Abstract
Aging is accompanied by the progressive decline in tissue regenerative capacity and functions of resident stem cells (SCs). Underlying mechanisms, however, remain unclear. Here we show that, during chronological aging, self-renewing mitoses of mammary SCs (MaSCs) are preferentially asymmetric and that their progeny divides less frequently, leading to decreased number of MaSCs and reduced regenerative potential. Underlying mechanisms are investigated in the p66Shc−/− mouse, which exhibits several features of delayed aging, including reduced involution of the mammary gland (MG). p66Shc is a mitochondrial redox sensor that activates a specific p53 transcriptional program, in which the aging-associated p44 isoform of p53 plays a pivotal role. We report here that aged p66Shc−/− MaSCs show increased symmetric divisions, increased proliferation and increased regenerative potential, to an extent reminiscent of young wild-type (WT) MaSCs. Mechanistically, we demonstrate that p66Shc, together with p53: (i) accumulates in the aged MG, (ii) sustains expression of the cell polarity determinant mInscuteable and, concomitantly, (iii) down-regulates critical cell cycle genes (e.g.,: Cdk1 and Cyclin A). Accordingly, overexpression of p53/p44 increases asymmetric divisions and decreases proliferation of young WT MaSCs in a p66Shc-dependent manner and overexpression of mInsc restores WT-like levels of asymmetric divisions in aged p66Shc−/− MaSCs. Notably, deletion of p66Shc has negligible effects in young MaSCs and MG development. These results demonstrate that MG aging is due to aberrant activation of p66Shc, which induces p53/p44 signaling, leading to failure of symmetric divisions, decreased proliferation and reduced regenerative potential of MaSCs.
Subject terms: Cell biology, Ageing
Introduction
In mammals, aging is accompanied by a decline in tissue homeostasis and regenerative capacity, suggesting a critical role for the functional decay of adult resident stem cells (SCs; [1]). Consistently, reduced SC number has been documented in various tissues, as the hippocampus [2], the skeletal muscle [3] and the mammary gland (MG; [4]). Underlying cellular and molecular mechanisms, however, are not yet fully elucidated.
Evidence in different models suggests that increased activity of p16INK4a or p53 tumor suppressors induces SC senescence or apoptosis and, thus, decreased SC number. Accordingly, mouse targeted-mutations which attenuate p16INK4a or p53 signaling diminish age-related functional decline of neuronal [5], hematopoietic [6] and mammary SCs (MaSCs; [7]), whereas constitutive activation of p53 leads to early SC exhaustion, reduced tissue regeneration and premature aging [8–10]. However, increased senescence or apoptosis in SCs during physiological aging remain poorly documented.
A further cellular mechanism that may affect SC number is the modality of the SC self-renewing mitosis. In the adult tissues, the prevailing modality of SC division is asymmetric, whereby daughter cells acquire distinct fates: one retains SC identity and the other undergoes commitment to differentiation. In certain conditions—e.g., tissue growth or injury-induced repair—SCs can instead divide symmetrically, where both daughter cells retain SC identity, an event that allows the expansion of the SC pool [11]. Thus, the binary choice between asymmetric and symmetric division is a critical mechanism to regulate SC number and the balance between self-renewal and differentiation. Analysis of self-renewing mitoses during physiological aging provided different results in different tissues: mathematical modelling of single neuronal SC-lineage dynamics suggests increased frequency of asymmetric divisions in the aged brain [12]; likewise, aged hair follicle SCs acquire an aberrant modality of division that generates two differentiating epidermal cells [13]. In the bone marrow and the epidermis SCs, instead, symmetric divisions increase during aging [14, 15].
Here, we investigated functional and molecular properties of self-renewing mitoses in aged MaSCs. As a model, we used the p66Shc knock-out (p66Shc−/−) mouse, which exhibits several features of delayed aging [16]. p66Shc is a mitochondrial redox sensor that uses reducing equivalents of the electron-transfer chain to generate reactive oxygen species [17] which, in turn, regulate specific intracellular targets, including phosphatases, kinases and membrane channels [18]. Deletion of p66Shc in mice induces oxidative-stress resistance at both systemic and cellular level, reducing penetrance of aging-associated diseases (e.g., obesity [19], atherosclerosis [20], ischemic injury [21] and diabetes [22]), ultimately delaying aging.
Results
Delayed age-related involution of the mammary gland (MG) in p66Shc−/− mice
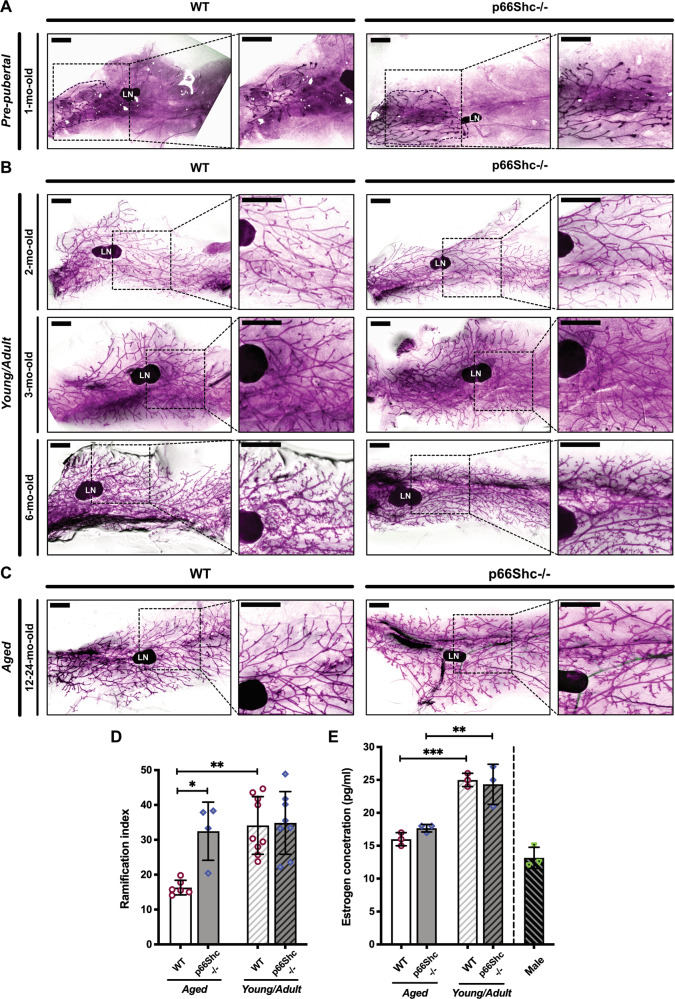
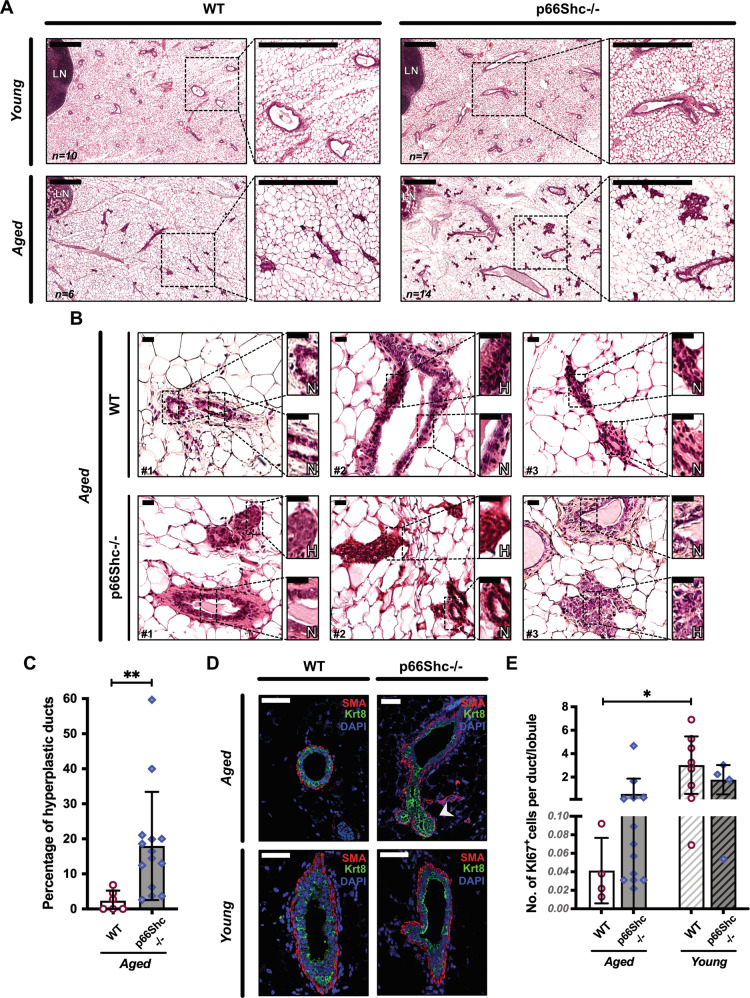
Inguinal MGs were removed from nulliparous C57BL/6 J WT or p66Shc−/− mice at different ages and analyzed by carmine-stained whole-mounts (Fig. 1A–C) or H&E-stained paraffin-embedded sections (Fig. 2A). In WT MGs, we observed typical features [23] of (i) post-natal development [small ductal trees with short-branched primary ducts in pre-pubertal glands (Fig. 1A)]; (ii) post-pubertal growth [multi-branched trees with extended ducts in young/adult glands (Fig. 1B)]; and (iii) age-related involution [few ducts and lobules, reduced ductal branching and increased fat/epithelium ratio in aged glands (Fig. 1C)]. Strikingly, MGs from aged p66Shc−/− mice showed little—if any—signs of age-related involution (Figs. 1C and 2A). Quantification of ductal branching confirmed that ramification of aged p66Shc−/− MGs was significantly higher than in WT counterparts (∼2-fold-increased, P = 0.016) and comparable to that of young/adult WT mice (Fig. 1D). Levels of circulating estrogen in aged WT and p66Shc−/− females were instead analogous (Fig. 1E). Aged p66Shc−/− mice showed multi-stratification of the normally bi-layered mammary epithelium, i.e., epithelial hyperplasia (Fig. 2B). Indeed, quantitative analyses of multilayered ducts on H&E-stained sections and of luminal and basal cells (marked by anti-Krt8 or anti-SMA antibodies, respectively) on confocal images showed increased multilayered ducts (∼10-fold; P = 0.003; Fig. 2B, C) and luminal overgrowth (∼1.7-fold-increased Krt8+/SMA+ ratio; P = 0.0028; Figs. 2D and S1A, B) in aged p66Shc−/− MGs. Accordingly, staining with anti-Ki67 antibody showed increased proliferating cells (∼14-fold-increased number of Ki67+ cells per duct or lobule, Figs. 2E and S1C). However, we never observed breast tumors in aged p66Shc−/− mice [24], thus confirming the benign nature of epithelial hyperplasia.
Fig. 1. Delayed age-related involution of mammary gland (MG) in p66Shc−/− mice.
A–C Carmine-stained whole-mounts of inguinal MGs from pre-pubertal (A), young/adult (B) and aged (C) WT or p66Shc−/− nulliparous mice, as indicated (see Materials and Methods for details). In each panel, the boxed area on the left is shown at higher magnification on the right. LN: lymph node. Scale bar = 1.5 mm. D Quantification of ductal branching in MGs from aged (12–24-mo-old) WT (n = 6) or p66Shc−/− (n = 4) mice and young/adult (2–6-mo-old) WT (n = 9) or p66Shc−/− (n = 8) mice. Data are represented as mean ± SD of ramification index (Sholl analysis; [63]) and analyzed by two-way ANOVA and Tukey post-hoc test; *: P = 0.016; **: P = 0.009. E Estrogen levels in aged (24-mo-old) and young/adult (3-mo-old) WT or p66Shc−/− nulliparous mice (n = 3 each, including control males), represented as mean ± SD of serum 17-β-estradiol concentrations analyzed by one-way ANOVA and Tukey post-hoc test; **: P = 0.006; ***: P = 0.0005.
