Abstract
Microbes are by far the dominant biomass in the world’s oceans and drive biogeochemical cycles that are critical to life on Earth. The composition of marine microbial communities is highly dynamic, spatially and temporally, with consequent effects on their functional roles. In part, these changes in composition result from viral lysis, which is taxon-specific and estimated to account for about half of marine microbial mortality. Here, we show that extracellular ribosomal RNA (rRNAext) is produced by viral lysis, and that specific lysed populations can be identified by sequencing rRNAext recovered from seawater samples. In ten seawater samples collected at five depths between the surface and 265 m during and following a phytoplankton bloom, lysis was detected in about 15% of 16,946 prokaryotic taxa, identified from amplicon sequence variants (ASVs), with lysis occurring in up to 34% of taxa within a water sample. The ratio of rRNAext to cellular rRNA (rRNAcell) was used as an index of taxon-specific lysis, and revealed that higher relative lysis was most commonly associated with copiotrophic bacteria that were in relatively low abundance, such as those in the genera Escherichia and Shigella spp., as well as members of the Bacteriodetes; whereas, relatively low lysis was more common in taxa that are often relatively abundant, such as members of the Pelagibacterales (i.e., SAR11 clade), cyanobacteria in the genus Synechococcus, and members of the phylum Thaumarchaeota (synonym, Nitrososphaerota) that comprised about 13–15% of the 16 S rRNA gene sequences below 30 m. These results provide an explanation for the long-standing conundrum of why highly productive bacteria that are readily isolated from seawater are often in very low abundance. The ability to estimate taxon-specific cell lysis will help explore the distribution and abundance of microbial populations in nature.
Subject terms: Microbial ecology, Water microbiology
Introduction
Although microorganisms drive carbon and nutrient cycles in the world’s oceans, and constitute more than 90% of its living biomass, they have turnover times from hours to days [1]. A major contributor to such high turnover is cell lysis, particularly by viral infection that is estimated to kill about 20% of this biomass each day [2], thereby directing cellular organic matter into dissolved and particulate organic matter in a process termed the viral shunt [3, 4]. Moreover, viral infection is taxon-specific, contributing to bacterial communities that are highly dynamic in taxonomic composition [5]. Other processes, such as programmed cell death [6] or infection by predatory bacteria [7, 8], may also contribute to prokaryotic cell lysis, but have not been shown to contribute substantially to cell lysis in natural waters.
Despite the significance of cell lysis to marine ecosystem processes, differences in relative lysis among taxa are unknown, and estimates of daily viral lysis are bulk estimates that provide no information on how lysis is distributed among thousands of microbial taxa. Being able to resolve taxon-specific cell lysis in natural microbial communities is one of the most pressing questions in microbial ecology, and being able to identify taxa that are permissive or resistant to cell lysis will allow the exploration of fundamental paradigms in microbial ecology such as Kill the Winner [9], the Seed Bank Model [10], and the relationship between potential growth rate and susceptibility to viral infection [11]. For example, it has been argued that the most abundant taxa are largely resistant to viral infection, while rare species that are capable of fast growth are highly susceptible to viral lysis [11]. Moreover, such data will help inform the relationship between cell lysis and community structure in microbial communities.
This study was motivated by large amounts of ribosomal RNA (rRNA) “contamination” that was consistently present in 0.22-µm filtered seawater collected to investigate the diversity of RNA viruses. We wondered if the source of the extracellular rRNA (rRNAext) was cell lysis and, if so, could the rRNAext be sequenced to reveal the taxonomic composition of the cells that had died? The premise of the argument is simple: if the source of the rRNAext is cell lysis, then sequencing this RNA will reveal the cells from which it was derived, and thus those taxa in which lysis occurred. Preliminary investigations using quantitative reverse-transcription PCR (qRT-PCR) indicated that there were typically millions of copies of rRNAext in each mL of 0.22-µm-filtered seawater, and that it was stable for days when incubated on the benchtop. Moreover, it was possible to sequence the rRNAext and taxonomically profile the community from which it was derived.
Here, we detail and expand on these initial observations to show that rRNAext is produced by cell lysis of prokaryotes, and that by sequencing and quantifying rRNAext, cellular rRNA (rRNAcell), and the genes encoding rRNA (rRNAgenecell), we can estimate the taxon-specific cell lysis of prokaryotes in complex natural communities.
Results and discussion
Extracellular rRNA (rRNAext) is produced by cell lysis but not flagellate grazing
We hypothesized that extracellular ribonucleoprotein-bound rRNAs, including free ribosomes, are produced in seawater as the result of cell lysis. To test this hypothesis, we infected the marine heterotrophic proteobacterium, Vibrio natriegens strain PWH3a, with the lytic phage, PWH3a-P1 [12], and used qRT-PCR to quantify the rRNAext in 0.22-µm filtrate. About 1.75 h after adding viruses to a culture of V. natriegens PWH3a, the concentration of rRNAext was about 100-fold higher than in an uninfected control culture (Fig. 1A), confirming that viral lysis releases rRNAext. The rRNAext is likely in the form of ribosomes that have been shown by transmission electron microscopy to be released from marine cyanobacteria in the genus Synechococcus as the result of viral lysis [13]. The presence of rRNAext in the uninfected control likely resulted from prophage induction in a small proportion of the cells, as the genome of V. natriegens strain PWH3a has multiple prophage elements. Nevertheless, any process that causes cell lysis will release rRNA. For example, RNA can be released in vesicles by some bacteria [14], although we are unaware of data showing that vesicles are a source of “free” rRNA.
Fig. 1. Production and persistence of free extracellular rRNA (rRNAext) in water.
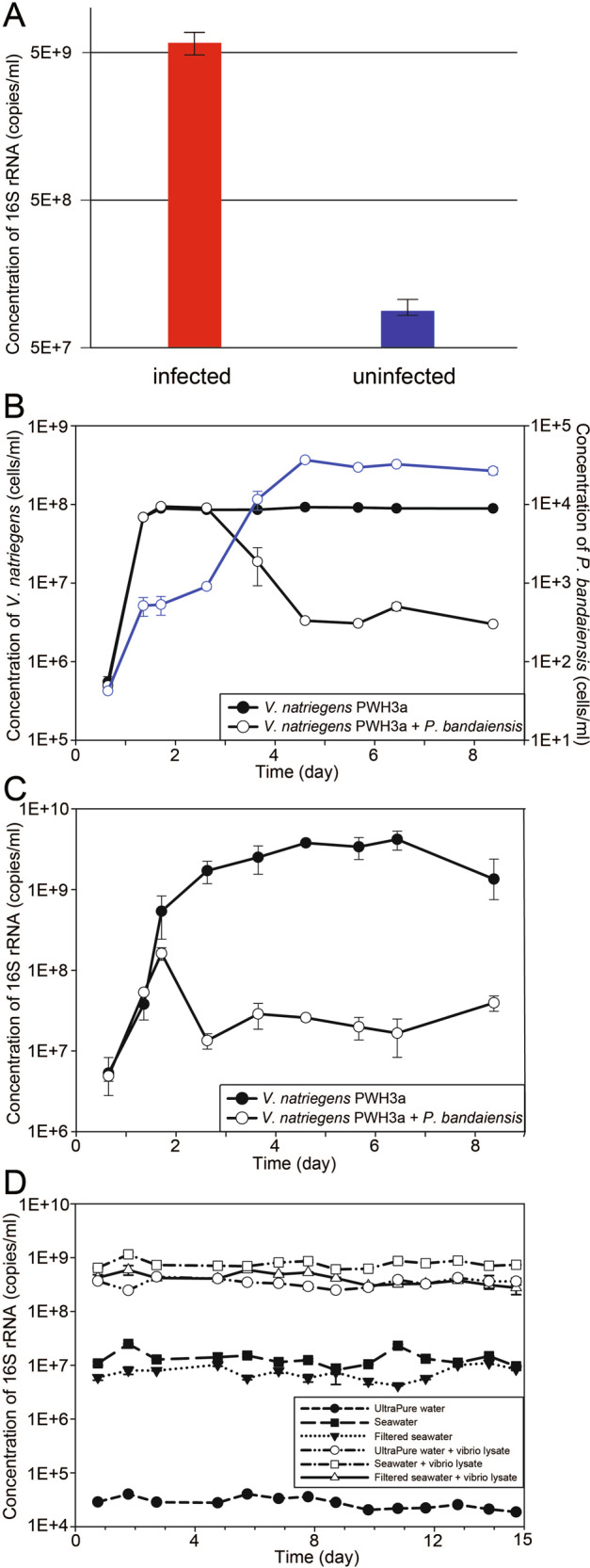
A Concentration of 16 S rRNAext in cultures of V. natriegens strain PWH3a, 1.75 h after the addition of the lytic phage PWH3a-P1 (red bars) and in control cultures to which the phage was not added (blue bars). Error bars represent the standard deviation of three biological replicates. B, C Bacteria, protists, and 16 S rRNAext concentrations in cultures of V. natriegens PWH3a, with or without the addition of the microflagellate grazer, Paraphysomonas bandaiensis. Panel (B) shows changes in the concentration of bacteria (black lines and left axis) and the grazer (blue line and right axis) during eight days of incubation. Panel (C) shows changes in the concentration of 16 S rRNAext, with and without the addition of the grazer. D 16 S rRNAext concentrations in ultrapure water, untreated seawater, and 0.22-µm-filtered seawater, measured over two weeks. Incubations were performed in the dark at 21 °C, with and without the addition of 0.22-µm-filtered V. natriegens PWH3a-P1 lysate (vibrio lysate).
