Abstract
Introduction
HLA-G plays a central role in immune tolerance at the maternal-fetal interface. The HLA-G gene is characterized by low allelic polymorphism and restricted tissue expression compared with classical HLA genes. HLA-G polymorphism is associated with HLA-G expression and linked to pregnancy complications. However, the association of parental HLA-G polymorphisms with soluble HLA-G (sHLA-G) expression and their roles in recurrent implantation failure (RIF) is unclear. The study aims to systematically review the association of HLA-G polymorphisms with RIF, the association of sHLA-G expression with RIF, and the association of HLA-G polymorphisms with sHLA-G expressions in patients attending in-vitro fertilization (IVF) treatment.
Methods
Studies that evaluated the association of HLA-G polymorphisms with RIF, the association between sHLA-G expression with RIF, and the association between HLA-G polymorphisms with sHLA-G expressions in patients attending IVF treatment were included. Meta-analysis was performed by random-effect models. Sensitivity analysis was performed by excluding one study each time. Subgroup analysis was performed based on ethnicity.
Results
HLA-G 14bp ins variant is associated with a lower expression of sHLA-G in seminal or blood plasma of couples attending IVF treatment. The maternal HLA-G*010101 and paternal HLA-G*010102 alleles are associated with RIF risk compared to other alleles. However, single maternal HLA-G 14bp ins/del polymorphism, HLA-G -725 C>G/T polymorphism, or circulating sHLA-G concentration was not significantly associated with RIF in the general population. HLA-G 14bp ins/ins homozygous genotype or ins variant was associated with a higher risk of RIF in the Caucasian population.
Discussion
Specific HLA-G alleles or HLA-G polymorphisms are associated with sHLA-G expression in couples attending IVF treatment. Several HLA-G polymorphisms may be related to RIF, considering different ethnic backgrounds. A combined genetic effect should be considered in future studies to confirm the association of HLA-G polymorphisms and sHLA-G expressions in relation to RIF.
Keywords: Human leukocyte antigen G, immune tolerance, recurrent implantation failure, genetic polymorphism, meta-analysis
Introduction
Human leukocyte antigen (HLA)-G is a non-classical HLA class Ib molecule that plays a vital role in the maternal acceptance of the semi-allogenic fetus. The HLA-G gene, locates on chromosome 6p21.3, consists of seven introns and eight exons. Alternative splicing of HLA-G mRNA generates seven HLA-G isoforms, including four membrane isoforms (G1-G4) and three soluble isoforms (G5-G7) (1). In addition, soluble HLA-G (sHLA-G) can be generated by shedding or proteolytic cleavage of membrane-anchored HLA-G through matrix metalloproteinases (MMPs) activity, such as shed HLA-G1 (2, 3). HLA-G gene is characterized by low allelic polymorphism and restricted tissue expression compared with highly polymorphic classical HLA Ia genes (HLA-A, B, C). HLA-G’s expression is mainly restricted to the maternal-fetal interface and immune tissues. HLA-G is detectable in body fluids as secreted soluble molecules despite the restricted tissue expression (4–6). Essential functions of HLA-G at the fetal-maternal interface include the inhibition of natural killer (NK) cells mediated cytolysis, enrichment of regulatory T (Treg) cells, and promotion of a shift from a T-helper (Th)1 to a Th2 cytokine profile (7). HLA-G polymorphisms are associated with abnormal HLA-G levels and linked to reproductive disorders such as implantation failure, recurrent miscarriage, preeclampsia, and placental abruption (8–14). One of the most studied HLA-G polymorphisms is the 14bp insertion/deletion (ins/del) polymorphism located on exon eight at the 3’ untranslated region (3’UTR). HLA-G 14bp ins/del affects the stability of HLA-G mRNA and leads to abnormal HLA-G expression (15, 16), which is associated with recurrent miscarriage (11). HLA-G -725C>G polymorphism located at the 5’upstream regulator region (5’URR) or promoter region is reported to change the methylation profile of CpG dinucleotide, resulting in a modification of HLA-G expression and also linked to miscarriage (17). Besides, the other HLA-G polymorphism such as HLA-G -964G>A at 5’ URR, HLA-G allele variation at exon 2, 3, 4, intron 2, and specific HLA-G haplotypes/diplotypes are associated with sHLA-G expression and may be linked to reproductive outcomes (8, 18–21). However, the role of HLA-G polymorphism on RIF has been investigated in only a few studies with contradictory results.
Recurrent implantation failure (RIF) is a complication following in-vitro fertilization and embryo transfer (IVF-ET), with an incidence rate of approximately 10~15%. RIF is defined as good-quality embryos repeatedly failing to implant. It is generally diagnosed based on the number of unsuccessful ET cycles, the number of transferred embryos, female age, or a combination of these factors (22). The causes of RIF include decreased quality of gametes or embryos, decreased endometrial receptivity, uterine anomalies, immune diseases, thrombophilia conditions, endocrine disorders, metabolic disorders, and genetic abnormalities (22). Genetic factors contribute to RIF susceptibility as several genetic polymorphisms have been reported to be associated with RIF (23–26). Investigating the role of genetic polymorphisms on RIF susceptibility can help to promote our understanding of the pathogenesis underlying RIF and contributes to the prediction and prevention of RIF. Increasing evidence underlines the essential role of immune factors on embryo implantation as pregnancy remains an immune challenge for the uterus. The key to successful implantation and pregnancy maintenance is the immune tolerance of the uterus to the semi-allogeneic fetus (27). HLA-G plays a central role in immune tolerance at the maternal-fetal interface. Interactions between sHLA-G and uterine lymphocytes induce maternal immune tolerance for the invading extravillous trophoblasts, which is the critical factor affecting embryo implantation. Soluble HLA-G is essential for embryo implantation. The embryo-secreted sHLA-G in the culture medium served as a promising predictor for a successful pregnancy (28–32). However, the role of parental sHLA-G expression before pregnancy is less studied. The sHLA-G expression in circulating blood is significantly increased in pregnant women compared to that of unpregnant women. sHLA-G level is dynamically changed during pregnancy. Recent studies indicate that HLA-G may be involved in preparing for an immune environment before embryo implantation because sHLA-G can be detected in the genital tract, endometrium, and circulating blood of unpregnant women and is also present in male semen. Though maternal sHLA-G levels before pregnancy have been measured, their relationship with RIF has not yet been well established. Therefore, the current study aims to investigate the association of HLA-G polymorphisms with RIF, the association of sHLA-G expression with RIF, and the association of HLA-G polymorphisms with sHLA-G expression in patients attending IVF treatment.
Materials and methods
The authors performed this meta-analysis following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guideline (33).
Searching strategy
We searched EMBASE, Pubmed, and CNKI (China National Knowledge Infrastructure) for related studies from their inception to 19 September 2022. The grey literature was searched in OpenGrey (http://www.opengrey.eu/). The references of the included studies were also hand-searched. For searching studies that evaluate the association between HLA-G polymorphism with RIF, the searching syntax in PubMed involves the following text words: “HLA-G” or “HLAG” or “human leukocyte antigen G” in combination with “polymorphism” or “mutation” or “allele” or “genotype” or “genetic” or “variant” or “haplotype” or “diplotype” and in combination with “implantation” or “in vitro fertilization” or “IVF” or “ICSI” or “Intracytoplasmic sperm injection” or “embryo transfer”. For searching studies that evaluate the association between sHLA-G expression with RIF, the searching syntax in Pubmed involves the following text words: “HLA-G” or “HLAG” or “human leukocyte antigen G” or “sHLA-G” or “sHLAG” in combination with “expression” or “level” or “concentration” and in combination with “implantation” or “in vitro fertilization” or “IVF” or “ICSI” or “Intracytoplasmic sperm injection” or “embryo transfer”. For searching studies that evaluate the association between HLA-G polymorphism with sHLA-G expression in patients attending IVF treatment, the searching syntax in Pubmed involves the following text words: “HLA-G” or “HLAG” or “human leukocyte antigen G” or “sHLA-G” or “sHLAG” in combination with “expression” or “level” or “concentration” and in combination with “implantation” or “in vitro fertilization” or “IVF” or “ICSI” or “Intracytoplasmic sperm injection” or “embryo transfer” in combination with “polymorphism” or “mutation” or “allele” or “genotype” or “genetic” or “variant” or “haplotype” or “diplotype” The detailed searching strategies and searching results in Pubmed, Embase, and CNKI were listed in the Supplementary Material .
Inclusion criteria and exclusion criteria
For evaluating the association between HLA-G polymorphism with RIF or the association between sHLA-G expression with RIF, the inclusion criteria were: (1) case-control studies; (2) the cases were RIF patients; (3) the control patients were fertile women with ≥1 normal pregnancy and lived birth or infertile women with ≥1 normal pregnancy following IVF; (4) genotype frequencies or HLA-G concentrations are eligible for calculation. The exclusion criteria were: (1) non-case-control studies (cohort studies, reviews, case reports, or meta-analyses); (2) the case group was not RIF patients. For evaluating the association between HLA-G polymorphism with sHLA-G expression, the inclusion criteria were: (1) case-control studies or cohort studies; (2) the study population was patients attending IVF treatment; (3) sHLA-G expression was compared in each HLA-G genetic group. The exclusion criteria were: (1) non-case-control or non-cohort studies (reviews, case reports, or meta-analysis); (2) the study population was not patients attending IVF treatment; (3) patients with pregnancy complications (recurrent miscarriage, pre-eclampsia) or other immune diseases. Studies in all languages were included. Conference literature was included if the data was eligible for analysis and did not overlap with the published papers. Two authors (Lian Hu and Dongmei He) independently performed the study selection. A meta-analysis was performed for each HLA-G polymorphism with two or more published studies.
Data collection and quality assessment
Two authors (Lian Hu and Dongmei He) independently extracted the data from each study. We extracted the following information from studies that evaluate associations between HLA-G polymorphism or sHLA-G expression with RIF: first author, publication year, country, ethnicity, age of cases and controls, number of cases and controls, sample origin, genotyping method, and the definition of RIF and control. We extracted the following information from studies that evaluate the association between HLA-G polymorphism with sHLA-G expression in patients attending IVF treatment: first author, publication year, country, ethnicity, study population, number of patients tested, the sample tested, the timing of sample collection, type of assay, sHLA-G isoform, detection limit, and result. We evaluated study quality following the modified Newcastle-Ottawa scale (NOS). The scores of NOS ranged from 0 points (worst) to 9 points (best). We defined scores ≥ 7 as high quality. All the other scores indicate low quality. Two authors (Lian Hu and Dongmei He) assessed the study quality independently. Disagreement in study selection, data extraction, and quality assessment was dependent on the third author (Hong Zeng).
Statistical analysis
We performed the meta-analysis using the random-effects model due to each study’s heterogenous definition of RIF. The odds ratios (ORs) with 95% confidence intervals (CIs) or standardized difference in means (SMD) with 95% CIs were reported to evaluate the associations. The heterogeneity of the included studies was analyzed using the Q test and quantified using the I2 test. I2<25%, I2= 25-50%, I2= 50-75%, and I2>75% indicated no, moderate, large, and extreme heterogeneity, respectively. We examined the Hardy-Weinberg equilibrium (HWE) in the control group by the “GWASExactHW” R package (https://CRAN.R-project.org/package=GWASExactHW). Deviation from HWE was confirmed if the p-value<0.05. We assessed the publication bias using Begg’s test and Egger’s test. Publication bias was confirmed if the p-value of Begg’s or Eggers’ test was < 0.05. We performed subgroup analysis based on ethnicity. We performed sensitivity analysis by excluding one study each time. Though we only reported the result of the random-effect model in the manuscript, both the fixed-effects model and the random-effect model were performed in each meta-analysis with the results listed in the Supplementary Materials . All statistical analyses were performed using the R software (The R Foundation for Statistical Computing, version 4.1.1, https://www.r-project.org). A p-value < 0.05 was considered statistically significant.
Results
Study characteristics
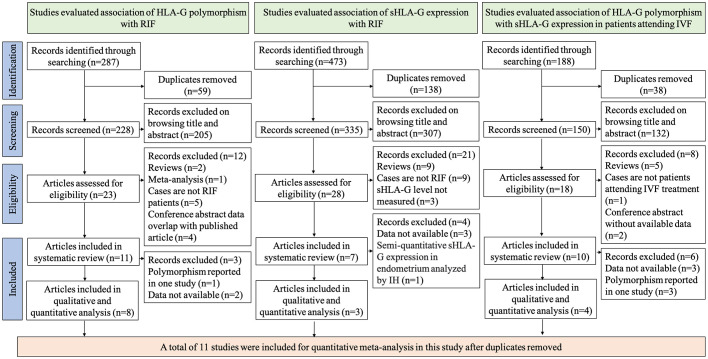
To investigate the association of HLA-G polymorphism with RIF, 287 records were identified through literature searching. We excluded 59 duplicates and excluded 205 records after browsing titles or abstracts. 23 articles were assessed for eligibility (8, 9, 18–21, 34–50). 12 articles were excluded from systematic review and meta-analysis [two are reviews (38, 40), one is a meta-analysis (9), five cases are not RIF patients (18, 21, 34, 48, 49), and four are conference abstracts whose data overlap with published articles (44–47)]. 11 articles were included for systematic review. However, three articles were excluded from the quantitative meta-analysis for reasons [two articles’ data are not available to extract for synthesis (41, 43), one study reported the polymorphisms only in their research thus could not be synthesized (36)]. Finally, eight studies were included in the quantitative analysis (8, 19, 20, 35, 37, 39, 42, 50). To investigate the association of sHLA-G expression with RIF, 473 records were identified through literature searching. We excluded 138 duplicates and excluded 307 records after browsing titles or abstracts. 28 articles were assessed for eligibility (8, 19, 35, 38, 39, 41, 42, 50–70). 21 articles were excluded from systematic review and meta-analysis [nine are reviews (38, 51–58), nine cases are not RIF patients (59–67), and three studies did not measure the sHLA-G levels (39, 42, 50)]. Four studies are excluded from quantitative analysis for reasons [three articles’ data are not available to extract for synthesis (8, 68, 69), one study measured sHLA-G levels in the endometrium by semi-quantitative method (41)]. Three articles were included in the quantitative synthesis (19, 35, 70). To investigate the association of HLA-G polymorphism with sHLA-G expression in patients attending IVF treatment, 188 records were identified through literature searching. We excluded 38 duplicates and excluded 132 records after browsing titles or abstracts. 18 articles were assessed for eligibility. Eight articles were excluded from the systematic review [five were reviews (57, 58, 71–73), two were conference abstracts without data (74), and cases in one study were not IVF patients (59)]. Six studies were excluded from the quantitative meta-analysis [three studies’ data are not available to extract for synthesis (19, 41, 75), three articles reported the polymorphism in only one study thus cannot be synthesized (8, 70, 76)]. Finally, four studies were included in the quantitative meta-analysis (35, 77–79). The flow diagram of selecting studies for systematic review and meta-analysis is shown in Figure 1 .
