Abstract
Based on the pharmaceutical potentials of coumarins, which have antitumor activity, we synthesized new coumarin derivatives and evaluated their biological activities. The new coumarin derivatives were chemically synthesized from 4-hydroxycoumarin, and their structures were confirmed by nuclear magnetic resonance data. Ten of the synthesized compounds were investigated for antimetastatic activity against lung carcinoma cells. Several of the tested compounds showed good to mild inhibitory effects on lung cancer cell motility. There were no cytotoxic effects related to the use of these compounds. 4-Hydroxycoumarin derivatives, 4h and 4i, elicited the significant inhibitory effect on lung cancer cell motility by suppressing expression of the epithelial–mesenchymal transition markers N-cadherin, Snail, and Twist.
Subject terms: Cancer, Chemical biology
Introduction
Lung adenocarcinoma is the most malignant diagnosed cancer and is the leading cause of cancer death in men worldwide, followed by colorectal and liver cancer1,2. Lung cancer has various preferential sites for metastasis, such as the liver, brain, and bones3. Patients with metastatic lung adenocarcinoma have a poor prognosis and usually survive for fewer than 5 years4. The epithelial–mesenchymal transition (EMT) is a common mechanism underlying lung cancer cell motility and progression5. It facilitates metastasis by promoting lung cancer cell invasion, migration, and resistance to apoptotic stimuli. Therefore, inhibition of the EMT suppresses metastasis and progression of lung cancer6.
Coumarins, which are well-known condensed benzoheterocycles, are used as medicinal chemicals because they are easily synthetically transformed into a large variety of functionalized structures7. They have various purposes, such as synthesis of medicines8; laser dyes; and compounds with antifungal9, antibacterial10, anti-inflammatory11, and anticancer activities11,12. Recently, coumarin derivatives were reported to have inhibitory effects on prostate cancer, colon cancer, gastric cancer, and pancreatic cancer, mainly by regulating cell proliferation and apoptosis. Coumarin derivative esculetin inhibits the proliferation of human prostate cancer cells by inducing apoptosis and arresting the cell cycle13. 5-Geranyloxy-7-methoxycoumarin inhibits colon cancer cell growth via induction of apoptosis14. Coumarin-indole derivative MY-413 induces gastric cancer cell apoptosis and inhibits proliferation through MAPK signaling pathway15. Coumarin derivatives (12a–x) functions as NEDD8 activating enzyme inhibitors and induces apoptosis in pancreatic cancer cells16. Due to their many uses, it has become important to synthesize coumarins or their derivatives with conventional drugs by various reactions17,18. Various synthetic methods for coumarins and their derivatives have been used to insert various substituents into specific areas of the coumarin structure, thereby allowing mechanistic exploration of their anticancer activities. Herein, we summarize the inhibitory effects of coumarin derivatives on lung cancer cell motility.
Results and discussion
Chemistry
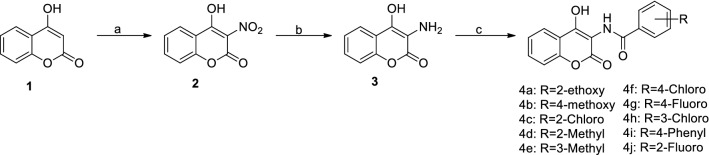
4-Hydroxycoumarin is an effective drug for small cell lung cancer15. Therefore, we used 4-hydroxycoumarin as a material to chemically synthesize 3-nitro-4-hydroxycoumarin (2) in the presence of HNO3 at 60 °C in an oil bath and then 3-amino-4-hydroxycoumarin (3) in the presence of Pd/C, and in a H2 atmosphere for 8 h (Fig. 1).
Figure 1.
Synthesis pathway of coumarin analogues. Reagents and conditions: (a) HNO3, NaNO2, AcOH, 1 h, 60 °C; (b) H2, Pd/C, EtOH, 8 h, rt; (c) Benzoyl chloride, Pyridine, CH2Cl2, 8 h, rt.
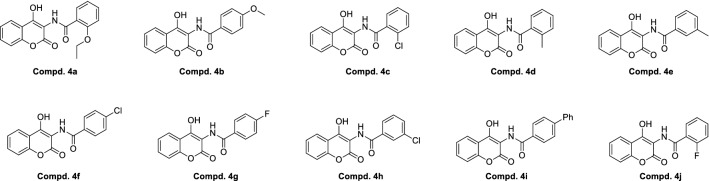
3-Amino-4-hydroxycoumarin (3) has an amino group (NH2), which is a stable functional site. Variously substituted acid chlorides (4a–4j) were selected as the linker via a carboxyl group (–COOH). Ten 3-amidocoumarins (4a–4j) were synthesized by a reaction involving formation of a peptide bond (–CO–NH–). A peptide bond was used due to its resonance stabilization and readiness to undergo chemical reactions. The structures of different aminocoumarin derivatives are presented in Fig. 2.
Figure 2.
Structure of Aminocoumarin derivatives (4a–4j).
