Abstract
Insects can assemble defensive microbiomes on their body surfaces to defend against fungal parasitic infections. The strategies employed by fungal pathogens to combat host cuticular microbiotas remains unclear. Here, we report the identification and functional characterization of the defensin-like antimicrobial gene BbAMP1 encoded by the entomopathogenic fungus Beauveria bassiana. The mature peptide of BbAMP1 can coat fungal spores and can be secreted by the fungus to target and damage Gram-positive bacterial cells. Significant differences in insect survival were observed between the wild-type and BbAMP1 mutant strains during topical infection but not during injection assays that bypassed insect cuticles. Thus, BbAMP1 deletion considerably reduced fungal virulence while gene overexpression accelerated the fungal colonization of insects compared with the wild-type strain in natural infections. Topical infection of axenic Drosophila adults evidenced no difference in fly survivals between strains. However, the gnotobiotic infections with the addition of Gram-positive but not Gram-negative bacterial cells in fungal spore suspensions substantially increased the survival of the flies treated with ∆BbAMP1 compared to those infected by the wild-type and gene-overexpression strains. Bacterial colony counts and microbiome analysis confirmed that BbAMP1 could assist the fungus to manipulate insect surface bacterial loads. This study reveals that fungal defensin can suppress the host surface defensive microbiomes, which underscores the importance to extend the research scope of fungus-host interactions.
Subject terms: Fungal ecology, Microbiome
Introduction
Studies have shown that defensive microbiomes are formed on animal body surfaces to defend against diseases [1]. For example, human skin microbiotas can mediate barrier functions against dermatoses and beyond [1, 2]. The microbiotas assembled on amphibian skins can assist the hosts to defend against chytrid infections [3, 4]. Likewise, the ectosymbiotic bacteria inhabited the cuticles of insects, such as ants, bees, beetles, and flies, play important roles in battling fungal parasitic infections [5–9]. The dominant bacteria, such as Lactiplantibacillus plantarum and Acetobacter persici, isolated from the body surface of Drosophila melanogaster could effectively inhibit the spore germination and therefore infection of fungal entomopathogens such as Beauveria bassiana and Metarhizium robertsii [9]. The actinobacterial Pseudonocardia installed on the cuticle of leaf-cutting ants [5] and Streptomyces on beetles [7] can produce antifungal chemicals to protect insects from fungal parasite infections. Thus, apart from the production of cuticle-degrading enzymes and generation of appressorium turgor for cuticle penetration [10–12], entomopathogenic fungi might have evolved the ability to outcompete host cuticle microbiotas to facilitate parasite infection.
Antimicrobial peptides (AMPs) are produced by different organisms including fungi [13]. Based on their specific activities, AMPs can be broadly classified as antibacterial peptides, antifungal peptides (AFPs) and antiviral peptides [14]. AMPs have also been classified according to their secondary structures, such as the α(-helix), β(-sheet), αβ and non-αβ families [14]. The largest group of AMPs are defensins, which are cysteine-rich, amphipathic, positively charged and have a mature length ranging from 34 to 54 amino acid residues [15]. Cysteine-stabilized αβ defensins are also encoded by different fungi and classified into different families based on sequence similarity and the presence/absence of αβ-motifs within peptides [16–18]. A few fungal defensin-like peptides (fDLPs), such as plactasin [19], eurocin [20], micasin [21], and afusinC [22], have been characterized with potent antibacterial activities. Plactasin and eurocin can bind to the bacterial cell-wall (CW) component lipid II to inhibit CW biosynthesis [19, 20]. Fungal production of fDLPs may assist the producers to outcompete bacteria for nutrients, niches and/or hosts [23]; however, this effect has not been well studied.
Here, we report the identification of the fDLP-type gene BbAMP1 from B. bassiana which contributes to the fungal infection of insects through dysbiosis of the host surface microbiomes. Mature BbAMP1 can effectively target and damage different Gram-positive (G+) bacteria isolated from insect body surfaces. The identification of this new type of fungal virulence factor broadens the understanding of fungus-insect interactions.
Materials and methods
Fungal strains, insects, and growth conditions
The wild-type (WT) and mutant strains of B. bassiana ARSEF 2860 were cultured on potato dextrose agar (PDA; BD Difco) at 25 °C for two weeks for harvesting conidial spores. For liquid growth, conidial spores were grown in Sabouraud dextrose broth (SDB; BD Difco) at 25 °C and 200 rpm on a rotatory shaker. The yeast strains of the Saccharomyces cerevisiae AH109 and Candida albicans SC5314 were grown in the YPD (yeast extract, 10 g L−1; peptone, 20 g L−1, and dextrose, 20 g L−1) broth for antifungal tests. For stress response assays, conidial suspensions (1 × 105 conidia mL−1) were prepared in 0.05% Tween 20 and inoculated on PDA plates amended with FeSO4 (2 mM), CaCl2 (5 mM), CuSO4 (100 mM), ZnSO4 (30 mM, 50 mM), sorbitol (1 M), farnesol (50 μM), Congo red (50 μg mL−1), sodium dodecyl sulfate (0.01%) and Calcofluor white (100 μg mL−1; Sigma-Aldrich) at 25 °C for 3 days.
The stocks of fruit fly D. melanogaster W1118 line were maintained at 25 °C and 12 h of light:dark cycles on the Bloomington formulation of cornmeal agar medium [24]. Axenic flies were prepared as described before by repeated washes of fly eggs with the diluted Walch disinfectant solution (1:30, v v−1), hypochlorite (1:1, v v−1), 75% ethanol and phosphate buffer (pH, 7.4) containing 0.1% Tween 20 [25]. The eggs were hatched and insects reared on the autoclaved medium. Prior to infection assays, the obtained adult flies were homogenized and checked by PCR with the universal bacterial 16S rRNA gene primers 27F/1492R before uses [9].
Phylogenetic analysis of fungal AMPs
The defensin-like AMP homologs of Aspergillus fumigatus afusinC [22], and the proteins containing the antifungal protein domain (Pfam11402) were retrieved from the genome of B. bassiana [26] by BLASTP search. In total, four proteins were obtained: the defensin-like peptides BBA_01785 (termed BbAMP1) and BBA_09303 (BbAMP2), and the AFP-like peptides BBA_08528 (BbAMP3) and BBA_08974 (BbAMP4, also called BbAFP1) [27]. Each sequence was then used for BLASTP search against the genomes of the sequenced entomopathogenic fungi [28, 29]. Otherwise, the AMPs of fungal origin reported elsewhere were obtained. The proteins were aligned with the program Clustal X [30]. A maximum-likelihood tree was generated using a Jones-Taylor-Thornton matrix-based model with 500 bootstrap replicates using the software MEGA X [31].
Gene deletion and fungal transformation
The individual deletions of BbAMP1-BbAMP4 genes were performed via Agrobacterium-mediated transformation as we described before [32]. Briefly, the 5′- and 3′-flanking regions of each gene were amplified using the genomic DNA as a template with different primer pairs (Table S1). The obtained fragments were purified, and cloned into the binary vector pDHt-Bar (conferring resistance to ammonium glufosinate) for transformation of the WT strain of B. bassiana. The drug-resistance colonies were used for single spore isolation and verification of gene deletion by PCR. For complementation of the BbAMP1 null mutant, the open reading frame of this gene together with its promoter region was amplified, purified and cloned into the binary vector pDHt-Ben (conferring benomyl resistance) for transformation of the null mutant. For overexpression of BbAMP1, the gene was amplified and made under the control of the constitutive GpdA promoter [33]. Each mutant was verified by PCR and or reverse transcription (RT)-PCR. In addition, BbAMP1 was fused in-frame at its 3′-terminus with a green fluorescent protein (GFP) gene and the cassette was controlled by GpdA promoter to transform the WT strain. The obtained overexpression strain OE::BbAMP1-GFP was used for examination of protein localization. The strain only transformed with the GpdA-controlled GFP gene (i.e., the obtaining of OE::GFP) was used as a control [34]. The WT and obtained mutants were inoculated in SDB for 36 h and fungal cell walls were stained with 0.01% (w v−1) Calcofluor white for 1 min and rinsed with the phosphate-buffered saline (PBS, pH 8.0) prior to the observations under a fluorescent microscope (Nikon, CX21) [35].