Figure 1.
Flow chart of study selection.
There are several HLA-G polymorphisms reported in different studies. Meta-analysis is performed if at least two studies evaluate the same HLA-G polymorphism with RIF, and the data can be extracted. Table 1 summarizes the different HLA-G polymorphisms that have been reported in RIF. Association of HLA-G 14bp ins/del polymorphism, HLA-G -725 C>G polymorphism, HLA-G alleles distribution at exon2-4 (HLAG*010101, HLAG*010102, HLAG*010103, HLAG*010106, HLAG*010107, HLAG*010108, HLAG*010401, HLAG*010403, HLAG*010404, HLAG*0106, HLAG*0105N) were reported in more than two studies; therefore, meta-analyses were performed in these HLA-G polymorphisms. Meta-analysis is also performed based on three studies that evaluated the association between sHLA-G levels and RIF. The characteristics of included studies in the meta-analysis that evaluate the association between HLA-G polymorphism with RIF or the association between sHLA-G expression with RIF are presented in Table 2 . The studies were conducted in Poland, Denmark, Brazil, Iran, and the Netherlands. The HLA-G 14bp ins/del genotype frequency in the control group has deviated from HWE in Hviid’s study (p-value=0.042) (39). The other studies’ HLA-G 14bp ins/del genotype frequencies did not deviate from HWE. The HLA-G -725C>G genotype frequencies in the included studies did not deviate from HWE.
Table 1.
Summary of the HLA-G polymorphisms that have been reported in RIF patients.
| HLA-G polymorphisms | Location | No. studies | Publication | Included in meta-analysis |
|---|---|---|---|---|
| HLA-G genotypes | ||||
| HLA-G 14-bp ins/del (rs66554220/rs371194629) | 3′UTR | 6 | Hviid 2004 (39) Sipak-Szmigie 2009 (42) Enghelabifar 2014 (37) Lashley 2014 (50) Nardi 2016 (35) Nowak 2019 (8) |
Yes |
| HLA-G -725 C>G(T) (rs1233334) | 5′URR | 2 | Sipak-Szmigie 2009 (42) Nowak 2019 (8) |
Yes |
| HLA-G -964G>A (rs1632947) | 5′URR | 1 | Nowak 2019 | No |
| HLA-G haplotypes | ||||
| Haplotypes of rs1632947-rs1233334-rs371194629 | – | 1 | Nowak 2019 (8) | No |
| Haplotypes of HLA-G alleles and rs371194629 | – | 1 | Nardi 2012b (36) | No |
| HLA-G diplotypes | ||||
| Diplotypes of rs1632947–rs1233334–rs371194629 | – | 1 | Nowak 2019 (8) | No |
| HLA-G allele distribution | ||||
| HLA-G*010101 | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*01010106 | Exon2-4 | 1 | Kuroshli 2015 (20) | No |
| HLA-G*010102 | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*010103 | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*010105 | Exon2-4 | 1 | Kuroshli 2015 (20) | No |
| HLA-G*010106 | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*010107 | Exon2-4 | 2 | Kuroshli 2015 (20) Nardi 2012a (19) |
Yes |
| HLA-G*010108 | Exon2-4 | 3 | Kuroshli 2015 Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*010109 | Exon2-4 | 1 | Sipak-Szmigie 2009 (42) | No |
| HLA-G*010112 | Exon2-4 | 1 | Nardi 2012a (19) | No |
| HLA-G*010114 | Exon2-4 | 1 | Nardi 2012a (19) | No |
| HLA-G*010120 | Exon2-4 | 1 | Nardi 2012a (19) | No |
| HLA-G*0103 | Exon2-4 | 1 | Kuroshli 2015 (19) | No |
| HLA-G*010301 | Exon2-4 | 1 | Nardi 2012a (19) | No |
| HLA-G*010401 | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*010403 | Exon2-4 | 2 | Kuroshli 2015 (20) Nardi 2012a (19) |
Yes |
| HLA-G*010404 | Exon2-4 | 2 | Kuroshli 2015 Nardi 2012a (19) |
Yes |
| HLA-G*0106 | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G*01:05N | Exon2-4 | 3 | Kuroshli 2015 (20) Nardi 2012a (19) Sipak-Szmigie 2009 (42) |
Yes |
| HLA-G +482T/C | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +485G/T | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +494A/C | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +505/+506 -/CC | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +506 -/C | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +615 A/- | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +636C/T | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +644G/T | Intron2 | 1 | Kuroshli 2015 (20) | No |
| HLA-G +685G/A | Intron2 | 1 | Kuroshli 2015 (20) | No |
3’UTR, 3 prime untranslated region; 5’URR, 5 prime upstream regulatory region.
Table 2.
Characteristics of studies included in the meta-analysis that reported the associations between HLA-G polymorphisms or sHLA-G expressions with RIF.
| Study(year) | Period | Country | Race | N Case/Ctr | Age of Case/Ctr | Sample | Item analyzed | HWE(P-value) | Definition of RIF | Definition of control |
|---|---|---|---|---|---|---|---|---|---|---|
| Hviid (2004) (39) |
NR | Denmark | Caucasian | 14/108 | NR | Blood | HLA-G 14-bp ins/del | 0.042 | Patients with ≥3 or more unsuccessful IVF treatments | Women with twin pregnancies after the first IVF treatment and fertile women with ≥2 uncomplicated pregnancies and births. |
| Sipak-Szmigiel (2007) (70) |
NR | Poland | Caucasian | 20/20 | NR | Blood | HLA-G concentration | – | Patients with ≥3 or more unsuccessful IVF treatments. | Fertile women with ≥2 uncomplicated pregnancies and births. |
| Sipak Szmigiel (2009) (42) |
NR | Poland | Caucasian | 50/71 | NR | Blood | HLA-G 14-bp ins/del HLA-G -725 C>G(T) HLA-G allele distribution |
0.449 0.520 - |
Patients with ≥3 or more unsuccessful IVF treatments. | Fertile women with ≥2 uncomplicated pregnancies and births. |
| Nardi (2012) (19) | NR | Brazil | Mixed | 41/60 | 31.6 ± 5.6/44.2 ± 11.7 | Blood | HLA-G concentration HLA-G allele distribution |
– | Patients with ≥2 or more unsuccessful IVF treatments. | Fertile women with ≥2 normal pregnancies. |
| Enghelabifar (2014) (37) |
NR | Iran | Caucasian | 40/40 | NR | Blood | HLA-G 14-bp ins/del | 0.752 | Women with at least two failed IVF-embryo transfers, using at least 6 appropriate cleaved embryos. | Women with pregnancy following IVF |
| Lashley (2014) (50) |
2005-2009 | Netherlands | Caucasian | 24/96 | 35 (26–40)/33 (25–38) | Blood | HLA-G 14-bp ins/del | 0.667 | Patients with ≥3 or more unsuccessful IVF treatments with high quality embryos transferred. | Women with live birth after one IVF/ICSI treatment and fertile women with ≥1 uncomplicated pregnancies and births. |
| Kuroshli (2015) (20) | NR | Iran | Caucasian | 100/50 | 32.3/NR | Blood | HLA-G alleles distribution | – | Patients with ≥2 unsuccessful IVF treatments | Healthy unrelated Iranian individuals |
| Nardi (2016) (35) |
NR | Brazil | Mixed | 49/34 | 36.04 ± 0.5/36.1 ± 1 | Blood | HLA-G 14-bp ins/del HLA-G concentration |
0.471 - |
Patients with ≥2 or more unsuccessful IVF treatments. | Fertile women with ≥2 pregnancies. |
| Nowak (2020) (8) |
NR | Poland | Caucasian | 239/437 | 34.63 ± 4/31.97 ± 3.63 | Blood | HLA-G 14-bp ins/del HLA-G -725 C>G(T) HLA-G concentration |
0.843 0.616 - |
Patients with ≥3 or more unsuccessful IVF treatments with good quality embryos transferred. | Women with live birth after one IVF/ICSI treatment and fertile women with ≥1 uncomplicated pregnancies and births. |
NR, not reported; HWE, Hardy-Weinberg equilibrium; RIF, recurrent implantation failure; PCR, polymerase chain reaction; hCG, human chorionic gonadotrophin; ctr, control.
Ten studies are included in the systematic review to evaluate the associations between HLA-G polymorphism with sHLA-G expressions in patients attending IVF treatment. Characteristics of the ten studies are shown in Table 3 . Four of the ten studies that reported the same HLA-G polymorphism with sHLA-G expression were subjected to meta-analysis. In summary, the studies were performed in Denmark, Poland, the Netherlands, and Brazil. Most of the study’s ethnicity is Caucasian. Seven studies reported the association of parental HLA-G polymorphism with peripheral blood plasma or serum sHLA-G expressions. Three studies reported the association of paternal HLA-G polymorphisms with seminal plasma sHLA-G expressions (77–79). One study detected the sHLA-G expression in the endometrium by IH (41). The other studies detected the sHLA-G expression in body fluid by ELISA with the detection limit range from 0.6U/ml to 3.9U/ml. Most studies detect sHLA-G isoforms of HLA-G1 and HLA-G5 by ELISA, except for one study that detected the sHLA-G isoforms of HLA-G5 and HLA-G6 (41). The quality assessment of all 11 studies for meta-analysis is listed in Table 4 . Ten of the studies were assessed as high-quality, and one study assessed 6 points was defined as low-quality.
Table 3.
Summary of the studies that reported the association of HLA-G polymorphisms with sHLA-G expression in patients attending IVF treatment.
| Study | Country | Ethnicity | Study population | Number of patients tested | Sample tested | Timing of sample collection | Type of assay | sHLA-G isoform | Detection limit | Result | Included in meta-analysis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Papúchová 2022 (41) | Denmark | Caucasian | Women attending IVF treatment | 121 | Endometrium | the day equivalent to embryo transfer | IH | HLA-G5 and HLA-G6 | NR | No significant differences were observed in sHLA-G expression between the different HLA-G genotypes | No |
| Piekarska 2021 (76) | Poland | Caucasian | Men attending IVF treatment | 183 | Seminal plasma | 2-7 days of sexual abstinence | ELISA | HLA-G1 and HLA-G5 | 0.6 IU/ml | Certain HLA-G haplotypes and diplotypes of rs1632947-rs1233334-rs371194629 are associated with seminal sHLA-G expression | No |
| Nilsson 2020 (79) | Denmark | Caucasian | Couples attending IVF treatment | 127 couples and 4 single women | Periphery blood plasma and seminal plasma | NR | ELISA | HLA-G1 and HLA-G5 | 0.6 IU/ml | HLA-G 14bp genotype or HLA-G 3’UTR diplotype is significantly associated with seminal sHLA-G levels, while is not significantly associated with female blood plasma sHLA-G levels | Yes |
| Nowak 2020 (8) | Poland | Caucasian | Women attending IVF | 234 before IVF-ET and 185 after IVF-ET | Periphery blood plasma | Before and after IVF-ET | ELISA | HLA-G1 and HLA-G5 | 0.6 IU/ml | Certain HLA-G haplotypes of rs1632947-rs1233334-rs371194629 are associated with sHLA-G expression in blood plasma before IVF-ET; whereas such observations were not detected after ET | No |
| Craenmehr 2019 (77) | Netherlands | Caucasian | Men attending IVF treatment | 156 | Seminal plasma | NR | ELISA | HLA-G1 and HLA-G5 | 0.6 IU/ml | Seminal plasma sHLA-G levels are associated with HLA-G SNPs (HLA-G 14 bp ins/del, +3003 C/T, +3010 C/G, +3142 C/G, +3187 A/G, +3196 C/G, +3496 A/G and + 3509 G/T) and haplotypes at the 3’UTR | Yes |
| Nardi 2016 (35) | Brazil | Mixed | RIF and control patients | 58 | Periphery blood serum | NR | ELISA | HLA-G1 and HLA-G5 | 0.25ng/ml | HLA-G 14bp del variant is associated with higher sHLA-G levels | Yes |
| Dahl 2014 (78) | Denmark | Caucasian | Couples attending IVF treatment | 43 men for seminal test; 53 women and 47 men for blood test | Seminal plasma and periphery blood plasma | NR | ELISA | HLA-G1 and HLA-G5 | 0.6 IU/ml | Male HLA-G 14bp del/del genotype is associated with higher sHLA-G levels in seminal plasma | Yes |
| Nardi 2012 (19) | Brazil | Mixed | RIF and control patients | 41 RIF women and 60 controls | Periphery blood serum | NR | ELISA | HLA-G1 and HLA-G5 | 3.9U/ml | HLA-G allele distribution in exon 2,3,4 is not associated with serum sHLA-G levels | No |
| Sipak-Szmigiel 2007 (70) | Poland | Caucasian | Women attending IVF treatment | 80 | Periphery blood plasma | NR | ELISA | NR | 2U/ml | Allele HLA-G10101, 10102, and 10108 was related to higher plasma sHLA-G levels than other alleles | No |
| Hviid 2004 (75) | Denmark | Caucasian | Couples attending IVF treatment | 43 women and 42 male partners | Periphery blood serum | Before pregnancy | ELISA | HLA-G1 and HLA-G5 | 1ng/ml | Detectable sHLA-G in serum is restricted to HLA-G genotypes with the 14bp del | No |
IH, Immunohistochemical staining; ELISA, enzyme-linked immunosorbent assay; IVF, in vitro fertilization; ET, embryo transfer; 3’UTR, 3’ untranslated region; 5’ URR, 5’ upstream regulatory region; NR, not reported.
Table 4.