Biological evaluation
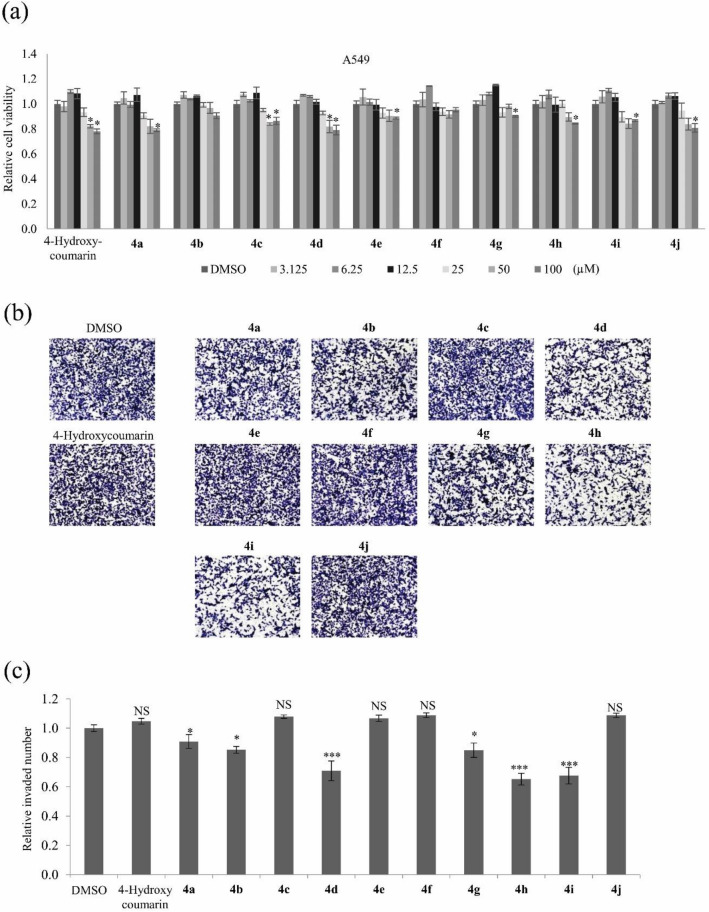
Cancer metastasis accounts for much of the morbidity and is associated with ~ 90% of the cancer-associated deaths19. According to the GLOBOCAN analysis, lung cancer is one of the most malignant types with metastasis, with a higher mortality rate of around 18.4%20. The motility of cancer cells contributes to tumor metastasis with several stages, including invading across the basement membranes, migration to blood, intravasation and movement into distant organs21. Cell migration and invasion are relevant phenotypes when studying the effect of novel therapeutic drugs against metastasis in cancer development and progression22. This study aimed to elucidate whether chemically synthesized coumarin derivatives affect lung cancer cell motility. To determine the cytotoxic effects of all the compounds on lung cancer cells, viability of A549, H460, and H1975 cells were evaluated via the MTT assays. Treatment with 3.125–20 μM of 4-hydroxycoumarin and its derivatives 4a–4j did not affect the viabilities of A549, H460, and H1975 cells. Treatment with 50–100 μM of 4-hydroxycoumarin and its derivatives 4a–4j inhibited the cell viabilities by ~ 25% in A549, H460, and H1975 cells (Fig. 3a and Supplementary Fig. S1). An invasion assay was performed to examine their effects on A549 cell motility. Cells were treated with 4-hydroxycoumarin and its derivatives 4a–4j at nontoxic concentration of 5 μM for 24 h. Treatment with compounds 4d, 4h, and 4i significantly inhibited A549 cell invasion by ~ 40%, ~ 50%, and ~ 40% (p < 0.0001), respectively (Fig. 3b,c).
Figure 3.
The effects of 4-hydroxycoumarin and its derivatives 4a–4j on invasion of A549 cells. (a) Relative viabilities of A549 cells after treatment with 4-hydroxycoumarin and its derivatives 4a–4j for 48 h at concentrations ranging from 3.125 to 100 μM measured by the MTT assay. (b) Representative images of invasion assays of A549 cells treated with 4-hydroxycoumarin and its derivatives 4a–4j at a concentration of 5 μM. (c) Quantitative analysis of invaded cell numbers after each treatment. Representative images from three independent experiments are shown (n = 3). Data represent the mean ± SD. *p < 0.05; ***p < 0.001; NS, no significant difference compared with dimethyl sulfoxide (DMSO)-treated A549 cells.
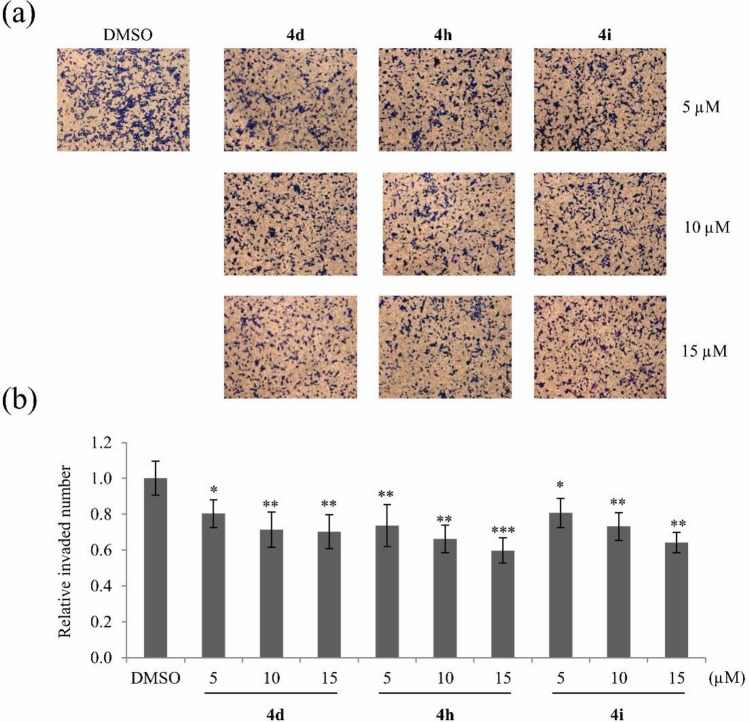
To elucidate how these three compounds inhibit cancer cell motility at noncytotoxic concentrations, cells were mainly treated with nontoxic concentrations at 5, 10, or 15 μM of compounds 4d, 4h, and 4i in subsequent experiments. Treatment with compounds 4d, 4h, and 4i dose-dependently decreased the number of invaded A549 cells (Fig. 4).
