Abstract
Hybrid insulin peptides (HIPs) form in pancreatic β-cells through the formation of peptide bonds between proinsulin fragments and other peptides. HIPs have been identified in pancreatic islets by mass spectrometry and are targeted by CD4 T cells in patients with type 1 diabetes (T1D) as well as by pathogenic CD4 T-cell clones in nonobese diabetic (NOD) mice. The mechanism of HIP formation is currently poorly understood; however, it is well established that proteases can drive the formation of new peptide bonds in a side reaction during peptide bond hydrolysis. Here, we used a proteomic strategy on enriched insulin granules and identified cathepsin D (CatD) as the primary protease driving the specific formation of HIPs targeted by disease-relevant CD4 T cells in T1D. We also established that NOD islets deficient in cathepsin L (CatL), another protease implicated in the formation of disease-relevant HIPs, contain elevated levels of HIPs, indicating a role for CatL in the proteolytic degradation of HIPs. In summary, our data suggest that CatD may be a therapeutic target in efforts to prevent or slow the autoimmune destruction of β-cells mediated by HIP-reactive CD4 T cells in T1D.
Introduction
In type 1 diabetes (T1D), autoreactive CD4 T cells mediate the destruction of insulin-producing β-cells. Hybrid insulin peptides (HIPs), which form in β-cells through peptide bond formation between proinsulin fragments and other β-cell peptides, represent a type of posttranslationally modified antigen targeted by autoreactive CD4 T cells in T1D (1–9). HIPs contain nongenomically encoded amino acid sequences, making them plausible targets for T cells that drive the destruction of β-cells. Indeed, several diabetes-triggering CD4 T-cell clones in nonobese diabetic (NOD) mice react to HIPs as antigens (1,2,4). In the human disease, presence of HIP-reactive CD4 T cells has been verified in residual pancreatic islets of deceased organ donors with T1D (2,3). Significantly elevated levels of various HIP-reactive T cells have also been detected in the peripheral blood of patients with recent-onset T1D, but not in control subjects without diabetes (8). Furthermore, the presence of various disease-relevant HIPs has been validated by mass spectrometry in pancreatic islets of humans and mice (5,10).
A number of HIPs identified in mouse and human islets by mass spectrometry share a common C-peptide fragment on the N-terminal side (left-peptide) of the HIP junction. Various diabetogenic NOD mouse–derived CD4 T-cell clones, including BDC-2.5, BDC-6.9, and NY4.1, target HIPs with this C-peptide fragment linked to natural cleavage products of chromogranin A (ChgA; 2.5HIP), islet amyloid polypeptide (IAPP; 6.9HIP), or insulin C-peptide (HIP11), respectively (1,2,4). T cells that target the human form of HIP11 were also detected at elevated frequencies in peripheral blood mononuclear cells (PBMCs) of patients with recent-onset T1D, but not in control subjects without diabetes (8). A CD4 T-cell clone (E2) was isolated from the PBMCs of one of these patients, and it was established that E2 recognizes HIP11 at low nanomolar concentrations (8). An HIP11-reactive CD4 T-cell receptor was also detected in residual pancreatic islets of an organ donor with T1D (5). The C-peptide fragment (left-peptide) shared by HIP11 and other HIPs, which is termed Des-(27-31)C-peptide (11), lacks the five C-terminal amino acid residues of the intact C-peptide (see Table 1). Previous analyses by Verchere et al. (11) had identified Des-(27-31)C-peptide as a major β-cell secretory product, cosecreted from β-cells with insulin and C-peptide upon glucose stimulation. These data indicate that a highly specific proteolytic mechanism exists within β-cell secretory granules driving the formation of Des-(27-31)C-peptide. Notably, of the HIPs so far identified by mass spectrometry in human and murine islets, the HIPs containing Des-(27-31)C-peptide as a left-peptide are the ones for which disease-relevant T-cell specificities have also been identified (1,2,4,5,7). Proteases (enzymes that cleave proteins into peptides) have been known to mediate not only the hydrolytic cleavage of peptide bonds but also the formation of new peptide bonds through transpeptidation reactions (12,13). Water thereby competes with peptides for a specific peptide bond within the target peptide in transpeptidation reactions. Therefore, protease-mediated hydrolysis or transpeptidation reactions typically occur at the same peptide bond within the peptide. Here, we hypothesized that a specific protease drives the primary formation of Des-(27-31)C-peptide as well as the corresponding HIPs in insulin granules.
Table 1.
Defined relevant peptide and HIP sequences
| Peptide sequence | Peptide origin | HIP-reactive T cells |
|---|---|---|
| Homo sapiens | ||
| EAEDLQVGQVELGGGPGAGSLQPLALEGSLQ | C-peptide | |
| EAEDLQVGQVELGGGPGAGSLQPLAL | Des-(27-31)-C-peptide | |
| EAEDLQVGQVELGGGPGAGSLQPLAL-EAEDLQVGQVELGGG… | HIP11 (human) | E2, GSE.8E3 |
| NOD mice | ||
| EVEDPQVAQLELGGGPGAGDLQTLALEVAQQ | C-peptide (insulin 2) | |
| EVEDPQVAQLELGGGPGAGDLQTLAL | Des-(27-31)C-peptide | |
| EVEDPQVAQLELGGGPGAGDLQTLAL-EVEDPQVAQLELGGG… | HIP11 (mouse) | NY4.1 |
| EVEDPQVAQLELGGGPGAGDLQTLAL-WSRMDQLAKELTAE | 2.5HIP | BDC-2.5, BDC-10.1, |
| WSRMDQLAKELTAE | ChgA: WE14 | BDC-9.46 |
| WSRMDQ | ChgA: WQ6 | |
| EVEDPQVAQLELGGGPGAGDLQTLAL-NAARDPNRESLDFLLV | 6.9HIP | BDC-6.9, BDC-9.3 |
| NAARDPNRESLDFLLV | IAPP: IAPP2 |
Research Design and Methods
Mice
Female and male NOD and NOD.RIP-TAg mice were bred and housed at University of Colorado Denver in specific pathogen-free conditions, as described previously (1,2). Female and male NOD, NOD.cathepsin L (CatL)+/−, and NOD.CatL−/− were obtained from The Jackson Laboratory and housed in the University of Alabama at Birmingham Research Support Building in specific pathogen-free conditions (14). Islets and RIP-TAg tumors were isolated from male and female mice. Mouse sex was not considered a factor in the statistical analysis of the data. All experiments were conducted under a protocol approved by the University of Alabama at Birmingham and the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committees.
