Abstract
The gastrointestinal (GI) environment plays a critical role in shaping enteric infections. Host environmental factors create bottlenecks, restrictive events that reduce the genetic diversity of invading bacterial populations. However, the identity and impact of bottleneck events on bacterial infection are largely unknown. We used Citrobacter rodentium infection of mice, a model of human pathogenic Escherichia coli infections, to examine bacterial population dynamics and quantify bottlenecks to host colonization. Using Sequence Tag-based Analysis of Microbial Populations (STAMP) we characterized the founding population size (Nb′) and relatedness of C. rodentium populations at relevant tissue sites during early- and peak-infection. We demonstrate that the GI environment severely restricts the colonizing population, with an average Nb′ of only 12–43 lineages (of 2,000+ inoculated) identified regardless of time or biogeographic location. Passage through gastric acid and escape to the systemic circulation were identified as major bottlenecks during C. rodentium colonization. Manipulating such events by increasing gastric pH dramatically increased intestinal Nb′. Importantly, removal of the stomach acid barrier had downstream consequences on host systemic colonization, morbidity, and mortality. These findings highlight the capability of the host GI environment to limit early pathogen colonization, controlling the population of initial founders with consequences for downstream infection outcomes.
Subject terms: Bacteriology, Intestinal diseases, Population dynamics, DNA sequencing, Microbial ecology
Introduction
Enteric infections are shaped by the dynamic interplay between pathogen and host [1–3]. The context of these interactions is determined by the gastrointestinal (GI) environment, a series of complex, distinct, and inter-connected ecosystems where enteric pathogens must quickly adapt to overcome variations in physical, immune, and microbial features for successful colonization [1]. It is important to consider how regional environmental factors create barriers that impact initial colonization events and ultimately determine the overall infection outcome.
The impact of the host environment on pathogen colonization and dissemination can be investigated using population dynamics, the study of how and why populations change in size and structure over time [4]. Of major interest are bottleneck events, which represent sharp reductions in the size and genetic diversity of a population due to an environmental stimulus [5]. In the GI tract, potential causes of bottlenecks include colonization resistance by the resident microbiota, thickness of the mucus layer, or immune cell infiltration [1, 3]. An important consequence of bottlenecks is founder effects, in which a loss of genetic variation occurs when a new population is established by a small number of individuals (the founding population). Bottlenecks can significantly impact the course of infection, as reducing genetic diversity can increase the rate at which mutations become fixed within the population, leading to consequences such as altered virulence or antibiotic resistance [5].
Enteropathogenic and enterohemorrhagic Escherichia coli (EPEC; EHEC) are enteric pathogens causing diarrheal disease that, in developing countries, is the second leading cause of death in children under age five [6, 7]. As neither EPEC nor EHEC colonize mice, a mouse model of EPEC/EHEC infection has been well-established using the natural murine pathogen Citrobacter rodentium [8, 9]. C. rodentium is the causative agent of transmissible murine colonic hyperplasia, a form of enteric colitis [10–12], and shares many hallmarks of infection with EPEC and EHEC, including the use of a Type III Secretion System (T3SS) for intimate attachment to the host epithelium and injection of protein effectors into host cells [13–16]. Transmitted through contaminated food or water, they pass from the external environment through the acidic stomach and navigate the changing architecture of the small and large intestines before shedding in the feces and passing to a new host [1].
To date, our understanding of EPEC, EHEC, and C. rodentium population dynamics, as with most other pathogens, has relied on the use of colony-forming units (CFU) as a read-out of bacterial colonization. However, in using CFU counts, any early genetically restrictive events are masked by the rapid expansion of colonizing bacteria. Instead, a technique called Sequence Tag-based Analysis of Microbial Populations (STAMP) has emerged as a tool to study bacterial population dynamics [17]. STAMP involves infecting a host with a library of individually genetically barcoded bacteria of equal fitness, to collect and sequence the colonizing populations of barcodes at desired locations and timepoints post-infection (Fig. 1A) [17]. Ecological formulas are then used to calculate the estimated founding population size and the degree of relatedness (chord distance) between samples [17, 18]. STAMP has been used to characterize enteric infections by Vibrio cholerae and Listeria monocytogenes in mice, demonstrating differences to founding population size by tissue site and route of administration [17, 19]. A modified barcoding protocol has also been used to measure systemic colonization by extraintestinal pathogenic E. coli (ExPEC), revealing the importance of clonal expansion to bacterial persistence [20]. It is clear from these studies that founding population size differs across pathogens and reflects key barriers to infection.
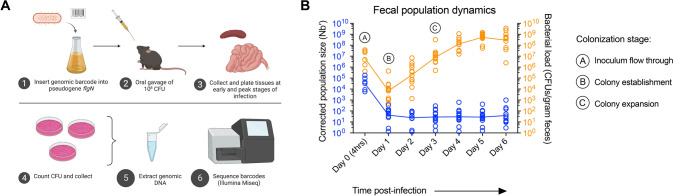
Fig. 1. Evidence for bottlenecks during C. rodentium infection.
A STAMP experimental design. In brief, a library is made of individually barcoded C. rodentium DBS100 of otherwise equal fitness. Mice are inoculated with the library by oral gavage and samples are taken at early- (day 3) and peak- (day 7) infection timepoints, in addition to fecal samples taken daily post-infection (p.i.). Samples are homogenized and plated in their entirety for enumeration of colony forming units (CFU) before collection and sequencing of barcode regions on a MiSeq instrument (Illumina). B Founding population size and bacterial load of C. rodentium shed in the feces p.i. (N = 6–12). Annotation illustrates the overall trend of C. rodentium colonization illustrating high shedding immediately post-infection, representing the inoculum flowing through, followed by colony establishment, and finally the expansion of successful colonizers.
Our study uses STAMP to directly quantify bottleneck events during C. rodentium infection. The measurement of founding population size across ten GI regions, in both mucosal and lumenal pathogen subpopulations, and two major systemic organs at early- and peak-infection timepoints provides a comprehensive picture of C. rodentium population dynamics. We identify major bottlenecks to colonization and evaluate how the manipulation of gastric pH, a key environmental barrier, impacts colonization. We further demonstrate that altering intestinal pathogen population size has downstream effects on disease severity and overall host susceptibility. Together, these results improve our understanding of colonization dynamics during intestinal infection.
Methods
Generation of the barcoded STAMP library
A library of tagged C. rodentium was created by inserting a 30-nucleotide barcode into the flagellar pseudogene flgN of wild-type (WT) C. rodentium strain DBS100 using suicide plasmid vector pRE112 by single cross-over to preserve chloramphenicol resistance [21]. The barcode region is flanked by 18-nucleotide constant regions for identification by sequencing (Fig. S1). Following triplicate bi-parental matings of E. coli MFDpir and C. rodentium DBS100, colonies were scraped and pooled before expansion in lysogeny broth (LB) broth at 37 °C overnight, sub-cultured 1:20 to prevent in vitro bottlenecks. This subculture was then aliquoted and frozen at −70 °C until use, with a new aliquot used for each experiment. Primers used to create the library are listed in Table S2. Bacterial fitness was evaluated by comparing 10 uniquely barcoded colonies to WT C. rodentium, and calculated population size (Nb) was calibrated to CFU (Supplemental Materials and Data; Figs. S1–S3).
Mouse infections and pre-treatments
Unless otherwise stated 7-week-old female C57BL/6 J mice were gavaged orally with 108 CFU of wild-type (WT) or barcoded C. rodentium DBS100 (100 µL volume). In the case of infection with the barcoded library of C. rodentium, the inoculum was prepared by culturing a 1 mL frozen aliquot of the barcoded library for 3 h 1:20 in LB supplemented with chloramphenicol at 30 µg/mL (+Cam30) before being spun down and washed to remove antibiotic, and resuspended in phosphate-buffered saline (PBS). Mice infected with the barcoded library were singly housed post-infection to prevent mixing of barcodes by coprophagy. Mice were monitored daily throughout the 3–12 day infection for weight loss and clinical symptoms. Mice were euthanized at experimental endpoint by isoflurane anesthesia followed by carbon dioxide inhalation.