Figure 4.
Suppression of A549 cell invasion by compounds 4d, 4h, and 4i. (a) Representative images of invaded A549 cells after treatment with various nontoxic concentrations (5, 10, and 15 μM) of compounds 4d, 4h, and 4i for 24 h. (b) Quantitative analysis of invaded cell numbers in each group. Representative images from three independent experiments are shown (n = 3). Data represent the mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001 compared with DMSO-treated A549 cells.
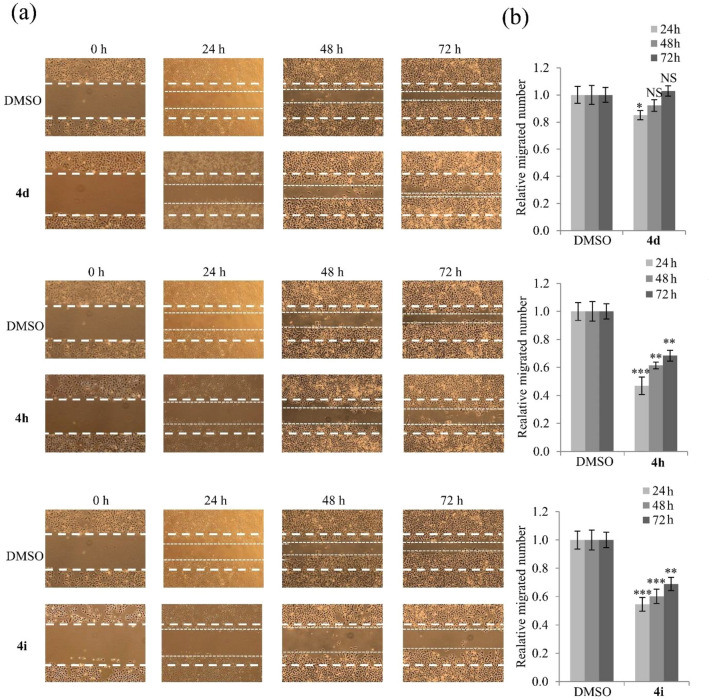
Wound healing assays are a commonly used method to evaluate cell migration ability23. The wound healing assay was performed by treating A549 cells with 15 μM compounds 4d, 4h, and 4i (Fig. 5). Cell migration was inhibited by treatment with compounds 4h and 4i in a time-dependent manner but was unaffected by compounds 4d treatment.
Figure 5.
Inhibition of A549 cell migration by compounds 4d, 4h, and 4i. (a) Representative images and (b) quantitative analysis of migration assays of A549 cells treated with 15 μM compounds 4d, 4h, and 4i for 24, 48, and 72 h. Representative images from three independent experiments are shown (n = 3). Data represent the mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; NS, no significant difference compared with DMSO-treated A549 cells.
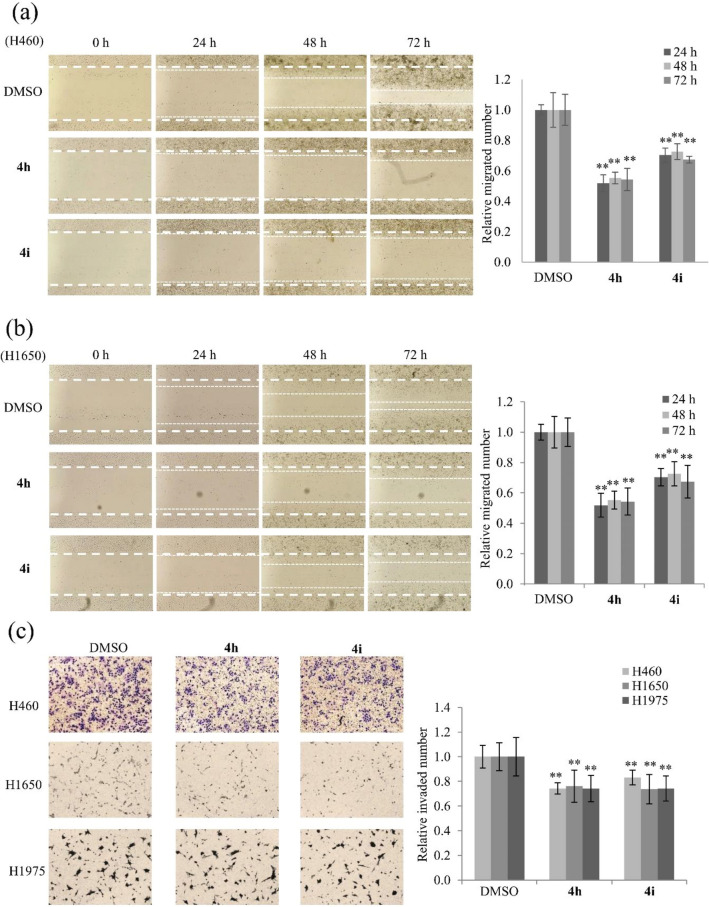
Moreover, the migration assay was performed using the other lung cancer cell lines H460, and H1650. Treatment with compounds 4h and 4i significantly decreased abilities of migrated cells in both cell lines (Fig. 6a,b). However, cell migration abilities of H460 and H1650 were also unaffected by treatment with compound 4d (Supplementary Fig. S2), indicating that it might not be a selection against lung cancer motility. The invasion assays were performed using H460, H1650, and H1975 cells. Treatment with compounds 4h and 4i significantly decreased numbers of invaded cells of all three cell lines by more than 25% (p < 0.005) (Fig. 6c). Taken together, these results indicate that compounds 4h and 4i significantly suppress lung cancer cell motility.