Insulin Secretory Granule Enrichment From β-Cells
NOD.RIP-TAg mice produce β-cell tumors via transduction of the rat insulin promoter with the large T antigen transgene. Approximately 800 mg of harvested tumor tissue was used as starting material for each experiment. In the absence of protease inhibitors, the tumor tissue was homogenized through a 70-μm filter and sheared through 22-, 27-, and 30-gauge needles consecutively. Secretory granules were then enriched from the homogenate through differential centrifugation, as published previously (2,15).
Fractionation of Proteases From β-Cell Granules
Enriched secretory granules underwent lysis with 2% octyl β-glucoside in buffer containing 80 mmol/L NaCl, 10 mmol/L Na2HPO4, and 10 mmol/L acetic acid adjusted to pH 7.0 with NaOH. Undissolved contents were pelleted at 17,000g for 5 min at 4°C, and the supernatant was collected and separated using size exclusion chromatography (SEC) (BioLogic Fast Protein Liquid Chromatography System, Bio-Rad) with the same buffer at a flow rate of 0.8 mL/min. Eight fractions (2 mL each) were collected starting at 7.8 min (6.2 mL).
Islet Isolation and Preparation
Islets used as an antigen source were prepared from whole-mouse pancreata at the University of Colorado Diabetes Research Center Tissue Procurement and Processing Core (K. Scott Beard) and in the laboratory of Dr. Hubert Tse at the University of Alabama at Birmingham, as described elsewhere (16). We observed no differences in the sex of mice used for islet isolation.
Cathepsin Reactions
Sodium acetate buffer (100 mmol/L) at the indicated pH values was used for the following reactions. Various concentrations of mouse cathepsin D (CatD; Sino Biological cat no. 50127-M08H) was incubated in the presence of 0.2 μmol/L mouse insulin 2 C-peptide (SynPeptide, >95% purity), with or without 0.2 μmol/L of the mouse ChgA peptide WQ6 (SynPeptide, >95% purity). Cysteine protease reactions with CatL (Sino Biological, cat no. 50015-M08H), cathepsin Z (Sino Biological cat no. 50089-M08H), cathepsin B (Sino Biological cat no. 50084-M08H), cathepsin S (Sino Biological cat no. 50769-M08H), and cathepsin H (Sino Biological cat no. 50619-M08H) were incubated in the presence of insulin 2 C-peptide and WQ6 peptide at the indicated concentrations (pH 5.0). Indicated concentrations of human CatD (Sino Biological cat no. 12517-H08H) were incubated with 0.8 μmol/L full-length human insulin C-peptide (SynPeptide, >95% purity) at the indicated pH values.
Maintenance of Mouse and Human T cells
Murine T-cell clones (BDC 2.5, BDC 9.3) were restimulated every 2 weeks, as previously described (17), with enriched β-cell granules (an antigenic preparation referred to as “beta membrane”) (15), antigen-presenting cells (APCs), and interleukin 2. The human T-cell clone E2 was grown and maintained as described elsewhere (5).
Mouse and Human T-Cell Antigen Assays
Murine T-cell clone assays (18) contained responder T cells (20,000/well), NOD peritoneal exudate cells (25,000/well) as APCs, and antigen (HIP-formation reactions, peptides, islet cells). Antigen samples in medium were added to the T-cell clones/APCs and incubated for 48 h in a 96-well tissue culture–treated U-bottom plate (Falcon cat no. 35-3077). Murine supernatants were harvested, and the concentration of interferon-γ (IFN-γ) was measured by ELISA (2,18). The hybrid insulin peptides 2.5HIP, 6.9HIP, and HIP14 (SLQPLAL-NAVEVLK) (GenScript, >90% purity) were used as positive and negative controls at a concentration of 10 μmol/L for mouse antigen assays.
The human E2 T-cell clone (50,000 cells/well) was incubated with APCs (irradiated autologous human Epstein-Barr virus cell line, 50,000/well) (8). Antigen samples in medium were added to the T-cell clones/APCs as described above. IFN-γ harvested from the human E2 clone was measured by an Invitrogen Human IFN-γ Uncoated ELISA kit (cat no. 88-7316-88). HIP11 and HIP14 (GenScript, >90% purity) were used as positive and negative controls at a concentration of 10 μmol/L for human antigen assays.
Liquid Chromatography–Tandem Mass Spectrometry Analysis
NOD, NOD.CatL+/−, and NOD.CatL−/− islets were prepared for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses, as described elsewhere (10). Digested SEC fractions (Trypsin, AspN, and GluC are the designated names used for these reagents in research), in vitro reactions, and prepared NOD islets were analyzed by LC-MS/MS using an Agilent 1200 series ultra-high-performance liquid chromatography system with a nanoflow adapter and an Agilent 6550 Quadrupole Time-of- Flight (Q-TOF) equipped with a nano-electrospray ionization source. Online separation was accomplished by reversed-phase liquid chromatography using a Thermo Acclaim Pepmap 100 C18 trap column (75 μm × 2 cm; 3-μm particles; 100-Å pores) and Thermo Acclaim Pepmap RSLC C18 analytical column (75-μm inner diameter; 2-μm particles; 100-Å pores) in a trap forward-elute configuration using a water/acetonitrile gradient (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid and 90% acetonitrile in water). A detailed description of the mass spectrometry data collection was previously published (19). Data were analyzed using the Spectrum Mill software with the SwissProt human/mouse database and HIP sequence databases as described below. Search settings were as follows: instrument = Agilent ESI Q-TOF; precursor mass tolerance = +/− 10 ppm; product ion mass tolerance = +/− 20 ppm; digest = no enzyme. Matches were considered valid if the following thresholds were satisfied: score >10, percentage scored peak intensity (SPI) >70%, and rank 1 minus rank 2 (R1 − R2) score >2.5.
HIP Sequence Databases
Databases were generated using an in-house computer algorithm. The mouse HIP database contained each possible C-terminal truncation of insulin C-peptide linked to predicted as well as identified naturally occurring cleavage products of insulin (1 and 2) (11), ChgA (20), islet amyloid polypeptide (21), secretogranin 1, and neuropeptide Y, making a total of 3,000 peptides. More detail of this database and our established set of confidence criteria are provided elsewhere (10). The human HIP database contained all 30 HIP sequences that can form between C-peptide fragments on the left side linked to intact C-peptide on the right side.
