Abstract
The stress response protein regulated in development and DNA damage response 1 (REDD1) has been implicated in visual deficits in patients with diabetes. The aim here was to investigate the mechanism responsible for the increase in retinal REDD1 protein content that is observed with diabetes. We found that REDD1 protein expression was increased in the retina of streptozotocin-induced diabetic mice in the absence of a change in REDD1 mRNA abundance or ribosome association. Oral antioxidant supplementation reduced retinal oxidative stress and suppressed REDD1 protein expression in the retina of diabetic mice. In human retinal Müller cell cultures, hyperglycemic conditions increased oxidative stress, enhanced REDD1 expression, and inhibited REDD1 degradation independently of the proteasome. Hyperglycemic conditions promoted a redox-sensitive cross-strand disulfide bond in REDD1 at C150/C157 that was required for reduced REDD1 degradation. Discrete molecular dynamics simulations of REDD1 structure revealed allosteric regulation of a degron upon formation of the disulfide bond that disrupted lysosomal proteolysis of REDD1. REDD1 acetylation at K129 was required for REDD1 recognition by the cytosolic chaperone HSC70 and degradation by chaperone-mediated autophagy. Disruption of REDD1 allostery upon C150/C157 disulfide bond formation prevented the suppressive effect of hyperglycemic conditions on REDD1 degradation and reduced oxidative stress in cells exposed to hyperglycemic conditions. The results reveal redox regulation of REDD1 and demonstrate the role of a REDD1 disulfide switch in development of oxidative stress.
Introduction
Oxidative stress plays a central role in the development of diabetic complications, including diabetic retinopathy (1). Hyperglycemic conditions cause oxidative stress by both increasing the production of reactive oxygen species (ROS) and reducing antioxidant capacity of the cell (2,3). ROS, such as superoxide and hydrogen peroxide, mediate cytotoxic effects through the oxidation of proteins, lipids, and nucleic acids. In addition, ROS act as critical signaling molecules through the covalent modification of intracellular redox sensors (4,5). The role of ROS as second messengers is mediated in part by oxidation of reactive cysteine thiol side chains, resulting in altered function, localization, or expression of the target protein.
The stress response protein regulated in development and DNA damage response 1 (REDD1, also known as DDIT4/RTP801) has long been linked with oxidative stress (6,7), and evidence supports a causal role for REDD1 in diabetes-induced oxidative stress in the retina (8–10). REDD1 acts as a dominant governor of mammalian target of rapamycin complex 1 (mTORC1) (11), an effect that is mediated at least in part by promoting the association of protein phosphatase 2A with Akt, leading to site-specific dephosphorylation of the kinase at T308 and altered substrate specificity (12). Retinal REDD1 protein expression is enhanced in preclinical rodent models of diabetes, and REDD1 deletion prevents the development of retinal neurodegeneration and functional deficits in vision (8,9,13–15). Intravitreal administration of a siRNA targeting the REDD1 mRNA (PF-04523655) has also demonstrated promise in improving the visual function of patients with diabetic macular edema (16). Surprisingly, REDD1 mRNA abundance in the retina of diabetic rats is similar to that observed in nondiabetic controls (17). This suggests the possibility that diabetes acts through a posttranscriptional mechanism to promote retinal REDD1 protein expression.
Protein degradation is a highly regulated process that is heavily influenced by diabetes (18). In particular, the expression of stress response proteins like REDD1 is often controlled at the level of protein stability (19). In the absence of cellular stress, REDD1 is rapidly degraded, as the protein exhibits a half-life of ∼5 min (20). The most well-studied mechanism for the rapid degradation of specific proteins is the ubiquitin-proteasome pathway. Indeed, prior studies support that REDD1 is ubiquitinated and degraded by the proteasome (21,22). While less well explored than the ubiquitin-proteasome pathway, selective protein degradation may also be mediated by autophagy. In particular, chaperone-mediated autophagy (CMA) facilitates lysosomal degradation of specific KFERQ-like motif-bearing proteins in response to changes in cellular environment (23).
The aim here was identify the mechanism responsible for the increase in retinal REDD1 protein content that is observed with diabetes. We found that hyperglycemic conditions promoted formation of a disulfide bond between cysteine residues on adjacent antiparallel β-strands of REDD1. The disulfide bond suppressed REDD1 degradation by CMA through allosteric regulation of a KFERQ-like motif that was required for REDD1 interaction with the cytosolic chaperone heat shock cognate 71-kDa protein (HSC70). The findings identify REDD1 as a molecular ROS sensor and delineate the mechanism of redox-sensitive REDD1 degradation.
Research Design and Methods
Animals
At ∼5 weeks of age, wild-type (WT) and REDD1-knockout (KO) B6;129 (24) and Pdgfra-cre; HA-Rpl22 mice (25) were administered 50 mg/kg (males) or 75 mg/kg (females) streptozotocin (STZ; MilliporeSigma) dissolved in sodium citrate buffer for 5 consecutive days to induce diabetes. Nondiabetic control mice received sodium citrate buffer vehicle. Mice were maintained at ∼18–23°C with 12-h light/12-h dark and had access to standard chow (Teklad) and water ad libitum. Diabetic phenotype was assessed as fasting blood glucose levels >250 mg/dL. Retinas were analyzed after 6 weeks of diabetes. In some studies, diabetic mice were orally administered ∼2 g/kg of N-acetyl-l-cysteine (NAC) daily via drinking water, as previously described (8). Thickness of retinal layers was evaluated by optical coherence tomography, as recently described (15). All procedures were approved by the Penn State College of Medicine Institutional Animal Care and Use Committee (Protocol No. 202101862) and were in accordance with the Association for Research in Vision and Ophthalmology statement for the ethical use of animals in ophthalmic and vision research.
Terminal TUNEL
Mice were euthanized by administration of 300 mg ketamine/30 mg xylazine/kg body wt, followed by decapitation. Retinas were isolated, immersion-fixed in 4% paraformaldehyde, and whole-mounted on microscope slides. Retinas were processed with an ApopTag Peroxidase in situ Apoptosis detection kit (EMD Millipore), as previously described (26). We used 3-amino-9-ethylcarbazole (Sigma-Aldrich) in 50 mmol/L sodium acetate buffer (pH 5.0) as the substrate.
Cell Culture
MIO-M1 human Müller cells were obtained from the University College London Institute of Ophthalmology (London, U.K.). Generation and culture of REDD1-KO MIO-M1 and human embryonic kidney (HEK) 293 tetracycline-on (Tet-On) HA-REDD1 cells was previously described (12,14). Cells were transfected using Lipofectamine 2000 (Life Technologies) with the following plasmids: pCMV5 empty vector, pCMV-HA-REDD1 (human), pCMV-Myc-FLAG-Lamp2A (OriGene), and pCMV-Myc-FLAG-REDD1 (OriGene). Plasmid variants were generated by site-directed mutagenesis using a QuikChange Lightning Kit (Agilent) and the appropriate primers (Supplementary Table 1). For studies on the effects of hyperglycemic conditions, cell culture medium contained 30 mmol/L glucose or 5 mmol/L glucose with 25 mmol/L mannitol as an osmotic control. In specific studies, culture medium was supplemented with 5 μmol/L cycloheximide (Dot Scientific), 20 μmol/L MG-132 (EMD Millipore), 1 mmol/L H2O2 (MilliporeSigma), 10 mmol/L NAC (MilliporeSigma), 20 μmol/L AR7 (MedChemExpress), or 10 nmol/L bafilomycin (MilliporeSigma). Where indicated, cells were preexposed to H2O2 (2 h) or MG-132 (30 min) before AR7 or cycloheximide.
Protein Analysis
Retinas were harvested, flash frozen in liquid nitrogen, and later homogenized in 250 µL extraction buffer, as previously described (27). Lysosomes were purified using a Lysosome Isolation Kit (Abcam) according to the manufacturer’s instructions. Retinal homogenates, cell lysates, and protein isolates were fractionated using Criterion Precast 4–20% gels (Bio-Rad Laboratories). Proteins were transferred to polyvinylidene fluoride, blocked in 5% milk in Tris-buffered saline Tween 20, washed, and incubated overnight at 4°C with the appropriate antibodies (Supplementary Table 2). The antigen-antibody interaction was visualized with enhanced chemiluminescence Clarity Reagent (Bio-Rad Laboratories) using a ProteinSimple Fluorochem E.