Figure 6.
Inhibition of migration and invasion migration of different lung cancer cell lines by compounds 4h and 4i. (a,b) Representative images and quantitative analysis of migration assays of H460, and H1650 cells treated with 15 μM of compounds 4h and 4i for 24, 48, and 72 h. (c) Representative images and quantitative analysis of invasion assays of H460, H1650, and H1975 cells treated with 15 μM of compounds 4h and 4i for each cell line. Representative images from three independent experiments are shown (n = 3). Data represent the mean ± SD. *p < 0.05, **p < 0.01 compared with DMSO-treated cells.
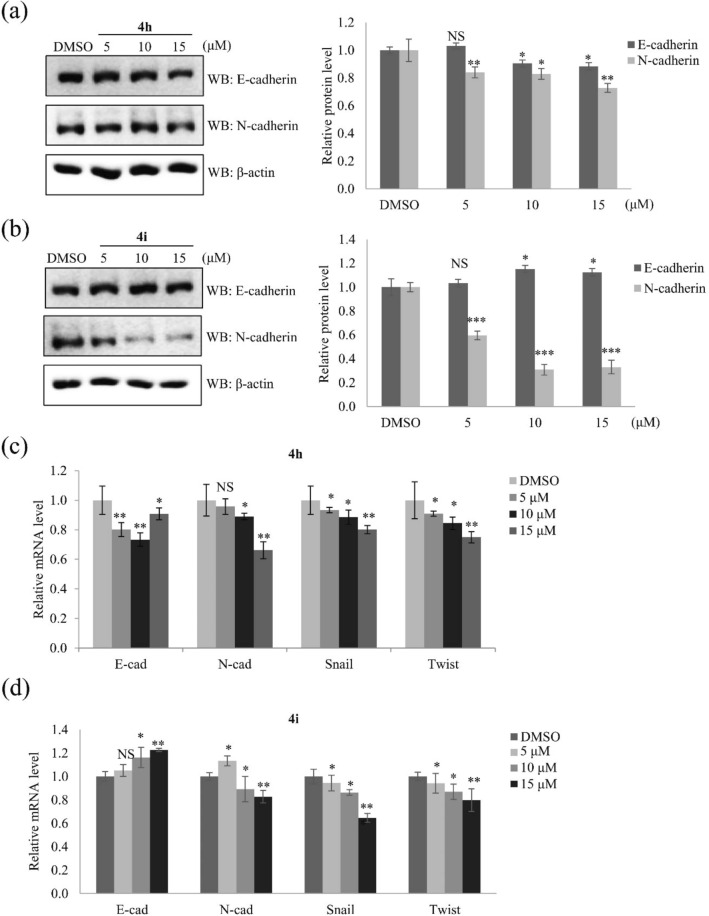
The EMT plays a role in lung cancer cell motility and metastasis24. In the EMT process, epithelial cells lose cell–cell adhesion and decrease the expression of epithelial cells marker such as E-cadherin, inversely, the expression of mesenchymal cell markers such as N-cadherin is increased, associated with invasive phenotype25. Several reports have proved that loss of E-cadherin expression or induction of N-cadherin led to poor prognosis in lung cancer26–28. E-cadherin and N-cadherin, as well as their regulators Snail and Twist, are indicators of the EMT24. The protein and mRNA expression levels of these EMT markers and their regulators were examined to determine how compounds 4h and 4i suppress lung cancer cell motility. Treatment with 15 μM compounds 4h decreased expression of not only N-cadherin but also E-cadherin and suppressed mRNA expression of Snail and Twist (Fig. 7a,c). Treatment with compound 4i significantly increased the expression of E-cadherin and downregulated the expression of N-cadherin, Snail, and Twist at a dose-dependent manner (Fig. 7b,d). These results indicate that compound 4i inhibits expression of EMT markers more than compound 4h. However, compound 4d did not affect E/N-cadherin expression as shown in Supplementary Fig. S2c. Taken together, our data demonstrate that compounds 4h and 4i inhibit lung cancer cell motility by downregulating EMT markers.
Figure 7.
Compounds 4h and 4i suppress A549 cell motility by modulating expression of EMT markers and their regulators. (a,b) Western blotting and quantification of the relative protein levels of E-cadherin and N-cadherin in A549 cells treated with nontoxic concentrations (5, 10, and 15 μM) of compounds 4h and 4i. β-Actin served as a loading control. (c,d) Relative mRNA levels of EMT effectors (E-cadherin and N-cadherin) and their transcription factors (Snail and Twist) in A549 cells treated with nontoxic concentrations (5, 10, and 15 μM) of compounds 4h and 4i. The mRNA levels were normalized against that of the housekeeping gene GAPDH. Quantitative data were obtained from at least three independent experiments. Data represent the mean ± SD. Statistical analysis was performed by a one-way analysis of variance. *p < 0.05; **p < 0.01; ***p < 0.001; NS, no significant difference compared with DMSO-treated A549 cells.
Conclusion
Ten amidocoumarin derivatives (4a–4j) were newly synthesized by a chemical reaction, and their inhibitory effects on lung carcinoma were evaluated. Compounds 4h and 4i inhibited invasion and migration of lung cancer cells by modulating expression of EMT effectors. Moreover, the inhibition of EMT markers by compound 4i was highest, suggesting that it is a potential inhibitor of lung cancer motility. A further biological study is required to elucidate the detailed underlying molecular mechanism.