Fig. 2. Benign ductal hyperplasia and increased epithelial proliferation in aged p66Shc−/− mammary glands (MGs).
A H&E staining of inguinal MGs from young (2-mo-old) WT (n = 10) or p66Shc−/− (n = 7) and aged (24-mo-old) WT (n = 6) or p66Shc−/− (n = 14) mice. In each panel, the boxed area on the left is shown at higher magnification on the right. LN: lymph node. Scale bar = 0.5 mm. B H&E images showing areas of normal (N) or hyperplastic (H) epithelium. Scale bar = 20 µm. C Quantification of epithelial hyperplasia in aged WT or p66Shc−/− mice (same as in B), represented as mean ± SD of percentages of hyperplastic ducts and analyzed by two-tailed Mann–Whitney test; **: P = 0.003. D Confocal images of basal (SMA, red) and luminal (Krt8, green) markers in MGs from aged WT (n = 3) or p66Shc−/− (n = 5) and young WT or p66Shc−/− mice (n = 3 each). Nuclei are stained by DAPI (blue). Scale bar = 50 µm; arrow-head: accumulation of Krt8+-cells. E Quantification of epithelial proliferation, represented as mean ± SD of number of Ki67+cells per duct or lobule in MGs from aged WT (n = 4) or p66Shc−/− (n = 13) and young WT (n = 8) or p66Shc−/− (n = 4) mice (same MGs as in A, B and D). Data are analyzed by two-way ANOVA and Tukey post-hoc test; *: P = 0.031.
Together, these data demonstrate a significant delay of age-related involution in the p66Shc−/− gland.
Increased number of mammary stem cells (MaSCs) in the aged p66Shc−/− MG
We then investigated the cellular composition of aged WT or p66Shc−/− MGs. Multipotent MaSCs were identified functionally, by limiting-dilution heterochronic transplantations of decreasing numbers of primary mammary epithelial cells (from 100,000 to 500) into the cleared fat-pads of 3-week-old syngeneic mice. Strikingly, MaSC frequency was markedly higher in aged p66Shc−/− mice (1:31,141), as compared to age-matched WT (1:236,701; P = 0.00004; Table 1). Uni-potent MaSCs (i.e., lineage-restricted progenitors) and differentiated cells were instead identified by FACS: staining of primary mammary epithelial cells with antibodies against basal (CD49f) or luminal (CD24) lineage-markers [25] showed comparable frequencies of luminal (CD24hiCD49fmed) or basal (CD24medCD49fhi) progenitors and of luminal (CD24hiCD49flow/neg) or basal (CD49fmed/hiCD24low/neg) differentiated cells (Fig. S2). Thus, numbers of MaSCs, but not of progenitors, is higher in the aged p66Shc−/− MG.
Table 1.
Increased number of mammary stem cells (MaSCs) in aged p66Shc−/− mice.
| Aged WT donors | Aged p66Shc−/− donors | |||
|---|---|---|---|---|
| No. of injected cells | No. of injected mice | No. of outgrowths | No. of injected mice | No. of outgrowths |
| 100,000 | 4 | 2 | 10 | 8 |
| 50,000 | 8 | 1 | 14 | 10 |
| 5000 | 8 | 0 | 16 | 6 |
| 500 | 6 | 0 | 14 | 4 |
| Total | 26 | 3 | 54 | 28 |
| Estimated SC frequency = 1:236,701 | Estimated SC frequency = 1:31,141 | |||
| Limits: lower = 1:726,359 - upper = 1:77,134 | Limits: lower = 1:50,840 - upper = 1:19,075 | |||
| P = 0.00004 | ||||
Limiting-dilution heterochronic transplantation of mammary epithelial cells from aged WT or p66Shc−/− mice into the cleared fat-pads of 3-week-old syngeneic mice. Numbers of injected cells, injected mice and obtained outgrowths are indicated, as well as estimated MaSC frequencies and confidence intervals. MaSC frequencies and statistical difference between the two groups are calculated with ELDA software [71] and indicated.
Increased regenerative potential of aged p66Shc−/− MaSCs
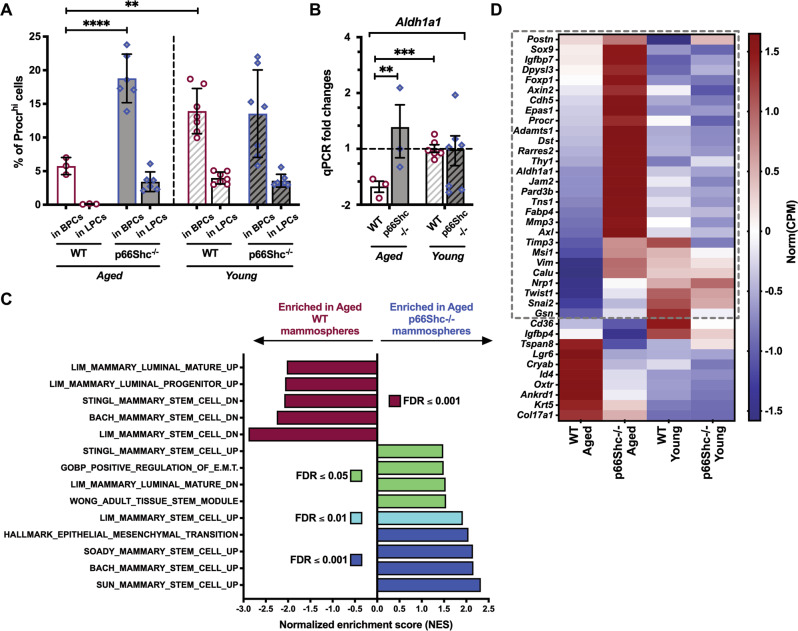
We then functionally investigated aged MaSCs using 3D cultures of primary epithelial cells (mammospheres), which allow for the enrichment and propagation of SCs and progenitors [26, 27]. Analysis of the MaSC markers Procr and Aldh1a1 [28, 29] showed higher MaSC frequency in aged p66Shc−/− mammospheres, as compared to the age-matched WT (∼19% vs ∼6% of Procrhi basal cells by FACS, P < 0.0001; ∼2-fold-increased Aldh1a1 expression by qRT-PCR, Q = 0.003; Figs. 3A, B and S3A).
Fig. 3. Increased expression of mammary stem cell (MaSC) markers in aged p66Shc−/− mammospheres.
A FACS-quantification of Procrhi cells among basal (BPCs) and luminal (LPCs) progenitors in M2-mammosphere independent preparations (n ≥ 3 each, as indicated) from aged and young, WT or p66Shc−/− mice. BPCs and LPCs are identified as CD24medCD49fhi and CD24hiCD49fmed, respectively. Data are represented as mean ± SD of Procrhi cell-percentages and analyzed by two-way ANOVA and Tukey post-hoc test; **: P = 0.005; ****: P < 0.0001. B Aldh1a1 mRNA levels in M2-mammosphere independent preparations (n ≥ 3 each) from aged and young, WT or p66Shc−/− mice. Data are represented in semi-logarithmic axis as mean ± SEM of qPCR fold-changes (FCs); 0 < FC < 1 are expressed as (−1/FC); “young WT” group is used as calibrator. Statistical analysis: multiple t-test performed on Table S3 (see Materials and Methods for further details); FDR = 5%; **: Q = 0.003; ***: Q = 0.0008. C Histogram representation of top-enriched, MaSC-related datasets obtained by Gene Set Enrichment Analysis [30] of aged p66Shc−/− (blue) vs aged WT (red) mammosphere RNA preparations (n = 3 each), analyzed by bulk RNA-sequencing. Raw expression levels were compared for enrichment in gene-sets listed in Dataset 1 (see Materials and Methods for further details); FDR: False Discovery Rate. D Heat-map representation of normalized expression levels of 38 MaSC-genes differentially expressed in aged p66Shc−/− vs aged WT mammosphere RNA preparations (n = 3 each), analyzed by bulk RNA-sequencing. Blue means low expression levels and red high expression levels. CPM: Counts Per Million mapped reads. The dashed gray box indicates genes upregulated in aged p66Shc−/− mammospheres (28 out of 38; see Dataset 2for details).
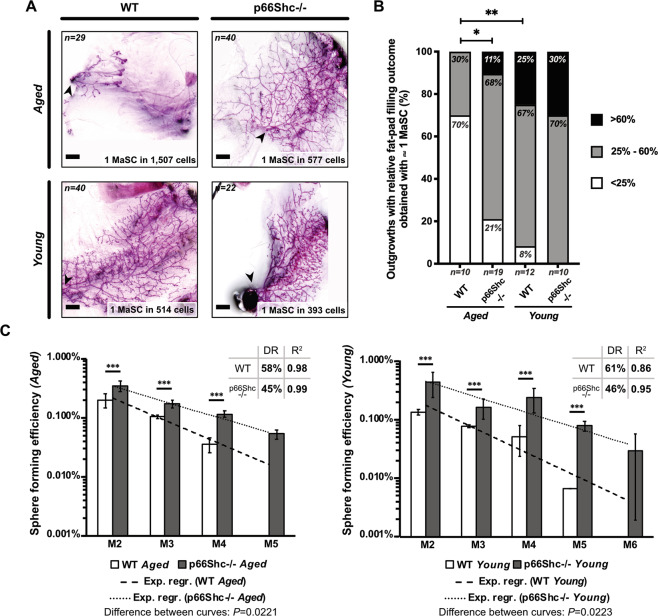
Bulk RNA-sequencing and Gene Set Enrichment Analysis (GSEA; [30]) revealed enrichment of: (i) gene-sets upregulated (UP) in normal MaSCs (Dataset 1; [31–35]) and (ii) genes involved in epithelial-to-mesenchymal transition [i.e., a distinguishing feature of normal MaSCs ([36, 37]; Figs. 3C and S3B) in aged p66Shc−/− mammospheres. Aged WT mammospheres showed, instead, enrichment of gene-sets downregulated (DN) in MaSCs ([31–34], Figs. 3C and S3C). Strikingly, among differentially expressed genes between aged mammospheres, we identified 28 MaSC-markers up-regulated in the aged p66Shc−/− (FDR ≤ 0.05; Fig. 3D and Dataset 2). Definitive evidence was obtained by limiting-dilution heterochronic transplantations, which showed a ∼3-fold-increased MaSC frequency in mammospheres from old p66Shc−/− mice (P = 0.006; Fig. 4A and Table S1).
Fig. 4. Increased mammary stem cell (MaSC) frequency and regenerative potential in aged p66Shc−/− mammospheres.
A Outgrowths obtained by limiting-dilution transplantation of M2-mammosphere cell suspensions. MaSC frequencies were calculated with ELDA [71] and indicated. Scale bar = 1 mm; arrow-head: injection site. B Frequencies of outgrowths obtained by transplantation of ∼1 MaSC (same experiment as in A), categorized according to the percentage of repopulated area (<25%, 25–60% and >60%) and analyzed by pairwise Chi-square test; *: P = 0.03; **: P = 0.008. C Representation in semi-logarithmic axis of sphere forming efficiency upon serial re-plating of mammary epithelial cells from aged (left) and young (right) WT or p66Shc−/− mice. Data are represented as mean ± SEM of three independent experiments. Statistical analysis: differences at each passage are calculated by two-way ANOVA and Tukey post-hoc test; ***: P < 0.001. Differences between fit-lines are calculated by simple linear regression after transformation of exponential curves into straight lines; P values are indicated. DR: decrease rate; R2: coefficient of determination.