Quantitative gene expression analysis
To investigate the pattern of gene expressions, we extracted the total RNA from the mycelia harvested from the two-day-old SDB cultures, conidial spores harvested from the 2-week-old PDA plates, and hyphal bodies harvested from the insect body cavity after infections [33]. In addition, conidial spores and mycelial samples were collected from the insect cadavers for RNA extraction and gene expression analysis. The qRT-PCR analysis of BbAMP1-BbAMP4 expression was performed using a SYBR mix (Yeasen, Shanghai) with the primer pairs for different genes (Table S1). The β-tubulin gene of B. bassiana was used as an internal reference [33]. For bacterial induction assays, L. plantarum was inoculated into the flasks (50 mL in size) containing 20 mL of the de Man, Rogosa and Sharpe medium (MRS, Oxoid) and incubated at 30 °C and 200 rpm; Staphylococcus aureus and Erwinia carotovora carotovora 15 (Ecc15) were grown in Luria-Bertani (LB) at 37 °C and 200 rpm for 18 h. The cells were then collected by centrifugation and washed twice with sterile water. The WT strain of B. bassiana was inoculated in LB (at a concentration of 5 × 106 conidia mL−1) and incubated at 28 °C and 200 rpm for 14 h, the bacterial cells (each at a final OD600 of 0.01) were then added individually. The cultures were kept for incubation for up to 3 h. The samples were collected at different times for RNA extraction and qPCR analysis of BbAMP1 expression.
BbAMP1 secretion assay
The yeast secretion trap system was used to verify the secretion feature of BbAMP1 [36]. To this end, the signal peptide (SP) region of the secreted Blys2 effector [33] was used as a positive control. The SPs of both Blys2 and BbAMP1 (predicted) were amplified and fused in-frame with an invertase into the plasmid pSUC2 to generate the vectors pSUC2-SPBlys2 and pSUC2-SPBbAMP1 for transformation of the yeast strain YTK12 (invertase negative). The empty vector pSUC2 was also transformed as a negative control. Positive transformants were obtained on the CMD-W (without Trp) plates (0.67% yeast nitrogen base without amino acids, 0.075% Trp drop-out supplement, 2% sucrose, 0.1% glucose and 2% agar) at 30 °C and verified by PCR with the primers pSUC2-F/pSUC2-R (Table S1). To assay the secretion and activity of invertase, we also inoculated yeast cells on the YPRAA plates [1% yeast extract, 2% peptone, 2% raffinose and 2 μg mL−1 antimicyn A (Coolaber, Beijing)] for the reduction of tetrazolium chloride (TTC, SinoPharm) to insoluble red pigment triphenylformazan [36]. Reduction assays were also conducted in liquid cultures. In addition, the WT, OE::BbAMP1-GFP and OE::GFP strains of B. bassiana were grown in SDB for three days. The culture supernatants were obtained by filtration and concentrated for protein precipitation with ammonium sulfate. Western blot analysis was then conducted using an anti-GFP antibody (Beyotime, China).
Mature peptide expression and antimicrobial activity assays
The cDNA region corresponding to the deduced mature BbAMP1 (mBbAMP1) was cloned into the pGEX-6P-3 plasmid by fusion with a glutathione S-transferase (GST) tag at the C-terminus for transformation of the Escherichia coli Rosetta cells (Weidi Biotech, Shanghai). The mature region of BbAMP1 was used for structure prediction with the program Phyre2 in comparison to the defensins phormicin and micasin [37]. The expression of mBbAMP1-GST was induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Sigma-Aldrich) in LB medium at 16 °C for overnight. The fusion protein was purified through the glutathione affinity chromatography and analyzed by SDS-PAGE analysis. The anti-GST antibody (Abcam, Shanghai) was used for Western blot verification. On-column cleavage of the GST-tag was conducted using enterokinase (Yeasen, Shanghai). The purified mBbAMP1 peptide was analyzed by Tricine-SDS-PAGE. The minimum inhibitory concentration (MIC) values of mBbAMP1 were examined against different bacteria (i.e., the G+ bacteria L. plantarum, Corynebacterium nuruki, Enterococcus faecalis and S. aureus; the G− bacteria Ecc15 and E. coli) and yeast cells (C. albicans and S. cerevisiae) by microplate assays. The antibiotic ampicillin and the antifungal drug fluconazole (SinoPharm, Beijing) were used as positive controls.
Insect bioassays
Both topical infection and injection assays were conducted to determine the virulence difference between the WT and mutants against two insect species, i.e., the last instar larvae of wax moth (Galleria mellonella; Keyun, China) and the females of D. melanogaster W1118 line (two to 3 days post eclosion) as described [38]. Briefly, the spore concentrations of each strain were adjusted to 3 × 107 conidia mL−1 in 0.05% Tween 20 for topical infections of the wax-moth larvae and flies by immersion in spore suspension for 30 s. The treated insects were kept at a high moisturizing condition (relative humidity > 95%) for 24 h and then maintained in the growth chamber at 25 °C. For injection assays, the wax-moth larvae were individually injected with 20 μL of spore suspension (1 × 106 conidia mL−1) at the second proleg with a hand microapplicator (Burkard, Hertfordshire, UK). The female flies were individually injected with 50 nL of spore suspension (5 × 105 conidia mL−1) with a microinjector (Nanoject III, Drummond, Broomall, PA).
The sterile flies obtained above were also used two to three days post eclosion for gnotobiotic assays with and without the addition of L. plantarum and Acetobacter persici (each at a final OD600 of 0.01 after trial assays) cells in each spore suspension (3 × 107 conidia mL−1 each) of WT and mutants. There were more than 70 flies and 45 caterpillar larvae used for each treatment. The experiments were repeated at least twice. Insect mortality was recorded every 12 h. The median lethal time (LT50) value of each strain was calculated by Kaplan–Meier analysis and the survival distributions were compared by log-rank test [34].
Cross-antagonism assays between fungi and bacteria
Different bacteria were used to test the inhibition effect on spore germination of the WT and mutant strains of B. bassiana, including those isolated from Drosophila cuticles, such as L. plantarum, C. nuruki, E. faecalis [9] and the common experimental bacteria, such as S. aureus, Ecc15, and E. coli. As indicated above, L. plantarum was grown in the flask containing the MRS media (20 mL) and incubated at 30 °C and 200 rpm. The other bacterial species were incubated in LB broth at 37 °C and 200 rpm for 14 h. The bacterial cells were collected by centrifugation, washed twice with sterile water, and re-suspended in fresh LB medium. Fungal spores were harvested from 2-week-old PDA plates and suspended in 0.05% Tween 20. The LB medium (20 mL) was inoculated with the adjusted value of bacterial cells (each at a final OD600 of 0.01) and fungal spores at the final concentration of 5 × 106 conidia mL−1. The mixed samples were incubated in a rotary shaker at 28 °C and 200 rpm for 12 h and the spore germination rate was determined for each strain after microscopic examinations. There were three replicates for each strain against each bacterium. More than 100 spores were counted for each replicate. Dual-culture overlay assay was conducted to determine the antifungal activity of L. plantarum as being described [39]. In brief, bacterial cells were inoculated on MRS agar plates at 30 °C for 24 h. The agar strips (2 × 0.5 cm) were then carefully cut and transferred upside down to the middle of a new plate (two strips each). Each plate was then overlaid with 10 mL of malt extract soft agar (BD Difco) containing the spores of each strain at the final concentration of 5 × 104 conidia mL−1. The plates were incubated at 30 °C for 3 days. There were three replicates for each treatment.
