Abstract
Patients presenting with metastatic hepatoblastoma have limited treatment options and survival rates as low as 25%. We previously demonstrated that Proviral Integration site in Maloney murine leukemia virus 3 (PIM3) kinase promotes tumorigenesis and cancer cell stemness in hepatoblastoma. In this study, we assessed the role of PIM3 kinase in promoting hepatoblastoma metastasis. We utilized a tail vein injection model of metastasis to evaluate the effect of CRISPR/Cas9-mediated PIM3 knockout, stable overexpression of PIM3, and pharmacologic PIM inhibition on the formation of lung metastasis. In vivo studies revealed PIM3 knockout impaired the formation of lung metastasis: 5 out of 6 mice injected with wild type hepatoblastoma cells developed lung metastasis while none of the 7 mice injected with PIM3 knockout hepatoblastoma cells developed lung metastasis. PIM3 overexpression in hepatoblastoma increased the pulmonary metastatic burden in mice and mechanistically, upregulated the phosphorylation and cell surface expression of CXCR4, a key receptor in the progression of cancer cell metastasis. CXCR4 blockade with AMD3100 decreased the metastatic phenotype of PIM3 overexpressing cells, indicating that CXCR4 contributed to PIM3’s promotion of hepatoblastoma metastasis. Clinically, PIM3 expression correlated positively with CXCR4 expression in primary hepatoblastoma tissues. In conclusion, we have shown PIM3 kinase promotes the metastatic phenotype of hepatoblastoma cells through upregulation of CXCR4 cell surface expression and these findings suggest that targeting PIM3 kinase may provide a novel therapeutic strategy for metastatic hepatoblastoma.
Keywords: hepatoblastoma, metastasis, PIM3 kinase, CXCR4, CRISPR/Cas9 knockout
INTRODUCTION
Hepatoblastoma is the most common primary malignant liver tumor in children [1]. Over half of patients initially present with advanced or metastatic disease [2]. The prognosis for these children remains dismal with survival rates less than 25% [3], primarily due to limited treatment options and chemotherapeutic resistance [4]. Current research efforts are focused on understanding tumor biology with the goal of identifying new targets for therapy.
Proviral Integration site for Moloney murine leukemia virus 3 (PIM3) kinase is overexpressed in hepatoblastoma tissue [5] but not normal liver [5, 6]. PIM3 kinase plays a pivotal role in hepatoblastoma tumorigenesis by promoting cell proliferation, survival, and cell-cycle progression [5-7]. The observation that higher PIM3 kinase expression in human tumors correlated with worse patient survival in many cancer types [8-10], including hepatoblastoma [11], suggests that PIM3 kinase may play a role in promoting hepatoblastoma metastasis. Furthermore, PIM3 knockout (KO) and PIM kinase inhibition significantly impaired hepatoblastoma cell migration and invasion in vitro [5], which are early steps in metastasis.
The mechanism by which PIM3 kinase enhances the migratory and invasive capabilities of cancer cells is not fully understood. One proposed mechanism is regulation of C-X-C chemokine receptor 4 (CXCR4), which interacts with C-X-C chemokine ligand 12 (CXCL12) to promote migration, invasion, and metastasis of tumor cells [12]. All three members of the PIM kinase family regulate surface expression of CXCR4 [13, 14].
Our aim was to assess the role of PIM3 kinase in promoting hepatoblastoma metastases. The findings presented in this study identify CXCR4 as a plausible mechanism for PIM3’s promotion of hepatoblastoma metastasis and provide evidence that targeting PIM3 may serve as a novel therapeutic strategy for metastatic hepatoblastoma.
MATERIALS AND METHODS
Cells and Cell Culture
Cells were cultured under standard conditions at 37 °C and 5% CO2. The human long-term passage hepatoblastoma cell line, HuH6, was obtained from Thomas Pietschmann (Hannover, Germany) [15] and maintained in Dulbecco’s Modified Eagle’s Medium (HyClone, GE Healthcare Life Sciences, Logan, UT, #D5030) supplemented with 10% fetal bovine serum (HyClone, GE Healthcare Life Sciences), 1 μg/mL penicillin/streptomycin (Gibco, Carlsbad, CA, #15140122), and 2 mmol/L L-glutamine (Thermo Fisher Scientific, Waltham, MA, #25035081). Short tandem repeat analysis (Genomics Core, University of Alabama at Birmingham (UAB), Birmingham, AL) validated the HuH6 cell line. The cells were free of mycoplasma.