Methodology
Chemistry
Synthesis of 4-hydroxy-3-nitrocoumarin (2)
Commercially available 4-hydroxycoumarin (1, 3.24 g, 20 mmol, Aldrich) was dissolved in AcOH (60 mL). One portion of sodium nitrite (14 mg, 0.2 mmol) was added to this solution before HNO3 (3 mL) dropwise. The reaction was allowed to stir 10 min at room temperature, followed by heating at 60 °C for 1 h in an oil bath. The brown solution was allowed to attain to the room temperature and the pure compound crystallized out from the solution. The suspension was filtered and washed with hexane, and then dried to afford 3-nitro-4-hydroxycoumarin (2) as off-yellow crystals. 3.5 g; yield 85%.
Synthesis of 3-amino-4-hydroxycoumarin (3)
4-Hydroxy-3-nitrocoumarin (2, 3.1 g, 15 mmol) was dissolved in EtOH, and a catalytic amount of Pd/C (30 mg) was added to the mixture at room temperature. The resulting solution was stirred at room temperature under H2 atmosphere for 5 h. After completion of the reaction, the mixture was filtered to remove the catalyst with celite pad, washed with MeOH and ethyl acetate, and concentrated to afford an off-white solid. 2.3 g; yield 87%.
General procedure for the preparation of 3-aminocoumarins (4a–4j)
3-Amino-4-hydroxycoumarin (3, 90 mg, 0.5 mmol) was dissolved in CH2Cl2. Pyridine (120 uL, 1.5 mmol) was then added, and the mixture was cooled to 0 °C. Variously substituted acid chloride (4a–4j) was added dropwise at 0 °C, and the mixture was stirred overnight at room temperature. The reaction mixture was evaporated and purified by column chromatography (Hexane/EtOAc, 9:1) to give the compounds 3-aminocoumarins (4a–4j). The yield of 10 compounds is over 80%. NMR and LC–MS spectra were obtained using a JEOL 400 MHz NMR spectrometer (JEOL, Ltd., Tokyo, Japan) and LC–MS spectrometer (Agilent Technologies, Santa Clara, CA, USA), respectively (Supplementary Fig. S5).
Compd. 4a: 2-ethoxy-N-(4-hydroxy-2-oxo-2H-chromen-3-yl)benzamide. 1H NMR (400 MHz, CDCl3) δ: 11.06 (s, NH), 8.27 (d, J = 7.6 Hz, 1H), 8.04 (d, J = 7.6 Hz, 1H), 7.569–7.50 (m, 2H), 7.35–7.30 (m, 2H), 7.13–7.05 (m, 2H), 4.37–4.32 (m, 2H), 1.72–1.68 (m, 3H). 13C NMR (100 MHz, CDCl3) δ: 165.35, 161.11, 157.80, 153.41, 150.79, 134.73, 132.64, 131.52, 124.54, 121.33, 118.70, 117.56, 116.22, 112.46, 105.57, 65.81, 14.73. LC–MS (ESI+): Calc. for C18H15NO5 [M + H]+: 326.10, Found: 326.2.
Compd. 4b: N-(4-hydroxy-2-oxo-2H-chromen-3-yl)-4-methoxybenzamide. 1H NMR (400 MHz, CDCl3) δ: 8.80 (s, NH), 8.03 (d, J = 6.8 Hz, 1H), 7.94 (d, J = 6.8 Hz, 1H), 7.55–7.53 (t, 2H), 7.38–7.33 (t, 2H), 7.01–6.99 (d, J = 6.8 Hz, 2H), 3.90 (s, 3H). 13C NMR (100 MHz, CDCl3) δ: 166.98, 163.75, 161.41, 152.72, 150.54, 131.71, 129.78, 124.80, 124.52, 123.60, 117.33, 116.34, 114.42, 104.95, 55.70. LC–MS (ESI+): Calc. for C17H14NO5 [M + H]+: 312.09, Found: 312.2.
Compd. 4c: 2-chloro-N-(4-hydroxy-2-oxo-2H-chromen-3-yl)benzamide. 1H NMR (400 MHz, CDCl3) δ: 9.14 (s, NH), 8.05 (d, J = 8.4 Hz, 1H), 7.87 (d, J = 8.4 Hz, 1H), 7.59–7.54 (t, 1H), 7.53–7.47 (m, 2H), 7.44–7.40 (m, 1H), 7.38–7.34 (m, 2H). 13C NMR (100 MHz, CDCl3) δ: 166.57, 162.75, 160.98, 150.75, 133.08, 132.05, 131.91, 131.64, 131.06, 130.94, 127.51, 124.86, 124.62, 117.07, 116.40, 104.91. LC–MS (ESI+): Calc. for C16H11ClNO4 [M + H]+: 316.04, Found: 316.2.
Compd. 4d: N-(4-hydroxy-2-oxo-2H-chromen-3-yl)-2-methylbenzamide. 1H NMR (400 MHz, CDCl3) δ: 8.50 (s, NH), 8.04 (d, J = 8.0 Hz, 1H), 7.62 (d, J = 8.4 Hz, 1H), 7.58–7.56 (t, 1H), 7.490–7.30 (m, 4H), 2.61 (s, 3H). 13C NMR (100 MHz, CDCl3) δ: 170.52, 161.12, 152.87, 150.65, 137.33, 133.00, 131.90, 131.86, 131.82, 127.66, 126.38, 124.86, 124.55, 117.16, 116.37, 105.08, 20.39. LC–MS (ESI+): Calc. for C17H14NO4 [M + H]+: 296.09, Found: 296.2.