Liquid Chromatography–Tandem Mass Spectrometry Analysis
HEK293 cells were transfected to express FLAG-REDD1 and then exposed to 1 mmol/L H2O2. Cells were collected in buffer A (20 mmol/L HEPES [pH 7.4], 2 mmol/L EGTA, 50 mmol/L NaF, 100 mmol/L KCl, 0.2 mmol/L EDTA, 50 mmol/L β-glycerophosphate, 1 mmol/L dithiothreitol, 1 mmol/L benzamidine, 50 mmol/L sodium vanadate, 1 mmol/L microcystin, 10 μL/mL protease inhibitor mixture [MilliporeSigma], and 2.5% Triton-X-100), and 1,000g supernatant fractions were collected. FLAG-REDD1 was affinity purified using EZview Red Anti-FLAG M2 Affinity Gel (MilliporeSigma) and eluted with 3xFLAG peptide (MilliporeSigma). FLAG-REDD1 was gel purified and subject to in-gel trypsin digestion, as previously described (28). Peptides were extracted, dried in a speed-vac, and reconstituted in 10 µL of solvent A (2.5% acetonitrile, 0.1% formic acid). A nanoscale reverse-phase high-performance liquid chromatography capillary column was created by packing 2.6-µm C18 spherical silica beads into a fused silica capillary. Peptides were loaded onto the column and eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). Eluted peptides were subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion trap mass spectrometer (Thermo Fisher Scientific). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions. Peptide sequences were determined by matching protein databases with the acquired fragmentation pattern using SEQUEST (Thermo Fisher Scientific) (29). Data were filtered to between a 1% and 2% peptide false discovery rate.
Biotin Switch Assay
Biotin labeling was performed as previous described (30). Briefly, three fully confluent 10-cm dishes of MIO-M1 cells were lysed on ice using a Dounce homogenizer with biotin labeling lysis buffer (BLLB, 50 mmol/L Tris-HCl [pH 7.0], 5 mmol/L EDTA, 120 mmol/L NaCl, and 0.5% IGEPAL-630) with 10 μL/mL protease inhibitor mixture (MilliporeSigma) and 100 mmol/L maleimide (MilliporeSigma) to alkylate all unmodified cysteines. Lysates were centrifuged at 20,000g for 10 min at 4°C to remove debris, and the protein content of the clarified supernatant was measured by DC Protein Assay (Bio-Rad). The concentration of each sample was then adjusted to 1 mg/mL with BLLB, and SDS was added to a final volume of 1%. Samples were incubated at room temperature for 2 h. Proteins from each sample were precipitated using 5 volumes of acetone prechilled at −20°C, and samples were incubated at −20°C for 20 min. The preparations were then centrifuged at 20,000g for 10 min at 4°C, and the protein pellets were air dried. Precipitants were resuspended in 200 µL BLLB with 1% SDS, 10 mmol/L dithiothreitol, and 0.1 mmol/L biotin-maleimide (MilliporeSigma) to reduce oxidized sulfhydryl groups and allow their reaction with biotin-malemide. Proteins were precipitated with 5 volumes of methanol, resuspended in 500 µL of BLLB, and incubated with 10 µL of 50% streptavidin-Sepharose bead slurry (GE Healthcare) for 2 h at 4°C. Beads were washed with BLLB, and proteins were eluted in SDS sample buffer.
Immunoprecipitations
Supernatant fractions of cell lysates were prepared in buffer A supplemented with protease and phosphatase inhibitors, as described above. Lysates were incubated at 4°C for 30 min. Anti-FLAG affinity gel (10 µL per sample) was washed thrice with low-salt buffer (20 mmol/L Tris-HCl [pH 7.4], 150 mmol/L NaCl, 5 mmol/L EDTA, 0.5% Triton X-100, and 0.1% β-mercaptoethanol) and blocked with low-salt buffer plus 1% BSA for 1 h at 4°C. Lysates were incubated with the affinity gel for 2 h at 4°C and washed thrice with buffer A. Samples were eluted in SDS sample buffer and boiled for 5 min. Müller cell-specific ribosomes were isolated from Pdgfra-cre; HA-Rpl22 mice as previously described (25). Ribosome-associated RNA was processed by adding 375 µL of Buffer RLT (QIAGEN) to the remaining beads and using a RNeasy Micro kit (QIAGEN).
RNA Analysis
Total RNA was extracted from whole retina with TRIzol (Invitrogen). RNA (1 μg) was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Rpl22HA-ribosomes were immunoprecipitated from the retina of Pdgfra-cre; HA-Rpl22 mice, and RNA was isolated as previously described (25). RNA was subjected to quantitative real-time PCR (QuantStudio 12K Flex Real-Time PCR System) using QuantiTect SYBR Green master mix (QIAGEN), as previously described (14). Primer sequences are listed in Supplementary Table 1. Mean cycle threshold (CT) values were determined for control and experimental samples. Changes in mRNA expression were normalized to GAPDH mRNA expression using the 2−ΔΔCT calculations.
Computational Investigation of REDD1 Allostery
A crystal structure of REDD1 (31) was obtained from Protein Data Bank (PDB ID 3LQ9). Discrete molecular dynamics (DMD) (32–34) was used to identify residues in REDD1 whose positions were correlated. Mutations at V178 were evaluated using the Eris web server (35) for overall increase in protein Gibbs free energy, with higher values of ΔG indicating greater destabilization of the crystal structure. All DMD simulations were performed for several million time steps (∼100 µs of real time). The disulfide bond was modeled as a hard distance constraint between the sulfurs of C150 and C157, with a range of 1.5 to 3.0 Å (36). The trajectories were examined in PyMol to ensure the protein did not unfold significantly during simulation. Trajectories were processed in Python 3.9. Each snapshot of the trajectory was aligned to the starting configuration using the Kabsch algorithm, an unweighted average position was calculated for each residue based on all the atoms in that residue, and Pearson correlation coefficients between all residue pairs were calculated for each dimension using NumPy 1.21.1 (37). The correlation coefficients were then averaged over the x, y, and z dimensions. Contact maps were plotted using Matplotlib 3.5.0. The Ohm web server (38) was used to estimate the allosteric contacts in WT REDD1. A Fisher r-to-z transformation was performed when comparing the contact maps. A two-sided z test was performed, with significance set at α = 0.01, applying a Bonferroni correction for multiple comparisons of n × (n − 1)/2, where n was the number of residues.
ROS Detection
Retinas were homogenized in lysis buffer as previously described (8). Lysates were centrifuged at 1,500g for 3 min, and the supernatant was exposed to 10 μmol/L 2′,7′-dichlorofluorescein (DCF). ROS were assessed in cells in culture using a DCF diacetate (DCFDA) Cellular ROS Detection Assay kit (Abcam). Fluorescence was measured using a SpectraMax M5 plate reader (Molecular Devices).
Statistical Analysis
Western blots were quantified using ImageJ software (National Institutes of Health). Results are expressed as mean ± SD. Statistical analyses were performed using GraphPad Prism software, with P value <0.05 defined as statistically significant. Data were analyzed overall with the Student t test or one-way or two-way ANOVA. Trend test and pairwise comparisons were conducted with the Tukey test for multiple comparisons.
Data and Resource Availability
Data sets, code to generate Figs. 4A–C and 6D and E, DMD trajectories, Ohm contact maps, and resources generated during this study are available from the corresponding author upon reasonable request.
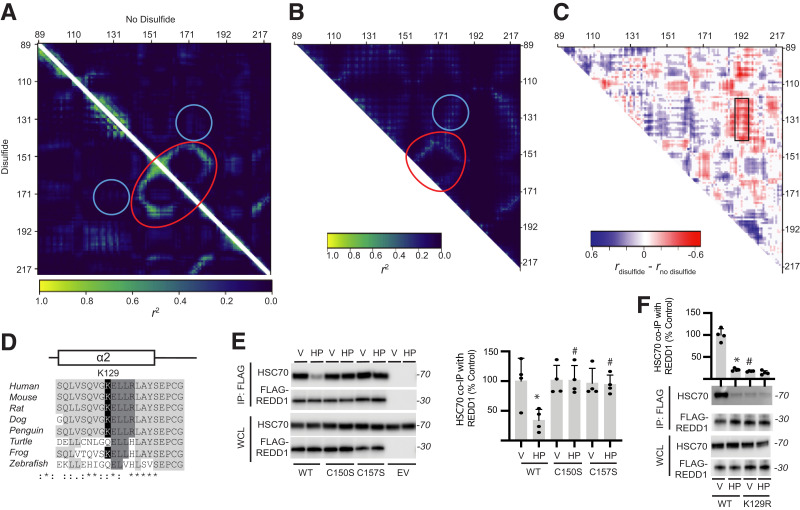
Figure 4.
