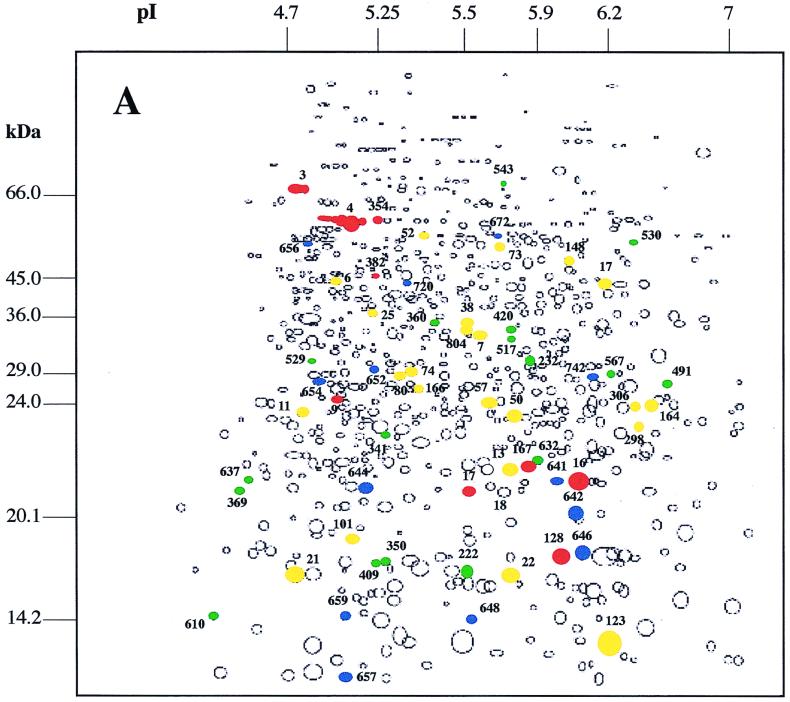
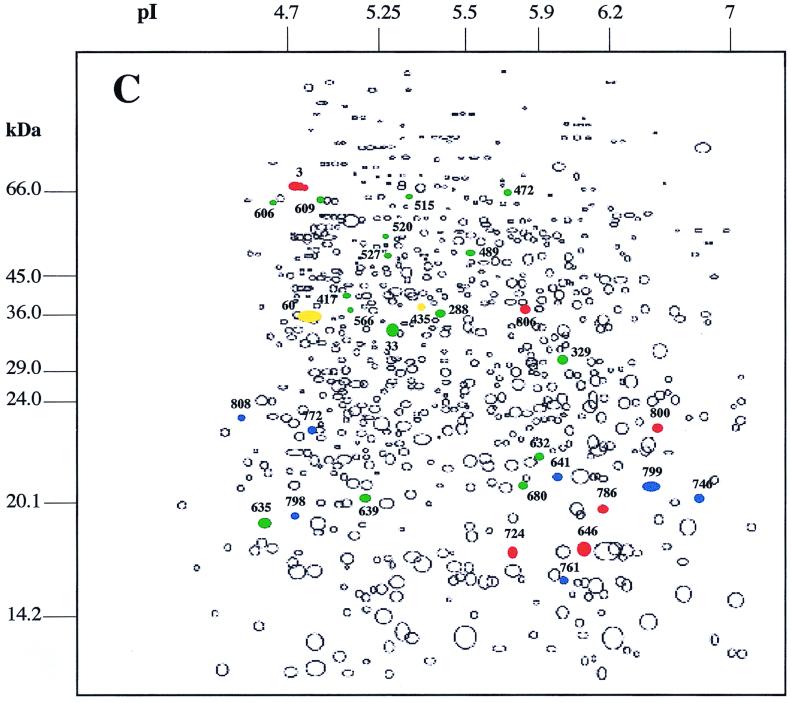
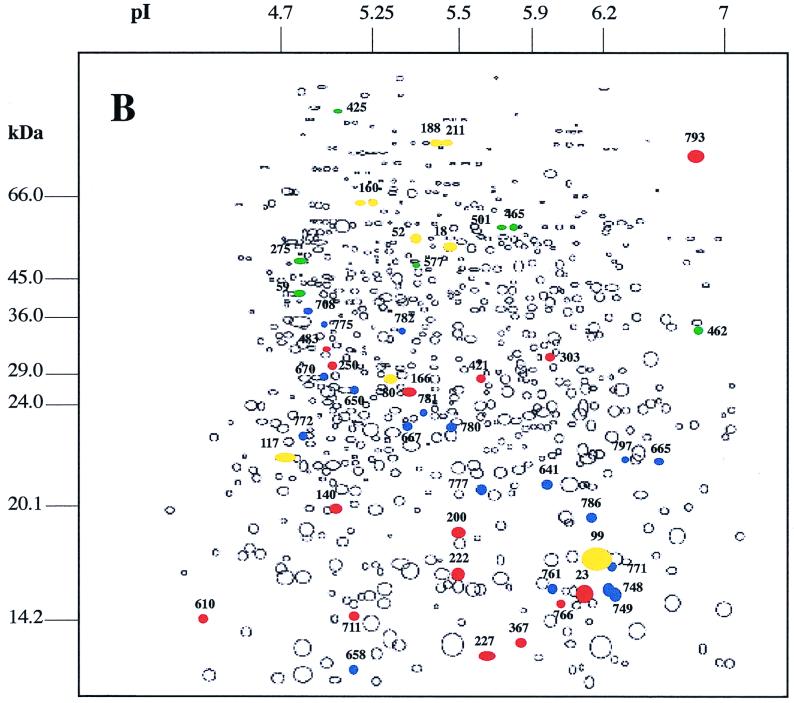
The comparison of the protein synthesis patterns of exponentially growing B. melitensis 16M cultures exposed or not to various environmental stresses allowed us to distinguish alterations in the expression of several proteins (Fig. 1). Proteins marked in each panel were found to have an altered synthesis relative to their synthesis in the unstressed control. Five subsets of proteins could be observed: (i) proteins synthesized de novo in response to the stress, (ii) proteins synthesized at an enhanced rate, (iii) proteins whose synthesis was undetectable, (iv) proteins synthesized at a reduced rate, and (v) proteins whose synthesis rate remained unchanged. In fact, only a small proportion of the total proteins have been the target of mechanisms of regulation. Depending on the stress, 6.4 to 12% of total proteins showed reproducible differences in expression level.
FIG. 1.
Schematic maps of autoradiograph produced by use of Kepler software and obtained after exposure of B. melitensis 16M to heat shock (A), H2O2 (B), and acid pH (C). Colored circles represent protein spots with altered expression under the indicated conditions. Proteins whose expression has been increased or induced during environmental stress are shown in red or blue, respectively. Proteins whose expression has decreased or that have not been detected in response to stress are shown in yellow or green, respectively. The numbers are the protein gel spot numbers as discussed in the text and in the tables.
Protein synthesis in response to heat shock.
An upshift of culture temperature from 37 to 42°C for 3 h resulted in an increase of synthesis of 31 protein spots. Among these proteins, 13 were synthesized de novo (Fig. 1A).
As we have previously shown, on a 2-D protein profile one protein can be present as a group of heterogeneous spots, which can reflect charge differences (60). Among the proteins synthesized at an enhanced rate, we detected clusters of protein spots previously identified as DnaK and GroEL proteins (60) (Table 1). Other heat shock proteins (Hsps) were identified by N-terminal microsequencing: the ribosome releasing factor (RRF) homologous protein (65), a protein showing homology to either Fe or Mn SOD from other bacteria, and a protein spot whose N-terminal amino acid sequence showed 85.7% identity with an amino acid ATP binding cassette-type transporter from Rhizobium leguminosarum, the AapJ protein (60, 66) (Table 1).
TABLE 1.