To neutralize the stomach acid pH, mice were pretreated with either sodium bicarbonate or proton pump inhibitor, Lansoprazole. Sodium bicarbonate was administered as a 5% w/v solution (100 µL volume) by oral gavage 30 min before infection with C. rodentium. Lansoprazole (Sigma-Aldrich; CAS Number 103577-45-3) was administered intraperitoneally daily for 8 days at a concentration of 8 mg/kg before C. rodentium gavage 12 h after the final injection [22].
To determine disease susceptibility, 7-week-old female C3H/HeJ mice were inoculated with 106 CFU of WT DBS100 (100 µL volume) from overnight culture after antacid treatment, as above. Mice were monitored twice daily for weight loss and clinical symptoms, and euthanized upon reaching humane endpoint.
Sample collection and processing
Fecal samples were collected daily post-infection (p.i.). At experimental endpoint, the GI tract, liver, gall bladder, and spleen were collected (Fig. 2A). To isolate mucosal and lumenal populations, GI samples were opened longitudinally and gently scraped to collect lumenal content. The remaining tissue was washed twice in PBS for collection to represent the mucosal population. All samples were collected in 1 mL PBS and homogenized in a FastPrep-24 (MP Biomedicals) at 5.5 m/s for 2 min. 250 μL of sample was plated in triplicate on MacConkey (Mac) agar +Cam30, capturing barcodes from 75% of the total sample. 100 µL of the remaining sample was diluted for enumeration of CFU. After 18–20 h of growth at 37 °C bacterial colonies were counted and plates containing at least 10 colonies were scraped to collect barcoded C. rodentium, pooling triplicate plates, and genomic DNA was extracted (Qiagen DNeasy Blood & Tissue Kit).
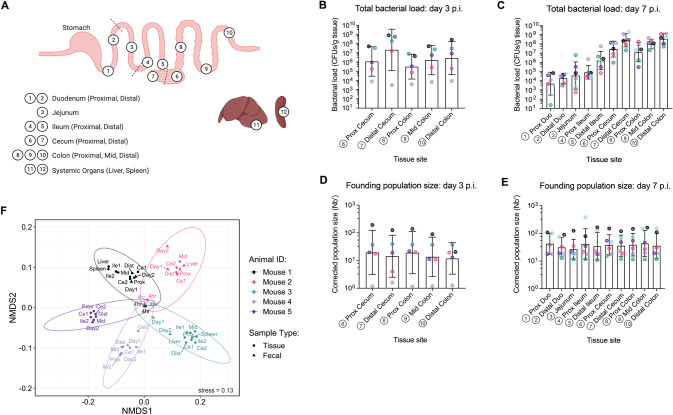
Fig. 2. A small population size exists across gut regions despite a high overall CFU burden.
A Map of the ten gastrointestinal sites sampled, as well as two systemic organs (liver and spleen). At each site bacterial load of C. rodentium was determined at B day 3 (N = 5), and C day 7 (N = 6) post-infection (p.i.). Corresponding founding population size is shown to be low across the entire gut despite the high overall CFU burden at both D day 3, and E day 7 p.i. Colours represent samples from individual mice, matched by timepoint. Error bars represent geometric mean ± geometric SD. F Non-metric Multi-dimensional Scaling (NMDS) analysis of sequenced day 3 samples shows clustering by mouse despite initial similarity at 4 h p.i. Colours represent samples from individual mice. Triangles = fecal samples. Circles = tissue samples.
Sequencing index tags were added by triplicate PCR to reduce PCR bias (Table S3). Cycling conditions were as follows: initial 30 s denaturation at 98 °C followed by 30 cycles of 10 s at 98 °C, 10 s at 52 °C, and 28 s at 72 °C with a 5 min final extension at 72 °C. Triplicate PCR reactions were pooled and purified using the Qiagen MinElute PCR Purification Kit before DNA quantification using a Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). Equal concentrations of each purified sample were then pooled and sequenced.
Amplicon sequencing and data analysis
Sequencing was performed at the UBC Biomedical Research Centre Sequencing Core, on a MiSeq (Illumina) using a 50-cycle V2 MiSeq reagent kit with 40% PhiX and a custom read 1 sequencing primer (Table S3). Demultiplexed single-end reads were processed in QIIME2, trimming off the conserved regions and filtering for barcodes of exactly 30 bp in length. Denoising was done using DADA2 to algorithmically determine the difference between sequencing errors and true biological variability, and thereby report exact amplicon sequence variants. DADA2 was chosen as it is applicable to any genetic locus, and reports fewer false positive sequence variants than other methods [23]. Data were then filtered to include barcodes present in a minimum of 5 reads in at least one tissue or fecal sample, and present in the inoculum in at least 2 reads. An average of 2062 unique barcodes were confirmed in the library. Nb, allele frequency, genetic relatedness, and directionality index were calculated using previously described formulas [18, 24, 25], substituting individual barcodes as alleles as described [5]. Generation time (g) was set to 1 for all calculations. Calculated Nb values were corrected using an in vitro calibration curve (IVCC; Supplemental Materials and Data) and plotted as Nb′. In directionality index calculations, the number of unique inoculated barcodes was used to determine population size (n).
Bacterial growth and survival at gastric pH
Bacterial growth was assessed over 20 h by diluting an overnight culture 1:200 in LB adjusted to neutral pH 7 or gastric pH 3.5. Cultures were grown at 37 °C with agitation and OD600 readings were taken every 10 min in a Synergy H1 plate reader (Biotek). Bacterial survival at gastric pH 3.5 was assessed by inoculating C. rodentium from overnight culture 1:50 into LB adjusted to neutral pH (pH 7 control) or gastric pH. At 30 min intervals bacteria were washed and resuspended in PBS before dilution and plating on neutral LB agar to determine viability. Time points were chosen to reflect average reported gastric emptying times of 30–120 min [26].
Statistical analysis
Statistical analysis was performed in GraphPad Prism (www.graphpad.com). All statistical tests used are described in the figure legends. Unless otherwise stated, analysis of non-normally distributed data was performed using Mann-Whitney tests to compare two groups and Friedman tests for more than two groups with Dunn’s multiple comparisons test. Unless otherwise stated, analysis of normally distributed data was performed using two-way ANOVA with Dunnett’s multiple comparisons test, or a Mixed-effects analysis with Holm-Šidák’s multiple comparisons test. Aggregate results represent the mean ± SD, and statistical significance is represented by *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Spearman correlation analysis between variables was performed in Graphpad Prism (p- and rho- values representing Spearman correlation values; lines representing linear regression ± 95% confidence intervals). Non-metric Multi-Dimensional Scaling (NMDS) plots were created by calculating the Bray-Curtis distance matrix using phyloseq (1.38.0) [27].
Results
Barcoding C. rodentium provides a robust tool for exploring population dynamics
To identify and quantify bottleneck events within the mouse gut, we generated a library of isogenic but uniquely-tagged C. rodentium DBS100 [17]. Barcodes consisting of 30 random nucleotides were inserted into the pseudogene flgN [21, 28] and the library, with an average of 2,062 uniquely-tagged lineages, was assessed for fitness. Individual lineages from the barcoded population showed similar growth to the WT C. rodentium (Fig. S1B, C). Type III Secretion (T3S) assays showed uniform banding patterns across 10 barcoded and WT C. rodentium strains, indicating similar ability to produce the T3SS and secrete protein effectors (Fig. S1D). In vivo colonization during single-strain infections resulted in no significant differences in fecal shedding, intestinal, and systemic bacterial load (Fig. S1E, F) and, in direct in vivo competition with the WT strain, average competitive index values (cecum = 1.09; colon = 0.98; spleen = 1.12; Fig. S1G) indicated no significant fitness defects. The barcode insert was also found to be stable both in vitro and in vivo (Fig. S2). Altogether, these data indicate that the flgN barcode insert does not affect bacterial fitness.