To investigate MaSC functionality in aged mice, we analyzed percentages of recipients’ replenished fat-pad areas in the same transplantation experiment (Table S1), focusing on outgrowths obtained with rate-limiting dilution of injected cells (i.e., containing ∼1 MaSC). Strikingly, the fat-pad filling capacity of aged p66Shc−/− MaSCs resulted significantly higher than that of age-matched WT (∼79% vs ∼30% of outgrowths filling >25% of recipient’s fat-pad, (P = 0.031; Figs. 4B and S4), showing that MaSCs from old p66Shc−/− mice are endowed with increased regenerative potential.
Increased self-renewal of aged p66Shc−/− MaSCs
To investigate cellular mechanisms underlying increased MaSC-frequency in p66Shc−/− mice, we first examined MaSC replicative potential, i.e., the number of consecutive divisions they can undergo during serial re-plating prior to functional exhaustion [26, 27]. Consistently with previous reports [27, 38, 39], sphere-forming efficiency of mammospheres from aged WT mice showed a constant decrease rate (DR = 58%, R2 = 0.98), which led to culture exhaustion after four passages (Fig. 4C). Likewise, mammospheres from aged p66Shc−/− mice showed progressive exhaustion upon serial re-plating, which occurred at the 5th passage (Fig. 4C), demonstrating that differences in replicative potential between aged WT and p66Shc−/− MaSCs are negligible. Nevertheless, p66Shc−/− mammospheres showed a significantly higher capacity of generating new mammospheres at each passage (DR = 45%, R2 = 0.99, P = 0.022; P < 0.001 for each passage; Fig. 4C), suggesting that they are endowed with increased self-renewal potential.
Increased symmetric divisions of aged p66Shc−/− MaSCs
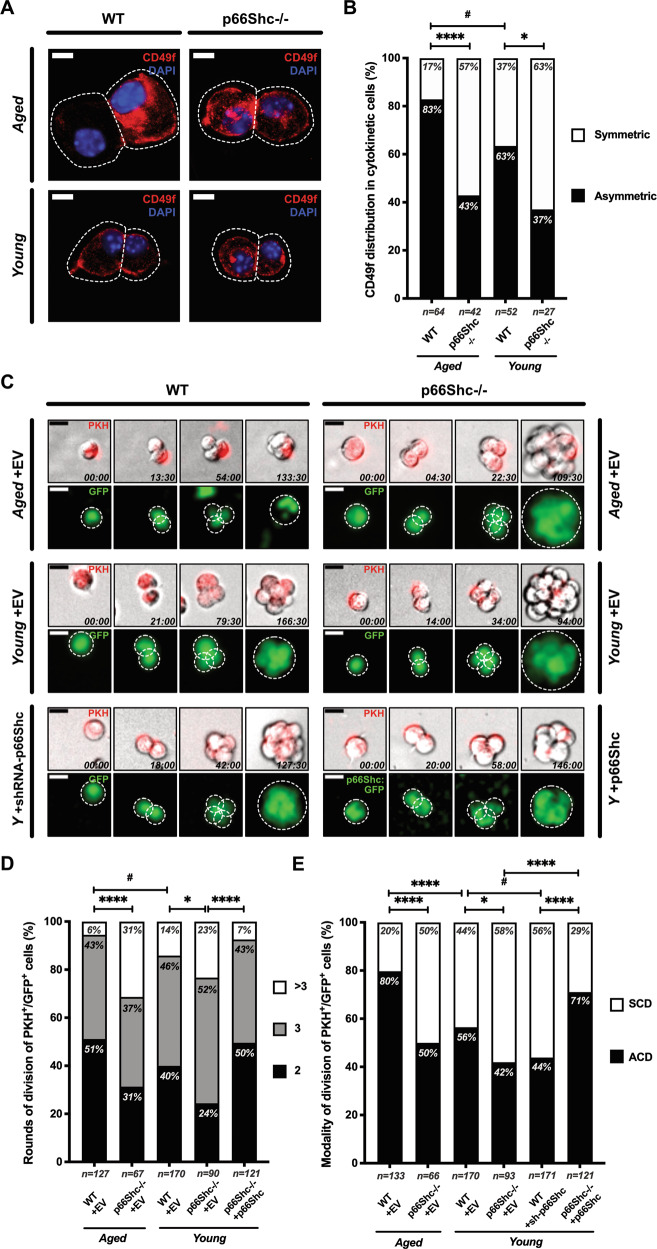
To investigate cellular mechanisms underlying increased self-renewal of p66Shc−/− MaSCs, we measured frequencies of asymmetric and symmetric divisions by immunofluorescence analysis of the MaSC marker CD49f [26, 27, 31, 40]. Modality of MaSC division was based on the identification of CD49f double-positive (symmetric) or single-positive (asymmetric) cytokinetic cell-doublets in mammosphere cell-suspensions [39, 41]. Results showed a predominantly asymmetric modality of division in MaSCs from aged WT mice (∼83%; Fig. 5A, B). Strikingly, MaSCs from old p66Shc−/− mice showed, instead, predominantly symmetric divisions (∼57%, P < 0.0001; Fig. 5A, B). Consistently, distribution of the fate-determinant Numb [27, 42] in the same cell-pairs was prevalently asymmetric in aged WT MaSCs (∼64%) and symmetric in the age-matched p66Shc−/− (∼73%, P = 0.006; Fig. S5A, B).
Fig. 5. Increased symmetric divisions and proliferation rate of p66Shc−/− aged mammary stem cells (MaSCs).
A CD49f distribution in cytokinetic cell-doublets from M2-mammospheres of aged and young, WT or p66Shc−/− mice (n ≥ 3 for each preparation). CD49f (red) is either asymmetrically or symmetrically distributed. Cells and cleavage sites are outlined. Nuclei are shown by DAPI (blue). Scale bar = 10 µm. B Frequencies of either asymmetrical (black) or symmetrical (white) CD49f distribution (same as in A). Numbers of scored doublets are indicated. Statistical analysis: pairwise Chi-square test with Yates’ correction. #: P = 0.031; *: P = 0.046; ****: P < 0.0001. C Time-lapse microscopy images of PKH+/GFP+ cells from M2-mammospheres infected with either empty- (+EV), or p66Shc-silencing- (+shRNA-p66Shc), or p66Shc:GFP-expressing- (+p66Shc) vectors. Early and late divisions are shown. Shooting-times are indicated as hours:minutes. Cells and spheroids are outlined. Scale bar = 20 µm. D Proliferation rates of PKH+GFP+ cells (same as in C), expressed as frequencies of rounds of division (2, 3 or >3) entered over 7 days. Numbers of scored cells are indicated. Statistical analysis: pairwise Chi-square test; #: P = 0.026; *: P = 0.024; ****: P < 0.0001. E Frequencies of either asymmetrical (ACD, black) or symmetrical (SCD, white) cell division (same experiment as in C). Numbers of scored cells are indicated. Statistical analysis: pairwise Chi-square test with Yates’ correction. *: P = 0.034; #: P = 0.026; ****: P < 0.0001.
Increased proliferation of aged p66Shc−/− MaSCs
To analyze their proliferation rate, we monitored MaSCs by time-lapse imaging for 7 days. MaSCs were identified using the PKH26 label-retaining protocol, which enables to distinguish PKH+ quiescent SCs from PKH− proliferating progenitors [27, 38, 42]. The vast majority of PKH+-MaSCs from aged WT mice underwent 2–3 division rounds, with only ∼6% undergoing >3 divisions. PKH+-MaSCs from aged p66Shc−/− mice, instead, showed a significantly higher proliferation rate, with ∼31% entering >3 division rounds in the same time-window (P < 0.0001; Fig. 5C, D). In the same experiments, PKH+-cell divisions were scored as asymmetric when one of the daughter cells did not divide further and symmetric when they both divided. Asymmetric divisions were largely prevalent in MaSCs from old WT mice, as compared to the age-matched p66Shc−/− (∼80% vs ∼50%, P < 0.0001; Fig. 5E). Within aged p66Shc−/− MaSCs, ∼30% of asymmetrically-dividing and ∼33% of symmetrically-dividing cells entered >3 division rounds. Likewise, within aged WT MaSCs, ∼6% of asymmetrically-dividing and ∼4% of symmetrically-dividing cells entered >3 division rounds (Table S2), indicating that the modality of division does not affect MaSC proliferation rate.
Notably, p66Shc re-expression in p66Shc−/− mammospheres from young mice increased MaSC asymmetric divisions (P < 0.0001; Fig. 5C, E), decreasing their proliferation rate (P < 0.0001; Fig. 5D and Table S2), to extents comparable to those of old WT mice. Thus, p66Shc promotes asymmetric division in aged MaSCs and restricts their proliferation, either they are dividing asymmetrically or symmetrically.
Increased symmetric divisions in aged p66Shc−/− MaSCs is due to de-regulation of the cell-polarity determinant mInscuteable
We then investigated molecular mechanisms underlying increased proliferation and symmetric divisions in aged p66Shc−/− MaSCs.
In MaSCs, cell-polarity is regulated by the tumor suppressor p53, as demonstrated by the prevailing symmetric modality of division of p53−/− MaSCs [27, 38, 41, 42]. Notably, p53 stabilization induces progeria and reduces lifespan [8, 9], two phenotypes that are rescued by concomitant p66Shc-deletion [43], suggesting that p66Shc is a critical determinant of p53 pro-aging effects. Mechanistically, we previously demonstrated that p66Shc is indispensable for the downregulation of ∼200 p53 target-genes critical for cell cycle progression (p53/p66Shc mitotic signature, [43]).
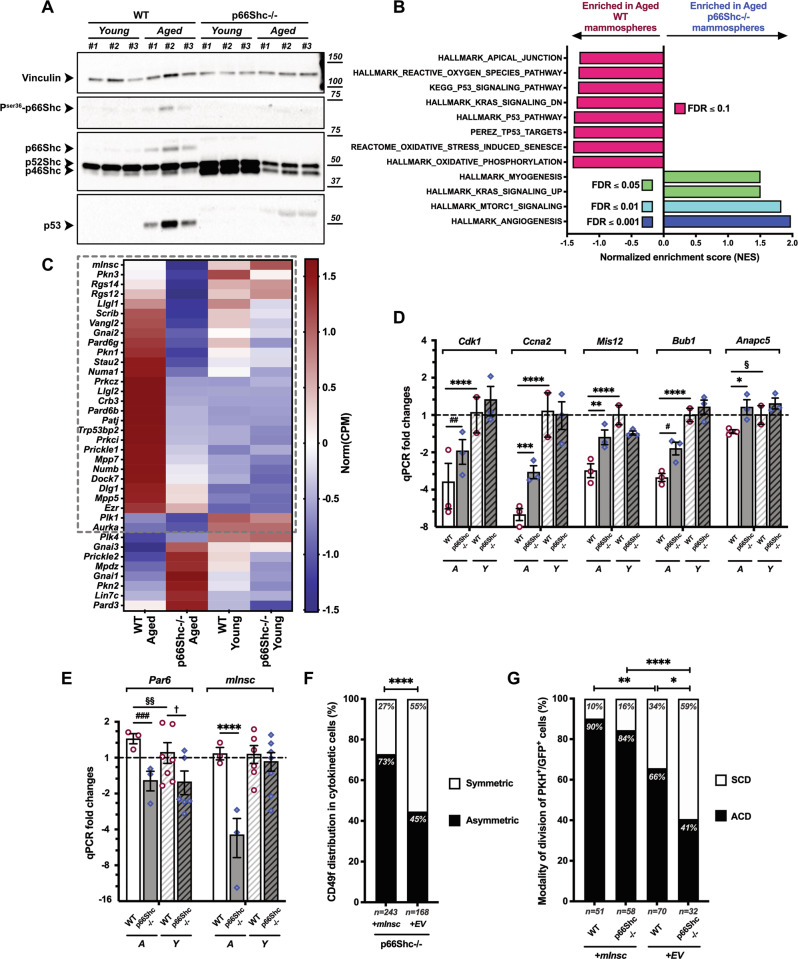
Western blotting on mammary epithelial cells showed that p66Shc, its activated (Ser36-phosphorylated) form and p53 were readily detectable in WT aged mice (Fig. 6A). p53, instead, was undetectable in age-matched p66Shc−/− samples, thus demonstrating that p66Shc is critical to achieve proper levels of p53 protein in the aged MG. This is consistent with findings in other aged tissues showing that p66Shc is indispensable for p53 stabilization upon oxidative stress [43]. Accordingly, RNA-sequencing and GSEA [30] showed enrichment of p53-signaling and oxidative stress response in aged WT mammospheres (Fig. 6B).