Potentially, grazing by protists could also produce rRNAext. We tested this by adding the bacterivorous flagellate Paraphysiomonas bandaiensis to cultures of V. natriegens PWH3a and quantifying rRNAext (Fig. 1B, C). Rather than production, in the presence of grazers, the concentration of rRNAext decreased relative to that of controls, implying that grazers consume rRNAext (Fig. 1C). Although we only tested a single grazer-prey system, the decrease in rRNAext is consistent with observations that phagotrophic protists consume virus particles [15], which are in the same size range as ribosomes. Moreover, because protistan grazers consume bacterial prey in their entirety [16, 17], the release of bacterial rRNA was not a result of protistan grazing.
Stability of extracellular rRNA (rRNAext) in seawater
The concentration of rRNAext in seawater is a balance of production and removal. Thus, we tested the stability of rRNAext, produced by lysis of cultures of V. natriegens PWH3a with phage PWH3a-P1, by adding it to molecular-grade ultrapure water, untreated seawater, and 0.22-µm-filtered seawater. After at least ten days of incubation in the dark at 21 °C, there were no detectable changes in the concentrations of rRNAext in any of the treatments (Fig. 1C). Collectively, these data demonstrate that, even in the presence of the natural microbial community, rRNAext detected by qRT-PCR, is stable in seawater. This stability suggests that rRNAext is in the form of ribosome complexes that prevent degradation of the RNA, consistent with observations that rRNA complexed with ribonucleoproteins is protected from nucleases [18, 19].
The concentration of rRNAext was stable over at least 10 days in laboratory incubations; however, in situ turnover rates would be faster because of other factors contributing to the decay of rRNAext. For example, solar radiation damages organic molecules [20], viral infectivity [12, 21], and DNA [22]. Exposure to UV also produces hydroxide radicals that damage rRNA, and creates covalent crosslinks between rRNA and protein, and between proteins. Thus, the conformation of the ribosome complex is changed, making it easier for nucleases to access the ribonucleoprotein complex and degrade rRNA [23]. Studies have shown nuclease [24] and protease [25] activity in seawater. As a result, despite the stability of rRNAext in the laboratory, in situ turnover rates will be higher. Moreover, regardless of the decay mechanism of the rRNAext, there is no prior evidence suggesting that the decay rate would be taxon-specific.
Although we did not measure turnover rates of rRNAext in field samples, decay and production of rRNAext must be balanced, on average. Because viral lysis is estimated to kill about 20 to 30 % of the standing stock of bacteria in the oceans each day [11, 26], viral lysis likely accounts for most of the production of rRNAext. Consequently, in situ turnover rates of rRNAext should also be about 20–30% per day, similar to the turnover rates of bacteria imposed by viral lysis. Relatively high turnover rates are also consistent with the high temporal and spatial variability seen in the taxonomic profiles of rRNAext (Fig. 2). Below, we use rRNAext as a window to investigate taxon-specific lysis of prokaryotes in seawater.
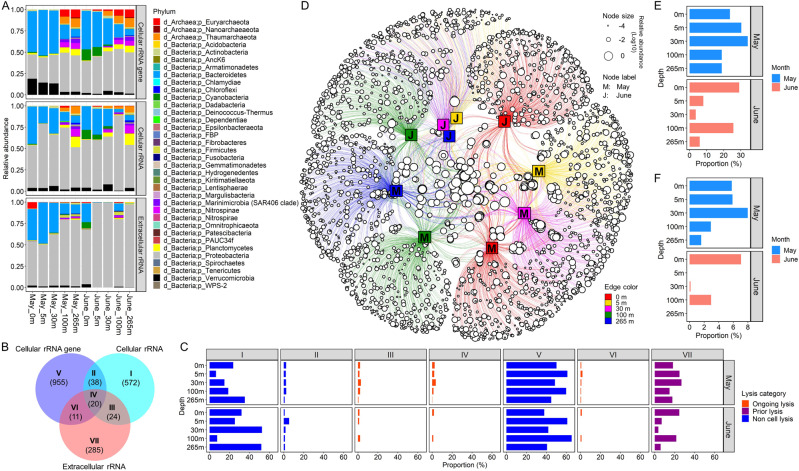
Fig. 2. Detection of cell lysis in coastal seawater as inferred from extracellular rRNA (rRNAext).
A Relative abundance of 16 S amplicons assigned to phyla based on amplicon sequence variants (ASVs) for cellular rRNAgene and rRNA (rRNAgenecell and rRNAcell, respectively) and rRNAext. B Venn diagram showing seven possible lysis groups, where I, V, and VII represent ASVs found in the rRNAgenecell, rRNAcell, and rRNAext pools, respectively; the overlap represents ASVs shared between groups. The numbers shown in parentheses represent the average number of ASVs detected for each lysis group across the ten seawater samples. C Each panel shows the proportion of ASVs in the rRNAgenecell, rRNAcell, and rRNAext pools across the seven lysis groups for each seawater sample. The orange bars represent taxa with ongoing lysis (Groups III, IV and VI) as indicated by the presence of the same ASVs in both the cellular rRNAgene and rRNA fractions, as well as in the extracellular rRNA fraction. The purple bars represent taxa with prior-lysis (group VII) as inferred from the presence of ASVs in the rRNAext pool, but which were undetectable in the rRNAgenecell or rRNAcell pools. The blue bars indicate taxa without cell lysis (group I, II, and V), as indicated by the absence of ASVs in the rRNAext fraction. D A network analysis showing the taxonomic distribution of ASVs detected in the extracellular rRNA, across seawater samples collected from five depths. Each open circle (circular node) represents a ribosomal ASV, while the size indicates its relative abundance; The line (edge) links ASVs to the seawater samples (square node) in which they were detected. The network shows that a high proportion of the taxa in which lysis was detected were relatively rare and confined to a single month and depth; whereas, there were relatively few taxa in which lysis was detected between months and across depths. E Proportion of all ASVs from each sample in the rRNAgenecell, rRNAcell, and rRNAext pools, in which rRNAext was detected, indicating cell lysis. F The proportion of ASVs detected in the rRNAgenecell pool from each sample in which rRNAext was also detected, consistent with ongoing lysis.
Cell lysis occurs across phyla
We analyzed seawater samples collected from five depths in the Strait of Georgia, British Columbia, Canada, during and after a phytoplankton bloom. To infer taxon-specific lysis, we used deep sequencing of a 412-bp amplicon to infer the taxonomic distribution of 16 S rRNAext, as well as rRNAcell, and rRNAgenecell. A total of 16,946 amplicon sequence variants (ASVs), belonging to 33 phyla of bacteria and three phyla of archaea (Fig. 2A) were detected across all ten seawater samples.
For each sample, we binned assigned taxa into seven different “lysis groups” based on their detection as rRNAcell, rRNAext, and rRNAgenecell (Fig. 2B), with their presence as rRNAext (Groups III, IV, VI, and VII) indicating lysis. Across samples, the taxa in which lysis was detected varied widely, ranging from 3.7% to 34.1% of taxa at 30 m in June and May, respectively (Fig. 2C–E). Lysis was associated with 28 bacterial and three archaeal phyla, including 19 phyla for which viruses have not been reported (i.e., Acidobacteria, AncK6, Armatimonadetes, Chloroflexi, Epsilonbacteria, FBP group, Fibrobacteres, Fusobacteria, Gemmatimonadetes, Hydrogenedentes, Kiritimatiellaeota, Lentisphaerae, Margulisbacteria, Nanoarchaeaeota, Nitrospinae, Planctomycetes, PAUC34f, Patescibacteria, and Spirochaetae) (Fig. S1). Moreover, the proportion of taxa within a phylum in which lysis occurred varied across phyla. In ten of the 31 phyla in which lysis occurred (i.e., Armatimonadetes, Chlamydiae, Cyanobacteria, Deinococcus-Thermus, FBP group, Fibrobacteres, Firmicutes, Fusobacteria, Nanoarchaeaeota, and Spirochaetae), more than half of the ASVs in those phyla were associated with cell lysis, and in some cases included all the taxa within a phylum (Fig. S1). Our results show that cell lysis occurred in taxa across a broad range of prokaryotic phyla; yet, for a given sample the maximum proportion of ASVs found in the rRNAext relative to the rRNAgenecell (Groups IV and V) was only ~8% (Fig. 2F), indicating that lysis was not detectable for the vast majority of taxa in the cellular fraction (i.e., rRNAgenecell). In fact, lysis was only detected for 14 of the 28 phyla identified from 16 S rRNA gene sequences in the cellular fraction, and within those phyla, lysis was detected for less than half of the ASVs, except for ASVs in the archaeal phylum Euryarchaeota (Fig. S2).