Sequencing of the barcode inserts within the bacterial population allowed us to calculate estimated population size (Nb), representing the number of unique founders in a given sample. To calibrate the Nb values determined from barcode sequencing back to recoverable bacterial load (as determined by CFU), an in vitro calibration curve (IVCC) was created. Here we imposed artificial bottlenecks through serial dilutions for both plating and sequencing analysis [17]. We found a strong correlation between CFU and Nb, allowing for the calculation of a corrected population size (Nb′), or bottleneck size, with a high degree of accuracy for Nb values of up to ~104 using the standard curve (R2 = 0.99; Fig. S3). As such, we concluded that this barcoded library is an appropriate tool for measuring C. rodentium population size across GI sites.
Bottleneck events occur during C. rodentium colonization
Studies monitoring fecal shedding and gut colonization during C. rodentium infection of mice hint that severe bottlenecks occur early during infection, restricting the number of bacteria able to colonize the GI tract, and reducing the genetic diversity of the invading population [29–32]. To establish the presence of bottleneck events during C. rodentium infection, we infected mice with the barcoded library by oral gavage and plated their feces daily post-infection (p.i.) to enumerate CFU burden and founding population size. Patterns of C. rodentium fecal shedding indicate a potential bottleneck event. We observed high shedding of viable bacteria at 4 h post-infection, the average GI transit time in adult C57BL/6 J mice as determined using Carmine Red (data not shown); this is followed by low-level shedding by remaining C. rodentium (days 2–4 p.i.), and expansion of this successfully colonizing population (days 6–10 p.i.; Fig. 1B). Following plating of large intestinal tissues p.i. we observed a similar pattern in the colonization of both the cecum and colon as determined by CFU burden (Fig. S4). In contrast, while fecal founding population size (Nb′) mimicked patterns of CFU burden at 4 h and day 1 p.i., this population did not recover, remaining at low levels throughout the infection time course (Fig. 1B). We propose a model in which much of the inoculating population flushes through the gut without colonizing, followed by low-level colonization by successful bacteria, and their later expansion to peak infection (Fig. 1B). Together, this evidence indicates that C. rodentium encounters bottlenecks during host colonization, enabling further STAMP analysis.
Limited founding populations of C. rodentium succeed in seeding the GI tract
To characterize the population dynamics of C. rodentium within a murine host, we chose to investigate early- and peak-infection timepoints (day 3 and 7 p.i. respectively), sectioning the GI tract into 10 regions for collection, along with the liver and spleen (Fig. 2A). At day 3, C. rodentium was reliably found to colonize the large intestine (LI; cecum-colon), colonizing the distal cecum (housing the cecal patch, an area of high C. rodentium colonization) at similar levels to peak infection, while approximately ten-fold lower in other regions (Fig. 2B) [33]. While some CFU were detected in the small intestine (SI; proximal duodenum-distal ileum), colonization was variable (Fig. S5). At day 7, CFU were detected in all GI regions, including the upper SI where C. rodentium colonization has not been well-characterized (Fig. 2C). Colonization across upper SI sites was similar from the proximal duodenum to proximal ileum, at roughly 100-fold lower than the LI, increasing in the distal ileum. As previously described [33], C. rodentium CFU burden is highest in the distal cecum and the distal colon towards the rectum. Immediately following the cecum we found that CFU burden decreases at the proximal colon (upper 1/3rd of the colon). This indicates that small-scale regional differences exist in pathogen burden across the intestine, even along the length of the colon, which is understood to be a preferred niche of C. rodentium.
To quantify C. rodentium population size across host tissues, we sequenced the barcodes within regional populations at 3- and 7-days post-infection. Despite an inoculating population of 2000+ unique barcodes, a founding population of only 1-385 individual lineages was found at each site in the GI tract, regardless of time post-infection (Fig. 2D, E), and all GI sites had a similar average Nb′ (day 3 = 15.76; day 7 = 36.74) regardless of pathogen burden, even at established niches of C. rodentium. Both the Nb′ and CFU values showed a high degree of variation between mice. Indeed, despite initial similarity at 4 h p.i., non-metric multi-dimensional scaling (NMDS) analysis of sequenced samples revealed clustering by mouse, indicating stochastic colonization of individuals by different barcode lineages (Fig. 2F). Spearman correlation between Nb′ values and C. rodentium burden showed a significant positive correlation at day 3 (rho = 0.705, p = <0.0001), but no correlation at day 7 p.i. (rho = 0.12, p = 0.35; Fig. S5). The loss of relationship between Nb′ and CFU by peak-infection indicates both a saturation of the gut environment, and a loss of diversity within the infecting population over time. Together, these data reveal a significant founder effect during C. rodentium infection of the mouse GI tract, with only a small portion of the initial population ultimately contributing to infection.
Diversity within the infecting population is lost during early infection
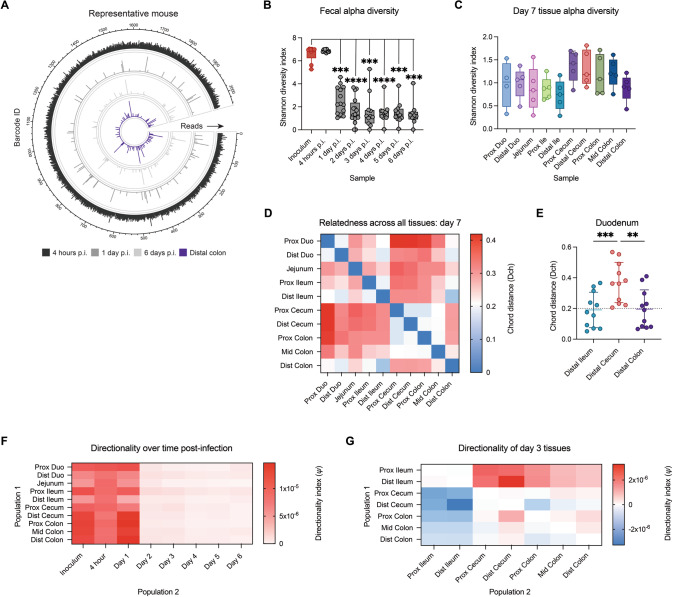
Given the low population size across the gut, it is clear that a significant loss of diversity occurs during infection, and so we aimed to determine when this loss occurs. We first plotted the number of reads per barcode within individual mice over time. While there was a high number of barcodes shed at 4 h p.i., representing the population of C. rodentium flushing through immediately after gavage, we found that most of these barcodes were lost by day 1 p.i., and were not represented in the GI tissues at day 7 (Fig. 3A). Alpha diversity measurements confirm a significant loss of diversity between fecal populations at 4 h and day 1 p.i. (Fig. 3B). As the diversity of the shed C. rodentium population likely mirrors events within the gut, these data indicate that a severe bottleneck event occurs within one day of gavage.
Fig. 3. Diversity within the infecting population is lost within the first two days of infection.