Fig. 6. Increased symmetric divisions of aged p66Shc−/− mammary stem cells (MaSCs) are due to impaired expression of mInsc and other cell polarity genes.
A Western blot on mammary epithelial cells from young and aged, WT or p66Shc−/− mice (n = 3 each). Vinculin is used as loading control. B Histogram representation of top-enriched, aging-related datasets obtained by Gene Set Enrichment Analysis [30] of aged p66Shc−/− (blue) vs aged WT (red) mammosphere RNA preparations (n = 3 each), analyzed by bulk RNA-sequencing. Raw expression levels were compared for enrichment in gene-sets listed in Dataset 1 (see Materials and Methods for further details); FDR: False Discovery Rate. C Heat-map representation of normalized expression levels of 36 cell polarity-genes differentially expressed in aged p66Shc−/− vs aged WT mammosphere RNA preparations (n = 3 each), analyzed by bulk RNA-sequencing. Blue means low expression levels and red high expression levels. CPM: Counts Per Million mapped reads. The dashed gray box indicates genes upregulated in aged p66Shc−/− vs aged WT mammospheres (28 out of 36; see Dataset 3for details). D, E mRNA levels of 5/30 p53/p66Shc mitotic signature-genes (D) and 2/6 cell polarity-genes (E) differentially regulated between young (Y) or aged (A), WT and p66Shc−/− M2-mammosphere independent preparations (n ≥ 3 for each group, see Table S3). Data are represented in semi-logarithmic axis as mean ± SEM of qPCR FCs; 0 < FC < 1 are expressed as (−1/FC); “young WT” group is used as calibrator. Statistical analysis: multiple t-test performed on Table S3 (see Materials and Methods for details); FDR = 5%; *: Q = 0.0347; #: Q = 0.0136; §: Q = 0.0393; †: Q = 0.0234; **: Q = 0.0044; ##: Q = 0.0023; §§: Q = 0.0087; ***: Q = 0.0003; ###: Q = 0.0001; ****: Q < 0.000001. F, G Effects of mInsc expression on MaSC division. Mammospheres from aged p66Shc−/− (F) and from young WT or p66Shc−/− (G) mice (n ≥ 3 for each preparation) were infected at M1-passage with either empty- (+EV) or mInsc- (+mInsc) expressing vectors. PKH+GFP+-MaSCs were analyzed for frequencies of asymmetrical (black) vs symmetrical (white) CD49f distribution in cytokinetic cell-doublets (F), or asymmetrical (ACD, black) vs symmetrical (SCD, white) cell division by time-lapse microscopy (G). Numbers of scored cells are indicated. Statistical analysis: pairwise Chi-square test with Yates’ correction. *: P = 0.038; **: P = 0.004; ****: P < 0.0001.
To investigate whether p66Shc regulates p53-dependent transcription in MaSCs/progenitors, we analyzed mRNA levels of 30 p53/p66Shc mitotic signature-genes (Tables S3), finding upregulation of key cell cycle-determinants (namely, Cdk1, Cyclin A, Anapc5, Mis12 and Bub1 [44–46]); Fig. 6D and Table S3]. Strikingly, RNA-sequencing showed downregulation of 28 genes involved in cell polarity-establishment and/or asymmetric division in aged p66Shc−/− mammospheres (FDR ≤ 0.05; Fig. 6C and Dataset 3). Among them, we found mInscuteable, a critical determinant of asymmetric division [47], which functions downstream to p53 and it is sufficient to restore normal levels of asymmetric divisions in p53−/− MaSCs [41]. qRT-PCR confirmed the marked downregulation of mInsc in aged p66Shc−/− mammospheres, as compared to age-matched controls (∼5-fold; Q < 0.000001; Fig. 6E and Table S3). These data suggest that mInsc is a downstream effector of the p53/p66Shc axis and that its downregulation in aged p66Shc−/− MaSCs is responsible for their increased frequency of symmetric divisions.
We therefore investigated whether mInsc-overexpression restores levels of asymmetric divisions in aged p66Shc−/− MaSCs. p66Shc−/− mammospheres were transduced with a mInsc-expressing lentivirus [41] and analyzed by anti-CD49f staining (Fig. S6A) and time-lapse imaging of PKH+-cells (Fig. S6B). CD49f distribution showed increased asymmetric divisions in mInsc-overexpressing aged p66Shc−/− cell-doublets (from ∼45% to ∼73%, P < 0.0001; Fig. 6F). Similarly, ∼84% of PKH+-MaSCs divided asymmetrically in time-lapse imaging analysis (P < 0.0001; Fig. 6G). Strikingly, mInsc-overexpression further increased asymmetric divisions in MaSCs from young WT mice, to an extent reminiscent of aged MaSCs (from ∼66% to ∼90%, P = 0.0037; Fig. 6G). Consistently, silencing of mInsc in aged WT mammospheres resulted in the significant increase of CD49f double-positive cell-doublets (from ∼32% to ∼67% or ∼57%, with two mInsc-targeting shRNAs, P = 0.013 and P = 0.038, respectively; Fig. S6C, D). Thus, increased frequency of symmetric divisions in the aged p66Shc−/− MaSCs is mediated by downregulation of the p53/p66Shc-target mInsc.
Effects of p66Shc on mInsc and asymmetric cell division are mediated by the p44 isoform of p53
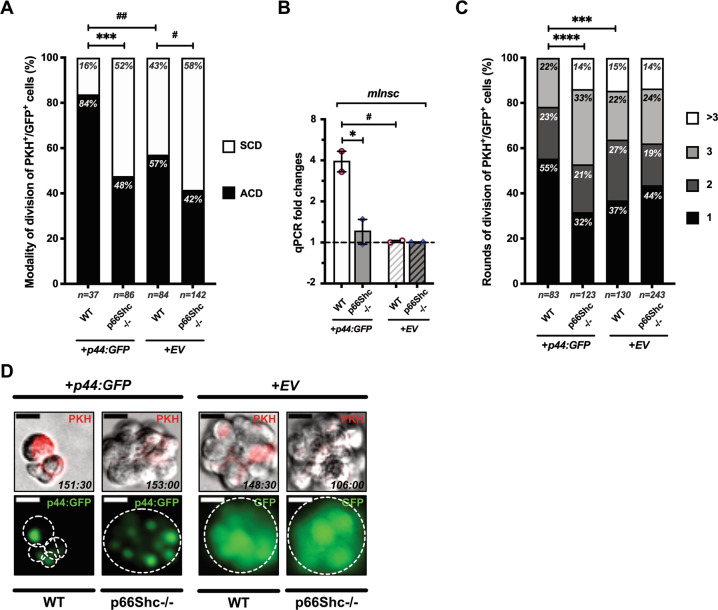
In cultured fibroblasts, p66Shc regulates p53 transcriptional response to oxidative stress by activating p44, an N-terminally truncated p53 isoform which induces p53 stabilization and favors its binding to specific promoters [9, 43, 48]. Unfortunately, western blotting with different anti-p53 antibodies did not produce reliable p44-specific signals in our model, probably due to low expression levels, as observed in other primary cells [43, 49]. Thus, we investigated the effects of lentivirus-mediated overexpression of p44:GFP fusion protein on the modality of MaSC division in young mice. Overexpressed p44:GFP induced p66Shc-independent stabilization of p53 (Fig. S7A), exerting also a modest effect on p66Shc stability or phosphorylation (Fig. S7B). In young WT mammospheres, p44:GFP induced a marked increase in asymmetric divisions (∼84%), reminiscent of aged MaSCs (P = 0.009; Figs. 7A and S7C). Strikingly, p44:GFP did not significantly affect the frequency of asymmetric divisions in p66Shc−/− MaSCs (from ∼42% to ∼58%; P = 0.444; Figs. 7A and S7C). Accordingly, p44-overexpression was associated with increased mInsc expression, which was, however, completely dependent on p66Shc (∼4-fold-increase in WT vs ∼1.2-fold in p66Shc−/− spheroids; P = 0.019; Fig. 7B). Likewise, p44-overexpression significantly impaired proliferation of WT MaSCs (from ∼37% to ∼22% of cells undergoing ≥3 division rounds; P = 0.0009; Fig. 7C), in a p66Shc-dependent manner (∼38% vs ∼47%; P = 0.129; Fig. 7C). Accordingly, p44:GFP-transduced WT mammospheres showed a considerably reduced size, which was completely dependent on p66Shc expression (Fig. 7D).
Fig. 7. Effects of p44 overexpression on mammary stem cell (MaSC) division.
A Mammospheres from young WT or p66Shc−/− mice (n ≥ 3 for each preparation) were infected at M1-passage with either empty- (+EV) or p44:GFP- (+p44:GFP) expressing vectors. PKH+GFP+-MaSCs were analyzed for frequencies of asymmetrical (ACD, black) vs symmetrical (SCD, white) cell division by time-lapse microscopy. Numbers of scored cells are indicated. Statistical analysis: pairwise Chi-square test with Yates’ correction. #: P = 0.033; ##: P = 0.009; ***: P = 0.0004. B mInsc mRNA levels in either p44:GFP-overexpressing- (+p44) or empty- (+EV) vector-infected WT or p66Shc−/− M2-mammospheres (n = 2 independent infection experiments). Data are represented as mean ± SEM of qPCR FCs; 0 < FC < 1 are expressed as (−1/FC); Statistical analysis: two-way ANOVA and Tukey post-hoc test; *: P = 0.019; #: P = 0.015. C Proliferation rate of PKH+GFP+-cells (same as in A), expressed as frequencies of rounds of division (1, 2, 3 or >3) entered over 7 days. Numbers of scored cells are indicated. Statistical analysis: pairwise Chi-square test; ***: P = 0.0009; ****: P < 0.0001. D Mammospheres obtained in the same experiment as in (A and C). PKH is shown in red. Spheroids are outlined. Shooting-times are indicated as hours:minutes. Note p44:GFP nuclear localization.
Thus, p44 increases mInsc expression and decreases symmetric divisions and proliferation in young WT MaSCs, to levels comparable to those of aged MaSCs, in a p66Shc-dependent manner.
p66Shc regulates chronological aging but it is marginally involved in MG post-natal development
Western blotting showed very low—if any—expression of p66Shc and p53 in young MGs, while they were both abundant in aged glands (Fig. 6A). Thus, we investigated whether p66Shc regulates MG chronological aging.
We first analyzed whether the same functions that are de-regulated in p66Shc−/− MaSCs (i.e., proliferation rate and modality of division) are modified in WT MaSCs during aging. Anti-CD49f staining and time-lapse imaging showed decreased symmetric divisions in aged vs young WT mammospheres (∼17% vs ∼37%, P = 0.031; Fig. 5A, B and ∼20% vs ∼44%, P < 0.0001; Fig. 5C, E). Likewise, proliferation was significantly reduced in aged vs young WT PKH+-MaSCs (∼6% vs ∼14% of cells entering >3 division rounds, P = 0.026; Fig. 5D). MaSCs from old WT mammospheres were ∼3-time-less frequent than in young counterparts (P = 0.009, Fig. 4A and Table S2; ∼5% vs ∼14% of Procrhi basal cells, P = 0.005, Figs. 3A and S3A), showing also decreased regenerative potential (from ∼25% to 0% of outgrowths filling >60% of recipients’ fat-pad; P = 0.008; Figs. 4B and S4). Consistently, both complexity and Ki67-positivity of ductal epithelium were reduced in old WT MGs (∼2-fold-decreased ramification, P = 0.001, Fig. 1D; ∼70-fold-decreased Ki67-positivity, P = 0.031; Figs. 2E and S1C).
