Abstract
Ever since coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, was declared a pandemic on March 11, 2020, by the WHO, a concerted effort has been made to find compounds capable of acting on the virus and preventing its replication. In this context, researchers have refocused part of their attention on certain natural compounds that have shown promising effects on the virus. Considering the importance of this topic in the current context, this study aimed to present a critical review and analysis of the main reports of plant-derived compounds as possible inhibitors of the two SARS-CoV-2 proteases: main protease (Mpro) and Papain-like protease (PLpro). From the search in the PubMed database, a total of 165 published articles were found that met the search patterns. A total of 590 unique molecules were identified from a total of 122 articles as potential protease inhibitors. At the same time, 114 molecules reported as natural products and with annotation of theoretical support and antiviral effects were extracted from the COVID-19 Help database. After combining the molecules extracted from articles and those obtained from the database, we identified 648 unique molecules predicted as potential inhibitors of Mpro and/or PLpro. According to our results, several of the predicted compounds with higher theoretical confidence are present in many plants used in traditional medicine and even food, such as flavonoids, carboxylic acids, phenolic acids, triterpenes, terpenes phytosterols, and triterpenoids. These are potential inhibitors of Mpro and PLpro. Although the predictions of several molecules against SARS-CoV-2 are promising, little experimental information was found regarding certain families of compounds. Only 45 out of the 648 unique molecules have experimental data validating them as inhibitors of Mpro or PLpro, with the most frequent scaffold present in these 45 compounds being the flavone. The novelty of this work lies in the analysis of the structural diversity of the chemical space among the molecules predicted as inhibitors of SARS-CoV-2 Mpro and PLpro proteases and the comparison to those molecules experimentally validated. This work emphasizes the need for experimental validation of certain families of compounds, preferentially combining classical enzymatic assays with interaction-based methods. Furthermore, we recommend checking the presence of Pan-Assay Interference Compounds (PAINS) and the presence of molecules previously reported as inhibitors of Mpro or PLpro to optimize resources and time in the discovery of new SARS-CoV-2 antivirals from plant-derived molecules.
1. Introduction
Coronavirus disease 2019, shortened to COVID-19, is caused by a severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2). It was first reported in Wuhan, China, in late 2019, and declared a pandemic on March 11, 2020, by the WHO.1 From that moment on, a frantic search was unleashed with the aim of identifying molecules, natural or synthetic, that would act on the virus in order to treat the disease and reduce its spread. Among the many studies written, some focused on studying and validating certain proteins of the SARS-CoV-2 virus as possible molecular targets for antiviral agents. Amid those already validated, scientists identified the inhibition of virus type 3C (main) or papain-type proteases as promising strategies for the development of drug candidates against COVID-19.2
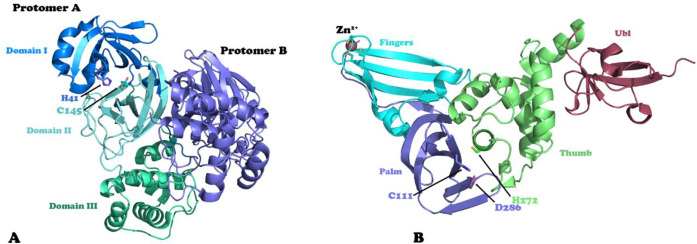
Structurally, the SARS-CoV-2 has two cysteine proteases involved in the proteolytic processing of the viral polyproteins: the papain-like protease (PLpro) and the main protease or 3C-like protease (Mpro/3CLpro). PLpro cleaves the viral polyproteins precursors pp1a and pp1ab at three sites to produce nonstructural proteins Nsp1, Nsp2, and Nsp3, while Mpro cleaves the same polyproteins precursors at 11 different sites to produce 12 viral proteins.3 Both proteases are essential for generating functional viral particles, making them attractive drug targets for the development of new antiviral drugs. In addition, the absence of human homologues of Mpro is considered an advantage in the development of antiviral drugs with high specificity for the viral protease. MPro has a chymotrypsin-like fold with three domains and is active as a homodimer, like other coronavirus MPros.4 The active site is located in a deep cleft between domains I and II of each monomer (Figure 1A).5,6 Dimerization is necessary for higher catalytic efficiency because the N-terminus of each monomer helps to stabilize the S1 pocket of the other monomer. Meanwhile, PLpro is part of the multidomain membrane-associated protein nsp3 and its structure has four distinct structural domains. Three of these domains form an extended right-hand architecture with distinct palm, thumb, and finger domains (Figure 1B). The N-terminal domain is well separated from the other three domains and adopts a fold-like ubiquitin domain.7 The active site of this enzyme is in the cleft between the thumb and palm domains, and contains a catalytic triad of cysteine, histidine, and aspartic acid.4 The conformation of a flexible β-loop present near the entrance of the active site is an important element for the binding of ligands into the active site. In addition to the proteolytic processing of the viral polyproteins precursors, PLpro can also bind and cleave ISG15 (interferon-stimulated gene product 15) and ubiquitin from ISGylated or ubiquitinated proteins.8,9 Both activities are vital for the coronavirus to counteract the host immune responses.
Figure 1.
Three-dimensional structures of the SARS-CoV-2 proteases Mpro (A) and PLpro (B). The structural domains and catalytic residues are highlighted.
There are several three-dimensional structures for both proteases in the Protein Data Bank (PDB), which are very useful for drug discovery using in silico approaches. In fact, the use of computer-assisted drug discovery tools has become an efficient way of discovering successful molecules that could enter the drug development pipeline against this disease. Various investigations have focused on the search for SARS-CoV-2 inhibitors using computational approaches, such as molecular docking and structure-based virtual screening. Thus, for SARS-CoV-2 Mpro, there are almost 495 entries reported in the PDB while for PLpro there are 54 entries. Inhibitors, mostly synthetic molecules, of these viral proteases have proven to be effective as antiviral agents. Currently, there are almost 2500 molecules in the databases PostEra (https://covid.postera.ai/covid/activity_data) and COVID-19 (https://covid19-help.org/) reported as Mpro inhibitors. However, most of these compounds are not from natural sources making the search for natural products as SARS-CoV-2 protease inhibitors an underexploited field. In particular, the plant-derived molecules are a diversified source of compounds in terms of chemical diversity and pharmacological activities.10,11 Against this backdrop, the aim of this study was to review and analyze the main reports available to date that propose plant-derived compounds as possible inhibitors of SARS-CoV-2. There are some previous review papers about the use of plant natural products as SARS-CoV-2 protease inhibitors;12−16 therefore, in this review, we focus on the analysis of the structural diversity among the plant-derived predicted inhibitors of both proteases of SARS-CoV-2 (Mpro and PLpro) and their structural similarity with those compounds that have been experimentally tested against these proteases.
2. Methodology
2.1. Dataset
A PubMed database search with the following query was carried out: (“plant″[Title/Abstract] OR “phyto″[Title/Abstract] OR “herbal″[Title/Abstract]) AND (“inhibitor″[Title/Abstract] OR “inhibition″[Title/Abstract] OR “binding″[Title/Abstract] OR “inhibitors″[Title/Abstract]) AND (“COVID-19″[Title/Abstract] OR “SARS-CoV2”) AND (“protease″[Title/Abstract] OR “CL-pro″[Title/Abstract] OR “M-pro″[Title/Abstract] OR “CLpro″[Title/Abstract] OR “Mpro″[Title/Abstract]). The database PubMed was chosen for the search as it allows the contents of the MEDLINE medical library, life sciences journals, and books to be consulted. In this way, access to the largest possible number of indexed articles related to the search topics was guaranteed. As a result of this search, a total of 165 published articles were downloaded and manually curated. From these 165 published articles, we excluded (a) review articles, (b) articles that do not exactly explore plant-derived compounds, and (c) articles that were not fully accessed. With the manual curation, the distribution was as follows: 122 articles were further processed, 3 articles analyzed full plant extracts (not chemical compounds at a theoretical or experimental level), 29 articles were excluded because of being reviews or articles that do not explore chemical compounds at a theoretical or experimental level, and 10 articles carried out experimental analysis including chemical compounds derived from plants (1 article was not analyzed because the full manuscript was not accessible during the analysis).
2.2. Dataset of Predicted Molecules as Inhibitors of Mpro and PLpro
A total of 590 unique molecules were identified from the 122 articles. Additionally, we extracted all molecules from the COVID-19 Help database reported in natural products and specifically with theoretical support annotation and antiviral effects (a total of 216 molecules excluding peptides or microRNAs). However, our study focuses on the SARS-CoV-2 proteases (Mpro and PLpro), and not all 216 molecules from the database were predicted as protease inhibitors. After filtering for only these two targets, 114 molecules remained. After combining the 590 molecules extracted from articles and the 114 obtained from the database, we finally identified 648 unique molecules predicted as potential inhibitors of Mpro and/or PLpro (Table S1). For the computation of the “evidence”, we used the number of articles referring to the interaction prediction in the group of the 590 identified by our literature curation and the number of articles reported in the database for the remaining molecules out of the 648 (this information is also presented in Table S1 under the column titled “evidence”). Nonetheless, each author’s affiliation (country), as well as the type of theoretical strategy used (docking and/or molecular dynamics) were only computed in the 590 molecules derived from our literature curation. Each of the 648 molecules was classified based on its chemical structure using NPClassifier.17 The final list with the 648 smiles and their classifications is presented in Tables S1 and S2 (Supporting Information).