Allosteric regulation of REDD1 KFERQ-like motif controls HSC70 binding. A: DMD was used to examine the correlation of pairwise residues in REDD1 with (lower triangle) and without (upper triangle) the C150/C157 disulfide bond. Strong interactions are indicated near residues C150/C157 and V178 (red circle). Interactions between V178 and α-helix 2 near K129 (blue circle) were also observed. B: An Ohm contact map was generated to identify allosteric communication networks within REDD1. C: Fisher r-to-z transformation and, subsequently, a z test was performed on the difference between correlation coefficients for the DMD simulations in A. Statistically significant differences between DMD positional coefficients for REDD1, with and without the disulfide, are shown, with nonsignificant results set to 0. Changes in coordination of residues with α-helix 2 are indicated (black box). D: Sequence conservation of an acetylation-generated KFERQ-like motif (129KELLR133) in α-helix 2 was observed among vertebrates. E: FLAG-tagged WT, C150S, and C157S FLAG-REDD1 variants were expressed in MIO-M1 REDD1 KO cells. Cells were exposed to vehicle (V) or H2O2 (HP) for 2 h. FLAG-REDD1 was immunoprecipitated from cell lysates. REDD1 and HSC70 were evaluated in the immunoprecipitate (IP) and whole-cell lysates (WCL) by Western blotting. Representative blots are shown. Protein molecular mass in kDa is indicated at right of blots. HSC70 binding with FLAG-REDD1 was quantified. F: FLAG-tagged WT and K129R REDD1 variants were expressed in cells, and REDD1 binding with HSC70 was evaluated by FLAG-tag immunoprecipitation. Data are represented as mean ± SD. *P < 0.05 vs. V; #P < 0.05 vs. WT.
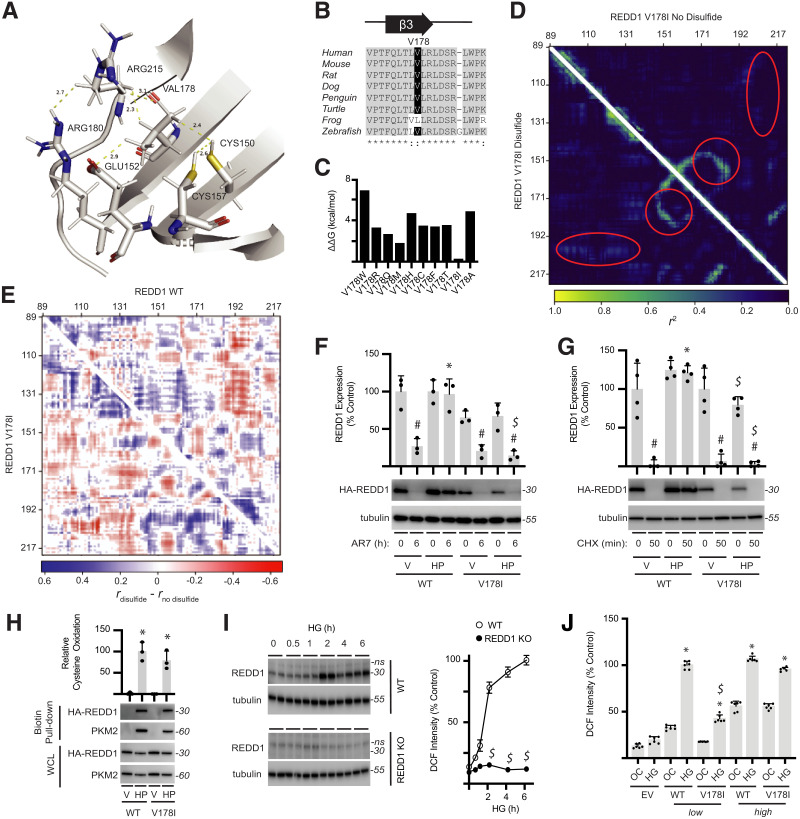
Figure 6.
Disruption of REDD1 allosteric regulation prevents REDD1 stabilization and hyperglycemia-induced oxidative stress. A: Side chains of C150/C157 in REDD1 coordinate with V178. B: REDD1 V178 exhibits sequence conservation among vertebrates. C: Eris web server was used to predict the stability of REDD1 V178 variants. D: DMD simulations were performed on REDD1 V178I in the absence (top triangle) and presence (bottom triangle) of the C150/C157 disulfide. E: Comparison of DMD simulations for REDD1 WT vs. V178I, with and without the disulfide, supported disruption of protein-wide allostery in the V178I mutant. F and G: HA-tagged REDD1 WT or V178I variants were expressed in REDD1-KO MIO-M1 cells. Cells were exposed to vehicle (V) or H2O2 (HP) for 2 h prior to administering AR7 (F) or cycloheximide (CHX) (G), and REDD1 expression was evaluated by Western blotting. Representative blots are shown. Protein molecular mass in kDa is indicated at right of blots. H: REDD1 oxidation was evaluated by biotin switch assay. PKM2 was evaluated in biotin pull-downs as a positive control. HA-REDD1 and PKM2 expression in whole-cell lysate (WCL) was determined by Western blotting. I: WT and REDD1-KO MIO-M1 cells were exposed to medium containing 30 mmol/L glucose (HG) for up to 6 h, and ROS were visualized with DCFDA. REDD1 and tubulin expression were evaluated by Western blotting, and DCF fluorescent intensity was quantified (n = 3). J: WT and V178I REDD1 variants were expressed at a low (12.5 ng of plasmid) or high (100 ng of plasmid) level in REDD1-KO MIO-M1 cells. Cells were exposed to medium containing 30 mmol/L glucose (HG) or 5 mmol/L glucose plus 25 mmol/L mannitol as an osmotic control (OC) for 24 h. DCFDA was used to quantify ROS in cells. Data are represented as mean ± SD. *P < 0.05 vs. V; #P < 0.05 vs. no treatment; $P < 0.05 vs. WT.
Results
Diabetes Enhances REDD1 Protein Expression in the Retina of Diabetic Mice
To determine the impact of diabetes on retinal REDD1 expression, REDD1+/+ and REDD1−/− mice were administered STZ to promote hyperglycemia (Fig. 1A). In support of prior studies demonstrating a role for REDD1 in diabetes-induced retinal pathology (39), diabetic REDD1+/+ mice exhibited reduced retinal thickness (Fig. 1B and Supplementary Figs. 1 and 2A) and increased retinal apoptosis (Figs. 1C and Supplementary Fig. 2B), whereas diabetic REDD1−/− mice did not. Consistent with prior studies (8,9,13,15,39), REDD1 protein expression in retinal lysates from diabetic mice was enhanced compared with nondiabetic controls (Fig. 1D). However, a corresponding increase in REDD1 mRNA abundance was not observed in the retina of diabetic mice (Fig. 1E). REDD1 expression in the retina localizes to Pdgfra-positive Müller glia (15). REDD1 mRNA association with Rpl22HA-ribosomes isolated from Müller glia of Pdgfra-cre; HA-Rpl22 RiboTag mice was not altered by diabetes (Fig. 1F). To determine whether oxidative stress influenced retinal REDD1 protein expression, diabetic mice were administered the antioxidant NAC. NAC reduced ROS in the retina of diabetic mice (Fig. 1G) and suppressed REDD1 protein expression (Fig. 1H). Together, the data support that diabetes promotes REDD1 protein content in the retina via a posttranslational mechanism that depends on augmented ROS levels.
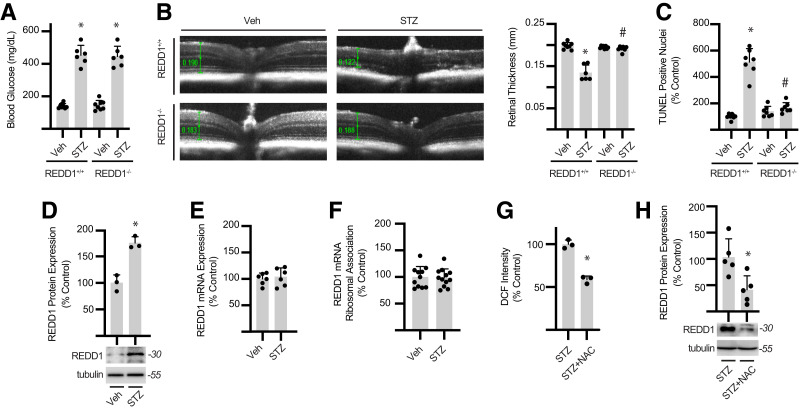
Figure 1.