The stable CRISPR/Cas9-mediated PIM3 KO hepatoblastoma cells have been extensively described and characterized in a previous publication [9]. PIM3 KO cells were cultured and validated as described above for parent HuH6 cell line. HuH6 EV and PIM3 OE cells have also been described and characterized in a prior publication [13]. HuH6 EV and PIM3 OE cells were cultured in media described above with the addition of G418 (2 mg/mL, Geneticin®, Sigma-Aldrich, St. Louis, MO) to maintain selection. HuH6 EVLuc and PIM3 OELuc cells were cultured in media containing puromycin (1 μg/mL, Sigma-Aldrich, St. Louis, MO, #P9620) and G418 (2 mg/mL, Sigma-Aldrich, #10131035) to establish stable overexpression of the PIM3 protein, which was confirmed by Western blotting (Supplementary Information, Fig. S1b). Short tandem repeat analysis (Genomics Core, UAB) validated all cell lines on a regular basis and cells were tested and determined to be free of mycoplasma.
The HuH6 WTLuc and PIM3 KOLuc cells were generously provided by the Hjelmeland laboratory and were established by stable transfection of HuH6 WT and PIM3 KO cells with the luciferase reporter that was cloned into the pCDH-CMV-MCS-EF1a-Puro lentiviral vector (System Biosciences, Palo Alto, CA, #CD510B-1). HuH6 WTLuc and PIM3 KOLuc cells were cultured in the media described above with the addition of puromycin (1 μg/mL, Sigma-Aldrich) to maintain selection.
Antibodies and Reagents
Antibodies utilized included rabbit monoclonal anti-PIM3 (4165), anti-CXCR4 (D4Z7W, #97680), rabbit polyclonal anti-phospho-CXCR4 (Ser339, #59028) from Cell Signaling Technology (Beverly, MA), and mouse monoclonal anti-β-actin (A1978) from Sigma Aldrich (St. Louis, MO). The pan-PIM inhibitor AZD1208 and the CXCR4 antagonist AMD3100 (plerixafor) were purchased from Selleck Chemicals (Houston, TX, #S7104 and #S8030).
Anchorage-Independent Growth
Soft agar colony formation assessed anchorage-independent growth as previously described [5]. A 3 mL mixture of 1% noble agar (BD Biosciences, #DF0142-17-0) and 2× culture media (in a 1:1 ratio) was poured into 60 × 15 mm petri dishes and allowed to cool. After solidification of the first layer, a second 1.5 mL layer containing the same ratio of agar and culture media, but also HuH6 wild-type (WT) or PIM3 KO cells (1 x 104 cells per dish), was added. Similarly, HuH6 EV or PIM3 OE cells (1 x 104 cells per dish) were added in a separate experiment to assess the effect of PIM3 overexpression on anchorage-independent growth. Culture media (1 mL) was added every 3-4 days for 6 weeks. Images of the dishes were taken using the Bio-Rad ChemiDoc™ MP Imager (Bio- Rad, Hercules, CA), colony growth was quantified using ImageJ software (https://imagej.nih.gov/ij), and reported as mean number of colonies ± standard error of the mean (SEM).
Formation of Lung Metastases In Vivo
The UAB Institutional Animal Care and Use Committee (IACUC-021420) approved all animal studies, which were conducted within institutional, national, and NIH guidelines. Animals were maintained in the specific pathogen-free facility with standard 12-hour light/dark cycles, allowed chow and water ad libitum, and euthanized humanely in their home cages with CO2 followed by cervical dislocation.
Four murine studies were performed using the tail vein injection metastasis model. In all studies, the cells were injected into the tail vein of female 6 week old athymic nude mice. In the first study, HuH6 WTLuc or PIM3 KOLuc (2.5 x 106) cells in 100 μl of phosphate-buffered saline (PBS) were injected (n=7 per group, Charles River, Frederick, MD). Four weeks later and then weekly, the mice received an intraperitoneal injection of d-luciferin and imaged 10 minutes post-injection using an IVIS® Lumina III in vivo imaging system (PerkinElmer, Waltham, MA) to monitor for lung metastasis. Animals were weighed three times a week and humanely euthanized at 8 weeks or when they met IACUC parameters. Upon euthanasia, lungs were harvested, imaged ex vivo, and fixed in 10 % formalin for histology. Investigators were not blinded to the group assignment. Bioluminescence was quantified as average radiance (photons/sec/cm2/sr; number of photons per second that leave a cm2 of tissue and radiate into a solid angle of one steradian) using the Living Image software version 3.0 (Caliper Life Sciences, Waltham, MA) and the region of interest (ROI) tool. Bioluminescence was utilized to confirm the comparable luciferase expression between the two cell lines prior to tail vein injections as well as immediately after injections to i) visualize the Luc-tagged cells within the lung microvasculature, confirming successful tail vein injections, and ii) obtain an initial baseline reading. A single photo of lung metastases was used to quantify the background signal serving as the standard for which the subsequent bioluminescence of each image was standardized.