2.3. Dataset of Experimentally Evaluated Molecules as Inhibitors of Mpro and PLpro
The experimental evidence was compiled from three different sources: (1) the 10 articles reporting experimental evidence of compounds that inhibit PLpro and Mpro proteases, (2) the COVID-19 Help database (https://covid19-help.org/), and (3) the PostEra Mpro activity data (https://covid.postera.ai/covid/activity_data). The molecules with experimental evidence were not restricted only to natural products. The vast majority of molecules with experimental evidence had been evaluated in Mpro rather than PLpro. Therefore, we decided to only continue our analysis with the Mpro inhibitors. From the 10 articles, we extracted 82 compounds; from the COVID-19 Help database, we extracted 719 records (restricting the search for antivirals and removing those compounds with only in silico evidence), and from PostEra, we found 2062 molecules. Of the 719 records obtained from the COVID-19 Help database, many are recombinant proteins, antibodies, and molecules that do not have any chemical structure reported in PubChem or even the COVID-19 Help database. Therefore, after removing molecules without smiles, 456 remained. After joining all the molecules, a total of 2600 molecules with experimental evidence as Mpro inhibitors were included. The full list of molecules with experimental evidence regarding Mpro is presented in Table S3 (Supporting Information).
2.4. Molecular Similarity, Scaffolds, and Fragment Identification
In order to compare all molecules between each other or between groups (i.e., experimental versus theoretical analysis), the EFCP4 fingerprint (1024 bits) was computed, and the similarity was calculated using the Tanimoto metric (using RDKit library in python). For identity matching (i.e., removal of duplicates), we used a Tanimoto similarity equal to or higher than 0.95. The comparison between molecules with theoretical predictions and those having experimental evidence was carried out using the Tanimoto similarity computed as previously described but using 0.75 and 0.95 similarity cutoffs.
For scaffold identification, we used the Murcko scaffold identification algorithm implemented in RDKit.18 However, even when this strategy is helpful, the resulting molecule can be too generic, and their use in medicinal chemistry, or as the initial point for chemical modifications, is questionable.19 We decided to additionally use a retrosynthetic combinatorial analysis (Recap) implemented in RDKit.20 Recap fragment molecules function according to 11 types of bonds and simple rules. These rules will keep intact rings and conjugated rings. Recap will produce much more structural diversity, considering linkers, core structures, and even different substituent variations, as compared to the Murcko algorithm. Each scaffold/fragment was weighted by frequency of appearing across the 648 predicted molecules. All scaffolds and fragments with their corresponding frequency are presented in Table S4 (Supporting Information).
3. Results and Discussion
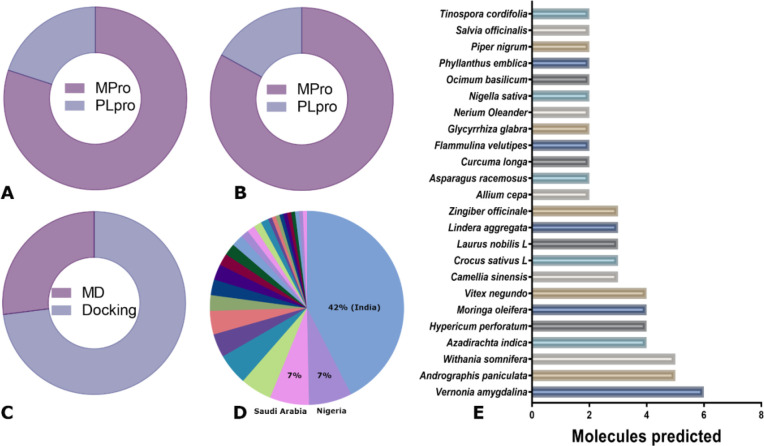
Most of the studies (and predicted molecules) retrieved in our search address the inhibition of the protease Mpro instead of PLpro (Figure 2A,B). The same pattern was observed for those molecules tested experimentally. Indeed, from the 648 unique molecules, 147 were predicted as potential inhibitors of PLpro and only 34 of them were predicted exclusively for this protease (Table S1). On the other hand, even when all molecules were evaluated using different docking strategies, molecular dynamics simulation was only performed in a small fraction of the molecular space (Figure 2C). Molecular dynamic simulation is a computationally expensive approach, and it was usually used only with the best candidates obtained from docking experiments.
Figure 2.
(A) Percentage of molecules predicted to bind each target protease. (B) Percentage of articles reporting molecules as inhibitors of each target protease. (C) Percentage of molecules identified as inhibitors of the target proteases using molecular docking or molecular dynamics simulations (MD). (D) Distribution of articles per country. (E) Plants with more than two compounds proposed as a SARS-CoV-2 protease inhibitor.
The analysis of the authors’ affiliations across all analyzed publications clearly indicates that more than half of all published articles were produced in India, Nigeria, and Saudi Arabia (Figure 2E). A total of 134 plants were reported as the source of molecules with the potential to inhibit one or both proteases of SARS-CoV-2. The plants with the highest number of articles were Vernonia amygdalina, Andrographis paniculata, Withania somnifera, Azadirachta indica, Hypericum perforatum, Moringa oleifera, and Vitex negundo (Figure 2D). Those plants with higher levels of evidence, for instance, Vermonia amygdalina and Azadirachta indica, are commonly used in several African countries as medicinal plants as well as for alimentary purposes. Andrographis paniculata, Withania somnifera, and Azadirachta indica are commonly used in India in Ayurvedic and traditional medicine. Hypericum perforatum and Moringa oleifera are used in natural medicine in several countries around the world, while Vitex negundo is native to several African and Asian countries.
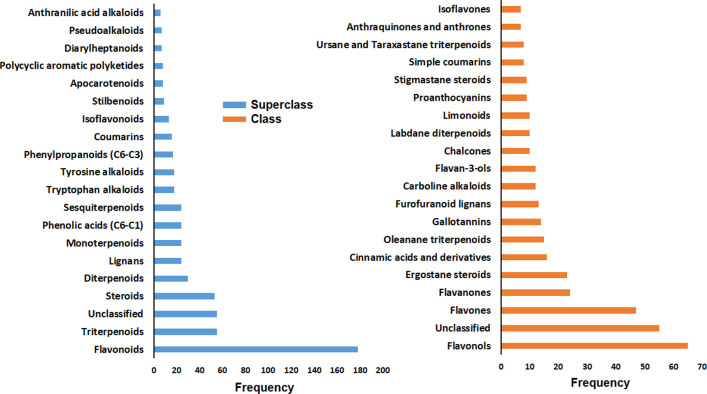
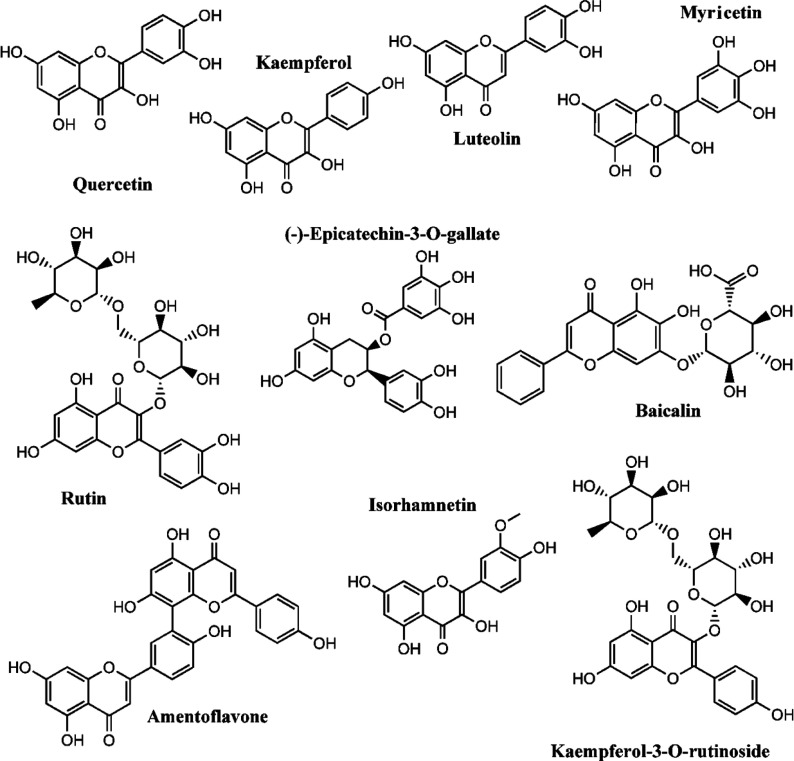
The top 20 most abundant superclasses and classes of the 648 molecules are presented in Figure 3. Most of the molecules are classified as belonging to the flavonoid superclass, specifically flavonols and flavones (Figure 3). It is important to note that 55 molecules (8.5%) are unclassified. A manual inspection of the unclassified molecules reveals that some of them are anthocyanins and charged compounds. Flavonoids are major constituents of plants and therefore, as expected, this group as well as triterpenoids and steroids were found to have the highest representation. The top 10 molecules with the highest number of articles predicting them as potential inhibitors of SARS-CoV-2 proteases are presented in Figure 4. All of these molecules appear in more than 8 papers and have been predicted as Mpro inhibitors as well as PLpro. The molecule with the highest number of reports is quercetin, followed by kaempferol, rutin, luteolin, baicalin, myricetin, isorhamnetin, (−)-epicatechin-3-O-gallate, amentoflavone, and kaempferol-3-O-rutinoside. We can clearly notice that among these top molecules, the structural diversity is actually very low. Almost all molecules share the same structural core of quercetin, luteolin, or galangin (7-hydroxyflavonol). Moreover, some of them had been proposed in glycosylated form with one or two sugar units. The role of these sugar substituents will be discussed later.
Figure 3.
Distribution of the molecular superclasses and classes across the 648 molecules using NPClassifier.
Figure 4.
Structures of the top ten molecules more frequently reported in the studies analyzed. These molecules are predicted as potential inhibitors of Mpro and PLpro.