Fungal penetration and colonization assays
To determine the bacterial blocking effect, we used the sterile cellophane membranes for examining fungal penetrations [40]. Spore suspension (5 × 106 conidia mL−1) of the WT and mutant strains of B. bassiana was mixed with the cells of different bacteria used above at a final amount of 0.01 OD600. The aliquots of 2 μL were then inoculated on the middle of membranes placed on the minimum medium agar plates for five days. The cellophanes were then carefully removed, and the plates were kept for incubation for four additional days to examine the cases of fungal penetration or not. Fungal loads within insects were also examined after topical infections. After trial assays, the sampling time was set at the half LT50 value of the WT strain. Thus, female flies were treated as indicated above for topical infections. The insects were then collected ca. 110 h post treatment and frozen in liquid nitrogen for fine homogenizations. Total DNA of each sample (five flies each) was extracted using the DNeasy Blood & Tissue kit (QIAGEN), and used for qPCR analysis with the primers (Table S1) designed for the 18S rRNA gene of B. bassiana and the Drosophila Rpl32 gene (used as a reference) [41]. There were six replicates for each strain.
Scanning electron microscopy analysis
Following overnight growth, the cells of L. plantarum and S. cerevisiae were collected by centrifugation and washed twice with sterile water. The cells were then treated with the BbAMP1-GFP and GFP proteins (each at a final concentration of 100 μg mL−1) for 0, 1 and 6 h. The cells (20 μL each) were then transferred onto the poly-L-lysine coated glass slides for one hour prior to being visualized using a Field-Emission Scanning Electron Microscope (Merlin Compact VP, Zeiss) [23]. For observation of microorganisms on fly surfaces, the flies were immersed with 0.05% Tween 20 and the WT spore suspension (3 × 107 conidia mL−1) for 30 s. After treatments for 18 h, the flies were freeze killed and used for scanning electron microscopy (SEM) observations of surface microorganisms as described before [9].
Microbiome analysis
To determine the effect of fungal inoculation on fly surface microbiome structures, we used female flies for immersions with the WT, ∆BbAMP1, and OE strains similar to the above topical infection treatments. The flies treated with 0.05% Tween 20 were included as a mock control. The treated flies were maintained at a high humidity condition (RH > 95%) for 12 h. The flies were then anesthetized with CO2 and washed (10 flies per replicate) with the sterile PBS buffer (pH, 7.4) [9]. The wash buffers were plated on LB medium for counting colony-forming units (CFUs) and comparison between treatments. The samples were also used for microbiome analysis as we described before [9]. In brief, after DNA lysis of the collected bacteria, each sample (1 μL) was used as the template for PCR amplification with the primers 515F/806R (Table S1). After quality checks, the products were used for the construction of paired-end libraries (2 × 250 bp) and sequencing (HiSeq 2500, Illumina) by the service company Biozeron (Shanghai, China). After normalization, sequencing reads were analyzed for estimation of operational taxonomic units (OTUs) at a cutoff of 97% identity using the program Mothur ver. 1.45.3 [42]. Different α-diversity indices: number of bacterial families (Nbf) per sample, number of bacterial species (Nbs) per sample, Shannon H and Simpson, were estimated based on the detected OTUs with the program Mothur. There were eight independent replicates for each sample.
Statistical analysis
The difference in insect survival was compared by log-rank test. One-way ANOVA analysis was conducted to determine the difference in gene expression, fungal spore germination, and CFU formation between different samples. Two-tailed Student’s t-tests were conducted to determine the α-diversity index difference between samples.
Results
Identification of a virulence-related antimicrobial gene
Our genome survey followed by a phylogenetic analysis revealed that BbAMP1 and BbAMP2 are defensin-like peptides while BbAMP3 and BbAMP4 are grouped with the characteristic AFPs (Fig. S1A). These genes were mainly expressed by the fungus at the conidiation stage with the exception that BbAMP1 could also be transcribed by the fungus growing in insect body cavity and during saprophytic growth (Fig. S2A). We performed the individual deletions of these four genes (Fig. S2B). The obtained null mutants had no growth defects compared with the WT strain (Fig. S2C). Topical infections of Drosophila adults revealed that the deletion of BbAMP1 (log-rank test: χ2 = 42.69; p = 0) but not the other genes could significantly increase insect survivals compared with the WT strain (Table S2). The function of this virulence-related BbAMP1 was thus further explored.
BbAMP1 can coat fungal cell walls and is inducible by different bacteria
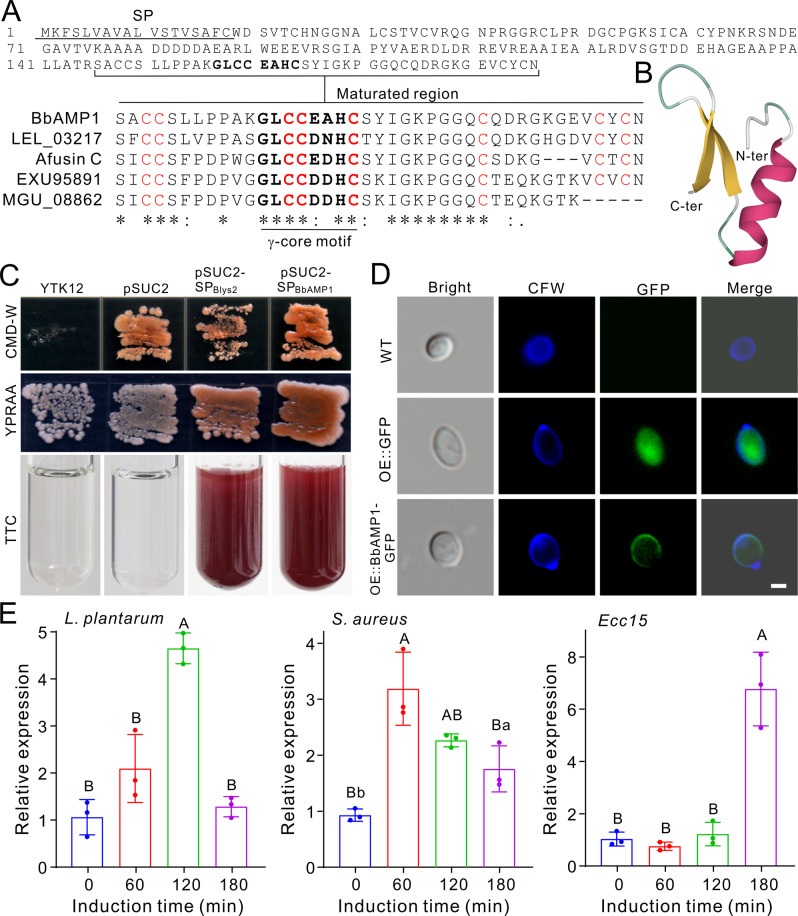
BbAMP1 contains a putative SP and is similar (44% identity at the amino acid level) to the afusinC produced by A. fumigatus, which is a cysteine-stabilized α-helix/β-sheet (CSαβ) defensin containing a γ-core motif GXCX3-9C [16, 43]. The putative mature peptide of BbAMP1 with 41 amino acid residues is cysteine-rich and harbors the conserved γ-core motif, which is highly similar to that of afusinC (Fig. 1A). The predicted peptide structure showed that mBbAMP1 contains one α-helix and two β-sheets, which is a typical αβ-structure (Fig. 1B). The predicted structure is highly similar to that of phormicin (defensin A, at a confidence level of 89.1%) of the blowfly Phormia terranovae [44], and the fDLP micasin (at a confidence level of 70.7%) of the dermatophytic fungus Microsporum canis [21] (Fig. S1B).
Fig. 1. BbAMP1 protein characteristics and inductive expression.
A BbAMP1 sequence feature and conservative relationships with other homologous proteins. SP, signal peptide. The residues highlighted in bold represent the γ-core motif. B Structural modeling of mature BbAMP1. C Yeast assay of BbAMP1 secretion. The signal peptide of the Blys2 effector was included as a positive control. The CMD-W (Trp) medium was used to select the yeast strain YTK12 carrying the pSUC2 plasmid, and YPRAA medium was used to indicate that the secreted invertase reduced tetrazolium chloride (TTC) to red formazan. D Localization of BbAMP1 on the cell wall of spores. CFW, Calcofluor white to stain cell wall chitin. Bar, 2 μm. E Inductive expression of BbAMP1 in B. bassiana after challenge with different bacteria. Values are the mean ± SD (standard deviation). Letters labeled above each column indicate the difference level of p < 0.01 (capital) and p < 0.05 (lowercase) between different inductive times after one-way ANOVA analysis.