Newcastle-Ottawa Scale assessment of studies included in the meta-analysis.
| Study (year) | Selection | Comparability | Exposure | Total Score | Quality | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case definition adequate | Representativeness of the cases | Selection of controls | Definition of controls | Adjustment for age | Adjustment for other factors | Ascertainment of exposure | Uniform method of ascertainment | Non-response rate | |||
| Hviid (2004) | * | * | * | * | – | – | * | * | * | 7 | High |
| Szmigiel (2007) | * | * | * | * | – | – | * | * | * | 7 | High |
| Szmigiel (2009) | * | * | * | * | – | * | * | * | * | 8 | High |
| Nardi (2012) | * | * | * | * | – | – | * | * | – | 6 | Low |
| Enghelabifar (2014) | * | * | * | * | – | – | * | * | * | 7 | High |
| Lashley (2014) | * | * | * | * | – | * | * | * | * | 8 | High |
| Dahl (2014) | * | * | * | * | – | – | * | * | * | 7 | High |
| Nardi (2016) | * | * | * | * | – | – | * | * | * | 7 | High |
| Craenmehr (2019) | * | * | * | * | – | – | * | * | * | 7 | High |
| Nowak (2020) | * | * | * | * | – | – | * | * | * | 7 | High |
| Nilsson (2020) | * | * | * | * | – | – | * | * | * | 7 | High |
*denotes one score.
Meta-analysis of association between HLA-G 14bp ins/del polymorphism and RIF
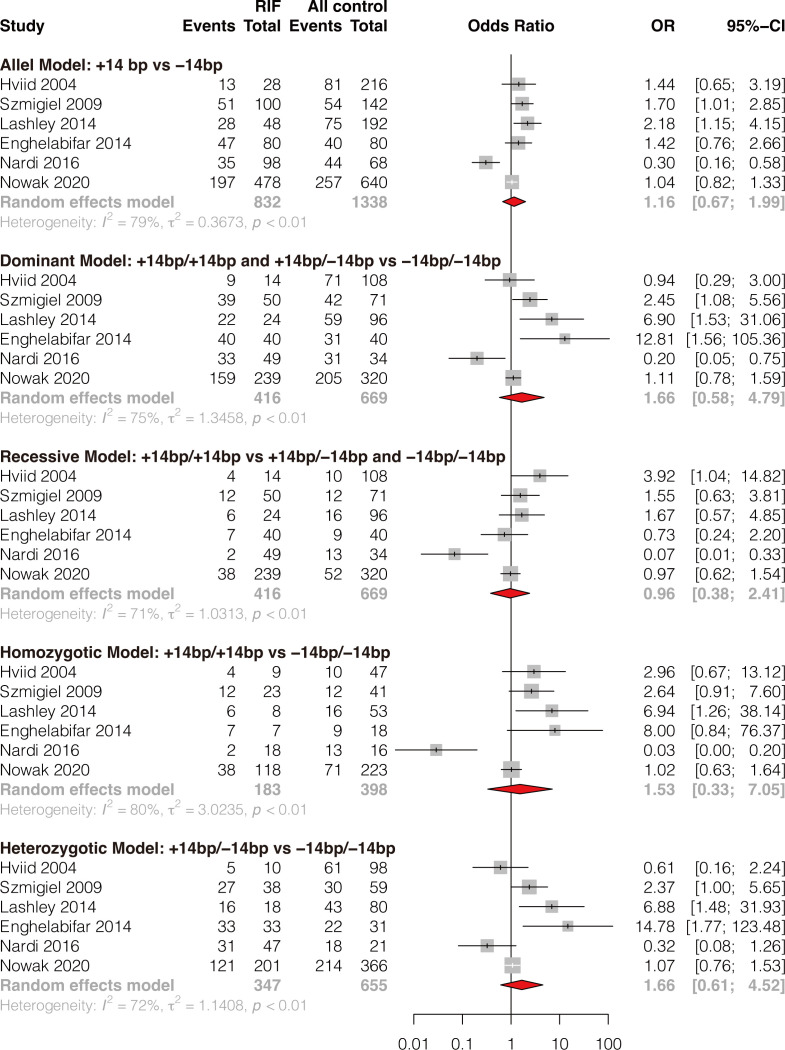
A total of six studies comprising 416 RIF cases and 669 controls were included (8, 35, 37, 39, 42, 50). Results showed that HLA-G 14bp ins/del polymorphism was not significantly associated with RIF in the general population under all genetic models (allele model: OR 1.16, 95%CI 0.67-1.99, p-value=0.599; dominant model: OR 1.66, 95%CI 0.58-4.79, p-value=0.344; recessive model: OR 0.96, 95%CI 0.38-2.41, p-value=0.925; homozygotic model: OR 1.53, 95%CI 0.33-7.05, p-value=0.584; heterozygotic model: OR 1.66, 95%CI 0.61-4.52, p-value=0.318) ( Figure 2 and Supplementary Table 1 ). Begg’s and Eggers’ tests showed no significant publication bias ( Supplementary Table 2 ). Considering the effect of different genetic backgrounds on the results, we performed a subgroup meta-analysis based on different ethnicities. The subgroup analysis showed that HLA-G 14bp ins/del polymorphism was significantly associated with RIF in the Caucasian population under the allele model (OR 1.41, 95%CI 1.04-1.90, p-value=0.028) and the homozygotic model (OR 2.48, 95%CI 1.09-5.62, p-value=0.030) ( Figure 3 and Supplementary Table 3 ). Sensitivity analysis was performed by excluding one study each time; results of the sensitivity analysis showed that the result changed from non-significant to significant under the allele model (OR 1.41, 95%CI 1.04-1.90, p-value=0.028) and the homozygotic model (OR 2.48, 95%CI 1.09-5.62, p-value=0.030) after excluding Nardi’s study conducted in 2016 ( Figure 3 ). It is worth noting that the study population in Nardi’s study conducted in 2016 was mixed ethnicity, while the population included in the other studies is Caucasian ethnicity. The subgroup analysis and sensitivity analysis consistently indicate that ethnic background plays an essential role in genetic susceptibility to RIF.
Figure 2.
Forest plot showing the association of HLA-G 14bp ins/del polymorphism with RIF under 5 genetic models.
Figure 3.
Forest plot showing subgroup analyses based on ethnicity and sensitivity analysis by omitting one study each time.
Meta-analysis of association between HLA-G -725 C>G(T) polymorphism and RIF
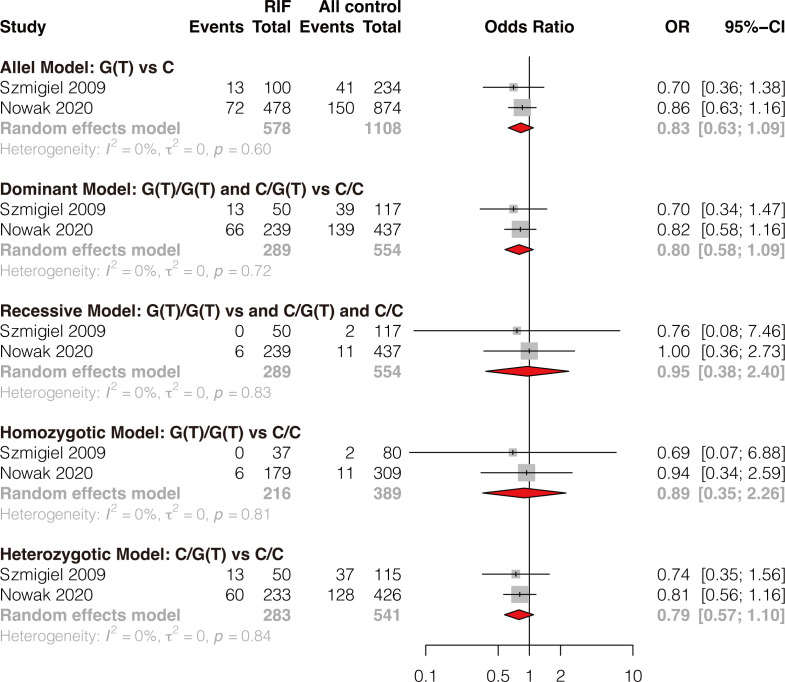
Two studies reported the association of HLA-G -725C>G(T) polymorphism and RIF (8, 42). The results showed that single HLA-G -725C>G(T) polymorphism was not significantly associated with RIF under all genetic models (allele model: OR 0.83, 95%CI 0.63-1.09, p-value=0.183; dominant model: OR 0.80, 95%CI 0.58-1.09, p-value=0.155; recessive model: OR 0.95, 95%CI 0.38-2.40, p-value=0.920; homozygotic model: OR 0.89, 95%CI 0.35-2.26, p-value=0.813; heterozygotic model: OR 0.79, 95%CI 0.57-1.10, p-value=0.163) ( Figure 4 and Supplementary Table 4 ). Publication bias and sensitivity analysis were not performed due to the small number of included studies. Therefore, the results should be explained with caution because only two studies evaluated HLA-G -725 C>G(T) polymorphism and RIF, and the sample size is small.
Figure 4.
Forest plot showing the association of HLA-G -725C>G/T polymorphism with RIF under 5 genetic models.
Meta-analysis of association between HLA-G alleles variants and RIF
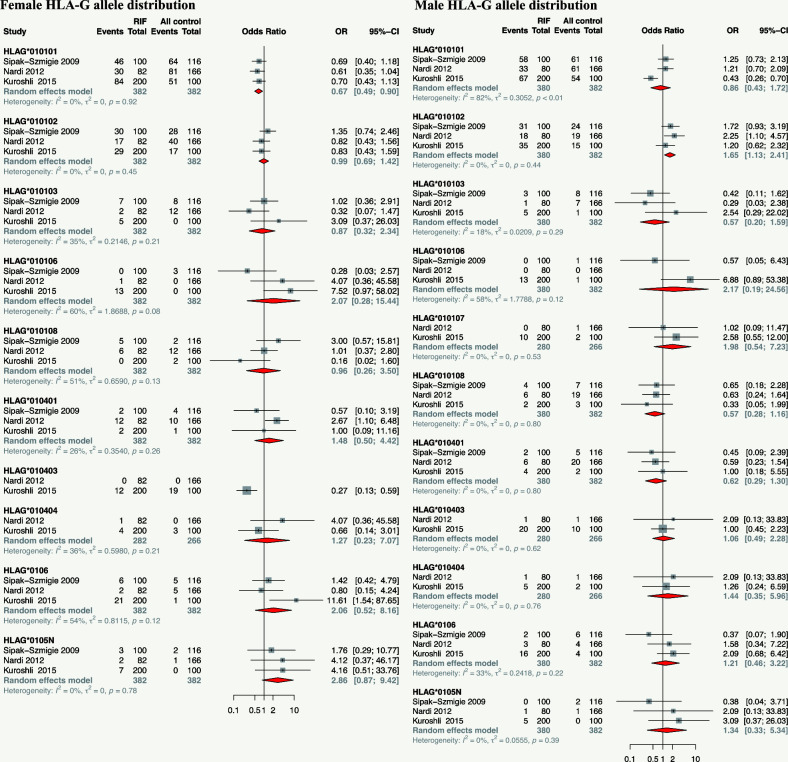
Three studies reported the association of HLA-G alleles distribution at exon 2-4 (HLA-G*010101, HLA-G*010102, HLA-G*010103, HLA-G*010106, HLA-G*010107, HLA-G*010108, HLA-G*010401, HLA-G*010403, HLA-G*010404, HLA-G*0106, and HLA-G*0105N) with RIF were included for meta-analysis. Results showed that the maternal HLA-G*010101 allele is associated with a lower risk of RIF (OR 0.67, 95%CI 0.49-0.90, p-value=0.008), maternal HLA-G*0105N tends to increase the risk of RIF though without statistical significance (OR 2.86, 95%CI 0.87-9.42, p-value=0.083). The paternal HLA-G*010102 allele is associated with a higher risk of RIF in their female partner (OR 1.65, 95%CI 1.13-2.41, p-value=0.010) ( Figure 5 and Supplementary Table 5 ).
Figure 5.
Forest plot showing the association of parental HLA-G allele distribution with RIF.
Meta-analysis of association between maternal circulating sHLA-G concentration and RIF
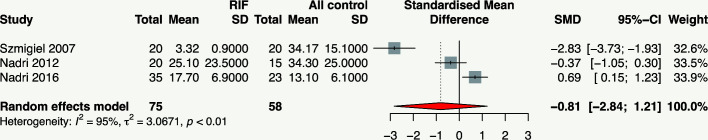
Three studies investigated the association of maternal circulating sHLA-G concentration with RIF (19, 35, 70). Meta-analysis showed that maternal circulating sHLA-G concentration was not significantly associated with RIF (SMD -0.81, 95%CI -2.84~-1.21, p-value=0.432) with extremely significant heterogeneity (I2= 95.4%, p-value<0.01) ( Figure 6 and Supplementary Table 6 ). The results should be explained cautiously due to the considerable heterogeneity and the small number of included studies.
Figure 6.
Forest plot showing the association of circulating sHLA-G concentration with RIF.
Meta-analysis of association between HLA-G polymorphism and sHLA-G expression in patients attending IVF treatment
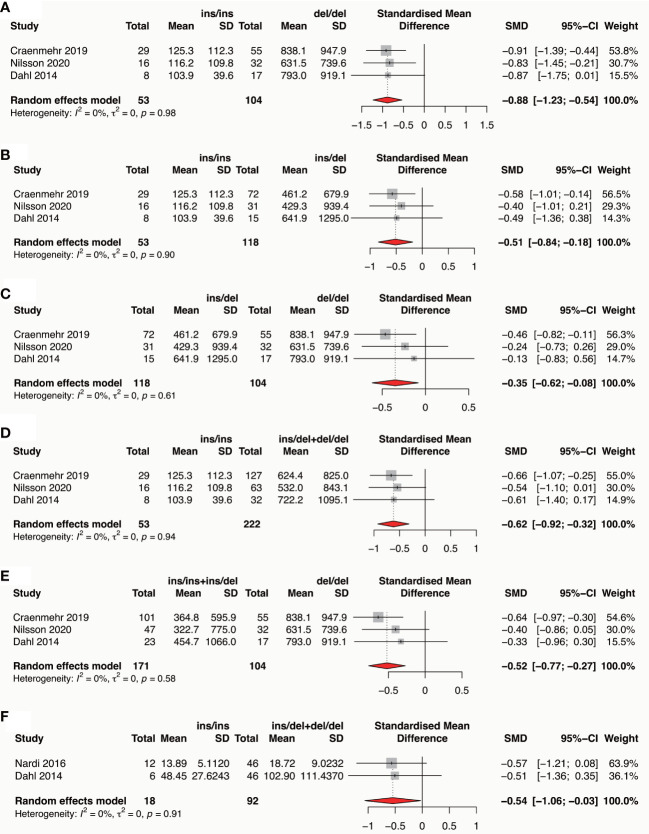
Three studies reported the association of paternal HLA-G polymorphisms with seminal plasma sHLA-G expression (77–79), and two studies reported the association of maternal HLA-G polymorphisms with blood plasma sHLA-G expression (35, 78) were included for meta-analysis, respectively. Results showed that the paternal HLA-G 14bp ins variant is associated with a lower seminal sHLA-G expression in all comparisons (ins/ins vs del/del: SMD -0.88, 95%CI -1.23~-0.54, p-value<0.0001; ins/ins vs ins/del: SMD -0.51, 95%CI -0.84~-0.18, p-value=0.002; ins/del vs del/del: SMD -0.35, 95%CI -0.62~-0.08, p-value=0.010; ins/ins vs ins/del+del/del: SMD -0.62, 95%CI -0.92~-0.32, p-value<0.0001; ins/ins+ins/del vs del/del: SMD -0.52, 95%CI -0.77~-0.27, p-value<0.0001) ( Figure 7A-E and Supplementary Table 7 ). Maternal HLA-G 14bp ins/ins genotype is associated with a lower blood plasma sHLA-G level than ins/del and del/del genotypes (SMD -0.54, 95%CI -1.06~-0.03, p-value=0.038) ( Figure 7F and Supplementary Table 8 ). Due to incomplete data, we cannot conduct a meta-analysis based on other maternal 14bp genotype comparisons.