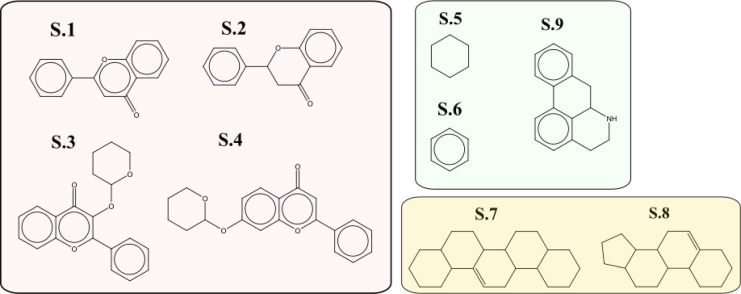
A total of 384 scaffolds were identified using the Murcko algorithm. Those present in more than 5 molecules are presented in Figure 5, where we can identify the flavones (S.1) and flavonones (S.2) cores, as well as two common glycosylated variants (S.3 and S.4). Some of these glycosylated S.1 variants are not only monosaccharides but also di, trisaccharides, or glucuronic acid, like in baicalin. The scaffolds S.5 and S.6 represent a major group of carboxylic acids, phenolic acids, and even lignans (i.e., quinic acid, gallic acid, and caffeic acid). The scaffolds presented in S.7 and S.8 are generic cores for triterpenes (i.e., ursane, oleanane, and steroid skeletons). Finally, S.9 is a generic core for aporphine alkaloids (which are a subclass of quinoline alkaloids). These scaffolds clearly represent the main superclasses and classes found in the 648 molecules (Figure 3).
Figure 5.
Top scaffolds identified using the Murcko algorithm in the 648 molecules. Only scaffolds found in more than 5 molecules are present.
The scaffold analysis is very helpful, but groups used in substitution are also of high relevance for chemical modification. We can see, for example, in S.4, a glycosylated substituent, but we ignore those that are more common. The fragment analysis of all the unique molecules using Recap generates a total of 1042 different fragments. Because of the large number of fragments (and also the wide chemical diversity), we included the most frequent fragments (with more than 4 non-hydrogen atoms and with an abundance from 0.25 to 24%) in Supporting Information (Figure S1). The fragments were arranged in groups, and we can easily identify the fragments related to gallic and caffeic acids, fragments, or even portions of the double ring fragments associated with the molecules with higher evidence (quercetin, luteolin, kaempferol, etc.). We also found several monosaccharides, disaccharides, and glucuronic fragments, which are consistent with glycosylated flavonoids. On the other hand, we found fragments present in molecules, such as piperine, coumaperine, piperundecalidi, and quinic acids, along with different aliphatic substituents commonly found in the dataset.
We previously described the most frequent plants found in our literature research. However, if we look at the predicted molecules with higher evidence (Figure 4), molecules including quercetin, kaempferol, rutin, luteolin, and others are very frequent across many different plants that can be found worldwide. In fact, after a quick consultation of the Lotus Database, quercetin (ID: LTS0004651) had been found in 1399 species and rutin (ID: LTS0042292) in 365 species. A large number of species can be found for other flavonoids, for example kaempferol, luteolin, and baicalin.
3.1. Molecules with Experimental Evidence as Inhibitors of SARS-CoV-2 Mpro and PLpro
As previously expressed, most data consulted with experimental evidence comprise the inhibition of Mpro. Therefore, our comparison between predicted molecules and those with experimental evidence focuses on Mpro. Nevertheless, we did not separate molecules predicted as inhibitors of Mpro from PLpro from our list of 648 (only 34 were predicted exclusively as inhibitors of PLpro). We will discuss our findings and indicate when the prediction was originally for Mpro, PLpro, or both.
Out of the 648 molecules predicted as inhibitors of the SARS-CoV-2 Mpro protease, 33 were directly found in our dataset of experimentally validated molecules (Table 1).21,22,31,32,23−30 We only find 10 molecules with experimental evidence of inhibition against SARS-CoV-2 PLpro: anacardic acid,33 baicalein,28 cyanidin-3-O-glucoside,34 ginkgolic acid,33 hypericin,34 isoforsythiaside,28 rutin,34 scutellarin,28 procyaninidin,28 and wogonin.28 From these 45 molecules predicted as Mpro inhibitors, 27 had been validated experimentally in 6 out of the 10 articles collected in our literature search.23−27,35 The other 6 molecules were found in the additional experimental databases used in this work. The compounds curcumin, (−)-epigallocatechin gallate, acteoside, hyperoside, silibinin, chlorogenic acid, isochlorogenic acid B, liquiritin, pectolinarin, ginkgetin, andrographiside, emodin, isoginkgetin, isoschaftoside, procyanidin, theaflavin, wogonoside, isochlorogenic acid A, and tannic acid were exclusively predicted as Mpro inhibitors. Additionally, many of the predicted molecules with experimental evidence of inhibiting Mpro are those with higher numbers of evidence (across computational studies; see Figure 3), and 14 of them have also been studied using molecular dynamics simulations. Some of these compounds with experimental evidence as SARS-CoV-2 Mpro inhibitors have been previously identified as inhibitors of the SARS-CoV Mpro. Quercetin, epigallocatechin gallate, amentoflavone, apigenin, luteloin, and curcumin are some of these compounds able to inhibit the main protease of both SARS-CoVs.36,37
Table 1. Molecules Predicted as SARS-CoV Mpro Inhibitors with Experimental Evidence.
| compound name | source | ref | compound name | source | ref |
|---|---|---|---|---|---|
| (+)-dihydromyricetin | COVID-19 DB | (21,22) | isoginkgetin | article | (25) |
| (−)-epigallocatechin gallate | article, COVID-19 DB | (23) | isorhamnetin (3-methylquercetin) | article | (25) |
| isochlorogenic acid A | article | (24) | isoschaftoside | article | (27) |
| acteoside | article | (24) | liquiritin | article, COVID-19 DB | (24) |
| amentoflavone (didemethyl-ginkgetin) | article | (25) | luteolin | article | (25) |
| andrographoiside | article | (24) | luteoloside | article | (24) |
| andrographolide | article, COVID-19 DB | (24) | myricetin | COVID-19 DB | (21) |
| apigenin | article | (25) | narcissin | article | (27) |
| baicalin | article, COVID-19 DB | (24) | pectolinarin | article, COVID-19 DB | (24) |
| chlorogenic acid | article, COVID-19 DB | (24) | procyanidin | COVID-19 DB | (28) |
| curcumin | article | (23) | quercetin | article | (23) |
| ellagic acid | article | (23) | rutin (quercetin 3-rutinoside) | article, PostEra DB, COVID-19 DB | (29) |
| emodin | article | (26) | silibinin | COVID-19 DB | (30) |
| ginkgetin | article | (25) | tannic acid | COVID-19 DB | (31) |
| glycyrrhizin | article, COVID-19 DB | (24) | theaflavin | COVID-19 DB | (31,32) |
| hyperoside (quercetin 3-galactoside) | article | (27) | wogonoside | article | (24) |
| isochlorogenic acid B | article | (24) |
Several flavonoids have been validated as able to bind SARS-CoV-2 Mpro using methods other than enzymatic assays. For instance, baicalein, baicalin (baicalein 7-O-glucuronide), scutellarein, ganhuangenin, wogonoside, oroxylin A-7-O-β-d-glucuronide, and luteoloside have been validated as Mpro inhibitors using a native mass spectrometry affinity method.24 From this group of molecules, baicalein, scutellarein, and ganhuangenin showed KD values against SARS-CoV-2 Mpro of 0.94, 3.02, and 0.84 μM, respectively.38 Furthermore, baicalein, wogonin, scutellarin, and isoforsythiaside have been identified as inhibitors of PLpro.38 Baicalein is one of two natural flavones with a three-dimensional structure in complex with the SARS-CoV-2 Mpro (PDB ID: 6M2N), and it binds to the active site of the protease. Baicalein does not appear in our database of predicted compounds but baicalin does, opening a question regarding the possible effects of glycosides and/or glucuronide derivatives in the inhibitory activity. The presence of glucuronic acid in baicalin results in a higher value of KD with respect to baicalein, and this difference in affinity for Mpro has been validated using interaction-based methods, like isothermal calorimetry and native mass spectrometry, as well as enzymatic assays.38 Another interesting finding with baicalein and its derivative baicalin is that only the first can inhibit PLpro, suggesting that the presence of glucuronic acid in baicalin is an impediment to the inhibition of this protease.
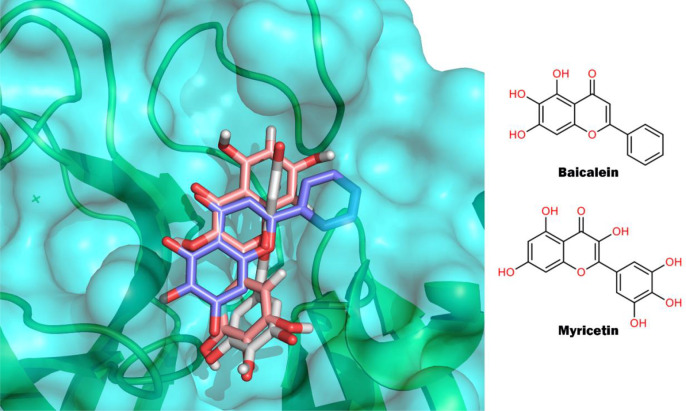
Myricetin is the other flavone with two three-dimensional structures with the SARS-CoV-2 Mpro (PDB IDs: 7DPP and 7B3E).22,39 In these structures, myricetin is covalently linked to the catalytic C145, but the chromone group has different orientations, as shown in Figure 6) This plasticity in the binding mode of flavones can also be seen when we compare the binding modes of baicalein and myricetin. Despite both molecules having the same scaffold (Figure 4), their binding modes in the active site of Mpro are quite different (Figure 6). The pyrogallol group of myricetin is oriented to the catalytic C145 and is covalently linked to it, while in baicalein the chromone group is the one that is close to the C145 and is not covalently connected to Mpro. These crystallographic structures have revealed that even molecules with the same scaffold can bind very differently to the active site of Mpro. Therefore, the assertion that molecules sharing a scaffold should bind in the same way to a protein target is not necessarily valid in this case. This plasticity in the binding modes is usually seen in molecules with affinities in the micromolar range for a target and has been reported for other families of structurally related compounds.40−43 We also found a third three-dimensional structure of SARS-CoV-2 with a natural plant product (shikonin), which is classified as a quinone (PDB ID: 7CA8).44 However, the analysis of this structure revealed negative electron density in the FO–FC electron density map (3σ contour) in almost the entire region where shikonin was modeled, indicating low confidence in the binding mode of the modeled ligand. Because of this, we do not include this structure in our analysis. The epigallocatechin gallate is a flavonol that showed inhibitory activity against Mpro in enzymatic assays. However, in the same study, the authors suggested a nonspecific binding between epigallocatechin gallate and Mpro, based on the analysis of SPR data.23 This could be an example of compounds that show inhibitory activity in enzymatic assays, but interaction-based assays revealed nonspecific interaction with the target.