A Circos plot of the number of reads per barcode in a representative mouse at 4 h, day 1, day 6, and in the distal colon at day 7 p.i., demonstrating that barcode diversity is lost during early infection. B Shannon alpha diversity within the shed fecal population over time. Statistics represent a Mixed-effects model with Geisser-Greenhouse correction and Dunnett’s multiple comparisons test (N = 9–16). C Shannon alpha diversity across intestinal sites at day 7 p.i. D Heat map showing the mean chord distance (Dch), a measure of genetic relatedness, between of all intestinal samples at day 7 p.i., where Dch values greater than 0.2 indicate that populations are distinct (N = 5–6). E Relatedness of the duodenal sites compared to key tissue regions (N = 11). Statistics represent a Friedman test with Dunn’s multiple comparisons test. F Directionality index calculations between intestinal tissues at day 7 p.i. and C. rodentium shed daily in the feces, where negative (blue) values indicate that population 1 is closer to the input population compared to population 2, and positive (red) values indicate that population 1 is farther from the input population. G Directionality index (ψ) calculations between intestinal tissues at day 3 p.i. (population 1 – population 2), where negative (blue) values indicate that population 1 is closer to the input population compared to population 2, and positive (red) values indicate that population 1 is farther from the input population.
To investigate compositional differences between founding populations across the gut, we used barcode sequencing data to determine the relatedness of regional C. rodentium populations within a single animal infection. Relatedness is measured as chord distance (Dch), which compares the relative frequency of individual barcodes between samples; populations with low Dch values share more similar frequencies of the same barcodes and are considered more closely related [17, 18]. In the study of mammalian populations Dch values >0.2 have been considered as distinct populations. Though an equivalent cut-off has not been established for bacterial populations, y = 0.2 has been indicated as a reference point in subsequent figures. We investigated Dch between fecal C. rodentium populations shed on sequential days p.i. (Fig. S6). While Dch values were initially high between shed populations, they reached a high degree of relatedness by day 2 p.i., again suggesting dynamic changes to the infecting population within the first two days.
Next, we wanted to investigate both the diversity and relatedness of C. rodentium populations across the gut. At day 7 p.i. we found a trend towards increased Shannon diversity in the proximal cecum to mid colon regions, as compared to both the small intestine and distal colon (Fig. 3C). We also investigated chord distance relationships between all tissues at day 7. We observed a general pattern of relatedness amongst small intestinal tissues and amongst large intestinal tissues, with separation between these two groups (Fig. 3D). Indeed, we found that adjacent tissues were related across sections (≤0.2) indicating movement or exchange of barcodes between sites, with the exception of the distal ileum-proximal cecum which has an increased chord distance (Fig. 3D, Fig. S6). While this may suggest the presence of a bottleneck event upon entry to the large intestine, overall, we found that regional C. rodentium populations show a high degree of relatedness. In contrast, we found that adjacent tissue sections at day 3 p.i. vary in genetic relatedness (Fig. S6), likely because populations are not well-established during early infection.
Coprophagy is considered a major source of transmission of C. rodentium between co-housed hosts [2], and may also play a role in normalizing colonizing populations within the gut. In particular, seeing little to no colonization of the upper SI at day 3 p.i., we hypothesized that coprophagy is important in seeding the SI later during infection. Comparing the chord distance of the duodenal samples to key tissue sites (Fig. 3E) revealed a higher degree of relatedness to all other SI regions, and to the distal colon. Though C. rodentium is thought to re-seed the colon from the cecal patch region [33], at peak infection we found a higher average Dch between the distal cecum and all SI regions, as well as the distal colon (Fig. S6E, F). These data point to the distal colon as a main site of shedding, rather than the cecum, determining the composition of the transmitted population.
To further investigate the possibility of range expansion of C. rodentium and serial founder effects across intestinal sites, we calculated the directionality index (ψ) [25]. This statistic allows for the detection of asymmetries in allele, or barcode, frequencies that result from population expansion. The calculation of ψ assumes the loss of rare barcode lineages as expansion occurs, and the subsequent increase in the frequency of abundant lineages within the newly expanded population when compared to the original. The ψ values, which account for differences in the frequency of shared lineages between two populations, will increase in the direction of expansion; as a result, a positive ψ value indicates a likely expansion from an initial founder population. We found that all ψ values comparing intestinal tissues to shed C. rodentium populations over time were positive, with the greatest expansion when compared to populations shed at 4 h and 1 day p.i. (Fig. 3F), corresponding to the significant loss of diversity which occurs at this time. Within the gut, the ψ values of small intestinal tissues at day 3 p.i. were positive as compared to large intestinal tissues, indicating further expansion from the starting population, and supporting the hypothesis that the small intestine is seeded later during infection through coprophagy (Fig. 3G).
Lumenal and mucosal subpopulations are closely related
C. rodentium exists in the large intestine as both a tissue-adherent and lumenal population [34]. Considering the apparent exchange of members between adjacent C. rodentium populations, we examined dynamics of founders in these two subpopulations by performing STAMP analysis separately on mucosal and lumenal GI populations (Fig. S7A). We hypothesized that colonization of the mucosa could present a bottleneck and expected to see differences in both CFU burden and Nb′ between subpopulations at the same GI site. We found a non-significant trend towards higher CFU in the lumenal population compared to corresponding mucosal populations, in the colon at day 3 and in all sections downstream of the jejunum at day 7 p.i. (Fig. S7B, C). However, barcode sequencing showed no difference in Nb′ values between lumenal and mucosal subpopulations (Fig. S7D, E). Instead, we found a strong positive correlation between lumenal and mucosal Nb′, as well as CFU burdens across sites (Fig. S7F, G). These data indicate a strong relationship between mucosal and lumenal populations such that restrictions to one subpopulation may influence the other.
We next calculated chord distance to measure the genetic relatedness between mucosal and lumenal subpopulations. Genetic relatedness of adjacent mucosal populations show that regions are more distinct at day 3 compared to day 7 p.i. (Fig. S7H). Adjacent lumenal populations in the small intestine were all highly related to each other, suggesting bacterial movement between regions (Fig. S7I). By comparing the mucosal and lumenal populations at each site, we found that the proximal and distal colon had the lowest Dch values, indicating that these may be possible sites of either increased shedding of the mucosal population or increased mucosal seeding by the lumenal population (Fig. S7J). These data indicate a high amount of relatedness between C. rodentium populations in the mucosa, intimate to the epithelial layer, and in the intestinal lumen.
Escape to systemic circulation represents a major barrier to colonization
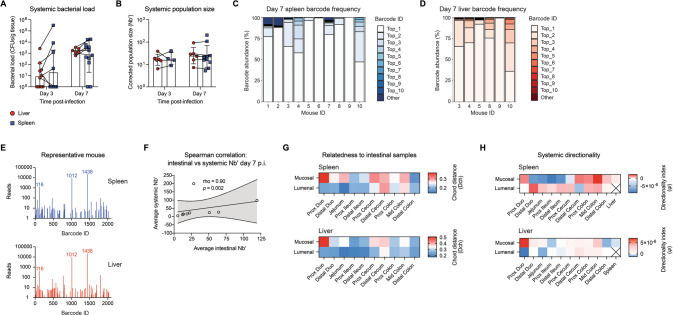
In our model of C. rodentium infection, only 81.8% of singly housed mice demonstrated systemic colonization by day 7 p.i. despite robust intestinal colonization, measured as those mice with recoverable CFU in the spleen. As intestinal colonization did not guarantee systemic colonization, bottlenecks likely exist to systemic spread. To determine whether escape from the GI tract represents a bottleneck to systemic infection by C. rodentium, we collected spleens (N = 10) and livers (N = 6) for CFU and sequencing analysis. Mice with detectable spleen CFU always had colonized livers, while the reverse was not true, suggesting the liver and spleen are colonized sequentially (Fig. 4A). Despite this, Nb′ was similar across organs (Fig. 4B). We found that one barcode lineage represented 36-99% of the bacterial populations in the spleen and liver in all mice sequenced (Fig. 4C, D), with dominant barcodes matching in both systemic organs (Fig. 4E). The dominating barcode was different in each mouse, confirming this was due to priority effects rather than a pre-existing fitness advantage by any one barcoded lineage. Per mouse, systemic and intestinal Nb′ was also similar, with a strong positive correlation between intestinal and spleen Nb′ (Fig. 4F).