For the second study, HuH6 EVLuc or PIM3 OELuc cells (2 x 106 cells in 100 μl of PBS) were injected (n=6 per group) and the mice monitored for 8 weeks. For the third study, PIM3 OELuc cells (2 x 106 cells in 100 μl of PBS) were injected and animals randomized the following day to receive vehicle (100 μl of PBS) or AMD3100 (5 mg/kg in 100 μl of PBS), via subcutaneous injections 5 days a week for 6 weeks (n=7 per group). AMD3100 dosing was based upon previous publication [16].
In a supplemental study, HuH6 WTLuc cells (2 x 106 cells in 100 μl of PBS) were injected and the following day animals were randomized to receive vehicle (100 μL of ORAPlus®, Perrigo, Allegan, MI) or AZD1208 (30 mg/kg in 100 μL of ORAPlus®), daily by oral gavage for 6 weeks (n=7 per group). Animals were imaged as described above to detect gross pulmonary metastasis (Supplementary Information Fig. S2).
Histology and Quantification of Metastatic Burden
Microscopic pulmonary metastases were confirmed using hematoxylin and eosin (H&E) staining. Lungs were fixed in 10% formalin, embedded in paraffin, sectioned into 5 μm slices at 100 μm intervals through the entire lung of each animal, and stained with H&E. A board-certified pediatric pathologist (E.M.M.) blinded to the treatment groups examined sections at six levels from each lung to confirm the presence or absence of pulmonary metastasis.
To quantify the metastatic burden in the second and third in vivo studies, we imaged H&E-stained lung sections at three different levels from each mouse using 10x objective of a light microscopy (Photometrics CoolSNAP HQ2 CCD camera (Tucson, AZ) attached to a Nikon Eclipse Ti microscope (Tokyo, Japan)). We quantified the metastatic burden using ImageJ software by determining the total area of lung metastasis (in pixel squared). Results were reported as mean metastatic burden ± SEM.
RNA Extraction, Library Preparation, and RNA Sequencing
RNA was extracted from HuH6 EV or PIM3 OE cells using the RNeasy kit (Qiagen Sciences Inc., Germantown, MD, #74106) per manufacturer's protocol. The UAB Genomics Core determined RNA sample quality control (QC), prepared the library, and performed sequencing as described [8]. The quality of total RNA was assessed using the Agilent 2100 Bioanalyzer followed by 2 rounds of poly A+ selection and conversion to cDNA. The NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® library generation kit (New England Biolabs, Ipswich, MA, #E7420L) was used per manufacturer’s instructions. The libraries were quantitated using qPCR in a Roche LightCycler 480 with the Kapa Biosystems kit (Kapa Biosystems, Woburn, MA, #07959362001). Sequencing was performed on the Illumina NextSeq500 using the latest versions of the sequencing reagents and flow cells with single-end 75 bp reads. We deposited raw and processed data in Gene Expression Omnibus (GEO), Accession #GSE164082 for HuH6 WT and PIM3 KO cells and #GSE176152 for HuH6 EV and PIM3 OE cells.
RNA Sequencing Analysis
The raw RNA-Seq fastq reads were aligned to the human reference genome (GRCh38 p13 Release 32) from Gencode using STAR (version 2.7.3a) and the parameters --outReadsUnmapped Fastx --outSAMtype BAM SortedBy Coordinate -- outSAMattributes All --outFilterIntronMotifs RemoveNoncanonicalUnannotated [17]. Following alignment, Cufflinks (version 2.2.1) was used to assemble transcripts, estimate their abundances, and test for differential expression and regulation using parameters--library-type fr-firststrand -G –L [18, 19]. Cuffmerge, a part of Cufflinks, merged the Cufflinks transcripts across multiple samples using default parameters. Finally, Cuffdiff found significant changes in transcript expression, splicing and promoter usage using default parameters.
For generating network analysis of canonical pathways, a data set containing gene identifiers and corresponding expression values was uploaded into Ingenuity Pathway Analysis (IPA) [20]. Each identifier was mapped to its corresponding object in Ingenuity’s Knowledge Base. A fold change cutoff of ± 2 was set to identify molecules whose expression was significantly differentially regulated. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in Ingenuity’s Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity. The Functional Analysis identified the canonical pathways that were most significant to the entire data set. The analysis included molecules from the dataset that met the fold change cutoff of ± 2 and were associated with biological functions and/or diseases in Ingenuity’s Knowledge Base. Right-tailed Fisher’s exact test calculated the p value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
Transcriptome Analysis of Published Hepatoblastoma Patient Cohorts
Gene expression and correlation analyses for hepatoblastoma patients were conducted using the R2: Genomics Analysis and Visualization Platform (http://r2platform.com). Normalized and log2-transformed clinically annotated microarray data was obtained from the GEO GSE131329, consisting of 53 hepatoblastoma samples [21], and PIM3 gene expression evaluated. RNA sequencing data of hepatoblastoma tumors (n=34 samples) was obtained from GSE133039 [22].