Figure 6.
Binding modes of baicalein (blue, PDB ID: 6M2N) and myricetin (white, PDB ID: 7DPP; salmon PDB ID: 7B3E) in the active site of SARS-CoV-2 Mpro.
Among the inhibitors of Mpro, there are also biflavonoids like ginkgetin, amentoflavone (didemethyl-ginkgetin), and isoginkgetin. It can be noticed from their names that their structures are very similar, as well as their IC50 against Mpro with values of 2.98, 2.33, and 8.65 μM, respectively.25 Even so, some of these flavonoids and biflavonoids, such as quercetin, amentoflavone, apigenin, and luteloin, which inhibit SARS-CoV-2 Mpro, were identified previously as inhibitors of the SARS-CoV Mpro.36 Therefore, in some sense, these molecules appear to be obvious potential inhibitors of SARS-CoV-2 Mpro, considering the high similitude between the main proteases of both SARS-CoVs.
The compounds glycyrrhizin, andrographolide, and andrographiside are terpenes identified as SARS-CoV-2 Mpro inhibitors.24 The structures of andrographolide and andrographiside are very similar, to the extent that the latter can be considered a glycosylated variant of the former. Andrographolide can inhibit Mpro from both SARS-CoVs through a covalent linkage with the C145.45 Although the binding mode of andrographiside has not been identified, it is very likely that it also binds to the Mpro C145 through a Michael addition reaction, considering the presence of an α,β-unsaturated γ-lactone moiety in its structure. In addition, it has been demonstrated that andrographolide is able to inhibit the production of infectious virions in SARS-CoV-2-infected human lung epithelial cells with low to no cytotoxic effects on human cell lines.46 These results make this molecule a promising hit (scaffold) for the development of new SARS-CoV-2 antiviral drugs.
Among the inhibitors of the SARS-CoV-2 Mpro, there are also some carboxylic acid derivatives, including chlorogenic acid (3-O-caffeoylquinic acid), isochlorogenic acid A (3,5-dicaffeoylquinic acid), and isochlorogenic acid B (3,4-dicaffeoylquinic acid). It is very possible that the interaction detected by native mass spectrometry for these three compounds and Mpro could be related to the presence of α,β-unsaturated ester moiety, which could react as a Michael acceptor with the thiol groups of the cysteines residues of Mpro, as was proposed for the forsythoside A.24 However, in the same study, it was suggested that the interaction between this molecule and Mpro, to which it is able to bind covalently, seems to be a nonspecific binding with every accessible free cysteine.
In the case of phenolic compounds, which include subgroups such as flavonoids, phenolic acids, and quinones, their aggregation tendency could lead to obtaining false positive results in the inhibition enzymatic assays.47 The inhibition of SARS-CoV Mpro of kaempferol and amentoflavone is lower when 0.1% of Triton X-100 is present in the enzymatic assays, suggesting that previous inhibition observed could be due to compound aggregation.48 This finding is quite interesting considering that the structural difference between quercetin and kaempferol is hydroxyl. On the other hand, the interaction between kaempferol and Mpro was detected by surface plasmon resonance (SPR) with a KD of 24.65 μM.27 Of course, SPR allows us to know that there is an interaction between two molecules but is not able to specify if this interaction has any effect on the enzymatic activity. In the same study, the authors found that the interaction of a glycosylated form of kaempferol (kaempferol 3-O-gentiobioside) with Mpro is 10 times stronger (KD: 2.7 μM), which again raises questions regarding the role of glycosylation in the inhibitory properties of molecules with this scaffold. The experimental setup is something to consider when you want to categorize a molecule as a hit or not. For instance, ellagic acid showed a 10% inhibition at 30 μM in one report,35 but more than 50% inhibition at 10 μM was also reported.23 These apparently contradictory results are very likely related to the concentration of Mpro or substrate used in both assays, but for inexperienced readers this could be difficult to understand. In this specific example, one enzymatic assay uses 0.1 μM of Mpro while the other uses 11 mM, which corresponds to more than 1000 times more enzyme.
3.2. Exploring Similarity and Scaffold Variations
Looking at the molecules with higher theoretical evidence (Figure 4), the classification of the molecules (Figure 3), as well as the scaffold frequency (Figure 5), a question emerges: How diverse is the chemical space of the predicted natural products? Moreover, how much of the chemical space is covered by experimental evidence? The quantification of the chemical space is a complex problem and instead of looking for a hard number, we decided to follow a graphical approach.
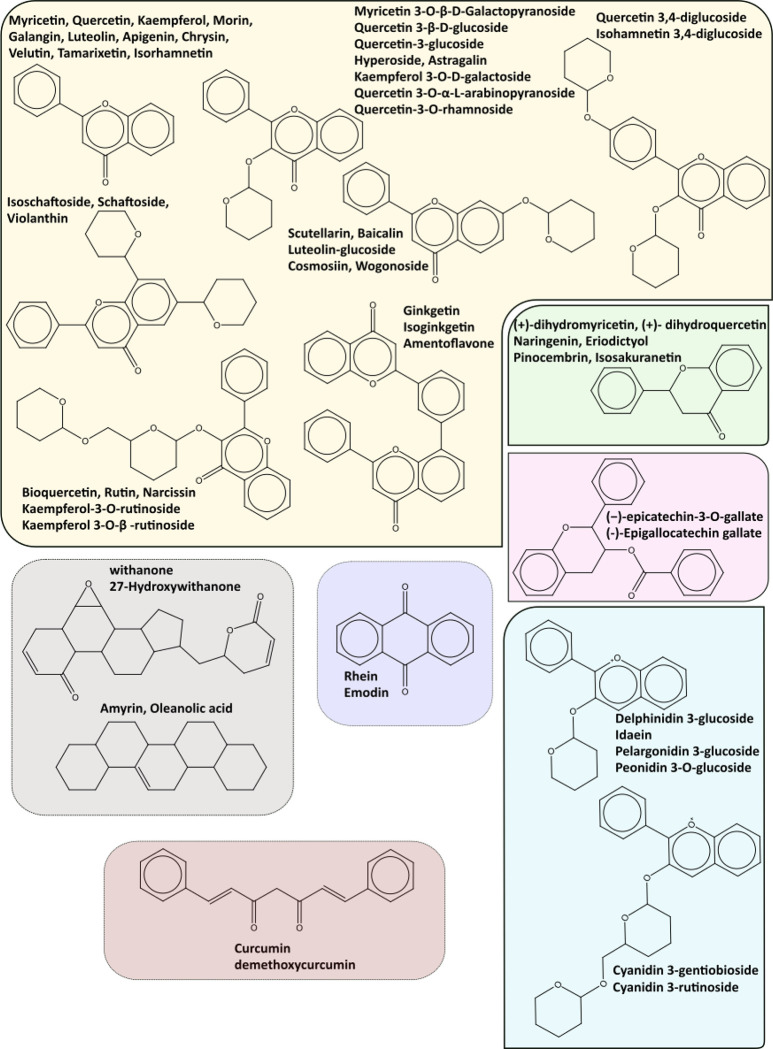
All 648 molecules were compared to all those with experimental evidence (similarity of ≥0.75). Of the 648 molecules, 100 (15.4%, including those with direct experimental evidence) are similar or equal to some of the molecules with experimental data. Of the 100 molecules showing some similarity (≥0.75) with respect to compounds with experimental evidence, a total of 50 scaffolds were identified. However, out of these 50 scaffolds, 15 of them comprise 60 of the 100 molecules (Figure 7). In addition, 6 of the 15 more abundant scaffolds contain the core flavone (comprising flavones, flavonols, and isoflavones) and diflavones with and without sugar derivatives (Figure 7, yellow panel). Six structures are represented in the flavanone core (including the flavanonols) (Figure 7, green panel), six others in the anthocyanin scaffold (Figure 7, blue panel), and two flavans (Figure 7, violet panel). This distribution where flavonoids are the most representative group of molecules is consistent with Figure 3A. Of course, these are not the only flavonoids in the 100 molecules, but rather are the scaffolds with the highest representations (i.e., there are flavans, flavones, anthocyanins, and others with more than two sugar derivatives and glucuronic moieties).
Figure 7.
Top 15 scaffolds representing the most abundant molecules in the 100 with high similarity (similarity is ≥0.75) to those with experimental evidence. The scaffolds were grouped according to flavone with and without sugar derivatives (yellow panel, including flavones, flavonols, isoflavones and diflavones), flavanone (green panel, including flavanonols), anthocyanins (blue panel), flavan-3-ols (red panel), anthraquinone (dark blue), steroids and triterpenoids (gray panel), and finally curcumin (brown panel).