Confidence Criteria for Putative HIP Identifications
1) Spectrum cannot be confidently matched to any peptides in a traditional protein database. 2) Spectrum matches with high confidence and little ambiguity to a peptide in the HIP database. 3) Peptide match corresponds to the proteolytic digest performed in the sample preparation. 4) The left peptide and right peptide regions each contain at least two amino acid residues. 5) The spectrum contains sufficient b-/y-ion coverage of both the left and right peptide region of the HIP (10).
Validation of HIPs Using Peptide-Spectrum Matches Validation With Internal Standards Approach
HIP peptides in our samples were digested with AspN to obtain a shorter peptide sequence. A synthetic version of the AspN-digested HIP sequence was obtained for validation purposes. This validation peptide and the biological/reaction sample were each spiked with a panel of internal standard peptides (ISPs). Peptide-spectrum matches (PSM)_validator was then used to compare the fragmentation spectrum of the biological/reaction peptide to the spectrum of the validation peptide by calculating the Pearson correlation coefficients (PCCs), which were also calculated for each of the ISPs. The PCCs of the ISPs were then used to calculate a 95% CI. If the PCC of the peptide of interest falls within the 95% CI, then it is considered a fully validated peptide. A detailed description of the PSM validation approach with the PSM_validator algorithm and internal standards (P-VIS) was previously published (19).
Synthetic Peptides and ISPs
PROCAL (22) (JPT Peptide Technologies, Berlin, Germany) was used as ISPs. Other synthetic peptides were obtained from GenScript or SynPeptide at chromatographic purities of ≥90%.
Data and Resource Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identifications (PRIDE) partner repository with the data set identifier PXD032664. The PSM_validator analyzed during the current study is available freely at https://github.com/Delong-Lab/PSM_validator/releases under the Creative Commons Attribution 4.0 International Public License.
Results
CatD From Enriched β-Cell Granule Lysates Drives Formation of 2.5HIP
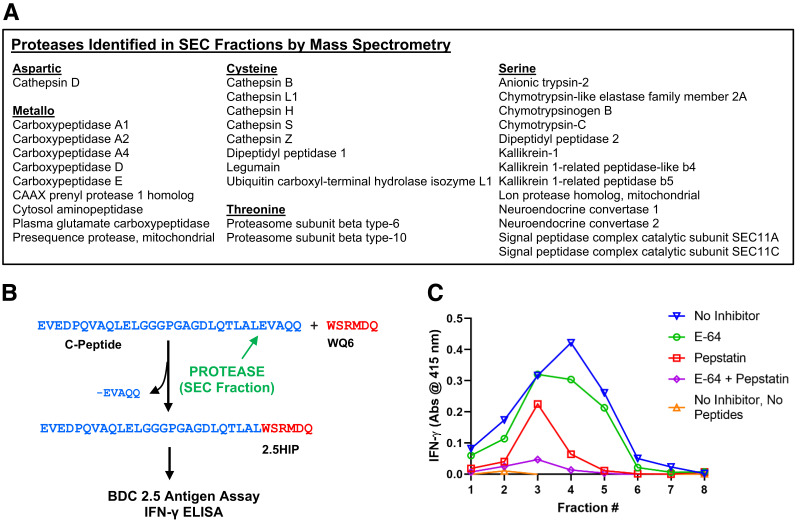
We used an untargeted proteomic strategy to identify a granular protease that specifically drives the formation of HIPs between Des-(27-31)C-peptide and other natural β-cell peptides. Insulin granules were enriched from β-cell tumors of NOD.RIP-TAg mice as previously described (2). Granular lysates were then fractionated by SEC in the absence of protease inhibitors. Mass spectrometric analyses of these fractions led to the identification of various proteases belonging to the five functional protease families (Fig. 1A). Proteolytic transpeptidation reactions require the presence of a protease as well as precursor proteins or peptides (Fig. 1B). To determine whether SEC fractions of granular lysates contain a protease that drives the formation of 2.5HIP, we incubated the fractions in the presence and absence of intact murine C-peptide as well as the ChgA peptide WQ6, which contains the N-terminal amino acid sequence of the naturally occurring ChgA cleavage product WE14 (20) (see Table 1). Unlike WE14, WQ6 does not stimulate the CD4 T-cell clone BDC-2.5 in an antigen assay, allowing us to add WQ6 to the SEC fractions without altering the endogenous antigen content of these fractions. We predicted that in the presence of our protease of interest a covalent bond would form between the C-terminus of Des(27-31)C-peptide and the N-terminus of the WQ6 peptide, leading to the formation of 2.5HIP (Fig. 1B). As shown in Fig. 1C (orange triangle line), in the absence of precursor peptides, BDC-2.5 did not respond to any fraction following a 2-h incubation. Although the presence of HIPs in granule lysates has been previously established (2,23), in these lysate SEC fractions, the 2.5HIP content was below the detection limit, possibly due to the proteolytic degradation of 2.5HIP in the absence of protease inhibitors. In the presence of precursor peptides, BDC-2.5 responded strongly to several fractions, peaking at fraction 4, (blue inverted triangle line), and indicating that a 2.5HIP-forming protease was present in these fractions. We also incubated the fractions with precursor proteins in the presence of cysteine and/or aspartic protease inhibitors, to determine whether formation of the 2.5HIP could be blocked. Synthesis of the 2.5HIP was reduced when the cysteine protease inhibitor E-64 (8 μmol/L) was used (Fig. 1C, green circle line). In contrast, the majority of 2.5HIP formation was blocked in the presence of the aspartic protease inhibitor pepstatin (220 μmol/L) (Fig. 1C, red square line). When both pepstatin and E-64 were used, almost all 2.5HIP formation was prevented (Fig. 1C, purple diamond line). We conclude that while one or more cysteine proteases partially contribute to the formation of 2.5HIP, it is CatD, the only aspartic protease identified in our fractions (see Fig. 1A), that is the primary catalyst in this reaction.
Figure 1.
Identification of HIP-forming proteases in β-cell lysates. A: Enriched insulin secretory granules were fractionated by SEC. Mass spectrometric analyses of the fractions led to the identification of 30 distinct proteases. B: Schematic of a proteolytic transpeptidation reaction leading to the formation of 2.5HIP between Des-(27-31)C-peptide and the ChgA fragment WQ6. C: BDC-2.5 T-cell responses to fractions incubated with or without 2.5HIP precursor peptides in the presence and absence of various inhibitors. See control experiments in Supplementary Fig. 7A. Experiment was done in triplicate. Above data show one representative experiment.