Immunoblotting
Cells were lysed using radio-immunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors (Sigma-Aldrich, #P8340), phosphatase inhibitors (Sigma-Aldrich), and phenyl-methane-sulfonyl-fluoride (Sigma-Aldrich, #P0044). Immunoblotting, gel transfer, and immunodetection was performed as previously described [5]. Precision Plus Protein Kaleidoscope molecular weight marker (Bio-Rad, #1610395) confirmed the expected size of target proteins and antibodies were used according to the manufacturers' recommendations. β-actin confirmed equal protein loading.
CXCR4 Cell Surface Expression
Cells (1 x 106) were labeled with a phycoerythrin (PE)-conjugated mouse immunoglobulin G2aκ (IgG2aκ) anti-human CXCR4 antibody (Miltenyi Biotec, Waltham, MA, #1-30-124-017), which competes with AMD3100 in binding to epitope 12G5. Unlabeled cells comprised negative controls. Flow cytometry detected the cell surface expression of CXCR4 by quantifying the percentage of cells positive for PE using the Attune™ NxT Flow Cytometer (Invitrogen™, Thermo Fisher) and FlowJo software (FlowJo, LLC).
Quantitative Real-Time PCR (qPCR)
Quantitative real-time PCR, as previously described [8], assessed the mRNA abundance of CXCR4. cDNA was synthesized using an iScript cDNA Synthesis kit (Bio-Rad, #1708891) according to supplier's instructions. For qPCR, SsoAdvanced™ SYBR® Green Supermix (Bio-Rad, #1725271) was utilized according to manufacturer's protocol. Primers specific for CXCR4 (Forward: 5’-CCCTCCTGCTGACTATTCCC-3', Reverse: 5'-TAAGGCCAACCATGATGTGC-3') were designed using Primer3 web version 4.1.0 [23] and checked for non-specific binding using the basic local alignment search tool (BLAST, NCBI), and β-actin primers were obtained from Applied Biosystems (Foster City, CA, #4326315E). qPCR was performed with 10 ng cDNA in 20 μL reaction volume with an Applied Biosystems 7900HT cycler (Applied Biosystems) with cycling conditions of 95 °C for 2 min, followed by 39-cycle amplification at 95 °C for 5 s and 60 °C for 30 s. Samples were analyzed in triplicate with β-actin as an internal control. The ΔΔCt method [24] calculated the gene expression, reported as mean fold change ± SEM.
Migration and Invasion Assays
Migration and invasion assays were performed as described [8]. For migration, the bottom of 8 μm micropore Transwell® inserts (Corning Life Sciences, Corning, NY, #3463) were coated with collagen I (10 μg/mL, MP Biomedicals, Santa Ana, CA, # 150026) overnight at 37 °C and then washed with PBS. For invasion, the top of the inserts was coated with 50 μL of Matrigel™ (1 mg/mL, BD Biosciences, San Jose, CA, #356234) overnight at 37 °C. For both assays, the inserts were placed in 24-well culture plates containing 350 μL media. HuH6 EV or PIM3 OE cells were treated with increasing concentrations of AMD3100 (0-30 μM) for 24 hours, plated into each insert (3 x 104 cells) and allowed to migrate or invade for 24 hours. The inserts were fixed with 4% paraformaldehyde and stained with 1% crystal violet. Insert images were obtained using light microscopy and the number of cells in seven random fields per insert were counted using ImageJ. Migration and invasion were reported as mean fold change in number of cells migrating or invading ± SEM.
Cell Viability
AlamarBlue® Cell Viability Assay (Thermo Fisher Scientific, #88952) was used to assess viability following treatment with AMD3100. HuH6 EV or PIM3 OE cells (5 x 103 cells per well) were plated in 96-well plates, allowed to attach overnight, and treated with AMD3100 (0-100 μM). Following 24 hours of treatment, the alamarBlue® reagent was added (10 μL) and absorbance read at 562 nm (reduced reagent) and 595 nm (oxidized reagent) using a microplate reader (BioTek Gen5, BioTek Instruments, Winooski, VT). Results were reported as mean fold change viability ± SEM.
Statistical Analysis
Data were generated with a minimum of three biologic replicates and results reported as mean ± SEM [25] of separate experiments. Student's t-test or analysis of variance (ANOVA) was used to compare means between groups and chi-square test was used for categorical variables as appropriate, with p≤0.05 considered statistically significant.