Lysis was detected in about 15% of the 16,946 ASVs, and of these most were only in the rRNAext fraction (Group VII; Fig. 2C), indicating that lysis likely occurred prior to sampling, because the corresponding sequences were not detected in either the rRNAgenecell or rRNAcell fractions. Ongoing or recent lysis, implied by the presence of ASVs in both the rRNAgenecell and rRNAext fractions (Groups IV and VI) was detected for 202 taxa, and within this group, 0.03% to 1.85% of the total taxa across samples were detected in the rRNAcell, rRNAext, and rRNAgenecell fractions (Group IV), making it possible to speculate on the relative mortality among taxa. Taxa that were detected in the rRNAgenecell and rRNAext pools (Group VI) occurred in 0.03% to 3.6% of taxa, implying lysis of taxa with little growth or activity at the time of sampling. For taxa in Groups I and III, rRNAcell was detected but rRNAgenecell was undetectable, suggesting they are active but in very low abundance, or dormant but harbour high numbers of ribosomes [27]. Among all taxa, up to 2.9% were in Group III, with detectable rRNAcell and rRNAext even though rRNAgenecell was undetectable, implying that these taxa were in very low abundance but undergoing lysis. The discrepancy in ASV numbers among the rRNAgenecell, rRNAcell, and rRNAext fractions is a reflection of the composition and activity of the microbial communities being highly dynamic, with consequent effects on the relative contribution of each taxon to each of the three pools.
Our observation that lysis was associated with specific ASVs is consistent with viral lysis, which is typically taxon-specific and would be expected to kill cells within a taxon, while leaving members of other taxa unaffected. These observations are congruent with the seed-bank theory of viral infection, in which only a small proportion of viruses are active at any given time [10]. Furthermore, as viral lysis is estimated to cause about 50% of marine prokaryotic mortality [28, 29] and kills about 20% of the standing stock each day [30], lysis must be high in taxa that are affected. Of the 31 phyla for which rRNAext was detected, there are only a few for which viruses have been isolated (Euryarchaeota, Thaumarchaeota (synonym, Nitrososphaerota), Proteobacteria (synonym, Pseudomonadota), Firmicutes (synonym, Bacillota), Bacteriodetes (Bacteriodota), Actinobacteria (synonym, Actinomycetota), Cyanobacteria, Chlamydiae (synonym, Chlamydiota), Tenericutes (synonym, Mycoplasmatota), and Deinococcus-Thermus (synonym, Deinococcota)) [31] or are known from single amplified genomes (Verrucomicrobia and Marinimicrobia) [32]. Thus, sequencing of rRNAext can also provide insights into taxa to be targeted for virus isolation. Although programmed cell death [6], predation by Bdellovibrio and like organisms (BALOs) [33], and bacteriocins [34, 35] can also cause lysis of prokaryotes, there is no evidence that they are a major source of prokaryotic mortality in the sea.
Cell lysis varies widely across taxa and seawater samples
We detected 2561 ASVs in the rRNAext pool from ten seawater samples, implying lysis in ~15.1% of the 16,946 ASVs detected across the rRNAcell, rRNAext, and rRNAgenecell fractions (Fig. 2). Overall, cell lysis was evident in members of the phyla Bacteroidetes (genera Aureispira, Lewinella, Marinoscillum, Fluviicola, Flavicella, Formosa, and marine groups NS4, NS5, and NS7), Cyanobacteria (genus Synechococcus), Firmicutes (genus Paenibacillus), Nitrospinae (members of the LS-NOB group), Proteobacteria (class Alphaproteobacteria: genera Methylobacterium, Ochrobactrum, Amylibacter, Planktomarina, Sulfitobacter, and clades PS1, SAR116, and SAR11 Ia; class Gammaproteobacteria: genera Alcaligenes, Delftia, Luminiphilus, Escherichia-Shigella, Acinetobacter, Pseudomonas, and Stenotrophomonas, and clades OM60 (NOR5) and SAR92), and Verrucomicrobiota (genus Roseibacillus) (Figs. 2A and 3A).
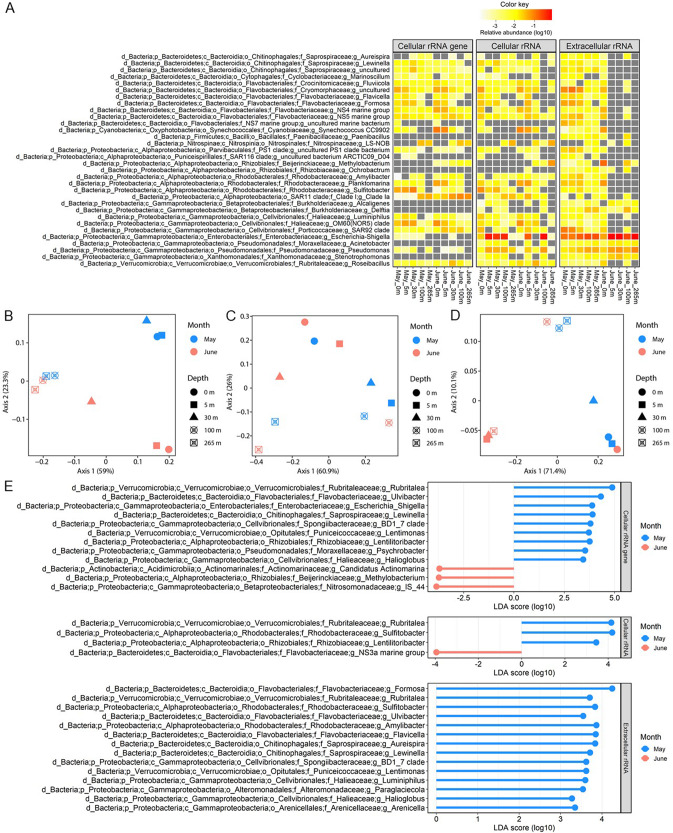
Fig. 3. Variability in the prokaryotic taxa in which lysis was detected in coastal seawater samples from the Strait of Georgia.
A Prokaryotic genera for which the relative abundance of rRNAext was >1% in at least one sample. The panels from left to right indicate the relative abundances of these genera in the 16 S rRNAgenecell, rRNAcell, and rRNAext pools, respectively. B–D Principal Coordinate Analyses (PCoA) of the community structure of the prokaryotic ASVs for 16 S rRNAgenecell (B), rRNAcell (C), and rRNAext (D). The PCoA ordinates the weighted Unifrac metrics that are based on the presence/absence and the relative abundance of ASVs detected in each fraction. E Differences in the relative abundance of taxa at the genus level between May and June revealed by Linear discriminant analysis Effect Size (LEfSe). Panels from top to bottom showed the data for the 16 S rRNAgenecell, rRNAcell, and rRNAext fractions, respectively. The absolute value of the logarithmic LDA score indicates the degree of difference in the relative abundance of taxa between the two months.
The taxonomic composition of the rRNAext varied greatly among seawater samples, implying different prokaryotic taxa were lysed across depths and times (Figs. 2D and 3). There was a significant difference in the taxonomic composition of the rRNAext between May and June (PERMANOVA: R2 = 0.24, p value = 0.027), but not across depths (PERMANOVA: R2 = 0.44, p value = 0.132). Linear discriminant analysis Effect Size (LEfSe) revealed differences in relative abundances of the rRNAext between May and June samples for the following taxa: members of the phylum Bacteroidetes (genera Aureispira, Lewinella, Flavicella, Formosa, and Ulvibacter), the classes Alphaproteobacteria (genera Amylibacter and Sulfitobacter) and Gammaproteobacteria (genera Luminiphilus, Paraglaciecola, Halioglobus and Arenicella, and the clade BD1-7), and the phylum Verrucomicrobia (genera Rubritalea and Lentimonas) (Fig. 3E), consistent with higher lysis of these taxa during the bloom-period in May. In contrast, there are no significant differences in the relative abundances of rRNAext taxa across depths using LEfSe.
Taxa with ongoing or recent lysis
Sequencing of the rRNAext reveals the taxa in which lysis has occurred; if the rRNAext is corrected for differences in “rRNA copy number per cell” that occur among taxa and with physiological state [36], the ratio of rRNAext to cellular rRNA (rRNAcell) provides an index of the relative amount of lysis in each taxon. Thus, a cell-lysis index (CLI) can be calculated for individual taxa from the ratio of the relative abundance of rRNAext to rRNAcell. A CLI > 1 indicates that there is more free rRNA in the water than within cells for a specific taxon; hence, the higher the CLI the greater the lysis relative to the size of the co-occurring population. If rRNAgenecell is undetectable for an ASV, it indicates the concentration of cells is below the detection limit, and is consistent with the lysis of the population. In contrast, if an ASV occurs in the rRNAext, rRNAcell, and rRNAgenecell pools (Group IV), this suggests that lysis of that taxon is ongoing, or has occurred very recently (Fig. 2B, C).
There were relatively few taxa in Group IV, and they were highly variable across samples. Of the 16,946 ASVs detected across all samples, the number of taxa in Group IV ranged between zero for the June 5- and 265-m sample and 46 for the surface sample in May (Fig. 4B). The CLI for these taxa ranged between 0.13 and 15.92, with the highest values for ASV in the genus Amylibacter from the Alphaproteobacteria family Rhodobacteraceae. Overall, we detected a relative high lysis index (CLI > 1) in members of the phylum Bacteroidetes (order Chitinophagales, Flavobacteriales, and Sphingobacteriales), Cyanobacteria (genus Synechococcus), Marinimicrobia, Nitrospinae (genus Nitrospina), Planctomycetes (genus Blastopirellula), Proteobacteria (family Beijerinckiaceae, Rhodobacteraceae, PS1 clade, SAR11 clade I, SAR86 clade, Halieaceae, Porticoccaceae, Enterobacteriaceae, and Woeseiaceae), and Verrucomicrobia (genus Roseibacillus and Rubritalea), which occurred mostly during the phytoplankton bloom in May (Fig. 4C). Many of the bacteria are putative copiotrophs (e.g., Bacteroidetes, Enterobacteriaceae, Rhodobacteraceae, and Verrucomicrobia) and known for breaking down high molecular weight dissolved organic material associated with phytoplankton blooms [37].