Together, these data demonstrate that loss of MaSC functionality (i.e., decreased proliferation, symmetric divisions and regenerative potential) are critical features of chronological aging and they suggest that p66Shc-deletion delays the aging process by preserving the same functions.
Our histological and quantitative analyses of pre-pubertal, young and adult MGs (Fig. 1A–D) did not reveal, instead, differences in post-natal development between WT and p66Shc−/− MGs. Likewise, in young p66Shc−/− mammospheres we did not observe relevant de-regulation of cell cycle- (Fig. 6D and Table S3) or cell-polarity genes (Fig. 6C, E), except for a modest downregulation of Par6 (∼1.8-fold, Q = 0.023; Fig. 6E and Table S3). Comparison of specific MaSC functions in young WT vs p66Shc−/− mammospheres, however, provided apparently contradictory results. Some of them were analogous, including (i) expression of the MaSC-markers Procr and Aldh1a1 (Figs. 3A, B and S3A), (ii) MaSC frequency (Table S1), and (iii) regenerative potential (Fig. 4A, B). Other functions, instead, were slightly enhanced in p66Shc−/− young mice, though differences with young WT were, by far, less extended than those between aged mice: (i) frequency of symmetric divisions (∼1.3-fold-increased, P = 0.034; Fig. 5B–E) and (ii) proliferation rate (∼1.6-fold-increase in MaSCs undergoing >3 division rounds, P = 0.024; Fig. 5D). Finally, the weak effect of p66Shc on the modality of division of young MaSCs was confirmed by p66Shc knock-down in mammospheres from young mice (∼1.3-fold-increased symmetric divisions, P = 0.026; Fig. 5C, E).
Discussion
Asymmetric division in SCs represents a critical mechanism to modulate the balance between self-renewal and differentiation. Seminal studies in Drosophila revealed that loss-of-function of several components of the asymmetric division machinery (e.g., Miranda, Prospero, Numb and Inscuteable) lead to SC over-proliferation, genomic instability and tumor-like behavior [50]. Whether aberrant regulation of symmetric SC division contributes to organismal aging, instead, is still controversial.
We show here that the self-renewing mitoses of aged MaSCs are more frequently asymmetric and that their progeny divides less frequently. This correlates with reduced MaSC number and decreased regenerative-potential, as evaluated by limiting-dilution transplantations. Notably, transplants were performed in young recipients, thus providing direct evidence that MG age-related involution is due to cell-intrinsic mechanisms. Other properties of self-renewing divisions are not altered in aged MaSCs, e.g., their replicative potential or the capacity of committed daughter cells to maintain physiological levels of basal and luminal progenitors, although single-cell RNA-sequencing revealed subtle changes in proportions of epithelial populations during aging [51]. Together, our data demonstrate that increased asymmetric divisions and decreased proliferation of aged MaSCs are markers of MG physiological aging and they suggest that these two mechanisms contribute significantly to reduce MG regenerative potential.
Genetic support to this hypothesis comes from p66Shc−/− mice: MGs of old p66Shc−/− mice exhibit little—if any—morphological involution. Aged p66Shc−/− MaSCs show increased propensity to divide symmetrically, while daughter cells display increased proliferation, as compared to age-matched controls. Accordingly, both number and regenerative potential of aged p66Shc−/− MaSCs are increased. These properties are reminiscent of young WT MaSCs, suggesting that p66Shc-deletion attenuates physiological mechanisms of chronological aging.
Mechanistically, p66Shc regulates the expression of specific p53-dependent genes. We and others have previously shown that p53/p66Shc signaling induces senescence or apoptosis in either cultured cells or in vivo, in response to oxidative stress [43, 52–54]. Upon oxidative stress, p66Shc is activated [16, 55, 56] and, specifically in this context, it is critical for the expression of the p44 isoform of p53 (through an alternative p53 initiation-site; [9, 48, 57]). p44, in turn, induces stabilization of p53 and modulates its binding to target promoters [43, 48]. Consistently, we do not detect p53 protein in aged p66Shc−/− MGs and p53 transcriptional activity is attenuated in aged p66Shc−/− MaSCs/progenitors, where we find downregulation of cell polarity-genes (e.g., aPKCζ/ι, Lgl1/2 and mInsc), up-regulation of critical cell cycle-genes (e.g., Cdk1, Cyclin A and Bub1) and enrichment in epithelial-to-mesenchymal transition and SC transcriptional signatures.
We focused on mInsc since it functions downstream to p53 as an asymmetric division determinant in MaSCs [41]. Impaired mInsc expression inhibits asymmetric divisions in the murine developing neocortex [58] and rat retina [59], while its overexpression increases asymmetric divisions in the epidermis [60] and p53−/− MaSCs [41]. Accordingly, we show that mInsc-overexpression is sufficient to restore WT-like levels of asymmetric divisions in aged p66Shc−/− MaSCs. Furthermore, p44-overexpression (and, thus, p53 stabilization) promotes mInsc expression and asymmetric divisions and it restricts proliferation in a p66Shc-dependent manner. These findings suggest that p53-p44/p66Shc-mediated de-regulation of mInsc and cell cycle-genes is responsible for increased asymmetric divisions and decreased proliferation of aged MaSCs.
Finally, we show that young/adult p66Shc−/− MGs are histologically undistinguishable from age-matched WT organs. Accordingly, we find similar expression levels of cell cycle-genes or mInsc and similar MaSC numbers or regenerative potential. Consistently, p66Shc/p53 expression is barely detectable in young MGs, suggesting that p66Shc has a negligible role in MG development. Expression of p66Shc (and p53) in the old MG may be due to the progressive accumulation of oxidative stress, which is a universal and proximal mechanism of aging, upstream to both p66Shc and p53 [18, 61]. Most notably, mInsc is not regulated by p66Shc in young MaSCs and it is not up-regulated during chronological aging. Nevertheless, its expression is near-completely supported by p66Shc in aged MaSCs, thus indicating that critical MaSC functions, which are under p66Shc control in aging, are regulated through p66Shc-independent mechanisms during development. Consistently, deletion of p66Shc does not increase breast cancer formation in aged mice [24]. In conclusion, targeting specific cell-fate or -cycle regulators that act near-exclusively in aging, as p66Shc, may represent a novel and safe approach to retard tissue degeneration.
Materials and methods
Animals
Generation of p66Shc−/− mice by targeted mutation of Shc1 gene (MGI:98296) has been described in [16]; further information can be found on the MGI_dedicated_page. C57BL/6 J WT and C57BL/6 J p66Shc−/− mice were housed in SPF conditions at Cogentech Mouse Genetics facility (Milan, MI, Italy), in individually ventilated cages (Tecniplast, Buguggiate, VA, Italy) with a light/dark cycle of 12 h; food and water were provided ad libitum. All the procedures on mice have been communicated to and approved by the Italian Ministry of Health (Project number 603/2016-PR).
Carmine-stained whole-mounts
Whole-mount analyses (Fig. 1A–C) were performed on a total of 15 WT and 18 p66Shc−/− female mice. Of each genotype, 3 pre-pubertal (1-month-old), 6 post-pubertal (2-, 3- and 6-month-old), 3 or 5 12-month-old and 3 or 4 24-month-old mice were analyzed. Inguinal MGs were removed, flattened onto glass slides and fixed over-night (ON) in 4% formalin at room-temperature (RT). MGs were then washed in ethanol, rinsed in water and stained ON in 0.2% carmine powder (Cat#C1022, Merck, Darmstadt, HE, Germany) dissolved in in 0.5% aluminum potassium sulfate dodecahydrate (Cat#A7167, Merck). MGs were then de-stained in ethanol and soaked in BABB solution [3-(N-Acetylamino)−5-(N-decyl-N-methylamino)benzyl alcohol/Benzyl benzoate, 1:2 – Cat#A4813 and Cat#B6630, Merck] until fat-pads were bleached and only ductal epithelium was clearly visible.
Quantification of ductal branching
Carmine-stained whole-mount images, acquired by stereomicroscopy (Table S5), were used to calculate ramification indexes (Fig. 1D) in Fiji software [62] as already described [63]. Statistical analysis was performed by comparing post-pubertal mice (“Young/Adult” in Fig. 1D) with aged mice (12- and 24-month-old) by ordinary two-way ANOVA and Tukey’s multiple comparison test, with individual variances computed for each comparison. Sample size and data representation are detailed in Fig. 1 legend.
Histopathological analyses on FFPE mammary glands (MGs)
Formalin-fixed MGs were paraffin-embedded with Logos J Processor (Milestone Medical, Sorisole, BG, Italy); 3 µm sections were cut and incubated ON at 37 °C before hematoxylin & eosin staining. Analyses represented in Fig. 2A were performed on 10 WT and 7 p66Shc−/− 2-month-old mice and 6 WT and 14 p66Shc−/− 24-month-old mice. Analysis of ductal hyperplasia (Fig. 2B) was performed on the same aged mice: ducts showing multilayered epithelium were scored as hyperplastic. Difference between genotypes (Fig. 2C) was computed using two-tailed Mann–Whitney test, assuming non-Gaussian distribution of percentages of hyperplastic ducts. Sample size and data representation are detailed in Fig. 2 legend. Images were acquired using an Aperio ScanScope XT system (Leica Biosystems, Nussloch, BW, Germany; Table S5) and analyzed with Fiji [62].
Immunofluorescence on FFPE mammary glands (MGs)
Immunofluorescence staining (Figs. 2D and S1A) was performed on dewaxed and rehydrated sections exposed for 20′ at 95 °C to sodium citrate, pH = 6, for antigen-retrieval. Slides were permeabilized in 0.1% Triton X-100 for 20′ and blocked in 5% BSA for 60′ in a humid chamber. Primary antibodies (Table S4) were incubated ON at 4 °C. Secondary antibodies (Table S4) were incubated for 60′ at RT. Experiment presented in Fig. 2D and S1A was performed on at least three mice per group of the same used for Fig. 2A. Images were acquired by confocal microscopy (Table S5) and analyzed with Fiji [62]. Quantification of luminal hyperplasia in aged p66Shc−/− mice was performed by manually counting luminal (Krt8+) and basal (SMA+) cells on each duct/lobule Z-stack-image and by computing the ratio of Krt8+ to SMA+ cells. Statistical analysis was performed by comparing post-pubertal mice (“Young/Adult” in Fig. S1B) with aged mice (24-month-old) by ordinary two-way ANOVA and Tukey’s multiple comparison test, with individual variances computed for each comparison. Sample size and data representation are detailed in Fig. S1 legend.
Immunohistochemistry on FFPE mammary glands (MGs)
Immunohistochemistry of Ki67 (Fig. S1C) was performed using a Bond-III fully-automated IHC/ISH stainer (Leica Biosystems). Antigen-retrieval was performed by exposing slides for 20′ at 95 °C to Bond Epitope Retrieval Solution 1 (Cat#AR9961, Leica Biosystems). Primary antibody (Table S4) was diluted in Bond Primary Antibody Diluent (Cat#AR9352, Leica Biosystems) and chromogenic reaction was obtained with BOND IHC Polymer Detection Kit (Cat#DS9800, Leica Biosystems); hematoxylin was used as counterstaining. Experiment presented in Fig. S1C was performed on same mice used for Fig. 2A. Ki67-positivity was scored as no. of Ki67+-cells per duct or lobule and differences between groups (Fig. 2E) were calculated by ordinary two-way ANOVA and Tukey’s multiple comparison test, with individual variances computed for each comparison. Sample size and data representation are detailed in Fig. 2 legend. Images were acquired using an Aperio ScanScope XT system (Leica Biosystems; Table S5) and analyzed with Fiji [62].
