Abstract
DNA sequencing upstream of the Salmonella enterica serovar Typhi pilV and rci genes previously identified in the ca. 118-kb major pathogenicity island (X.-L. Zhang, C. Morris, and J. Hackett, Gene 202:139–146, 1997) identified a further 10 pil genes apparently forming a pil operon. The product of the pilS gene, prePilS protein (a putative type IVB structural prepilin) was purified, and an anti-prePilS antiserum was raised in mice. Mutants of serovar Typhi either lacking the whole pil operon or with an insertion mutation in the pilS gene were constructed, as was a strain in which the pilN to pilV genes were driven by the tac promoter. The pil+ strains synthesized type IVB pili, as judged by (i) visualization in the electron microscope of thin pili in culture supernatants of one such strain and (ii) the presence of PilS protein (smaller than the prePilS protein by removal of the leader peptide) on immunoblotting of material pelleted by high-speed centrifugation of either the culture supernatant or sonicates of pil+ strains. Control pil mutants did not express the PilS protein. A pilS mutant of serovar Typhi entered human intestinal INT407 cells in culture to levels only 5 to 25% of those of the wild-type strain, and serovar Typhi entry was strongly inhibited by soluble prePilS protein (50% inhibition of entry at 1.4 μM prePilS).
Earlier, it was reported that the major pathogenicity island of Salmonella enterica serovar Typhi, which is ca. 118 kb in size (11), contained pilV and rci genes, which were cloned and sequenced (22). The Rci gene product was shown to be a site-specific recombinase, active to invert DNA in the C-terminal region of the pilV gene, so that two PilV proteins could be synthesized. Comparisons with database sequences indicated that the two possible pilV genes might code for pilus-tip adhesins, as the serovar Typhi PilV sequence was similar to that of PilV proteins encoded by the Escherichia coli R64 plasmid. In R64-bearing strains, different PilV proteins, borne on type IV pili, select various recipients in liquid mating (the R64-bearing cell is the donor) (10). Both serovar Typhi PilV proteins were seen when the two pilV genes were transcribed from the T7 promoter. The discovery of the serovar Typhi pilV and rci genes in the ca. 118-kb pathogenicity island (henceforth in this work termed the large pathogenicity island) suggested that serovar Typhi might synthesize thin pili belonging to the type IV pilin family (9). As type IV pili, encoded in a Vibrio cholerae pathogenicity island (7, 8) are used by V. cholerae as mediators of adhesion to human cells (13, 18), it was of interest to ask (i) if serovar Typhi also synthesizes type IV pili and (ii) if such pili are important in adherence to or invasion of human intestinal cells. These topics are the subject of this paper.
MATERIALS AND METHODS
Materials.
All reagents were of molecular biology grade. Enzymes active on DNA were obtained from either GibcoBRL or Boehringer Mannheim and were used as directed by the suppliers. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside and isopropyl-β-d-thiogalactopyranoside were purchased from Amersham. Anti-mouse immunoglobulin G (from sheep), conjugated with horseradish peroxidase, was from Amersham. Phosphatase-labeled goat anti-mouse immunoglobulin G (heavy and light chains) was purchased from KPL Laboratories. p-Nitrophenyl phosphate tablets were from Sigma. Bio-Rad was the supplier of polyvinylidene difluoride membrane. Freund's adjuvant was from GibcoBRL.
Strains and vectors.
Serovar Typhi J341 (Ty2 Vi−) (22) was the source of DNA for a cosmid bank (partially Sau3AI-cut DNA in BamHI-cut pHC79), which was probed with 32P-labeled total DNA (including the virulence plasmid pSLT) of (wild-type, mouse-virulent) serovar Typhimurium C5, to yield cosmids such as pUST90 (22) containing DNA from the large pathogenicity island of serovar Typhi (such cosmids did not probe with the total serovar Typhimurium DNA). Serovar Typhi A21-6 (our laboratory strain) is serovar Typhi J341 with a ca. 20-kb deletion in the DNA of the large pathogenicity island, but with the deletion commencing ca. 10 kb downstream of the pilV gene (so pil DNA is not affected by this deletion). A Kmr cassette was inserted in place of the deleted DNA. Serovar Typhimurium J357 [leu trpD2 ilv452 metE551 lys metA22 hsdA (r− m+), hsdB(r− m+), hsdL(r− m+) rpsL120 thyA] cured of virulence plasmid pSLT served as a modifying strain prior to transformation of recombinant plasmids to serovar Typhi. E. coli DH5α [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was the host for recombinant plasmids. E. coli JM101 [supE thi Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] was the host for recombinant phage M13mp18 propagation. E. coli BL21(DE3) (E. coli B; F− dcm ompT hsdS (rB− mB−) gal; λ(DE3))[pLysS] was obtained from Stratagene for the expression of glutathione-S-transferase (GST) fusion proteins (the plasmid expresses chloramphenicol resistance). Plasmids pUC18 and pUC19 were used in subclonings. Plasmid pUC4K, which carries a kanamycin resistance cassette, was obtained from Pharmacia Biotechnology.
Media.
Luria-Bertani broth (LB) was prepared as described by Miller (14). Solid medium contained 1.5% (wt/vol) agar. Antibiotics were added, when appropriate, to 30 μg/ml (chloramphenicol) or 50 μg/ml (kanamycin and ampicillin). The 2× YT medium was prepared with Difco Bacto Tryptone (16 g/liter), Difco Bacto Yeast Extract (10 g/liter), and NaCl (5 g/liter).
Analytical methods.
