Abstract
Two protein pairs in Vibrio cholerae, ToxRS and TcpPH, are necessary for transcription from the toxT promoter and subsequent expression of cholera virulence genes. We have previously shown that transcription of tcpPH in classical strains of V. cholerae is activated at mid-log-phase growth in ToxR-inducing conditions, while transcription of tcpPH in El Tor strains is not. In this study, we showed that while transcription of tcpPH differs at mid-log-phase growth in ToxR-inducing conditions between the biotypes, transcription is equivalently high during growth in AKI conditions. We used tcpPH::gusA transcriptional fusions to quantitate expression of tcpPH in each biotype throughout growth in ToxR-inducing conditions and showed that although transcription of tcpPH is reduced at mid-log-phase growth in an El Tor strain, transcription is turned on later in growth to levels in excess of those in the classical strain (although cholera toxin is not produced). This suggests that the difference in expression of cholera virulence factors in response to ToxR-inducing conditions between the El Tor and classical biotypes of V. cholerae may be related to the timing of transcription of tcpPH rather than the absolute levels of transcription.
Vibrio cholerae is a gram-negative bacterium that causes the watery-diarrheal illness cholera. Strains of V. cholerae are classified serotypically based on their O antigen, and V. cholerae O1 is the predominant cause of epidemic cholera. V. cholerae O1 is divided into two biotypes, classical and El Tor, which are distinguished by a variety of phenotypic markers (13). The major virulence factors for V. cholerae are cholera toxin and the toxin-coregulated pilus (TCP). Cholera toxin is a heterodimeric protein exotoxin which consists of an enzymatically active A subunit, which acts as an ADP-ribosyl transferase and elevates intracellular cyclic AMP levels, and a pentamer of B subunits, which bind holotoxin to its receptor, the ganglioside GM1, on eukaryotic cells. The genes for cholera toxin are encoded as an operon, ctxAB, and are contained within the genome of a filamentous phage, CTXφ (16, 29). The major subunit of the TCP is encoded by tcpA (26). This gene is transcribed with a larger operon of 12 genes which are involved in processing and assembly of TCP on the surface of V. cholerae.
In classical strains of V. cholerae, expression of cholera toxin and TCP is strongly regulated by environmental signals, such as pH, temperature, amino acid concentration, and osmolarity (19, 20). Coordinate regulation of expression of these virulence factors depends on a transmembrane DNA-binding protein, ToxR, which is encoded in an operon with a second regulatory protein, ToxS (6, 17, 18, 20). ToxR and ToxS are necessary for transcription of a gene encoding an important outer membrane protein of V. cholerae, ompU (5), as well as a gene encoding a second regulatory protein, toxT (7, 10, 22). ToxT belongs to the AraC class of transcriptional regulatory proteins and activates transcription of the ctxAB and tcpA operons. The toxT gene is contained within the tcpA gene cluster itself, and transcription of toxT occurs both from a promoter immediately upstream of toxT and also as part of the tcpA operon, from a longer transcript which originates at the tcpA promoter (1, 9).
Several groups have recently shown that two additional regulatory proteins, TcpP and TcpH, are necessary for transcription from the toxT promoter and have presented a model in which ToxRS and TcpPH interact to activate toxT transcription (4, 8). The genes for tcpP and tcpH are located immediately upstream of tcpA and are transcribed together as an operon (4, 23, 27). The tcpI gene is transcribed divergently from tcpPH, and TcpI has previously been suggested to be a negative regulator of cholera virulence gene expression (27), although the mechanism of this negative regulation has not yet been elucidated. Expression of the tcpPH operon in classical strains of V. cholerae is regulated by the same environmental conditions that regulate expression of cholera virulence factors, suggesting that the expression of tcpPH couples these environmental signals to transcription of toxT and expression of the virulence genes (4). The environmental conditions that induce expression of cholera virulence genes in classical strains of V. cholerae (30°C and pH 6.5) are termed ToxR-inducing conditions, whereas the environmental conditions of 37°C and pH 8.5 repress expression of the ToxR regulon (19).