Brucella proteins assigned to the 2-D maps
|
Brucella protein characteristics
|
Most closely matching protein from databases (% identity, accession no. in OWL database) | Change in protein expression aftera:
|
|||||
|---|---|---|---|---|---|---|---|
| Spot no. | N-terminal sequence | pI | Molecular mass (kDa) | Heat shock | Oxidative stress | Acid pH | |
| 3 | AKVIGIDLGT | 4.88–4.98 | 70 | DnaK (100, DNAK_BRUOV) | ↑ | − | ↑ |
| 4 | AAKDVKFGRT | 5.1–5.2 | 57 | GroEL (100, CH60_BRUAB) | ↑ | − | − |
| 6 | ADLPGKFEGVTVNAK | 5.12 | 45 | Unknown | ↓ | − | − |
| 7 | KTSFVLPT (Y) DT (D) MV (Y) | 5.7 | 33 | Unknown | ↓ | − | − |
| 9 | KTLSDVKAKGFLQXG | 5.15 | 31 | AapJ (85.7, AAPJ_RHILV) | ↑ | − | − |
| 11 | AILLIAEHDNAQLSDQTA | 4.7 | 24 | Alpha-ETF (80, ETFA_BRAJA) | ↓ | − | − |
| 13 | NLVPMVVEQTNRGER | 5.84 | 22 | ClpP (85.7, CLPP_ECOLI) | ↓ | − | − |
| 16 | SDAFDIBBLK | 6.09 | 21 | RRF (80, RRF_BRUME) | ↑ | − | − |
| 17 | AFGLPALPYD | 5.6 | 21 | Fe and/or Mn SOD (73.1, SODF_SALTY) | ↑ | − | − |
| 21 | MKGEPKVIER | 4.7 | 18 | Bacterioferritin (100, BMU19760) | ↓ | − | − |
| 22 | KEIPIGKPQLLGGME | 5.84 | 18 | Unknown | ↓ | − | − |
| 23 | ESTTVKMYEALPTGP | 6.2 | 17 | Cu-Zn SOD (100, SODC_BRUAB) | − | ↑ | − |
| 33 | ARNKIALIGSGMIG | 5.28 | 36 | Malate dehydrogenase (100, MDH_RHILV) | − | − | ↓ |
| 50 | MKEASATQTIALVDD | 5.75 | 23 | BvrR (100, AF005157) | ↓ | − | − |
| 99 | KRHVNISP | 6.18 | 18.2 | Unknown | − | ↓ | − |
| 123 | ASLPGGASTL | 6.2 | 14 | IalB (60, INVB_BARBA) | ↓ | − | − |
| 160 | SQSNPTRADFEXLLA | 5.2 | 64 | 30S ribosomal protein S1 (64.3, RS1_RHIME) | − | ↓ | − |
| 222 | AYKDPENTLVLETTK | 5.53 | 18 | Unknown | ↓ | ↑ | − |
| 804 | XXMTMIEAIQ | 5.53 | 35.7 | Pyruvate dehydrogenase E1 component beta subunit (66.7, ODPB_MYCGE) | ↓ | − | − |
Symbols: ↑, increase; ↓, decrease; −, no change.
An appreciable reduction in synthesis of about 40 protein spots was observed in response to heat shock (Fig. 1A). For 17 of them, the expression was completely repressed. Ten proteins whose expression appeared to be reduced in 2-D gels were identified by N-terminal microsequencing (Table 1). Four proteins were previously reported for Brucella: the electron transfer flavoprotein alpha subunit (alpha-ETF), the bacterioferritin, the ClpP, and the BvrR protein (56, 60). Two protein spots not yet reported for Brucella showed homology to proteins of other bacteria: protein spot 804 with the pyruvate dehydrogenase E1 component beta subunit (20) and protein spot 123 with an invasion-associated protein of Bartonella bacilliformis, IalB (35). The N-terminal sequences of four protein spots whose synthesis rate was also reduced, i.e., protein spots 6, 7, 22, and 222, did not show any significant sequence homology with proteins found in databases. The synthesis of protein spot 222 had the distinction of being totally blocked by heat shock.
Protein synthesis in response to oxidative stress.
Oxidative stress was created by increasing the medium concentration of H2O2 to 50 mM. 2-D gel electrophoretic analysis showed that, among the 34 induced protein spots, 19 were synthesized de novo. Sixteen protein spots were repressed, while seven of them became undetectable (Fig. 1B). In response to oxidative stress, the level of expression of Cu-Zn SOD was increased, confirming previous results (46) (Table 1). Two proteins not yet reported for Brucella were repressed by this stress: the 30S ribosomal protein S1 (52) (Table 1) and the protein spot 99.
Protein synthesis in response to acid pH.
When B. melitensis 16M was subjected to pH 5.5, 15 protein spots were induced. Among these proteins, seven were synthesized de novo (Fig. 1C). DnaK was identified within the group of induced proteins as previously reported in other work (28, 46) (Table 1). The expression of about 18 protein spots was repressed in response to acid pH, while two of them became undetectable (Fig. 1C). One of these two proteins corresponded to malate dehydrogenase (Table 1).
General stress proteins.