The other relevant group is the terpenoids. In this group, we have four molecules comprising two scaffolds. However, a total of 9 steroids and triterpenoids were identified in the 100 molecules, while 108 (steroids and triterpenoids) were predicted. Phytosterols are a diverse family of compounds presenting several challenges for extraction and identification.49 The high number of predictions compared to those experimentally validated (or similar to) suggests that more research needs to be performed concerning these compounds, especially at an experimental level. A similar underrepresentation is evident for anthraquinone and curcumin derivates (phenolic classification). Curcumin derivatives need to be carefully considered because they are well-known interfering compounds (pan-assay interference compounds). Curcumin and its derivatives have been predicted as Mpro inhibitors by several authors using docking and molecular dynamics.50−54 It was also proven to have an antiviral effect in Vero E6 cells55 and an IC50 = 11.9 μM, determined using an enzymatic assay, but was not analyzed by surface plasmon resonance because of its low solubility.23 Even though few experimental studies had been carried out exploring curcumin interaction with Mpro of SARS-CoV-2, we found two ongoing clinical trials (NCT05150782 and NCT05008003) and one completed trial (NCT05130671) that evaluated the benefits of a compound’s mixture (curcumin, vitamin D, quercetin, etc.) as a dietary supplement in COVID-19 patients. So, although we know it is an interfering molecule (interfering molecules are usually avoided in medical chemistry) and even with its limitations in terms of pharmaceutical properties (bioavailability, poor absorption, and rapid elimination),56 further research needs to be carried out, especially regarding the physical interaction between curcumin and SARS-CoV-2 main protease.
We can see from the scaffolds (Figure 7) that several of them comprise substituents in various positions that correspond with sugar moieties and glucuronic derivatives. We previously discussed that glucuronic acid in baicalin results in a higher value of KD than the same acid in baicalein, and this difference in affinity for Mpro has been validated by different experimental methods. This finding highlights the problem of glycosylation and its effects on Mpro (and PLpro) inhibition. An early computational study57 found that the presence of sugar units improves the binding energy with respect to Mpro. However, this study has several limitations, for instance the lack of a negative control.58 Moreover, the experimental evidence is markedly more complex mainly because (1) several studies have explored the effect of glycosylated flavonoids in several problems59,60 but few studies have used glycosylated and nonglycosylated compounds under the same experimental conditions in Mpro or PLpro inhibition; (2) few studies allow us to define the precise binding site (to be discussed later). A few cases need to be addressed: rutin/quercetin and baicalin/baicalein. Quercetin has been initially proven to bind Mpro, having a Ki = 7.4 μM,61 and rutin was found to bind in a similar way with a Ki = 11.0 μM,29 suggesting a very similar binding affinity for glycosylated and nonglycosylated flavonoids. It is important to mention that the glycosylation of quercetin improves some pharmaceutical properties (solubility and bioavailability). Both baicalin and baicalein showed SARS-CoV-2 antiviral effect in vitro (Vero CCL-81 cells),62,63 and several pharmacodynamic evaluations have been performed in various organisms.63 These results cannot allow us to generalize that the glycosylation of flavonoids can improve inhibition potential but clearly indicate the need for more research in this area, particularly with Mpro and PLpro interactions.
3.3. Allosteric Binding Sites in Mpro and PLpro
Allosteric binding sites have been identified in both proteases of SARS-CoV-2 through the screening of compound libraries. Two allosteric binding sites have been identified in SARS-CoV-2 Mpro using X-ray crystallography. One of these sites is a hydrophobic pocket to the C-terminal of one protomer and is close to the active site of the other protomer. Five molecules that bind to this binding site were identified, all five with different scaffolds: pelitinib, RS-102895, ifenprodil, PD-168568, and tofogliflozin.64 Despite binding to the same pocket, the binding modes of pelitinib and tofogliflozin are different from the other three molecules. The second allosteric binding site is formed by a deep groove between the dimerization and catalytic domains. One molecule, AT7519, was found in this binding site, and its allosteric modulator inhibition of Mpro was related to its interaction with R298, which is a crucial amino acid for dimerization and proper stabilization of the active site.64 To date, no natural plant products or derivatives have been identified as being able to bind these allosteric binding sites.
In the case of SARS-CoV-2 PLpro, three natural molecules from plants have been identified as allosteric ligands in a screening study of a library of 500 molecules using protein X-ray crystallography: 4-(2-hydroxyethyl)phenol, 4-hydroxybenzaldehyde, and methyl 3,4-dihydroxybenzoate.65 The former two compounds bind at the same allosteric binding site, located between the domains Ubl and thumb, while methyl 3,4-dihydroxybenzoate binds to a binding site on the surface of the thumb domain. In all three cases, the inhibitor interacts with a helix (S2 helix), which is important for the PLpro interaction with the ISG15 molecule and inhibits the deISGylating enzymatic activity of PLpro.
The identification of these allosteric binding sites represents an opportunity for the discovery of potential allosteric inhibitors of Mpro and PLpro using a combination of in silico and experimental approaches, as have been used in the search for active-site binding molecules.
3.4. Challenges and Future Perspectives
In the context of a pandemic, there is an urgent need to find therapeutic agents to control it. In the rush to discover treatment alternatives for the SARS-CoV-2 virus, many research reports have focused on plant-based compounds and their potential and confirmed bioactivity. Given that this is a “hot” and urgent topic, some published works solely containing computational predictions may contain pitfalls that affect their quality and hence the reliability of their predictions. Perhaps the most common misinterpretation of computational studies related to SARS-CoV-2 is taking molecular docking results as a demonstration of ligand binding.66 Examples of this can be found in the scientific literature, particularly in reports where a few plant-derived compounds are docked into the Mpro and/or PLpro enzymes, and authors claimed that the top-scoring ones are enzyme inhibitors.50,67−71 We understand that not all research groups have the capability of experimentally testing their predictions. However, some theoretical options could add additional support to computational studies, making them attractive for groups who do have the capacity for performing experimental evaluations. For example, mixing the natural products with known enzyme inhibitors and evaluating how the studied compounds rank among these known inhibitors and noninhibitors. Other options could be to perform additional MD simulations and accurate binding energy estimations from MD simulations.
Another important aspect to consider when analyzing the confirmed anti-SARS-CoV-2 activity of the plant-based compounds here discussed is their possible interference with the outcome of the assays. PAINS are defined as compounds with substructural motifs associated with increased chances of being registered as a hit in different assays.72,73 This issue has been previously discussed in the context of natural products.74 We used the Filter program75,76 to identify PAINS alerts in the set of 648 plant-based molecules collected for our investigation. The results of this analysis are provided in Table S5 (Supporting Information).
We found that 248 of the compounds are identified as PAINS, a result to consider before proceeding to the experimental evaluation of any of these chemicals. Strikingly, 26 out of 45 compounds with experimental evidence of SARS-CoV-2 activity are identified as PAINS. Extensive scrutiny and experimentation must be undertaken to fully clarify if the activity of all these 26 compounds is because they are real PAINS or protease inhibitors. Among these 26 compounds, 20 are identified as containing a catechol A motif that can react and form covalent adducts with the catalytic cysteine of Mpro. A clear example of this reactivity is myricetin, which has been cocrystallized with this enzyme. The fact that myricetin has different orientations within the active site of Mpro in the two structures, together with the multiple cysteine-containing targets associated with it in the ChEMBL database, strongly suggests that myricetin is indeed a PAINS. The presence of PAINS in several common scaffolds of plant-derived molecules is something that should be consider in the selection of the assay to be used to validate the binding/inhibition of the target. Additionally, the use of a standardized enzymatic assay is very important to obtain straightforward data about whether a specific compound can inhibit or not the target, and to compare with other studies, Interaction-based methodologies like biolayer interferometry, surface plasmon resonance, native mass spectrometry, and isothermal titration calorimetry should be used to confirm or refute the inhibition detected by enzymatic assays.
Certainly, extensive experimental work is essential to further validate the plant-based compounds with either confirmed or predicted SARS-CoV-2 inhibition activity discussed above, especially those scaffolds with not a single compound experimentally validated. This work has shown that most of the plant-based compounds predicted as potential SARS-CoV-2 inhibitors lack experimental evaluation. The validation of some of these predictions, perhaps those based on the most solid computational approaches, could be among the next steps to take. Another important issue to clarify is whether or not many plant-based compounds with confirmed activity against the SARS-CoV-2 virus predicted are actual PAINS. The latter is a highly relevant issue since the detection of PAINS compounds at the early stages of the drug discovery process can save time and economic resources. This will allow the scientific community to prioritize available resources for the optimization of the antiviral activity of the most promising and specific plant-based compounds.
4. Conclusions
This study presents a critical review of the main reports in the literature on plant-derived compounds as possible inhibitors of SARS-CoV-2, as well as a comparison with the drugs reported in clinical trials for the same purpose. According to the analyses carried out on 648 molecules predicted as potential inhibitors of Mpro and/or PLpro, the main conclusions are as follows.
-
(1)
Several of the predicted compounds with higher theoretical confidence are present in many plants used in traditional medicine and even food. However, we need to be cautious about further research reporting the same compounds in other plants and only addressing computational models. These studies will not increase the validation of the already identified molecules or provide more comparable information regarding the already reported ones.
-
(2)
Given that several plants can share the same compounds already identified with computational or even experimental information, further research into the inhibition of Mpro or PLpro using plant crude extracts or semipurified fractions needs to be complemented along with chemical characterization.
-
(3)
The most effective or attractive compounds are baicalein, scutellarin, and myricetin considering the availability of 3D-structures in complex with Mpro and/or evidence of binding from different techniques.
-
(4)
Several phytosterols and triterpenoids have been predicted as potential inhibitors of Mpro and PLpro. However, little experimental information was found for those families of compounds. It is necessary to perform further chemical characterization and experimental evaluation, focusing on these bioactive molecules.
-
(5)
We need more experimental data to assess the effect of glycosylation in the flavonoid inhibition of Mpro or PLpro, using standardized methodologies.
-
(6)
Future studies exploring the natural compound inhibition of Mpro and PLpro should address the identified and potential allosteric binding sites.
Acknowledgments
This work was supported by Universidad de Las Américas (Research Grant BIO.YGB.21.02 (Y. Guerra)). We also want to thank Universidad de Las Américas for supporting E. Tejera and Y. Castillo, and Universidad San Francisco de Quito for supporting J.M. Alvarez-Suarez.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05766.