In El Tor strains of V. cholerae, expression of cholera virulence factors does not occur in vitro in ToxR-inducing conditions but rather requires special growth conditions termed AKI conditions (11). In AKI conditions, V. cholerae are grown statically at 37°C for 4 h and then shifted to overnight shaking at 37°C; these conditions lead to expression of cholera virulence factors in both classical and El Tor strains. Both toxT and tcpPH are necessary for expression of cholera virulence factors in El Tor strains of V. cholerae, as for classical strains.
We have recently shown that transcription of tcpPH in El Tor strains of V. cholerae cannot be detected by Northern blotting at mid-log-phase growth in ToxR-inducing conditions but that expression of tcpPH from a constitutive promoter in an El Tor strain leads to expression of cholera virulence factors independent of environmental signals (21). Two other proteins, AphA and AphB, have recently been shown to be necessary for transcription of tcpPH in both classical and El Tor strains, and these proteins may help couple environmental signals to expression of cholera virulence factors (14, 25). We wished to examine more carefully differences in transcription of tcpPH between classical and El Tor strains of V. cholerae.
Mapping the tcpPH and tcpI transcription start sites in classical and El Tor biotypes and in different environmental conditions.
The sequences of the intergenic regions between tcpPH and tcpI are quite homologous between the classical and El Tor strains of V. cholerae (Fig. 1A). We investigated the transcriptional start sites of the tcpPH operon and tcpI in each of these biotypes, utilizing primer extension analysis and oligonucleotide primers spanning the tcpI-tcpP intergenic region. The oligonucleotides used for primer extension analysis included YM2 (CCGGCTTCATTGGATCTTGTGCATAATAGA), YM10 (GGGTAAGCCAAACATTGGATAGATTACCTTGATAA), YM11 (GCATTCGTTCCACCAAAGGTTATCGGGAAATT), YM8 (GCCCCAAACGGAAGGGGCAAAGTGTCACAGGAAA), YM13 (GGGGCAAAGTGTCACAGGAAAGATAATGTAACCAA), and YM14 (CGTTTTAAATAGTATTTTTTTTCTTTAGGAAAAT). V. cholerae RNA was purified from cells grown in ToxR-inducing, ToxR-repressing, or AKI conditions as described previously (21), and primer extension was done with 20 μg of RNA with the Primer Extension System (Promega Corporation, Madison, Wis.) according to the manufacturer's instructions with the following modifications: primer annealing was carried out at 58°C for 1 h, and primer extension was carried out at 42°C for 1 h. Samples were ethanol precipitated and resuspended in 5 μl of water and 5 μl of loading dye. DNA sequencing was done with the Sequenase quick-denature plasmid sequencing kit (Amersham Life Sciences, Cleveland, Ohio) according to the manufacturer's instructions, using the same oligonucleotide used to generate the primer extension product. Five microliters of primer extension product and 2.5 μl of each DNA sequencing reaction were resolved on a 6% denaturing polyacrylamide gel and visualized by autoradiography (24).
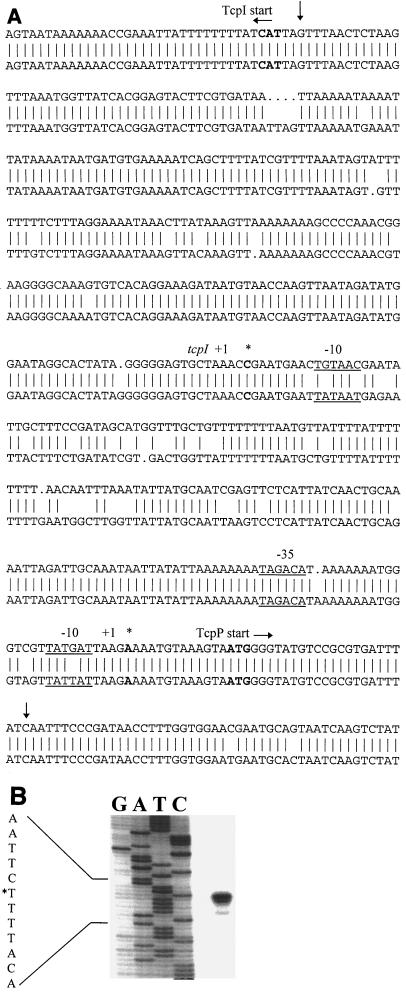
FIG. 1.