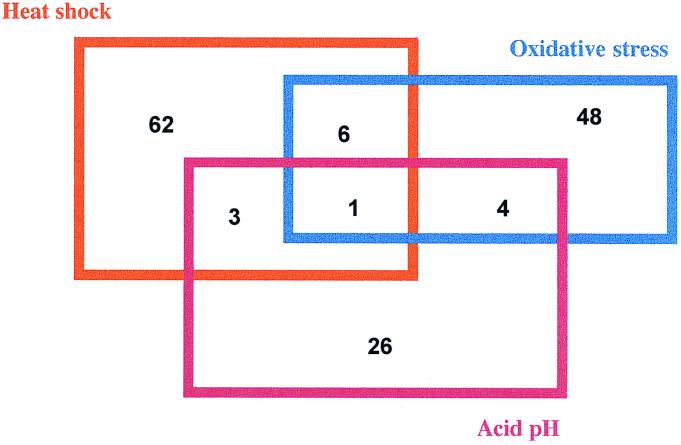
All patterns obtained in response to the various stress conditions showed that 12 proteins were regulated by at least two distinct stresses (Fig. 2). For example, DnaK was up-regulated by heat shock and acid stress at the same cellular level, as also described by Köhler et al. (28). Protein spots 166, 222, and 610 were differently regulated in response to different stresses. They were strongly induced in response to oxidative stress, whereas their expression was stopped during heat shock. Only one protein spot, at 21 kDa (spot 641), showed an enhanced expression in response to all stress conditions examined.
FIG. 2.
The number of B. melitensis 16M stress proteins whose expression was altered by various environmental stresses.
DISCUSSION
Survival and replication of brucellae in host phagocytes are key components of their virulence (17, 55). In order to understand the mechanisms of intracellular survival employed by virulent B. melitensis 16M, we envisaged a proteomic approach for a comprehensive description of protein expression in response to various environmental conditions relevant to the environment encountered during bacterial dissemination. This approach had been already initiated by Rafie-Kolpin et al. (46) with B. abortus but without identification of the proteins implicated.
The present study revealed that environmental stresses rapidly modified the gene expression in B. melitensis. In fact, as observed for other bacteria, such as Escherichia coli or Salmonella enterica serovar Typhimurium (18, 23, 63), the modifications in protein expression concerned about 10% on average of the detected proteins. 2-D gel electrophoresis detected at least 128 proteins whose expression level was modulated by at least one environmental stress. Some of them were identified by N-terminal microsequencing.
In E. coli, from which detailed data about the stress response have been obtained, the induction of the majority of up-regulated cytoplasmic proteins in response to heat shock occurs through the activation of ς32 while the periplasmic response is coordinated by ςE (8, 33, 69). In Brucella, the only Hsps described up to now were the cytoplasmic molecular chaperones DnaK and GroEL and the periplasmic HtrA protease (6, 28, 30, 46). Increased cellular levels of the two cytoplasmic molecular chaperones in Brucella during a heat shock also seem to be controlled by ς32, suggesting a fairly well conserved regulation (6, 30). Our results confirmed the heat shock induction in B. melitensis of both chaperones, DnaK and GroEL, previously described for B. abortus (6, 30, 46) and B. suis (28). As reported for the latter Brucella species (28), an induction of DnaK expression was also observed for B. melitensis in response to low pH. Thus, the Brucella DnaK protein could be subjected to multiple controls or be controlled by a single regulon which would partially overlap heat and acid shock proteins. As a consequence, as reported for S. enterica serovar Typhimurium (19), the Brucella DnaK protein might be involved in the adaptive acid tolerance response.
However, according to some of the stress proteins identified, some mechanisms of regulation of the stress response could be specific to Brucella. This concerns in particular regulation of expression of the RRF, the ClpP protease, the Fe- and/or Mn-SOD, and the bacterioferritin.
In E. coli, the RRF promoter contains two regions showing homology to the conserved −35 (TTGACA) and −10 (TATAAT) regions of E. coli ς70 promoters (53). There was no such homologous region downstream of the gene coding for the RRF of B. melitensis (65). However, we found putative −35 and −10 regions homologous to the E. coli ς32-specific promoter at nucleotide positions 110 to 116 (CCATGAA) and 127 to 134 (TGGCCGAT), respectively (GenBank accession no. U53133) (Table 2). The presence of this putative promoter could explain why the RRF was up-regulated in response to heat shock and therefore is part of the heat shock regulon in B. melitensis. This result supports also the notion that the rapid and efficient synthesis of Hsps is achieved as a result of changes in the translational capacity of the cell (54). To our knowledge, no other rrf genes with heat shock promoters have been reported. Interestingly, the elevated expression of RRF has also been reported for Staphylococcus aureus upon infection in animals (31). We have also previously shown that the B. melitensis RRF was immunogenic in infected sheep (65). Therefore, possibly for some pathogens the RRF could participate in bacterial pathogenesis. This could be demonstrated for B. melitensis by gene deletion, provided that the rrf gene is not essential for growth. In some bacteria, the RRF appears indeed to be an essential protein for bacterial growth (24).
TABLE 2.