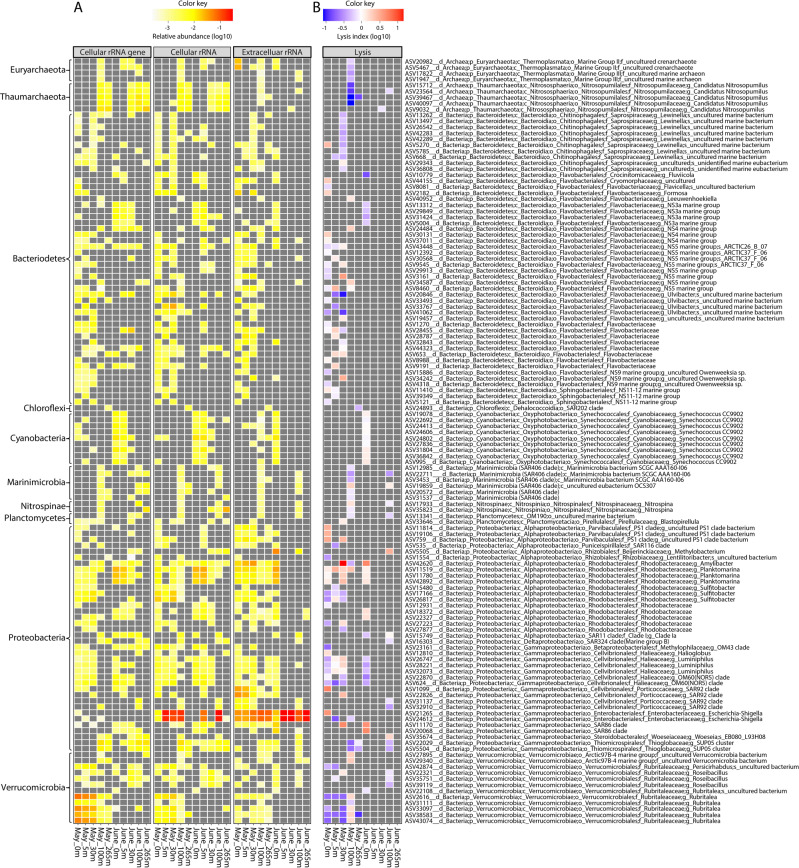
Fig. 4. Estimates of taxon-specific cell lysis in coastal seawater samples from the Strait of Georgia.
A Panels from left to right show the relative abundance of the 128 ASVs for which 16 S rRNAgenecell, rRNAcell, and rRNAext (Group IV) data were available. B The taxon-specific cell-lysis index (CLI) is the ratio of the relative abundance of rRNAext to rRNAcell, for each of these 128 ASVs. In the heatmap, grey indicates that a value required to make the calculation was undetectable. Red indicates that for the specific ASV the relative abundance of rRNAext was greater than for rRNAcell (CLI > 1), suggesting relatively high relative lysis. In contrast, blue indicates that the relative abundance of rRNAext is less than for rRNAcell (CLI < 1), suggesting relatively low relative lysis. White indicates that the relative abundance of rRNAext and rRNAcell were very similar (CLI = 1), suggesting an intermediate level of cell lysis.
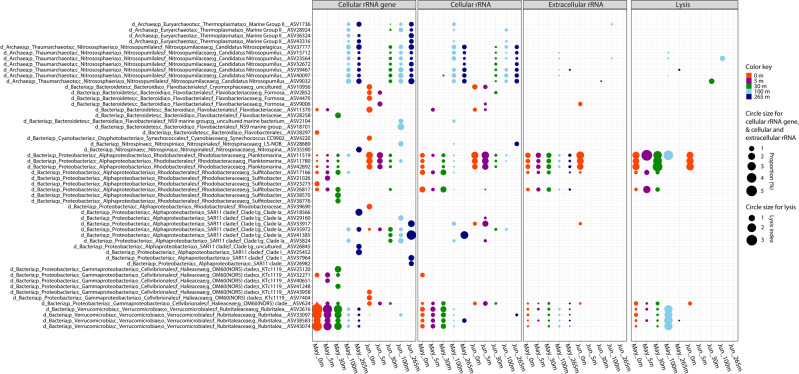
Cyanobacteria in the genus Synechococcus were relatively abundant, especially at 0 and 5 m in June (Fig. 1A); yet, only one of the 138 ASVs assigned to Synechococcus comprised >1% of the total rRNAgenecell, although we found no evidence of cell lysis for this taxon (Fig. 5; Fig. S3). In contrast, during June at 0 and 5 m, lysis was detected in nine of the less-abundant ASVs assigned to the genus Synechococcus (Fig. 4B; Fig. S3). These results are consistent with observations of high host specificity for most cyanophages [38, 39].
Fig. 5. Relative abundance of 16 S rRNAgenecell, rRNAcell, and rRNAext for different taxa and the taxon-specific cell-lysis index (CLI) for the dominant ASVs with a relative abundance of rRNAgenecell >1%.
The CLI is the ratio of the relative abundances of rRNAext to rRNAcell for each ASV. A CLI > 1 indicates that the relative abundance of rRNAext is greater than that for rRNAcell, and suggests relatively high lysis.
Only a small proportion of taxa were associated with ongoing or recent lysis (Groups III, IV, and VI); thus, most of the phyla represented in the rRNAext pool were the result of prior lysis (Fig. S1). It is noteworthy that rRNAext could be produced by microbial communities that were not sampled in our study, such as those particles-associated microbes that were removed by the 120-µM prefiltration step during seawater treatment. Yet, this will not affect the CLI estimation, because the 120-µM prefiltration would remove the rRNAgenecell and rRNAcell of those particle-associated microorganisms while leaving their free rRNAext detected in the rRNAext pool in case of cell lysis occurred.
High lysis is associated with low abundance
An enduring puzzle in marine microbial ecology is the relationship between viral infection and the structure of prokaryotic communities, and in particular the relationship between the abundance of specific taxa and viral lysis. It has been proposed that lysis rates are relatively low for cells in the most abundant taxa, and higher in relatively rare bacteria that are capable of fast growth [11]. This view is congruent with “Kill the Winner”, a model, in which viral lysis prevents the most ecologically “fit” cells from dominating the community [9, 40]. Conversely, cells associated with rare taxa may be subject to significant viral lysis [41].
Here, we examined taxa for which we could measure rRNAext, rRNAcell, and rRNAgenecell (Group IV), and demonstrate that the CLI was negatively correlated with relative abundance (Fig. 6), consistent with experimental evidence that rare groups of marine bacteria are more susceptible to virus-induced mortality [41]. For example, an ASV assigned to the phylum Proteobacteria in the family Rhodobacteraceae (genus Amylibacter) occurred in low abundance at 30 m in May (Fig. 4A), and yet the estimate of cell lysis was the highest measured (Fig. 4B). In contrast, ASVs assigned to the phylum Verrucomicrobia in the family Rubritaleaceae (genus Rubritalea) were the most abundant prokaryotes at 0–30 m, while being relatively rare (<1% in relative abundance) at 100 m, in May (Figs. 4A and 5); yet, estimates of cell lysis were higher at 100 m and lower at 0–30 m, where they were relatively abundant (Fig. 4B and 5). Similarly, at 0–5 m in June, bacteria in the Rhodobacteraceae (genus Planktomarina) were also among the most abundant bacteria, along with members of the phylum Bacteroidetes in the family Flavobacteriaceae (genus Formosa), as well as other ASVs assigned to OM60 (Fig. 5). Again, although members of these taxa were relatively abundant, the CLI was low (Fig. 5).
Fig. 6. Linear regression (red line) between the relative abundance of 16 S rRNAgenecell and lysis index for individual ASVs across all samples.
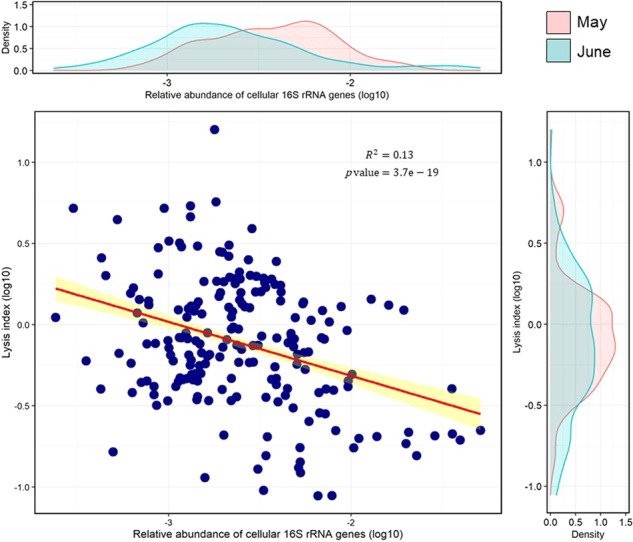
These represent the ASVs that belong to Group IV in which rRNAgenecell, rRNAcell, and rRNAext were detectable, allowing calculation of the cell-lysis index (CLI). Yellow indicates the 95% confidence interval for the slope of the regression line. The equation of the trendline (red) for linear regression is: Y = −0.416X–2.665. The density plots above and to the right of the data show the distribution of values for May (pink) and June (green).