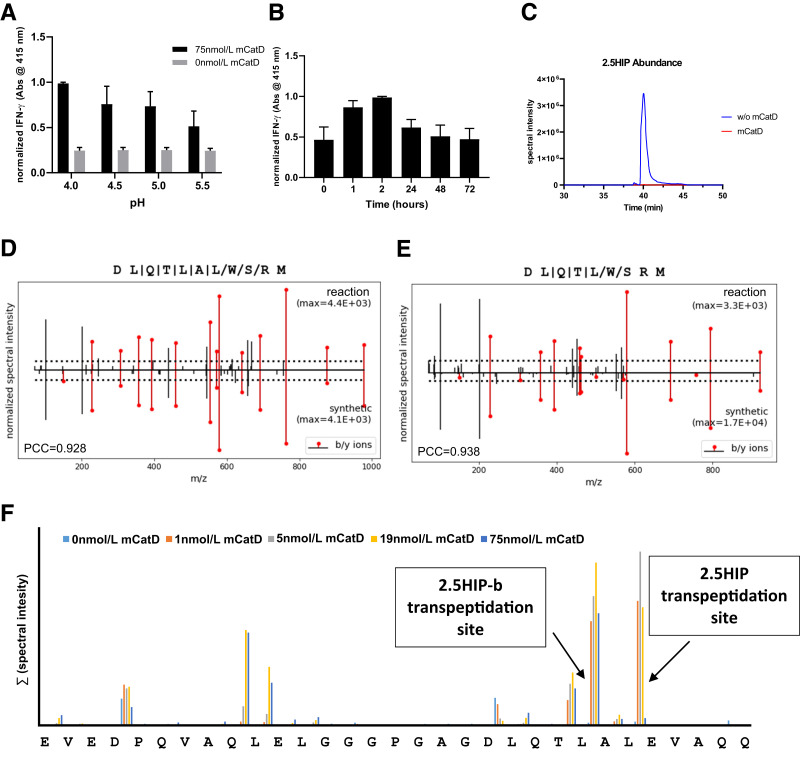
Formation of 2.5 HIP by Recombinant Murine CatD
To better characterize CatD-mediated HIP formation, we incubated C-peptide and the ChgA fragment WQ6 in the presence of recombinant murine CatD (see Fig. 1B) and then assessed 2.5HIP formation using the BDC-2.5 T-cell assay system. We tested reactions containing CatD and precursor peptides at pH 4.0, 4.5, 5.0, and 5.5, encompassing the hydrolytic pH-optimum of CatD (pH 4.5–5.0) (24) and the physiological pH of various β-cell organelles, such as insulin granules (pH 5.0–6.0) (25), lysosomes (pH 4.5–5.0) (26), or crinosomes, which form through the fusion between lysosomes and insulin granules (27–29). Following a 2-h incubation in presence of CatD, the yield of 2.5HIP was highest at pH 4.0 and declined to the minimal yield at pH 5.5 (Fig. 2A). In the absence of CatD, T-cell responses remained at background levels at all the pH values tested, indicating that 2.5HIP did not form at levels that could elicit an IFN-γ response from the BDC-2.5 T-cell clone. This also indicates that the presence of precursor peptides, C-peptide and WQ6, alone do not elicit a T-cell response. During long incubations with high CatD concentrations, intact C-peptide substrate may be depleted due to proteolytic processing by CatD and therefore would no longer be available for the formation of new 2.5HIP. To determine whether prolonged incubations in the presence of CatD can lead to the proteolytic degradation of newly formed 2.5HIP, we performed a time course analysis of 2.5HIP formation at pH 4.0 for up to 72 h. Following incubation, yield of 2.5HIP was the highest after 2 h, but with additional time, the yield declined to background levels (Fig. 2B), indicating that CatD can mediate not only the formation but also the degradation of the 2.5HIP antigen upon depletion of the C-peptide substrate. To confirm CatD is responsible for the degradation of 2.5HIP over time, we incubated a synthetic version of the 2.5HIP (DLQTLAL-WSRM) in the presence and absence of CatD for 24 h (Fig. 2C). The 2.5HIP was degraded only in the presence of CatD after a 24-h incubation. In addition to monitoring the IFN-γ production of BDC-2.5 in antigen assays, we also verified the presence of 2.5HIP in our samples by mass spectrometry. For this, we stopped the CatD reactions after 2 h through the addition of the aspartic protease inhibitor pepstatin and then digested the samples with the metalloprotease AspN, allowing us to obtain a peptide spanning the 2.5HIP junction that has an adequate length for mass spectrometric validation (DLQTLAL-WSRM). Following mass spectrometric analyses, we used our established P-VIS validation method (19) and verified the presence in these samples of 2.5HIP (Fig. 2D and Supplementary Fig. 1) as well as another HIP (2.5HIP-b) with the amino acid sequence DLQTL-WSRM (Fig. 2E and Supplementary Fig. 2). We previously reported that 2.5HIP-b is recognized by some, but not all, 2.5HIP-reactive T-cell clones (2) from the BDC panel, leading us to define 2.5HIP-b as a secondary T cell target in NOD mice. Using our established set of confidence criteria (10) for the discovery of HIPs (see Research Design and Methods), no other HIPs were detected that may have formed at lower yields between C-peptide and WQ6 in these samples.
Figure 2.
Cleavage and transpeptidation site-specificity of murine (m)CatD. A: BDC-2.5 T-cell responses to in vitro reactions (pH 4.0–5.5) of 2.5HIP precursor peptides (insulin 2 C-peptide and WQ6) (see Table 1) incubated for 2 h in the presence and absence of mCatD. B: BDC-2.5 T-cell responses to precursor peptides incubated in the presence of mCatD (pH 4.0) for 0, 1, 2, 24, 48, and 72 h. Results from A and B are the mean ± SEM from three independent experiments. Data were normalized to the maximal IFN-γ signal for each experiment. See control experiments in Supplementary Fig. 7B. C: Incubation for 24 h of AspN-digested synthetic 2.5HIP sequence at pH 4.0 in the presence and absence of 75 nmol/L murine CatD. Following AspN digestion, the in vitro formation of 2.5HIP (D) and 2.5HIP-b (E) was validated using the P-VIS protocol (19). Figure shows mirror plots generated during peptide validation. F: Following 24-h treatment of murine C-peptide with variable concentrations of mCatD, the spectral intensities of peptides resulting from cleavages at individual peptide bonds were summarized and graphed.