Common fragments in the 648 predicted molecules (Figure S1) (PDF)
List of unique predicted inhibitors retrieved from articles and databases (Table S1); chemical classification of the unique molecules using NPClassifier (Table S2); list of molecules with experimental validation as SARS-CoV-2 Mpro inhibitors (Table S3); scaffolds and fragments identified for each unique molecule (Table S4); predicted PAINS alerts in compounds with either predicted or confirmed activity against SARS-CoV-2 proteases (Table S5) (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- WHO . WHO Timeline - COVID-19; https://www.who.int/news/item/27-04-2020-who-timeline—covid-19 (accessed August 25, 2022).
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. Structure of Mpro from SARS-CoV-2 and Discovery of Its Inhibitors. Nature 2020, 582 (7811), 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Harcourt B. H.; Jukneliene D.; Kanjanahaluethai A.; Bechill J.; Severson K. M.; Smith C. M.; Rota P. A.; Baker S. C. Identification of Severe Acute Respiratory Syndrome Coronavirus Replicase Products and Characterization of Papain-Like Protease Activity. J. Virol 2004, 78 (24), 13600–13612. 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R.; Kumari S.; Pandey B.; Mistry H.; Bihani S. C.; Das A.; Prashar V.; Gupta G. D.; Panicker L.; Kumar M. Structural Insights into SARS-CoV-2 Proteins. J. Mol. Biol. 2021, 433 (2), 166725. 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.; Ratia K.; Johnston R. E.; Mesecar A. D.; Baric R. S. Severe Acute Respiratory Syndrome Coronavirus Papain-Like Protease Ubiquitin-Like Domain and Catalytic Domain Regulate Antagonism of IRF3 and NF-KB Signaling. J. Virol 2009, 83 (13), 6689–6705. 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.; Mukherjee R.; Grewe D.; Bojkova D.; Baek K.; Bhattacharya A.; Schulz L.; Widera M.; Mehdipour A. R.; Tascher G.; Geurink P. P.; Wilhelm A.; van der Heden van Noort G. J.; Ovaa H.; Müller S.; Knobeloch K.-P.; Rajalingam K.; Schulman B. A.; Cinatl J.; Hummer G.; Ciesek S.; Dikic I. Papain-like Protease Regulates SARS-CoV-2 Viral Spread and Innate Immunity. Nature 2020, 587 (7835), 657–662. 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K.; Saikatendu K. S.; Santarsiero B. D.; Barretto N.; Baker S. C.; Stevens R. C.; Mesecar A. D. Severe Acute Respiratory Syndrome Coronavirus Papain-like Protease: Structure of a Viral Deubiquitinating Enzyme. Proc. Natl. Acad. Sci. 2006, 103 (15), 5717–5722. 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H. A.; Fotouhi-Ardakani N.; Lytvyn V.; Lachance P.; Sulea T.; Ménard R. The Papain-Like Protease from the Severe Acute Respiratory Syndrome Coronavirus Is a Deubiquitinating Enzyme. J. Virol 2005, 79 (24), 15199–15208. 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N.; Jukneliene D.; Ratia K.; Chen Z.; Mesecar A. D.; Baker S. C. The Papain-Like Protease of Severe Acute Respiratory Syndrome Coronavirus Has Deubiquitinating Activity. J. Virol 2005, 79 (24), 15189–15198. 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov A. G.; Waltenberger B.; Pferschy-Wenzig E.-M.; Linder T.; Wawrosch C.; Uhrin P.; Temml V.; Wang L.; Schwaiger S.; Heiss E. H.; Rollinger J. M.; Schuster D.; Breuss J. M.; Bochkov V.; Mihovilovic M. D.; Kopp B.; Bauer R.; Dirsch V. M.; Stuppner H. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33 (8), 1582–1614. 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance H.; Wetzel S.; Kumar K.; Waldmann H. Charting, Navigating, and Populating Natural Product Chemical Space for Drug Discovery. J. Med. Chem. 2012, 55 (13), 5989–6001. 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- Remali J.; Aizat W. M. A Review on Plant Bioactive Compounds and Their Modes of Action Against Coronavirus Infection. Front. Pharmacol 2021, 10.3389/fphar.2020.589044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarba B.; Pandiella A. Medicinal Plants as Sources of Active Molecules against COVID-19. Front. Pharmacol 2020, 10.3389/fphar.2020.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio A. D. S.; Wiedemann L. S. M.; Veiga-Junior V. F. Natural Products’ Role against COVID-19. RSC Adv. 2020, 10 (39), 23379–23393. 10.1039/D0RA03774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saakre M.; Mathew D.; Ravisankar V. Perspectives on Plant Flavonoid Quercetin-Based Drugs for Novel SARS-CoV-2. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10 (1), 21. 10.1186/s43088-021-00107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M.; Jaganathan S. K. Antiproliferative Effects of Honey and of Its Polyphenols: A Review. J. Biomed. Biotechnol 2009, 2009, 830616. 10.1155/2009/830616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. W.; Wang M.; Leber C. A.; Nothias L.-F.; Reher R.; Kang K. B.; van der Hooft J. J. J.; Dorrestein P. C.; Gerwick W. H.; Cottrell G. W. NPClassifier: A Deep Neural Network-Based Structural Classification Tool for Natural Products. J. Nat. Prod 2021, 84 (11), 2795–2807. 10.1021/acs.jnatprod.1c00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis G. W.; Murcko M. A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39 (15), 2887–2893. 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Stumpfe D.; Bajorath J. Computational Exploration of Molecular Scaffolds in Medicinal Chemistry. J. Med. Chem. 2016, 59 (9), 4062–4076. 10.1021/acs.jmedchem.5b01746. [DOI] [PubMed] [Google Scholar]
- Sharma S.; Karri K.; Thapa I.; Bastola D.; Ghersi D. Identifying Enriched Drug Fragments as Possible Candidates for Metabolic Engineering. BMC Med. Genomics 2016, 9 (S2), 46. 10.1186/s12920-016-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Ye F.; Sun Q.; Liang H.; Li C.; Li S.; Lu R.; Huang B.; Tan W.; Lai L. Scutellaria Baicalensis Extract and Baicalein Inhibit Replication of SARS-CoV-2 and Its 3C-like Protease in Vitro. J. Enzyme Inhib. Med. Chem. 2021, 36 (1), 497–503. 10.1080/14756366.2021.1873977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.; Yao S.; Zhao W.; Zhang Y.; Liu J.; Shao Q.; Wang Q.; Li M.; Xie H.; Shang W.; Ke C.; Feng L.; Jiang X.; Shen J.; Xiao G.; Jiang H.; Zhang L.; Ye Y.; Xu Y. Identification of Pyrogallol as a Warhead in Design of Covalent Inhibitors for the SARS-CoV-2 3CL Protease. Nat. Commun. 2021, 12 (1), 3623. 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahun M.; Jukić M.; Oblak D.; Kranjc L.; Bajc G.; Butala M.; Bozovičar K.; Bratkovič T.; Podlipnik Č.; Poklar Ulrih N. Inhibition of the SARS-CoV-2 3CLpro Main Protease by Plant Polyphenols. Food Chem. 2022, 373, 131594. 10.1016/j.foodchem.2021.131594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D.; Su H.; Ke C.; Tang C.; Witt M.; Quinn R. J.; Xu Y.; Liu J.; Ye Y. Efficient Discovery of Potential Inhibitors for SARS-CoV-2 3C-like Protease from Herbal Extracts Using a Native MS-Based Affinity-Selection Method. J. Pharm. Biomed. Anal 2022, 209, 114538. 10.1016/j.jpba.2021.114538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y.; Zhu G.-H.; Wang H.-N.; Hu Q.; Chen L.-L.; Guan X.-Q.; Li H.-L.; Chen H.-Z.; Tang H.; Ge G.-B. Discovery of Naturally Occurring Inhibitors against SARS-CoV-2 3CLpro from Ginkgo Biloba Leaves via Large-Scale Screening. Fitoterapia 2021, 152, 104909. 10.1016/j.fitote.2021.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrot-Hadzik I.; Zmudzinski M.; Matkowski A.; Preissner R.; Kęsik-Brodacka M.; Hadzik J.; Drag M.; Abel R. Reynoutria Rhizomes as a Natural Source of SARS-CoV-2 Mpro Inhibitors–Molecular Docking and In Vitro Study. Pharmaceuticals 2021, 14 (8), 742. 10.3390/ph14080742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q.; Chen Z.; Tao Y.; Zhang B.; Wu X.; Yang L.; Wang Q.; Wang Z. An Integrated Method for Optimized Identification of Effective Natural Inhibitors against SARS-CoV-2 3CLpro. Sci. Rep 2021, 11 (1), 22796. 10.1038/s41598-021-02266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-J.; Chao T.-L.; Kao H.-C.; Tsai Y.-M.; Liu Y.-K.; Wang L. H.-C.; Hsieh M.-C.; Chang S.-Y.; Liang P.-H. Kinetic Characterization and Inhibitor Screening for the Proteases Leading to Identification of Drugs against SARS-CoV-2. Antimicrob. Agents Chemother. 2021, 65 (4), e02577-20. 