Abstract
Haemophilus influenzae lacks most of the biosynthetic enzymes for hemin synthesis. However, the organism has retained ferrochelatase activity, which we identified to be encoded by a hemH-homologous gene. In this report we characterize the growth physiology conferred by hemH mutations under infection and laboratory conditions.
Haemophilus influenzae is a gram-negative bacterium that is responsible for significant morbidity and mortality in young children (4, 20). Under normal growth conditions H. influenzae needs two essential growth factors: NAD and hemin (2). Hemin can serve as a source of both iron and porphyrin, and protoporphyrin IX (PPIX) can substitute for hemin if exogenous iron is available (5, 21). The activity of a putative ferrochelatase was found in whole-cell extracts of H. influenzae (11), indicating that Fe2+ and PPIX are chelated into heme. Iron-citrate or ferric ions can be utilized by an uptake system, which was found to be encoded by the genes hitABC (18). Since free iron is limited under in vivo conditions and no siderophores are synthesized, H. influenzae has evolved strategies to scavenge host iron-binding proteins as sources of iron (19). Human transferrin has been shown, for example, to be a suitable substrate and is specifically recognized by two outer-membrane-located receptor proteins encoded by tbpAB (6). Free hemin or PPIX may also be utilized by H. influenzae if it is present in growth media, and they are also scavenged from hemin-containing host proteins by specific hemopexin- and hemoglobin- or haptoglobin-binding protein complexes (1, 7–9, 12, 13, 16, 22).
H. influenzae has only a rudimentary hemin biosynthetic pathway, in which no other enzymes except that encoded by the hemH homologue are known to exist. In this study, we establish that the hemH gene identified by Fleischmann et al. (3) and designated HI1160 encodes a ferrochelatase and that defined mutations in the gene region corresponding to hemH confer a profound growth phenotype.
Construction of hemH mutants.
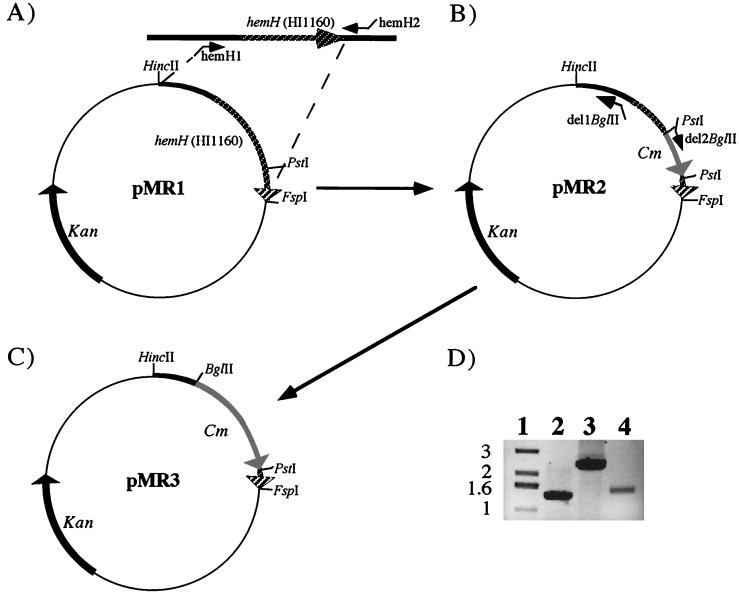
Utilizing the genome sequence provided by Fleischmann et al. (3) (http://www.tigr.org), hemH (HI1160) was PCR amplified from H. influenzae strain Rd chromosomal DNA with oligonucleotides containing flanking EcoRV restriction sites, i.e., hemH1 (AAGATATCAGTGGATCATCGTACTATGC) and hemH2 (AAGATATCGCTGATTTTAGCAAAGTGCG) (synthesized by MWG-Biotech, Ebersberg, Germany). The resulting 1,315-bp product of HI1160 (Fig. 1A) was cleaved with EcoRV and subcloned into HincII- and FspI-linearized pACYC177 (17), resulting in plasmid pMR1 (Fig. 1A). A PCR-generated chloramphenicol acetyltransferase-encoding gene (cat) with flanking PstI restriction sites (10) was inserted into a unique PstI site, resulting in pMR2 (Fig. 1B). An 837-bp segment of the 5′ hemH sequence was deleted by PCR amplification of pMR2 with oligonucleotides del1BglII (GAAGATCTCAGGCGTTTAAGGGCACC) and del2BglII (GAAGATCTTTTGCCAAACTTGGATATT), containing flanking BglII restriction sites. Subsequent ligation of the BglII sites resulted in plasmid pMR3 (Fig. 1C). The hemH:: cat gene from pMR2 and the ′hemH(Δ837)::cat gene from pMR3 were amplified again by PCR, using oligonucleotides hemH1 and hemH2, and the DNA fragments were retransformed into H. influenzae strain Rd and H. influenzae type b strain Eagan (Hib), respectively. Chloramphenicol-resistant (Cmr) colonies were obtained on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.) supplemented with hemin (20 μg/ml) (Sigma), NAD (10 μg/ml) (Sigma), and chloramphenicol (2 μg/ml) (Sigma). Transformants were designated SCH01 (Rd hemH::cat) and SCH02 [Hib; hemH(Δ837)::cat]. Correct gene replacement in both strains was verified by PCR (Fig. 1D) and Southern blot analysis (data not shown).