Comparison of the putative heat shock promoter −35 and −10 regions of the Brucella RRF operon to the corresponding heat shock promoter sequences in Brucellaa
| Promoter | −35 region | Spacer (bp) | −10 region |
|---|---|---|---|
| Consensus | CCCTTGAAA | TCCCCAT | |
| RRF | tCCaTGAAA | 10 | ggCCgAT |
| GroEL | ggCTTGAAc | 12 | gCCCCAg |
| DnaK | aCCTTGAAg | 26 | TgCCCAT |
The ClpP protein is described as an Hsp with a ς32-specific promoter in E. coli (29). In contrast, as shown in the present study, in B. melitensis this protein is strongly down-regulated in response to heat stress. This discrepancy could result from a different clpP gene promoter and/or the occurrence of a negative repressor in B. melitensis. The physiological function of the ClpP protein in Brucella is yet unknown. In prokaryotes, most ATP-dependent ClpP proteases are involved in protein catabolism under both optimal and stress conditions (44). As heat shock leads to severe down-regulation of its expression in B. melitensis, it is possible that alternative proteases may compensate for its absence.
Of particular interest is also the Fe and/or Mn SOD (protein spot 17) identified in our study as an Hsp. In E. coli, the control of Mn SOD induction during a heat shock is independent of ς32 regulation (62). It has been postulated that reactive oxygen induced in the onset of the heat shock response may be involved in the induction of this antioxidant metalloenzyme, resulting in protection of the cell (45). In a previous study (60), we had identified another protein spot (spot 18 in Fig. 1A) showing the same molecular mass and the same N-terminal amino acid sequence but a different pI (5.8). The lack of up-regulation of the latter protein in response to heat stress suggests that at least two homologous Fe and/or Mn SODs occur in B. melitensis and that their expression is under the control of different promoters.
Finally, expression of the B. melitensis bacterioferritin previously identified by Denoel et al. (11) appeared reduced in response to heat shock but was not increased in response to oxidative stress. In Pseudomonas aeruginosa, bacterioferritin limits labile iron levels and assists in the protection of the cell against oxidative stress (32). According to our results, the bacterioferritin production in B. melitensis is not altered in the face of exposure to oxidative stress, and this is in accordance with the results of Denoel et al. (12), who reported that the bacterioferritin gene was not at all required for survival in the macrophage.
Besides the four proteins mentioned above, there are three other proteins identified in our study that merit at present further attention, namely, BvrR and the IalB and AapJ homologous proteins. BvrR is a protein homologous to a regulator found in bacterial genera of the same family as Brucella, such as Rhizobium, and which has been recently found to be essential in virulence of B. abortus, in particular in invasion and survival in the macrophage (56). We have found this protein perhaps surprisingly down-regulated in response to heat shock. This regulator is of particular interest since the mutant for this protein was selected on the basis of increased sensitivity to polycations that are known to interact with surface molecules of the outer membrane such as lipopolysaccharide and possibly outer membrane protein-lipopolysaccharide complexes. Thus, the BvrR regulator in Brucella is probably at least partially responsible for regulation of expression of important surface molecules of Brucella possibly required for full virulence. IalB is an invasion-associated protein described for B. bacilliformis, also a bacterium closely genetically related to Brucella (35). The expression of this protein appeared also reduced in B. melitensis in response to heat shock. Little is known about surface proteins required for invasion of Brucella, and thus this protein merits also further studies. AapJ is a periplasmic protein and a component of an l-amino acid permease found in R. leguminosarum (66). The control of AapJ expression is presumed to aid in adjusting the cellular amino acid pool. This protein was actually up-regulated in B. melitensis in response to heat shock, and interestingly, it has also been found previously to be immunogenic in infected sheep (61).
Many bacterial stress proteins are targets of the host immune response in a broad spectrum of infections (26, 27, 38, 68). It has been reported that their immunodominance is probably related to their abundance within antigen-presenting cells under conditions of stress (5, 27, 38). Previously, we identified some proteins important in the humoral immune responses of B. ovis- and B. melitensis-infected sheep combining 2-D gel electrophoresis, N-terminal microsequencing, and immunoblotting results (61). Among them, we identified DnaK, GroEL, and Cu-Zn SOD as highly immunogenic proteins which have the potential to be required or to participate in intracellular survival (28, 46, 57). The new proteins identified in the present study as stress proteins, such as the B. melitensis RRF and the AapJ homologous proteins, merit further studies with regard to their potential role in virulence since they were also identified as interesting immunogenic proteins in infected sheep.
ACKNOWLEDGMENTS
Ana P. Teixeira-Gomes was supported by a grant from the Ministère de l'Education Nationale, de la Recherche et de la Technologie.
We are grateful to Guy Bézard for microsequencing assistance.