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Expression profile data analyzed in this study were obtained from GEO GSE131329 and GSE133039. The sequence data generated in this study are publicly available in GEO at GSE164082 and GSE176152.
RESULTS
PIM3 Kinase Promotes a Metastatic Phenotype of Hepatoblastoma Cells In Vitro and In Vivo
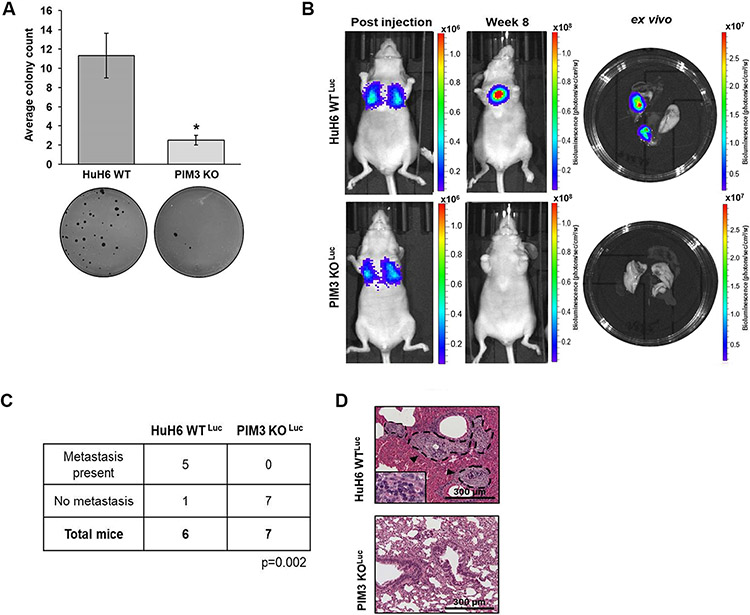
We previously demonstrated that PIM3 KO decreased hepatoblastoma cell migration and invasion [7]. We utilized colony formation assays to examine whether PIM3 contributes to anchorage-independent growth, an indicator of metastatic ability [26]. PIM3 KO diminished colony formation (11 ± 3 vs. 3 ± 0.5 colonies, HuH6 WT vs. PIM3 KO cells, p≤0.05, Fig. 1a). We advanced to an in vivo model of metastasis. HuH6 WTLuc or PIM3 KO Luc cells were injected, and bioluminescence imaging used to detect gross metastasis (Fig. 1b). Representative images of mice were taken immediately post tail vein injections and at the end of the 8-week study period, along with images of the ex vivo lungs (Fig. 1b). PIM3 KO significantly impaired the formation of lung metastasis. Five of 6 mice injected with HuH6 WTLuc cells developed lung metastases compared to none of the animals injected with PIM3 KOLuc cells (p≤0.01, Fig. 1c). Finally, representative photomicrographs of H&E staining demonstrating metastatic lesions in the HuH6 WTLuc group (Fig. 1d, upper panel, arrowheads, dotted lines), compared to lungs from the PIM3 KOLuc group which showed no histologic evidence of metastases (Fig 1d, lower panel).
Fig. 1. PIM3 kinase knockout decreased hepatoblastoma cell metastasis.
(a) Soft agar assays assessed anchorage-independent growth in HuH6 wild-type (WT) or PIM3 knockout (KO) cells. PIM3 KO significantly diminished colony formation. Representative images of soft agar dishes are shown (panels below graph). (b) Tail vein injections were used for an in vivo model of metastasis. HuH6 WTLuc (2.5 × 106 cells, n=6 animals) or PIM3 KOLuc cells (2.5 × 106 cells, n=7 animals) were injected and mice were monitored for formation of lung metastasis using bioluminescence imaging. Each figure demonstrates representative imaging compared to a single, standardized, background signal. Scales are thus unique to each experiment to better visualize the changes in bioluminescence. Representative images of mice immediately post tail vein injection (left panel) and at the end of the 8-week study period (middle panel). Representative bioluminescence images of ex vivo lungs shown at completion of the 8-week study period (right panel). (c) PIM3 KO significantly impaired the formation of lung metastasis. Five out of 6 mice injected with HuH6 WTLuc cells developed lung metastases while no lung metastases were seen in any of the 7 mice injected with PIM3 KOLuc cells (p=0.002). (d) Representative H&E stained lung sections from both groups confirmed the presence (in HuH6 WTLuc) or absence (in PIM3 KOLuc) of pulmonary metastases. Metastatic lesions (upper panel, arrowheads and dotted lines) were visualized outside the vessels in the lung parenchyma and were composed of cancer cells (inset showing tumor cells at higher magnification). Lungs from PIM3 KOLuc injected mice showed no histologic evidence of metastases (lower panel). Scale bars represent 300 μm. Data from in vitro studies represent at least three biologic replicates. Data reported as mean ± SEM. Data compared with Student’s t-test or chi-square. *p≤0.05.