Figure 7.
Forest plot showing the association of parental HLA-G 14bp ins/del polymorphism with sHLA-G expression in seminal plasma or in blood plasma. (A) Meta-analysis of sHLA-G expression in male seminal plasma in comparison of ins/ins genotype versus del/del genotype. (B) Meta-analysis of sHLA-G expression in male seminal plasma in comparison of ins/ins genotype versus ins/del genotype. (C) Meta-analysis of sHLA-G expression in male seminal plasma in comparison of ins/del genotype versus del/del genotype. (D) Meta-analysis of sHLA-G expression in male seminal plasma in comparison of ins/ins genotype versus ins/del+del/del genotype. (E) Meta-analysis of sHLA-G expression in male seminal plasma in comparison of ins/ins+ins/del genotype versus de/del genotype. (F) Meta-analysis of sHLA-G expression in female blood plasma in comparison of ins/ins genotype versus ins/del+del/del genotype.
Discussion
Summary of this study
RIF is a multi-factorial complication following embryo transfer. In addition to the embryo factor, immune dysfunction is one of the leading factors contributing to implantation failure, which has been a focus of interest. HLA-G plays a central role in inducing immune tolerance through interactions between HLA-G and its receptors, including CD8, ILT-2/LILRB1/CD85j, ILT-4/LILRB2/CD85d, KIR2DL4, and NKG2A/CD94 (58, 80). Besides inducing immune tolerance, HLA-G controls trophoblast invasion, regulates vascular remodeling, and facilitates fetal growth, allowing for successful embryo implantation and pregnancy maintenance (81–85). HLA-G is predominantly produced by the extravillous trophoblasts (EVTs). The embryo-secreted sHLA-G increases during embryo development, from a relative 35% in the cleavage stage to 100% in the morula or blastocyst stage (86). The sHLA-G level in the embryo culture medium is a promising predictor of pregnancy outcome (28–32). Besides, immune cells in the endometrium and maternal peripheral antigen-presenting cells (APC) can be a source of sHLA-G (87). Decidualization by progesterone and cAMP can increase HLA-G expression (88). Immunohistochemistry (IH) experiments validated that sHLA-G is located in the endometrial stroma and glandular epithelium of pre-implantation and peri-implantation endometrium. sHLA-G expression is correlated with CD56+ uNK cell abundance and associated with pregnancy outcomes (41, 63). Several studies reported that abnormal sHLA-G expression is associated with pregnancy complications such as preeclampsia, recurrent miscarriage (RM), and recurrent implantation failure (RIF), and may be further linked to HLA-G polymorphisms (89, 90). Some meta-analyses have confirmed the association between HLA-G polymorphisms with susceptibility to preeclampsia (91, 92) and recurrent miscarriage (RM) (93, 94). The current systematic review and meta-analysis focused on the implication of the genetic variants responsible for altered HLA-G expression in relation to RIF. Our study indicates that specific HLA-G alleles or HLA-G polymorphisms are associated with sHLA-G expression in couples attending IVF treatment. Parental HLA-G*010101 and HLA-G*010102 alleles distribution is associated with RIF risk. However, single maternal HLA-G 14bp ins/del polymorphism, HLA-G -725 C>G/T polymorphism, or circulating sHLA-G concentration is not significantly associated with RIF in the general population. Whereas the HLA-G 14bp insertion variant is associated with RIF under a homozygotic genetic model in the Caucasian population.
HLA-G 14bp ins/del polymorphism with reproductive disorders
HLA-G 14bp ins/del is the most commonly studied polymorphism of HLA-G. Although maternal HLA-G 14bp ins/del polymorphism was not significantly associated with RIF in the general population, the sensitivity analysis and the subgroup analysis consistently suggested that HLA-G 14bp ins/del polymorphism was significantly associated with RIF in the Caucasian population under the allele and homozygotic models. The subgroup analyses should be explained cautiously due to the small number of studies in the subgroup. Consistent with our study, a meta-analysis in 2017 reported similar results that the HLA-G 14bp ins/del polymorphism is related to RIF in Caucasian patients (9). However, compared to Fan’s meta-analysis in 2017, which only assessed the association between HLA-G 14bp ins/del polymorphism with RIF, our meta-analysis adds the associations between other HLA-G polymorphisms (including HLA-G -725C>G/T, multiple allele distributions at HLA-G exon2~4, specific haplotypes and diplotypes of HLA-G) with RIF. It adds the association between HLA-G polymorphism with sHLA-G expression as well as the association between sHLA-G expression with RIF susceptibility. Besides, the sample size in this systematic review and meta-analysis is the largest so far. Further studies are needed to confirm whether an association exists between HLA-G 14bp ins/del polymorphism with RIF in patients of other ethnicities. In addition to RIF, maternal HLA-G 14bp ins/del polymorphism is also reported to be associated with preeclampsia (95), gestational diabetes mellitus (96), and recurrent miscarriage (97–99). Almeida et al. combined implantation failure, preeclampsia, recurrent miscarriage, and spontaneous miscarriage as reproductive disorders and performed a meta-analysis; they found that the HLA-G 14bp ins variant is associated with reproductive disorders (92). Moreover, the HLA-G 14bp ins/ins genotype is associated with insulin resistance (100), birth weight, and placental weight (101). Nevertheless, other studies have observed contrary results (102–104). The contradictory findings may be explained by differences in ethnic background, sample size, and genotyping methodology.
HLA-G -725 polymorphism with reproductive disorders
HLA-G -725C>G/T polymorphism at the promoter region is reported to change the methylation profile of CpG dinucleotide resulting in a modification of HLA-G expression (17). HLA-G -725C>G/T polymorphism is reported to be associated with male fertility (76), endometriosis progression (105), and miscarriage (17). Our study found no significant association of HLA-G -725C>G/T polymorphism with RIF under all genetic models. Sipak et al. reported that HLA-G 725 C>G/T polymorphism is not associated with pregnancy complications, including antiphospholipid syndrome, preeclampsia, intrauterine growth restriction, and recurrent spontaneous abortion (106). In summary, an association of HLA-G -725C>G/T polymorphism with pregnancy-related complications or pregnancy outcomes could not be confirmed.
Other HLA-G polymorphisms or HLA-G haplotypes or diplotypes with reproductive disorders
Other HLA-G polymorphisms include HLA-G -964G>A (rs1632947), HLA-G haplotype of rs1632947–rs1233334–rs371194629, diplotypes of rs1632947–rs1233334–rs371194629, and HLA-G alleles distribution at exon2-4 and intron2 are potentially associated with RIF or reproductive outcomes following IVF-ET. HLA-G polymorphisms such as -716 G/T rs2249863 (107) and 3142C/G rs1063320 (108) have been reported to play a role in spontaneous abortion but have not been reported in RIF. Our results showed that the maternal HLA-G*010101 allele is associated with a lower RIF risk than other HLA-G alleles, whereas the paternal HLA-G*010102 allele is associated with a higher RIF risk. HLA-G*010101 and HLA-G*010102 are the most prevalent alleles compared to other HLA-G alleles and are associated with sHLA-G expressions. Whereas Warner et al. reported that the HLA-G*01011 allele has a statistically significant association with an enhanced chance of reproductive success following IVF-ET in Caucasian women (109). The HLA-G*0105N null-allele contains a deletion in exon 3, which leads to a frameshift, and no functional full-length HLA-G1 and -G5 protein can be expressed (110). Our meta-analysis observed a tendency that the HLA-G*0105N allele increased the RIF risk, while without statistical significance, possible because the sample size is too small to reach significance. Nonetheless, HLA-G1 and HLA-G5 are reported to be not essential for fetal survival, indicating that other HLA-G isoforms or other HLA molecules may compensate for the lack of HLA-G1 and HLA-G5 in immune modulation.
Soluble HLA-G expression with reproductive disorders
Soluble HLA-G can be secreted into circulating blood, amniotic fluid, umbilical cord blood, and semen; it can be detected in non-pregnant/pregnant women, men, embryo/fetus, and the maternal-fetal interface. It is well-studied that the embryo-secreted sHLA-G level in the culture medium is a promising predictor for embryo implantation (28–31). However, the role of maternal sHLA-G expression on embryo implantation was less studied. The sHLA-G expression is different before pregnancy or during pregnancy. The blood sHLA-G level is higher in pregnant women than in non-pregnant women, and the sHLA-G level is higher in the first trimester of pregnancy than in the second and third trimesters (60, 111–113). It is reported that lower serum sHLA-G levels at the pre-ovulatory stage increase the risk of early miscarriage (61). A reduced frequency of HLA-G expressing CD4+T and CD8+ T cells in the peripheral blood is associated with RM and RIF (69). However, results concerning whether the maternal sHLA-G expression is associated with RIF are conflicting. This meta-analysis showed that single maternal sHLA-G expression was not significantly associated with RIF, while the heterogeneity is extremely significant (I2= 98%, p-value<0.01). The considerable heterogeneity may be caused by: (1) Each clinic applied a different ELISA system for detecting sHLA-G with different detection sensitivity or limits (114); (2) The timing of measurement during preimplantation development could be critical. Because sHLA-G expression before or after ET is different (8), it dynamically changes over the pregnancy weeks. Therefore, the timing of sHLA-G measurement may significantly affect the result; (3) Maternal circulating sHLA-G concentration might not be correlated with sHLA-G levels at the fetal-maternal interface. Whether maternal circulating sHLA-G levels are associated with fetal circulating sHLA-G levels is conflicting (111, 115); (4) Other pathological conditions or diseases also affect sHLA-G levels. Crohn’s disease, Behçet’s disease, multiple sclerosis, or organ transplant are associated with sHLA-G expression (116–119); (5) IVF factors such as maternal age, cycle type (frozen cycles or fresh cycles), or exogenous hormone supplementation may change sHLA-G levels (8). It is reported that HLA-G expression was positively associated with progesterone supplementation but negatively with estradiol (120) and maternal age; (6) The detection of sHLA-G level in most cases is limited to sHLA-G1/HLA-G5 isotype measured by ELISA due to restriction in antibodies. Whereas the expression pattern of the other isoforms is rarely analyzed. Different HLA-G isoforms may interact differently with the receptors; for example, ILT-2 only binds β2m-associated HLA-G1/G5 isotypes, while ILT-4 preferably binds β2m-free isoforms. How these isoforms of the HLA-G protein differ in function is poorly understood. A recent study detected endometrium sHLA-G5 and sHLA-G6 isotypes by IH; they found that endometrial sHLA-G5 and sHLA-G6 levels are higher in RIF patients compared to controls (41). Besides, sHLA-G levels are reported to be associated with oocyte competence (121, 122), endometriosis progression (123), pregnancy-related conditions such as SGA neonates (124), GDM (125), advanced labor (126), preterm premature rupture of membranes (127), intrauterine growth retardation (IUGR) (112), and preeclampsia (113, 128–130). Moreover, maternal circulating sHLA-G levels in the second trimester were significantly lower in pregnant women with 18-trisomy fetuses (T18) and significantly higher in those with 21-trisomy fetuses (T21) compared to the normal controls (131), and it is inversely correlated with fetal microchimerism levels (132). Also, there are contrary results that the HLA-G expression is similar between samples of normal and abnormal karyotypes, and there is no association between the HLA-G polymorphisms and altered expression in reduced abortion and miscarriage groups (133). Schallmoser et al. reported no association of sHLA-G expression with female reproductive outcomes following IVF-ET (134). More evidence is needed to determine whether maternal circulating sHLA-G expression is a predictor for RIF and whether a combination of maternal-, paternal- and embryo-derived sHLA-G levels have more clinical significance than single detection.
Association of HLA-G polymorphism with sHLA-G expression
Multiple HLA-G polymorphisms are reported to be associated with sHLA-G expression. HLA-G 14bp ins/del polymorphism is the most commonly reported to be related to sHLA-G levels. Most studies indicate a significant association between HLA-G 14bp ins variant and reduced sHLA-G levels in maternal circulating blood or paternal semen (35, 75, 77, 78, 96, 104, 135–139). HLA-G 14bp del/del genotype is also associated with higher HLA-G on the trophoblast membrane (101). However, there are different results; two studies report that the HLA-G 14bp ins variant is associated with higher circulating sHLA-G levels (140, 141). The contradictory observations may have the following explanations: the correlation of sHLA-G expression and HLA-G polymorphism is affected by genetic backgrounds, pregnancy state, and diverse pathophysiologies or diseases. The expression pattern of HLA-G is different under various complications. Therefore, in this study, we review the associations of HLA-G polymorphism with sHLA-G expression in patients attending IVF treatment. Our meta-analyses observed that the parental HLA-G 14bp ins variant is associated with lower sHLA-G expression in female blood and male semen. The result indicates that parental HLA-G polymorphism may affect sHLA-G expressions in body fluid. However, further studies must confirm whether sHLA-G levels in parental body fluid affect pregnancy outcomes.