Plasmid preparation, restriction enzyme digestion, agarose gel electrophoresis of DNA, nick translation, Southern transfer, and hybridization followed the methods of Sambrook et al. (17). DNA sequence (from clones in either pUC18, pUC19, or M13mp18) was determined using AutoCycle sequencing kits (purchased from Pharmacia), or ABI Prism BigDye terminator kits (purchased from PE Applied Biosystems), and run on Pharmacia ALF or ABI Prism 310 Genetic Analyzer sequencing systems, respectively. Development of immunoblots used the Enhanced Chemiluminescence system (Amersham). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was usually performed on 5% (wt/vol) (stacking gel)–18% (wt/vol) (separating gel) polyacrylamide. The GST fusion system was purchased from Pharmacia Biotechnology. The Expand Long Template PCR System (ELTPS) was from Boehringer Mannheim.
prePilS-GST fusion protein expression and use of protein as immunogen.
The prePilS protein was used as an immunogen, an SDS-PAGE standard, and an inhibitor of serovar Typhi association with human intestinal epithelial cells. To amplify the pilS gene before fusion to the GST gene, two primers, prpilSL and prpilSR, which contained BamHI and EcoRI sites, respectively (the pilS gene has no BamHI or EcoRI sites), were synthesized, with sequences as follows: prpilSL, 5′-GTTAAGCAATGGATCCAAAAATGAAACAGAGGG-3′; prpilSR, 5′-CACACATCCCGCGAATTCAGCCGTTGTAGT-3′. A 0.65-kb pilS gene fragment was amplified from a 12.6-kb SacI-SnaBI fragment from pUST90 (22) and cloned into pUC19. The pilS PCR product was digested with EcoRI and BamHI, purified, and ligated with similarly digested pGEX-2T vector, so that pilS was in the same translational reading frame as the GST gene in the vector. The ligation mixture was transformed to E. coli BL21(DE3)[pLysS], and after selection of recombinants, purified prePilS protein was prepared by affinity purification of the GST-prePilS fusion protein and release of the prePilS protein by thrombin cleavage. Some 400 μg of lyophilized prePilS protein was resuspended in 1.894 ml of 0.1% (vol/vol) trifluoroacetic acid, to a calculated concentration of 10 pmol/μl, and N-terminal protein sequencing proceeded on an Applied Biosystems 476A Protein Sequencer. To prepare an antiserum against the prePilS protein, BALB/c male mice, 2 months old, were immunized. Some 50 to 200 μg of prePilS protein/mouse, in 0.3 ml of 1× phosphate-buffered saline (PBS), was emulsified with an equal volume of Freund's adjuvant (complete for the first inoculation, incomplete for later injections) for intraperitoneal injection. The mice received five injections in all over 2 months; serum was collected and stored. In an enzyme-linked immunosorbent assay using alkaline phosphatase-conjugated secondary antibody, an anti-prePilS titer of 102,400 was obtained.
Construction of serovar Typhi pil mutants.
In general, a pil mutation was constructed in a plasmid bearing all or part of the pil operon DNA and was accompanied by introduction of a selectable antibiotic resistance marker. The modified plasmids were transferred to serovar Typhi via the serovar Typhimurium modifying strain J357 and with selection for both the vector-encoded antibiotic resistance and that introduced into the pil DNA. Individual clones were then grown in 5 ml of LB with antibiotic selection for the mutation marker alone at 42°C for periods of ca. 24 h before transfer of a 0.1-ml aliquot into 5 ml of fresh medium. After three to four transfers, aliquots were plated onto LB with selection for only the antibiotic resistance introduced with the pil mutation (thus, the antibiotic resistance marker of the vector was not selected). Resulting colonies were checked for sensitivity to the vector-encoded antibiotic resistance. Invariably, such colonies had lost the transformed plasmids, and the pil mutations, with associated antibiotic resistance cassette insertions, had recombined into the serovar Typhi genome. The mutants were checked by use of PCR involving primers which were in DNA either (i) adjacent to or (ii) inserted by the recombination events.
To construct serovar Typhi J341 pilS::Kmr, a SacI-SnaBI fragment of pUST90 encoding part of pilO and all of pilP through pilV was cloned into pUC19 digested with SacI and HincII. The resulting plasmid was further digested with StuI, which recognizes a single site within pilS. The linearized molecule was then ligated with a Kmr cassette obtained by HincII digestion of pUC4K. Transformants of E. coli DH5α were selected for Kmr and Apr; in the resulting plasmid the Kmr cassette is transcribed in the direction opposite to that of pil operon transcription. The mutation was recombined into the serovar Typhi chromosome, and ELTPS was used to confirm the presence of the mutation.
To construct serovar Typhi J341 Δpil (a deletion mutant lacking the entire pil operon and adjacent DNA), cosmid pUST90 (22) was digested with XhoI and religated to produce a deletion derivative. A 1.3-kb Kmr cassette isolated from pUC4K by SalI digestion was then cloned into the unique XhoI site. Homologous recombination between the resulting plasmid and the serovar Typhi chromosome introduced the Kmr gene-bearing cassette into the serovar Typhi Δpil chromosome with simultaneous deletion of DNA between the XhoI sites of pUST90 (Fig. 1).