Similar observations were made for archaea, which were some of the most abundant cells at 100 m and 265 m, but the CLI was low (Fig. 5). These included archaea in Marine Group I (phylum Thaumarchaeota), which are ubiquitous in the ocean and important in nitrification [42]. Archaea classified as Candidatus Nitrosopumilus, which has the potential to oxidize ammonia to nitrite, were estimated to account for up to 12.9% of the 16 S rRNA gene sequences below 30 m in May, and up to 14.5% of the sequences in June, but consistently had a low CLI.
Another example is proteobacteria in the order Pelagibacterales (a.k.a. SAR11 clade), which are among the most abundant bacteria in the open ocean. There were 612 ASVs assigned to the SAR11 clade, which encompassed three putative families (Clades I, II, and III). Members of Clade I were relatively abundant in our samples from 100 m and 265 m in May, and at all depths in June (Fig. 5); yet, with the exception of one ASV at 100 m in June (Fig. 4), extracellular rRNA from the SAR11 group was undetectable, indicating negligible cell lysis. Even though bacteria in the SAR11 clade were relatively abundant in several samples, and are known to be infected by a variety of viruses [43], we could detect little to no lysis (Fig. 5). Collectively, the CLI data support the idea that relatively high lysis and release of rRNA can be attributed to prokaryotic cells that are typically present in relatively low abundance; whereas, populations of cells in relatively high abundance typically have low rates of cell lysis.
A caveat to using the CLI to infer the magnitude of cell lysis is that it is based on the relative contribution of the rRNAcell for each taxon at the time of sampling, which may differ from the rRNAcell at the time of lysis. For example, the rRNA in a cell can be very sensitive to growth rate, particularly in copiotrophs [44, 45]. Variability in the RNA per cell would not be expected to increase bias in the data, but would increase the variability in the CLI. More importantly, any taxon in which lysis is occurring or has recently undergone lysis will have released a significant amount of rRNAext at the expense of rRNAcell, thus, would be expected to have a high CLI. Although a number of factors may contribute to the significant negative relationship between the CLI and estimated cell abundance, it is striking that the taxa in which the CLI is high are taxa that are typically in relatively low abundance, while a low CLI is associated with taxa that are often dominant.
As a final caveat, there is no reason to expect that the mechanisms causing decay of rRNAext, such as solar radiation, temperature, and protistan grazing, are taxon-specific. Thus, turnover of rRNAext should not affect the ratio of the relative abundance of rRNAext to rRNAcell, or calculations of relative taxon-specific lysis (CLI). Hence, the conclusion that high lysis is coupled with low abundance should not be affected by the turnover of rRNAext.
Enterobacteria represent a large portion of the rRNAext in seawater
Sequences that were assigned to enterobacteria in the genera Escherichia and Shigella, mostly clustered into Groups I and III, and comprised a large portion of the rRNAext in all ten samples, with the highest relative abundance (83%) in the June 5-m sample (Fig. 3A). These enterobacterial taxa were rare, accounting for at most 0.07% of the relative abundance of the rRNAgenecell fraction, but were very active, accounting for up to 66% of the relative abundance in the rRNAcell fraction. Although there were only two out of 156 Escherichia and Shigella ASVs in which cell lysis was measurable, they displayed higher lysis compared to other prokaryotic taxa (Figs. 3A and 4B; Fig. S4), indicative of fast-growing r-selected taxa with high lysis rates. It is unlikely that the detection of bacteria in the genera Escherichia and Shigella was the result of contamination, as they were present across samples, the equipment and containers used for sample collecting and processing were fastidiously cleaned or sterile, and members of these genera are found in coastal [e.g. [46–48],] and oceanic seawater (Fig. S5) [49], as well as marine zooplankton [50]. Given that the Strait of Georgia is subject to large freshwater inflows from the Fraser River, as well as other freshwater inputs, which are a source of coliform contamination [51], it is possible that these taxa were from terrestrial sources and were lysed when exposed to seawater.
Taxon-specific lysis rates
The determination of absolute lysis rates of different taxa requires estimates of turnover rates of rRNAext (Fig. S6). Although we do not have absolute estimates of turnover rates, ultimately rRNAext decay must be balanced by production. For example, if the decay and production rates of rRNAext is 1 d−1, which is a typical bacterial production rate for coastal seawater, then the taxon-specific lysis rate will be equal to the ratio of the concentration of rRNAext to rRNAcell for each taxon multiplied by the rRNAext turnover rate (Fig. S6).
Conclusion
In summary, we showed for representative model systems that cell lysis of bacteria caused by viral infection releases extracellular ribosomal RNA (rRNAext) into seawater, but not protistan grazing. Although our approach cannot distinguish among different causes of cell lysis, given that lysis by viruses accounts for about half of the mortality of prokaryotes in seawater [28, 29], and kills about 20–30% of the standing stock of bacteria each day [30], most rRNAext likely stems from viral lysis. Moreover, the rRNAext can be sequenced to taxonomically profile the cells in which lysis has occurred. We demonstrate that although lysis is widespread across prokaryotic phyla, it is only detected in a low proportion of taxa within a sample. As well, relative taxon-specific lysis (i.e., CLI), which indicates the taxon-specific mortality rate, can be estimated using the ratio of the relative abundance of rRNAext to rRNAcell. Our results indicate that high lysis is significantly related to low abundance, suggesting that, overall, rare taxa are associated with high lysis. Conversely, low lysis is associated with high abundance, indicating that in general the dominant taxa are subject to low relative rates of lysis. MoRS provides a powerful new approach for understanding how taxon-specific lysis shapes microbial communities and processes, and provides an important tool in our efforts to explain the distribution and abundance of specific microbial taxa in nature.
Although not considered here, 18 S rRNAext from eukaryotic cells was also abundant and easily sequenced. However, the causes of community production of rRNAext from eukaryotic cells is more difficult to constrain and may include sloppy feeding by crustacean zooplankton, or cells bursting during filtration. Nonetheless, the production of 18 S rRNAext may also provide insights into the mortality of eukaryotic microbes.
Materials and methods
Culturing conditions
The heterotrophic marine bacterium Vibrio natriegens strain PWH3a and its phage PWH3a-P1 [12] were grown in CPM medium (0.05% Casamino Acids [Difco], 0.05% Bacto Peptone [Difco] in ultrafiltered ~25 psu seawater) [12]. A culture of the phagotrophic flagellate Paraphysomonas bandaiensis (courtesy David Caron, University of Southern California) was grown with its associated bacterial community in F/2-enriched seawater [52] to which a rice grain was added [53]. All media were sterilized using autoclave; cultures were maintained at room temperature (~21 °C) in the dark.
Seawater sampling and filtration
Seawater samples were collected from Hakai Oceanographic station QU#39 (50.0307 N, 125.0992 W) in the Strait of Georgia near Quadra Island, British Columbia, Canada, during (May 5, 2015) and after (June 4, 2015) a dinoflagellate phytoplankton bloom. One-liter seawater samples were collected from five depths (0, 5, 30, 100, and 265 m) using Niskin bottles.
Filtration was used to separate extracellular rRNA (rRNAext) from the cells. Briefly, 100 mL of seawater was pre-filtered through a 120-μm mesh-size Nitex screen to remove large particles, followed by gentle vacuum filtration through a 47-mm 0.22-µm pore-size PVDF filter (Millipore, GVWP). The rRNAext was collected in the <0.22-µm filtrate, and the cellular rRNA (rRNAcell) was retained on the filter. Both the filter and filtrate were flash-frozen in liquid nitrogen, and kept at −80 °C prior to nucleic acid extraction. The filtrate was divided into several aliquots prior to freezing, so that subsamples could be thawed for downstream applications. The absence of cells in the filtrate was verified by flow cytometry (FCM) and quantitative PCR (qPCR) using primers that target the 16 S rRNA gene. See below for further details.
Assay to examine rRNAext production by bacteria infected with viruses
To assess whether viral lysis resulted in rRNAext production, V. natriegens strain PWH3a was cultured alone (control) and with its phage PWH3a-P1. The bacterium and its phage were originally isolated from the Gulf of Mexico [12], and was previously classified as V. natriegens PWH3a [54, 55]. Briefly, exponentially growing cultures of V. natriegens PWH3a were inoculated into 50 mL of CPM medium in a 250-mL Erlenmeyer flask. After 1.75 h the cells were split into two cultures; one was infected with phage PWH3a-P1 at a multiplicity of infection (MOI) of ~10, while the other served as an untreated control. Ten is the minimum MOI to ensure that all cells are infected simultaneously, but is well below the minimum MOI of 100 that can result in lysis from without in some T4-like phages [56]. Approximately 1.75 h after infection, the cultures were filtered through 0.22-µm pore-size syringe filters (Durapore, PVDF, 33 mm diameter, Millipore) to collect the filtrate containing the rRNAext. Filtrate samples were flash-frozen in liquid nitrogen and stored at −80 °C until the rRNAext was quantified in triplicate samples by reverse-transcription qPCR (RT-qPCR).