Estradiol measurement
Vaginal smears were collected every day from WT and p66Shc−/− mice to determine their estrous cycle phase, according to standard procedures [64]. When only epithelial cornified cells were found, it was inferred that young/adult mice (3-month-old) were in their estrous phase and aged mice (24-month-old) in acyclic phase, characterized by extended cornification and prolonged estrogen secretion [64], as expected. Female mice, together with control males, were then subjected to blood sampling to measure serum 17-β-estradiol levels by ELISA (Cat#EK7003, BosterBio, Pleasanton, CA, USA). Sample size and data representation are detailed in Fig. 1 legend. Due to the presence of the control male group, data were analyzed by ordinary one-way ANOVA followed by Tukey’s multiple comparison test. Statistical significance between the control male group and female groups is not shown in figure.
Primary mammary epithelial cell preparation
Mammary glands were removed from female mice and enzymatically and mechanically digested as previously described [27, 42]. After 1 week of culture in ultra-low adherence conditions, M1-spheroids were collected, dissociated by pipetting ∼200 times and re-plated on Poly-HEMA- (Cat#P3932, Merck, 1.2% in ethanol) -coated 6-multiwell plates.
FACS analyses
Cells collected from primary mammary epithelial cell preparations or M2-mammospheres were resuspended in blocking solution (10% BSA in PBS; ∼10ˆ6 cells/sample) for 30′, stained on ice for 60′ with specific antibody combinations, fixed for 30′ with Cytofix/Cytoperm™ solution (50 µl/sample, Cat#554714, BD Biosciences, Franklin Lakes, NJ, USA) and washed in PBS prior to acquisition on a FACSAria™ Fusion Flow Cytometer (BD Biosciences). In experiments performed in primary cells (Fig. S2), n is intended as number of mice (one mouse = one preparation). In experiments performed in M2-mammosphere cell-suspensions (Figs. 3A and S3A), n is intended as independent preparations, where one preparation was obtained from ≥3 mice. Aged mice of both genotypes were 12-month-old, while young mice were 2-month-old. Staining and exclusion of endothelial and haemopoietic cells was done using a combination of Lin antibodies (anti-CD45/Ter-119/CD31; Table S4). Same experimental conditions were applied for primary cells and M2-mammospheres to avoid differences in acquisition parameters and gating among experiments. Epithelial cell populations were identified by combinations of specific markers ([25]; Table S4) as described in Figs. 3, S2 and S3 legends. Gates were first set on “Young WT” group and kept for all the samples within the same experiment; representative gating images are reported in Figs. S2 and S3. Sample size and data representation are detailed in Figs. 3 and S2 legends. Differences between groups were calculated by ordinary two-way ANOVA with main effects only (“cell population” and “age+genotype”), followed by Tukey’s multiple comparison test with a single pooled variance. Flow cytometry data were analyzed with FlowJo™ v10 Software (FlowJo, Ashland, OR, USA).
RNA extraction
Total RNAs were extracted from M2-mammospheres (∼100,000 cells/sample) with PicoPure™ RNA isolation kit (Cat#KIT0204, Applied Biosystems™, Bedford, MA, USA), according to manufacturer’s instructions. RNA quality was checked with 2100 Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA) and RNAs with RIN (RNA Integrity Number) >7 were subjected to in vitro reverse-transcription (Cat#A3800, ImProm-II™ Reverse Transcription System, Promega Corporation, Madison, WI, USA), according to manufacturer’s instructions.
Bulk RNA-sequencing
Total RNAs were extracted as detailed above from three independent M2-mammosphere preparations derived from ≥3 mice each. WT and p66Shc−/− aged mice were 12-month-old, while young mice were 2-month-old. RNA libraries were prepared from 200 ng of total RNA using the Illumina TruSeq Stranded Total RNA kit (Cat#20020597, Illumina, Inc., San Diego, CA, USA), according to manufacturer’s instructions. cDNA libraries were then amplified by PCR and checked with 2100 Bioanalyzer Instrument. Libraries with distinct TruSeq RNA UD Indexes (Cat#20022371, IDT for Illumina) were multiplexed and sequenced for 51 bp in paired-end mode, with a coverage of 60*10ˆ6 reads, using the NovaSeq™6000 sequencer (Illumina, Inc). Quality-control-checks on raw sequences was done using the tool FastQC (Babraham Institute). Samples were then aligned on the murine reference genome (Mus Musculus UCSC, mm10), using TopHat software [65], which implements the Bowtie2 aligner [66]. Mapped sequences were processed with HTSeq software [67] to count reads per gene, as a first raw measure of gene expression levels. Pairwise differential gene expression analysis was performed with the R package edgeR [68], using exact test for negative binomial distribution on TMM-normalized gene counts.
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA; [30]) was performed using the GSEA v4.2.3 Mac App, available at MSigDB (Broad Institute) website. Briefly, the average number of Counts Per Million mapped reads (AvgCPM) was computed for each gene, for each condition (three replicates per condition); only genes with AvgCPM > 0 in all conditions were considered for further analysis. GSEA was then performed on a list of 16,150 genes whose expression levels were indicated with CPMs, remapping mouse gene symbols to human orthologs. Results presented in Figs. 3C, 6B and S3B, C were obtained by comparing p66Shc−/− aged replicates vs WT aged replicates with gene-sets listed in Dataset 1. Gene-sets were chosen among those available at MSigDB collections, for being involved in cellular senescence or growth, oxidative stress response, p53 signaling pathway and adult MaSCs. The seven gene sets listed below:
STINGL_MAMMARY_STEM_CELL_UP [31]
STINGL_MAMMARY_STEM_CELL_DN [31]
SOADY_MAMMARY_STEM_CELL_UP [33]
SOADY_MAMMARY_STEM_CELL_DN [33]
BACH_MAMMARY_STEM_CELL_UP [34]
BACH_MAMMARY_STEM_CELL_DN [34]
SUN_MAMMARY_STEM_CELL_UP [35]
were manually curated starting from the supplemental material provided by the Authors of the cited articles, to generate lists of genes UP or DN in murine MaSCs, purified according to different methods. To integrate these sets with the others downloaded from MSigDB, mouse gene symbols were converted to human orthologs using the g:Orth (Orthology Research) tool [69].
Heat maps
Heat maps were generated using Prism v9 Software for macOS (GraphPad, San Diego, CA, USA). Averages of normalized gene expression levels [Norm(CPM), CPM = Counts Per Million mapped reads] were used to build matrixes represented as heat maps in Figs. 3D and 6C. Genes were manually clustered to prioritize mammary stem cell-genes UP in aged p66Shc−/− vs WT mammospheres (Fig. 3D, Dataset 2) or, vice versa, cell-polarity genes DN in aged p66Shc−/− vs WT mammospheres (Fig. 6C, Dataset 3). Datasets 2, 3 were generated starting from differentially expressed genes between aged p66Shc−/− vs WT mammospheres, setting the significance threshold at false discovery rate (FDR) ≤ 0.05. Gene lists were then manually curated as follows.
Dataset 2: genes were categorized as “mammary stem cell-markers” if satisfying one of the two following criteria: (i) being present in at least 3 out of 5 datasets of genes UP in normal MaSCs [31–35] and/or (ii) being object of a functional study; a total of 38 genes met at least one of these requirements.
Dataset 3: genes were categorized as “cell polarity- or asymmetric division-markers” if object of a functional or descriptive study in vertebrates; a total of 36 genes met this condition.
All the papers used as references were cited with their respective Digital Object Identifier in Datasets 2, 3.
Limiting dilution heterochronic transplantations
Primary mammary epithelial cells or M2-mammospheres were mechanically dissociated to obtain single-cell suspensions and diluted in PBS at different concentrations, to inject identical volumes into cleared inguinal fat-pads of syngeneic 3-week-old female recipient mice. The surgical procedure on recipient mice was done as previously described [70]. Recipient mice were euthanized 8 weeks later and outgrowths were observed by whole-mount carmine-staining, as described above. Outgrowth extensions were quantified as percentage of fat-pad area repopulated by transplanted cells. Outgrowths repopulating <15% were considered as unsuccessful engraftment. Aged donor mice were 12-month-old, while young donor mice were 2-month-old. Amounts of transplanted cells, as numbers of injected mice and numbers of outgrowths observed are indicated in Table 1 and S1. In experiments presented in Figs. 4A, B and S4, outgrowths were categorized according to the percentage of repopulated area, as indicated in Table S1. In the same experiment, the putative number of injected MaSCs was retrospectively calculated as the ratio of injected cells to the estimated MaSC frequencies in young or aged mice. Limiting-dilution transplantation data were analyzed with Extreme_Limiting_Dilution_Analysis (ELDA) software [71]. To estimate MaSC regenerative potential, only outgrowths obtained with ∼1 MaSC were considered: in detail, for young mice of both the genotypes, injection of 500, 100, and 50 cells resulted as extreme dilutions, i.e., when ∼1 MaSC was transplanted in case of successful outgrowth. For WT aged mice, same conditions were obtained with 2000, 1000, 500, 100, and 50 injected cells and for p66Shc−/− aged mice with 1000, 500, 100, and 50 injected cells. Outgrowths obtained with ∼1 MaSC were categorized as filling <25%, 25–60% or >60% of recipient’s fat-pad area. Differences in regenerative potential between genotypes or between ages were calculated pairwise by two-sided Chi-square test.
Mammosphere serial re-plating
Mammary glands were extracted and pooled from ≥3 mice and processed as described above. Serial re-plating experiments were performed seeding 5000 cells from M1-spheroids in 10% MethoCult™ (Cat#M3236, StemCell™ Technologies, Vancouver, BC, Canada). After 7 days, M2-spheroids were counted using an EVOS® FL Cell Imaging System (Thermo Fisher Scientific, Waltham, MA, USA) and then collected, dissociated and re-plated. This procedure was repeated every 7 days until ∼0 mammospheres were detected. Biological replicates (n) indicated in Fig. 4 legend are intended as independent serial re-plating experiments performed in three biological replicates (three primary mammary epithelial cell preparations), each of them plated in three technical replicates. Sphere forming efficiency (SFE) was calculated at each passage (M) as follows:
where no. of plated cells was 5000 at each passage. In these conditions, SFE data fitted a curve of the type:
where k represents the growth rate (GR), which is constant at each passage. Since cultures of both WT and p66Shc−/− mammospheres underwent progressive exhaustion and k resulted <0, GR was expressed, more intuitively, as DR, which represents the percentage of spheres lost at each passage. DR was defined as follows:
In Fig. 4C, data were represented in semi-logarithmic scale and, as a consequence, the exponential regression curve was represented as:
where k became the slope of the line. Differences between WT and p66Shc−/− mammosphere exhaustion-rates were analyzed by testing whether the slopes of the fitting lines were different. Differences between WT and p66Shc−/− SFEs at each passage were instead calculated by ordinary two-way ANOVA with main effects only (“genotype” and “passage”), followed by Tukey’s multiple comparison test with individual variances computed for each comparison. Data representation is detailed in Fig. 4 legend.
Immunofluorescence on cytokinetic cell-doublets
Mammospheres were obtained from young (2-month-old) or aged (12-month-old) WT or p66Shc−/− mice. At passage 2, spheroids were mechanically dissociated to obtain single-cell suspensions and cells were plated in 50% MethoCult™ (Cat#M3236, StemCell™ Technologies). After ON incubation, cells were checked for doublet-formation, collected by gentle centrifugation, fixed for 10′ in 2% para-formaldehyde and moved onto poly-D-lysine-coated glass slides (Cat#P6407, Merck). Cells were then permeabilized for 10′ in 0.1% Triton X-100 in PBS, blocked for 30′ in 2% BSA and stained ON at 4 °C with specific primary antibodies (Table S4; anti-Numb was gifted by Ivan Colaluca [72]) and for 1 h at RT with secondary antibodies (Table S4) and, finally, for 5′ with 4 µg/ml DAPI (Cat#32670, Merck). To select only cell-doublets with bright fluorescent emission (i.e., dividing MaSCs), we used Fiji [62] to calculate mean fluorescence of both the cytokinetic cell-doublet and the background. When at least one of the two cells resulted ∼1000 times brighter than the background, the cell-doublet was scored as asymmetric (when one cell was evidently brighter than the other) or symmetric (when fluorescence levels of both the cells were similar). In experiment presented in Fig. S6A, C, cells were previously infected with lentiviruses at M1-passage, as described below. Images presented in Figs. 5A, S5 and S6A, C were collected by wide-field microscopy (Table S5). Sample size and data representation are detailed in Figs. 5, S5 and S6 legends. Differences in frequency of asymmetric or symmetric cell division were analyzed pairwise between groups by two-sided Chi-square test with Yates’ correction for continuity. Only comparisons yielding a statistically significant difference (P < 0.05) were indicated in figures.