HLA-G -725C>G/T polymorphism is reported to be associated with sHLA-G levels in an in-vitro study, which found that JEG-3 cells with HLA-G -725G allele produces higher levels of sHLA-G compared to HLA-G -725C/T allele (142). Specific HLA-G allele distribution at exon2-4 is also reported to be associated with sHLA-G levels; for example, the HLA-G10101 allele is reported to be associated with higher sHLA-G levels in circulating blood (59, 70), while the HLA-G*01013 allele, HLA-G*0105N allele, or 1597ΔC null allele is associated with lower sHLA-G levels (143, 144). Three other SNPs in the 3’UTR are associated with HLA-G mRNA stability and sHLA-G levels: +3142 (rs1063320) substituting a C to a G, +3187 (rs9380142) substituting an A to a G, and +3196 (rs1610696) substituting a C to a G (15, 57, 145). Moreover, some polymorphisms in the 3’UTR can act as targets for miRNAs and control HLA-G mRNA stability and expression levels (146). Unfortunately, we cannot perform a meta-analysis based on those HLA-G polymorphisms in patients attending IVF treatment because of insufficient studies. Whether there is a combined effect of multiple HLA-G polymorphisms on sHLA-G expressions needs further investigation.
Interaction of HLA-G with other HLA Ia and Ib genes in relation to reproductive disorders
We speculate that single maternal HLA-G polymorphism or circulating sHLA-G concentration is not a single major cause of implantation failure. Whereas combined genetic effect would have been more potent than a single polymorphism analysis (147). The association of HLA-G with RIF is more likely to depend on the combined HLA-G genetic effect rather than single polymorphisms in the 3’-UTR, 5’URR, or coding regions. The 14bp ins, in combination with the +3187A/A and +3142G/G SNP, plays a significant role in HLA-G mRNA regulation in human endometrial stromal cells (148). One single maternal genetic polymorphism or circulating sHLA-G level could not become an adequate independent cause of RIF because: (1) The mechanisms regulating maternal-fetal tolerance are complex. There are many immune mechanisms and other compensatory processes of maternal-fetal tolerance; (2) More than two HLA-G genotypic effects (maternal, paternal, and embryo) may participate in immune regulation during embryo implantation. Therefore, a single maternal or paternal HLA-G genotype cannot wholly reflect the immune state at the maternal-fetal interface; (3) The influence of clinical variables such as maternal age, gestational age, embryo factors, other diseases, or pathophysiological conditions were not controlled in the previous studies and can cause bias; (4) One gene has a variety of polymorphisms. The interaction of different polymorphisms of the same gene, the interaction of various genetic polymorphisms, and their combined effects on gene expression and molecular functions are not entirely understood. Except for HLA-G 14bp polymorphism, the other HLA-G genotypes/haplotypes/diplotypes at the 3’UTR, 5’URR, and exon regions, HLA-C polymorphism, HLA-F polymorphism, or HLA molecule receptor polymorphism as KIR is also associated with RIF (41, 149–151). Certain HLA-G variations are in linkage disequilibrium with three HLA-F locus SNPs that influence reproduction (152). HLA-G expression and function are under the control of miRNA via the miRNA binding site at HLA-G genes (146). Other cytokines such as IF-10, IFN-γ, LIF, PIF, and Galectin-1 can induce the production of HLA-G (88, 153–155). Rizzo et al. found that endometrium and uterine flushing fluid with high LIF, HB-EGF, Glycodelin-A, MCP1, IP10, HLA-G, and HLA-E, but low MUC-1 expression presented a higher permittivity to embryo implantation by an endometrial 3D in vitro model (156). In summary, multiple genetic factors, non-coding molecules, and cytokines together generate a pro-tolerance milieu network to regulate embryo implantation. Considering all of these factors, it is doubtful that one single molecule (sHLA-G) or a small genetic region could be uniquely associated with reproductive outcomes. At least a single HLA-G gene mutation, while it may contribute, is not a significant independent cause of RIF. The present challenge is to find the correct combination of genetic factors that define the susceptibility of RIF. More studies involving a careful selection of strictly-defined RIF patients and control patients, good-quality embryo transfers, and other essential molecules involved in embryo implantation (such as VEGFA, PAI-1, and MTHFR), studies controlling confounding factors are needed to determine whether the presence of HLA-G polymorphism have a crucial role in an immune imbalance during embryo implantation. And to what extent this could affect maternal-fetal tolerance and be linked to RIF.
Limitations
As far as we know, this is the first systematic review and meta-analysis to investigate the association of HLA-G polymorphisms and sHLA-G expression in relation to RIF. The following potential limitations should be considered: (1) The validity of the meta-analysis depends on the internal validity of the included studies. We could only use the information provided at the study level. We could not analyze the unmeasured or unreported factors of the patient, such as maternal age, underlying causes of infertility, varying infertility treatment, or exogenous hormone supplementation; (2) The generalizability of our result is another limitation. Patients included in this study were mainly Caucasian. ‘RIF’ is a broad term, including heterogeneous causes and diagnoses; (3) HLA-G expression is multifactorial and can be influenced by many other factors such as splice variants, DNA methylation, miRNA-mediated post-transcriptional regulation, etc., which were not explored in this study.
Future expectations and conclusions
The immuno-genetics of infertility is complex and might depend on different genes involved in embryo implantation. A better understanding of HLA-G allele structure and how the genetic diversity at regulatory sites shared by different alleles and haplotypes could affect its expression might shed further light on the comprehension of immuno-genetics mechanisms acting at the feto-maternal interface. The primary source of sHLA-G at the maternal-fetal interface is the embryo-derived trophoblasts. In contrast, most studies assessing the role of HLA-G in pregnant diseases have considered only the maternal genotype and ignored the contribution of the fetus and paternal partner. Ideally, mother-father-fetus genotypes should be tested. The significant association between HLA-G 14bp ins/del polymorphism, HLA-G -725 C>G/T polymorphism, or circulating sHLA-G concentration with RIF could not be confirmed in the general population. However, if and to what extent the use of the multiple polymorphisms combined with the sHLA-G test from parental body fluid and the culture medium might increase accuracy in the RIF prediction remains to be elucidated. In the future, testing multiple genetic variances or biomarkers at a time by a high-throughput method combined with machine learning may screen the best predictive model for RIF. Animal studies have shown that recombinant sHLA-G or synthetic HLA-G may have a therapeutic effect on arthritis disease or prolong the acceptance of skin grafts (157, 158). Whether recombinant sHLA-G or synthetic HLA-G can be used to treat reproductive disorders needs far more studies.
In conclusion, our study indicates that specific HLA-G alleles or HLA-G polymorphisms are associated with sHLA-G expression in couples attending IVF treatment. Several HLA-G polymorphisms may be associated with RIF, considering different ethnic backgrounds. A combined genetic effect should be considered in future studies to confirm the association of HLA-G polymorphisms and sHLA-G expressions in relation to RIF. Our findings will hopefully stimulate further research to identify whether multiple HLA-G genetics combined with the sHLA-G test of parental body fluid and the culture medium is of clinical relevance to implantation success.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author. The extracted data and statistical R scripts are uploaded in our Github (https://github.com/minizenghong/HLA-G_sHLA-G_RIF).
Author contributions
HZ designed the study, performed the statistical analyses, drafted and revised the manuscript, tables, and figures. LH and DH searched the studies, extracted the data, and performed the quality assessment. All authors approved the submission of the manuscript.
Funding
The study is supported by funds from Guangdong Basic and Applied Basic Research Foundation (grant number: 2021A1515110601). The study is supported by China Postdoctoral Science Foundation (grant number: 2022M711521).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.988370/full#supplementary-material
References
- 1. Arnaiz-Villena A, Juarez I, Suarez-Trujillo F, López-Nares A, Vaquero C, Palacio-Gruber J, et al. HLA-G: Function, polymorphisms and pathology. Int J Immunogenet (2021) 48(2):172–92. doi: 10.1111/iji.12513 [DOI] [PubMed] [Google Scholar]
- 2. Park GM, Lee S, Park B, Kim E, Shin J, Cho K, et al. Soluble HLA-G generated by proteolytic shedding inhibits NK-mediated cell lysis. Biochem Biophys Res Commun (2004) 313(3):606–11. doi: 10.1016/j.bbrc.2003.11.153 [DOI] [PubMed] [Google Scholar]
- 3. Rizzo R, Trentini A, Bortolotti D, Manfrinato MC, Rotola A, Castellazzi M, et al. Matrix metalloproteinase-2 (MMP-2) generates soluble HLA-G1 by cell surface proteolytic shedding. Mol Cell Biochem (2013) 381(1-2):243–55. doi: 10.1007/s11010-013-1708-5 [DOI] [PubMed] [Google Scholar]
- 4. Morales PJ, Pace JL, Platt JS, Langat DK, Hunt JS. Synthesis of beta(2)-microglobulin-free, disulphide-linked HLA-G5 homodimers in human placental villous cytotrophoblast cells. Immunology (2007) 122(2):179–88. doi: 10.1111/j.1365-2567.2007.02623.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, et al. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-e, f, and G in maternal-placental immune recognition. J Immunol (2003) 171(3):1376–84. doi: 10.4049/jimmunol.171.3.1376 [DOI] [PubMed] [Google Scholar]
- 6. Gonen-Gross T, Goldman-Wohl D, Huppertz B, Lankry D, Greenfield C, Natanson-Yaron S, et al. Inhibitory NK receptor recognition of HLA-G: regulation by contact residues and by cell specific expression at the fetal-maternal interface. PloS One (2010) 5(1):e8941. doi: 10.1371/journal.pone.0008941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Khunaizi NR, Tabbara KS, Farid EM. Is there a role for HLA-G in the induction of regulatory T cells during the maintenance of a healthy pregnancy? Am J Reprod Immunol (2020) 84(2):e13259. doi: 10.1111/aji.13259 [DOI] [PubMed] [Google Scholar]
- 8. Nowak I, Wilczyńska K, Radwan P, Wiśniewski A, Krasiński R, Radwan M, et al. Association of soluble HLA-G plasma level and HLA-G genetic polymorphism with pregnancy outcome of patients undergoing in vitro fertilization embryo transfer. Front Immunol (2019) 10:2982. doi: 10.3389/fimmu.2019.02982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan W, Huang Z, Li S, Xiao Z. The HLA-G 14-bp polymorphism and recurrent implantation failure: a meta-analysis. J Assist Reprod Genet (2017) 34(11):1559–65. doi: 10.1007/s10815-017-0994-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amodio G, Canti V, Maggio L, Rosa S, Castiglioni MT, Rovere-Querini P, et al. Association of genetic variants in the 3'UTR of HLA-G with recurrent pregnancy loss. Hum Immunol (2016) 77(10):886–91. doi: 10.1016/j.humimm.2016.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monti M, Lupoli R, Sosa Fernandez LM, Cirillo F, Di Minno MND. Association of human leukocyte antigen-G 14 bp polymorphism with recurrent pregnancy loss in European countries: a meta-analysis of literature studies. Fertil Steril (2019) 112(3):577–85.e573. doi: 10.1016/j.fertnstert.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 12. Hylenius S, Andersen AM, Melbye M, Hviid TV. Association between HLA-G genotype and risk of pre-eclampsia: A case-control study using family triads. Mol Hum Reprod (2004) 10(4):237–46. doi: 10.1093/molehr/gah035 [DOI] [PubMed] [Google Scholar]
- 13. Larsen MH, Hylenius S, Andersen AM, Hviid TV. The 3'-untranslated region of the HLA-G gene in relation to pre-eclampsia: revisited. Tissue Antigens (2010) 75(3):253–61. doi: 10.1111/j.1399-0039.2009.01435.x [DOI] [PubMed] [Google Scholar]
- 14. Steinborn A, Rebmann V, Scharf A, Sohn C, Grosse-Wilde H. Placental abruption is associated with decreased maternal plasma levels of soluble HLA-G. J Clin Immunol (2003) 23(4):307–14. doi: 10.1023/a:1024592901663 [DOI] [PubMed] [Google Scholar]
- 15. Hviid TV, Hylenius S, Rorbye C, Nielsen LG. HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics (2003) 55(2):63–79. doi: 10.1007/s00251-003-0547-z [DOI] [PubMed] [Google Scholar]
- 16. Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P. The 14 bp deletion-insertion polymorphism in the 3' UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol (2003) 64(11):1005–10. doi: 10.1016/j.humimm.2003.08.347 [DOI] [PubMed] [Google Scholar]