Compd. 4e: N-(4-hydroxy-2-oxo-2H-chromen-3-yl)-3-methylbenzamide. 1H NMR (400 MHz, CDCl3) δ: 8.88 (s, NH), 8.04 (d, J = 6.8 Hz, 1H), 7.76–7.74 (m, 2H), 7.58–7.54 (m, 1H), 7.43–7.41 (m, 2H), 7.40–7.341 (m, 2H), 2.45 (s, 3H). 13C NMR (100 MHz, CDCl3) δ: 170.60, 167.76, 161.32, 150.61, 139.24, 134.17, 131.86, 131.52, 129.08, 128.20, 124.87, 124.78, 124.75, 116.37, 110.16, 104.86, 21.47. LC–MS (ESI+): Calc. for C17H14NO4 [M + H]+: 296.09, Found: 296.2.
Compd. 4f.: 4-chloro-N-(4-hydroxy-2-oxo-2H-chromen-3-yl)benzamide. 1H NMR (400 MHz, CDCl3) δ: 8.86 (s, NH), 8.04 (d, J = 6.4 Hz, 1H), 7.92 (d, J = 8.4 Hz, 2H), 7.59–7.55 (t, 1H), 7.53–7.50 (m, 2H), 7.39–7.34 (m, 2H). 13C NMR (100 MHz, CDCl3) δ: 166.40, 161.27, 152.96, 150.72, 141.04, 139.93, 132.03, 129.53, 129.07, 124.93, 124.59, 117.11, 116.42, 104.66. LC–MS (ESI+): Calc. for C16H11ClNO4 [M + H]+: 316.04, Found: 316.2.
Compd. 4g: 4-fluoro-N-(4-hydroxy-2-oxo-2H-chromen-3-yl)benzamide. 1H NMR (400 MHz, CDCl3) δ: 9.66 (s, NH), 8.18–8.14 (t, 1H), 8.04 (d, J = 7.2 Hz, 1H), 7.64–7.60 (t, 1H), 7.58–7.54 (m, 1H), 7.38–7.33 (m, 2H), 7.26–7.23 (m, 2H). 13C NMR (100 MHz, CDCl3) δ: 163.39, 161.00, 153.07, 150.78, 135.35, 135.26, 132.20, 131.93, 125.33, 124.77, 124.59, 117.16, 116.86, 116.62, 116.40, 105.08. LC–MS (ESI+): Calc. for C16H11FNO4 [M + H]+: 300.07, Found: 300.2.
Compd. 4h: 3-chloro-N-(4-hydroxy-2-oxo-2H-chromen-3-yl)benzamide. 1H NMR (400 MHz, CDCl3) δ: 8.87 (s, NH), 8.05 (d, J = 8 Hz, 1H), 7.96 (m, 1H), 7.83 (d, J = 8 Hz, 1H), 7.61–7.55 (m, 2H), 7.50–7.46 (m, 1H), 7.9–7.35 (m, 2H). 13C NMR (100 MHz, CDCl3) δ: 166.15, 161.22, 153.03, 150.65, 135.60, 133.39, 132.08, 130.46, 128.12, 125.49, 124.94, 124.61, 116.43. LC–MS (ESI+): Calc. for C16H11ClNO4 [M + H]+: 316.04, Found: 316.2.
Compd. 4i: N-(4-hydroxy-2-oxo-2H-chromen-3-yl)-[1,1′-biphenyl]-4-carboxamide. 1H NMR (400 MHz, CDCl3) δ: 8.95 (s, NH), 8.05–8.02 (m, 3H), 7.76 (d, J = 8.8 Hz, 2H), 7.65 (d, J = 8.8 Hz, 2H), 7.59–7.54 (m, 1H), 7.50–7.47 (m, 2H), 7.44–7.35 (m, 3H). 13C NMR (100 MHz, CDCl3) δ: 167.19, 161.35, 152.93, 150.61, 146.23, 139.52, 131.88, 130.07, 129.14, 128.58, 128.25, 127.78, 127.38, 124.87, 124.58, 117.24, 116.39, 104.87. LC–MS (ESI+): Calc. for C22H16NO4 [M + H]+: 358.11, Found: 358.2.
Compd. 4j: 2-fluoro-N-(4-hydroxy-2-oxo-2H-chromen-3-yl)benzamide. 1H NMR (400 MHz, CDCl3) δ: 8.05–8.01 (m, 2H), 7.60–7.54 (m, 2H), 7.25–7.14 (m, 4H). 13C NMR (100 MHz, CDCl3) δ: 169.89, 164.01, 161.41, 135.66, 135.57, 132.83, 124.20, 124.16, 117.87, 117.79, 117.35, 117.12. LC–MS (ESI+): Calc. for C16H11FNO4 [M + H]+: 300.07, Found: 300.2.
Biological evaluation
Cell culture and material
The lung cancer cell lines A549, H1650, H1975, and H460 were obtained from the Korean Cell Line Bank (Seoul, South Korea). Cells were cultured in RPMI 1640/DMEM culture medium (Gen Depot, TX, USA) supplemented with 10% fetal bovine serum (Gen Depot, TX, USA) and 1% Penicillin–Streptomycin solution. Cells were cultured in 5% CO2 in a humidified atmosphere at 37 °C. 4-Hydroxycoumarin was purchased from Sigma-Aldrich (H23805).
MTT assay
Cells (2 × 104 cells/well) were seeded on a 96-well plate, grown overnight, and then treated with 4-hydroxycoumarin and its derivatives 4a–4j at concentrations from 3.125 to 100 μM for 48 h. After incubation with MTT at 37 °C, cells were lysed with 150 μL of DMSO (Sigma-Aldrich, St. Louis, USA) and absorbance was measured spectrophotometrically at 540 nm.