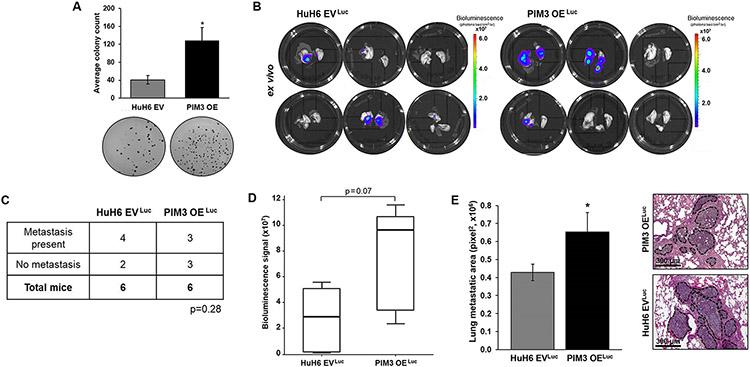
Since PIM3 KO decreased formation of hepatoblastoma lung metastasis, we aimed to explore whether PIM3 OE could promote metastasis. PIM OE cells exhibited increased anchorage-independent growth compared to HuH6 EV controls (3 ± 0.7 fold change, p≤0.05, Fig. 2a). These findings, in conjunction with those in Fig. 1a, indicate that PIM3 kinase plays a role in the in vitro metastatic phenotype of hepatoblastoma cells. We injected animals with HuH6 EVLuc or PIM3 OELuc cells into the tail vein and monitored them with bioluminescence imaging. At 8 weeks, there was a trend toward increased gross metastasis in animals injected with PIM3 OELuc compared to HuH6 EVLuc cells (p=0.28, Fig. 2b-d). Average bioluminescence signal measured in the ex vivo lungs of PIM3 OELuc injected mice was over 2.6 times higher than in the control group lungs (p=0.07, Fig. 2b-d). When we examined microscopic metastatic burden, we found animals injected with PIM3 OELuc cells had significantly increased metastatic burden compared to animals injected with HuH6 EVLuc cells (p≤0.05, Fig. 2e). Representative H&E images are shown with dotted lines encircling metastatic lesions (Fig. 2e, right panels).
Fig. 2. PIM3 kinase promoted hepatoblastoma cell metastasis.
(a) Soft agar assays assessed anchorage-independent growth in HuH6 empty vector (EV) or PIM3 overexpression (OE) cells. Representative images of the soft agar dishes are shown (panels below). PIM OE cells exhibited increased anchorage-independent growth compared to HuH6 EV controls cells. (b) Tail vein injections of 2 x 106 HuH6 EVLuc or PIM3 OELuc cells were performed (n=6 per group). Bioluminescence imaging was used to monitor the mice weekly for development of lung metastases. At 8 weeks, lungs were imaged ex vivo. (c) Gross pulmonary metastases were detected by bioluminescence in 4 out of 6 mice injected with HuH6 EVLuc cells and in 3 out of 6 mice injected with PIM3 OELuc cells. (d) Bioluminescence was quantified in the ex vivo lungs. Average bioluminescence signal measured in the lungs of PIM3 OELuc mice was higher than in lungs of HuH6 EVLuc injected animals. (e) Lung sections from three different levels from each animal were imaged using light microscopy. Metastatic burden was quantified using ImageJ software and by determining the total area of lung metastases (in pixel squared). PIM3 OELuc cells led to a greater microscopic metastatic burden compared to HuH6 EVLuc cells. Representative H&E lung sections from both groups are shown with metastatic lesions marked (dotted lines). Scale bars represent 300 μm. Data from in vitro studies represent at least three biologic replicates. Data reported as mean ± SEM. Data compared with Student’s t-test or chi-square. *p≤0.05.
Pharmacologic Inhibition of PIM3 Kinase Impaired Formation of Hepatoblastoma Lung Metastasis In Vivo
Since pharmacological inhibition is more amenable to clinical translation, we examined the effect of PIM kinase inhibition with the pan PIM inhibitor, AZD1208, on the formation of hepatoblastoma lung metastasis in vivo. Mice injected with HuH6 EVLuc cells were randomized to receive AZD1208 (30 mg/kg/day, n=7) or vehicle control (ORAPlus®, n=7) daily by oral gavage. At 6 weeks, animals treated with AZD1208, did not develop lung metastasis compared to 4 out of 7 control mice which did (p≤0.05, Supplemental Information Fig. S2a-c). Animal weights, a surrogate measure of treatment toxicity, were not affected by AZD1208 (Supplemental Information Fig. S3a).