- 17. Ober C, Aldrich CL, Chervoneva I, Billstrand C, Rahimov F, Gray HL, et al. Variation in the HLA-G promoter region influences miscarriage rates. Am J Hum Genet (2003) 72(6):1425–35. doi: 10.1086/375501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Costa CH, Gelmini GF, Wowk PF, Mattar SB, Vargas RG, Roxo VM, et al. HLA-G regulatory haplotypes and implantation outcome in couples who underwent assisted reproduction treatment. Hum Immunol (2012) 73(9):891–7. doi: 10.1016/j.humimm.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 19. Nardi Fda S, Slowik R, Wowk PF, da Silva JS, Gelmini GF, Michelon TF, et al. Analysis of HLA-G polymorphisms in couples with implantation failure. Am J Reprod Immunol (2012) 68(6):507–14. doi: 10.1111/aji.12001 [DOI] [PubMed] [Google Scholar]
- 20. Kuroshli Z, Gourabi H, Bazrgar M, Sanati M, Zamani Esteki M. The relationship between HLA-G gene polymorphisms and repeated implantation failure in infertile couples undergoing assisted reproductive technique. Iran J Allergy Asthma Immunol (2015) 14(5):535–42. [PubMed] [Google Scholar]
- 21. Costa CH, Gelmini GF, Nardi FS, Roxo VM, Schuffner A, da Graça Bicalho M. HLA-G profile of infertile couples who underwent assisted reproduction treatment. Hum Immunol (2016) 77(12):1179–86. doi: 10.1016/j.humimm.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 22. Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, Gurgan T, et al. Recurrent implantation failure: definition and management. Reprod BioMed Online (2014) 28(1):14–38. doi: 10.1016/j.rbmo.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 23. Zhu Y, Wu T, Ye L, Li G, Zeng Y, Zhang Y. Prevalent genotypes of methylenetetrahydrofolate reductase (MTHFR) in recurrent miscarriage and recurrent implantation failure. J Assist Reprod Genet (2018) 35(8):1437–42. doi: 10.1007/s10815-018-1205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee JY, Ahn EH, Kim JO, Park HS, Ryu CS, Kim JH, et al. Associations between microRNA (miR-25, miR-32, miR-125, and miR-222) polymorphisms and recurrent implantation failure in Korean women. Hum Genomics (2019) 13(1):68. doi: 10.1186/s40246-019-0246-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeng H, He D, Hu L, Ma W, Quan S. PAI-1 4G/4G genotype is associated with recurrent implantation failure: A systematic review and meta-analysis. Reprod Sci (2021) 28(11):3051–60. doi: 10.1007/s43032-021-00623-1 [DOI] [PubMed] [Google Scholar]
- 26. Zeng H, Hu L, Xie H, Ma W, Quan S. Polymorphisms of vascular endothelial growth factor and recurrent implantation failure: a systematic review and meta-analysis. Arch Gynecol Obstet (2021) 304(2):297–307. doi: 10.1007/s00404-021-06072-0 [DOI] [PubMed] [Google Scholar]
- 27. Alecsandru D, Garcia-Velasco JA. Is there a role for human leukocyte antigen-G typing in infertility treatment? Fertil Steril (2020) 114(3):515–6. doi: 10.1016/j.fertnstert.2020.06.048 [DOI] [PubMed] [Google Scholar]
- 28. Vercammen MJ, Verloes A, Van de Velde H, Haentjens P. Accuracy of soluble human leukocyte antigen-G for predicting pregnancy among women undergoing infertility treatment: meta-analysis. Hum Reprod Update (2008) 14(3):209–18. doi: 10.1093/humupd/dmn007 [DOI] [PubMed] [Google Scholar]
- 29. Rebmann V, Switala M, Eue I, Grosse-Wilde H. Soluble HLA-G is an independent factor for the prediction of pregnancy outcome after ART: A German multi-centre study. Hum Reprod (2010) 25(7):1691–8. doi: 10.1093/humrep/deq120 [DOI] [PubMed] [Google Scholar]
- 30. Kotze D, Kruger TF, Lombard C, Padayachee T, Keskintepe L, Sher G. The effect of the biochemical marker soluble human leukocyte antigen G on pregnancy outcome in assisted reproductive technology–a multicenter study. Fertil Steril (2013) 100(5):1303–9. doi: 10.1016/j.fertnstert.2013.07.1977 [DOI] [PubMed] [Google Scholar]
- 31. Niu Z, Wang L, Pang RTK, Guo Y, Yeung WSB, Yao Y. A meta-analysis of the impact of human leukocyte antigen-G on the outcomes of IVF/ICSI. Reprod BioMed Online (2017) 34(6):611–8. doi: 10.1016/j.rbmo.2017.03.002 [DOI] [PubMed] [Google Scholar]
- 32. Vani V, Vasan SS, Adiga SK, Varsha SR, Sachdeva G, Kumar P, et al. Soluble human leukocyte antigen-G is a potential embryo viability biomarker and a positive predictor of live-births in humans. Am J Reprod Immunol (2021) 86(6):e13499. doi: 10.1111/aji.13499 [DOI] [PubMed] [Google Scholar]
- 33. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Bmj (2009) 339:b2535. doi: 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Costa CH, Gelmine GF, Nardi FS, Schuffner A, Da Graça Bicalho M. HLA-g∗01:03 and HLA-G∗01:04:01 higher frequencies in couples who conceived naturally compared to couples undergoing assisted reproduction treatment. Tissue Antigens (2015) 85(5):388–9. doi: 10.1002/phar.1497 [DOI] [Google Scholar]
- 35. Nardi Fda S, Slowik R, Michelon T, Manvailer LF, Wagner B, Neumann J, et al. High amounts of total and extracellular vesicle-derived soluble HLA-G are associated with HLA-G 14-bp deletion variant in women with embryo implantation failure. Am J Reprod Immunol (2016) 75(6):661–71. doi: 10.1111/aji.12507 [DOI] [PubMed] [Google Scholar]
- 36. Da Silva-Nardi F, Slowik R, Da Silva JS, Gelmini GF, Hernandes-Costa C, Wowk PF, et al. Haplotypes and genotypes of HLA-G alleles and 14-bp Del/Ins in the 3'UTR region in implantation failure couples. Tissue Antigens (2012) 80(1):82. doi: 10.1111/j.1399-0039.2012.01899.x [DOI] [Google Scholar]
- 37. Enghelabifar M, Allafan S, Khayatzadeh J, Shahrokh Abadi K, Hasanzadeh Nazarabadi M, Moradi F, et al. Association of the maternal 14-bp insertion/deletion polymorphism in the histocompatibility leukocyte antigen G gene with recurrent implantation failure. Iran J Reprod Med (2014) 12(9):641–6. [PMC free article] [PubMed] [Google Scholar]
- 38. Hviid TV. HLA-G in human reproduction: Aspects of genetics, function and pregnancy complications. Hum Reprod Update (2006) 12(3):209–32. doi: 10.1093/humupd/dmi048 [DOI] [PubMed] [Google Scholar]
- 39. Hviid TV, Hylenius S, Lindhard A, Christiansen OB. Association between human leukocyte antigen-G genotype and success of in vitro fertilization and pregnancy outcome. Tissue Antigens (2004) 64(1):66–9. doi: 10.1111/j.1399-0039.2004.00239.x [DOI] [PubMed] [Google Scholar]
- 40. Nowak I, Wilczyńska K, Wilczyński JR, Malinowski A, Radwan P, Radwan M, et al. KIR, LILRB and their ligands' genes as potential biomarkers in recurrent implantation failure. Arch Immunol Ther Exp (Warsz) (2017) 65(5):391–9. doi: 10.1007/s00005-017-0474-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Papúchová H, Saxtorph MH, Hallager T, Jepsen IE, Eriksen JO, Persson G, et al. Endometrial HLA-f expression is influenced by genotypes and correlates differently with immune cell infiltration in IVF and recurrent implantation failure patients. Hum Reprod (2022) 37(8):1816–34. doi: 10.1093/humrep/deac118 [DOI] [PubMed] [Google Scholar]
- 42. Sipak-Szmigiel O, Cybulski C, Wokolorczyk D, Lubinski J, Kurzawa R, Baczkowski T, et al. HLA-G polymorphism and in vitro fertilization failure in a polish population. Tissue Antigens (2009) 73(4):348–52. doi: 10.1111/j.1399-0039.2008.01205.x [DOI] [PubMed] [Google Scholar]
- 43. Wilczyńska K, Jasek M, Wagner M, Niepiekło-Miniewska W, Wiśniewski A, Radwan P, et al. The study of genetic polymorphisms for KIR2DL4, LILRB1, LILRB2 receptors and their ligand–HLA-G in recurrent implantation failure. J Reprod Immunol (2017) 122:43. [Google Scholar]
- 44. Kuroshli Z, Gourabi H, Bazrgar M, Sanati MH. Parental contribution of HLA-G∗0106 and G∗0105N to repeated implantation failure. Int J Fertility Sterility (2013) 7:131–2. [Google Scholar]
- 45. Kuroshli Z, Gourabi H, Bazrgar M, Sanati MH. Association between HLA-G genotypes and repeated implantation failure in Iranian couples. Iranian J Reprod Med (2013) 11:52. [Google Scholar]
- 46. Da Silva Nardi F, Gelmine GF, Costa CH, Roxo VS, Da Graca Bicalho M. Comparative analysis of allele frequencies of HLA-G gene in embryo implantation failure, assisted reproduction treatment, spontaneous recurrent miscarriage and fertile control couples. Tissue Antigens (2013) 81(5):380–1. doi: 10.1111/tan.12108 [DOI] [Google Scholar]
- 47. Kuroshli Z, Gourabi H, Bazrgar M, Sanati MH. Investigating association of HLA-G gene polymorphisms and failed implantation in human embryos. Int J Fertility Sterility (2012) 6:123–4. [Google Scholar]
- 48. Costa CH, Gelmini GF, Wowk PF, Mattar SB, Vargas RG, Sperandio VMM, et al. Comparative analysis of ins/del 14 bp at the 3UTR region of HLA-G gene in couples under assisted reproduction. Tissue Antigens (2011) 77(5):467. doi: 10.1111/j.1399-0039.2011.01671.x [DOI] [Google Scholar]
- 49. Costa CH, Gelmini GF, Wowk PF, Mattar SB, Vargas RG, Silva JS, et al. SNPs in the HLA-G 5UTR region in couples submitted to assisted reproduction. Tissue Antigens (2011) 77(5):466–7. doi: 10.1111/j.1399-0039.2011.01671.x [DOI] [Google Scholar]
- 50. Lashley LE, van der Westerlaken LA, Haasnoot GW, Drabbels JJ, Spruyt-Gerritse MJ, Scherjon SA, et al. Maternal HLA-C2 and 14 bp insertion in HLA-G is associated with recurrent implantation failure after in vitro fertilization treatment. Tissue Antigens (2014) 84(6):536–44. doi: 10.1111/tan.12452 [DOI] [PubMed] [Google Scholar]
- 51. Persson G, Melsted WN, Nilsson LL, Hviid TVF. HLA class ib in pregnancy and pregnancy-related disorders. Immunogenetics (2017) 69(8-9):581–95. doi: 10.1007/s00251-017-0988-4 [DOI] [PubMed] [Google Scholar]
- 52. González A, Rebmann V, Lemaoult J, Horn PA, Carosella ED, Alegre E. The immunosuppressive molecule HLA-G and its clinical implications. Crit Rev Clin Lab Sci (2012) 49(3):63–84. doi: 10.3109/10408363.2012.677947 [DOI] [PubMed] [Google Scholar]
- 53. Hviid TVF. HLA-G polymorphism, soluble HLA-G and early and late pregnancy complications. J Reprod Immunol (2011) 90(2):137. doi: 10.1016/j.jri.2011.06.015 [DOI] [Google Scholar]
- 54. Fanchin R, Gallot V, Rouas-Freiss N, Frydman R, Carosella ED. Implication of HLA-G in human embryo implantation. Hum Immunol (2007) 68(4):259–63. doi: 10.1016/j.humimm.2006.11.002 [DOI] [PubMed] [Google Scholar]
- 55. Hviid TVF. Non-classical HLA genes: From implantation to transplantation. Ugeskrift Laeger (2006) 168(5):461–6. [PubMed] [Google Scholar]
- 56. Mosaferi E, Majidi J, Mohammadian M, Babaloo Z, Monfaredan A, Baradaran B. HLA-G expression pattern: Reliable assessment for pregnancy outcome prediction. Adv Pharm Bull (2013) 3(2):443–6. doi: 10.5681/apb.2013.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lynge Nilsson L, Djurisic S, Hviid TV. Controlling the immunological crosstalk during conception and pregnancy: HLA-G in reproduction. Front Immunol (2014) 5:198. doi: 10.3389/fimmu.2014.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nilsson LL, Hviid TVF. HLA class ib-receptor interactions during embryo implantation and early pregnancy. Hum Reprod Update (2022) 28(3):435–54. doi: 10.1093/humupd/dmac007 [DOI] [PubMed] [Google Scholar]
- 59. Sipak O, Rył A, Grzywacz A, Laszczyńska M, Zimny M, Karakiewicz B, et al. The relationship between the HLA-G polymorphism and sHLA-G levels in parental pairs with high-risk pregnancy. Int J Environ Res Public Health (2019) 16(9):1546. doi: 10.3390/ijerph16091546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rizzo R, Andersen AS, Lassen MR, Sørensen HC, Bergholt T, Larsen MH, et al. Soluble human leukocyte antigen-G isoforms in maternal plasma in early and late pregnancy. Am J Reprod Immunol (2009) 62(5):320–38. doi: 10.1111/j.1600-0897.2009.00742.x [DOI] [PubMed] [Google Scholar]
- 61. Pfeiffer KA, Rebmann V, Passler M, van der Ven K, van der Ven H, Krebs D, et al. Soluble HLA levels in early pregnancy after in vitro fertilization. Hum Immunol (2000) 61(6):559–64. doi: 10.1016/s0198-8859(00)00123-3 [DOI] [PubMed] [Google Scholar]
- 62. Ouji-Sageshima N, Yuui K, Nakanishi M, Takeda N, Odawara Y, Yamashita M, et al. sHLA-G and sHLA-I levels in follicular fluid are not associated with successful implantation. J Reprod Immunol (2016) 113:16–21. doi: 10.1016/j.jri.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 63. Kofod L, Lindhard A, Bzorek M, Eriksen JO, Larsen LG, Hviid TVF. Endometrial immune markers are potential predictors of normal fertility and pregnancy after in vitro fertilization. Am J Reprod Immunol (2017) 78(3):e12684. doi: 10.1111/aji.12684 [DOI] [PubMed] [Google Scholar]
- 64. Parvanov D, Ganeva R, Todorova M, Nikolova K, Vasileva M, Rangelov I, et al. Hla-G levels in follicular fluid are not associated with follicular G-CSF concen-trations in women undergoing in vitro fertilization. Fertility Sterility (2020) 114(3 SUPPL):e313. doi: 10.1016/j.fertnstert.2020.08.853 [DOI] [Google Scholar]
- 65. Filippini-Cattaneo G, Bortolotti D, Spalvieri S, Rotola A, Jemec M, Suter T, et al. Soluble HLA-G as a non-invasive biomarker from ovulation to early pregnancy in assisted reproduction. Hum Reprod (2015) 30:i183. doi: 10.1093/humrep/30.Supplement-1.1 [DOI] [Google Scholar]
- 66. Wunder DM, Birkhäuser MH, Bersinger NA. Soluble human leukocyte antigen-G (sHLA-G) in follicular fluid and embryo culture medium and its impact on pregnancy prediction in IVF-ICSI treatment. Immuno-Analyse Biologie Specialisee (2013) 28(1):43–50. doi: 10.1016/j.immbio.2012.10.004 [DOI] [Google Scholar]