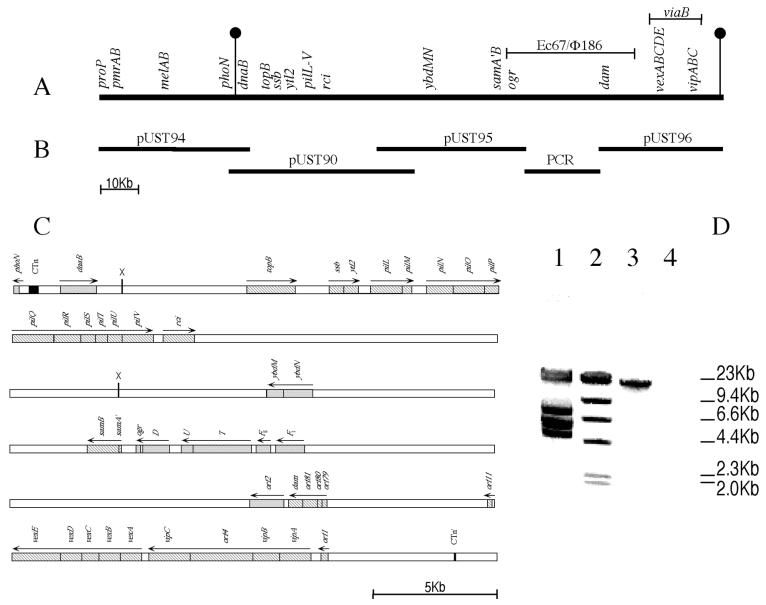
FIG. 1.
The large pathogenicity island of serovar Typhi. (A) A linkage map of the serovar Typhi chromosome from proP to the viaB region, as deduced from partial sequence analysis within the cosmid inserts shown in panel B. The viaB gene cluster extends from the orf1 gene to the orf11 gene, inclusive, and thus contains the vip and vex genes. The DNA of the large pathogenicity island lies between the two vertical markers. A line above the chromosome indicates DNA with similarity to coliphages P2 and φ186, and the E. coli retron Ec67. (B) Overlapping inserts of cosmids pUST90, pUST94, and pUST95 which extend from the serovar Typhi proP to the P2 ogr homolog, the ELTPS fragment (see panel D) linking pUST95 and the insert of cosmid pUST96 (which carries the Ec67 dam homolog and the serovar Typhi viaB region). (C) Detailed linkage map of the large pathogenicity island between the residual sequences (black boxes) of the duplications created by entry of a conjugative transposon. Hatched boxes indicate sequenced genes (4, 22; also this study), and shaded boxes represent genes identified through partial sequence homology to representatives in the GenBank database. Regions where no genes have yet been identified are left blank. Arrows above the map indicate directions of transcription only; they do not imply identification of transcription units. The two sites marked with an X are XhoI site between which DNA was removed in the construction of the Δpil mutant. (D) Agarose (0.7%, wt/vol) gel electrophoresis of the ELTPS product (ca. 20 kb in size) linking pUST95 and pUST96. Lane 1: molecular weight standards from λDNA cut with EcoRI; lane 2: molecular weight standards from λDNA cut with HindIII; lane 3: PCR product synthesized with primers from the 3′ end of pUST95 and the 5′ end of pUST96, using total serovar Typhi chromosomal DNA as the template; lane 4: absence of such a PCR product when total serovar Typhimurium DNA was used as template. The primers were 5′-GGTTCAGGTGTTTTAACTTGCGTGGTAAAGC-3′ within pUST95 and 5′-GCTGCTGGTACTCCCTGAAATGAATCAG-3′ within pUST96.
Strain serovar Typhi J341 pilM(::Cmr-tac)pilN contains a tac promoter inserted before the pilN gene. To detect the recombination event, a Cmr cassette was inserted simultaneously, just upstream of the tac promoter. In this construction, the 575-nucleotide (nt) pilM-pilN intergenic gap was removed. The tac promoter drives transcription of the pilN through pilV genes of the pil operon. In this strain, the last base of the −10 sequence of the tac promoter is 76 nt away from the A of the ATG start codon of the pilN gene.
Preparation and analysis of pili from serovar Typhi.
In early experiments, cultures of serovar Typhi J341 (pil+), serovar Typhi A21-6 (pil+), serovar Typhi Δpil (pil), and serovar Typhimurium C5 (wild type) were grown in 100 ml of 2× YT medium, with shaking at 37°C for 22 h. The cultures were centrifuged at 7,800 × g for 10 min. The supernatants were saved and centrifuged for 30 min at 12,000 × g. The supernatants from this centrifugation step were spun in an ultracentrifuge for 3 h at 100,000 × g. The pellets were solubilized and analyzed by SDS-PAGE and immunoblotting. In later experiments, cells of serovar Typhi J341 (pil+), serovar Typhi pilM(::Cmr-tac)pilN (a pil+ strain), and serovar Typhi pilM(::Cmr-tac)pilN pilS::Kmr (pilS) were grown in stationary culture in 250 ml of LB at 30°C for 14 to 16 h. The culture was centrifuged at 4,000 × g for 10 min, and the cells were resuspended in 1 ml of PBS. Sonication (three bursts of 18 s, with 12-s intervals between bursts; microtip probe; output control no. 5; 4°C) with a model 250 Branson Sonifier followed. After sonication, the cell lysate was spun at 1,000 × g for 5 min. The supernatant was saved and diluted 10-fold before SDS-PAGE and immunoblotting.
Electron microscopy.
Serovar Typhi pilM(::Cmr-tac)pilN was grown in stationary culture in 250 ml of LB for 14 h at 30°C, to an optical density at 600 nm of 0.6. The culture was centrifuged twice at 9,200 × g for 10 min to remove the bacterial cells. The supernatant was again centrifuged at 140,000 × g for 1 h. The pellet was resuspended in 0.9% (wt/vol) NaCl and negatively stained with 2% (wt/vol) uranyl acetate after adsorption onto a 400-mesh carbon-coated copper grid. The sample was observed in a Philips CM20 electron microscope.
Experiments with human intestinal cells.