Invasion assay
Invasion assays were performed in Transwell chambers (Corning, New York, USA) with 8 μm pore size polycarbonate membrane coated with 1% gelatin. A549, H1650, H1975, and H460 cells were plated at 2 × 106 cells/mL in RPMI1640 containing 0.2% bovine serum albumin (BSA) in the upper compartment of the chamber. The lower chamber was filled with 400 μL RPMI containing 0.2% BSA and 10 μg/mL fibronectin (EMD MilliporeCorp., Billerica, MA, USA) as a chemoattractant. A549 cells were treated with 5 μM of 4-hydroxycoumarin and its derivatives (4a–4j) for 48 h. H1650, H1975, and H460 cells were treated with 5, 10, and 15 μM of derivatives 4h and 4i for 48 h. After incubation, the cells in the upper chamber were fixed with Diff Quik kit (Sysmex, Kobe, Japan). The numbers of cells in five fields per chamber were counted using a Nikon Eclipse 400fluorescence microscope (Nikon Instech, Co., Ltd, Kawasaki, Japan) and iSolution FL Auto × 64 software (IMT iSolution Inc., Vancouver, Quebec, Canada).
Wound healing assay
A549, H460, and H1650 cells were plated at a density of 2.5 × 105 cells/well and grown overnight to the confluence. Monolayer cells were scratched to create a wound. The cells were then washed twice and incubated in RPMI1640 culture medium supplemented with 2% FBS with 15 μM of derivatives 4d, 4h, and 4i. For the quantitation of relative migration ability, photographs of cells were taken at 0, 24, 48, and 72 h after wounding to measure the width of the wound. The distance migrated by the cells was calculated as the difference between the edges of the wound at time point 1 and at time point 2. For each sample, an average of five wound distances was taken to determine the average rate of migration at the time-course of 24, 48, and 72 h. The migrating rate was determined with the following formula: migrating rate [%] = (width t1 [mm] − width t2 [mm]) / width t1 × 100%.
Western blots
A549 cells were treated with 5, 10, or 15 μM concentrations of compounds 4d, 4h and 4i for 24 h. Cells were lysed and the proteins (25 μg) were separated by 10% polyacrylamide gel electrophoresis containing 0.1% SDS and electrophoretically transferred to nitrocellulose membranes. Anti-E-cadherin (#3195) (Cell signaling) and anti-N-cadherin(#13A9) (Cell signaling) were used as primary antibodies for the detection of EMT markers. Anti-ß-actin (Santa Cruz Biotechnology) was used as internal standards. All results are representative from at least three independent experiments. Bands were measured by Multi Gauge version 3.0 (Fuji photo film Co., Ltd., Tokyo, Japan) and their relative density calculated based on the density of the ß-actin bands in each sample. Values were expressed as arbitrary densitometric units corresponding to signal intensity. Cropped gels retain important bands, but whole gel images with membrane edges visible are available in Supplementary Figs. S3 and S4.
qRT-PCR
Briefly, total RNA was isolated from compounds 4h or 4i treated A549 cells using RNAiso Plus (TaKaRa, Otsu, Shiga 520-2193, Japan) and then total RNAs (1 μg) were used to synthesize cDNA using a M-MLV reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) and SYBR green (Enzynomics, Seoul, Korea). qRT-PCR reactions and analysis were performed using CFX (Bio-Rad, Hercules, CA, USA). The primers used for real-time PCR were as follows: E-cadherin (forward) 5′-cagaaagttttccaccaaag-3′ and (reverse) 5′-aaatgtgagcaattctgctt-3′; N-cadherin (forward) 5′-ctcctatgagtggaacaggaacg-3′ and (reverse) 5′-ttggatcaatgtcataatcaagtgctgta-3′; Snail (forward) 5′-tcccgggcaatttaacaatg-3′ and (reverse) 5′-tgggagacacatcggtcga-3′; Twist (forward) 5′-cgggagtccgcagtctta-3′ and (reverse) 5′-tgaatcttgctcagcttgtc-3′; and GAPDH (forward) 5′-atcaccatcttccaggagcga-3′ and (reverse) 5′-agttgtcatggatgaccttggc-3′. Target gene expression was normalized relative to the expression of GAPDH in the same sample.
Statistical analysis
All in vitro experiments were performed in triplicate. Data are presented as the mean standard deviation (SD) of 3 independent experiments with 3 replicates each. The student's t-test was utilized to determine statistical significance between two groups, and analysis of variance was utilized between three or more groups, respectively. Values of p < 0.05 were considered statistically significant. All statistical analyses were performed using Sigma Plot 12.5 software (Systat software, Erkrath, Germany).
Supplementary Information
Acknowledgements
This work was supported by the National Research Foundation of Korea Grant (NRF-2020R1C1C1007832) funded by the Korea government (MSIP).
Author contributions
H.K. and H.H.H. conceived and designed the experiments. R.Z. and Y.H.Y. performed the experiments. R.Z. and H.K. analyzed the data and wrote the manuscript. All authors read and approved the final manuscript. All authors read the M.S. and are agree with submission in its present for to the journal of Scientific Reports. The M.S. is original and is not considered for publication by other journals.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hangun Kim, Email: hangunkim@sunchon.ac.kr.
Hyung-Ho Ha, Email: hhha@sunchon.ac.kr.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-26212-z.
References
- 1.Roser, M. & Ritchie, H. Cancer. https://ourworldindata.org/cancer/ (2015).