Lentiviral constructs
Re-expression of p66Shc:GFP in p66Shc−/− mammospheres (Fig. 5C, lower-right panel) and overexpression of p44:GFP in both WT and p66Shc−/− primary mammary epithelial cells or mammospheres (Figs. 7 and S7) were obtained by sub-cloning the C-terminally GFP-tagged murine p66Shc or p44 cDNAs into pLVX-Puro lentiviral vector (Cat#632164, Takara Bio, Mountain View, CA, USA). Silencing of endogenous p66Shc expression in WT mammospheres (Fig. 5C, lower-left panel) was obtained by cloning a p66Shc-specific shRNA oligonucleotide (5′-TGAGTCTCTGTCATCGCTG-3′) into pRSI17-U6-sh-UbiC-TagGFP2-2A-Puro lentiviral vector (Cat#SVSHU617-L, Cellecta, Mountain View, CA, USA), according to manufacturer’s instructions. Overexpression of mInsc in WT or p66Shc−/− mammospheres (Figs. 6F, G and S6A, B) was obtained by sub-cloning human mInsc cDNA into pCDH-CMV-MCS-EF1α-copGFP lentiviral vector (Cat#CD511B-1, System Biosciences, Palo Alto, CA, USA), as previously described [41]. Silencing of endogenous mInsc expression in mammospheres from aged WT mice (Fig. S6C, D) was obtained using two mInsc-specific pGIPZ™ shRNA lentiviral constructs, designed by Horizon Discovery (Cambridge, UK) and provided as bacterial stabs. Plasmids were recovered, amplified and used to produce lentiviral particles according to manufacturer’s protocols.
Lentiviral infections
Lentiviral supernatants were obtained by transfecting plasmids into HEK293T cells (Cat#CRL-3216, ATCC®, Manassas, VA, USA), using standard calcium phosphate method. After transfection, HEK293T cells were cultured in MEBM (Cat#CC-3151, Lonza, Basel, BS, Switzerland) for 36 h; supernatant was then filtered and supplemented with other components of stem cell medium and 10 µg/ml Polybrene (Cat#TR1003, Merck). Primary mammary epithelial cells or mammospheres were dissociated thoroughly and plated at a density of 50,000/ml in viral supernatant for 24 h. Cells were then collected and resuspended in freshly prepared viral supernatant for a second infection cycle of 12–24 h, at the end of which primary cells were plated in stem cell medium for 48 h before being collected for further applications. M1-mammosphere cells, instead, were stained for 5′ on ice with 0.5 μM PKH26 (Cat#PKH26GL, Merck – final concentration adjusted to maximize cell viability) and cultured until M2-spheroid formation.
Time-lapse microscopy
M2-mammospheres were dissociated thoroughly and 5000 cells/300 µl were plated on a glass-bottom six-multiwell dish (Cat#P06G-1.0-10-F, MatTek Life Sciences, Ashland, MA, USA) in 80% MethoCult™ (Cat#M3236, StemCell™ Technologies). After ON incubation, cells were observed by time-lapse microscopy for 7 days, collecting auto-focused images every 1.5 h in both green (FITC) and red (TRITC) fluorescence and DIC (Differential Interference Contrast). At least two independent replicates (i.e., viral supernatants and cells prepared at different times) were performed for each of the presented time-lapse experiment; different instruments were used for acquisition (Table S5). Analysis was done with Fiji [62]: briefly, only cells with bright red fluorescence emission were selected (i.e., MaSCs, according to their lower proliferation rate and thus higher PKH26 label-retention), by comparing at t = 1 (i.e., initial time-point) cell mean fluorescence with background fluorescence. Only cells with mean fluorescence intensity ∼500 times brighter than the background were considered for further analysis. Cells with green mean fluorescence brighter than the background—regardless of its intensity—were all scored as GFP+.
Modality of division and proliferation rate of mammary stem cells
The modality of division was retrospectively assigned, as previously described [27, 42]. Rounds of mitotic divisions entered by PKH+GFP+ cells were scored on the basis of both the modality of division and the number of cells present at late time-points: i.e., a group of seven cells was categorized as deriving from four rounds of division when the modality was asymmetric, or from three rounds when the modality was symmetric. In Fig. 5E and Table S2, rounds of division were categorized as 2, 3 or >3. In Fig. 7C, also cells entering only one round of division were considered, since this was the prevalent category of one of the analyzed groups. Sample size (i.e., number of PKH+GFP+ cells) and data representations are detailed in Figs. 5 and 7 legends. Differences in frequencies of asymmetric or symmetric cell division were analyzed pairwise by two-sided Chi-square test with Yates’ correction for continuity. Differences in frequencies of rounds of divisions were analyzed pairwise by uncorrected two-sided Chi-square test. Only comparisons yielding a statistically significant difference (P < 0.05) were indicated in figures.
Quantitative RT-PCR
Relative mRNA expression levels were estimated using a custom TaqMan® Array Card (Thermo Fisher Scientific, Table S6), designed to perform simultaneous qRT-PCRs of chosen genes on multiple samples. qRT-PCRs on M2-mammospheres shown in Figs. 3B, 6D, E and Table S3 were performed in three technical replicates on at least three independent mammosphere preparations for each group. When n > 3, qRT-PCRs were repeated on biological replicates using single-gene TaqMan® assays. Data representation is detailed in Figs. 3 and 6 legends, while sample size (i.e., independent mammosphere preparation of at least three mice each) is detailed in Table S3. Data were analyzed with the 2−∆∆Ct method, as described [73]. Statistical analysis of qRT-PCRs obtained with the TaqMan® Array Card was performed pairwise between experimental groups on the entire set of data (Table S3) using multiple t-test with a single pooled variance, correcting for multiple comparisons with the two-stage linear step-up method of Benjamini, Krieger and Yekutieli, with FDR set to 5%. Only comparisons yielding a statistically significant difference (Q < 0.05) were indicated in figures. Differences in mInsc expression upon p44-overexpression (Fig. 7B) were analyzed by ordinary two-way ANOVA followed by Tukey’s multiple comparison test.
Western blotting
Total proteins were extracted from primary mammary epithelial cells or M2-mammospheres (at least 500,000 cells/sample) with RIPA buffer, supplemented with protease and phosphatase inhibitor cocktails [cOmplete™, Cat#11697498001 (Roche, Basel, BS, Switzerland) and PhosSTOP™, Cat#4906845001, (Roche)]. Total extracts were quantified by Pierce™ BCA Protein Assay Kit (Cat#23227, Thermo Fisher Scientific) and stored at −80 °C in SDS-containing Laemmli sample buffer, according to standard protocols and manufacturer’s instructions. Protein levels in Figs. 6A and S7A were analyzed on primary cells from 3 young (2-month-old) or aged (12-month-old) WT or p66Shc−/− mice, or lentivirus-infected primary cells from pools of at least three young WT or p66Shc−/− mice, respectively. For western blot analysis, at least 40 µg of total protein extracts were separated on a precast gradient gel (Cat#5678084, Bio-Rad Laboratories, Hercules, CA, USA) and transferred on a 0.2 µm nitrocellulose membrane (Cat#1704159, Bio-Rad Laboratories) using a Trans-Blot® Turbo™ Transfer System (Cat#1704150, Bio-Rad Laboratories), according to manufacturer’s instructions. Blocking was done in 5% non-fat milk, 1 h at RT. Primary antibodies (Table S4 were incubated ON at 4 °C. Secondary antibodies (Table S4) were incubated for 1 h at RT. Proteins of interest were then detected by chemiluminescent reaction (Cat#1705062, Bio-Rad Laboratories,) using ChemiDoc XRS+ and Image Lab software (Bio-Rad Laboratories). Uncropped western blot images are available as Original Data in the Supplemental Material.
Immunoprecipitation
Levels of p66Shc phosphorylation upon p44-overexpression (Fig. S7B) were determined after immunoprecipitating total protein extracts obtained from lentivirus-infected primary mammary epithelial cells, prepared from pools of at least three young WT or p66Shc−/− mice. Briefly, 100 µg of protein extracts were incubated ON at 4 °C with 0.25 µg of anti-Shc1 antibody (Cat#610081, BD Biosciences), under gentle rotation. Protein extracts were then incubated with 100 µl of Protein A-Sepharose bead slurry (Cat#GE17-0780-01, Merck) for 4 h at 4 °C, under gentle rotation. Beads were then washed extensively with RIPA buffer, supplemented with protease and phosphatase inhibitor cocktails. The antigen-antibody complex was eluted by boiling pelleted beads for 5′ in 2x SDS-containing Laemmli sample buffer. Samples were then analyzed by western blotting as described above.
Statistical analyses
All the measurements employed in this study are from independent samples. Variance between samples within the same experiment was similar, except for percentages of hyperplastic ducts in aged mice (Fig. 2C), which were therefore analyzed by a non-parametric test. Sample size (n), estimate of variation (SD or SEM) and age of the animals at euthanasia are noted in figure legends or below the bar-graphs, as in case of single-cell imaging experiments. No data were excluded in descriptive statistics or statistical test computation. Except for histopathological analysis on FFPE MG (Fig. 2A) and immunohistochemistry analysis of proliferating cells (Figs. 2E and S1C), which were performed in blind by an expert pathologist, other experiments were not analyzed in blind. All the statistical analyses presented in this study, except for RNA-sequencing, gene-set enrichment and mammary stem cell frequency analyses (computed with edgeR [68], GSEA [30] and ELDA [71], respectively), were done in Prism v9 software, performing tests always as two-tailed and correcting for multiplicity or continuity when needed. P < 0.05 and Q < 0.05 were considered as thresholds for statistical significance.
Supplementary information
Acknowledgements
We are grateful to Pier Paolo Di Fiore, Ivan Colaluca, Andrea Basile and Maria Grazia Filippone for sharing protocols and reagents, to Simona Rodighiero for her invaluable assistance with imaging experiments, to Chiara Soriani and Stefano Freddi for advice on image analysis, to Salvatore Pece, Francesca Montani, Micol Tillhon and Giovanna Jodice for assistance with histopathological analyses, to Giorgio Pacecchi for technical help, to Stefania Averaimo for reading the paper and to Cristina Bonvicini for secretarial work. We thank the IEO Genomic Unit and the Cogentech Mouse Genetics and Quantitative PCR facilities.
Author contributions
EM and PGP conceptualized and supervised the research project; CP, DM, GDM, and FR performed all the experiments; GBe curated histopathological analysis, GBi contributed to statistical analyses, AP and SR to FACS analyses, LL to bioinformatic analyses, VG, MM, and MG to reagents and interpretation of results. CP, EM, and PGP analyzed the data and wrote the paper.
Funding
European Research Council: 2013-ADG_341131; Fondazione Cariplo: 2016-1031; Associazione Italiana per la Ricerca sul Cancro: AIRC-IG-2014-20162 and AIRC-IG-2017-20162. CP was recipient of a FIRC (Fondazione Italiana per la Ricerca sul Cancro) fellowship and DM of a FUV (Fondazione Umberto Veronesi) fellowship.