Fig. 4. Escape to the systemic circulation is a major bottleneck during C. rodentium infection.
A Bacterial load in systemic organs at days 3 and 7 post-infection (p.i.) reveal sequential colonization of the liver and spleen (N = 6-12). Legend applies to A and B. B Founding population size (Nb′) in the liver and spleen (N = 4–10). Error represents geometric mean±geometric SD. C, D Frequency of colonizing barcodes in the spleen and liver show that the population is dominated by 1-2 barcodes (N = 6–10). Spleen and liver frequencies matched by mouse number. E Number of reads per barcode in spleen and liver populations are highly similar with dominant barcodes consistent between organs. F Spearman correlation of average intestinal Nb′ to average systemic Nb′ shows that systemic population size is positively correlated with the population size in the gut. Line represents linear regression ± 95% confidence interval. G Genetic relatedness of the spleen and liver to mucosal and lumenal populations across the gut (N = 3–5). H Directionality index calculations of the spleen and liver to mucosal and lumenal populations across the gut.
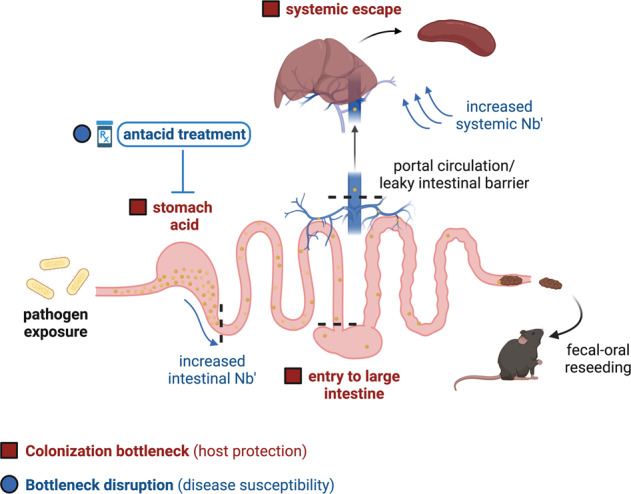
While dominant systemic barcodes were found across all gut regions, spleen and liver populations showed a higher degree of relatedness to small intestinal and distal colon populations (Fig. 4G). Considering the sequential colonization of liver and spleen, C. rodentium may exit the gut through the portal circulation, which drains from the SI and LI, as well as through a “leaky” colon resulting from intestinal damage, followed by entrance to the systemic circulation and subsequent spleen colonization. C. rodentium was not detected in the gall bladder (data not shown) and therefore cannot re-enter the GI tract through the bile. To investigate this further we calculated directionality index, which indicates that liver populations are founded earlier than spleen populations (a positive ψ value when comparing spleen to liver and a negative ψ value when comparing liver to spleen; Fig. 4H). We further found that liver populations were more distant from the expansion origin compared to various tissue sites in both the small and large intestines, supporting the possibility of spread through the portal circulation (Fig. 4H). Together, these data indicate that clonal expansion occurs during colonization of extra-intestinal sites, possibly due to chance escape of the GI tract through the portal circulation during early infection.
Gastric acid is a major early bottleneck to C. rodentium colonization
We have established that most diversity of colonizing bacterial lineages is lost within one day post-infection. Next, we wanted to identify the bottleneck event responsible. We hypothesized that passage through the acidic stomach environment is a major barrier to infection. Indeed, Dch values ~0.8 between the inoculum and intestinal populations at both timepoints indicate the populations are highly divergent (Fig. S8A). While viable C. rodentium shed at 4 h p.i. retains relatedness to the inoculum (mean Dch = 0.16), considering the short timeframe this may still indicate a large extinction event (Fig. S8B). These observations indicate an early stress experience by C. rodentium that results in a dramatic bottleneck immediately upon infection, which could be due to low stomach pH.
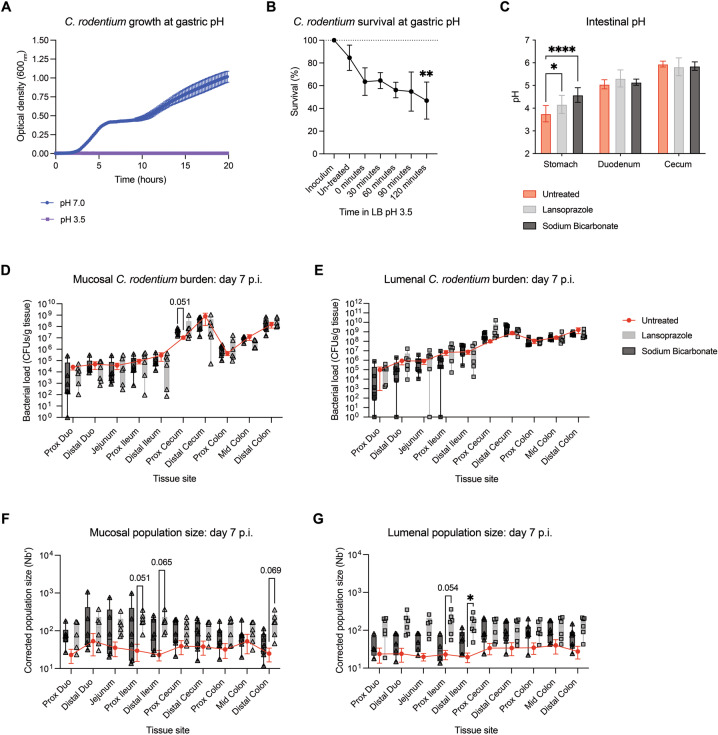
Stomach acid is a significant barrier to enteric infection [22, 35, 36]. We found that C. rodentium does not grow in LB at pH 3.5, representing the average gastric pH in mice (Fig. 5A) [37]. Plating after exposure to pH 3.5 revealed that over half (53.1%) of the C. rodentium population dies after a range of 30–120 min (the lower and upper limits of gastric emptying time) (Fig. 5B) [26]. The 0-minute control (inoculation into LB pH 3.5 before immediate processing) further shows that most cell death occurs upon immediate exposure to low pH, while processing after subculture at a neutral control pH did not result in significant death. Therefore, while C. rodentium is tolerant of gastric pH, a large portion of the population may die before reaching the intestines.
Fig. 5. Pre-treatment to neutralize the stomach acid bottleneck with either sodium bicarbonate or proton pump inhibitor Lansoprazole dramatically increases intestinal founding population size independent of CFUs.
A Growth of C. rodentium at gastric pH 3.5 and pH 7.0 over 20 h at 37 °C. Curve represents mean±SD of 3 biological replicates (average of 3 technical replicates). B Survival of C. rodentium in gastric pH 3.5 over the range of average gastric emptying time indicates an average loss of 53.1% of the population. Each point represents the average of 4–5 biological replicates (3 technical replicates per biological replicate). Error represents means ± SD. C Intestinal pH measurements in the stomach, small intestine, and cecum indicate that only gastric pH is altered after sodium bicarbonate and Lansoprazole pre-treatments (N = 5–8). D, E Mucosal and lumenal bacterial loads after antacid treatment at day 7 p.i. (N = 5–7). Legend applies to D–G. F–G Mucosal and lumenal Nb′ at day 7 p.i. (N = 5–7). Box plots show points with range. Laz, Lansoprazole. SB, sodium bicarbonate. Statistical analysis represents C) a two-way ANOVA with Dunnett’s multiple comparisons test, and D–G a Mixed-effects model with Dunnett’s multiple comparisons test.