FIG. 1.
Cloning of hemH and construction of hemH mutants. (A) Cloning of hemH PCR product, containing flanking EcoRV restriction sites, into a HincII- and FspI-linearized pACYC177 plasmid. (B) pMR1 was digested with PstI, and cat was ligated into hemH. (C) Plasmid pMR3 was derived from pMR2 by deletion of the 5′ end of hemH (see text). Antibiotic markers and relevant restriction sites are indicated. The hemH gene (hatched arrows), chloramphenicol resistance gene (cat) (light arrows), and kanamycin resistance marker (Kan) (black arrows) are shown. (D) A 0.7% agarose gel with PCR-generated fragments obtained by using oligonucleotides hemH1 and hemH2 and purified chromosomal DNAs of H. influenzae strains Hib (lane 2), SCH01 (lane 3), and SCH02 (lane 4). Lane 1, 1-kbp standard (Gibco Life Technologies).
Characterization of hemH growth phenotypes.
Strains SCH01 and SCH02 were tested for their abilities to grow on PPIX- or hemin-supplemented BHI medium. Both hemH mutants failed to grow on medium supplemented with PPIX (20 μg/ml) (Sigma) but grew well on medium containing hemin (20 μg/ml) (Table 1). Complementation of both strains with plasmid pMR1 resulted in growth of both strains on PPIX-containing medium (Table 1), indicating that hemH was responsible for utilization of PPIX. We further tested the ability of strain SCH01 to grow on hemin as an intracellular iron source. To establish iron-limiting conditions, BHI medium was supplemented with the iron chelator deferoxamine mesylate (DFX) (Sigma), which preferentially chelates extracellular iron. We observed no growth of the wild-type (wt) Rd strain on BHI medium with PPIX (20 μg/ml) supplemented with DFX (0.08 mM), indicating that under these conditions no iron source was available for hemin biosynthesis (Table 1). With hemin instead of PPIX, both the wt Rd and the hemH mutant SCH01 could grow in the presence of DFX. This finding demonstrates that both strains can utilize hemin as an iron source and that if HemH could release iron from hemin, as has previously been suggested (11), then an additional cytoplasmic hemin-utilizing and iron liberation system must coexist with hemH.
TABLE 1.
Growth abilities of H. influenzae hemH mutant and complemented strains on hemin and PPIX
| Supplement(s)a | Growth phenotype
|
||||
|---|---|---|---|---|---|
| wt | SCH01 | SCH01(pMR1) (hemH+) | SCH02 | SCH02(pMR1) (hemH+) | |
| Hemin | + | + | + | + | + |
| Hemin, DFX | + | + | NDb | ND | ND |
| PPIX | + | − | + | − | + |
| PPIX, DFX | − | − | ND | ND | ND |
BHI medium containing NAD (10 μg/ml) was supplemented with hemin (20 μg/ml) or PPIX (20 μg/ml), with or without DFX (0.08 mM).
ND, not done.
The in vivo relevance of hemH was assessed by intraperitoneal and intranasal inoculation of 5-day-old Sprague-Dawley infant rats (14, 15). Infant rats were inoculated intraperitoneally with 100 μl of 0.1% gelatin in phosphate-buffered saline containing 102 CFU of wt Hib (n = 5) or the hemH mutant Hib strain SCH02 (n = 5). At 48 h, there was no difference in bacteremia (2.78 × 106 ± 1.9 × 106 CFU/ml for wt Hib [mean ± standard deviation] versus 2.98 × 106 ± 2.1 × 106 CFU/ml for SCH02), and all animals died by 72 h, suggesting that the two strains had similar virulence. Infant rats were also inoculated intranasally with 10 μl of 0.1% gelatin–phosphate-buffered saline containing 107 CFU of wt Hib (n = 3) or SCH02 (n = 3). There was no difference in recovery (around 103 to 104 CFU) of either strain from 40-μl nasal washings at 48 h. These experiments indicate that HemH is not essential for bloodstream survival or nasopharyngeal colonization and suggest that PPIX is not a major in vivo source of factor X.