REFERENCES
- 1.Alton G G. Brucella melitensis. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 383–409. [Google Scholar]
- 2.Anderson T D, Cheville N F. Ultrastructural morphometric analysis of Brucella abortus-infected trophoblasts in experimental placentitis. Am J Pathol. 1986;124:226–237. [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson T D, Cheville N F, Meador V P. Pathogenesis of placentitis in the goat inoculated with Brucella abortus. Ultrastructural studies. Vet Pathol. 1986;23:227–239. doi: 10.1177/030098588602300302. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin C L, Jiang X, Fernandes D. Macrophage control of Brucella abortus: influence of cytokines and iron. Trends Microbiol. 1993;1:99–104. doi: 10.1016/0966-842x(93)90115-8. [DOI] [PubMed] [Google Scholar]
- 5.Buchmeier N A, Heffron F. Induction of Salmonella stress proteins upon infection of macrophages. Science. 1990;248:730–732. doi: 10.1126/science.1970672. [DOI] [PubMed] [Google Scholar]
- 6.Cellier M F M, Teyssier J, Nicolas M, Liautard J P, Marti J, Sri Widada J. Cloning and characterization of the Brucella ovis heat shock protein DnaK functionally expressed in Escherichia coli. J Bacteriol. 1992;174:8036–8042. doi: 10.1128/jb.174.24.8036-8042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherwonogrodzky J W, Dubray G, Moreno E, Mayer H. Antigens of Brucella. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 19–64. [Google Scholar]
- 8.Chuang S-E, Blattner F R. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbel M J, Brinley-Morgan W J. Genus Brucella Meyer and Shaw 1920, 173AL. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 377–388. [Google Scholar]
- 10.De Ley J, Mannheim W, Segers P, Lievens A, Denijn M, Vanhoucke M, Gillis M. Ribosomal ribonucleic acid cistron similarities and taxonomic neighborhood of Brucella and CDC group Vd. Int J Syst Bacteriol. 1987;37:35–42. [Google Scholar]
- 11.Denoel P A, Zygmunt M S, Weynants V, Tibor A, Lichtfouse B, Briffeuil P, Limet J N, Letesson J-J. Cloning and sequencing of the bacterioferritin gene of Brucella melitensis 16M strain. FEBS Lett. 1995;361:238–242. doi: 10.1016/0014-5793(95)00189-g. [DOI] [PubMed] [Google Scholar]
- 12.Denoel P A, Crawford R M, Zygmunt M S, Tibor A, Weynants V E, Godfroid F, Hoover D L, Letesson J-J. Survival of a bacterioferritin deletion mutant of Brucella melitensis 16M in human monocyte-derived macrophages. Infect Immun. 1997;65:4337–4340. doi: 10.1128/iai.65.10.4337-4340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detilleux P G, Deyoe B L, Cheville N F. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect Immun. 1990;58:2320–2328. doi: 10.1128/iai.58.7.2320-2328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detilleux P G, Deyoe B L, Cheville N F. Entry and intracellular localization of Brucella spp in Vero cells: fluorescence and electron microscopy. Vet Pathol. 1990;27:317–328. doi: 10.1177/030098589002700503. [DOI] [PubMed] [Google Scholar]
- 15.Dorsch M, Moreno E, Stackebrandt E. Nucleotide sequence of 16S rRNA from Brucella abortus. Nucleic Acids Res. 1989;17:1765. doi: 10.1093/nar/17.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elzer P H, Phillips R W, Robertson G T, Roop R M., II The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect Immun. 1996;64:4838–4841. doi: 10.1128/iai.64.11.4838-4841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright F M. The pathogenesis and pathobiology of Brucella infection in domestic animals. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 301–320. [Google Scholar]
- 18.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster J W. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit Rev Microbiol. 1995;21:215–237. doi: 10.3109/10408419509113541. [DOI] [PubMed] [Google Scholar]
- 20.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J L, Nguyen D T, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 21.Frenchick P J, Markham R J F, Cochrane A H. Inhibition of phagosome-lysosome fusion in macrophages by soluble extracts of virulent Brucella abortus. Am J Vet Res. 1985;46:332–335. [PubMed] [Google Scholar]