(A) DNA sequences of the classical (top strand; GenBank accession no. X64098, bases 1250 to 1792) and El Tor (bottom strand; GenBank X74730, bases 307 to 853) tcpI-tcpP intergenic regions. Transcriptional start sites for tcpI and tcpP, as determined by primer extension, are indicated by asterisks. Putative −10 and −35 boxes are underlined. Note that the transcriptional start sites and putative promoter regions differ from those that were reported previously (23). Down arrows denote the region amplified by PCR and cloned for the tcpP::uidA promoter fusion constructs. (B) Primer extension analysis of the tcpPH transcript prepared from classical strain O395 RNA purified from cells grown to mid-log phase in ToxR-inducing conditions; the corresponding DNA sequence is also shown. Primer extension and DNA sequencing were carried out with oligonucleotide YM2.
For classical V. cholerae strain O395, a 109-bp primer extension product was obtained when oligonucleotide YM2 was annealed to RNA purified from cells grown to mid-log phase in ToxR-inducing conditions (Fig. 1B). This corresponds to a transcriptional start site at an adenine residue 13 nucleotides upstream of the TcpP start codon (indicated by an asterisk in Fig. 1A). Putative −10 and −35 boxes were found upstream of the transcriptional start site (indicated by underlining in Fig. 1A). Oligonucleotides YM11 and YM12, which reside 50 nucleotides upstream and downstream of YM2, respectively, were used to confirm the primer extension results (data not shown). Using oligonucleotide YM2, identical primer extension products were obtained from RNA purified from O395 grown in AKI conditions (harvested at the end of 4 h of static culture) and from RNA purified from V. cholerae El Tor strain C6709 cells grown in both ToxR-inducing and AKI conditions (data not shown). No primer extension products were found in either biotype when grown in ToxR-repressing conditions (data not shown). Of note, the −10 boxes of the tcpPH promoters differed by 1 bp between the classical and El Tor strains, and the −35 box in the El Tor strain was spaced 1 bp further away from the −10 box than in the classical strain (Fig. 1A).
The tcpI transcriptional start site was similarly mapped. A 90-bp primer extension product was obtained from O395 RNA purified from cells grown to mid-log phase in ToxR-inducing conditions with oligonucleotide YM8 (data not shown). This corresponds to a transcriptional start site at a cytosine residue 241 bp upstream of the TcpI translational start codon (denoted by an asterisk in Fig. 1A). This start site was confirmed by using oligonucleotides YM13 and YM14, residing 50 nucleotides on either side of YM8. With YM8, identical products were obtained from O395 RNA purified from cells grown in AKI conditions as well as from C6709 RNA purified from cells grown in ToxR-inducing and AKI conditions (data not shown); no primer extension products were found in either biotype when grown in ToxR-repressing conditions. The putative −10 boxes of the tcpI promoters differed by 2 bp between the two biotypes, and we did not find an obvious −35 box in either biotype (Fig. 1A). Note that the transcription start sites and putative promoter regions for both tcpI and tcpPH differed from those reported previously (23).
Quantitating transcription of tcpPH in classical and El Tor strains by primer extension.
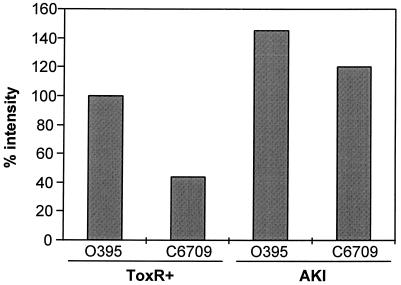
We used primer extension to quantitate transcription of tcpPH in the two biotypes in ToxR-inducing and AKI conditions (Fig. 2). Primer extension was carried out as described above, and the products were visualized by autoradiography. Densitometry analysis was performed by scanning the radiographs and performing analysis with a Power Macintosh G3 computer using the public-domain NIH Image Program (version 1.61) (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). At the mid-log phase of growth, the classical V. cholerae strain O395 had approximately twice as much tcpPH transcript as did the El Tor biotype, while both strains had equivalently high levels of transcription of tcpPH during growth in AKI conditions (Fig. 2).