Diabetes promotes retinal degeneration and increases REDD1 protein content. A: Diabetes was induced in WT (REDD1+/+) and REDD1-KO (REDD1−/−) transgenic mice by administration of STZ or a vehicle (Veh) control. Fasting blood glucose concentrations were evaluated. B: Retinal thickness was determined with optical coherence tomography. Retinal thickness was manually measured from the retinal nerve fiber layer to the photoreceptor outer segments as indicated by calipers (green). Representative images are shown. C: Retinal flat mounts were labeled with TUNEL-horseradish peroxidase, and TUNEL-positive nuclei were quantified. D: REDD1 and tubulin protein expression were evaluated in retinal lysates from REDD1+/+ mice by Western blotting. Representative blots are shown. Protein molecular mass in kDa is shown at right. E: REDD1 mRNA expression in retinal lysates was quantified by quantitative PCR analysis. F: Müller glia–specific expression of HA-tagged Rpl22 in the retina was achieved by crossing WT RiboTag mice to a PDGFRa-cre recombinase–expressing mouse, resulting in deletion of the WT exon 4 in the target cell population and replacement with the Rpl22HA exon. PDGFRa-Cre+; HA-Rpl22 mice were administered STZ or Veh. Ribosomes were isolated from retinal lysates of PDGFRa-Cre+; HA-Rpl22 mice by affinity purification. REDD1 mRNA association with ribosomes was determined by quantitative PCR. G: Oxidative stress was quantified by evaluating DCF fluorescence in retinal lysates. H: REDD1 and tubulin protein expression in retinal lysates was evaluated by Western blotting. Diabetic mice received ∼2 g/kg NAC daily via drinking water (G and H). Data are represented as mean ± SD. *P < 0.05 vs. Veh or no NAC treatment; #P < 0.05 vs. REDD1+/+.
ROS Inhibit Proteasome-Independent REDD1 Degradation
The impact of hyperglycemic conditions on REDD1 turnover was investigated by transiently expressing HA-tagged REDD1 in a previously characterized human retinal MIO-M1 Müller cell line (14). Hyperglycemic conditions reduced HA-REDD1 degradation in a cycloheximide-chase assay relative to an osmotic control (Fig. 2A). Consistent with the prior report (8), retinal cells exposed to hyperglycemic conditions exhibited increased cellular ROS levels (Fig. 2B). In cells exposed to hyperglycemic conditions, NAC addition to culture medium prevented enhanced ROS levels and restored HA-REDD1 degradation (Fig. 2C). The data support that hyperglycemic conditions suppress REDD1 degradation by promoting ROS levels. Indeed, exposure to the oxidant H2O2 reduced REDD1 degradation (Fig. 2D and E and Supplementary Fig. 3). To determine whether the suppressive effect of ROS on REDD1 degradation was mediated by reduced proteolysis, cells were exposed to the proteasome inhibitor MG-132. Surprisingly, H2O2 and proteasomal inhibition had an additive effect in reducing HA-REDD1 degradation in both HEK293 Tet-On (Fig. 2D) and MIO-M1 (Figs. 2E and Supplementary Fig. 3) cells. Consistent with a proteasome-independent effect of ROS, MG-132 was not sufficient to prevent reduced HA-REDD1 degradation in cells exposed to hyperglycemic conditions (Fig. 2F).
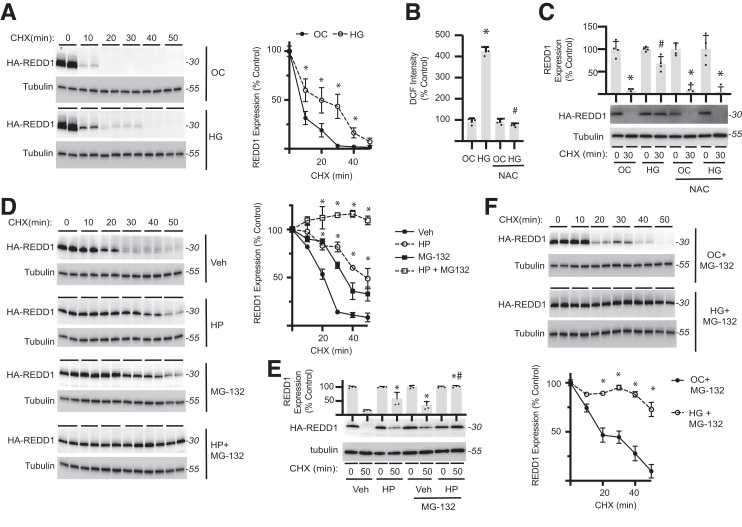
Figure 2.
Hyperglycemic conditions suppress proteasome-independent REDD1 degradation by promoting oxidative stress. A: HA-REDD1 was expressed in REDD1-KO MIO-M1 cells by transient transfection. Cells were exposed to medium containing 30 mmol/L glucose (HG) or 5 mmol/L glucose plus 25 mmol/L mannitol as an osmotic control (OC) for 4 h. Cycloheximide (CHX) was used to inhibit protein synthesis. REDD1 and tubulin protein expression were evaluated by Western blotting (n = 6). Representative blots are shown. Protein molecular mass in kDa is indicated at right of blots. B: ROS were visualized in WT MIO-M1 cells with DCFDA. DCF fluorescent intensity was quantified after exposure to HG or OC in the presence and absence of the antioxidant NAC. C: HA-REDD1 protein degradation was assessed in REDD1-KO MIO-M1 cells by CHX-chase assay. D: HEK293 Tet-On HA-REDD1 cells were exposed to culture medium containing H2O2 (HP) for 2 h, followed by CHX-chase in the presence/absence the proteasome inhibitor MG-132 (n = 4). E: HA-REDD1 was expressed in REDD1-KO MIO-M1 cells, and REDD1 protein degradation was evaluated by CHX-chase following exposure to HP and/or MG-132. F: HA-REDD1 was expressed in REDD1-KO MIO-M1 cells, and REDD1 protein degradation was evaluated by CHX chase in the presence of MG-132 following exposure to either HG or OC (n = 4). Data are represented as mean ± SD. *P < 0.05 vs. OC or Veh; #P < 0.05 vs. control.
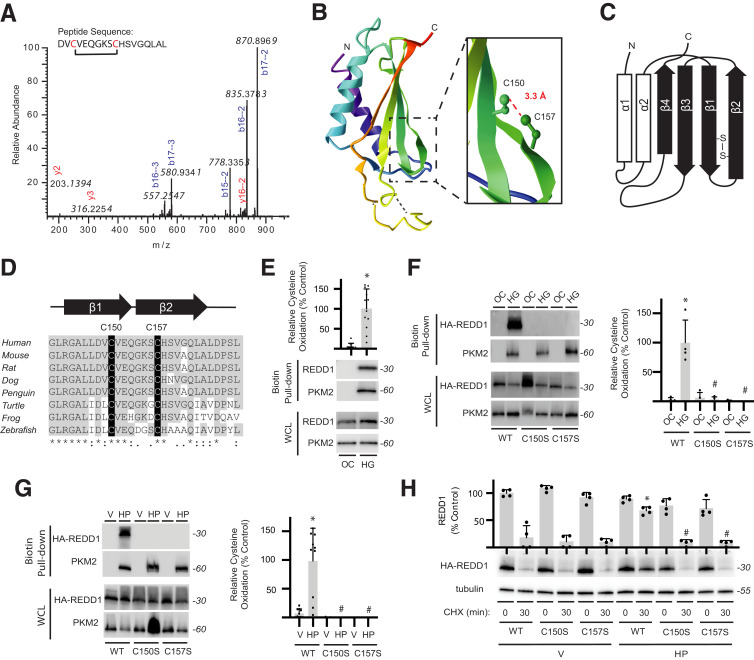
Formation of a Redox-Sensitive Cross-Strand Disulfide Inhibits REDD1 Degradation
To investigate redox modification of REDD1, purified recombinant FLAG-tagged REDD1 (Supplementary Fig. 4A) was exposed to H2O2 and subject to liquid chromatography–tandem mass spectrometry. A disulfide-linked peptide (148DVCVEQGKSCHSVGQLAL165) was observed in the spectra, and fragmentation revealed a cystine at C150/C157 (Fig. 3A). REDD1 C150 and C157 side chains are located superficially and in proximity to form an intramolecular disulfide (31) (Fig. 3B and Supplementary Fig. 4B). The two cysteines are on adjacent antiparallel β-strands (Fig. 3C) and well conserved (Fig. 3D). Redox modification of cysteines in REDD1 was examined by biotin switch assay (40). In cells exposed to hyperglycemic conditions, biotin-maleimide was readily incorporated into REDD1 (Fig. 3E). However, neither the HA-REDD1 C150S nor HA-REDD1 C157S exhibited an increase in reactive cysteines upon exposure to hyperglycemic conditions (Fig. 3F) or H2O2 (Fig. 3G). Moreover, the suppressive effect of H2O2 on HA-REDD1 degradation was not observed with HA-REDD1 C150S or HA-REDD1 C157S (Fig. 3H and Supplementary Fig. 4C and D). WT and C150S/C157S REDD1 variants acted similarly in the suppression of S6K1 phosphorylation (Supplementary Fig. 4E), supporting that the disulfide was not required for REDD1 to inhibit mTORC1.