To assess the C-peptide cleavage site specificity of CatD, we incubated murine C-peptide in the presence and absence of various CatD concentrations at pH 4.0 for 24 h and then analyzed these samples by mass spectrometry. Spectral intensities of detected peptides describing individual cleavage sites were then summarized. As shown in Fig. 2F, we identified two major CatD cleavage sites within the murine C-peptide. The first site is at the C-terminus of the Des-(27-31)C-peptide site. Another preferred cleavage site was at C-peptide amino acid residue 24, the site at which 2.5HIP-b forms. In the presence of high CatD concentrations, the detectable spectral intensities of peptides describing various peptide bonds declined. This is a likely result of further proteolytic processing of these peptides, leading to the formation of smaller, singly charged peptide ions, the analysis of which was not pursued by mass spectrometry. In sum, our data demonstrate that murine CatD selectively drives the formation of 2.5HIP and 2.5HIP-b and that the cleavage site specificity of murine CatD aligns with the transpeptidation site specificities.
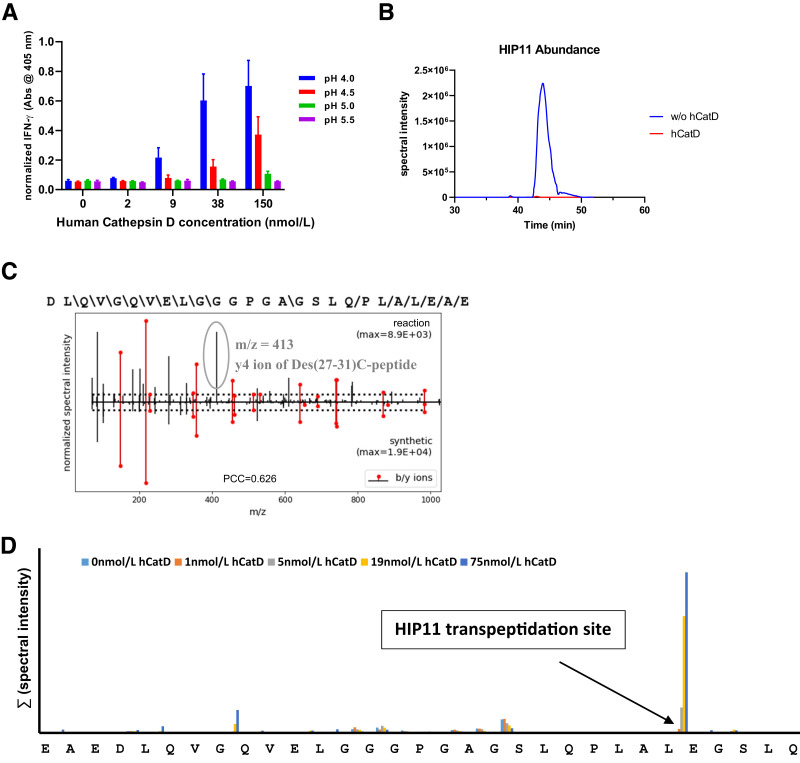
Recombinant Human CatD Selectively Forms HIP11
To test HIP formation with the human form of CatD, we incubated several concentrations of recombinant human CatD in the presence of human C-peptide at pH 4.0–5.5 with the expectation that HIP11 would form between the Des-(27-31)C-peptide fragment and the N-terminus of human C-peptide (see Table 1). In our previous studies, we isolated a HIP11-reactive CD4 T-cell clone (E2) from PBMCs of a patient with recent-onset T1D (8). To measure the transpeptidation yields of HIP11 from intact human C-peptide in the presence of human CatD we used our E2 T-cell assay system (see research design and methods) (8). As a negative control, samples were incubated with C-peptide in the absence of CatD. Based on the human E2 T cell assays, the highest yields of HIP11 were accumulated after 24 h of incubation in the presence of 150 nmol/L CatD at pH 4.0 (Fig. 3A). Lower or no yields were obtained when the reactions were performed at higher pH values or lower CatD concentrations. In the absence of CatD, HIP11 formation could not be detected, and T-cell responses remained at background levels. To determine whether human CatD is capable of HIP degradation, as was the case with murine CatD (Fig. 2C), we incubated a higher concentration of human CatD (225 nmol/L) in the presence of a synthetic version of HIP11 (DLQVGQVELGGGPGAGSLQPLAL-EAE). After a 24-h incubation, HIP11 degradation was evident and only in the presence of human CatD (Fig. 3B). Using Spectrum Mill analysis, we also verified presence of HIP11 by mass spectrometry in these samples (score: 16.0; SPI: 80%), with Spectrum Mill scores exceeding standard validation parameters (score: >10.0; SPI: >70%). However, we were unable to validate HIP11 formation in these samples using our rigorous P-VIS protocol due to HIP11 and Des-(27-31)C-peptide having identical masses and close coelution times during LC-MS/MS analyses (see sequences in Table 1). Because of identical masses, these peptides could not be separated by the instrument’s quadrupole, causing ions of both peptides to be concurrently present in the fragmentation chamber. As a result, fragmentation spectra of the low-abundance HIP11 ions were cross-contaminated with high-intensity ions of Des-(27-31)C-peptide. As indicated in Fig. 3C, the y4-ion of Des-(27-31)C-peptide is cross-contaminating the fragmentation spectrum of HIP11, making rigorous validation unfeasible. However, Spectrum Mill scoring and our data with the CD4 T-cell clone E2 confirm the formation of HIP11 in these samples. Using our confidence criteria (10), we did not identify other HIPs that may have formed in these reactions.
Figure 3.
Cleavage and transpeptidation site-specificity of human (h)CatD. A: E2 T-cell responses to in vitro reactions (pH 4.0–5.5) of HIP11 precursor peptide (C-peptide) incubated for 24 h in the presence of CatD. Results shown are the mean ± SEM from three independent experiments. Data were normalized to maximal IFN-γ signal for each experiment. See control experiments in Supplementary Fig. 7C. B: Incubation for 24 h of AspN-digested synthetic HIP11 sequence at pH 4.0 in presence and absence of 225 nmol/L human CatD. C: Mirror plot of synthetic HIP11 and HIP11 that formed in the presence of CatD and C-peptide. Data show the y4 ion of Des-(27-31)C-peptide that coisolated with the HIP11 ion during LC-MS/MS analyses. m/z, mass-to-charge. D: Following 24-h treatment of human C-peptide with variable concentrations of human (h)CatD, the spectral intensities of peptides resulting from cleavages at individual peptide bonds were summarized and graphed.