10.1128/AAC.02577-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuti B.; Grande F.; Conforti F.; Jimenez-Alesanco A.; Ceballos-Laita L.; Ortega-Alarcon D.; Vega S.; Reyburn H. T.; Abian O.; Velazquez-Campoy A. Rutin Is a Low Micromolar Inhibitor of SARS-CoV-2 Main Protease 3CLpro: Implications for Drug Design of Quercetin Analogs. Biomedicines 2021, 9 (4), 375. 10.3390/biomedicines9040375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardanelli A. M.; Isgrò C.; Palese L. L. SARS-CoV-2 Main Protease Active Site Ligands in the Human Metabolome. Molecules 2021, 26 (5), 1409. 10.3390/molecules26051409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T. H.; Jung J.-H.; Kim M.-K.; Lim S.; Choi J.-M.; Chung B.; Kim D.-W.; Kim D. The Inhibitory Effects of Plant Derivate Polyphenols on the Main Protease of SARS Coronavirus 2 and Their Structure–Activity Relationship. Molecules 2021, 26 (7), 1924. 10.3390/molecules26071924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.; Park Y.-I.; Cha Y.-E.; Park R.; Namkoong S.; Lee J. I.; Park J. Tea Polyphenols EGCG and Theaflavin Inhibit the Activity of SARS-CoV-2 3CL-Protease In Vitro. Evidence-Based Complement. Altern. Med. 2020, 2020, 1–7. 10.1155/2020/5630838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Cui Q.; Cooper L.; Zhang P.; Lee H.; Chen Z.; Wang Y.; Liu X.; Rong L.; Du R. Ginkgolic Acid and Anacardic Acid Are Specific Covalent Inhibitors of SARS-CoV-2 Cysteine Proteases. Cell Biosci 2021, 11 (1), 45. 10.1186/s13578-021-00564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsillou E.; Liang J.; Ververis K.; Lim K. W.; Hung A.; Karagiannis T. C.. Identification of Small Molecule Inhibitors of the Deubiquitinating Activity of the SARS-CoV-2 Papain-like Protease: In Silico Molecular Docking Studies and in Vitro Enzymatic Activity Assay Front. Chem. 2020, 8, 10.3389/fchem.2020.623971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito A.; Colantuono A.; Pirone L.; Pedone E.; Intartaglia D.; Giamundo G.; Conte I.; Vitaglione P.; Apone F. Pomegranate Peel Extract as an Inhibitor of SARS-CoV-2 Spike Binding to Human ACE2 Receptor (in Vitro): A Promising Source of Novel Antiviral Drugs. Front. Chem. 2021, 9, 638187. 10.3389/fchem.2021.638187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T. H.; Woo H.-J.; Kang H.-K.; Nguyen V. D.; Kim Y.-M.; Kim D.-W.; Ahn S.-A.; Xia Y.; Kim D. Flavonoid-Mediated Inhibition of SARS Coronavirus 3C-like Protease Expressed in Pichia Pastoris. Biotechnol. Lett. 2012, 34 (5), 831–838. 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C.-C.; Kuo Y.-H.; Jan J.-T.; Liang P.-H.; Wang S.-Y.; Liu H.-G.; Lee C.-K.; Chang S.-T.; Kuo C.-J.; Lee S.-S.; Hou C.-C.; Hsiao P.-W.; Chien S.-C.; Shyur L.-F.; Yang N.-S. Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007, 50 (17), 4087–4095. 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- Su H.; Yao S.; Zhao W.; Li M.; Liu J.; Shang W.; Xie H.; Ke C.; Hu H.; Gao M.; Yu K.; Liu H.; Shen J.; Tang W.; Zhang L.; Xiao G.; Ni L.; Wang D.; Zuo J.; Jiang H.; Bai F.; Wu Y.; Ye Y.; Xu Y. Anti-SARS-CoV-2 Activities in Vitro of Shuanghuanglian Preparations and Bioactive Ingredients. Acta Pharmacol. Sin 2020, 41 (9), 1167–1177. 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzikov M.; Costanzi E.; Reinshagen J.; Esposito F.; Vangeel L.; Wolf M.; Ellinger B.; Claussen C.; Geisslinger G.; Corona A.; Iaconis D.; Talarico C.; Manelfi C.; Cannalire R.; Rossetti G.; Gossen J.; Albani S.; Musiani F.; Herzog K.; Ye Y.; Giabbai B.; Demitri N.; Jochmans D.; Jonghe S. De; Rymenants J.; Summa V.; Tramontano E.; Beccari A. R.; Leyssen P.; Storici P.; Neyts J.; Gribbon P.; Zaliani A. Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen. ACS Pharmacol. Transl. Sci. 2021, 4 (3), 1096–1110. 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert M.; Köster H.; Bartholomäus R.; Park A. Y.; Shahim A.; Heine A.; Steuber H.; Klebe G.; Diederich W. E. Tracing Binding Modes in Hit-to-Lead Optimization: Chameleon-Like Poses of Aspartic Protease Inhibitors. Angew. Chemie Int. Ed 2015, 54 (9), 2849–2853. 10.1002/anie.201411206. [DOI] [PubMed] [Google Scholar]
- Czodrowski P.; Hölzemann G.; Barnickel G.; Greiner H.; Musil D. Selection of Fragments for Kinase Inhibitor Design: Decoration Is Key. J. Med. Chem. 2015, 58 (1), 457–465. 10.1021/jm501597j. [DOI] [PubMed] [Google Scholar]
- Malamas M. S.; Manas E. S.; McDevitt R. E.; Gunawan I.; Xu Z. B.; Collini M. D.; Miller C. P.; Dinh T.; Henderson R. A.; Keith J. C.; Harris H. A. Design and Synthesis of Aryl Diphenolic Azoles as Potent and Selective Estrogen Receptor-β Ligands. J. Med. Chem. 2004, 47 (21), 5021–5040. 10.1021/jm049719y. [DOI] [PubMed] [Google Scholar]
- Kramer C.; Fuchs J. E.; Liedl K. R. Strong Nonadditivity as a Key Structure–Activity Relationship Feature: Distinguishing Structural Changes from Assay Artifacts. J. Chem. Inf. Model 2015, 55 (3), 483–494. 10.1021/acs.jcim.5b00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Zhou X.; Zhang Y.; Zhong F.; Lin C.; McCormick P. J.; Jiang F.; Luo J.; Zhou H.; Wang Q.; Fu Y.; Duan J.; Zhang J. Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Natural Product Inhibitor Shikonin Illuminates a Unique Binding Mode. Sci. Bull. 2021, 66 (7), 661–663. 10.1016/j.scib.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T.-H.; Huang Y.-L.; Chen C.-C.; Pi W.-C.; Hsu Y.-L.; Lo L.-C.; Chen W.-Y.; Fu S.-L.; Lin C.-H. Andrographolide and Its Fluorescent Derivative Inhibit the Main Proteases of 2019-NCoV and SARS-CoV through Covalent Linkage. Biochem. Biophys. Res. Commun. 2020, 533 (3), 467–473. 10.1016/j.bbrc.2020.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa-ngiamsuntorn K.; Suksatu A.; Pewkliang Y.; Thongsri P.; Kanjanasirirat P.; Manopwisedjaroen S.; Charoensutthivarakul S.; Wongtrakoongate P.; Pitiporn S.; Chaopreecha J.; Kongsomros S.; Jearawuttanakul K.; Wannalo W.; Khemawoot P.; Chutipongtanate S.; Borwornpinyo S.; Thitithanyanont A.; Hongeng S. Anti-SARS-CoV-2 Activity of Andrographis Paniculata Extract and Its Major Component Andrographolide in Human Lung Epithelial Cells and Cytotoxicity Evaluation in Major Organ Cell Representatives. J. Nat. Prod 2021, 84 (4), 1261–1270. 10.1021/acs.jnatprod.0c01324. [DOI] [PubMed] [Google Scholar]
- Pohjala L.; Tammela P. Aggregating Behavior of Phenolic Compounds — A Source of False Bioassay Results?. Molecules 2012, 17 (9), 10774–10790. 10.3390/molecules170910774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Kim S.; Shin D. H.; Kim M.-S. Inhibition of SARS-CoV 3CL Protease by Flavonoids. J. Enzyme Inhib. Med. Chem. 2020, 35 (1), 145–151. 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Llatas G.; Alegría A.; Barberá R.; Cilla A. Current Methodologies for Phytosterol Analysis in Foods. Microchem. J. 2021, 168, 106377. 10.1016/j.microc.2021.106377. [DOI] [Google Scholar]
- Babaeekhou L.; Ghane M.; Abbas-Mohammadi M. In Silico Targeting SARS-CoV-2 Spike Protein and Main Protease by Biochemical Compounds. Biologia (Bratisl) 2021, 76 (11), 3547–3565. 10.1007/s11756-021-00881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R.; Singh S.; Singh A.; Misra K. Two Pronged Approach for Prevention and Therapy of COVID-19 (Sars-CoV-2) by a Multi-Targeted Herbal Drug, a Component of Ayurvedic Decoction. Eur. J. Integr. Med. 2021, 43, 101268. 10.1016/j.eujim.2020.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat J.; Singh P.; Patel R. A Computational Approach for the Screening of Potential Antiviral Compounds against SARS-CoV-2 Protease: Ionic Liquid vs Herbal and Natural Compounds. J. Mol. Liq. 2021, 326, 115298. 10.1016/j.molliq.2021.115298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teli D. M.; Shah M. B.; Chhabria M. T. In Silico Screening of Natural Compounds as Potential Inhibitors of SARS-CoV-2 Main Protease and Spike RBD: Targets for COVID-19. Front. Mol. Biosci 2021, 7, 599079. 10.3389/fmolb.2020.599079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adem Ş.; Eyupoglu V.; Ibrahim I. M.; Sarfraz I.; Rasul A.; Ali M.; Elfiky A. A. Multidimensional in Silico Strategy for Identification of Natural Polyphenols-Based SARS-CoV-2 Main Protease (Mpro) Inhibitors to Unveil a Hope against COVID-19. Comput. Biol. Med. 2022, 145, 105452. 10.1016/j.compbiomed.2022.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Palma D.; Tabares-Guevara J. H.; Zapata-Cardona M. I.; Flórez-Álvarez L.; Yepes L. M.; Rugeles M. T.; Zapata-Builes W.; Hernandez J. C.; Taborda N. A. Curcumin Inhibits In Vitro SARS-CoV-2 Infection In Vero E6 Cells through Multiple Antiviral Mechanisms. Molecules 2021, 26 (22), 6900. 10.3390/molecules26226900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-S.; Chen T.-H.; Weng L.; Huang L.; Lai D.; Weng C.-F. Pharmacological Properties and Underlying Mechanisms of Curcumin and Prospects in Medicinal Potential. Biomed. Pharmacother 2021, 141, 111888. 10.1016/j.biopha.2021.111888. [DOI] [PubMed] [Google Scholar]