Figure 3.
REDD1 degradation is reduced by a redox-sensitive disulfide bond. A: Liquid chromatography–tandem mass spectrometry fragmentation of AspN-digested FLAG-REDD1 identified a cystine-linked peptide by loss of 2.01565 Da that included C150/C157. B: REDD1 protein structure (PDB ID 3LQ9) includes an α/β sandwich with two antiparallel α-helices and four β-strands. Inset highlights 3.3-Å distance between the C150 and C157 side chains. C: Two-dimensional graphic of REDD1 secondary structure illustrates location of C150/C157 cross-strand disulfide. D: C150/C157 of REDD1 exhibit sequence conservation. E: REDD1 cysteine oxidation in MIO-M1 cells exposed to medium supplemented with 30 mmol/L glucose (HG) or an osmotic control (OC) for 4 h was evaluated by biotin switch assay. Western blotting was used to evaluate REDD1 and PKM2 in biotin pull-downs and whole-cell lysates (WCL). Representative blots are shown. Protein molecular mass in kDa is indicated at right of blots. REDD1 in biotin pull-down was quantified. PKM2 was evaluated as a positive control. F and G: WT, C150S, and C157S HA-REDD1 variants were expressed in REDD1-KO MIO-M1 cells. Cells were exposed to OC vs. HG for 4 h (F) or to vehicle (V) or H2O2 (HP) for 2 h (G). REDD1 cysteine oxidation was evaluated by biotin switch assay. H: WT, C150S, and C157S HA-REDD1 variants were expressed in REDD1-KO MIO-M1 cells. HA-REDD1 variant degradation was evaluated by cycloheximide (CHX)-chase assay. Data are represented as mean ± SD. *P < 0.05 vs. OC or Veh; #P < 0.05 vs. WT.
REDD1 Oxidation Disrupts Recognition by HSC70
To understand the potential impact of the C150/C157 disulfide on REDD1 structure/function, we performed DMD simulations in the absence and presence of the disulfide bond, modeled as a distance constraint between the sulfurs (1.5–3.0 Å). As an approximation to allostery, we visualized correlation of pairwise residue positions over the course of each simulation (Fig. 4A). These correlation coefficients revealed whether two residues were prone to traveling in the same direction at any given time, with a higher correlation coefficient indicating a stiffer allostery. Our results agreed with the allosteric pathways predicted by Ohm (Fig. 4B), in which V178 was seen to interact with C150/157 as well as α-helix 2 (Fig. 3B, teal), distant from the disulfide. Global allosteric differences were observed in the presence and absence of the disulfide bond, indicating that the disulfide significantly affects REDD1 structure (Fig. 4C). Examination of the α-helix 2 sequence for a putative degron identified an acetylation-activated KFERQ-like motif (129KELLR133, KFERQ finder [23]) (Fig. 4D). WT REDD1 was recognized by an anti–acetyl-lysine antibody, but not the REDD1 K129R variant (Supplementary Fig. 5A). H2O2 did not alter REDD1 lysine acetylation, nor was there a difference in lysine acetylation between WT and the C150S/C157S variants of REDD1 (Supplementary Fig. 5A and B). The chaperone HSC70 binds KFERQ-like motifs to direct substrate proteins to the lysosome for CMA (41). HSC70 coimmunoprecipitated with FLAG-tagged REDD1, and the interaction was reduced upon exposure to H2O2 (Fig. 4E). HSC70 coimmunoprecipitation with FLAG-REDD1 C150S or C157S was not altered by H2O2. In support of a role for the KFERQ-like motif, REDD1 K129R failed to associate with HSC70 (Fig. 4F) but was oxidized upon H2O2 exposure (Supplementary Fig. 5C and D).
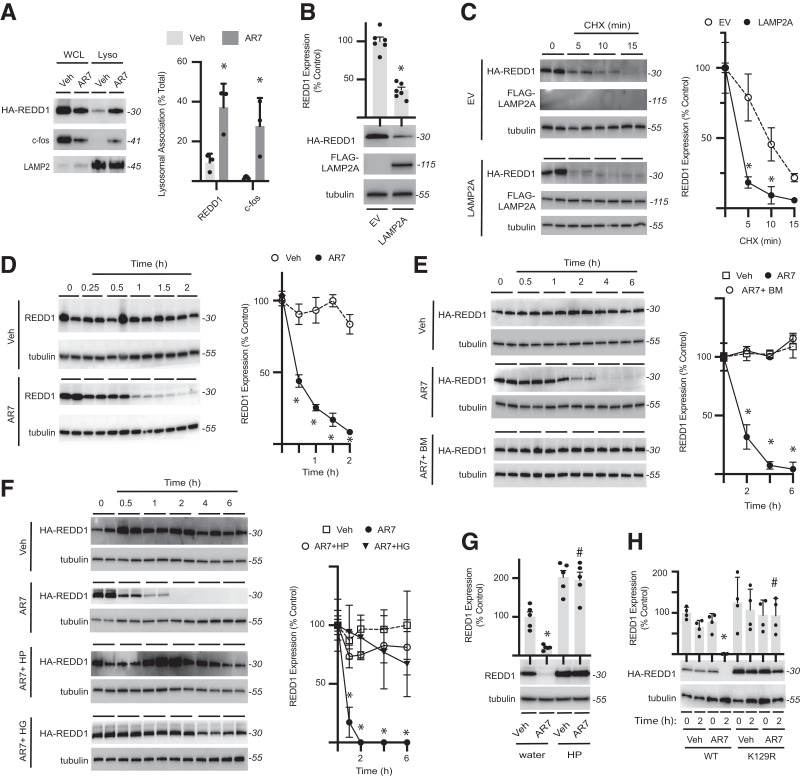
Hyperglycemia-Induced ROS Prevent REDD1 Degradation by CMA
Lysosomal proteolysis of REDD1 was investigated by exposing cells to the CMA-activator/retinoic acid receptor-α antagonist AR7 (42). HA-REDD1 localization to lysosomes was enhanced by AR7 in a manner that was similar to the CMA target c-fos (43) (Fig. 5A). Previous studies have shown that expression of LAMP2A is sufficient to promote CMA (44). In the current study, LAMP2A expression reduced HA-REDD1 expression (Fig. 5B) and accelerated its degradation in a cycloheximide-chase assay (Fig. 5C). Endogenous REDD1 expression was also reduced in cells exposed to AR7 compared with vehicle (Fig. 5D). The lysosomal proton-pump inhibitor bafilomycin prevented the suppressive effect of AR7 on HA-REDD1 expression (Fig. 5E and Supplementary Fig. 6A and B). Similarly, exposure to H2O2 or hyperglycemic conditions blunted the reduction in HA-REDD1 expression by AR7 (Fig. 3F and Supplementary Fig. 6C and D). H2O2 also prevented a reduction in endogenous REDD1 expression after exposure to AR7 (Fig. 5G). Unlike WT REDD1, REDD1 K129R expression was not reduced in cells exposed to AR7 (Fig. 5H). Moreover, hyperglycemic conditions failed to prevent the reduction in expression of REDD1 C150S and C157S in cells exposed to AR7 (Supplementary Fig. 6E).