The human embryonic intestine cell line INT407 was obtained from the American Type Culture Collection. Cells were maintained in liquid nitrogen and cultivated in basal medium Eagle's (BME). Cells were seeded in 24 multiwell plates with BME until monolayers (ca. 105 cells/well) were obtained (48 to 72 h). Bacterial strains were grown in stationary culture in LB for 14 to 16 h at 30°C to reach an optical density at 650 nm of 0.5 to 0.7. Immediately before the bacterium-cell association assays, old cell growth medium was removed, cells were washed once with 10 mM PBS, and fresh BME (at 37°C) was added. Added bacteria (either single strains or equal mixtures of two strains) were centrifuged onto the cells for 10 min at 2,164 × g. Bacterium-cell interaction proceeded for 2 h at 37°C in a 5% (vol/vol) CO2 atmosphere. In some tests, prePilS protein at various concentrations was added to the bacterium-containing media. Then, either washing alone or washing followed by gentamicin treatment was performed. Cells were washed six times with 10 mM PBS, and, in some tests, residual bacteria were then enumerated (after cell lysis with detergent; see below). When gentamicin treatment was to follow, fresh BME with gentamicin (100 μg/ml) was added and cells were incubated for 1 h at 37°C in a 5% (vol/vol) CO2 atmosphere. Cells were washed three times with 10 mM PBS to remove gentamicin and lysed with 0.02% (wt/vol) Triton X-100. Released bacteria were enumerated on both LB agar plates or LB agar plates supplemented with Km (both the A21-6 strain and the pilS mutant are Kmr). Basic controls (three of them) for each experiment included bacteria only (no INT407 cells) with either the washing procedure or the washing and gentamicin treatment (to confirm no bacterial background) and INT407 cells only (to confirm absence of bacterial contamination).
Nucleotide sequence accession number.
The pil sequence presented in this paper has been submitted to GenBank under accession no. AF000001, which also contains the pilV and rci sequences and sequence upstream (Fig. 1) of the pil operon.
RESULTS
Cloning and mapping of the large pathogenicity island of serovar Typhi.
Earlier, part of the large pathogenicity island of serovar Typhi was cloned in five overlapping cosmids and partially mapped (22). The 5′ end of the large pathogenicity island was defined as the boundary between serovar Typhimurium-homologous phoN sequence and DNA showing no homology to DNA of serovar Typhimurium. The 5′ end of the large pathogenicity island was contained in cosmid pUST94 (Fig. 1), the insert DNA of which overlapped with that of cosmid pUST90. Use of the 3′ end of cosmid pUST90 as a probe identified pUST95 as containing adjacent DNA (Fig. 1). When the insert DNA of cosmid pUST96 was probed with DNA from the viaB region, it was clear that the cosmid contained the viaB operon (4), which is known to be within the large pathogenicity island (11). Attempts to link the inserts of pUST95 and pUST96 by chromosomal walking 3′ from pUST95 insert DNA or 5′ from pUST96 insert DNA were not successful (see Discussion) (Fig. 1). Instead, the ELTPS, with primers based on the ends of pUST95 DNA and pUST96 DNA, was used to link the insert DNAs of the two cosmids (Fig. 1D).
Sequencing just downstream of phoN yielded the sequence 5′-TGGTGCCCGGACTCGGAATCGAACCACGGACACG GGGATTTTCAATCCCC-3′, in which only 1 nt (boldface type) differs from the reverse complement of the duplication element present at either end of the conjugative transposon CTnscr94 of serovar Senftenberg (6). Use of the above sequence to probe pUST96 subclones, followed by sequencing of a region of reactive DNA identified the sequence 5′-TGGTGCCCGGACTCGG-3′ ca. 4.5 kb 3′ of the end of the orf1 gene of the viaB region (Fig. 1). The continuation of the sequence 3′ to this region bore no homology to the first (near phoN) presumptive CTn sequence identified above. The distance between the short duplicated sequences (full and partial) identified above is ca. 118 kb (Fig. 1). The presence of the duplicated elements (full and partial) suggests the means whereby serovar Typhi may have acquired the large pathogenicity island (see Discussion).
A type IVB pilin operon is located upstream of pilV of serovar Typhi.
The discovery of pilV and rci genes in the large pathogenicity island of serovar Typhi (22) suggested that serovar Typhi might synthesize thin pili belonging to the type IV pilin family (9). Upstream of serovar Typhi pilV, a type IV pil operon with 11 genes in all (pilL through pilV) was fully sequenced in both directions (GenBank accession no. AF000001).
The serovar Typhi pil operon has no pilI, pilJ, or pilK genes, all of which are present in R64 (21), although the pilI and pilJ genes are not required for type IV pilus synthesis by R64-bearing cells (21). For the biogenesis of the R64 type IV pili, the pilS (prepilin structural) and pilV (pilus-tip adhesin) genes, the accessory genes traB and traC, and 10 pil genes (pilK through pilR, pilT, and pilU) are required (21). Between the serovar Typhi pilM and pilN genes, a sequence of 575 nt appears to represent noncoding sequence. In R64, there are only 16 nt between the end of pilM and the start of pilN (9).
The remainder of the serovar Typhi pil operon is similar, in gene arrangement, size, and sequence, to that of R64. As with the pil operon of the R64 plasmid (9), it appears that the serovar Typhi pil genes form part of the same transcriptional unit, as evidenced by the absence of independent promoter sequences for each gene. The presumptive start codons of the pilM, pilO, pilR, pilS, pilT, and pilU genes either overlap with the coding sequences for the preceding pil genes or are within 15 nt of the end of the previous genes. With the exception of the presumptive PilN protein, all the possible serovar Typhi Pil proteins are comparable in size to their R64 homologs. The serovar Typhi PilN homolog has 378 amino acid (aa) residues, while the R64 homolog has 560 aa. The high homology between serovar Typhi PilN and that of R64 reflects the fact that the smaller serovar Typhi presumptive PilN protein is very similar to the C-terminal portion of the R64 homolog. The serovar Typhi pilM-pilN intergenic region bears no similarity to the N-terminal R64 pilN sequence. The serovar Typhi presumptive PilS protein has 45% identity and 67% identity or similarity with R64 PilS (CLUSTAL W alignment [19]).