Assay to examine rRNAext production by protist grazing on bacteria
To determine if grazing by protozoa resulted in rRNAext production, we added exponentially growing V. natriegens PWH3a to a culture of the phagotrophic flagellate P. bandaiensis growing in 100 mL of 20% CPM medium (bacteria:grazers = 12,500:1 at T0). Vibrio natriegens PWH3a grown in the absence of the grazer was used as a control. The experiment was conducted in triplicate in the dark at room temperature (21 °C) in 250-mL polycarbonate Erlenmeyer flasks (Corning) for nine days. The cultures were subsampled (250 µL) once or twice a day to count the bacteria and protists by flow cytometry (FCM). Another 1 mL was taken from each flask and filtered through a 0.22-µm pore-size syringe filter (Durapore, PVDF, 33 mm diameter, Millipore), and the filtrate flash-frozen in liquid nitrogen and stored at −80 °C until the concentration of rRNAext was determined by qRT-PCR.
Stability of rRNAext in water
The stability of rRNAext in water was assayed by adding 100 µL of 0.22-μm-filtered PWH3a-P1 lysate of V. natriegens PWH3a to 35 mL of Ultrapure water (Invitrogen), unfiltered seawater or 0.22-μm-filtered seawater. The seawater sample was collected from nearby Wreck Beach (49.262 N, 123.262 W; British Columbia, Canada) about an hour before starting the experiment, and transported to the laboratory in the dark in a sterile polypropylene bottle. The seawater was then pre-filtered through 120-μm mesh Nitex screening followed by gentle vacuum filtration through a 47-mm diameter 0.22-µm pore-size PVDF filter (Millipore, GVWP).
Each treatment was conducted in duplicate and incubated in the dark at room temperature (21 °C) in 50-mL polypropylene tubes (BD Falcon). Every day for two weeks, 500 µL was taken from each tube, 0.22-µm filtered, flash-frozen in liquid nitrogen and stored at −80 °C until the concentration of rRNAext was determined by qRT-qPCR. All filtrations were done using syringe filters containing a Durapore, PVDF, 33-mm diameter membrane (Millipore).
Flow cytometry (FCM) to count bacteria, viruses and protists
Bacteria, virus and protist numbers were determined using a FACSCalibur flow cytometer (Becton Dickinson) equipped with an air-cooled laser (15 mW, 488 nm) following established methods [57, 58]. The flow cytometer list mode files were analyzed using WEASEL v3.1 (The Walter and Eliza Hall Institute of Medical Research).
Samples of bacteria and viruses were fixed with EM-grade glutaraldehyde (0.5% final concentration) for 15 min in the dark at 4 °C, then flash-frozen in liquid nitrogen and stored at −80 °C until analysis. Prior to analysis, samples were diluted in 0.02-µm-filtered TE buffer (10 mM Tris-HCL and 1 mM EDTA, pH 8), and stained with SYBR Green I (at a final 5 × 10−5 dilution of the commercial stock solution, Molecular Probes), for 15 min at room temperature (21 °C) for bacteria counts, or at 80 °C for virus counts.
Protist numbers were determined after staining live cells with LysoTracker Green (Molecular Probes) [59]. Briefly, flagellates were stained by adding 25 µl of 1 mM dye to 250 µl of culture and incubated for 15 min in the dark at room temperature.
Nucleic acids extraction
Total (cellular) nucleic acids (DNA and RNA) were extracted from the 0.22-µm pore-size filters using a MasterPure™ Complete DNA and RNA Purification Kit (Epicentre) by following the manufacturer’s directions for tissue samples. Half of each total nucleic acid sample was treated with 1 µL of RNase A (Epicentre) at 37 °C for 30 min to obtain cellular DNA; the other half of the sample was treated with DNase I (Amplification Grade, Invitrogen) to obtain cellular RNA. Briefly, the DNase I treatment was conducted at room temperature (21 °C) for 15 min in a 20-µL reaction mixture that included 16 µL of total nucleic acids (<2 µg) and 2 µL of DNase I (1 U/µL). The reaction was stopped by adding 2 µL of 25 mM EDTA solution and heating at 65 °C for 10 min. The purified cellular RNA was immediately used for cDNA synthesis step.
Extracellular nucleic acids (DNA and RNA) were extracted from 400 µL of the <0.22-µm filtrate using a PureLink Viral RNA/DNA mini Kit (Invitrogen) by following the manufacturer’s directions and eluted in 25 µL of molecular grade water. To obtain the RNA fraction only, DNA was removed by DNase I (Amplification Grade, Invitrogen) treatment by following the manufacturer’s instructions as shown above. The removal of DNA was verified by qPCR to quantify the 16 S rRNA gene before and after DNase I treatment. The purified extracellular RNA was immediately applied to the following cDNA synthesis.
cDNA synthesis
The DNA-free cellular and extracellular RNA samples were subsequently reverse-transcribed to the complementary DNA (cDNA) using SuperScriptTM III Reverse Transcriptase (Invitrogen) following the manufacturer’s directions. The first-strand cDNA synthesis reaction was primed using random hexamers (approximately 50 ng of random hexamers per 5 μg of RNA). The cDNA samples were then used as the template in quantitative PCR (qPCR) to estimate rRNA copies, and in PCR for deep sequencing of amplicons to obtain the taxonomic distribution of cellular and extracellular rRNAs.
Quantitative PCR (qPCR)
We used quantitative PCR (qPCR) with the primer set 331 F/518 R (Table S1) to target the conserved V3 region of the 16 S rRNA gene [60, 61]. When qPCR was applied to cDNA, it is referred to as quantitative reverse-transcription PCR (qRT-PCR). The amplification of 16 S rRNA sequences was done as follows: (1) The number of 16 S rRNA genes (rRNAgenecell) was estimated by amplifying DNA from the 0.22-µm pore-size filter; (2) cellular 16 S rRNA (rRNAcell) was estimated by amplifying cDNA from reverse-transcribed rRNA on the 0.22-µm pore-size filter; (3) the extracellular 16 S rRNA (rRNAext) was estimated by amplifying cDNA from reverse-transcribed rRNA in the <0.22-µm filtrate. The amplification was carried out as described below.
Briefly, the 10 µL qPCR reaction contained 1X SsoFast™ EvaGreen Supermix (Bio-Rad), 0.5 µM of each primer, and 1 µL of cDNA or DNA template. Thermal cycling was conducted in a CFX96 real-time PCR detection system (Bio-Rad) with the following program: 3 min denaturation at 95 °C, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing and extension at 62.8 °C for 30 s. Nine, 10-fold serially diluted standards (ranging from 5 × 100 to 5 × 109 molecules per mL) were run in duplicate along with two no-template control reactions containing 1 µL of nuclease-free water. The amplicon standards were made from a cloned 16 S rRNA gene amplified from V. natriegens PWH3a using primer set 331 F/518 R, purified using a MiniElute PCR Purification Kit (Qiagen), and quantified using a Qubit dsDNA High Sensitivity Assay Kit (Invitrogen). The size of the amplicon (i.e., 187 bp) was verified using gel-electrophoresis, and the qPCR melting curves confirmed that the fluorescence signal corresponds to a DNA fragment of a single size. The qPCR amplification efficiency was between 0.95 and 1.05 for the cloned amplicons (R2 > 0.98, n = 9).
PCR, amplicon sequencing library construction and sequencing
PCR was conducted to amplify the 16 S rRNA gene sequences from DNA or cDNA among samples of each fraction, including cellular DNA and cDNA from the 0.22-µm pore-size filter, and extracellular cDNA from <0.22-µm filtrate, to examine the taxonomic distribution of 16 S rRNAgenecell, rRNAcell, and rRNAext, respectively, using high-throughput sequencing.
The preparation of 16 S rRNA gene amplicon libraries was adapted from the online Illumina protocol [62] with several modifications. Briefly, two successive runs of PCR were performed. The first PCR generated amplicons of the 412-bp 16 S rRNA gene between the V4 and V5 regions using the modified primers 515F-Nxt and 926R-Nxt (Table S1). Compared to 515 F and 926 R [63], the modified primers incorporate overhanging adapter sequences (Table S1) that are compatible with Illumina index and sequencing adapters. These modifications enabled the use of Illumina Nextera XT indexes as forward and reverse primers to create the dual-indexed amplicon libraries in the second PCR.
First amplicon PCR
The 25 μL reaction mix consisted of 1X PCR buffer, 4 mM MgCl2, 50 µg of Bovine Serum Albumin (Invitrogen), 200 mM of each dNTP (Invitrogen), 0.4 µM of each primer, 0.5 U of Q5 high fidelity polymerase (NEB), and approximately 5 ng of DNA or 0.5 ng of cDNA template. Each sample was amplified in triplicate.
The PCR program [63] used 25 cycles for rRNAgenecell and rRNAcell samples but increased the number of cycles to 34 for the rRNAext samples. Briefly, PCR amplifications were subject to an initial denaturation at 95 °C for 3 min, followed by 25 to 34 cycles of denaturation at 95 °C for 45 s, annealing at 50 °C for 45 s, and elongation at 68 °C for 90 s, and a final elongation step at 68 °C for 5 min to ensure complete amplification. Triplicate first PCR products were pooled and then purified using magnetic Agencourt AMPure XP beads (Beckman Coulter) with a ratio of 1:1 for beads:product to remove fragments less than 200 bp (e.g., dimers). Purified amplicons from the first PCR were then used as the template for the index PCR (second PCR).
Second PCR (index-PCR)
PCR amplifications followed the Illumina protocol [62], but substituted Q5 high fidelity polymerase (NEB). The 25 μL reaction mix consisted of 1X PCR buffer, 4 mM MgCl2, 200 mM of each dNTP (Invitrogen), 2.5 µL of each index primer (N7XX and S5XX of Nextera XT Index Kit), 1 U of Q5 high-fidelity polymerase (NEB), and 2.5 µL of purified DNA product from the first PCR.