Data availability
RNA-sequencing data have been deposited at NCBI Gene Expression Omnibus [GSE201253]. The Authors declare that all the data supporting their study are available within the paper or in its Supplemental Material file.
Competing interests
The authors declare no competing interests.
Ethics approval
All the procedures on mice have been communicated to and approved by the Italian Ministry of Health (Project number 603/2016-PR).
Footnotes
Edited by: G Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chiara Priami, Daniela Montariello.
Contributor Information
Enrica Migliaccio, Email: enrica.migliaccio@ieo.it.
Pier Giuseppe Pelicci, Email: piergiuseppe.pelicci@ieo.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01029-5.
References
- 1.Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nat Cell Biol. 2011;13:506–12.. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Gotz M, et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–56. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Hwang AB, Brack AS. Muscle Stem Cells and Aging. Curr Top Dev Biol. 2018;126:299–322. doi: 10.1016/bs.ctdb.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Gross KM, Zhou W, Breindel JL, Ouyang J, Jin DX, Sokol ES, et al. Loss of Slug Compromises DNA Damage Repair and Accelerates Stem Cell Aging in Mammary Epithelium. Cell Rep. 2019;28:394–407.e6. doi: 10.1016/j.celrep.2019.06.043. [DOI] [PubMed] [Google Scholar]
- 5.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–52.. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–6. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 7.Bazarov AV, Lee WJ, Bazarov I, Bosire M, Hines WC, Stankovich B, et al. The specific role of pRb in p16 (INK4A) -mediated arrest of normal and malignant human breast cells. Cell Cycle. 2012;11:1008–13. doi: 10.4161/cc.11.5.19492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 9.Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–19. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatza CE, Dumble M, Kittrell F, Edwards DG, Dearth RK, Lee AV, et al. Altered mammary gland development in the p53+/m mouse, a model of accelerated aging. Dev Biol. 2008;313:130–41.. doi: 10.1016/j.ydbio.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santoro A, Vlachou T, Carminati M, Pelicci PG, Mapelli M. Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep. 2016;17:1700–20.. doi: 10.15252/embr.201643021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bast L, Calzolari F, Strasser MK, Hasenauer J, Theis FJ, Ninkovic J, et al. Increasing Neural Stem Cell Division Asymmetry and Quiescence Are Predicted to Contribute to the Age-Related Decline in Neurogenesis. Cell Rep. 2018;25:3231–40.e8. doi: 10.1016/j.celrep.2018.11.088. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura H, Liu N, Nanba D, Ichinose S, Takada A, Kurata S, et al. Distinct types of stem cell divisions determine organ regeneration and aging in hair follicles. Nat Aging. 2021;1:190–204. doi: 10.1038/s43587-021-00033-7. [DOI] [PubMed] [Google Scholar]
- 14.Florian MC, Klose M, Sacma M, Jablanovic J, Knudson L, Nattamai KJ, et al. Aging alters the epigenetic asymmetry of HSC division. PLoS Biol. 2018;16:e2003389. doi: 10.1371/journal.pbio.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charruyer A, Weisenberger T, Li H, Khalifa A, Schroeder AW, Belzer A, et al. Decreased p53 is associated with a decline in asymmetric stem cell self-renewal in aged human epidermis. Aging Cell. 2021;20:e13310. doi: 10.1111/acel.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–13. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 17.Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–33. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Mir HA, Ali R, Mushtaq U, Khanday FA. Structure-functional implications of longevity protein p66Shc in health and disease. Ageing Res Rev. 2020;63:101139. doi: 10.1016/j.arr.2020.101139. [DOI] [PubMed] [Google Scholar]
- 19.Berniakovich I, Trinei M, Stendardo M, Migliaccio E, Minucci S, Bernardi P, et al. p66Shc-generated oxidative signal promotes fat accumulation. J Biol Chem. 2008;283:34283–93. doi: 10.1074/jbc.M804362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spescha RD, Glanzmann M, Simic B, Witassek F, Keller S, Akhmedov A, et al. Adaptor protein p66(Shc) mediates hypertension-associated, cyclic stretch-dependent, endothelial damage. Hypertension. 2014;64:347–53. doi: 10.1161/HYPERTENSIONAHA.113.02129. [DOI] [PubMed] [Google Scholar]
- 21.Carpi A, Menabo R, Kaludercic N, Pelicci P, Di Lisa F, Giorgio M. The cardioprotective effects elicited by p66(Shc) ablation demonstrate the crucial role of mitochondrial ROS formation in ischemia/reperfusion injury. Biochim Biophys Acta. 2009;1787:774–80. doi: 10.1016/j.bbabio.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Albiero M, Ciciliot S, Tedesco S, Menegazzo L, D’Anna M, Scattolini V, et al. Diabetes-Associated Myelopoiesis Drives Stem Cell Mobilopathy Through an OSM-p66Shc Signaling Pathway. Diabetes. 2019;68:1303–14.. doi: 10.2337/db19-0080. [DOI] [PubMed] [Google Scholar]
- 23.Radisky DC, Hartmann LC. Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia. 2009;14:181–91.. doi: 10.1007/s10911-009-9123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltrami E, Valtorta S, Moresco R, Marcu R, Belloli S, Fassina A, et al. The p53-p66Shc apoptotic pathway is dispensable for tumor suppression whereas the p66Shc-generated oxidative stress initiates tumorigenesis. Curr Pharm Des. 2013;19:2708–14. doi: 10.2174/1381612811319150005. [DOI] [PubMed] [Google Scholar]
- 25.Smalley MJ, Kendrick H, Sheridan JM, Regan JL, Prater MD, Lindeman GJ, et al. Isolation of mouse mammary epithelial subpopulations: a comparison of leading methods. J Mammary Gland Biol Neoplasia. 2012;17:91–7. doi: 10.1007/s10911-012-9257-1. [DOI] [PubMed] [Google Scholar]
- 26.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70.. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–95.. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–4. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67.. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 32.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soady KJ, Kendrick H, Gao Q, Tutt A, Zvelebil M, Ordonez LD, et al. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res. 2015;17:31. doi: 10.1186/s13058-015-0539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat Commun. 2017;8:2128. doi: 10.1038/s41467-017-02001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun H, Miao Z, Zhang X, Chan UI, Su SM, Guo S, et al. Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J Biol Chem. 2018;293:8315–29.. doi: 10.1074/jbc.RA118.002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen QH, Pervolarakis N, Blake K, Ma D, Davis RT, James N, et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat Commun. 2018;9:2028. doi: 10.1038/s41467-018-04334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Insinga A, Cicalese A, Faretta M, Gallo B, Albano L, Ronzoni S, et al. DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc Natl Acad Sci USA. 2013;110:3931–6. doi: 10.1073/pnas.1213394110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santoro A, Vlachou T, Luzi L, Melloni G, Mazzarella L, D’Elia E, et al. p53 Loss in Breast Cancer Leads to Myc Activation, Increased Cell Plasticity, and Expression of a Mitotic Signature with Prognostic Value. Cell Rep. 2019;26:624–38.e8. doi: 10.1016/j.celrep.2018.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 41.Culurgioni S, Mari S, Bonetti P, Gallini S, Bonetto G, Brennich M, et al. Insc:LGN tetramers promote asymmetric divisions of mammary stem cells. Nat Commun. 2018;9:1025. doi: 10.1038/s41467-018-03343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tosoni D, Zecchini S, Coazzoli M, Colaluca I, Mazzarol G, Rubio A, et al. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. J Cell Biol. 2015;211:845–62. doi: 10.1083/jcb.201505037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gambino V, De Michele G, Venezia O, Migliaccio P, Dall’Olio V, Bernard L, et al. Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell. 2013;12:435–45. doi: 10.1111/acel.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 45.Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–77.. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goto GH, Mishra A, Abdulle R, Slaughter CA, Kitagawa K. Bub1-mediated adaptation of the spindle checkpoint. PLoS Genet. 2011;7:e1001282. doi: 10.1371/journal.pgen.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–5. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- 48.Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias MM, Marullo S, et al. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol Cell. 2010;38:78–88. doi: 10.1016/j.molcel.2010.01.041. [DOI] [PubMed] [Google Scholar]
- 49.Pehar M, Ko MH, Li M, Scrable H, Puglielli L. P44, the ‘longevity-assurance’ isoform of P53, regulates tau phosphorylation and is activated in an age-dependent fashion. Aging Cell. 2014;13:449–56.. doi: 10.1111/acel.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–60. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li CM, Shapiro H, Tsiobikas C, Selfors LM, Chen H, Rosenbluth J, et al. Aging-Associated Alterations in Mammary Epithelia and Stroma Revealed by Single-Cell RNA Sequencing. Cell Rep. 2020;33:108566. doi: 10.1016/j.celrep.2020.108566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomita K, Teratani T, Suzuki T, Oshikawa T, Yokoyama H, Shimamura K, et al. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J Hepatol. 2012;57:837–43.. doi: 10.1016/j.jhep.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Vashistha H, Singhal PC, Malhotra A, Husain M, Mathieson P, Saleem MA, et al. Null mutations at the p66 and bradykinin 2 receptor loci induce divergent phenotypes in the diabetic kidney. Am J Physiol Ren Physiol. 2012;303:F1629–40. doi: 10.1152/ajprenal.00246.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Qiu B, Li X, Zhang H, Liu W. p53-p66(shc)/miR-21-Sod2 signaling is critical for the inhibitory effect of betulinic acid on hepatocellular carcinoma. Toxicol Lett. 2015;238:1–10. doi: 10.1016/j.toxlet.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 55.Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–63.. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 56.Khalid S, Drasche A, Thurner M, Hermann M, Ashraf MI, Fresser F, et al. cJun N-terminal kinase (JNK) phosphorylation of serine 36 is critical for p66Shc activation. Sci Rep. 2016;6:20930. doi: 10.1038/srep20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Candeias MM, Powell DJ, Roubalova E, Apcher S, Bourougaa K, Vojtesek B, et al. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene. 2006;25:6936–47.. doi: 10.1038/sj.onc.1209996. [DOI] [PubMed] [Google Scholar]
- 58.Postiglione MP, Juschke C, Xie Y, Haas GA, Charalambous C, Knoblich JA. Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron. 2011;72:269–84. doi: 10.1016/j.neuron.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zigman M, Cayouette M, Charalambous C, Schleiffer A, Hoeller O, Dunican D, et al. Mammalian inscuteable regulates spindle orientation and cell fate in the developing retina. Neuron. 2005;48:539–45. doi: 10.1016/j.neuron.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 60.Poulson ND, Lechler T. Robust control of mitotic spindle orientation in the developing epidermis. J Cell Biol. 2010;191:915–22. doi: 10.1083/jcb.201008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi T, Dansen TB. Reactive Oxygen Species Induced p53 Activation: DNA Damage, Redox Signaling, or Both? Antioxid Redox Signal. 2020;33:839–59.. doi: 10.1089/ars.2020.8074. [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanko JP, Easterling MR, Fenton SE. Application of Sholl analysis to quantify changes in growth and development in rat mammary gland whole mounts. Reprod Toxicol. 2015;54:129–35. doi: 10.1016/j.reprotox.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson JF, Felicio LS, Randall PK, Sims C, Finch CE. A longitudinal study of estrous cyclicity in aging C57BL/6J mice: I. Cycle frequency, length and vaginal cytology. Biol Reprod. 1982;27:327–39.. doi: 10.1095/biolreprod27.2.327. [DOI] [PubMed] [Google Scholar]
- 65.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–8.. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faraldo MM, Glukhova MA, Deugnier MA. The transplantation of mouse mammary epithelial cells into cleared mammary fat pads. Methods Mol Biol. 2015;1293:161–72. doi: 10.1007/978-1-4939-2519-3_9. [DOI] [PubMed] [Google Scholar]
- 71.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 72.Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data have been deposited at NCBI Gene Expression Omnibus [GSE201253]. The Authors declare that all the data supporting their study are available within the paper or in its Supplemental Material file.