- 22.Geisow M J, D'Arcy Hart P, Young M R. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: studies by fluorescence spectroscopy. J Cell Biol. 1981;89:645–652. doi: 10.1083/jcb.89.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickey E W, Hirshfield I N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl Environ Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janosi L, Shimizu I, Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Natl Acad Sci USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann S H E. Heat shock proteins and the immune response. Immunol Today. 1990;11:129–136. doi: 10.1016/0167-5699(90)90050-j. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann S H E. Introduction: heat shock proteins in protection, surveillance and pathogenesis. Semin Immunol. 1991;3:1–3. [Google Scholar]
- 28.Köhler S, Teyssier J, Cloeckaert A, Rouot B, Liautard J-P. Participation of the molecular chaperone DnaK intracellular growth of Brucella suis within U937-derived phagocytes. Mol Microbiol. 1996;20:701–712. doi: 10.1111/j.1365-2958.1996.tb02510.x. [DOI] [PubMed] [Google Scholar]
- 29.Kroh H E, Simon L D. The ClpP component of Clp protease is the ς32-dependent heat shock protein F21.5. J Bacteriol. 1990;172:6026–6034. doi: 10.1128/jb.172.10.6026-6034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, Adams L G, Ficht T A. Characterization of the heat shock response in Brucella abortus and isolation of the genes encoding the GroE heat shock proteins. Infect Immun. 1992;60:2425–2431. doi: 10.1128/iai.60.6.2425-2431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 32.Ma J-F, Ochsner U A, Klotz M G, Nanayakkara V K, Howell M L, Johnson Z, Posey J E, Vasil M L, Monaco J J, Hassett D J. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mecas J, Rouviere P E, Erickson J W, Donohue T J, Gross C A. The activity of ςE, an Escherichia coli heat-inducible ς-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 34.Miller R A, Britigan B E. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell S J, Minnick M F. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect Immun. 1995;63:1552–1562. doi: 10.1128/iai.63.4.1552-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno E. Evolution of Brucella. In: Plommet M, editor. Prevention of brucellosis in the mediterranean countries. Wageningen, The Netherlands: Pudoc Scientific; 1992. pp. 197–218. [Google Scholar]
- 38.Murray P J, Young R A. Stress and immunological recognition in host-pathogen interactions. J Bacteriol. 1992;174:4193–4196. doi: 10.1128/jb.174.13.4193-4196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 40.Pearce J H, Williams A E, Harris-Smith P W, Fitzgeorge R B, Smith H. The chemical basis of the virulence of Brucella abortus. II. Erythritol, a constituent of bovine foetal fluids which stimulates the growth of Brucella abortus in bovine phagocytes. Br J Exp Pathol. 1962;43:31–36. [Google Scholar]
- 41.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips R W, Elzer P H, Roop R M., II A Brucella melitensis high temperature requirement A (htrA) deletion mutant demonstrates a stress response defective phenotype in vitro and transient attenuation in the BALB/c mouse model. Microb Pathog. 1995;19:277–284. [PubMed] [Google Scholar]
- 43.Pizarro-Cerda J, Moreno E, Sanguedolce V, Mege J-L, Gorvel J-P. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun. 1998;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porankiewicz J, Wang J, Clarke A K. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- 45.Privalle C T, Fridovich I. Induction of superoxide dismutase in Escherichia coli by heat shock. Proc Natl Acad Sci USA. 1987;84:2723–2726. doi: 10.1073/pnas.84.9.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rafie-Kolpin M, Essenberg R C, Wyckoff J H., III Identification and comparison of macrophage-induced proteins and proteins induced under various stress conditions in Brucella abortus. Infect Immun. 1996;64:5274–5283. doi: 10.1128/iai.64.12.5274-5283.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riley L K, Robertson D C. Ingestion and intracellular survival of Brucella abortus in human and bovine polymorphonuclear leukocytes. Infect Immun. 1984;46:224–230. doi: 10.1128/iai.46.1.224-230.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riley L K, Robertson D C. Brucellacidal activity of human and bovine polymorphonuclear leukocyte granule extracts against smooth and rough strains of Brucella abortus. Infect Immun. 1984;46:231–236. doi: 10.1128/iai.46.1.231-236.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson G T, Elzer P H, Roop R M., II In vitro and in vivo phenotypes resulting from deletion of the high temperature requirement A (htrA) gene from the bovine vaccine strain Brucella abortus S19. Vet Microbiol. 1996;49:197–207. doi: 10.1016/0378-1135(96)84554-8. [DOI] [PubMed] [Google Scholar]