We performed a yeast secretion trap assay using an invertase reporter gene [36], and the results confirmed that BbAMP1 is a secreted protein (Fig. 1C). After the growth of OE::BbAMP1-GFP strain on solid PDA and in liquid SDB media, the GFP signal could be detected on the spore surfaces (Fig. 1D) as well as the mycelial cell surfaces (Fig. S3). After challenging B. bassiana with different bacteria, it was confirmed that BbAMP1 expression could be highly induced by L. plantarum in 2 h, by S. aureus in 60 min and by Ecc15 in 3 h (Fig. 1E). Taken together, these results indicate that BbAMP1 is a secreted fDLP that can coat fungal cells and its expression is inducible by both G + and G− bacteria.
Mature BbAMP1 can inhibit different G+ bacteria
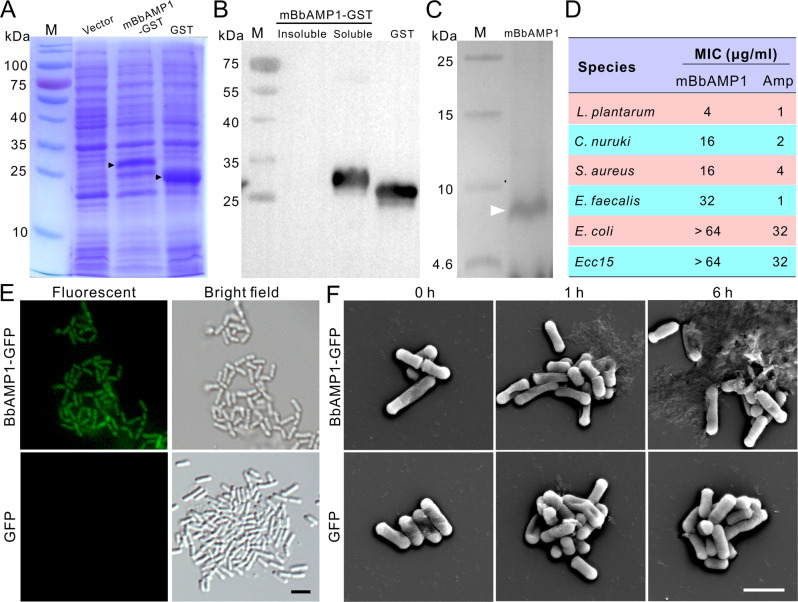
We next performed the heterologous expression of mBbAMP1 in E. coli (Fig. 2A–2C). The purified mBbAMP1 peptide was used to test the MIC values against different bacteria. The results indicated that mBbAMP1 could effectively inhibit G+ but not G− bacteria, especially the examined G+ bacterium L. plantarum isolated from fly surfaces (Fig. 2D).
Fig. 2. BbAMP1 expression and antimicrobial activity assays.
A Heterologous expression of the GST-fused form of mature BbAMP1 (mBbAMP1-GST). The targeted protein bands are arrowed. B Western blot analysis of the fused protein with an anti-GST antibody. C Tricine-gel analysis of the purified mBbAMP1 (arrowed). D MIC analysis of mBbAMP1 against different bacteria. Ampicillin (Amp) was used as a positive control. E Targeting of bacterial cells by BbAMP1. The GFP-fused protein was concentrated and incubated with the cells of L. plantarum for 1 h. GFP treatment was included as a control. The treated cells were washed twice with sterile water prior to being imaged. Bar, 1 μm. F Lysis of L. plantarum cells by BbAMP1. The cells of L. plantarum were treated with BbAMP1-GFP and GFP proteins (100 μg mL−1) for different times as indicated and then fixed on the poly-lysine coated slides for SEM observations. Bar, 1 μm.
We also harvested the mBbAMP1-GFP fusion protein from the culture filtrate of the OE::BbAMP1-GFP strain, and the concentrated protein was used to treat bacterial cells. A green fluorescent signal could be detected on the cells of L. plantarum, which was in contrast to the cells treated with the GFP protein (Fig. 2E). Further SEM analysis revealed the lysis of bacterial cells after treatment with BbAMP1-GFP but not with GFP for 1 or 6 h (Fig. 2F). The mature peptide could also inhibit the propagation of yeast cells such as C. albicans and S. cerevisiae (Fig. S4A). Yeast cells could be similarly targeted and damaged by BbAMP1-GFP (Fig. S4B, C). Thus, consistent with the reported dual functions of CSαβ defensins [15], BbAMP1 can target the cells of both G+ bacteria and yeasts.
BbAMP1 contributes to topical fungal infection of insects
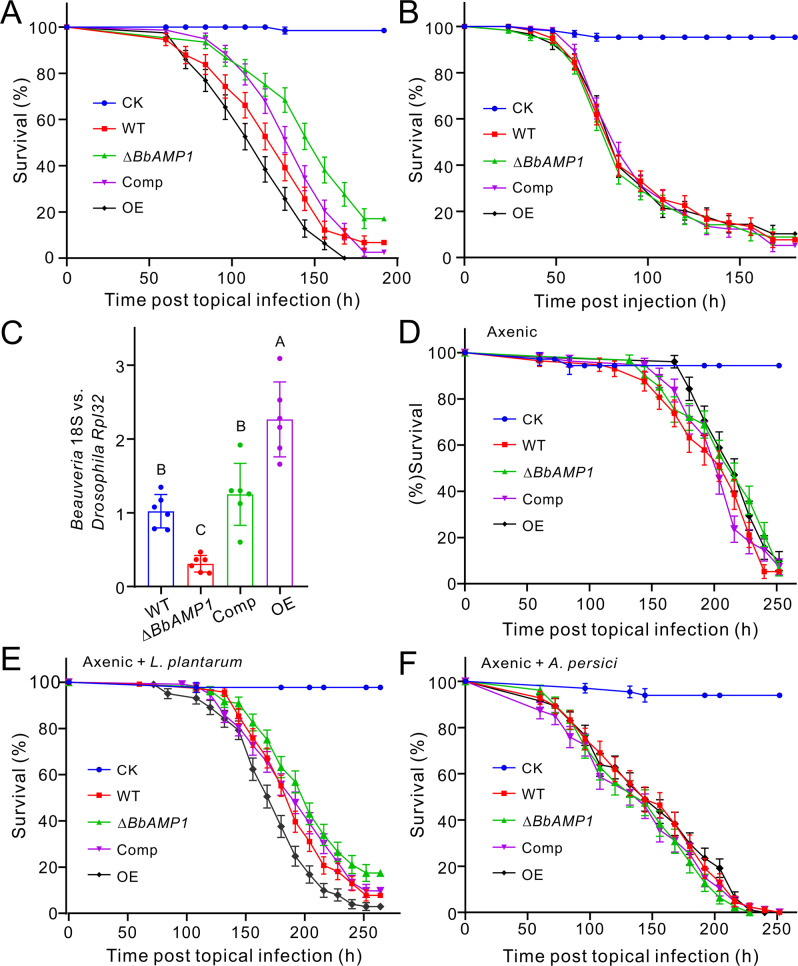
For further functional analysis, the null mutant of BbAMP1 was complemented (Comp), and the overexpression mutant (OE) was also generated on the WT strain background (Fig. S5A, B). The obtained mutants had no obvious differences in growth or stress responses compared with the WT strain (Fig. S5C). We then performed topical infection and injection assays against two insect species. After the topical infection of the female flies, significant differences in survival were evident between the flies treated with WT and ∆BbAMP1 (χ2 = 13.93; p = 0.0002), and between the WT and OE mutant treatments (χ2 = 6.17, p = 0.013). The estimation of LT50 values revealed an approximately 18% increase of fly survival time for the treatment with ∆BbAMP1 compared with that of the WT whereas the OE infection reduced fly survival times by 30% in reference to that of ∆BbAMP1. The difference between the WT and Comp strains was not significant (χ2 = 0.24, p = 0.88) (Fig. 3A). In fly injection assays, no obvious difference (p > 0.05) in survival was evident in response to the WT and mutant strains (Fig. 3B). Similar results were obtained in the topical and injection assays of wax-moth larvae (Fig. S6A, B). We quantified fungal loads after natural infection of flies. Consistent with the results of topical infections, deletion of BbAMP1 substantially impaired the ability of the fungus to colonize flies whereas the OE mutant could more quickly (p < 0.05) infect and proliferate in insects than the WT strain (Fig. 3C). The results indicated therefore that BbAMP1 could facilitate natural fungal infection of insects.