C-peptide cleavage products that formed in the presence of various CatD concentrations were also detected by mass spectrometry. To assess C-peptide cleavage site specificity of human CatD, we summarized spectral intensities of peptides describing individual cleavage sites for C-peptide (Fig. 3D). The highest spectral yields of CatD-mediated cleavages were detected on the C-terminal side of Des-(27-31)C-peptide. Unlike the CatD-mediated cleavage of murine C-peptide, human C-peptide was not significantly truncated by human CatD at the peptide bond two amino acid residues N-terminally adjacent to the Des-(27-31)C-peptide bond (see 2.5HIP-b for comparison). However, we previously identified various HIPs that formed at this adjacent site with high confidence in human islets (10), indicating that another protease or mechanism may be responsible for the formation of these N-terminally adjacent HIPs. In sum, our data demonstrate that human CatD selectively drives the formation of HIP11 and that the cleavage site specificity of human CatD aligns with its transpeptidation site specificity.
Absence of CatL in NOD Islets Leads to an Increased Content of Disease-Relevant HIPs
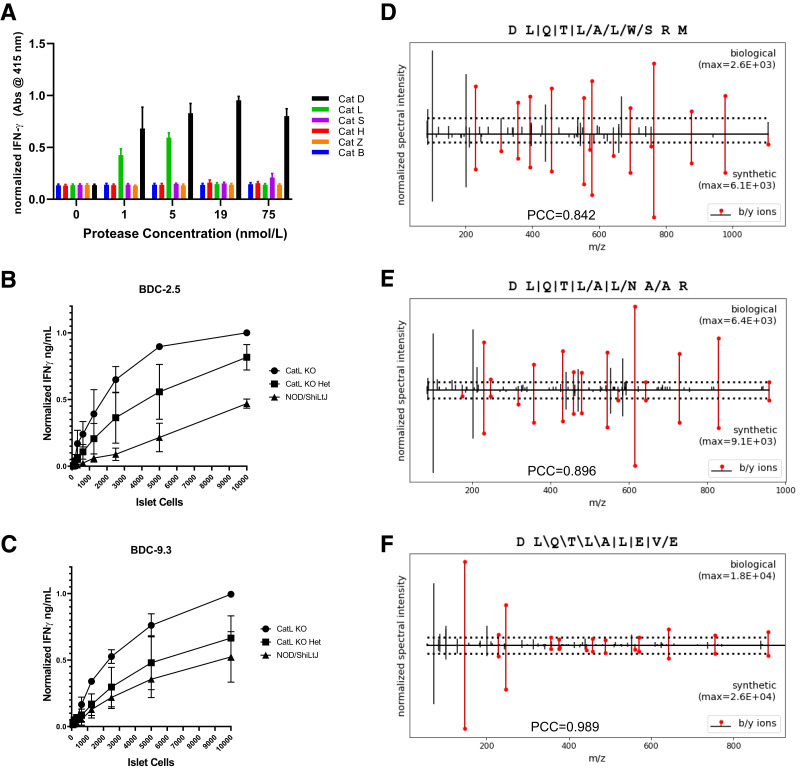
Our data indicate a minor role of cysteine proteases (such as CatL) in the formation of the 2.5HIP within enriched murine β-cell granule lysates (see Fig. 1C). To study the role of cysteine proteases in the formation of the 2.5HIP, we incubated murine C-peptide and WQ6 in the presence of various recombinant cysteine proteases (Fig. 4A) that were identified in SEC fractions of granule lysates (Fig. 1A). We excluded the proteases legumain and dipeptidyl peptidase 1 from these analyses as their established cleavage site specificities would not mediate 2.5HIP formation (30,31). We also omitted ubiquitin carboxyl-terminal hydrolase isozyme L1 from these experiments due to its established role in ubiquitin-dependent proteolysis (32). We used recombinant forms of the remaining cysteine proteases— CatL, cathepsin Z, cathepsin B, cathepsin S, and cathepsin H—and incubated each in the presence of 2.5HIP precursor peptides. For comparison, we included reactions with CatD. Samples were incubated at pH 5.0, at the interface of the lysosomal and insulin granular pH. Formation of 2.5HIP was monitored using the BDC-2.5 assay system. The results of this experiment are seen in Fig. 4A and show that cysteine protease CatL is capable of 2.5HIP formation, while the other cysteine proteases did not significantly contribute. CatL-mediated HIP formation was only observed at 1 and 5 nmol/L, but not at higher concentrations, suggesting that CatL can both form and degrade the 2.5HIP in vitro. To demonstrate that CatL is capable of HIP degradation, we incubated synthetic 2.5HIP (DLQTLAL-WSRM) in the presence and absence of CatL at pH 5.0. In the presence of CatL, 2.5HIP degradation is prevalent (Supplementary Fig. 3C). Although it has been proposed that CatL may contribute to the formation of 2.5HIP in NOD mice (33), the observed CatL cleavage site specificity (Supplementary Fig. 3) and transpeptidation site specificity (24) did not align with the HIPs that we identified in murine islets (10).
Figure 4.
CatL does not contribute significantly to HIP formation in murine islets. A: BDC-2.5 T-cell responses to in vitro reactions (pH 5.0) of 2.5HIP precursor peptides incubated for 24 h in the presence and absence of various cysteine proteases and CatD. Responses of BDC-2.5 (B) and BDC-9.3 (C) T-cell clones to dispersed islets from NOD, NOD.CatL+/−, and NOD.CatL−/− islets. KO, knockout. See control experiments in Supplementary Fig. 7D and E. Results from A, B, and C are the mean ± SEM from three independent experiments. Data were normalized to the maximal IFN-γ signal for each experiment. D–F: Mirror plots showing the fragmentation spectra of the AspN-digested 2.5HIP, 6.9HIP, and murine HIP11 sequences identified in NOD CatL−/− islets compared with the synthetic peptides. Presence of 2.5HIP, 6.9HIP, and mouse HIP11 in CatL-KO islets was fully validated through P-VIS (19).