- 50.Sangari F J, Agüero J. Molecular basis of Brucella pathogenicity: an update. Microbiol SEM. 1996;12:207–218. [PubMed] [Google Scholar]
- 51.Sangari F J, Grillo M J, Jiménez De Bagües M P, Gonzalez-Carrero M I, Garcia-Lobo J M, Blasco J M, Agüero J. The defect in the metabolism of erythritol of the Brucella abortus B19 vaccine strain is unrelated with its attenuated virulence in mice. Vaccine. 1998;16:1640–1645. doi: 10.1016/s0264-410x(98)00063-2. [DOI] [PubMed] [Google Scholar]
- 52.Schnier J, Thamm S, Lurz R, Hussain A, Faist G, Dobrinski B. Cloning and characterization of a gene from Rhizobium melilotii 2011 coding for ribosomal protein S1. Nucleic Acids Res. 1988;16:3075–3089. doi: 10.1093/nar/16.7.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu I, Kaji A. Identification of the promoter region of the ribosome-releasing factor cistron (frr) J Bacteriol. 1991;173:5181–5187. doi: 10.1128/jb.173.16.5181-5187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sierra J M, Zapata J M. Translational regulation of the heat shock response. Mol Biol Rep. 1994;19:211–220. doi: 10.1007/BF00986963. [DOI] [PubMed] [Google Scholar]
- 55.Smith L D, Ficht T A. Pathogenesis of Brucella. Crit Rev Microbiol. 1990;17:209–230. doi: 10.3109/10408419009105726. [DOI] [PubMed] [Google Scholar]
- 56.Sola-Landa A, Pizarro-Cerda J, Grillo M-J, Moreno E, Moriyon I, Blasco J-M, Gorvel J-P, Lopez-Goni I. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. 1998;29:125–138. doi: 10.1046/j.1365-2958.1998.00913.x. [DOI] [PubMed] [Google Scholar]
- 57.Tatum F M, Detilleux P G, Sacks J M, Halling S M. Construction of Cu-Zn superoxide dismutase deletion mutants of Brucella abortus: analysis of survival in vitro in epithelial and phagocytic cells and in vivo in mice. Infect Immun. 1992;60:2863–2869. doi: 10.1128/iai.60.7.2863-2869.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatum F M, Morfitt D C, Halling S M. Construction of a Brucella abortus RecA mutant and its survival in mice. Microb Pathog. 1993;14:177–185. doi: 10.1006/mpat.1993.1018. [DOI] [PubMed] [Google Scholar]
- 59.Tatum F M, Cheville N F, Morfitt D. Cloning, characterization and construction of htrA and htrA-like mutants of Brucella abortus and their survival in BALB/c mice. Microb Pathog. 1994;17:23–36. doi: 10.1006/mpat.1994.1049. [DOI] [PubMed] [Google Scholar]
- 60.Teixeira-Gomes A P, Cloeckaert A, Bézard G, Dubray G, Zygmunt M S. Mapping and identification of Brucella melitensis proteins by two-dimensional electrophoresis and microsequencing. Electrophoresis. 1997;18:156–162. doi: 10.1002/elps.1150180128. [DOI] [PubMed] [Google Scholar]
- 61.Teixeira-Gomes A P, Cloeckaert A, Bézard G, Bowden R A, Dubray G, Zygmunt M S. Identification and characterization of Brucella ovis immunogenic proteins using two-dimensional electrophoresis and immunoblotting. Electrophoresis. 1997;18:1491–1497. doi: 10.1002/elps.1150180824. [DOI] [PubMed] [Google Scholar]
- 62.Touati D. Transcriptional and posttranscriptional regulation of manganese superoxide dismutase biosynthesis in Escherichia coli, studied with operon and protein fusions. J Bacteriol. 1988;170:2511–2520. doi: 10.1128/jb.170.6.2511-2520.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.VanBogelen R A, Kelley P M, Neidhardt F C. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Velasco J, Romero C, Lopez-Goni I, Leiva J, Diaz R, Moriyon I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol. 1998;48:759–768. doi: 10.1099/00207713-48-3-759. [DOI] [PubMed] [Google Scholar]
- 65.Vizcaíno N, Cloeckaert A, Dubray G, Zygmunt M S. Cloning, nucleotide sequence, and expression of the gene coding for a ribosome releasing factor-homologous protein of Brucella melitensis. Infect Immun. 1996;64:4834–4837. doi: 10.1128/iai.64.11.4834-4837.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walshaw D L, Poole P S. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol Microbiol. 1996;21:1239–1252. doi: 10.1046/j.1365-2958.1996.00078.x. [DOI] [PubMed] [Google Scholar]
- 67.Yanagi M, Yamasato K. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol Lett. 1993;107:115–120. doi: 10.1111/j.1574-6968.1993.tb06014.x. [DOI] [PubMed] [Google Scholar]
- 68.Young D B, Mehlert A, Smith D F. Stress proteins and infectious diseases. In: Morimoto R I, Tissières A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 131–165. [Google Scholar]
- 69.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]