By homology with other type IV prepilins, the prePilS protein is also a structural prepilin. A putative signal sequence cleavage site is located between aa 25 (Gly) and aa 26 (Met) (Fig. 2). It appears that the peptide aa 1 to aa 25 represents a signal sequence which is hydrophilic in nature, while the region aa 26 to aa 52 is highly hydrophobic and may be required for efficient transmembrane transport of PilS. The length of the serovar Typhi PilS signal sequence (25 aa), and the presence of Met (and not Phe) as the first amino acid residue of the mature protein, identifies the serovar Typhi PilS protein as a member of the type IVB family, of which the other members are R64 PilS, TcpA of V. cholerae, and BfpA of enteropathogenic E. coli (Fig. 2) (9). The serovar Typhi PilT protein may possess a peptidoglycan-lytic activity, as has been suggested for its R64 homolog (9). The serovar Typhi PilU protein is proposed to be the prepilin peptidase; it is clear that the R64 homolog has such a function (9). The functions of the remaining Pil proteins of R64 and, by homology, serovar Typhi, are less clear (21). The R64 PilN protein is a lipoprotein, while the PilQ protein has a nucleotide-binding motif, and the PilR protein may be an inner membrane protein (21).
FIG. 2.
Comparison of signal sequences of prePilS proteins of the type IVB family. Signal sequences of the serovar Typhi prePilS, R64 prePilS, and V. cholerae preTcpA proteins are shown, as is the signal sequence of the preBfpA protein of enteropathogenic E. coli. Signal sequences of the prePilV proteins of serovar Typhi and R64 are also included. The cleavage sites are indicated by the vertical arrow. The glycine residue before the cleavage sites is indicated in boldface type. The first 10 aa residues of the mature proteins are also shown.
Serovar Typhi synthesizes type IVB pili encoded by the pil operon.
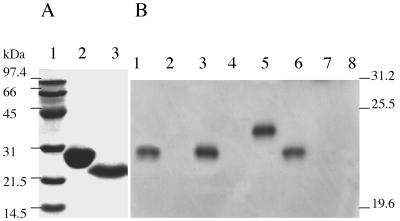
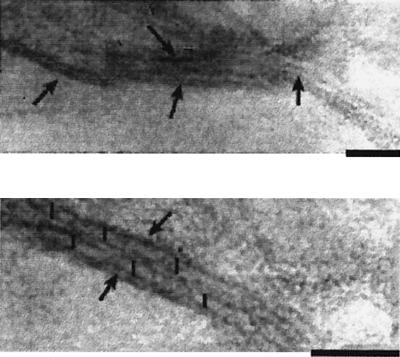
First, an antiserum against the prePilS protein was prepared. The pre-pilS gene was fused, from the second amino acid residue, in-frame to the C terminus of GST, and thrombin cleavage of cell lysates bearing the fusion protein yielded pure prePilS protein (Fig. 3A), which had the expected N-terminal sequence of GSKNE TEGKM MNEVS TLNPC NRPDR GM. The first two amino acid residues are encoded by DNA of the pGEX-2T vector; from the third amino acid K, the protein sequence is exactly as expected from the N-terminal region of prePilS. Mice were immunized with the protein, and an antiserum, with a titer of 1/102,400 against purified prePilS protein, was obtained. This antiserum was used in immunoblotting (Fig. 3B). Culture supernatants of serovar Typhi pil+ strains grown at 37°C with shaking contained PilS protein, which was absent in culture supernatants of serovar Typhi Δpil and serovar Typhimurium (Fig. 3B, lanes 1 to 4). The level of PilS expression appeared to be very low, however, at ca. 150 molecules PilS/cell (calculated from comparisons of immunoblotting densities of samples with that of purified prePilS standard), and type IVB pili could not be reliably found by electron microscopy of cells or culture supernatants. It was decided to first view sonicates of whole cells, in an effort to detect PilS protein not removed from the cells by the shearing action of shake flask growth. Also, a pilS insertion mutation was made (the Δpil strain used above lacks not only the entire pil operon but also adjacent DNA) to provide further evidence that the protein detected is the pilS gene product. When PilS production by the pilS::Kmr strain was assessed in this manner, PilS production was no better than that seen in Fig. 3B, lane 1. Finally, the tac promoter was inserted in the pilM-pilN intergenic region, in an effort to increase expression of pili. Insertion of the tac promoter greatly increased (by ca. 100-fold compared to the earlier tests) type IVB pilus expression, which was better when cells were grown at 30°C rather than 37°C (Fig. 3B, lanes 6 to 8). In the electron microscope, thin pili were visible in the culture supernatant of even a stationary culture of serovar Typhi J341 pilM(::Cmr-tac)pilN (Fig. 4). The pili were 6 nm in diameter and aggregated laterally, features also noted on examination of ColIb-P9 type IVB pili (20).
FIG. 3.