- Cherrak S. A.; Merzouk H.; Mokhtari-Soulimane N. Potential Bioactive Glycosylated Flavonoids as SARS-CoV-2 Main Protease Inhibitors: A Molecular Docking and Simulation Studies. PLoS One 2020, 15 (10), e0240653 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Avalos G.; Vargas-Ruiz A. P.; Delgado-Pease N. E.; Olivos-Ramirez G. E.; Sheen P.; Fernández-Díaz M.; Quiliano M.; Zimic M.; Agurto-Arteaga A.; Antiparra R.; Ardiles-Reyes M.; Calderon K.; Cauna-Orocollo Y.; de Grecia Cauti-Mendoza M.; Chipana-Flores N.; Choque-Guevara R.; Chunga-Girón X.; Criollo-Orozco M.; De La Cruz L.; Delgado-Ccancce E.; Elugo-Guevara C.; Fernández-Sanchez M.; Guevara-Sarmiento L.; Gutiérrez K.; Heredia-Almeyda O.; Huaccachi-Gonzalez E.; Huerta-Roque P.; Icochea E.; Isasi-Rivas G.; Juscamaita-Bartra R. A.; Licla-Inca A.; Montalvan A.; Montesinos-Millan R.; Núñez-Fernández D.; Ochoa-Ortiz A.; Páucar-Montoro E.; Pauyac K.; Perez-Martinez J. L.; Perez-M N.; Poma-Acevedo A.; Quiñones-Garcia S.; Ramirez-Ortiz I.; Ramos-Sono D.; Rios-Angulo A. A.; Rios-Matos D.; Rojas-Neyra A.; Romero Y. K.; Salguedo-Bohorquez M. I.; Sernaque-Aguilar Y.; Soto L. F.; Tataje-Lavanda L.; Ticona J.; Vallejos-Sánchez K.; Villanueva-Pérez D.; Ygnacio-Aguirre F. Comprehensive Virtual Screening of 4.8 k Flavonoids Reveals Novel Insights into Allosteric Inhibition of SARS-CoV-2 MPRO. Sci. Rep 2021, 11 (1), 15452. 10.1038/s41598-021-94951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slámová K.; Kapešová J.; Valentová K. Sweet Flavonoids”: Glycosidase-Catalyzed Modifications. Int. J. Mol. Sci. 2018, 19 (7), 2126. 10.3390/ijms19072126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronina T.; Strugała P.; Popłoński J.; Włoch A.; Sordon S.; Bartmańska A.; Huszcza E. The Influence of Glycosylation of Natural and Synthetic Prenylated Flavonoids on Binding to Human Serum Albumin and Inhibition of Cyclooxygenases COX-1 and COX-2. Molecules 2017, 22 (7), 1230. 10.3390/molecules22071230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abian O.; Ortega-Alarcon D.; Jimenez-Alesanco A.; Ceballos-Laita L.; Vega S.; Reyburn H. T.; Rizzuti B.; Velazquez-Campoy A. Structural Stability of SARS-CoV-2 3CLpro and Identification of Quercetin as an Inhibitor by Experimental Screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi K.; Musall K.; Oo A.; Cao D.; Liang B.; Hassandarvish P.; Lan S.; Slack R. L.; Kirby K. A.; Bassit L.; Amblard F.; Kim B.; AbuBakar S.; Sarafianos S. G.; Schinazi R. F. Baicalein and Baicalin Inhibit SARS-CoV-2 RNA-Dependent-RNA Polymerase. Microorganisms 2021, 9 (5), 893. 10.3390/microorganisms9050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.; Zhang L.; Xu Y.; Yang D.; Zhang L.; Yang S.; Zhang W.; Wang J.; Tian S.; Yang S.; Yuan T.; Liu A.; Lv Q.; Li F.; Liu H.; Hou B.; Peng X.; Lu Y.; Du G. The Comprehensive Study on the Therapeutic Effects of Baicalein for the Treatment of COVID-19 in Vivo and in Vitro. Biochem. Pharmacol. 2021, 183, 114302. 10.1016/j.bcp.2020.114302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther S.; Reinke P. Y. A.; Fernández-García Y.; Lieske J.; Lane T. J.; Ginn H. M.; Koua F. H. M.; Ehrt C.; Ewert W.; Oberthuer D.; Yefanov O.; Meier S.; Lorenzen K.; Krichel B.; Kopicki J.-D.; Gelisio L.; Brehm W.; Dunkel I.; Seychell B.; Gieseler H.; Norton-Baker B.; Escudero-Pérez B.; Domaracky M.; Saouane S.; Tolstikova A.; White T. A.; Hänle A.; Groessler M.; Fleckenstein H.; Trost F.; Galchenkova M.; Gevorkov Y.; Li C.; Awel S.; Peck A.; Barthelmess M.; Schlünzen F.; Lourdu Xavier P.; Werner N.; Andaleeb H.; Ullah N.; Falke S.; Srinivasan V.; França B. A.; Schwinzer M.; Brognaro H.; Rogers C.; Melo D.; Zaitseva-Doyle J. J.; Knoska J.; Peña-Murillo G. E.; Mashhour A. R.; Hennicke V.; Fischer P.; Hakanpää J.; Meyer J.; Gribbon P.; Ellinger B.; Kuzikov M.; Wolf M.; Beccari A. R.; Bourenkov G.; von Stetten D.; Pompidor G.; Bento I.; Panneerselvam S.; Karpics I.; Schneider T. R.; Garcia-Alai M. M.; Niebling S.; Günther C.; Schmidt C.; Schubert R.; Han H.; Boger J.; Monteiro D. C. F.; Zhang L.; Sun X.; Pletzer-Zelgert J.; Wollenhaupt J.; Feiler C. G.; Weiss M. S.; Schulz E.-C.; Mehrabi P.; Karničar K.; Usenik A.; Loboda J.; Tidow H.; Chari A.; Hilgenfeld R.; Uetrecht C.; Cox R.; Zaliani A.; Beck T.; Rarey M.; Günther S.; Turk D.; Hinrichs W.; Chapman H. N.; Pearson A. R.; Betzel C.; Meents A. X-Ray Screening Identifies Active Site and Allosteric Inhibitors of SARS-CoV-2 Main Protease. Science (80-.) 2021, 372 (6542), 642–646. 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V.; Brognaro H.; Prabhu P. R.; de Souza E. E.; Günther S.; Reinke P. Y. A.; Lane T. J.; Ginn H.; Han H.; Ewert W.; Sprenger J.; Koua F. H. M.; Falke S.; Werner N.; Andaleeb H.; Ullah N.; Franca B. A.; Wang M.; Barra A. L. C.; Perbandt M.; Schwinzer M.; Schmidt C.; Brings L.; Lorenzen K.; Schubert R.; Guaragna Machado R. R.; Candido E. D.; Leal Oliveira D. B.; Durigon E. L.; Yefanov O.; Lieske J.; Gelisio L.; Domaracky M.; Middendorf P.; Groessler M.; Trost F.; Galchenkova M.; Saouane S.; Hakanpää J.; Wolf M.; Turk D.; Pearson A. R.; Chapman H. N.; Hinrichs W.; Wrenger C.; Meents A.; Betzel C.. SARS-CoV-2 Papain-like Protease PLpro in Complex with Natural Compounds Reveal Allosteric Sites for Antiviral Drug Design bioRxiv 2021, 10.1101/2021.11.17.468943. [DOI] [Google Scholar]
- Medina-Franco J. L.; Martinez-Mayorga K.; Fernández-de Gortari E.; Kirchmair J.; Bajorath J. Rationality over Fashion and Hype in Drug Design. F1000Research 2021, 10, 397. 10.12688/f1000research.52676.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R.; Chen L.; Lan R.; Shen R.; Li P. Computational Screening of Antagonists against the SARS-CoV-2 (COVID-19) Coronavirus by Molecular Docking. Int. J. Antimicrob. Agents 2020, 56 (2), 106012. 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchi S.; Messasma Z. Exploring the Inhibitory Potential of Saussurea Costus and Saussurea Involucrata Phytoconstituents against the Spike Glycoprotein Receptor Binding Domain of SARS-CoV-2 Delta (B.1.617.2) Variant and the Main Protease (Mpro) as Therapeutic Candidates, Using. J. Mol. Struct. 2022, 1263, 133032. 10.1016/j.molstruc.2022.133032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T. Q.; Kim J. A.; Woo M. H.; Min B. S. SARS-CoV-2 Main Protease Inhibition by Compounds Isolated from Luffa Cylindrica Using Molecular Docking. Bioorg. Med. Chem. Lett. 2021, 40, 127972. 10.1016/j.bmcl.2021.127972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin S.; Calapoglu F.; Ozmen I. Didemnins Inhibit COVID-19 Main Protease (Mpro). Biointerface Res. Appl. Chem. 2020, 11 (1), 8204–8209. 10.33263/BRIAC111.82048209. [DOI] [Google Scholar]
- Snoussi M.; Redissi A.; Mosbah A.; De Feo V.; Adnan M.; Aouadi K.; Alreshidi M.; Patel M.; Kadri A.; Noumi E. Emetine, a Potent Alkaloid for the Treatment of SARS-CoV-2 Targeting Papain-like Protease and Non-Structural Proteins: Pharmacokinetics, Molecular Docking and Dynamic Studies. J. Biomol. Struct. Dyn 2021, 1–14. 10.1080/07391102.2021.1946715. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53 (7), 2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Nissink J. W. M. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations. ACS Chem. Biol. 2018, 13 (1), 36–44. 10.1021/acschembio.7b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. B. Feeling Nature’s PAINS: Natural Products, Natural Product Drugs, and Pan Assay Interference Compounds (PAINS). J. Nat. Prod 2016, 79 (3), 616–628. 10.1021/acs.jnatprod.5b00947. [DOI] [PubMed] [Google Scholar]
- OMEGA 4.2.0.1; https://docs.eyesopen.com/applications/omega/ (accessed August 31, 2022).
- Hawkins P. C. D.; Skillman A. G.; Warren G. L.; Ellingson B. A.; Stahl M. T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model 2010, 50 (4), 572–584. 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.