Figure 5.
REDD1 is degraded by chaperone-mediated autophagy. A: HA-REDD1 was expressed in REDD1-KO MIO-M1 cells. Lysosomes were isolated by subcellular fractionation from cells exposed to the CMA-activator AR7 or vehicle (Veh) for 1 h. REDD1, c-fos, and LAMP2 expression were evaluated in whole-cell lysate (WCL) vs. lysosomes (Lyso) by Western blotting. Representative blots are shown. Protein molecular mass in kDa is indicated at right of blots. REDD1 and c-fos lysosomal association was quantified. B: HEK293 Tet-On HA-REDD1 cells were transfected to express FLAG-tagged LAMP2A or an empty vector (EV) control. REDD1, LAMP2A, and tubulin protein expression was evaluated by Western blotting. C: REDD1 degradation was evaluated in HEK293 Tet-On HA-REDD1 cells expressing FLAG-LAMP2A or EV by cycloheximide (CHX)-chase (n = 4). D: HEK293 cells were exposed to tunicamycin for 4 h to promote endogenous REDD1 expression and subsequently exposed to AR7 or Veh (n = 5). E: HEK293Tet-On HA-REDD1 cells were exposed to culture medium containing AR7 in the presence/absence of the autophagy inhibitor bafilomycin (n = 4). F: HA-REDD1 was expressed in REDD1-KO MIO-M1 cells by transient transfection. Cells were exposed to medium containing 30 mmol/L glucose (HG) or H2O2 (HP) for 24 h prior to addition of AR7 or Veh (n = 4). G: Endogenous REDD1 expression was evaluated in HEK293 cells pretreated with HP or water as a control for 2 h prior to exposure to AR7 or Veh for 2 h. H: WT or K129R HA-REDD1 variants were expressed in REDD1-KO MIO-M1 cells by transient transfection (n = 4). Cells were exposed to Veh or AR7 as indicated. Data are represented as mean ± SD. *P < 0.05 vs. Veh or EV; #P < 0.05 vs. water or WT.
V178 Is Critical for Allosteric Regulation of REDD1 by the Disulfide Bond
To better understand the allosteric regulation of REDD1, we investigated residues that interacted with the C150/C157 disulfide bond. V178 is located in close proximity to the disulfide bond and surrounded by several charged side chains (i.e., E152, R180, R215) (Fig. 6A). V178 is located in β-strand 3, which exhibits high sequence conservation (Fig. 6B). Because of the close packing of these residues, we hypothesized that a minor mutation at V178 may disrupt the overall REDD1 allostery. To identify substitutions at V178 that would be favorable for REDD1 stability, ΔΔG calculations were performed using the Eris web server (35) (Fig. 6C). V178I mutagenesis of REDD1 resulted in the minimal increase in ΔΔG (Fig. 6C). As with the WT REDD1, DMD simulations were performed on the REDD1 V178I variant in the presence and absence of the disulfide (Fig. 6D). Residual differences identified in DMD simulations of WT REDD1, with and without the C150/C157 disulfide, differed from those in the REDD1 V178I variant (Fig. 6E), supporting the blunting of allosteric regulation.
Disruption of Allosteric Regulation Prevents REDD1 Stabilization and Oxidative Stress
To test the hypotheses generated from our computational work, we examined degradation of the REDD1 V178I variant. Whereas H2O2 prevented the suppressive effect of AR7 on WT REDD1 expression, a similar protective effect of H2O2 was not observed with REDD1 V178I (Fig. 6F). Similarly, H2O2 prevented the rapid degradation of WT REDD1, but not REDD1 V178I (Fig. 6G). The absence of a protective effect on REDD1 V178I was not due to disruption of the disulfide bond, as REDD1 WT and V178I exhibited a similar extent of cysteine oxidation following exposure to H2O2 (Fig. 6H). Consistent with the prior report (8), REDD1 expression in MIO-M1 cells exposed to hyperglycemic conditions was increased in coordination with cellular ROS levels (Fig. 6I). Importantly, REDD1 expression was required for hyperglycemia-induced oxidative stress. In REDD1-KO cells wherein REDD1 was rescued by HA-REDD1 expression, hyperglycemic conditions enhanced ROS levels (Fig. 6J). Exposure to hyperglycemic conditions also enhanced HA-REDD1 protein content in cells expressing low concentrations of the plasmid (Supplementary Fig. 7). However, an increase in HA-REDD1 expression upon exposure to hyperglycemic conditions was not observed when WT REDD1 was expressed at higher plasmid concentrations nor with HA-REDD1 V178I expression at low or high plasmid levels. In coordination with the failure to upregulate HA-REDD1 V178I protein expression at low plasmid levels, hyperglycemia-induced ROS were blunted compared with WT HA-REDD1 (Fig. 6J). However, rescuing REDD1 expression with WT or V178I HA-REDD1 was sufficient to restore hyperglycemic-induced oxidative stress when either protein was expressed at a high concentration. The data support a model wherein allosteric regulation of REDD1 upon formation of the disulfide bond leads to REDD1 accumulation and hyperglycemia-induced oxidative stress (Fig. 7).
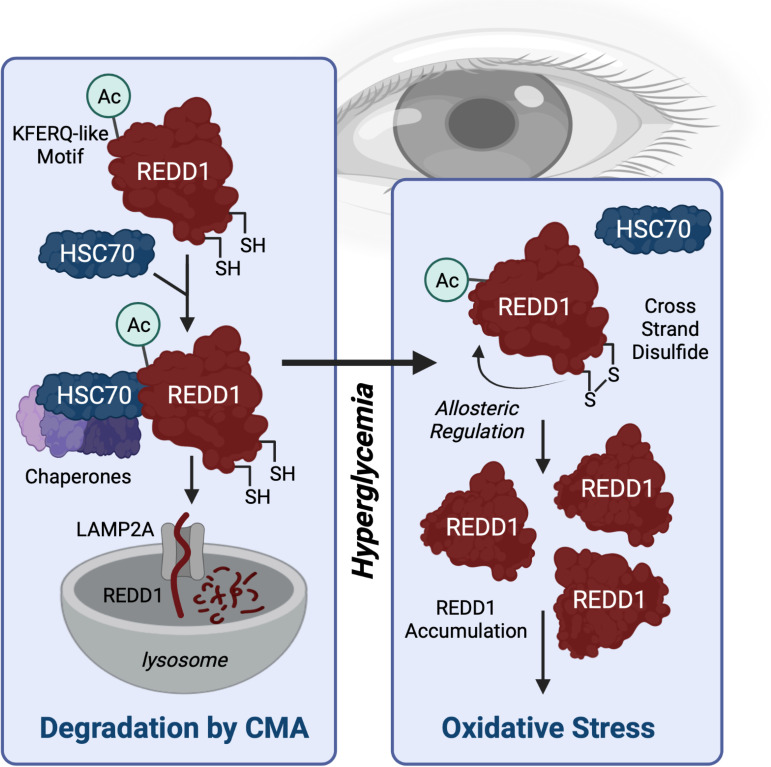
Figure 7.
Working model for a REDD1 redox switch in development of oxidative stress in DR. HSC70 interacts with REDD1 through a KFERQ-like motif that is created upon K129 acetylation (Ac) to facilitate degradation by CMA. In response to hyperglycemic conditions, a disulfide bridge (S-S bond) forms at REDD1 C150/C157, which acts allosterically to disrupt HSC70 binding to REDD1. The consequent reduction in REDD1 degradation leads to its accumulation and facilitates hyperglycemia-induced oxidative stress. Created with Biorender.com.
Discussion
An array of human pathologies results from the failure of regulatory proteins to be expressed appropriately. In rodent models of both type 1 and type 2 diabetes, REDD1 protein expression is increased in the retina (8,9,13,14), and positive effects of REDD1 suppression have been demonstrated in the context of diabetic retinopathy (13,16,39). Protein expression is the ultimate sum of events, including transcription, translation, and mRNA and protein degradation, that determine cellular protein content. In the retina of diabetic mice, REDD1 protein expression was enhanced in the absence of a corresponding change in either abundance or ribosome-association of REDD1 mRNA. We previously found that the increase in retinal REDD1 protein content was mediated by hyperglycemia per se, as the induction of glucosuria in diabetic mice to normalize blood glucose concentrations prevented the effect. Herein, we demonstrate that hyperglycemic conditions impair lysosomal proteolysis of REDD1 by promoting the formation of a redox-sensitive disulfide.