(A) Purification of the prePilS protein. A fusion protein, with prePilS attached to GST, was purified by affinity chromatography, and prePilS protein was then released by thrombin cleavage. The prePilS protein was viewed by SDS-PAGE with Coomassie blue staining. Lane 1: molecular mass standards, with sizes given on the left; lane 2: GST (60 μg); lane 3: prePilS protein (30 μg). (B) Immunoblotting to show that serovar Typhi synthesizes external PilS protein. Two pil+ strains of serovar Typhi—serovar Typhi J341 and serovar Typhi A21-6—and serovar Typhi Δpil were grown in a shaking liquid culture, as was serovar Typhimurium C5. The culture medium was subjected to ultrahigh-speed centrifugation (100,000 × g for 3 h), and the pellets were solubilized and subjected to SDS-PAGE. The proteins were transferred to a membrane, and immunoblotting, using a mouse anti-prePilS antiserum, was carried out. Lane 1: material pelleted from serovar Typhi J341 culture supernatant (from ca. 2 × 1010 cells); lane 2: material pelleted from serovar Typhi J341 Δpil culture supernatant (from ca. 2 × 1010 cells); lane 3: material pelleted from serovar Typhi A21-6 culture supernatant (from ca. 2 × 1010 cells); lane 4: material pelleted from serovar Typhimurium C5 culture supernatant (from ca. 2 × 1010 cells); lane 5: purified prePilS protein (100 ng); lane 6: material released by sonication from ca. 2 × 108 cells of serovar Typhi J341 pilM(::Cmr-tac)pilN; lane 7: material released by sonication from ca. 2 × 108 cells of serovar Typhi J341 pilM(::Cmr-tac)pilN pilS::Kmr; lane 8: material released by sonication from ca. 2 × 108 cells of serovar Typhi J341. The positions of some molecular mass standards are shown on the right. The prePilS and PilS proteins have apparent molecular masses of 24.5 and 21 kDa, respectively, although the amino acid sequence indicates that the molecular masses should be 21.1 and 18.3 kDa, respectively.
FIG. 4.
Electron microscopy of thin pili synthesized by serovar Typhi J341 pilM(::Cmr-tac)pilN. Material in the supernatant from stationary culture (30°C) was concentrated, stained with uranyl acetate, and viewed. Fibers with a diameter of 6 nm and with a tendency to laterally aggregate are visible. Fibers are indicated by arrows in the top micrograph. In the lower (high-magnification) picture, two parallel thin pili are indicated by arrows, and vertical black bars traverse the fibers. Bars, 50 nm.
Type IVB pili are involved in bacterial adherence to human intestinal epithelial cells.
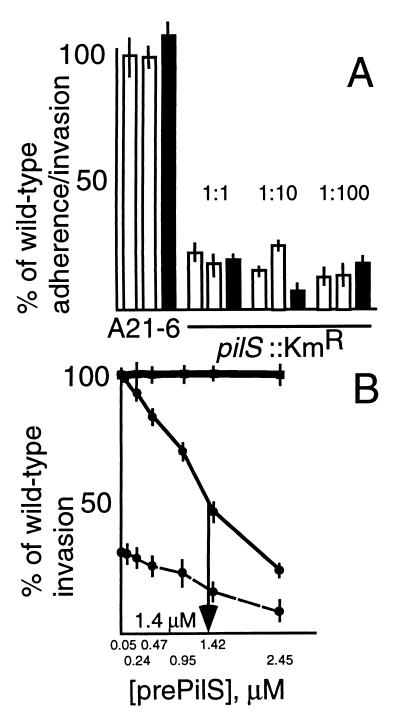
First, wild-type serovar Typhi and Kmr serovar Typhi mutants were mixed in equal numbers and exposed to human epithelial intestinal cells in culture (Fig. 5A). After adherence to and/or invasion of the bacteria, organisms which did not become cell associated were removed either by washing or with gentamicin, and residual bacteria were enumerated on solid media with or without kanamycin. The counts on the kanamycin-free medium minus the counts on the kanamycin-containing plates gave values for the entry levels of the wild-type (Kms) serovar Typhi. The Kmr counts were expressed as percentages of the Kms numbers, and these are the data shown in Fig. 5A.
FIG. 5.
(A) Adherence to and invasion of human intestinal INT407 cells by serovar Typhi J341, serovar Typhi A21-6, and serovar Typhi pilS::Kmr. The experimental details are given in Materials and Methods. Data from a total of 12 experiments is shown. In each test, serovar Typhi J341 and a Kmr strain were mixed in equal numbers before addition to the media overlying the intestinal cell monolayers. After a period during which the bacteria could adhere to or enter the INT407 cells, bacteria which did not associate with the cells were removed either by washing or by killing with gentamicin. Residual bacteria were enumerated on plates which either did or did not contain kanamycin. Bacterial counts on the kanamycin-free plates measured INT407 cell association and/or entry of both the wild-type (Kms) serovar Typhi, and the admixed Kmr strain. Counts on the kanamycin-containing medium measured INT407 cell association and/or entry of the Kmr strain only. The cell association and/or entry level of the wild-type strain was calculated by difference, and the cell association and/or entry level of the Kmr mutant was expressed as a percentage of this figure. The bars in Fig. 5A represent these percentages. In eight tests, bacteria which did not become cell associated (87 to 95% of added bacteria) were removed by washing (open bars), and in the remaining four tests, bacteria which did not invade the cells (95 to 97% of added bacteria) were killed with gentamicin (filled bars). Within each of the 12 experiments, the tests were performed in triplicate, and averages and standard errors (error bars) are shown. Two different Kmr bacterial strains were used. Strain serovar Typhi A21-6 is a Kmr control (pil+), and was used in three experiments. The other Kmr strain used is the pilS::Kmr mutant, and this was used in the remaining nine experiments. With this strain, three different bacterium/INT407 cell ratios (1:1, 1:10, and 1:100) were tested. (B) Inhibition by prePilS protein of human intestinal cell entry by wild-type serovar Typhi (thin line), serovar Typhimurium (thick line), and serovar Typhi pilS::Kmr (dashed line). The bacterium/cell ratio was 1:1, and gentamicin was used to kill bacteria external to the intestinal cells; residual bacteria were then enumerated. The tests were performed five to eight times; averages and standard errors (error bars) are shown.