To determine how removing the stomach acid barrier affects downstream population dynamics of C. rodentium, we increased gastric pH of the mice via pre-treatment with either sodium bicarbonate or the proton pump inhibitor Lansoprazole before C. rodentium infection [22, 38]. Both interventions effectively increased gastric pH (Fig. 5C). Although antacid pre-treatment did not significantly alter intestinal CFU counts (Fig. 5D, E; Fig. S8C, D), the Nb′ was increased at peak infection across all GI regions, with up to a 6.8-fold increase at day 7 in response to Lansoprazole in the lumenal populations of the upper SI, from the proximal duodenum to the proximal ileum (Fig. 5F, G). These data indicate that low stomach pH represents a major early bottleneck to C. rodentium colonization.
Widening intestinal colonization bottlenecks may alter systemic spread and overall disease susceptibility
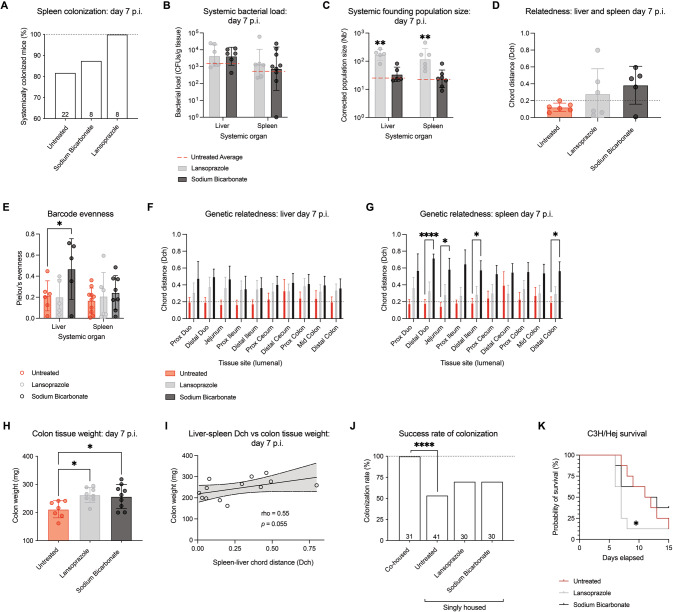
Next, we sought to determine whether manipulating intestinal founding population size had additional consequences to downstream infection. Compared to 81.8% in untreated controls, 100% of colonized mice pre-treated with Lansoprazole exhibited spleen colonization (Fig. 6A), with a modest increase also observed in response to sodium bicarbonate pre-treatment, indicating that systemic spread may be affected by increasing intestinal Nb′. While CFU burdens in the spleen and liver were not affected by either antacid pre-treatment (Fig. 6B), Lansoprazole significantly increased systemic Nb′ (Fig. 6C). While the genetic relatedness between the spleen and liver of untreated mice was high (mean Dch=0.12), chord distance values increased in both antacid treatments (Fig. 6D), indicating a difference in community composition. Indeed, in sodium bicarbonate pre-treated mice, barcodes within the liver populations were more evenly spread across colonizing barcodes, while the spleen population remained restrictive (Fig. 6E). This was reflected in changes to Dch values between the systemic and intestinal regions of antacid-treated mice (Fig. 6F–G). As antacid pre-treated mice were found to have higher colon weights than untreated mice, indicating increased tissue hyperplasia, loss of epithelial barrier integrity may allow for increased GI escape and an increased diversity of barcode lineages in the liver, though an additional bottleneck may exist to further colonization of the spleen. This is supported by a positive trend (rho = 0.55, p = 0.055) between liver-spleen Dch and colon weight (Fig. 6H–1).
Fig. 6. Antacid pre-treatment has unintended consequences to systemic colonization and susceptibility to enteric infection.
A Rate of spleen colonization across treatment groups (N = 8–22). B CFU burden in the liver and spleen in antacid-treated mice at day 7 post-infection (p.i.; N = 6–8). Legend applies to B and C. C Population size in liver and spleen sites at day 7 p.i. in antacid-treated mice (N = 5–7). D Genetic relatedness between the liver and spleen indicates that populations become more divergent as a result of antacid treatment (N = 5–6). E Barcode evenness within liver and spleen populations after antacid treatment (N = 5–10). Statistics represent a Mixed-effects model for matched samples with Dunnett’s multiple comparisons test. F, G Genetic relatedness of the liver and spleen compared to lumenal intestinal samples (N = 4–6). Statistics represent a Mixed-effects model for matched samples with Geisser-Greenhouse correction and Dunnett’s multiple comparisons tests. H Colon tissue weight at day 7 p.i. increases after antacid treatment (N = 7–9). I Spearman correlation of colon tissue weight compared to liver-spleen Dch. Line represents linear regression ± 95% confidence interval. p- and rho- values represent Spearman correlation values. J Success rate of C. rodentium colonization across co-housed, and singly housed untreated, Lansoprazole-treated, and sodium bicarbonate-treated groups (N = 30–41). Statistical analysis represents a Fisher’s exact test. K Morbidity and mortality of susceptible C3H/HeJ mice infected with low-dose C. rodentium in antacid pre-treated mice compared to untreated controls (N = 8). Statistical analysis represents a Gehan-Breslow-Wilcoxon test.
Given these changes to intestinal Nb′ and systemic colonization, we investigated whether antacid pre-treatment could increase overall host susceptibility to enteric infection. While singly housed untreated mice developed robust C. rodentium infection at an average rate of 53.6%, antacid pre-treatment increased this infection rate to 70% (Fig. 6J). Manipulating natural bottlenecks, such as acidity of the stomach, may therefore increase host susceptibility to infection. To test this further, susceptible C3H/HeJ mice were pre-treated with antacids before infection with a low infectious dose of C. rodentium [39]. We found that antacid pre-treated mice displayed increased morbidity and mortality compared to untreated animals, significantly in the Lansoprazole-treated group (Fig. 6K). These data indicate that manipulating early barriers to infection can have downstream consequences on host colonization and disease progression.
Discussion
STAMP allowed us to make several key observations about population dynamics during C. rodentium infection. The most striking is the small founding population size of C. rodentium across the GI tract, with an average of ~50 individually barcoded bacteria across any site or timepoint, indicating a severe restriction to the genetic diversity. Even areas of high C. rodentium density, such as the cecal patch and rectal regions, demonstrate a highly restrictive Nb′, suggesting they may be competitive for initial colonization while favouring the expansion of successful colonizers. This finding was in stark contrast to the reported population sizes of up to 105 individuals of enteric pathogen V. cholerae which also expresses a T3SS [17], and instead more similar to intracellular pathogen L. monocytogenes [19]. This is an intriguing result considering that C. rodentium is a natural murine pathogen, expected to be adapted for colonization of the mouse gut [17, 19].
Interestingly, we noted differences in Nb′ values at the same site between individual mice (Fig. 2). This may indicate that the carrying capacity for C. rodentium differs between individuals, even between those of the same genetic background [19]. For example, subtle differences between hosts, such as differences in host metabolism, have been shown to impact colonization [39]. Individual differences coupled with stochasticity of colonization may explain the inherent variability found in the infection model, with a dramatic decrease in colonization rate from 100% in co-housed animals to only 53.7% upon single-housing (Fig. 6J). Despite differences in Nb′ values between mice, by peak-infection C. rodentium CFU burdens were the same, even after dramatic changes to Nb′ following antacid-treatment (Fig. S5C, Fig. 5D, E). These data indicate a saturation of the gut environment by peak-infection.