Acknowledgments
This work was funded by BMBF grant 01KI8906.
REFERENCES
- 1.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans N M, Smith D D, Wicken A J. Hemin and nicotinamide adenine dinucleotide requirements of Haemophilus influenzae. J Med Microbiol. 1974;7:359–365. doi: 10.1099/00222615-7-3-359. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Frichman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 4.Funkhouser A, Steinhoff M C, Ward J. Haemophilus influenzae disease and immunization in developing countries. Rev Infect Dis. 1991;13:542–554. doi: 10.1093/clinids/13.supplement_6.s542. [DOI] [PubMed] [Google Scholar]
- 5.Granick S, Gilder H. The porphyrin requirements of Haemophilus influenzae and some functions of the vinyl and propionic acid chains of heme. J Gen Physiol. 1946;30:1–13. [PMC free article] [PubMed] [Google Scholar]
- 6.Gray-Owen S D, Loosmore S, Schryvers A B. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infect Immun. 1995;63:1201–1210. doi: 10.1128/iai.63.4.1201-1210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson M S, Pelzel S E, Latimer J, Muller-Eberhard U, Hansen E J. Identification of a genetic locus of Haemophilus influenzae type b necessary for the binding and utilization of heme bound to human hemopexin. Proc Natl Acad Sci USA. 1992;89:1973–1977. doi: 10.1073/pnas.89.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin H, Ren Z, Pozsgay J M, Elkins C, Whitby P W, Morton D J, Stull T L. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H, Ren Z, Whitby P W, Morton D J, Stull T L. Characterization of hgpA, a gene encoding a haemoglobin/haemoglobin-haptoglobin-binding protein of Haemophilus influenzae. Microbiology. 1999;145:905–914. doi: 10.1099/13500872-145-4-905. [DOI] [PubMed] [Google Scholar]
- 10.Kraiß A, Schlör S, Reidl J. In vivo transposon mutagenesis in Haemophilus influenzae. Appl Environ Microbiol. 1998;64:4697–4702. doi: 10.1128/aem.64.12.4697-4702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb M. Ferrochelatase activity and protoporphyrin IX utilization in Haemophilus influenzae. J Bacteriol. 1995;177:3613–3615. doi: 10.1128/jb.177.12.3613-3615.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maciver I, L. L J, Liem H H, Muller-Eberhard U, Hrkal Z, Hansen E J. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect Immun. 1996;64:3703–3712. doi: 10.1128/iai.64.9.3703-3712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morton D J, Whitby P W, Jin H, Ren Z, Stull T L. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect Immun. 1999;67:2729–2739. doi: 10.1128/iai.67.6.2729-2739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moxon E, Glode M, Sutton A, Robbins J. The infant rat model of bacterial meningitis. J Infect Dis. 1977;136:186–190. doi: 10.1093/infdis/136.supplement.s186. [DOI] [PubMed] [Google Scholar]
- 15.Moxon E R, Smith A L, Averill D R, Smith D H. Haemophilus influenzae menigitis in infant rats after intranasal inoculation. J Infect Dis. 1974;129:154–162. doi: 10.1093/infdis/129.2.154. [DOI] [PubMed] [Google Scholar]
- 16.Ren Z, Jin H, Morton D J, Stull T L. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect Immun. 1998;66:4733–4741. doi: 10.1128/iai.66.10.4733-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders J D, Cope L D, Hansen E J. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schryvers A B, Gray-Owen S. Iron acquisition in Haemophilus influenzae: receptor for human transferrin. J Infect Dis. 1992;165:103–104. doi: 10.1093/infdis/165-supplement_1-s103. [DOI] [PubMed] [Google Scholar]
- 20.Truk D C. The pathogenicity of Haemophilus influenzae. J Med Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 21.White D C, Granick S. Hemin biosynthesis in Haemophilus. J Bacteriol. 1963;85:842–850. doi: 10.1128/jb.85.4.842-850.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong J C Y, Patel R, Kendall D, Whitby P W, Smith A, Holland J, Williams P. Affinity, conservation, and surface exposure of hemopexin-binding proteins in Haemophilus influenzae. Infect Immun. 1995;63:2327–2333. doi: 10.1128/iai.63.6.2327-2333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]