FIG. 2.
Quantitation by densitometry of primer extension products of classical (O395) and El Tor (C6709) tcpPH transcripts at mid-log-phase growth in ToxR-inducing conditions (ToxR+) and at the end of the static phase in AKI conditions.
Construction of tcpPH::gusA transcriptional fusions and quantitation of expression of tcpPH in each biotype during growth in ToxR-inducing conditions.
To better quantitate expression of the tcpPH promoter in each biotype over the course of growth, we constructed transcriptional fusions between the tcpPH promoters and the gusA gene, encoding β-glucuronidase. We recovered the tcpI-tcpP intergenic region, containing the putative tcpPH promoters, from both biotypes by PCR (Fig. 1A) and cloned these fragments into plasmid pUJ10, then replaced the phoA gene in each with the uidA gene, encoding β-glucuronidase. This resulted in plasmids pYM2-25 (classical tcpPH promoter) and pYM2-24 (El Tor tcpPH promoter). The tcpPH promoter-uidA fragments were then recovered from these plasmids by digestion with XbaI and NotI and cloned into a variant of plasmid p6891MCS, which contains a multiple cloning site within a fragment of the V. cholerae lacZ gene (2) and the rrnB transcriptional terminator from plasmid pKK223-3 (positioned to be 5′ of the tcpPH promoter in each plasmid). This yielded plasmids pYM2-27 (classical tcpPH::uidA fusion) and pYM2-26 (El Tor tcpPH::uidA fusion). Each tcpPH::uidA fusion was then exchanged by double homologous recombination into the lacZ gene of the respective V. cholerae host, as previously described (3), resulting in strains YM2-34 (classical) and YM2-35 (El Tor).
β-Glucuronidase assays were performed as described previously with some modifications (12). Briefly, strains YM2-34 and YM2-35 were grown overnight and then back-diluted into 5 ml of LB medium and grown in ToxR-inducing conditions. At various time points, 500 μl of bacterial culture was centrifuged and washed once with 50 mM phosphate buffer (pH 7.0). Washed cells (50 μl) were lysed by adding 10 μl of toluene and vortexing for 20 s. Eight hundred fifty microliters of 50 mM phosphate buffer was added to the lysed cells, and samples were incubated for 10 min at 37°C. p-Nitrophenyl-β-d-glucuronide (100 μl; Sigma, St. Louis, Mo.) was added, and the samples were incubated at 37°C until they developed a yellow color, at which point reactions were stopped by the addition of 400 μl of 3 M 2-amino-2-methylpropanediol (Sigma). Samples were vortexed for 20 s and centrifuged for 5 min, and the absorbance of the supernatants at 420 nm was measured.
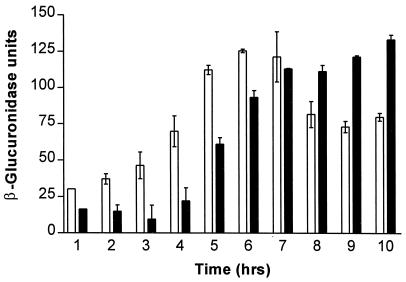
As shown in Fig. 3, the tcpPH::uidA fusion in the classical background demonstrated increased transcription from the tcpPH promoter by 4 h in ToxR-inducing conditions, peaking at 6 h and decreasing thereafter. In contrast, transcription from the tcpPH promoter in the El Tor background began later, increasing after 5 h and not peaking until 10 h. As seen previously (Fig. 2), transcription of tcpPH in the classical biotype was substantially higher than in the El Tor biotype at mid-log-phase growth in ToxR-inducing conditions. However, transcription of tcpPH in the El Tor biotype was greater than in the classical biotype during late-log-phase and stationary-phase growth, even though previous results suggest that little or no cholera toxin or TCP is expressed after overnight growth in ToxR-inducing conditions in the El Tor biotype (21).