We found that exit from the GI tract represents a significant bottleneck to systemic dissemination of C. rodentium. Spleen and liver populations were dominated by 1-2 barcode lineages, despite a high systemic Nb′, suggesting that colonization of systemic organs occurs by chance escape of the GI tract early during infection, allowing for clonal expansion before the host becomes overwhelmed at peak infection and additional barcodes enter the systemic circulation (Fig. 4). However, further studies are necessary to rule out within-host adaptation by members of the infecting population that facilitate systemic colonization. Another substantial bottleneck event was identified in the stomach. Enteric pathogens require acid tolerance to survive low gastric pH; EPEC has been demonstrated to tolerate simulated gastric fluid [36], whereas 20–80% of ingested EHEC may be lost in the gastric fluid [35]. We demonstrated that average gastric pH completely inhibits C. rodentium growth, and while recovery from this dormant state is possible after removal from low pH, almost half of the population was unable to survive the upper limits of gastric emptying time (Fig. 5). Gastric pH is therefore a substantial barrier to enteric colonization, from which C. rodentium must quickly recover to prevent being flushed through the GI tract and lost.
By increasing gastric pH we demonstrated how manipulating individual barriers can alter the course of infection, with an expansion of Nb′ across all intestinal sites compared to untreated mice (Fig. 5). Significantly, through infection of susceptible C3H/HeJ mice, we demonstrated that removal of early-infection barriers can increase host susceptibility, morbidity, and mortality (Fig. 6). Sodium bicarbonate pre-treatment, administered locally 30 min before C. rodentium gavage, represents a temporary reduction in gastric pH, while prolonged Lansoprazole exposure represents continual prescription administration. It is notable that the largest differences in population size and disease susceptibility were demonstrated in the context of this prolonged PPI administration, which allows for potential additional temporal effects on the host environment [22]. These data are relevant to human health outcomes as a human case-control study identified patient use of PPIs as a major risk factor for infection by Enterobacteriaceae [40]. Gastric acid suppressants are over-used both inside and outside of hospital settings [41]; between 40–60% of acute hospital inpatients are prescribed PPIs, of which Lansoprazole is one of the most common [41]. Our study shows that gastric acid suppressants may eliminate at least one critical early barrier to enteric infection. In the case of hospital inpatients, increasing gastric pH may therefore further predispose an already vulnerable population to hospital-acquired infection. As gastric pH is a significant barrier to even well-adapted pathogens such as C. rodentium, it may similarly prevent the effective colonization of beneficial microbes. It is currently unclear how efficiently commercial probiotic strains colonize the human gastrointestinal tract, though studies suggest their colonization to be largely transient, relying on the continual administration of strains to produce beneficial effects [42]. This is likely due to significant bottlenecks to niche establishment, which could be mitigated through the further study of bottlenecks during host colonization. It may be that a combination of encapsulation or neutralization of gastric acid could promote the maintenance of beneficial microbial communities upon administration.
In summary, our analysis of population dynamics during C. rodentium infection demonstrates how restrictive initial GI colonization is, and places further emphasis on the need for bacterial pathogens to sense and respond rapidly to their changing surroundings as they traverse the gut to establish their niche. Early infection events are important to determine the diversity and number of initial founders, with potential downstream consequences on infection outcome. We have demonstrated that bottlenecks result in stochastic colonization at the individual level, eliminating lineages regardless of fitness. It follows that, during natural infection by a non-uniform pathogen population under selective environmental pressure, bottlenecks could create selective sweeps that enrich for individuals with increased fitness. This suggests that adaptive mutations, for instance that increase virulence or antibiotic resistance, could be rapidly fixed in a population within the host gastrointestinal environment, and transmitted to new hosts. At the population level, this means that bottlenecks have important epidemiological impacts such as on the rate of horizontal gene transfer and acquisition of drug resistance genes, impacting the diversity of infection outbreaks and enriching adaptive mutations within the population [5]. As such, understanding the effects of bottlenecks on a genetically non-uniform population could greatly inform evolution of pathogen virulence or even adaptation to new hosts.
Supplementary information
Acknowledgements
We thank our colleagues in the Finlay laboratory for their support and assistance; particularly W Deng and L Thorson. We would also like to thank R Vander Werff, T Stach, and E Limber for technical support, as well as M Whitlock for early consultation on conceptual design. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to BB Finlay (FDN-159935). SE Woodward is a CIHR CGS-D Graduate Scholar, supported by a UBC Four Year Fellowship and Dmitry Apel Memorial Scholarship. RA Melnyk was a supported by Simons Foundation Fellowship through the Life Sciences Research Foundation. CH Haney was supported by a Canada Research Chairs salary award. Supporting images were created using Biorender.com.
Author contributions
SEW and SLV conceived the project with guidance from CHH and BBF, SEW and SLV designed experiments. SEW, SLV, JP-D, ASP, LMN, KEH, and MAW performed experiments. SEW, JP-D, RAM, and MC wrote bioinformatics pipelines and analyzed data. SEW wrote the original draft of the manuscript with input from all authors. BBF acquired funding for the project.
Data availability
The datasets generated during the current study are available in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA810564.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01321-9.
References
- 1.Woodward SE, Krekhno Z, Finlay BB. Here, There, and Everywhere: How Pathogenic Escherichia coli Sense and Respond to Gastrointestinal Biogeography. Cell Microbiol. 2019;21:e13107. doi: 10.1111/cmi.13107. [DOI] [PubMed] [Google Scholar]
- 2.Collins JW, Keeney KM, Crepin VF, Rathinam VAK, Fitzgerald KA, Finlay BB, et al. Citrobacter rodentium: Infection, Inflammation and the Microbiota. Nat Rev Microbiol. 2014;12:612–23. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 3.Mowat AM, Agace WW. Regional Specialization Within the Intestinal Immune System. Nat Rev Immunol. 2014;14:nri3738. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 4.Population Dynamics - Latest Research and News | Nature [Internet]. [cited 2017 Nov 5]. Available from: https://www.nature.com/subjects/population-dynamics
- 5.Abel S, Abel zur Wiesch P, Davis BM, Waldor MK. Analysis of Bottlenecks in Experimental Models of Infection. PLoS Pathog. 2015;11:e1004823. [DOI] [PMC free article] [PubMed]
- 6.Global Burden of Disease Study. 2013 Collaborators. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 301 Acute and Chronic Diseases and Injuries in 188 Countries, 1990-2013: A Systematic Analysis for the Global Burden of Disease Study 2013. Lancet Lond Engl. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO | Diarrhoeal Disease [Internet]. WHO. [cited 2017 Oct 22]. Available from: http://www.who.int/mediacentre/factsheets/fs330/en/.