Fig. 3. Insect survivals after being treated with different fungal strains.
The fungal strains used include the wild type (WT), BbAMP1 gene deletion (ΔBbAMP1), complementation (Comp), and overexpression (OE) mutants of B. bassiana. Survivals of female flies after topical infection (A) and injection (B) with the different strains of B. bassiana. Plotted values are the mean ± SEM (standard error of the mean). The flies treated with 0.05% Tween 20 were used as a control (CK). C Variations in the fungal loads of flies after treatment with different strains. After topical infections for 110 h, total DNA was extracted and used for qPCR analysis of the fungal 18S rRNA gene and the fly Rpl32 gene. Values are the mean ± SD. Different letters labeled on the columns indicate the difference level of P < 0.01 between strains after one-way ANOVA analysis. D, E Survivals of axenic female flies after topical infection with the spore suspensions without (D) and plus (E) the addition of L. plantarum (Lp) cells. F Fly survivals after topical infection with the spore suspensions plus the addition of A. persici cells. Plotted values are the mean ± SEM. The flies treated with 0.05% Tween 20 plus the same amount of bacterial cells were used as a control. More than 70 flies were used for each treatment.
We further obtained and used axenic files for gnotobiotic assays. The survivals of sterile files had no statistical difference (e.g., WT vs. ∆BbAMP1 at χ2 = 2.65; p = 0.104) after being topically treated by the WT and mutant strains (Fig. 3D). In contrast, significant differences in fly survival distributions were observed between the WT and ΔBbAMP1 groups (χ2 = 5.76; p = 0.016), and between the WT and OE mutant treatments (χ2 = 9.42; p = 0.002) after the addition of L. plantarum cells (Fig. 3E). Consistent with the ineffective activity of BbAMP1 against G− bacteria, the addition of the G− A. persici cells abrogated the difference in survival between the treatments with the WT and mutant strains (Fig. 3F). Unsurprisingly, the addition of bacterial cells could significantly delay the topical infection of sterile flies by both the WT and ∆BbAMP1 strains. For example, fly survival was significantly (χ2 = 68.95; p < 0.0001) delayed when the insects were infected with the WT strain in the presence of L. plantarum cells (Fig. S6C). Similar results were observed when the flies were infected with ∆BbAMP1 with or without the addition of bacterial cells (χ2 = 76.33; p < 0.0001) (Fig. S6D). Relative to the treatments with the pure fungal spores, the addition of bacterial cells increased fly survival times by 22% and 46.7% for WT and ∆BbAMP1, respectively. It is noteworthy that, disregarding the physiological differences from each other, the conventionally reared (CR) flies generally succumbed faster to fungal infections than the same age axenic flies did (Fig. 3A, D). For example, the WT infection of CR flies had a LT50 value of 132 ± 5.0 h while its infection of sterile flies resulted in a LT50 value of 216 ± 8.02 h. Likewise, the LT50 values of ∆BbAMP1 were 156 ± 4.4 h against the CR flies but 216 ± 7.8 h against the sterile flies.
BbAMP1 facilitates the fungus to counteract bacterial inhibition
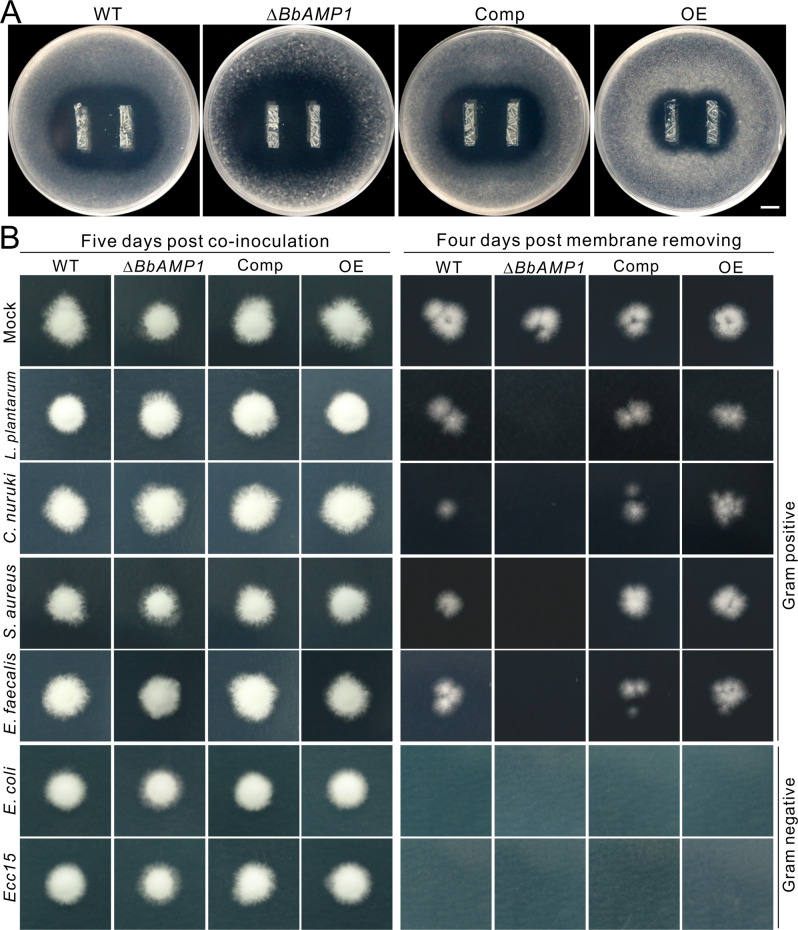
To verify whether the infection contribution of BbAMP1 was due to its antimicrobial activity against cuticular bacteria, we performed spore germination assays of WT and mutant strains in the presence of different bacteria. No difference was observed between strains in the absence of bacteria. However, relative to that of the WT strain, the spore germination rate of ∆BbAMP1 was significantly inhibited whereas those of the OE mutant were substantially increased after the addition of the G+ bacteria L. plantarum, C. nuruki, S. aureus and E. faecalis. The addition of the G− bacteria E. coli and Ecc15 could almost completely constrain the germination of the WT and mutant spores (Fig. S7A, B). The dual-culture overlay assays revealed that the OE mutant could effectively counteract the inhibition of L. plantarum whereas ∆BbAMP1 largely succumbed to bacterial suppression compared with the WT and Comp mutant of B. bassiana (Fig. 4A).
Fig. 4. Inhibition of fungal growth and membrane penetration by different bacteria.
A Dual overlay assays showing the variations between the WT and mutant strains of B. bassiana to counteract the antifungal activity of L. plantarum. The bacterial strips were overlaid with the malt-soft agars containing the spores of each strain (at a final concentration of 5 × 104 conidia mL−1) for three days. Bar, 1 cm. B Interference of different bacteria on the fungal penetration of cellophane membranes. The spore suspensions (2 μL each of 5 × 106 conidia mL−1) of each strain added with bacterial cells (each at a final OD600 = 0.01) were inoculated on the middle of membranes placed on PDA plates for 5 days (left panels). The membranes were then carefully removed and the plates were kept for incubation for another 4 days (right panels) to determine the penetration or non-penetration of each strain with or without bacterial cells.
To further corroborate these findings, we examined the fungal penetration of cellophane membranes in the presence or absence of different bacteria. Both the WT and mutants could penetrate the membranes in the absence of bacterial cells while the addition of G+ bacteria could impair the penetration of ∆BbAMP1 but not the other strains. Consistent with the inhibitory effects on spore germination, supplementation with the G− bacteria E. coli and Ecc15 completely blocked the penetration of both the WT and mutant strains (Fig. 4B). The results could thus explain the virulence defects of ∆BbAMP1 during natural infection of insects.