As a control, serovar Typhi strain A21-6 (Kmr but pil+) was shown to attach to or invade INT407 cells to the same extent as did wild-type serovar Typhi. This was true in each of three experiments (two used washing to remove bacteria; one test used gentamicin), as evidenced by the cell entry levels shown by all three A21-6 bars (Fig. 5A). Compared to the wild-type strain, however, the pilS::Kmr mutant was reduced in human intestinal cell association and/or entry (to 5 to 25% of wild-type levels). This pattern was seen in nine separate experiments, using three different bacterium/INT407 cell ratios (so, three experiments with each bacterium/cell ratio) (Fig. 5A). In six tests (two with each bacterium/cell ratio), washing was used to remove bacteria which did not become cell associated, while gentamicin killing was employed in the other three experiments (one with each bacterium/cell ratio). While the reduction in INT407 entry caused by the pilS::Kmr mutation was consistent, elimination of pilus production did not completely abrogate intestinal cell adherence and/or invasion.
Next, it was shown that prePilS protein, when present in the bacterium-containing medium overlaying the cell monolayers, inhibited the entry of serovar Typhi, with 50% inhibition noted at 1.4 μM prePilS (Fig. 5B). This was true not only for the wild-type serovar Typhi, but also for the pilS::Kmr mutant (Fig. 5B), suggesting that serovar Typhi expresses at least two adhesins active against INT407 cells. One adhesin is the PilS protein of the type IVB pili, while the other is unknown. Both adhesins may bind to the same cell receptor, however, since prePilS protein blocks adherence to and/or invasion of both wild-type and pilS::Kmr strains.
In contrast to the situation with serovar Typhi strains, INT407 cell entry by serovar Typhimurium was not notably affected by addition of prePilS (Fig. 5B), indicating that serovar Typhi and serovar Typhimurium enter INT407 cells by different mechanisms (see Discussion).
DISCUSSION
In this work, the large pathogenicity island of serovar Typhi was partially cloned in four cosmids, and the remaining DNA (linking two of the cosmid inserts) was amplified by ELTPS. This DNA contains sequences homologous to coliphages P2 and φ186 and the E. coli retron Ec67, and it may be suggested that a cloned prophage gene is lethal to E. coli K-12 cells, due to either (i) high copy number or (ii) separation from its control elements.
The short duplicated sequences (full and partial) identified in the large pathogenicity island are very similar to those at the end of the conjugative transposon CTnscr94 of serovar Senftenberg (6), and the distance between these elements is, indeed, ca. 118 kb (Fig. 1). The presence of these duplicated elements (full and partial) suggest how serovar Typhi may have acquired the large pathogenicity island. It is possible that the large pathogenicity island entered serovar Typhi as a conjugative transposon, with concomitant creation of a short duplicated sequence (one copy at either end of the transposon) and that the duplicated element nearest viaB was subsequently mutated by a DNA insertion which (i) split the element and (ii) may have eliminated the mobility of the transposon.
From near the left border of the large pathogenicity island, a type IVB pilus operon was subcloned and characterized; its closest known homolog is the type IVB pilus operon of the R64 plasmid. With R64 type IVB pili, the (PilV-mediated) binding of donor to recipient, in liquid, is a prelude to plasmid transfer. In enterotoxigenic E. coli and V. cholerae, which are the other two enteropathogens known to elaborate type IVB pili (2, 18), the pili are important virulence determinants. With enterotoxigenic E. coli, it is thought that the type IVB bundle-forming pili cause bacterial aggregation and that such aggregates may enter intestinal cells better than do single bacteria. The pili are thus not the direct mediators of adhesion between the bacteria and the eukaryotic cells; this role is performed by a different protein (intimin) (1, 5). The Tcp pili of V. cholerae are absolutely required, by both Classical and El Tor strains, for virulence in the infant mouse model (13).
The serovar Typhi pil operon includes a simplified “shufflon” (10, 22) in which two possible pilV gene products may be generated by recombinase-mediated DNA rearrangement in the C-terminal region of the pilV gene. In the R64-encoded type IVB pili, the PilV proteins are thought to be pilus-tip adhesins, and it is clear that different PilV proteins mediate association of potential donor bacteria (the R64-bearing strains) with different potential recipients (10). The serovar Typhi pil operon is not plasmid-encoded, and there is no indication that the type IVB pili are part of a system whereby serovar Typhi transfers DNA to other bacteria. The role, if any, of the PilV proteins in serovar Typhi pathogenesis remains to be determined.
Under the growth conditions used, expression of type IVB pili by serovar Typhi J341 appears to be weak, with ca. 150 molecules/cell detectable in the supernatant after shaking culture. When sonicates of 30°C-grown bacteria were examined, material from 2 × 108 serovar Typhi J341 pilM(::Cmr-tac)pilN organisms had an obvious PilS band on immunoblot, but this was not the case with an equivalent amount of material from the wild-type serovar Typhi J341. It appears that insertion of the tac promoter in the pilM-pilN intergenic region may have increased PilS production by ca. 100-fold. Thin pili were readily seen upon electron microscopy of culture supernatant from serovar Typhi J341 pilM(::Cmr-tac)pilN, but not from culture supernatant or in sonicate of serovar Typhi J341. If, indeed, the level of type IVB pilus production by serovar Typhi J341, under current laboratory culture conditions, averages 150 molecules of PilS/cell, this would correspond to ca. 1 conventional type IVB pilus/10 cells (12). This figure is derived by estimate of material spontaneously sheared from cells in shaking culture and will therefore be an underestimate of pilus expression. Also, a pilus effective in the mediation of serovar Typhi-eukaryotic cell adhesion may have 150 PilS subunits or fewer. The experiments with the INT407 cells (Fig. 5) suggest, indeed, that most or all of wild-type serovar Typhi cells express some PilS protein even when grown under laboratory conditions.