PIM3 Modulation Differentially Regulated CXCR4 Signaling Pathway
To evaluate whether PIM3 kinase correlates with advanced or metastatic disease in hepatoblastoma patients, we utilized clinically annotated microarray data from GEO GSE131329 [21] and evaluated PIM3 gene expression within 53 pretreatment, primary hepatoblastoma tumors. PIM3 gene expression was significantly higher in patients with progressive disease (event +, n=21), of which 12 patients developed lung metastasis, compared to patients with event-free survival (n=32, p≤0.05, Fig. 3a).
Fig. 3. PIM3 modulation differentially regulated CXCR4 signaling pathway.
(a) Microarray data was obtained from GSE131329 and PIM3 gene expression evaluated within primary hepatoblastoma tumors (n=53 samples). PIM3 gene expression was significantly higher in hepatoblastoma patients with recurrent or progressive disease (event +) compared to those with event-free survival (event free). (b) RNA sequencing was performed to identify differentially expressed genes (p value < 0.05 and fold change cut off of > or < 2) in PIM3 OE compared to HuH6 EV cells (GSE176152), which were analyzed in comparison to PIM3 KO and HuH6 WT cells (GSE164082). Comparative analysis of differentially expressed genes resulting from PIM3 modulation are illustrated in a heatmap. Associated canonical pathways identified by Ingenuity Pathway Analysis with their corresponding p value are also included. PIM3 OE upregulated expression of genes involved in pathways of cell movement, migration, and chemokine signaling, including CXCR4 signaling. (c) RNA sequencing data of hepatoblastoma patient tumors (n=34 samples) was obtained from GSE133039. Spearman rank correlation coefficient analysis evaluated correlation between expression of PIM3 and CXCR4. Gene expression levels of PIM3 correlated positively with those of CXCR4 in hepatoblastoma tumors.
Given that RNA sequencing of PIM3 KO cells identified chemokine signaling as one of the top downregulated pathways [7], we performed RNA sequencing on PIM3 OE cells to evaluate whether the differences in the transcriptome and associated pathways identified by IPA are consistent with a metastatic phenotype. Expression of genes involved in pathways of cell movement, invasion, and chemokine signaling, including CCR5 and CXCR4 signaling, were significantly upregulated (p<0.05 and fold change cutoff + 2) in the PIM3 OE cells compared to HuH6 EV controls. These changes are illustrated in a heat map along with the canonical pathways and their respective p values (Fig. 3b).
To verify the biological significance of CXCR4 signaling activation by PIM3, we utilized RNA sequencing data of hepatoblastoma patient samples from the publicly available GSE133039 dataset [21] and found a positive correlation between PIM3 and CXCR4 in hepatoblastoma tumors (r=0.6, p≤0.001, Fig. 3c), supporting the notion that PIM3 may affect the expression of CXCR4 in hepatoblastoma.
Overexpression of PIM3 Kinase Upregulated Cell Surface Expression of CXCR4
To understand the association of PIM3 kinase and CXCR4 expression in hepatoblastoma cells, we first used immunoblotting to evaluate CXCR4 expression at the protein level. We found that PIM3 OE cells had higher phosphorylated CXCR4 protein (at Ser339) compared to HuH6 EV control or HuH6 WT cells, while total CXCR4 protein remained unchanged (Fig. 4a). To explore the effect of PIM3 OE on the cell surface expression of CXCR4, we employed flow cytometry to evaluate the percentage of cells positive for CXCR4. CXCR4 cell surface expression was significantly increased with PIM3 OE (34 ± 0.5 vs. 22 ± 1.2%, PIM3 OE vs. HuH6 EV, p≤0.001, Fig. 4b).
Fig. 4. PIM3 kinase regulated cell surface expression of CXCR4.
(a) Immunoblotting evaluated CXCR4 expression and phosphorylation. PIM3 OE cells had more phosphorylated CXCR4 protein (at Ser339) compared to HuH6 EV control or HuH6 WT cells while the total amount of CXCR4 protein remained unchanged. β-actin was used as a loading control. (b) CXCR4 cell surface expression was determined using flow cytometry. PIM3 OE cells had significantly increased surface expression of CXCR4 compared to HuH6 EV cells. A representative histogram of each cell line along with the corresponding negative staining control are shown. (c) Quantitative real-time PCR assessed mRNA abundance of CXCR4. Gene expression was normalized to β-actin and calculated as fold change to HuH6 EV or HuH6 WT cells using the ΔΔCt method. PIM3 OE had no significant effect on abundance of CXCR4 mRNA. (d) PIM3 KO cells had significantly decreased surface expression of CXCR4 compared to HuH6 WT cells. A representative histogram of each cell line along with the corresponding negative staining control are shown. Results reported as mean ± SEM and represent data from at least three biologic replicates. Data compared using Student’s t-test. ***p≤0.001
To further examine the effect of PIM3 on CXCR4 cell surface expression, we compared PIM3 KO to HuH6 WT cells. PIM3 KO cells had significantly decreased percentage of cells positive for CXCR4 (13 ± 0.1 vs. 32 ± 0.4%, PIM3 KO vs. HuH6 WT cells, p≤0.001, Fig. 4d). Representative histograms of HuH6 EV and PIM3 OE as well as HuH6 WT and PIM3 KO cells, along with their corresponding negative staining controls, are shown (Fig. 4b,d).