BbAMP1 facilitates the fungus to combat the fly surface microbiomes
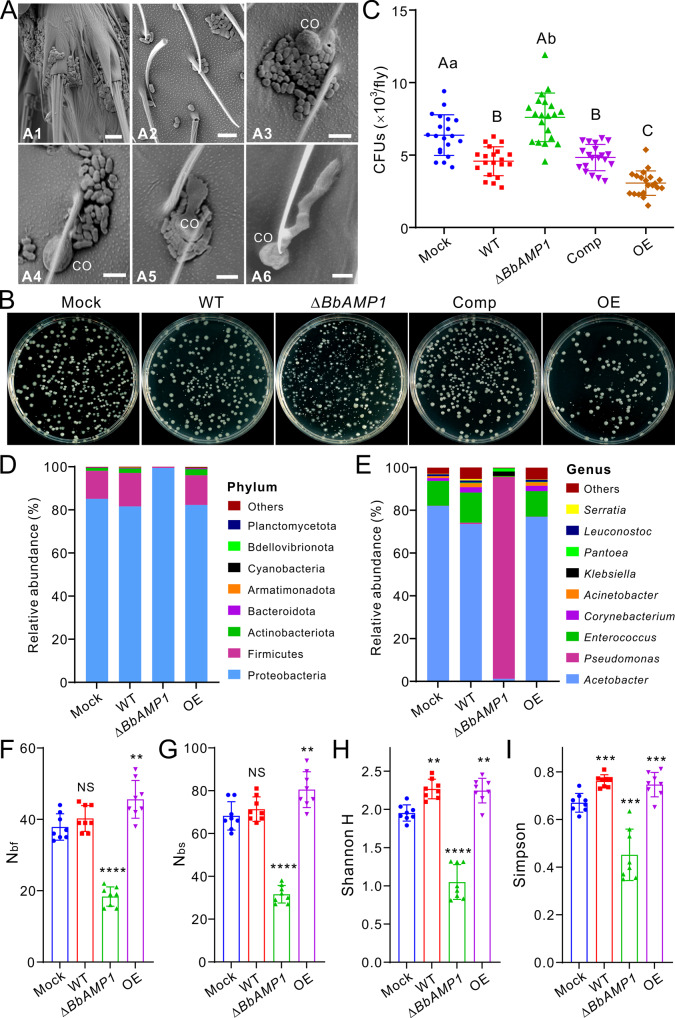
We next performed the SEM analysis of fly surfaces, CFU count, and microbiome structure analysis. Consistent with the previous observations [9], bacterial cells were observed on the flies especially on the tarsal segments of the fly legs, thorax, and abdomen surfaces (Fig. 5A). On fly surfaces, the inhibition of fungal spore germination was clearly evident once the spores were in contact with bacterial cells in reference to those without bacterial contacts (Fig. 5A). After the topical infection of flies with different strains, the insects were washed and the wash buffers were plated on LB agars for CFU counting (Fig. 5B). Statistically, the treatments with the spores of the WT, Comp and OE mutants could significantly (p < 0.01) lower the bacterial CFUs compared with the mock control. In comparison, the OE mutant could more significantly (p < 0.01) reduce bacterial numbers than the WT or Comp strains (Fig. 5C). In contrast, the deletion of BbAMP1 resulted in a substantial increase in bacterial CFU numbers (p < 0.01) (Fig. 5C). The results confirmed that BbAMP1 can bestow a competitive advantage on the fungus to combat fly surface bacteria.
Fig. 5. Differential manipulation of the fly surface microbiomes by different fungal strains.
A SEM observation of microorganisms on fly body surfaces. Panels A1 and A2 show the bacterial cells inhabiting the tarsus and thorax of flies. After topical infection of flies with WT spores for 18 h, conidium (CO) germination was inhibited once in contact with bacterial cells (Panels A3–A5) but not for those without contact with bacterial cells (Panel A6). Bar, 5 μm. B Colony-forming features of the bacteria washed off from the flies after the treatment with different strain spores (100× dilution). The LB plates were incubated at 37 °C for two days. C Variation in bacterial CFUs washed from fly surfaces after treatment with different strains. The letters above each sample represent the level of difference between treatments after one-way ANOVA analysis. Different capital letters indicate differences at the level of p < 0.01, and different lowercase letters represent the differences at the level of p < 0.05. Relative abundance of core bacterial phylum (D) and genera (E) detected on fly surfaces after fungal treatments by microbiome analysis. Variation in the α-diversity indices of Nbf (F, number of bacterial families), Nbs (G, number of bacterial species), Shannon H (H) and Simpson (I) between treatments. Values are the mean ± SD. Two-tailed Student’s t-test was performed to determine the significance of difference between the mock control and different strains: **p < 0.01; ***p < 0.001; ****p < 0.0001. NS not significant. The mock control was treated with 0.05% Tween 20. D–I: there were eight independent replicates for each treatment.
Further microbiome analysis of the flies after different treatments revealed the differences in relative bacterial abundance between strains, especially between ∆BbAMP1 and the other treatments at both the phylum and genus levels (Fig. 5D, E). Thus, the G− Pseudomonas bacteria of the Proteobacteria phylum dominated on the ∆BbAMP1-treated flies while the mock flies and those treated with the WT and OE strains were largely inhabited by Acetobacter (G−) and Enterococcus (G+) bacteria. Based on the detected OTUs (Table S3), the calculations of α-diversity indices revealed that the indices of Nbf and Nbs had similar patterns that were considerably increased (t-test, p < 0.01) after OE treatment but decreased after the inoculation of ∆BbAMP1 spores when compared with those of mock control (Fig. 5F, G). There were no statistical differences between WT and mock for these two indices. However, the Shannon H and Simpson indices were significantly (p < 0.01) increased by both the WT and OE spores whereas they were sharply (P < 0.001) reduced by the treatment of ∆BbAMP1 relative to those of mock control (Fig. 5H, I). These results revealed therefore that BbAMP1 could alter the fly surface microbiotas to facilitate fungal penetration of insect cuticles for systematic infections.
Discussion
We have recently shown that the microbiomes assembled on Drosophila body surfaces can defend the flies against fungal parasitic infections [9]. In this study, we revealed that the entomopathogenic fungus B. bassiana evolved a defensin-like BbAMP1 to battle the insect surface microbiotas to facilitate fungal infection of insect hosts. This peptide can be secreted by B. bassiana, coat fungal cells, bind and damage both bacterial and yeast cells. Likewise, mature afusinC can coat the spores of A. fumigatus [45]. In contrast to the wide distribution of AFP genes in different fungi [27], BbAMP1-like defensin is patchily distributed among fungal species and is largely absent from plant pathogens. Considering that mature BbAMP1 has a typical αβ-structure that is structurally similar to insect defensins, it is possible but remains to be determined whether BbAMP1 originated from insect hosts.
Functional genetic studies of entomopathogenic fungi such as Beauveria and Metarhizium species have identified an array of virulence-related genes involved in the regulation of infection structure differentiation, the penetration of cuticles and the evasion or invasion of host immunities [10, 12, 46]. In this study, we showed that BbAMP1 contributes to natural fungal infection of insects by suppressing the host surface microbiotas. In support of this observation, different bacteria, such as L. plantarum and E. faecalis, have been demonstrated to have broad antifungal activities through the production of bacteriocins and/or nutrient deprivation [47–49]. Considering that BbAMP1 also shows antifungal effects and the gene is inducible in saprophytic conditions, it remains to be determined whether this peptide is also involved in outcompeting other fungi or bacteria to facilitate B. bassiana to compete for insect hosts and/or micro-niches [34, 50]. A previous study has showed that the spore-coated antifungal protein BbAFP1 (BbAMP4 named in this study) of B. bassiana could inhibit the growth of competing fungi and has a potent potential to battle plant pathogens after transgenic expression in tomatoes [27]. It has been recently found that the soil-borne plant pathogen Verticillium dahliae can secrete AMPs to control soil and root surface microbiotas to facilitate fungal colonization of tomato roots [51] or to ward off fungal niche competitors [23]. Thus, AMP genes may enable the fungal producers to better survive in the diverse environments, including during the infection of hosts.