To assess the in vivo role of CatL in the formation of disease-relevant HIPs, we obtained islets from NOD mice deficient in CatL (NOD.CatL−/− and NOD.CatL+/−), and NOD mice (CatL+/+). Although CatL-deficient NOD mice are protected from diabetes (14), it was also shown that BDC-2.5 could be used to trigger diabetes in these mice, indicating that 2.5HIP antigen was still present in CatL-deficient β-cells. We titrated dispersed islet cells in the presence of APC and BDC-2.5 or the 6.9HIP-reactive CD4 T-cell clone, BDC-9.3 (see Table 1) as responding T cells. As shown in Fig. 4B and 4C, both T-cell clones responded to dispersed islet cells from wild-type NOD mice, verifying presence of endogenous HIPs. In comparison, the responses to islets from CatL+/− were increased, and responses to islets from CatL−/− mice were even higher, signifying elevated levels of 2.5HIP and 6.9HIP in these mice. We also validated the presence of 2.5HIP, 6.9HIP, and HIP11 in the CatL−/− islets using mass spectrometric analysis in combination with the P-VIS validation protocol (Fig. 4D–F and Supplementary Figs. 4, 5, and 6). In summary, although CatL is capable of mediating 2.5HIP formation in vitro, our data indicate that the primary role of CatL in islets is in the degradation of disease-relevant HIPs.
Discussion
In the work reported here, we used a combination of techniques from proteomics, immunology, and biochemistry to identify the aspartic protease CatD as the primary protease responsible for the specific formation of various disease-relevant HIPs in β-cells. We demonstrated that murine CatD, in alignment with its cleavage site specificity, drives the formation of 2.5HIP, a dominant antigen for diabetogenic CD4 T cells in NOD mice. We further established that human CatD, also in alignment with its cleavage site specificity, drives the formation of HIP11, which is targeted by CD4 T cells in PBMCs of patients with new-onset T1D as well as residual islets of organ donors with T1D (5,8). Using recombinant forms of human and murine CatD, we demonstrated that CatD-mediated cleavage at its primary C-peptide cleavage site yields Des-(27-31)C-peptide, a major granular peptide that is cosecreted with insulin upon glucose stimulation of β-cells (11). We further analyzed the role of additional cysteine proteases in HIP synthesis and confirmed data by others (33) that CatL is a protease that can mediate 2.5HIP formation. However, when analyzing the islets from NOD mice that are deficient of CatL, we established that they contain significantly elevated levels of disease-relevant HIPs (Fig. 4B and C). CatL has been established to mediate transpeptidation reactions leading to the formation of HIPs in vitro. However, unlike CatD, the broad specificity of CatL (5,33) does not lead to the selective formation of disease-relevant HIPs that we identified in islets (5,10,19). HIP content in organelles is mediated by various factors such as the presence of different proteases, the concentration of proteases and peptides, and the age of the organelle. While CatL can mediate the formation of HIPs in vitro, the observation that CatL-deficient islets contain an elevated content of HIPs supports the argument that CatL fulfills its primary function, the proteolytic degradation of proteins and peptides. This suggests that in vivo CatL is primarily involved in the autophagic degradation and not the formation of HIPs. Conversely, CatL-deficient NOD mice have previously been shown to be protected from diabetes (14). This, however, may be due to a lack of antigen processing mediated by CatL (34). As we have shown, HIPs still form in the absence of CatL, but may not be processed correctly for MHC presentation in the absence of CatL, therefore, evading T-cell reactivity.
HIP formation may take place within insulin granules or crinophagic bodies that form by fusion of lysosomes and insulin granules. Both types of subcellular organelles contain insulin as well as various proteases and peptides that can mediate the formation of HIPs. It is therefore plausible to assume that HIP formation takes place in both organelles. However, the yield of HIPs formed in these organelles may play a critical role in rendering HIPs disease-relevant or not. Our mass spectrometric analyses have so far led to the identification of a limited number of HIPs in human and murine islets. Of those HIPs, there are three in murine islets (2.5HIP, 6.9HIP, HIP11) and one in human islets (HIP11), for which disease-relevant T-cell specificities have also been verified. These HIPs share the common C-peptide fragment Des-(27-31)C-peptide as a left-peptide, implying that a highly specific protease, rather than a broad panel of proteases, is responsible for generation of these disease-relevant HIPs. Our analyses demonstrated that the specificity of CatD precisely mediates the formation of these disease-relevant HIPs, as well as Des-(27-31)C-peptide. In combination with previous analyses by others verifying the presence of CatD in insulin granules (35), our data imply that insulin granules are the primary site of disease-relevant HIP formation (35).
Reversal of insulin dependence in T1D may soon be achieved through implantation of β-cells derived from induced pluripotent stem cells. Yet, the presence of disease-driving antigens in such cells may require the use of antigen-specific tolerance induction strategies or immunosuppressant therapies to prevent their rejection. It may, however, be possible to interfere with this T-cell mediated destruction process by depriving β-cells of disease-relevant antigens. For example, stem cell-derived β-cells could be manipulated to lack disease-relevant HIPs by interfering with CatD-mediated transpeptidation reactions. With a reduced requirement for immunomodulation, such β-cells may be resistant to the destruction mediated by autoreactive memory T cells following transplantation into recipients with T1D. Overall, understanding the mechanism of HIP formation may allow us to block formation of disease-relevant autoantigens in T1D and further substantiate the role of HIPs in the pathogenesis of T1D.
Article Information
Acknowledgments. The authors thank Jeanine Forman-Ham for her generous support of research efforts in the Delong laboratory through the University of Colorado Foundation.
Funding. This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases R01 grants DK119529 (T.D.), DK081166 (K.H.), DK126456 (H.M.T.), and DK127497 (H.M.T.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. S.A.C., T.A.W., J.M.W., R.L.P., G.B., M.D., J.G., J.M.B., K.L.S.B., and A.C.H. conducted experiments. S.A.C., T.A.W., J.M.W., R.L.B., H.M.T., K.H., and T.D. contributed to discussion and reviewed and edited the manuscript. S.A.C., T.A.W., J.M.W., and T.D. analyzed the data. S.A.C., R.L.B., H.M.T., and T.D. designed experiments. S.A.C. and T.D. wrote the manuscript. T.D. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.20606019.