- 2.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X, Chen J, Yang F, Tang C. Multiple sites of soft-tissue metastases secondary to lung cancer: A case report. Medicine (Baltimore) 2019;98:e18162. doi: 10.1097/MD.0000000000018162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zappa C, Mousa SA. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016;5:288. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittal V. Epithelial mesenchymal transition in aggressive lung cancers. Adv. Exp. Med. Biol. 2016;890:37–56. doi: 10.1007/978-3-319-24932-2_3. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, et al. lncRNA PCBP1-AS1 correlated with the functional states of cancer cells and inhibited lung adenocarcinoma metastasis by suppressing the EMT progression. Carcinogenesis. 2021;42:931–939. doi: 10.1093/carcin/bgab047. [DOI] [PubMed] [Google Scholar]
- 7.Adimule VM, Nandi SS, Kerur S, Khadapure SA, Chinnam SJ. Recent advances in the one-pot synthesis of coumarin derivatives from different starting materials using nanoparticles: A review. Top. Catal. 2022 doi: 10.1007/s11244-022-01571-z. [DOI] [Google Scholar]
- 8.Khursheed A, Jain V. Medicinal research progress of natural coumarin and its derivatives. Nat. Prod. J. 2021;11:648–662. [Google Scholar]
- 9.Hu XL, Xu Z, Liu ML, Feng LS, Zhang GD. Recent developments of coumarin hybrids as anti-fungal agents. Curr. Top. Med. Chem. 2017;17:3219–3231. doi: 10.2174/1568026618666171215100326. [DOI] [PubMed] [Google Scholar]
- 10.Gao F, Xiao J, Huang G. Current scenario of tetrazole hybrids for antibacterial activity. Eur. J. Med. Chem. 2019;184:111744. doi: 10.1016/j.ejmech.2019.111744. [DOI] [PubMed] [Google Scholar]
- 11.Thomas V, Giles D, Basavarajaswamy G, Kumar Das A, Patel A. Coumarin derivatives as anti-inflammatory and anticancer agents. Anticancer Agents Med. Chem. 2017;17:415–423. doi: 10.2174/1871520616666160902094739. [DOI] [PubMed] [Google Scholar]
- 12.Song XF, Fan J, Liu L, Liu XF, Gao F. Coumarin derivatives with anticancer activities: An update. Arch. Pharm. (Weinheim) 2020;353:2000025. doi: 10.1002/ardp.202000025. [DOI] [PubMed] [Google Scholar]
- 13.Turkekul K, Colpan RD, Baykul T, Ozdemir MD, Erdogan SJ. Esculetin inhibits the survival of human prostate cancer cells by inducing apoptosis and arresting the cell cycle. J. Cancer Prev. 2018;23:10. doi: 10.15430/JCP.2018.23.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patil JR, et al. 5-Geranyloxy-7-methoxycoumarin inhibits colon cancer (SW480) cells growth by inducing apoptosis. Planta Med. 2013;79:219–226. doi: 10.1055/s-0032-1328130. [DOI] [PubMed] [Google Scholar]
- 15.Song J, et al. Discovery of novel coumarin-indole derivatives as tubulin polymerization inhibitors with potent anti-gastric cancer activities. Eur. J. Med. Chem. 2022;238:114467. doi: 10.1016/j.ejmech.2022.114467. [DOI] [PubMed] [Google Scholar]
- 16.Gong L, et al. Design, synthesis and biological evaluation of coumarin derivatives as NEDD8 activating enzyme inhibitors in pancreatic cancer cells. Med. Chem. 2022;18:679–693. doi: 10.2174/1573406418666211210163817. [DOI] [PubMed] [Google Scholar]
- 17.Lončarić M, Gašo-Sokač D, Jokić S, Molnar M. Recent advances in the synthesis of coumarin derivatives from different starting materials. Biomolecules. 2020;10:151. doi: 10.3390/biom10010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aslam K, Khosa MK, Jahan N, Nosheen S. Short communication: Synthesis and applications of coumarin. Pak. J. Pharm. Sci. 2010;23:449–454. [PubMed] [Google Scholar]
- 19.Ganesh K, Massagué J. Targeting metastatic cancer. Nat. Med. 2021;27:34–44. doi: 10.1038/s41591-020-01195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 21.Stuelten CH, Parent CA, Montell DJ. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pijuan J, et al. In vitro cell migration, invasion, and adhesion assays: From cell imaging to data analysis. Front. Cell Dev. Biol. 2019;7:107. doi: 10.3389/fcell.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinotti S, Ranzato E. Scratch wound healing assay. Methods Mol. Biol. 2019 doi: 10.1007/7651_2019_259. [DOI] [PubMed] [Google Scholar]
- 24.Loh CY, et al. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells. 2019;8:1118. doi: 10.3390/cells8101118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao D, He J. Epithelial mesenchymal transition and lung cancer. J. Thorac. Dis. 2010;2:154. doi: 10.3978/j.issn.2072-1439.2010.02.03.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LT, Zhu KB, Qu CF. Novel 4-hydroxycoumarin derivatives: Inhibiting growth of human lung cancer cells. Main Group Met. Chem. 2018;17:301–307. doi: 10.3233/MGC-180677. [DOI] [Google Scholar]
- 27.Sinkevicius KW, et al. E-cadherin loss accelerates tumor progression and metastasis in a mouse model of lung adenocarcinoma. Am. J. Respir. Cell Mol. Biol. 2018;59:237–245. doi: 10.1165/rcmb.2017-0210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quintanal-Villalonga Á, et al. The FGFR4-388arg variant promotes lung cancer progression by N-cadherin induction. Sci. Rep. 2018;8:1–13. doi: 10.1038/s41598-018-20570-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information file.