- 67. Alinejad Z, Shakib RJ, Yari F, Zahiri Z, Forghan-parast K, Roushan ZA, et al. The relationship between soluble serum HLA-G and ICSI success rates. J Reprod Infertility (2011) 12(2):93–9. [Google Scholar]
- 68. Wilczyńska K, Wiśniewski A, Krasiński R, Radwan P, Radwan M, Wilczyński J, et al. Recurrent implantation failure and soluble HLA-G plasma levels. J Reprod Immunol (2017) 122:46–7. doi: 10.1016/j.jri.2017.07.028 [DOI] [Google Scholar]
- 69. Liu S, Wei H, Lian R, Xu J, Zeng Y. Impaired peripheral blood HLA-g+ T cells are associated with recurrent miscarriage and recurrent implantation failure: Biomarkers in pregnancy complications–present knowledge and future direction. J Reprod Immunol (2018) 128:57. doi: 10.1016/j.jri.2018.05.043 [DOI] [Google Scholar]
- 70. Sipak-Szmigiel O, Ronin-Walknowska E, Cybulski C, Plonka T, Lubiński J. Antigens HLA-G, sHLA- G and sHLA- class I in reproductive failure. Folia Histochem Cytobiol (2007) 45 Suppl 1:S137–141. [PubMed] [Google Scholar]
- 71. Yeung HY, Dendrou CA. Pregnancy immunogenetics and genomics: Implications for pregnancy-related complications and autoimmune disease. Annual review of genomics and human genetics (2019) 20:73–97. doi: 10.1146/annurev-genom-083118-014943 [DOI] [PubMed] [Google Scholar]
- 72. Würfel FM, Winterhalter C, Trenkwalder P, Wirtz RM, Würfel W. European Patent in immunoncology: From immunological principles of implantation to cancer treatment. Int J Mol Sci (2019) 20(8):1830. doi: 10.3390/ijms20081830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Veit TD, Vianna P, Chies JAB. HLA-G - from fetal tolerance to a regulatory molecule in inflammatory diseases. Curr Immunol Rev (2010) 6(1):1–15. doi: 10.2174/157339510790231806 [DOI] [Google Scholar]
- 74. Dahl M, Perin T, Lindhard A, Djurisic S, Hviid TVF. Soluble human leukocyte antigen (HLA)-G and HLA-G genotype in couples undergoing treatment for infertility. J Reprod Immunol (2011) 90(2):172–3. doi: 10.1016/j.jri.2011.06.077 [DOI] [Google Scholar]
- 75. Hviid TV, Rizzo R, Christiansen OB, Melchiorri L, Lindhard A, Baricordi OR. HLA-G and IL-10 in serum in relation to HLA-G genotype and polymorphisms. Immunogenetics (2004) 56(3):135–41. doi: 10.1007/s00251-004-0673-2 [DOI] [PubMed] [Google Scholar]
- 76. Piekarska K, Radwan P, Tarnowska A, Wiśniewski A, Krasiński R, Radwan M, et al. The association of HLA-G gene polymorphism and its soluble form with Male infertility. Front Immunol (2021) 12:791399. doi: 10.3389/fimmu.2021.791399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Craenmehr MHC, Haasnoot GW, Drabbels JJM, Spruyt-Gerritse MJ, Cao M, van der Keur C, et al. Soluble HLA-G levels in seminal plasma are associated with HLA-G 3'UTR genotypes and haplotypes. Hla (2019) 94(4):339–46. doi: 10.1111/tan.13628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dahl M, Perin TL, Djurisic S, Rasmussen M, Ohlsson J, Buus S, et al. Soluble human leukocyte antigen-G in seminal plasma is associated with HLA-G genotype: possible implications for fertility success. Am J Reprod Immunol (2014) 72(1):89–105. doi: 10.1111/aji.12251 [DOI] [PubMed] [Google Scholar]
- 79. Nilsson LL, Hornstrup MB, Perin TL, Lindhard A, Funck T, Bjerrum PJ, et al. Soluble HLA-G and TGF-β in couples attending assisted reproduction - a possible role of TGF-β isoforms in semen? J Reprod Immunol (2020) 137:102857. doi: 10.1016/j.jri.2019.102857 [DOI] [PubMed] [Google Scholar]
- 80. Hò GT, Celik AA, Huyton T, Hiemisch W, Blasczyk R, Simper GS, et al. NKG2A/CD94 is a new immune receptor for HLA-G and distinguishes amino acid differences in the HLA-G heavy chain. Int J Mol Sci (2020) 21(12):4362. doi: 10.3390/ijms21124362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Guo Y, Lee CL, So KH, Gao J, Yeung WS, Yao Y, et al. Soluble human leukocyte antigen-g5 activates extracellular signal-regulated protein kinase signaling and stimulates trophoblast invasion. PloS One (2013) 8(10):e76023. doi: 10.1371/journal.pone.0076023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rajagopalan S. HLA-g-mediated NK cell senescence promotes vascular remodeling: implications for reproduction. Cell Mol Immunol (2014) 11(5):460–6. doi: 10.1038/cmi.2014.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee CL, Guo Y, So KH, Vijayan M, Guo Y, Wong VH, et al. Soluble human leukocyte antigen G5 polarizes differentiation of macrophages toward a decidual macrophage-like phenotype. Hum Reprod (2015) 30(10):2263–74. doi: 10.1093/humrep/dev196 [DOI] [PubMed] [Google Scholar]
- 84. Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, et al. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity (2017) 47(6):1100–1113.e1106. doi: 10.1016/j.immuni.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 85. Xu X, Zhou Y, Wei H. Roles of HLA-G in the maternal-fetal immune microenvironment. Front Immunol (2020) 11:592010. doi: 10.3389/fimmu.2020.592010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yao YQ, Barlow DH, Sargent IL. Differential expression of alternatively spliced transcripts of HLA-G in human preimplantation embryos and inner cell masses. J Immunol (2005) 175(12):8379–85. doi: 10.4049/jimmunol.175.12.8379 [DOI] [PubMed] [Google Scholar]
- 87. Alegre E, Diaz-Lagares A, Lemaoult J, Lopez-Moratalla N, Carosella ED, Gonzalez A. Maternal antigen presenting cells are a source of plasmatic HLA-G during pregnancy: longitudinal study during pregnancy. Hum Immunol (2007) 68(8):661–7. doi: 10.1016/j.humimm.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 88. Blanco O, Tirado I, Muñoz-Fernández R, Abadía-Molina AC, García-Pacheco JM, Peña J, et al. Human decidual stromal cells express HLA-G: Effects of cytokines and decidualization. Hum Reprod (2008) 23(1):144–52. doi: 10.1093/humrep/dem326 [DOI] [PubMed] [Google Scholar]
- 89. Xu Y, Ban Y, Ran L, Yu Y, Zhai S, Sun Z, et al. Relationship between unexplained recurrent pregnancy loss and 5,10-methylenetetrahydrofolate reductase) polymorphisms. Fertil Steril (2019) 111(3):597–603. doi: 10.1016/j.fertnstert.2018.11.011 [DOI] [PubMed] [Google Scholar]
- 90. Akdemir Y, Ayvaci H, Uludogan M. Effect of multiple thrombophilic gene mutations on uterine artery blood flow in nonpregnant recurrent pregnancy loss patients: are we searching enough? J Matern Fetal Neonatal Med (2019) 33(14):1–7. doi: 10.1080/14767058.2019.1569618 [DOI] [PubMed] [Google Scholar]
- 91. Pabalan N, Jarjanazi H, Sun C, Iversen AC. Meta-analysis of the human leukocyte antigen-G (HLA-G) 14 bp insertion/deletion polymorphism as a risk factor for preeclampsia. Tissue Antigens (2015) 86(3):186–94. doi: 10.1111/tan.12627 [DOI] [PubMed] [Google Scholar]
- 92. de Almeida BS, Muniz YCN, Prompt AH, Castelli EC, Mendes-Junior CT, Donadi EA. Genetic association between HLA-G 14-bp polymorphism and diseases: A systematic review and meta-analysis. Hum Immunol (2018) 79(10):724–35. doi: 10.1016/j.humimm.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 93. Shi X, Xie X, Jia Y, Li S. Maternal genetic polymorphisms and unexplained recurrent miscarriage: a systematic review and meta-analysis. Clin Genet (2017) 91(2):265–84. doi: 10.1111/cge.12910 [DOI] [PubMed] [Google Scholar]
- 94. Jalilvand A, Yari K, Heydarpour F. Role of polymorphisms on the recurrent pregnancy loss: A systematic review, meta-analysis and bioinformatic analysis. Gene (2022) 844:146804. doi: 10.1016/j.gene.2022.146804 [DOI] [PubMed] [Google Scholar]
- 95. Phoswa WN, Ramsuran V, Naicker T, Singh R, Moodley J. HLA-G polymorphisms associated with HIV infection and preeclampsia in south africans of African ancestry. BioMed Res Int (2020) p:1697657. doi: 10.1155/2020/1697657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Martinetti M, Beneventi F, Capittini C, Locatelli E, Simonetta M, Cavagnoli C, et al. The immunosignature of Mother/Fetus couples in gestational diabetes mellitus: Role of HLA-G 14 bp ins/del and PAPP-a A/C polymorphisms in the uterine inflammatory milieu. Dis Markers (2017) 2017:4254750. doi: 10.1155/2017/4254750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hviid TV, Hylenius S, Hoegh AM, Kruse C, Christiansen OB. HLA-G polymorphisms in couples with recurrent spontaneous abortions. Tissue Antigens (2002) 60(2):122–32. doi: 10.1034/j.1399-0039.2002.600202.x [DOI] [PubMed] [Google Scholar]
- 98. Aruna M, Sirisha PV, Andal Bhaskar S, Tarakeswari S, Thangaraj K, Reddy BM. Role of 14-bp insertion/deletion polymorphism in HLA-G among Indian women with recurrent spontaneous abortions. Tissue Antigens (2011) 77(2):131–5. doi: 10.1111/j.1399-0039.2010.01584.x [DOI] [PubMed] [Google Scholar]
- 99. Christiansen OB, Kolte AM, Dahl M, Larsen EC, Steffensen R, Nielsen HS, et al. Maternal homozygocity for a 14 base pair insertion in exon 8 of the HLA-G gene and carriage of HLA class II alleles restricting HY immunity predispose to unexplained secondary recurrent miscarriage and low birth weight in children born to these patients. Hum Immunol (2012) 73(7):699–705. doi: 10.1016/j.humimm.2012.04.014 [DOI] [PubMed] [Google Scholar]
- 100. Marzuillo P, Bellini G, Punzo F, Di Sessa A, Guarino S, Umano GR, et al. Association between 14 bp insertion/deletion HLA-G functional polymorphism and insulin resistance in a cohort of Italian children with obesity. Pediatr Diabetes (2018) 19(8):1357–61. doi: 10.1111/pedi.12768 [DOI] [PubMed] [Google Scholar]
- 101. Emmery J, Christiansen OB, Nilsson LL, Dahl M, Skovbo P, Møller AM, et al. Associations between fetal HLA-G genotype and birth weight and placental weight in a large cohort of pregnant women - possible implications for HLA diversity. J Reprod Immunol (2017) 120:8–14. doi: 10.1016/j.jri.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 102. Mandò C, Pileri P, Mazzocco MI, Lattuada D, Zolin A, Plebani M, et al. Maternal and fetal HLA-G 14 bp gene polymorphism in pregnancy-induced hypertension, preeclampsia, intrauterine growth restricted and normal pregnancies. J Matern Fetal Neonatal Med (2016) 29(9):1509–14. doi: 10.3109/14767058.2015.1052398 [DOI] [PubMed] [Google Scholar]
- 103. Iversen AC, Nguyen OT, Tømmerdal LF, Eide IP, Landsem VM, Acar N, et al. The HLA-G 14bp gene polymorphism and decidual HLA-G 14bp gene expression in pre-eclamptic and normal pregnancies. J Reprod Immunol (2008) 78(2):158–65. doi: 10.1016/j.jri.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 104. Kalotra V, Lall M, Verma IC, Kaur A, Kaur A. The HLA-G 14 bp insertion/deletion polymorphism and its association with soluble HLA-G levels in women with recurrent miscarriages. Hla (2018) 91(3):167–74. doi: 10.1111/tan.13198 [DOI] [PubMed] [Google Scholar]
- 105. Bylińska A, Wilczyńska K, Malejczyk J, Milewski Ł., Wagner M, Jasek M, et al. The impact of HLA-G, LILRB1 and LILRB2 gene polymorphisms on susceptibility to and severity of endometriosis. Mol Genet Genomics (2018) 293(3):601–13. doi: 10.1007/s00438-017-1404-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sipak O, Rył A, Grzywacz A, Laszczyńska M, Szymański S, Karakiewicz B, et al. Molecular analysis of HLA-G in women with high-risk pregnancy and their partners with regard to possible complications. Int J Environ Res Public Health (2019) 16(6):982. doi: 10.3390/ijerph16060982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nowak I, Malinowski A, Barcz E, Wilczyński JR, Wagner M, Majorczyk E, et al. Possible role of HLA-G, LILRB1 and KIR2DL4 gene polymorphisms in spontaneous miscarriage. Arch Immunol Ther Exp (Warsz) (2016) 64(6):505–14. doi: 10.1007/s00005-016-0389-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hashemi M, Mokhtari M, Khazaeian S, Bahari G, Rezaei M, Nakhaee A, et al. Evaluation of HLA-G 14-bp ins/del and +3142G>C polymorphisms with susceptibility to recurrent spontaneous abortion. Taiwan J Obstet Gynecol (2017) 56(3):276–80. doi: 10.1016/j.tjog.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 109. Warner CM, Tyas DA, Goldstein C, Comiskey M, Cohen J, Brenner CA. Genotyping: the HLA system and embryo development. Reprod BioMed Online (2002) 4(2):133–9. doi: 10.1016/s1472-6483(10)61930-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ober C, Aldrich C, Rosinsky B, Robertson A, Walker MA, Willadsen S, et al. HLA-G1 protein expression is not essential for fetal survival. Placenta (1998) 19(2-3):127–32. doi: 10.1016/s0143-4004(98)90000-5 [DOI] [PubMed] [Google Scholar]
- 111. Klitkou L, Dahl M, Hviid TV, Djurisic S, Piosik ZM, Skovbo P, et al. Human leukocyte antigen (HLA)-G during pregnancy part I: correlations between maternal soluble HLA-G at midterm, at term, and umbilical cord blood soluble HLA-G at term. Hum Immunol (2015) 76(4):254–9. doi: 10.1016/j.humimm.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 112. Steinborn A, Varkonyi T, Scharf A, Bahlmann F, Klee A, Sohn C. Early detection of decreased soluble HLA-G levels in the maternal circulation predicts the occurrence of preeclampsia and intrauterine growth retardation during further course of pregnancy. Am J Reprod Immunol (2007) 57(4):277–86. doi: 10.1111/j.1600-0897.2007.00475.x [DOI] [PubMed] [Google Scholar]