References
- 1. Wiles TA, Delong T, Baker RL, et al. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J Autoimmun 2017;78:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delong T, Wiles TA, Baker RL, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016;351:711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babon JAB, DeNicola ME, Blodgett DM, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes [published correction appears in Nat Med 2017;23:1004] Nat Med 2016;22:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parras D, Solé P, Delong T, Santamaría P, Serra P. Recognition of multiple hybrid insulin peptides by a single highly diabetogenic T-cell receptor. Front Immunol 2021;12:737428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiles TA, Hohenstein A, Landry LG, et al. Characterization of human CD4 T cells specific for a C-peptide/C-peptide hybrid insulin peptide. Front Immunol 2021;12:668680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arribas-Layton D, Guyer P, Delong T, et al. Hybrid insulin peptides are recognized by human T cells in the context of DRB1*04:01. Diabetes 2020;69:1492–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wiles TA, Delong T. HIPs and HIP-reactive T cells. Clin Exp Immunol 2019;198:306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baker RL, Rihanek M, Hohenstein AC, et al. Hybrid insulin peptides are autoantigens in type 1 diabetes. Diabetes 2019;68:1830–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker RL, Jamison BL, Wiles TA, et al. CD4 T cells reactive to hybrid insulin peptides are indicators of disease activity in the NOD mouse. Diabetes 2018;67:1836–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiles TA, Powell R, Michel R, et al. Identification of hybrid insulin peptides (HIPs) in mouse and human islets by mass spectrometry. J Proteome Res 2019;18:814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Verchere CB, Paoletta M, Neerman-Arbez M, et al. Des-(27-31)C-peptide: a novel secretory product of the rat pancreatic beta cell produced by truncation of proinsulin connecting peptide in secretory granules. J Biol Chem 1996;271:27475–27481 [DOI] [PubMed] [Google Scholar]
- 12. Cho YK, Northrop DB. Transpeptidation by porcine pepsin catalyzed by a noncovalent intermediate unique to its iso-mechanism. J Biol Chem 1998;273:24305–24308 [DOI] [PubMed] [Google Scholar]
- 13. Bergmann M, Fraenkel-Conrat H. The role of specificity in the enzymatic synthesis of proteins: syntheses with intracellular enzymes. J Biol Chem 1937;119:707–720 [Google Scholar]
- 14. Maehr R, Mintern JD, Herman AE, et al. Cathepsin L is essential for onset of autoimmune diabetes in NOD mice. J Clin Invest 2005;115:2934–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bergman B, McManaman JL, Haskins K. Biochemical characterization of a beta cell membrane fraction antigenic for autoreactive T cell clones. J Autoimmun 2000;14:343–351 [DOI] [PubMed] [Google Scholar]
- 16. Kozlovskaya V, Zavgorodnya O, Chen Y, et al. Ultrathin polymeric coatings based on hydrogen-bonded polyphenol for protection of pancreatic islet cells. Adv Funct Mater 2012;22:3389–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haskins K, Portas M, Bergman B, Lafferty K, Bradley B. Pancreatic islet-specific T-cell clones from nonobese diabetic mice. Proc Natl Acad Sci U S A 1989;86:8000–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stadinski BD, Delong T, Reisdorph N, et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol 2010;11:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wiles TA, Saba LM, Delong T. Peptide-spectrum match validation with internal standards (P-VIS): internally-controlled validation of mass spectrometry-based peptide identifications. J Proteome Res 2021;20:236–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Orr DF, Chen T, Johnsen AH, et al. The spectrum of endogenous human chromogranin A-derived peptides identified using a modified proteomic strategy. Proteomics 2002;2:1586–1600 [DOI] [PubMed] [Google Scholar]
- 21. Marzban L, Trigo-Gonzalez G, Verchere CB. Processing of pro-islet amyloid polypeptide in the constitutive and regulated secretory pathways of β cells. Mol Endocrinol 2005;19:2154–2163 [DOI] [PubMed] [Google Scholar]
- 22. Zolg DP, Wilhelm M, Yu P, et al. PROCAL: a set of 40 peptide standards for retention time indexing, column performance monitoring, and collision energy calibration. Proteomics 2017;17:1700263. [DOI] [PubMed] [Google Scholar]
- 23. Bergman B, Haskins K. Islet-specific T-cell clones from the NOD mouse respond to β-granule antigen. Diabetes 1994;43:197–203 [DOI] [PubMed] [Google Scholar]
- 24. Briozzo P, Morisset M, Capony F, Rougeot C, Rochefort H. In vitro degradation of extracellular matrix with Mr 52,000 cathepsin D secreted by breast cancer cells. Cancer Res 1988;48:3688–3692 [PubMed] [Google Scholar]
- 25. Hutton JC. The internal pH and membrane potential of the insulin-secretory granule. Biochem J 1982;204:171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeng J, Shirihai OS, Grinstaff MW. Modulating lysosomal pH: a molecular and nanoscale materials design perspective. J Life Sci (Westlake Village) 2020;2:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith RE, Farquhar MG. Lysosome function in the regulation of the secretory process in cells of the anterior pituitary gland. J Cell Biol 1966;31:319–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halban PA, Wollheim CB. Intracellular degradation of insulin stores by rat pancreatic islets in vitro. An alternative pathway for homeostasis of pancreatic insulin content. J Biol Chem 1980;255:6003–6006 [PubMed] [Google Scholar]
- 29. Weckman A, Di Ieva A, Rotondo F, et al. Autophagy in the endocrine glands. J Mol Endocrinol 2014;52:R151–R163 [DOI] [PubMed] [Google Scholar]
- 30. McDonald JK, Zeitman BB, Reilly TJ, Ellis S. New observations on the substrate specificity of cathepsin C (dipeptidyl aminopeptidase I). Including the degradation of beta-corticotropin and other peptide hormones. J Biol Chem 1969;244:2693–2709 [PubMed] [Google Scholar]
- 31. Dall E, Brandstetter H. Structure and function of legumain in health and disease. Biochimie 2016;122:126–150 [DOI] [PubMed] [Google Scholar]
- 32. Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry 1996;35:6735–6744 [DOI] [PubMed] [Google Scholar]
- 33. Reed B, Crawford F, Hill RC, et al. Lysosomal cathepsin creates chimeric epitopes for diabetogenic CD4 T cells via transpeptidation. J Exp Med 2021;218:e20192135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsieh CS, deRoos P, Honey K, Beers C, Rudensky AY. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J Immunol 2002;168:2618–2625 [DOI] [PubMed] [Google Scholar]
- 35. Brunner Y, Couté Y, Iezzi M, et al. Proteomics analysis of insulin secretory granules. Mol Cell Proteomics 2007;6:1007–1017 [DOI] [PubMed] [Google Scholar]