Second PCR amplifications were subject to an initial denaturation at 95 °C for 3 min, followed by 12–16 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and elongation at 72 °C for 30 s, and a final elongation step for at 72 °C for 10 min. Twelve cycles for rRNAgenecell and rRNAcell, and 16 cycles (for rRNAext) of index-PCR were used to append Illumina Nextera XT indexes to each side of the custom-designed amplicons.
Cleanup was conducted using magnetic Agencourt AMPure XP beads (Beckman Coulter) to purify amplicons >200 bp with a ratio of 1:1 for beads:product. Amplicon libraries were quantified using a Qubit dsDNA HS Assay Kit (Invitrogen), and the average fragment size was determined using an Agilent Bioanalyser and Agilent High Sensitivity DNA Kit. Equimolar amounts of purified amplicons from the second PCR were pooled for each library. The multiplexed pool was sequenced at the UCLA Sequencing and Genotyping Core facility, using MiSeq 2 × 300-bp paired-end chemistry (Illumina).
Sequence analysis
Amplicon sequences from each of the rRNAgenecell, rRNAcell, and rRNAext fractions were processed and analyzed using the QIIME pipeline version 2 (qiime2.2018.06) [64]. Briefly, adapters and low-quality reads were trimmed with Trimmomatic-0.36 [65] using the following settings (EADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36). Paired-end reads were then merged using PEAR [66] with the default setting. On average, 204,970 ± 95,344 merged reads of ~412 bp were obtained per library; only about 3% of low-quality reads needed to be discarded. These assembled sequences were then loaded into the QIIME pipeline version 2 and DADA2 [67] was used to filter noisy and chimeric sequences to obtain amplicon sequence variant (ASV) features. Taxonomy was assigned for these ASV features using a pre-built Naïve Bayes classifier that was trained based on the SILVA v132 database [68] at 99% nucleotide sequence similarity. ASV features were removed for downstream analysis for those with taxonomy-assignment confidence less than 80%, with no more than ten reads in either sample, unclassified at the kingdom level, or belonging to eukaryotes, mitochondria, and chloroplasts. To minimize stochastic effects resulting from rare ASV between three fractions (rRNAgenecell, rRNAcell, and rRNAext), we set ten reads as the cutoff value for the minimum number of reads for analysis: the number of reads ≥10 was considered and kept, otherwise the value was set as zero. To make the data on relative abundance comparable across samples, we normalized the ASV feature table to the lowest number of total reads per library by rarefication with the rarefy_even_depth() function of the Phyloseq package version 1.26.1 [69] in the R environment [70]. In the end, for each fraction (rRNAgenecell, rRNAcell, and rRNAext), samples were normalized by analyzing the relative abundance for each ASV as the proportion of all sequences within a sample. Statistical analysis was conducted in R version 4.0.3 [70] and figures were generated using ggplot2 version 3.3.3 [71]. The network showing the taxonomic distribution of rRNAext ASVs across seawater samples was plotted using R package igraph v1.2.6 [72]. Linear regression analysis to reveal the relative abundance of rRNAgenecell and lysis was conducted using R package methods v3.6.2 [70], ggplot2, and ggpmisc v0.3.7 [73].
To compare rRNAgenecell, rRNAcell, or rRNAext community structures among seawater samples, principal coordinate analyses (PCoA) were performed on the ordination of the weighted UniFrac metrics [74] that were based on the presence/absence and relative abundance of ASVs. The dissimilarity of the community composition among months or depths was examined using PERMANOVA analysis [75] with the adonis function and Bray–Curtis method in the R package, Vegan v.2.5 [76], and Microbiome version 1.13.12 [77].
Linear discriminant analysis Effect Size (LEfSe) [78] was conducted to identify prokaryotic taxa (rRNAgenecell, rRNAcell, or rRNAext) that were differentially abundant between months or depths, and was calculated with default settings using the Galaxy modules provided by the Huttenhower lab (https://huttenhower.sph.harvard.edu/galaxy/) [79]. Briefly, in each rRNAgenecell, rRNAcell, or rRNAext fraction, LEfSe used the two-tailed nonparametric Kruskal-Wallis test to evaluate the significance of differences in taxonomic abundance between months or depths. Then, a set of pairwise tests between months or depths was performed using the unpaired Wilcoxon test. In the end, the linear discriminant analysis (LDA) was conducted to estimate the effect size of each differentially abundant taxa for each fraction [78].
One of the challenges of working with the ASV data is the potential differences in detection limits for ASVs among the three bins (rRNAgenecell, rRNAcell, and rRNAext). We minimized this potential bias by normalizing the ASV feature table to the lowest number of total reads per library by rarefication using the rarefy_even_depth() function of the Phyloseq package [69]. Nonetheless, there are numerous counter-intuitive examples of ASVs that are found in the rRNAcell but are absent in the rRNAgenecell. These cases were largely unaffected by changing the cutoff value for the minimum number of reads per ASV (e.g., 1, 5, 10, 20, 50, and 100 reads). For those ASVs that are in the rRNAcell pool, but absent in the rRNAgenecell pool, the most parsimonious explanation is that rRNAgenecell is present, but below the limit of detection.
Another consideration is that the SILVA database that was used for taxonomically assigning the ASVs is periodically updated. We used SILVA v132 for taxonomically assigning the ASVs, which was current at the time of our analyses. Repeating the assignments with SILVA v138 that was released subsequent to our analysis produced very similar results. Regardless, of the version of the database that is used, because MoRS is based on ASVs, lysis is always assigned to an exact rRNA sequence, so the results across experiments are always comparable.
Calculation of the taxon-specific cell lysis index (CLI)
The taxon-specific cell lysis index (CLI) and identification of lysis groups were determined using a custom R package, tslysis (https://github.com/kevinzhongxu/tslysis). Briefly, for each taxon (ASV) in a given sample, a cell lysis index (CLI) was calculated as the ratio of the relative abundance of rRNAext to rRNAcell, as follows:
The CLI adjusts for the amount of rRNA present for each taxon at the time of sampling. The amount of rRNA per cell can vary greatly, particularly in copiotrophic bacteria [44, 45]; hence, the measured rRNAcell may not be an accurate reflection of the value at the time of cell lysis. The likely consequence of this would be to introduce more variability into the lysis-index data, rather than causing a systematic bias.
Because the CLI is based on the relative abundances of rRNAext and rRNAcell, changes in the abundance of a single taxon will not affect the relative abundances among other taxa. Thus, the CLI is quite robust to changes in the abundance of even rare taxa, and stochastic effects will be minor, allowing for a robust comparison of cell lysis among taxa. Furthermore, rarefaction curves were generated for each sample to ensure that the depth of sequencing was adequate, and ASVs with fewer than ten reads in a library were removed from the analysis to minimize potential stochastic errors because of detection limits. We also compared different cutoff values for the minimum number of ASV reads (i.e., 1, 5, 10, 20, 50, and 100 reads), and found that they did not affect our conclusions.
Supplementary information
Acknowledgements
We thank members of the Hakai Institute for facilitating the collection of seawater samples, particularly Brian Hunt, Kate Lansley, Alex Hare and Megan Foss. We are grateful to David Caron for providing Paraphysiomonas bandaiensis for the grazing studies. Comments from the editor and two anonymous reviewers are gratefully acknowledged and were instrumental in improving the manuscript. This work was supported by grants to CAS from the Tula Foundation, the Gordon and Betty Moore Foundation (grant: GBMF#5600), a Discovery grant from the Natural Sciences and Engineering Research Council of Canada, and infrastructure awards from the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund.
Author contributions
KXZ designed experimental approaches, conducted experimental work, analyzed the data, wrote the initial draft of the manuscript, and oversaw subsequent versions. JFW conducted initial experimental work and edited the manuscript. AMC provided technical support throughout the project and edited the manuscript. CAS conceived the project, contributed to experimental design and data interpretation, and helped write the paper.
Data availability
Sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under the accession numbers SRR14873150 to SRR14873179.
Code availability
The related codes for analyzing the taxon-specific lysis (lysis-index and lysis-rate) are included in the custom R package: tslysis (https://github.com/kevinzhongxu/tslysis).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kevin Xu Zhong, Email: xzhong@eoas.ubc.ca.
Curtis A. Suttle, Email: suttle@science.ubc.ca
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01327-3.