- 8.Crepin VF, Collins JW, Habibzay M, Frankel G. Citrobacter rodentium Mouse Model of Bacterial Infection. Nat Protoc. 2016;11:1851–76. doi: 10.1038/nprot.2016.100. [DOI] [PubMed] [Google Scholar]
- 9.Bhinder G, Sham HP, Chan JM, Morampudi V, Jacobson K, Vallance BA. The Citrobacter rodentium Mouse Model: Studying Pathogen and Host Contributions to Infectious Colitis. J Vis Exp. 2013;19:e50222. doi: 10.3791/50222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barthold SW, Coleman GL, Bhatt PN, Osbaldiston GW, Jonas AM. The Etiology of Transmissible Murine Colonic Hyperplasia. Lab Anim Sci. 1976;26:889–94. [PubMed] [Google Scholar]
- 11.Deng W, Vallance BA, Li Y, Puente JL, Finlay BB. Citrobacter rodentium Translocated Intimin Receptor (Tir) is an Essential Virulence Factor Needed for Actin Condensation, Intestinal Colonization and Colonic Hyperplasia in Mice. Mol Microbiol. 2003;48:95–115. doi: 10.1046/j.1365-2958.2003.03429.x. [DOI] [PubMed] [Google Scholar]
- 12.Schauer DB, Falkow S. Attaching and Effacing Locus of a Citrobacter freundii Biotype That Causes Transmissible Murine Colonic Hyperplasia. Infect Immun. 1993;61:2486–92. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott SJ, Yu J, Kaper JB. The Cloned Locus of Enterocyte Effacement From Enterohemorrhagic Escherichia coli O157:H7 is Unable to Confer the Attaching and Effacing Phenotype Upon E. coli K-12. Infect Immun. 1999;67:4260–3. doi: 10.1128/IAI.67.8.4260-4263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per Regulon of Enteropathogenic Escherichia coli: Identification of a Regulatory Cascade and a Novel Transcriptional Activator, the Locus of Enterocyte Effacement (LEE)-Encoded Regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis KG, Girón JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli Contains a Putative Type III Secretion System Necessary for the Export of Proteins Involved in Attaching and Effacing Lesion Formation. Proc Natl Acad Sci. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. Attaching and Effacing Activities of Rabbit and Human Enteropathogenic Escherichia coli in Pig and Rabbit Intestines. Infect Immun. 1983;41:1340–51. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abel S, Abel zur Wiesch P, Chang HH, Davis BM, Lipsitch M, Waldor MK. Sequence Tag–based Analysis of Microbial Population Dynamics. Nat Methods. 2015;12:223–6. doi: 10.1038/nmeth.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis. Models and Estimation Procedures. Am J Hum Genet. 1967;19:233–57. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Abel S, Abel Zur Wiesch P, Sasabe J, Davis BM, Higgins DE, et al. Deciphering the Landscape of Host Barriers to Listeria monocytogenes Infection. Proc Natl Acad Sci. 2017;114:6334–9. doi: 10.1073/pnas.1702077114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hullahalli K, Waldor MK. Pathogen Clonal Expansion Underlies Multiorgan Dissemination and Organ-Specific Outcomes During Murine Systemic Infection. eLife. 2021;10:e70910. doi: 10.7554/eLife.70910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petty NK, Feltwell T, Pickard D, Clare S, Toribio AL, Fookes M, et al. Citrobacter rodentium is an Unstable Pathogen Showing Evidence of Significant Genomic Flux. PLOS Pathog. 2011;7:e1002018. doi: 10.1371/journal.ppat.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasutomi E, Hoshi N, Adachi S, Otsuka T, Kong L, Ku Y, et al. Proton Pump Inhibitors Increase the Susceptibility of Mice to Oral Infection with Enteropathogenic Bacteria. Dig Dis Sci. 2018;63:881–9. doi: 10.1007/s10620-017-4905-3. [DOI] [PubMed] [Google Scholar]
- 23.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krimbas CB, Tsakas S. The Genetics of Dacus Oleae. v. Changes of Esterase Polymorphism in a Natural Population Following Insecticide Control-Selection or Drift? Evol Int J Org Evol. 1971;25:454–60. doi: 10.1111/j.1558-5646.1971.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 25.Peter BM, Slatkin M. Detecting Range Expansions From Genetic Data. Evol Int J Org Evol. 2013;67:3274–89. doi: 10.1111/evo.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz R, Kaspar A, Seelig J, Künnecke B. Gastrointestinal Transit Times in Mice and Humans Measured with 27Al and 19F Nuclear Magnetic Resonance. Magn Reson Med. 2002;48:255–61. doi: 10.1002/mrm.10207. [DOI] [PubMed] [Google Scholar]
- 27.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PloS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petty NK, Bulgin R, Crepin VF, Cerdeño-Tárraga AM, Schroeder GN, Quail MA, et al. The Citrobacter rodentium Genome Sequence Reveals Convergent Evolution with Human Pathogenic Escherichia coli. J Bacteriol. 2010;192:525–38. doi: 10.1128/JB.01144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mundy R, Pickard D, Wilson RK, Simmons CP, Dougan G, Frankel G. Identification of a Novel Type IV Pilus Gene Cluster Required for Gastrointestinal Colonization of Citrobacter rodentium. Mol Microbiol. 2003;48:795–809. doi: 10.1046/j.1365-2958.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- 30.Darwin AJ, Miller VL. Identification of Yersinia enterocolitica Genes Affecting Survival in an Animal Host Using Signature-Tagged Transposon Mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 31.Maroncle N, Balestrino D, Rich C, Forestier C. Identification of Klebsiella pneumoniae Genes Involved in Intestinal Colonization and Adhesion Using Signature-Tagged Mutagenesis. Infect Immun. 2002;70:4729–34. doi: 10.1128/IAI.70.8.4729-4734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiles S, Dougan G, Frankel G. Emergence of a ‘Hyperinfectious’ Bacterial State After Passage of Citrobacter rodentium Through the Host Gastrointestinal Tract. Cell Microbiol. 2005;7:1163–72. doi: 10.1111/j.1462-5822.2005.00544.x. [DOI] [PubMed] [Google Scholar]
- 33.Wiles S, Clare S, Harker J, Huett A, Young D, Dougan G, et al. Organ Specificity, Colonization and Clearance Dynamics In Vivo Following Oral Challenges with the Murine Pathogen Citrobacter rodentium. Cell Microbiol. 2004;6:963–72. doi: 10.1111/j.1462-5822.2004.00414.x. [DOI] [PubMed] [Google Scholar]
- 34.Kitamoto S, Nagao-Kitamoto H, Kuffa P, Kamada N. Regulation of Virulence: The Rise and Fall of Gastrointestinal Pathogens. J Gastroenterol. 2016;51:195–205. doi: 10.1007/s00535-015-1141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takumi K, de Jonge R, Havelaar A. Modelling Inactivation of Escherichia coli by Low pH: Application to Passage Through the Stomach of Young and Elderly People. J Appl Microbiol. 2000;89:935–43. doi: 10.1046/j.1365-2672.2000.01193.x. [DOI] [PubMed] [Google Scholar]
- 36.Pienaar JA, Singh A, Barnard TG. Acid-Happy: Survival and Recovery of Enteropathogenic Escherichia coli (EPEC) in Simulated Gastric Fluid. Micro Pathog. 2019;128:396–404. doi: 10.1016/j.micpath.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 37.McConnell EL, Basit AW, Murdan S. Measurements of Rat and Mouse Gastrointestinal pH, Fluid and Lymphoid Tissue, and Implications for In-Vivo Experiments. J Pharm Pharm. 2008;60:63–70. doi: 10.1211/jpp.60.1.0008. [DOI] [PubMed] [Google Scholar]
- 38.Tan A, Petty NK, Hocking D, Bennett-Wood V, Wakefield M, Praszkier J, et al. Evolutionary Adaptation of an AraC-Like Regulatory Protein in Citrobacter rodentium and Escherichia Species. Infect Immun. 2015;83:1384–95. doi: 10.1128/IAI.02697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez KK, Chen GY, Schieber AMP, Redford SE, Shokhirev MN, Leblanc M, et al. Cooperative Metabolic Adaptations in the Host Can Favor Asymptomatic Infection and Select for Attenuated Virulence in an Enteric Pathogen. Cell. 2018;175:146–.e15. doi: 10.1016/j.cell.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham R, Jones L, Enki DG, Tischhauser R. Proton Pump Inhibitor Use as a Risk Factor for Enterobacteriaceal Infection: A Case-Control Study. J Hosp Infect. 2018;100:60–4. doi: 10.1016/j.jhin.2018.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Kelly OB, Dillane C, Patchett SE, Harewood GC, Murray FE. The Inappropriate Prescription of Oral Proton Pump Inhibitors in the Hospital Setting: A Prospective Cross-Sectional Study. Dig Dis Sci. 2015;60:2280–6. doi: 10.1007/s10620-015-3642-8. [DOI] [PubMed] [Google Scholar]
- 42.Federici S, Suez J, Elinav E. Our Microbiome: On the Challenges, Promises, and Hype. Results Probl Cell Differ. 2020;69:539–57. doi: 10.1007/978-3-030-51849-3_20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA810564.