FIG. 3.
Assays of tcpPH::uidA promoter fusion activities during growth in ToxR-inducing conditions. Error bars represent standard errors of the mean for quadruplicate measurements. Results for the classical promoter fusion strain YM2-34 are shown as open bars, and those for the El Tor promoter fusion strain YM2-35 are shown as solid bars.
Assessment of tcpPH RNA stability in each biotype.
One possibility is that the tcpPH transcript is less stable in the El Tor strain, even though initiation of transcription is seen in late-log-phase or stationary-phase growth. The stability of the tcpPH message was compared between the classical and El Tor biotypes using rifampin as previously described (28). In brief, cultures were grown to mid-log phase in ToxR-inducing conditions, rifampin was added (200 μg/ml, final concentration), and RNA was harvested at various times following the addition of rifampin. RNA was quantitated spectrophotometrically and visualized by using ethidium bromide and agarose gel electrophoresis. Twenty micrograms of RNA was used for primer extension analysis as described above. One half of the total primer extension product was loaded onto a 6% denaturing polyacrylamide gel and separated by electrophoresis. Products were visualized by autoradiography, and band intensities were determined as above and compared with the zero timepoint.
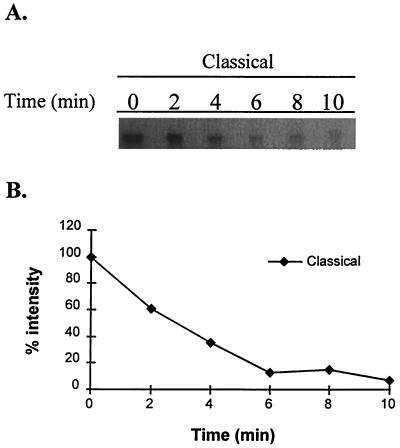
The intensity of the tcpPH transcripts at mid-log-phase growth in ToxR-inducing conditions was greater for the classical than the El Tor biotype strain prior to the addition of rifampin, as seen above (data not shown). The half-life of the tcpPH message in the classical biotype was approximately 2 min (Fig. 4). This short half-life is similar to that reported previously for the toxT transcript (30). Although the faint primer extension signal seen with the El Tor biotype prevented a comparably accurate measurement, the half-life of the tcpPH transcript in this biotype also approximated 2 min, no different from the classical strain (data not shown).
FIG. 4.
Estimation of tcpPH RNA stability after addition of rifampin, as measured by primer extension analysis and densitometry analysis. (A) Primer extension results after addition of rifampin at time zero for the classical strain. (B) Plot of densities of primer extensions results from panel A.
Yu et al. have previously shown in a classical strain of V. cholerae that TcpPH and ToxR are required for transcription from the promoter immediately upstream of toxT in early- to mid-log-phase growth in ToxR-inducing conditions but that transcription from this promoter begins to diminish 5 to 9 h after back-dilution of a culture and is undetectable after 9 h (30). Later in growth, they showed that transcription of toxT occurs from the upstream tcpA promoter as part of an autoregulatory loop primed by the initial transcription from the toxT promoter. If no toxT transcription occurred early in growth, then no cholera toxin was produced.
Medrano et al. examined transcription of toxT in an El Tor strain of V. cholerae in AKI conditions (15). They demonstrated that ToxR-dependent transcription from the toxT promoter was seen at the end of 4 h of static growth but was not seen after a shift to shaking growth conditions. During shaking growth, only the longer toxT transcript from the tcpA promoter was seen, as part of an autoregulatory loop. If the cultures were kept in static growth, the initial transcription from the toxT promoter disappeared, and no cholera toxin (and presumably no transcription from the tcpA promoter) was seen later in growth. This suggests that the temporal sequence of transcription from the toxT and tcpA promoters is important to the expression of cholera virulence genes.