References
- 1.Fuhrman JA, Cram JA, Needham DM. Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol. 2015;13:133–46. doi: 10.1038/nrmicro3417. [DOI] [PubMed] [Google Scholar]
- 2.Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea: Viruses play critical roles in the structure and function of aquatic food webs. BioScience. 1999;49:781–8. doi: 10.2307/1313569. [DOI] [Google Scholar]
- 4.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 5.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol. 2014;12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams HN, Lymperopoulou DS, Athar R, Chauhan A, Dickerson TL, Chen H, et al. Halobacteriovorax, an underestimated predator on bacteria: potential impact relative to viruses on bacterial mortality. ISME J. 2016;10:491–9. doi: 10.1038/ismej.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez J, Moraleda-Muñoz A, Marcos-Torres FJ, Muñoz-Dorado J. Bacterial predation: 75 years and counting! Environ Microbiol. 2016;18:766–79. doi: 10.1111/1462-2920.13171. [DOI] [PubMed] [Google Scholar]
- 9.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 10.Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–84. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Suttle CA. Marine viruses–major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 12.Suttle CA, Chen F. Mechanisms and rates of decay of marine viruses in seawater. Appl Environ Microbiol. 1992;58:3721–9. doi: 10.1128/aem.58.11.3721-3729.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502:707–10. doi: 10.1038/nature12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 15.González J, Suttle CA. Grazing by marine nanoflagellates on viruses and virus-sized particles: ingestion and digestion. Mar Ecol Prog Ser. 1993;94:1–10. doi: 10.3354/meps094001. [DOI] [Google Scholar]
- 16.Seong KA, Jeong HJ, Kim S, Kim GH, Kang JH. Bacterivory by co-occurring red-tide algae, heterotrophic nanoflagellates, and ciliates. Mar Ecol Prog Ser. 2006;322:85–97. doi: 10.3354/meps322085. [DOI] [Google Scholar]
- 17.Sherr BF, Sherr EB, Fallon RD. Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol. 1987;53:958–65. doi: 10.1128/aem.53.5.958-965.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta AK, Burma DP. Association of ribonuclease I with ribosomes and their subunits. J Biol Chem. 1972;247:6795–801. doi: 10.1016/S0021-9258(19)44656-5. [DOI] [PubMed] [Google Scholar]
- 19.Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–91. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- 20.Timko SA, Maydanov A, Pittelli SL, Conte MH, Cooper WJ, Koch BP, et al. Depth-dependent photodegradation of marine dissolved organic matter. Front Mar Sci. 2015;66:1–13.. [Google Scholar]
- 21.Noble RT, Fuhrman JA. Virus decay and its causes in coastal waters. Appl Environ Microbiol. 1997;63:77–83. doi: 10.1128/aem.63.1.77-83.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm SW, Jeffrey WH, Suttle CA, Mitchell DL. Estimation of biologically damaging UV levels in marine surface waters with DNA and viral dosimeters. Photochem Photobio. 2002;76:268–73. doi: 10.1562/0031-8655(2002)076<0268:EOBDUL>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul JH, Jeffrey WH, DeFlaun MF. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987;53:170–9. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bong CW, Obayashi Y, Suzuki S. Succession of protease activity in seawater and bacterial isolates during starvation in a mesocosm experiment. Aquat Micro Ecol. 2013;69:33–46. doi: 10.3354/ame01618. [DOI] [Google Scholar]
- 26.Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nat Microbiol. 2018;3:754–66. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 27.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061–8. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuhrman JA, Noble RT. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–42. doi: 10.4319/lo.1995.40.7.1236. [DOI] [Google Scholar]
- 29.Mojica KDA, Brussaard CPD. Significance of viral activity for regulating heterotrophic prokaryote community dynamics along a meridional gradient of stratification in the Northeast Atlantic Ocean. Viruses. 2020;12:1293. doi: 10.3390/v12111293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suttle CA. The significance of viruses to mortality in aquatic microbial communities. Micro Ecol. 1994;28:237–43. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- 31.Bibby K. Improved bacteriophage genome data is necessary for integrating viral and bacterial ecology. Micro Ecol. 2014;67:242–4. doi: 10.1007/s00248-013-0325-x. [DOI] [PubMed] [Google Scholar]
- 32.Labonté JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ, et al. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 2015;9:2386–99. doi: 10.1038/ismej.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sockett RE. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol. 2009;63:523–39. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 34.Chao L, Levin BR. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA. 1981;78:6324–8. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granato ET, Meiller-Legrand TA, Foster KR. The evolution and ecology of bacterial warfare. Curr Biol. 2019;29:R521–R537. doi: 10.1016/j.cub.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Bremer H, Dennis PP Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Ingraham JL, Magasanik B, Low KB, Schaechter M, Umbarger HE (eds). Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd ed. American Society for Microbiology, 1996. pp. 1559, Tab. 3.
- 37.Landa M, Cottrell MT, Kirchman DL, Blain S, Obernosterer I. Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquat Micro Ecol. 2013;69:157–68. doi: 10.3354/ame01632. [DOI] [Google Scholar]
- 38.Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–9. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan MB, Waterbury JW, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–51. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 40.Våge S, Storesund JE, Thingstad TF. SAR11 viruses and defensive host strains. Nature. 2013;499:E3–4. doi: 10.1038/nature12387. [DOI] [PubMed] [Google Scholar]
- 41.Bouvier T, del Giorgio PA. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ Microbiol. 2007;9:287–97. doi: 10.1111/j.1462-2920.2006.01137.x. [DOI] [PubMed] [Google Scholar]
- 42.Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing Archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, et al. Abundant SAR11 viruses in the ocean. Nature. 2013;494:357–60. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 44.Kerkhof L, Kemp P. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol Ecol. 1999;30:253–60. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- 45.Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA. 2011;108:12776–81. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alves MS, Pereira A, Araújo SM, Castro BB, Correia ACM, Henriques I. Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front Microbiol. 2014;5:426. doi: 10.3389/fmicb.2014.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen R, Paikin S, Rokney A, Rubin-Blum M, Astrahan P. Multidrug-resistant enterobacteriaceae in coastal water: an emerging threat. Antimicrob Resist Infect Control. 2020;9:169. doi: 10.1186/s13756-020-00826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorrasi S, Pasqualetti M, Franzetti A, González-Martínez A, González-López J, Muñoz-Palazon B, et al. Persistence of Enterobacteriaceae Drawn into a Marine Saltern (Saline di Tarquinia, Italy) from the Adjacent Coastal Zone. Water. 2021;13:1443. doi: 10.3390/w13111443. [DOI] [Google Scholar]
- 49.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 50.Fernandes V, Bogati K. Persistence of fecal indicator bacteria associated with zooplankton in a tropical estuary-west coast of India. Environ Monit Assess. 2019;191:420. doi: 10.1007/s10661-019-7531-z. [DOI] [PubMed] [Google Scholar]
- 51.Rocchini RJ, Bergerud WA, Drinna RW. FRASER RIVER ESTUARY STUDY,. WATER QUALITY,. SURVEY OF FECAL COLIFORMS IN 1978. APD Bulletin 21, Province of British Columbia, Ministry of Environment, Assessment and Planning Division. 1981.
- 52.Guillard RRL. Culture of Phytoplankton for Feeding Marine Invertebrates. In: Smith WL, Chanley MH (eds). Culture of Marine Invertebrate Animals: Proceedings - 1st Conference on Culture of Marine Invertebrate Animals Greenport. Springer US, 1975. pp. 29-60.
- 53.Caron DA Enrichment, Isolation, and Culture of Free-Living Heterotrophic Flagellates. In: Kemp PF, Sherr BF, Sherr EB, Cole JJ (eds). Handbook of Methods in Aquatic Microbial Ecology. CRC Press, 1993. pp. 77–89.
- 54.Mioni C, Poorvin L, Wilhelm S. Virus and siderophore-mediated transfer of available Fe between heterotrophic bacteria: characterization using an Fe-specific bioreporter. Aquat Micro Ecol. 2005;41:233–45. doi: 10.3354/ame041233. [DOI] [Google Scholar]
- 55.Hennes KP, Suttle CA, Chan AM. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl Environ Microbiol. 1995;61:3623–7. doi: 10.1128/aem.61.10.3623-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abedon ST. Lysis from without. Bacteriophage. 2011;1:46–49. doi: 10.4161/bact.1.1.13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marie D, Partensky F, Vaulot D, Brussaard CPD. Enumeration of phytoplankton, bacteria, and viruses in marine samples. Curr Protoc Cytom. 2001;Chapter 11:Unit 11.11. doi: 10.1002/0471142956.cy1111s10. [DOI] [PubMed] [Google Scholar]
- 58.Brussaard CPD. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–13. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose JM, Caron DA, Sieracki ME, Poulton N. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquat Micro Ecol. 2004;34:263–77. doi: 10.3354/ame034263. [DOI] [Google Scholar]
- 60.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiol Read Engl. 2002;148:257–66. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 61.Sekiguchi H, Watanabe M, Nakahara T, Xu B, Uchiyama H. Succession of bacterial community structure along the changjiang river determined by denaturing gradient gel electrophoresis and clone library analysis. Appl Environ Microbiol. 2002;68:5142–50. doi: 10.1128/AEM.68.10.5142-5150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anonymous. 16S Metagenomic Sequencing Library Preparation. Illumina Inc. 2013. Available at https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html.
- 63.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 64.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End read merger. Bioinformatics. 2014;30:614–20. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP, et al. DADA2: High resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, 2020. https://www.R-project.org/.
- 71.Wickham H ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York, 2016. https://ggplot2.tidyverse.org.
- 72.Csardi G, Nepusz T The igraph software package for complex network research. InterJournal, Complex Systems 1695, 1-9 (2006).
- 73.Aphalo PJ Learn R…as you learnt your mother tongue. Leanpub, Helsinki, 2017. https://leanpub.com/learnr.
- 74.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson MJ Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online. 2017. American Cancer Society, pp 1–15.
- 76.Oksanen FJ, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. R package Version 2.5-7. 2020. URL: https://CRAN.R-project.org/package=vegan
- 77.Lahti L, Shetty S Tools for microbiome analysis in R. Microbiome package version 1.13.12. 2017. URL: http://microbiome.github.com/microbiome
- 78.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–4. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544.. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under the accession numbers SRR14873150 to SRR14873179.
The related codes for analyzing the taxon-specific lysis (lysis-index and lysis-rate) are included in the custom R package: tslysis (https://github.com/kevinzhongxu/tslysis).