Besides for humans [1, 2], the microbiotas assembled on amphibian skins [3, 52] and plant phyllospheres [53–55] are also beneficial for hosts to fight pathogen infections. Taken together with the findings of the employment of surface microbiotas against fungal parasitic infections by insects [7, 9, 56], the results of this study highlight that the future investigations of fungus-host interactions have to be extended to the level of multiple interactions among fungi, surface microorganisms and hosts. Aside from AMP genes, small molecules with antibiotic activities may play similar roles in conferring fungal resistance to host surface microbiomes [5, 57]. In association with the insect habit of living, eating and excretion at the same environments, insect surface microbiomes are principally assembled from fecal bacteria [9]. Thus, insect ectomicrobiome structures are closely connected with their gut microbiotas. The latter are controlled by insect immune system [58, 59]. Fungal infections can invade and evade insect immunities [10], which leads to the dysbiosis of gut microbiotas to accelerate insect death [60, 61]. The findings would support our indirect evidence that, without considering the presence of physiological differences, the same age sterile flies survived a longer time than the CR flies upon the treatments with the same concentration of fungal spore suspensions. Thus, fungal infections may involve in manipulating both the ecto- and endo-microbiomes of insects. Obviously, further investigations are still required to determine the multi-interactive causes and effects.
Considering the wide occurrence of antagonism between microorganisms, it is not surprising to found that the topical infection of flies with B. bassiana could alter host surface microbiome structure and reduce bacterial loads. Likewise, the treatment of frogs with the chytrid fungus Batrachochytrium dendrobatidis could alter microbiome composition and the relative abundances of dominant bacterial taxa [62]. We found that BbAMP1 is largely effective against G+ bacteria, and the fungal reduction of surface bacterial loads was associated with BbAMP1 activity. Thus, consistent with our previous finding [9], the culturable G+ bacteria would dominate on Drosophila surfaces. The data also corroborated the finding that G− bacteria became expanded on the flies after treatment with the ∆BbAMP1 spores, which would lead to the reduction of α-diversity indices compared with other treatments. Likewise, the expansion of G− Proteobacteria on aging flies led to the substantial reduction of microbial diversity, i.e., the tradeoff between diversity and abundance [9]. This kind of negative correlations could also help explain that, despite of the fact that BbAMP1 has a bactericidal activity against G+ bacteria, the WT and especially OE treatments considerably increased the microbial diversities on fly surfaces in reference to those of mock control. The selective targeting mechanism of BbAMP1 against G+ but not G− bacteria requires further investigations.
In conclusion, we report the identification of a defensin-like peptide gene that can facilitate B. bassiana to infect insects by suppressing the host surface microbiotas. The findings of this study highlight that the outcome of the battle between pathogenic fungi and other microorganisms on host surfaces may essentially determine the fate of fungal infection or of host survival in response to pathogen attacks. Since B. bassiana has been developed and widely used as biocontrol agents against different insect pests [63], the results of this study suggest that the future development and application of mycoinsecticides might have to consider the influence of the defensive microbiotas formed on insect pest surfaces.
Supplementary information
Supplemental Figure S1-S7 and Table S1-S2
Author contributions
Conceptualization, supervision and funding acquisition: CW; performed the experiments: SH, YS, HC; analyzed the data: SH, HC; wrote the paper: SH, CW. All the authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32021001) and the Chinese Academy of Sciences (Nos. XDPB16 and QYZDJ-SSW-SMC028).
Data availability
Bacterial 16S rRNA sequencing fastq data have been deposited in the SRA (Sequencing Read Achieve) database with the BioProject accessions PRJNA751686 (SRX11638323-SRX11638354).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01323-7.
References
- 1.Ross AA, Rodrigues Hoffmann A, Neufeld JD. The skin microbiome of vertebrates. Microbiome. 2019;7:79. doi: 10.1186/s40168-019-0694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris-Tryon TA, Grice EA. Microbiota and maintenance of skin barrier function. Science. 2022;376:940–5. doi: 10.1126/science.abo0693. [DOI] [PubMed] [Google Scholar]
- 3.Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009;3:818–24. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- 4.Bletz MC, Perl RGB, Bobowski BT, Japke LM, Tebbe CC, Dohrmann AB, et al. Amphibian skin microbiota exhibits temporal variation in community structure but stability of predicted Bd-inhibitory function. ISME J. 2017;11:1521–34. doi: 10.1038/ismej.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batey SFD, Greco C, Hutchings MI, Wilkinson B. Chemical warfare between fungus-growing ants and their pathogens. Curr Opin Chem Biol. 2020;59:172–81. doi: 10.1016/j.cbpa.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruner-Montero G, Wood M, Horn HA, Gemperline E, Li L, Currie CR. Symbiont-mediated protection of Acromyrmex leaf-cutter ants from the entomopathogenic fungus Metarhizium anisopliae. mBio. 2021;12:e0188521. doi: 10.1128/mBio.01885-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pessotti RC, Hansen BL, Reaso JN, Ceja-Navarro JA, El-Hifnawi L, Brodie EL, et al. Multiple lineages of Streptomyces produce antimicrobials within passalid beetle galleries across eastern North America. Elife. 2021;10:e65091. doi: 10.7554/eLife.65091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DL, Smith EA, Newton ILG. A bacterial symbiont protects honey Bees from fungal disease. mBio. 2021;12:e0050321. doi: 10.1128/mBio.00503-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong S, Sun Y, Sun D, Wang CS. Microbiome assembly on Drosophila body surfaces benefits the flies to combat fungal infections. iScience. 2022;25:104408. doi: 10.1016/j.isci.2022.104408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CS, Wang SB. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu Rev Entomol. 2017;62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- 11.Ortiz-Urquiza A, Keyhani NO. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects. 2013;4:357–74. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Meng Y, Huang Y, Zhang D, Fang W. A novel cascade allows Metarhizium robertsii to distinguish cuticle and hemocoel microenvironments during infection of insects. PLoS Biol. 2021;19:e3001360. doi: 10.1371/journal.pbio.3001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: application informed by evolution. Science. 2020;368:eaau5480. doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol. 2015;1268:43–66. doi: 10.1007/978-1-4939-2285-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Oliverira Dias R, Franco OL. Cysteine-stabilized alphabeta defensins: from a common fold to antibacterial activity. Peptides. 2015;72:64–72. doi: 10.1016/j.peptides.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Zhu S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSalphabeta defensins. Mol Immunol. 2008;45:828–38. doi: 10.1016/j.molimm.2007.06.354. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Gao B, Zhu S. New fungal defensin-like peptides provide evidence for fold change of proteins in evolution. Biosci Rep. 2017;37:BSR20160438. doi: 10.1042/BSR20160438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Gao B, Zhu S. The fungal defensin family enlarged. Pharmaceuticals. 2014;7:866–80. doi: 10.3390/ph7080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science. 2010;328:1168–72. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 20.Oeemig JS, Lynggaard C, Knudsen DH, Hansen FT, Norgaard KD, Schneider T, et al. Eurocin, a new fungal defensin: structure, lipid binding, and its mode of action. J Biol Chem. 2012;287:42361–72. doi: 10.1074/jbc.M112.382028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu S, Gao B, Harvey PJ, Craik DJ. Dermatophytic defensin with antiinfective potential. Proc Natl Acad Sci USA. 2012;109:8495–500. doi: 10.1073/pnas.1201263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Contreras G, Braun MS, Schäfer H, Wink M. Recombinant AfusinC, an anionic fungal CSαβ defensin from Aspergillus fumigatus, exhibits antimicrobial activity against gram-positive bacteria. PLoS ONE. 2018;13:e0205509. doi: 10.1371/journal.pone.0205509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snelders NC, Petti GC, van den Berg GCM, Seidl MF, Thomma B. An ancient antimicrobial protein co-opted by a fungal plant pathogen for in planta mycobiome manipulation. Proc Natl Acad Sci USA. 2021;118:e2110968118. doi: 10.1073/pnas.2110968118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neyen C, Bretscher AJ, Binggeli O, Lemaitre B. Methods to study Drosophila immunity. Methods. 2014;68:116–28. doi: 10.1016/j.ymeth.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 25.Kietz C, Pollari V, Meinander A. Generating germ-free Drosophila to study gut-microbe interactions: Protocol to rear Drosophila under axenic conditions. Curr Protoc Toxicol. 2018;77:e52. doi: 10.1002/cptx.52. [DOI] [PubMed] [Google Scholar]