It remains true, however, that conditions for the natural induction of the pil operon require further investigation. In El Tor biotypes of V. cholerae, Tcp pilus expression is tightly controlled by a system involving the ToxR and ToxT activator proteins (3); this control is released in Classical biotypes due to higher expression (relative to El Tor biotypes) of the ToxT protein in standard growth medium.
Wild-type (pil+) serovar Typhi adhered to and entered human intestinal epithelial cells better than did a pilS::Kmr mutant, although the mutant retained 5 to 25% of the adherence and/or invasion ability of the wild-type strain. These results suggest that the type IVB pili might be used by serovar Typhi as an intestinal cell adhesin and also that a second, different adhesin is active, at least in tissue culture, to mediate serovar Typhi-intestinal cell attachment. Both adhesins may use the same eukaryotic cell receptor, as adherence and/or invasion mediated by either adhesin was inhibited by soluble prePilS protein (with 50% inhibition seen at ca. 1.4 μM prePilS) when the protein was added to bacterium-containing media overlying the intestinal cell monolayers. Soluble prePilS protein did not perceptibly inhibit the intestinal cell entry of serovar Typhimurium, indicating that serovar Typhi and serovar Typhimurium enter intestinal epithelial cells by different mechanisms. This has been shown previously (15); serovar Typhi uses the cystic fibrosis transmembrane conductance regulator (CFTR) for entry, while serovar Typhimurium does not.
The fact that the INT407 cell entry of serovar Typhi is inhibited by soluble prePilS protein suggests that the PilS protein of the type IVB pilus interacts directly with a receptor on the eukaryotic cell surface. If the type IVB pili were used for bacterial aggregation, resulting in more effective INT407 cell entry (compared with that shown by a pil mutant), soluble prePilS protein would not be expected to inhibit such clumping. Again, it would be interesting to seek a direct interaction between the prePilS protein and the CFTR protein; this work is in progress.
In vivo, it is possible that type IVB pilus-mediated intestinal cell attachment is an essential first step for subsequent invasion by serovar Typhi. This idea may be tested either in human volunteers or in mice expressing human CFTR (15). It is recognized that experiments identifying bacterial adhesins active on cells in tissue culture may not necessarily be perfect predictors of bacterium-eukaryotic cell interactions in vivo (16).
If, indeed, the type IVB pili of serovar Typhi are essential for the intestinal invasion of humans by ingested bacteria, then intestinal immunity against the prePilS protein should be protective against typhoid fever in humans.
ACKNOWLEDGMENTS
Xiao-Lian Zhang, Inez Tsui, and Cecilia Yip contributed equally to the work described in this paper.
This work was supported by the Hong Kong Government Research Grants Council.
We thank T. Komano for his constant and generous support. M. Popoff kindly provided probes for the S. enterica serovar Typhi viaB region.
REFERENCES
- 1.Bieber D, Ramer S W, Wu C-Y, Toru M W J T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 2.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiRita V J, Neely M, Taylor R K, Bruss P M. Differential expression of the ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci USA. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto Y, Li N, Yokohama H, Ezaki T. Complete nucleotide sequence and molecular characterization of ViaB region encoding Vi antigen in Salmonella typhi. J Bacteriol. 1993;175:4456–4465. doi: 10.1128/jb.175.14.4456-4465.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochhut B, Jahreis K, Lengeler J W, Schmid K. CTnscr94, a conjugative transposon found in enterobacteria. J Bacteriol. 1997;179:2097–2102. doi: 10.1128/jb.179.7.2097-2102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus, and a phage receptor in cholera bacteria. Nature (London) 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 9.Kim S-R, Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komano T, Kim S-R, Yoshida T, Nisioka T. DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J Mol Biol. 1994;243:6–9. doi: 10.1006/jmbi.1994.1625. [DOI] [PubMed] [Google Scholar]
- 11.Liu S-L, Sanderson K E. Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci USA. 1995;92:1018–1022. doi: 10.1073/pnas.92.4.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Low D, Braaten B, van der Woude M. Fimbriae. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 150–157. [Google Scholar]
- 13.Manning P A. The tcp gene cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/s0378-1119(97)00036-x. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 15.Pier G B, Grout M, Zaidi T, Meluleni G, Mueschenborn S S, Banting G, Ratcliff R, Evans M J, Colledge W H. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature (London) 1998;393:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- 16.Salyers A A, Whitt D D. Bacterial pathogenesis, a molecular approach. Washington, D.C.: ASM Press; 1994. pp. 75–79. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida T, Nobuhisa F, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. Purification and characterization of thin pili of IncI plasmids ColIb-P9 and R64: formation of PilV-specific aggregates by type IV pili. J Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida T, Kim S R, Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol. 1999;181:2038–2043. doi: 10.1128/jb.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X-L, Morris C, Hackett J. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major “pathogenicity island” of Salmonella typhi. Gene. 1997;202:139–146. doi: 10.1016/s0378-1119(97)00466-6. [DOI] [PubMed] [Google Scholar]