Kovacikova et al. used a tcpP::lacZ fusion in an El Tor strain of V. cholerae to show that transcription of tcpPH was minimal at mid-log-phase growth in ToxR-inducing conditions but was much more abundant in AKI conditions (14). We have previously shown an absence of tcpPH mRNA by Northern blot in an El Tor strain of V. cholerae at mid-log-phase growth in ToxR-inducing conditions (21).
Utilizing the more sensitive assays here, we now show that the tcpPH message is markedly reduced but not absent at mid-log-phase growth in an El Tor strain of V. cholerae in ToxR-inducing conditions but that transcription is turned on later in growth. This suggests the possibility that failure of tcpPH transcription early in growth in ToxR-inducing conditions in the El Tor biotype of V. cholerae (compared to the classical biotype) may fail to activate the autoregulatory loop from the tcpA promoter that is necessary for continued expression of toxT and that this may abrogate expression of cholera toxin and TCP. An early peak of tcpPH (and toxT) transcription may be necessary for the later expression of cholera toxin and TCP, and the transcription of tcpPH seen later in the El Tor biotype, after cells have entered the late log phase of growth, may not be sufficient to activate expression of cholera virulence factors. This model suggests that the difference in expression of cholera virulence factors between the El Tor and classical biotypes of V. cholerae may be related to the timing of transcription of tcpPH during ToxR-inducing conditions rather than the absolute levels of transcription.
Acknowledgments
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases, RO1 AI44487, to S.B.C. Y.M.M. was supported by a training grant from the National Institute of Allergy and Infectious Diseases, T32 AI07061.
REFERENCES
- 1.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 2.Butterton J R, Beattie D T, Gardel C L, Carroll P A, Hyman T, Killeen K P, Mekalanos J J, Calderwood S B. Heterologous antigen expression in Vibrio cholerae vector strains. Infect Immun. 1995;63:2689–2696. doi: 10.1128/iai.63.7.2689-2696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butterton J R, Ryan E T, Acheson D W, Calderwood S B. Coexpression of the B subunit of Shiga toxin 1 and EaeA from enterohemorrhagic Escherichia coli in Vibrio cholerae vaccine strains. Infect Immun. 1997;65:2127–2135. doi: 10.1128/iai.65.6.2127-2135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 5.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 6.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 7.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hase C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 12.Jefferson R A, Burgess S M, Hirsh D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacikova G, Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medrano A I, DiRita V J, Castillo G, Sanchez J. Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator ToxT in response to culture conditions. Infect Immun. 1999;67:2178–2183. doi: 10.1128/iai.67.5.2178-2183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, de Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 17.Miller V L, DiRita V J, Mekalanos J J. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by ToxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires ToxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 21.Murley Y M, Carroll P A, Skorupski K, Taylor R K, Calderwood S B. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect Immun. 1999;67:5117–5123. doi: 10.1128/iai.67.10.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogierman M A, Manning P A. Homology of TcpN, a putative regulatory protein of Vibrio cholerae, to the AraC family of transcriptional activators. Gene. 1992;116:93–97. doi: 10.1016/0378-1119(92)90634-2. [DOI] [PubMed] [Google Scholar]
- 23.Ogierman M A, Voss E, Meaney C, Faast R, Attridge S R, Manning P A. Comparison of the promoter proximal regions of the toxin-co-regulated tcp gene cluster in classical and El Tor strains of Vibrio cholerae O1. Gene. 1996;170:9–16. doi: 10.1016/0378-1119(95)00744-x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Skorupski K, Taylor R K. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor R K, Shaw C E, Peterson K M, Spears P, Mekalanos J J. Safe live Vibrio cholerae vaccines? Vaccine. 1988;6:151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- 28.von Gabain A, Belasco J G, Schottel J L, Chang A C Y, Cohen S N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 30.Yu R R, DiRita V J. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol. 1999;181:2584–2592. doi: 10.1128/jb.181.8.2584-2592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]