- 26.Xiao G, Ying SH, Zheng P, Wang ZL, Zhang S, Xie XQ, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong S, Li M, Keyhani NO, Liu Y, Yuan M, Lin D, et al. Characterization of a fungal competition factor: Production of a conidial cell-wall associated antifungal peptide. PLoS Pathog. 2020;16:e1008518. doi: 10.1371/journal.ppat.1008518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang YF, Xiao GH, Zheng P, Cen K, Zhan S, Wang CS. Divergent and convergent evolution of fungal pathogenicity. Genome Biol Evol. 2016;8:1374–87. doi: 10.1093/gbe/evw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu X, Xiao G, Zheng P, Shang Y, Su Y, Zhang X, et al. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc Natl Acad Sci USA. 2014;111:16796–801. doi: 10.1073/pnas.1412662111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang JM, Shang YF, Tang GR, Wang CS. Identification of a key G-protein coupled receptor in mediating appressorium formation and fungal virulence against insects. Sci China Life Sci. 2021;64:466–77. doi: 10.1007/s11427-020-1763-1. [DOI] [PubMed] [Google Scholar]
- 33.Cen K, Li B, Lu YZ, Zhang SW, Wang CS. Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 2017;13:e1006604. doi: 10.1371/journal.ppat.1006604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Yi W, Chen S, Wang CS. Empirical support for the pattern of competitive exclusion between insect parasitic fungi. J Fungi. 2021;7:385. doi: 10.3390/jof7050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang GR, Shang YF, Li SQ, Wang CS. MrHex1 is required for Woronin body formation, fungal development and virulence in Metarhizium robertsii. J Fungi. 2020;6:172. doi: 10.3390/jof6030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, et al. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat Plants. 2019;5:1167–76. doi: 10.1038/s41477-019-0527-4. [DOI] [PubMed] [Google Scholar]
- 37.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Song S, Wei X, Tang G, Wang CS. Activation of microlipophagy during early infection of insect hosts by Metarhizium robertsii. Autophagy. 2022;18:608–23. doi: 10.1080/15548627.2021.1943179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnusson J, Strom K, Roos S, Sjogren J, Schnurer J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol Lett. 2003;219:129–35. doi: 10.1016/S0378-1097(02)01207-7. [DOI] [PubMed] [Google Scholar]
- 40.Mei L, Wang X, Yin Y, Tang G, Wang CS. Conservative production of galactosaminogalactan in Metarhizium is responsible for appressorium mucilage production and topical infection of insect hosts. PLoS Pathog. 2021;17:e1009656. doi: 10.1371/journal.ppat.1009656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carboni AL, Hanson MA, Lindsay SA, Wasserman SA, Lemaitre B. Cecropins contribute to Drosophila host defense against a subset of fungal and Gram-negative bacterial infection. Genetics. 2022;220:iyab188. doi: 10.1093/genetics/iyab188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD. Reintroducing mothur: 10 years later. Appl Environ Microbiol. 2020;86:e02343–19. doi: 10.1128/AEM.02343-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonderegger C, Váradi G, Galgóczy L, Kocsubé S, Posch W, Borics A, et al. The evolutionary conserved γ-core motif influences the anti-Candida activity of the Penicillium chrysogenum antifungal protein PAF. Front Microbiol. 2018;9:1655. doi: 10.3389/fmicb.2018.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornet B, Bonmatin JM, Hetru C, Hoffmann JA, Ptak M, Vovelle F. Refined three-dimensional solution structure of insect defensin A. Structure. 1995;3:435–48. doi: 10.1016/S0969-2126(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 45.Dümig M, Binder J, Gaculenko A, Daul F, Winandy L, Hasenberg M, et al. The infectious propagules of Aspergillus fumigatus are coated with antimicrobial peptides. Cell Microbiol. 2021;23:e13301. doi: 10.1111/cmi.13301. [DOI] [PubMed] [Google Scholar]
- 46.St Leger RJ, Wang JB. Metarhizium: jack of all trades, master of many. Open Biol. 2020;10:200307. doi: 10.1098/rsob.200307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo P, Fares C, Longo A, Spano G, Capozzi V. Lactobacillus plantarum with broad antifungal activity as a protective starter culture for bread production. Foods. 2017;6:110. doi: 10.3390/foods6120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown AO, Graham CE, Cruz MR, Singh KV, Murray BE, Lorenz MC, et al. Antifungal activity of the Enterococcus faecalis peptide EntV requires protease cleavage and disulfide bond formation. mBio. 2019;10:e01334–19. doi: 10.1128/mBio.01334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Vuyst L, Leroy F. Bacteriocins from lactic acid bacteria: production, purification, and food applications. J Mol Microbiol Biotechnol. 2007;13:194–9. doi: 10.1159/000104752. [DOI] [PubMed] [Google Scholar]
- 50.Chen B, Sun YL, Li SQ, Yin Y, Wang CS. Inductive production of the iron-chelating 2-pyridones benefits the producing fungus to compete for diverse niches. mBio. 2021;12:e0327921. doi: 10.1128/mbio.03279-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snelders NC, Rovenich H, Petti GC, Rocafort M, van den Berg GCM, Vorholt JA, et al. Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat Plants. 2020;6:1365–74. doi: 10.1038/s41477-020-00799-5. [DOI] [PubMed] [Google Scholar]
- 52.Catenazzi A, Flechas SV, Burkart D, Hooven ND, Townsend J, Vredenburg VT. Widespread elevational occurrence of antifungal bacteria in Andean amphibians decimated by disease: A complex role for skin symbionts in defense against chytridiomycosis. Front Microbiol. 2018;9:465. doi: 10.3389/fmicb.2018.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Brettell LE, Singh B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020;25:841–4. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Wang J, Yang N, Wen Z, Sun X, Chai Y, et al. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat Commun. 2018;9:3429. doi: 10.1038/s41467-018-05683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogel CM, Potthoff DB, Schafer M, Barandun N, Vorholt JA. Protective role of the Arabidopsis leaf microbiota against a bacterial pathogen. Nat Microbiol. 2021;6:1537–48. doi: 10.1038/s41564-021-00997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chevrette MG, Carlson CM, Ortega HE, Thomas C, Ananiev GE, Barns KJ, et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun. 2019;10:516. doi: 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun YL, Hong S, Chen HM, Yin Y, Wang CS. Production of helvolic acid in Metarhizium contributes to fungal infection of insects by bacteriostatic inhibition of the host cuticular microbiomes. Microbiol Spectr. 2022;10:e0262022. doi: 10.1128/spectrum.02620-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marra A, Hanson MA, Kondo S, Erkosar B, Lemaitre B. Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. mBio. 2021;12:e00824–21. doi: 10.1128/mBio.00824-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 60.Wei G, Lai Y, Wang G, Chen H, Li F, Wang S. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc Natl Acad Sci USA. 2017;114:5994–9. doi: 10.1073/pnas.1703546114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu LT, Deng JD, Zhou FY, Cheng CH, Zhang LW, Zhang J, et al. Gut microbiota in an invasive bark beetle infected by a pathogenic fungus accelerates beetle mortality. J Pest Sci. 2019;92:343–51. doi: 10.1007/s10340-018-0999-4. [DOI] [Google Scholar]
- 62.Jani AJ, Bushell J, Arisdakessian CG, Belcaid M, Boiano DM, Brown C, et al. The amphibian microbiome exhibits poor resilience following pathogen-induced disturbance. ISME J. 2021;15:1628–40. doi: 10.1038/s41396-020-00875-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mei LJ, Chen M, Shang Y, Tang G, Tao Y, Zeng L, et al. Population genomics and evolution of a fungal pathogen after releasing exotic strains to control insect pests for 20 years. ISME J. 2020;14:1422–34. doi: 10.1038/s41396-020-0620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1-S7 and Table S1-S2
Data Availability Statement
Bacterial 16S rRNA sequencing fastq data have been deposited in the SRA (Sequencing Read Achieve) database with the BioProject accessions PRJNA751686 (SRX11638323-SRX11638354).