- 113. Yie SM, Taylor RN, Librach C. Low plasma HLA-G protein concentrations in early gestation indicate the development of preeclampsia later in pregnancy. Am J Obstet Gynecol (2005) 193(1):204–8. doi: 10.1016/j.ajog.2004.11.062 [DOI] [PubMed] [Google Scholar]
- 114. Ouji-Sageshima N, Geraghty DE, Ishitani A, Hatake K, Ito T. Establishment of optimized ELISA system specific for HLA-G in body fluids. Hla (2016) 88(6):293–9. doi: 10.1111/tan.12919 [DOI] [PubMed] [Google Scholar]
- 115. Shaikly VR, Morrison IE, Taranissi M, Noble CV, Withey AD, Cherry RJ, et al. Analysis of HLA-G in maternal plasma, follicular fluid, and preimplantation embryos reveal an asymmetric pattern of expression. J Immunol (2008) 180(6):4330–7. doi: 10.4049/jimmunol.180.6.4330 [DOI] [PubMed] [Google Scholar]
- 116. Zidi I, Ben Yahia H, Bortolotti D, Mouelhi L, Laaribi AB, Ayadi S, et al. Association between sHLA-G and HLA-G 14-bp deletion/insertion polymorphism in crohn's disease. Int Immunol (2015) 27(6):289–96. doi: 10.1093/intimm/dxv002 [DOI] [PubMed] [Google Scholar]
- 117. Sakly K, Maatouk M, Hammami S, Harzallah O, Sakly W, Feki S, et al. HLA-G 14 bp insertion/deletion polymorphism and its association with sHLA-G levels in behçet's disease Tunisian patients. Hum Immunol (2016) 77(1):90–5. doi: 10.1016/j.humimm.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 118. Darbas S, Yilmaz VT, Kocak H, Kisaoglu A, Demiryilmaz I, Aydinli B, et al. New markers for predictions of acute and chronic rejection and graft outcomes in kidney transplant recipients; HLA-G gene 3'UTR 14 bp polymorphism and sHLA-G. Gene (2021) 790:145712. doi: 10.1016/j.gene.2021.145712 [DOI] [PubMed] [Google Scholar]
- 119. Rizzo R, Bortolotti D, Fredj NB, Rotola A, Cura F, Castellazzi M, et al. Role of HLA-G 14bp deletion/insertion and +3142C>G polymorphisms in the production of sHLA-G molecules in relapsing-remitting multiple sclerosis. Hum Immunol (2012) 73(11):1140–6. doi: 10.1016/j.humimm.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 120. Nguyen LS, Rouas-Freiss N, Funck-Brentano C, Leban M, Carosella ED, Touraine P, et al. Influence of hormones on the immunotolerogenic molecule HLA-G: a cross-sectional study in patients with congenital adrenal hyperplasia. Eur J Endocrinol (2019) 181(5):481–8. doi: 10.1530/eje-19-0379 [DOI] [PubMed] [Google Scholar]
- 121. Jee BC, Suh CS, Kim SH, Moon SY. Soluble human leukocyte antigen G level in fluid from single dominant follicle and the association with oocyte competence. Yonsei Med J (2011) 52(6):967–71. doi: 10.3349/ymj.2011.52.6.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Rizzo R, Fuzzi B, Stignani M, Criscuoli L, Melchiorri L, Dabizzi S, et al. Soluble HLA-G molecules in follicular fluid: a tool for oocyte selection in IVF? J Reprod Immunol (2007) 74(1-2):133–42. doi: 10.1016/j.jri.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 123. Santoso B, Sa'adi A, Dwiningsih SR, Tunjungseto A, Widyanugraha MYA, Mufid AF, et al. Soluble immune checkpoints CTLA-4, HLA-G, PD-1, and PD-L1 are associated with endometriosis-related infertility. Am J Reprod Immunol (2020) 84(4):e13296. doi: 10.1111/aji.13296 [DOI] [PubMed] [Google Scholar]
- 124. Sehgal S, Vyawhare S, Bhatnagar S, Kshetrapal P. Longitudinal profile of sHLA-G during pregnancy and its association with small for gestational age births in north Indian pregnant females: A nested case-control study. Am J Reprod Immunol (2021) 87(1):e13504. doi: 10.1111/aji.13504 [DOI] [PubMed] [Google Scholar]
- 125. Beneventi F, Simonetta M, Locatelli E, Cavagnoli C, Badulli C, Lovati E, et al. Temporal variation in soluble human leukocyte antigen-G (sHLA-G) and pregnancy-associated plasma protein a (PAPP-a) in pregnancies complicated by gestational diabetes mellitus and in controls. Am J Reprod Immunol (2014) 72(4):413–21. doi: 10.1111/aji.12270 [DOI] [PubMed] [Google Scholar]
- 126. Knafel A, Basta P, Pitynski K, Mach P, Bednarek W, Klimek M, et al. Soluble HLA-G changes in maternal blood serum during the progression of labor. Neuro Endocrinol Lett (2009) 30(1):67–73. [PubMed] [Google Scholar]
- 127. Beneventi F, Locatelli E, De Amici M, Simonetta M, Cavagnoli C, Bellingeri C, et al. Soluble HLA-G concentrations in maternal blood and cervical vaginal fluid of pregnant women with preterm premature rupture of membranes. J Reprod Immunol (2016) 116:76–80. doi: 10.1016/j.jri.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 128. Yie SM, Li LH, Li YM, Librach C. HLA-G protein concentrations in maternal serum and placental tissue are decreased in preeclampsia. Am J Obstet Gynecol (2004) 191(2):525–9. doi: 10.1016/j.ajog.2004.01.033 [DOI] [PubMed] [Google Scholar]
- 129. He Y, Chen S, Huang H, Chen Q. Association between decreased plasma levels of soluble human leukocyte antigen-G and severe pre-eclampsia. J Perinat Med (2016) 44(3):283–90. doi: 10.1515/jpm-2015-0062 [DOI] [PubMed] [Google Scholar]
- 130. Eche S, Mackraj I, Moodley J. Circulating fetal and total cell-free DNA, and sHLA-G in black south African women with gestational hypertension and pre-eclampsia. Hypertens Pregnancy (2017) 36(4):295–301. doi: 10.1080/10641955.2017.1385794 [DOI] [PubMed] [Google Scholar]
- 131. Xu D, Zhu Y, Li L, Xu Y, Yan W, Dai M, et al. Evaluation of maternal serum sHLA-G levels for trisomy 18 fetuses screening at second trimester. Front Genet (2020) 11:497264. doi: 10.3389/fgene.2020.497264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Di Cristofaro J, Karlmark KR, Kanaan SB, Azzouz DF, El Haddad M, Hubert L, et al. Soluble HLA-G expression inversely correlates with fetal microchimerism levels in peripheral blood from women with scleroderma. Front Immunol (2018) 9:1685. doi: 10.3389/fimmu.2018.01685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. García-Láez V, Serra V, Bellver J, Ferro J, Vidal C, De Los Santos JM, et al. Gene polymorphisms and HLA-G expression in spontaneous abortions. Medicina Reproductiva y Embriol Clinica (2015) 2(3):82–92. doi: 10.1016/j.medre.2015.09.001 [DOI] [Google Scholar]
- 134. Schallmoser A, Sänger N. Quantitative analysis of the sHLA-G protein in seminal plasma. Hum Reprod (2020) 35(SUPPL 1):i145. doi: 10.1111/aji.13152 [DOI] [PubMed] [Google Scholar]
- 135. Rokhafrooz S, Ghadiri A, Ghandil P, Ghafourian M, Hossaini SH, Daraei N, et al. Association between HLA-G 14bp gene polymorphism and serum sHLA-G protein concentrations in preeclamptic patients and normal pregnant women. Immunol Invest (2018) 47(6):558–68. doi: 10.1080/08820139.2018.1467925 [DOI] [PubMed] [Google Scholar]
- 136. Persson G, Picard C, Marin G, Isgaard C, Stæhr CS, Molinari N, et al. Maternal HLA ib polymorphisms in pregnancy allo-immunization. Front Immunol (2021) 12:657217. doi: 10.3389/fimmu.2021.657217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Beneventi F, Badulli C, Locatelli E, Caporali R, Ramoni V, Cavagnoli C, et al. Soluble HLA-G in pregnancies complicated by autoimmune rheumatic diseases. J Reprod Immunol (2015) 110:67–73. doi: 10.1016/j.jri.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 138. Chen XY, Yan WH, Lin A, Xu HH, Zhang JG, Wang XX. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens (2008) 72(4):335–41. doi: 10.1111/j.1399-0039.2008.01107.x [DOI] [PubMed] [Google Scholar]
- 139. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PloS One (2013) 8(10):e71742. doi: 10.1371/journal.pone.0071742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Capittini C, Bergamaschi P, Sachetto S, Truglio M, Viola M, Marchesi A, et al. The plasma levels of soluble HLA-G molecules correlate directly with CD34+ cell concentration and HLA-G 14bp insertion/insertion polymorphism in cord blood donors. Blood Transfus (2014) 12 Suppl 1(Suppl 1):s361–366. doi: 10.2450/2012.0144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Gonzalez A, Alegre E, Torres MI, Díaz-Lagares A, Lorite P, Palomeque T, et al. Evaluation of HLA-G5 plasmatic levels during pregnancy and relationship with the 14-bp polymorphism. Am J Reprod Immunol (2010) 64(5):367–74. doi: 10.1111/j.1600-0897.2010.00855.x [DOI] [PubMed] [Google Scholar]
- 142. Ober C, Billstrand C, Kuldanek S, Tan Z. The miscarriage-associated HLA-G -725G allele influences transcription rates in JEG-3 cells. Hum Reprod (2006) 21(7):1743–8. doi: 10.1093/humrep/del036 [DOI] [PubMed] [Google Scholar]
- 143. Rebmann V, van der Ven K, Passler M, Pfeiffer K, Krebs D, Grosse-Wilde H. Association of soluble HLA-G plasma levels with HLA-G alleles. Tissue Antigens (2001) 57(1):15–21. doi: 10.1007/s00393-020-00783-6 [DOI] [PubMed] [Google Scholar]
- 144. Loisel DA, Billstrand C, Murray K, Patterson K, Chaiworapongsa T, Romero R, et al. The maternal HLA-G 1597ΔC null mutation is associated with increased risk of pre-eclampsia and reduced HLA-G expression during pregnancy in African-American women. Mol Hum Reprod (2013) 19(3):144–52. doi: 10.1093/molehr/gas041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Castelli EC, Mendes-Junior CT, Deghaide NH, de Albuquerque RS, Muniz YC, Simoes RT, et al. The genetic structure of 3'untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun (2010) 11(2):134–41. doi: 10.1038/gene.2009.74 [DOI] [PubMed] [Google Scholar]
- 146. Manaster I, Goldman-Wohl D, Greenfield C, Nachmani D, Tsukerman P, Hamani Y, et al. MiRNA-mediated control of HLA-G expression and function. PloS One (2012) 7(3):e33395. doi: 10.1371/journal.pone.0033395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Castelli EC, Ramalho J, Porto IO, Lima TH, Felício LP, Sabbagh A, et al. Insights into HLA-G genetics provided by worldwide haplotype diversity. Front Immunol (2014) 5:476. doi: 10.3389/fimmu.2014.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Verloes A, Spits C, Vercammen M, Geens M, LeMaoult J, Sermon K, et al. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res (2017) 19:118–27. doi: 10.1016/j.scr.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 149. Piekarska K, Radwan P, Tarnowska A, Wiśniewski A, Radwan M, Wilczyński JR, et al. ERAP, KIR, and HLA-c profile in recurrent implantation failure. Front Immunol (2021) 12:755624. doi: 10.3389/fimmu.2021.755624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Alecsandru D, Barrio A, Garrido N, Aparicio P, Pellicer A, Moffett A, et al. Parental human leukocyte antigen-c allotypes are predictive of live birth rate and risk of poor placentation in assisted reproductive treatment. Fertil Steril (2020) 114(4):809–17. doi: 10.1016/j.fertnstert.2020.05.008 [DOI] [PubMed] [Google Scholar]
- 151. Alecsandru D, Garrido N, Vicario JL, Barrio A, Aparicio P, Requena A, et al. Maternal KIR haplotype influences live birth rate after double embryo transfer in IVF cycles in patients with recurrent miscarriages and implantation failure. Hum Reprod (2014) 29(12):2637–43. doi: 10.1093/humrep/deu251 [DOI] [PubMed] [Google Scholar]
- 152. Nilsson LL, Scheike T, Langkilde CH, Jørgensen N, Hornstrup MB, Perin TL, et al. Examining extended human leukocyte antigen-G and HLA-f haplotypes: the HLA-G UTR-4 haplotype is associated with shorter time to pregnancy in an infertility treatment setting when both female and male partners are carriers. Fertil Steril (2020) 114(3):628–39. doi: 10.1016/j.fertnstert.2020.04.052 [DOI] [PubMed] [Google Scholar]
- 153. Bamberger AM, Jenatschke S, Schulte HM, Löning T, Bamberger MC. Leukemia inhibitory factor (LIF) stimulates the human HLA-G promoter in JEG3 choriocarcinoma cells. J Clin Endocrinol Metab (2000) 85(10):3932–6. doi: 10.1210/jcem.85.10.6849 [DOI] [PubMed] [Google Scholar]
- 154. Hakam MS, Miranda-Sayago JM, Hayrabedyan S, Todorova K, Spencer PS, Jabeen A, et al. Preimplantation factor (PIF) promotes HLA-G, -e, -f, -c expression in JEG-3 choriocarcinoma cells and endogenous progesterone activity. Cell Physiol Biochem (2017) 43(6):2277–96. doi: 10.1159/000484378 [DOI] [PubMed] [Google Scholar]
- 155. Tirado-González I, Freitag N, Barrientos G, Shaikly V, Nagaeva O, Strand M, et al. Galectin-1 influences trophoblast immune evasion and emerges as a predictive factor for the outcome of pregnancy. Mol Hum Reprod (2013) 19(1):43–53. doi: 10.1093/molehr/gas043 [DOI] [PubMed] [Google Scholar]
- 156. Rizzo R, D'Accolti M, Bortolotti D, Caccuri F, Caruso A, Di Luca D, et al. Human herpesvirus 6A and 6B inhibit in vitro angiogenesis by induction of human leukocyte antigen G. Sci Rep (2018) 8(1):17683. doi: 10.1038/s41598-018-36146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kuroki K, Hirose K, Okabe Y, Fukunaga Y, Takahashi A, Shiroishi M, et al. The long-term immunosuppressive effects of disulfide-linked HLA-G dimer in mice with collagen-induced arthritis. Hum Immunol (2013) 74(4):433–8. doi: 10.1016/j.humimm.2012.11.060 [DOI] [PubMed] [Google Scholar]
- 158. LeMaoult J, Daouya M, Wu J, Loustau M, Horuzsko A, Carosella ED. Synthetic HLA-G proteins for therapeutic use in transplantation. FASEB J (2013) 27(9):3643–51. doi: 10.1096/fj.13-228247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author. The extracted data and statistical R scripts are uploaded in our Github (https://github.com/minizenghong/HLA-G_sHLA-G_RIF).