Disulfide bonds have long been regarded as static structural features that occur in the oxidizing environment of the endoplasmic reticulum; however, the formation of reversible redox-sensitive disulfide bonds has more recently been appreciated to play a central role in the detection and response to oxidative stress (45,46). Strands β1–β3 of REDD1 form a Ψ-loop, wherein the antiparallel strands β2 and β3 are separated by the intervening β1 strand that hydrogen bonds with both. The disulfide at C150/C157 seemed unlikely to confer additional rigidity or stability to the protein, as cross-strand disulfides typically produce highly stressed torsional angles for the half-cystines (45). Such disulfides were dubbed “forbidden,” as the adjacent strands must adopt a high twist with respect to each other that is stoichiometrically prohibitive (46). The functional implications of these strained confirmations are now recognized in a number of important redox switches (45). We found that the strain induced by formation of the REDD1 C150/C157 disulfide allosterically regulated an acetylation-activated KFERQ-like motif on the opposite face of the protein.
REDD1 was recognized by an acetyl-lysine antibody, and K129 was required for both the modification and coimmunoprecipitation with HSC70. REDD1 K129 is preceded by a glycine at the −1 position and fits the consensus motif for diabetes-induced lysine acetylation sites identified in the kidney of diabetic rats (47). Sequence conservation of K129 was observed among vertebrates, with the exception of substitution to the acetyl-lysine mimetic glutamine in reptiles and fishes. The best characterized example of an acetylation-activated KFERQ-like motif is found in PKM2. Hyperglycemia-induced acetylation of PKM2 at K305 leads to activation of its KFERQ-like motif to facilitate HSC70 recruitment and CMA degradation (48). PKM2 also contains multiple oxidation-sensitive cysteines that become modified to inhibit activity of the kinase (49). Thus, PKM2 oxidation and acetylation act in concert to inhibit and degrade the protein. By contrast, REDD1 oxidation and acetylation were found to work in opposition, with acetylation of REDD1 favoring lysosomal degradation and oxidation preventing the effect. No variation in REDD1 lysine acetylation was observed in the presence/absence of the disulfide. Rather, the data are consistent with a model in which the C150/C157 disulfide of REDD1 disrupts HSC70 recognition of the K129 acetylation-activated KFERQ-like motif.
The synthesis of rapidly degraded proteins is energetically unfavorable but can be advantageous by allowing cellular concentrations of regulatory proteins to be quickly fine-tuned in response to stresses or environmental cues. REDD1 is ubiquitinated and targeted to the 26S proteasome by both CUL4A–DDB1–ROC–β-TRCP (21) and HUWE1 (50) E3 ligases. Initially, we suspected that the reduction in REDD1 turnover under hyperglycemic conditions was due to disrupted proteasomal degradation. However, we found that hyperglycemic/oxidative conditions were additive with proteasomal inhibition in suppressing REDD1 degradation. In a prior study, proteasomal inhibition slowed REDD1 degradation, but was insufficient to prevent rapid turnover of the protein (i.e., REDD1 half-life was only increased from ∼5 to 40 min) (21). We extend on the prior investigations of REDD1 turnover by demonstrating that the protein is also subject to rapid degradation via CMA.
A role for REDD1 C150/C157 was previously investigated when their superficial orientation and proximity was observed in the REDD1 crystal structure (31). Consistent with the prior report, C150S/C157S mutagenesis did not alter the suppressive effect of REDD1 on mTORC1-dependent phosphorylation of S6K1, demonstrating that C150/C157 was not required for REDD1 to act on its downstream targets. Importantly, we found little evidence of the C150/C157 disulfide without exposure to oxidants or hyperglycemic conditions. This supports that the disulfide bond is not generated in the endoplasmic reticulum during protein folding. Thus, it is likely that these studies compared mTORC1 suppression by WT REDD1 without the disulfide to C150S/C157S variants that could not form the disulfide, as cells were not manipulated to promote oxidation of cytoplasmic proteins (31). Regardless, the findings here do not support a role for the C150/C157 disulfide in the suppressive effect of REDD1 on Akt/mTORC1 signaling. Rather, the data demonstrate that REDD1 oxidation contributes to accumulation of the protein and increased oxidative stress in response to hyperglycemic conditions.
In both the retina of diabetic mice and in retinal cells in culture exposed to hyperglycemic conditions, REDD1 is necessary for the development of oxidative stress (8,9). Evidence supports that REDD1 contributes to the development of oxidative stress by both enhancing production of mitochondrial reactive oxygen species and undermining the Nrf2 antioxidant response (10). Hyperglycemic conditions suppressed degradation of REDD1 protein via formation of a C150/C157 disulfide that allosterically regulated interaction with HSC70. Surprisingly, either C150S or C157S substitution was sufficient to prevent cysteine oxidation on REDD1 in cells exposed to hyperglycemic conditions. This observation suggests that the local environment created by both cysteine thiols is necessary for formation of a highly reactive sulfenic acid intermediate that precedes the disulfide bond (51). These studies also support that allosteric regulation of REDD1 by the disulfide is mediated through V178, as V178I substitution prevented the suppressive effect of the disulfide on REDD1 degradation by CMA. In REDD1-KO cells where REDD1 expression was rescued, hyperglycemia-induced ROS accumulation was restored. In contrast to WT REDD1, REDD1 V178I expression was not increased upon exposure to hyperglycemic conditions, and the lower expression of REDD1 V178I was associated with blunted hyperglycemia-induced ROS. When the REDD1 V178I variant was expressed at a higher level by increasing the amount of plasmid used in transfection, hyperglycemia-induced oxidative stress was restored. This supports that the disulfide promotes REDD1 upregulation, which can be prevented by V178I mutagenesis. However, when REDD1 V178I expression was artificially enhanced by transfecting a higher concentration of the plasmid, glucose-induced oxidative stress returned. Overall, these proof-of-concept studies demonstrate a role for V178 in increased REDD1 expression and consequently development of oxidative stress in response to hyperglycemic conditions.
Diabetes promotes REDD1 expression in the retina, and the protein has been implicated in visual deficits in both preclinical models and in patients with diabetes. When considering the modest performance of PF-04523655 in improving visual acuity in patients with diabetic macular edema (16), it is important to note that effectiveness of the siRNA in suppressing REDD1 protein expression in the context of diabetes has never been determined. In fact, variation in rates of mRNA and protein turnover determines the suitability of a gene target for knockdown by siRNA. Thus, diabetes-induced stabilization of REDD1 protein would be counterproductive to reducing REDD1 protein expression in the retina by siRNA knockdown. Targeting a redox-sensitive molecular switch that causes a decline in REDD1 protein degradation in response to diabetes potentially represents a superior therapeutic strategy for preventing the pathological increase in REDD1 protein content.
Article Information
Acknowledgments. The authors thank Dr. Elena Feinstein (Quark Pharmaceuticals) for permission to use the REDD1−/− mice and the Harvard Medical School Taplin Biological Mass Spectrometry Facility for performing mass spectrometry analysis.
Funding. This research was supported by American Diabetes Association Pathway to Stop Diabetes grant 1-14-INI-04, National Institutes of Health National Eye Institute grants R01 EY029702, R01 EY032879 (to M.D.D.), and F31 EY031199 (to W.P.M.). Support was also provided by the National Institutes of Health National Institute of General Medical Sciences grant 1R35 GM134864, the National Science Foundation (2040667), and the Passano Foundation (to N.V.D.).
Duality of Interests. No potential conflicts of interest relevant to this article were reported.
Author Contributions. W.P.M., C.M.S., S.S., A.L.T., A.M.V., N.V.D., and M.D.D. contributed to data curation. W.P.M., C.M.S., S.S., A.L.T., A.M.V., and M.D.D. contributed to formal analysis. W.P.M., C.M.S., S.S., and A.M.V. contributed to the investigation. W.P.M., C.M.S., S.R.K., N.V.D., and M.D.D. contributed to conceptualization. W.P.M., C.M.S., S.R.K., N.V.D., and M.D.D. contributed to methodology. W.P.M., C.M.S., and M.D.D. contributed to visualization. W.P.M., C.M.S., and M.D.D. contributed to writing. W.P.M., A.L.T., S.R.K., N.V.D., and M.D.D. contributed to reviewing and editing. S.R.K., N.V.D., and M.D.D. contributed to resources. S.R.K., N.V.D., and M.D.D. supervised the research. W.P.M. and M.D.D. contributed to funding acquisition. M.D.D is guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 81st Scientific Sessions of the American Diabetes Association, virtual meeting, 25–29 June 2021.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.21183496.
References
- 1. Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res 2010;107:1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes 2005;54:1615–1625 [DOI] [PubMed] [Google Scholar]
- 3. Schulze PC, Yoshioka J, Takahashi T, He Z, King GL, Lee RT. Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. J Biol Chem 2004;279:30369–30374 [DOI] [PubMed] [Google Scholar]
- 4. Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE 2000;2000:pe1. [DOI] [PubMed] [Google Scholar]
- 5. Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012;48:158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellisen LW, Ramsayer KD, Johannessen CM, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell 2002;10:995–1005 [DOI] [PubMed] [Google Scholar]
- 7. Shoshani T, Faerman A, Mett I, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol 2002;22:2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller WP, Toro AL, Barber AJ, Dennis MD. REDD1 activates a ROS-generating feedback loop in the retina of diabetic mice. Invest Ophthalmol Vis Sci 2019;60:2369–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller WP, Sunilkumar S, Giordano JF, Toro AL, Barber AJ, Dennis MD. The stress response protein REDD1 promotes diabetes-induced oxidative stress in the retina by Keap1-independent Nrf2 degradation. J Biol Chem 2020;295:7350–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller WP, Sunilkumar S, Dennis MD. The stress response protein REDD1 as a causal factor for oxidative stress in diabetic retinopathy. Free Radic Biol Med 2021;165:127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004;18:2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dennis MD, Coleman CS, Berg A, Jefferson LS, Kimball SR. REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal 2014;7:ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dennis MD, Kimball SR, Fort PE, Jefferson LS. Regulated in development and DNA damage 1 is necessary for hyperglycemia-induced vascular endothelial growth factor expression in the retina of diabetic rodents. J Biol Chem 2015;290:3865–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dai W, Miller WP, Toro AL, et al. Deletion of the stress-response protein REDD1 promotes ceramide-induced retinal cell death and JNK activation. FASEB J 2018;32:6883–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller WP, Toro AL, Sunilkumar S, et al. Müller glial expression of REDD1 is required for retinal neurodegeneration and visual dysfunction in diabetic mice. Diabetes 2022;71:1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nguyen QD, Schachar RA, Nduaka CI, et al.; DEGAS Clinical Study Group . Dose-ranging evaluation of intravitreal siRNA PF-04523655 for diabetic macular edema (the DEGAS study). Invest Ophthalmol Vis Sci 2012;53:7666–7674 [DOI] [PubMed] [Google Scholar]
- 17. Rittenhouse KD, Johnson TR, Vicini P, et al. RTP801 gene expression is differentially upregulated in retinopathy and is silenced by PF-04523655, a 19-Mer siRNA directed against RTP801. Invest Ophthalmol Vis Sci 2014;55:1232–1240 [DOI] [PubMed] [Google Scholar]
- 18. James HA, O’Neill BT, Nair KS. Insulin regulation of proteostasis and clinical implications. Cell Metab 2017;26:310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flick K, Kaiser P. Protein degradation and the stress response. Semin Cell Dev Biol 2012;23:515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimball SR, Do AND, Kutzler L, Cavener DR, Jefferson LS. Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. J Biol Chem 2008;283:3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katiyar S, Liu E, Knutzen CA, et al. REDD1, an inhibitor of mTOR signalling, is regulated by the CUL4A-DDB1 ubiquitin ligase. EMBO Rep 2009;10:866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan CY, Hagen T. mTORC1 dependent regulation of REDD1 protein stability. PLoS One 2013;8:e63970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirchner P, Bourdenx M, Madrigal-Matute J, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol 2019;17:e3000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brafman A, Mett I, Shafir M, et al. Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Invest Ophthalmol Vis Sci 2004;45:3796–3805 [DOI] [PubMed] [Google Scholar]
- 25. Dierschke SK, Toro AL, Miller WP, Sunilkumar S, Dennis MD. Diabetes enhances translation of Cd40 mRNA in murine retinal Müller glia via a 4E-BP1/2-dependent mechanism. J Biol Chem 2020;295:10831–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gastinger MJ, Singh RS, Barber AJ. Loss of cholinergic and dopaminergic amacrine cells in streptozotocin-diabetic rat and Ins2Akita-diabetic mouse retinas. Invest Ophthalmol Vis Sci 2006;47:3143–3150 [DOI] [PubMed] [Google Scholar]
- 27. Miller WP, Mihailescu ML, Yang C, et al. The translational repressor 4E-BP1 contributes to diabetes-induced visual dysfunction. Invest Ophthalmol Vis Sci 2016;57:1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 1996;68:850–858 [DOI] [PubMed] [Google Scholar]
- 29. Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 1994;5:976–989 [DOI] [PubMed] [Google Scholar]
- 30. Anastasiou D, Poulogiannis G, Asara JM, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011;334:1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vega-Rubin-de-Celis S, Abdallah Z, Kinch L, Grishin NV, Brugarolas J, Zhang X. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry 2010;49:2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shirvanyants D, Ding F, Tsao D, Ramachandran S, Dokholyan NV. Discrete molecular dynamics: an efficient and versatile simulation method for fine protein characterization. J Phys Chem B 2012;116:8375–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Proctor EA, Dokholyan NV. Applications of discrete molecular dynamics in biology and medicine. Curr Opin Struct Biol 2016;37:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des 1998;3:577–587 [DOI] [PubMed] [Google Scholar]
- 35. Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods 2007;4:466–467 [DOI] [PubMed] [Google Scholar]
- 36. Sun MA, Wang Y, Zhang Q, Xia Y, Ge W, Guo D. Prediction of reversible disulfide based on features from local structural signatures. BMC Genomics 2017;18:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harris CR, Millman KJ, van der Walt SJ, et al. Array programming with NumPy. Nature 2020;585:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J, Jain A, McDonald LR, Gambogi C, Lee AL, Dokholyan NV. Mapping allosteric communications within individual proteins. Nat Commun 2020;11:3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller WP, Yang C, Mihailescu ML, et al. Deletion of the Akt/mTORC1 repressor REDD1 prevents visual dysfunction in a rodent model of type 1 diabetes. Diabetes 2018;67:110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li R, Kast J. Biotin switch assays for quantitation of reversible cysteine oxidation. Methods Enzymol 2017;585:269–284 [DOI] [PubMed] [Google Scholar]
- 41. Dice JF, Walker CD, Byrne B, Cardiel A. General characteristics of protein degradation in diabetes and starvation. Proc Natl Acad Sci U S A 1978;75:2093–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol 2013;9:374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aniento F, Papavassiliou AG, Knecht E, Roche E. Selective uptake and degradation of c-Fos and v-Fos by rat liver lysosomes. FEBS Lett 1996;390:47–52 [DOI] [PubMed] [Google Scholar]
- 44. Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy 2012;8:1643–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haworth NL, Wouters MJ, Hunter MO, Ma L, Wouters MA. Cross-strand disulfides in the hydrogen bonding site of antiparallel β-sheet (aCSDhs): forbidden disulfides that are highly strained, easily broken. Protein Sci 2019;28:239–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wouters MA, George RA, Haworth NL. “Forbidden” disulfides: their role as redox switches. Curr Protein Pept Sci 2007;8:484–495 [DOI] [PubMed] [Google Scholar]
- 47. Kosanam H, Thai K, Zhang Y, et al. Diabetes induces lysine acetylation of intermediary metabolism enzymes in the kidney. Diabetes 2014;63:2432–2439 [DOI] [PubMed] [Google Scholar]
- 48. Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell 2011;42:719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Irokawa H, Numasaki S, Kato S, et al. Comprehensive analyses of the cysteine thiol oxidation of PKM2 reveal the effects of multiple oxidation on cellular oxidative stress response. Biochem J 2021;478:1453–1470 [DOI] [PubMed] [Google Scholar]
- 50. Thompson JW, Nagel J, Hoving S, et al. Quantitative Lys-ϵ-Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J Biol Chem 2014;289:28942–28955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rehder DS, Borges CR. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 2010;49:7748–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]