Finally, we evaluated the mRNA abundance of CXCR4 following PIM3 modulation using qPCR. Abundance of CXCR4 mRNA was unchanged in PIM3 OE compared to HuH6 EV control cells (Fig. 4c), indicating that changes were at the protein level.
Chemokine Receptor CXCR4 Mediated PIM3-Induced Migration, Invasion, and Anchorage-Independent Growth in Hepatoblastoma Cells
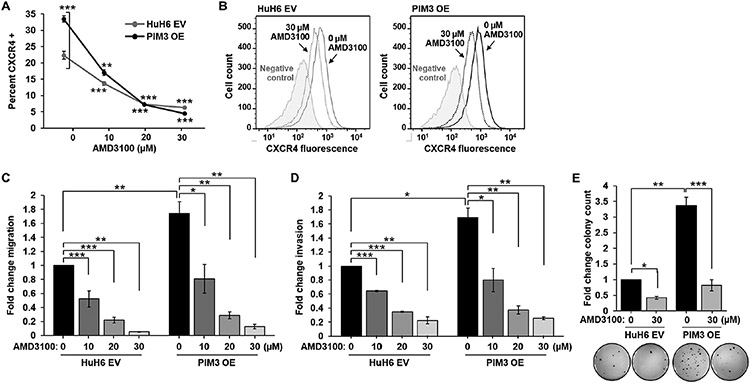
Because PIM3 enhanced CXCR4 cell surface expression and promoted hepatoblastoma metastasis, we hypothesized that CXCR4 may mediate the PIM3-induced metastatic phenotype of hepatoblastoma cells. We examined whether AMD3100 CXCR4 blockade would inhibit the migration, invasion, and anchorage-independent growth in PIM3 OE cells.
HuH6 EV or PIM3 OE cells were treated with AMD3100 (0-30 μM) for 24 h and flow cytometry assessed CXCR4 fluorescence signal. AMD3100 blocks binding of the anti-CXCR4 antibody to the 12G5 epitope of the CXCR4 receptor [27]. AMD3100 treatment decreased CXCR4 fluorescence signal in both HuH6 EV and PIM3 OE cells (Fig. 5a,b), implying blockade of the CXCR4 receptor. We then assessed the in vitro phenotype of HuH6 EV and PIM3 OE cells following CXCR4 blockade. PIM3 OE cells exhibited significantly increased migration and invasion compared to HuH6 EV cells (Fig. 5c,d, black bars). AMD3100 treatment of both cell lines decreased migration and invasion in a concentration-dependent manner (Fig. 5c,d). Treatment of HuH6 EV and PIM3 OE cells with AMD3100 (30 μM) significantly decreased colony formation in both cell lines (Fig. 5e). These findings were independent of viability, since AMD3100 did not affect viability of HuH6 EV or PIM3 OE cells at concentrations as high as 100 μM (Supplemental Information, Fig. S4a,b). Finally, there was no significant difference in the in vitro findings between PIM3 OE cells treated with AMD3100 and HuH6 EV cells treated with AMD3100 (Fig. 5c-e, light grey bars), implying that CXCR4 blockade abrogated the increase in the in vitro metastatic phenotype seen with PIM3 overexpression.
Fig. 5. CXCR4 blockade with AMD3100 abrogated the in vitro metastatic phenotype of PIM3 overexpressing hepatoblastoma cells.
(a, b) Following treatment of HuH6 EV or PIM3 OE cells with AMD3100 (0 to 30 μM) for 24 h, CXCR4 fluorescence signal was assessed using an anti-CXCR4 flow cytometry antibody that competes with AMD3100 in binding to the 12G5 epitope of CXCR4. AMD3100 treatment decreased CXCR4 fluorescence signal in both HuH6 EV and PIM3 OE cells. (b) Representative histograms of both cell lines, along with negative staining controls, are shown and demonstrate the decrease in CXCR4 fluorescence signal (shift to the left) following AMD3100 treatment (30 μM, dotted lines) compared to untreated cells (solid lines). (c) Migration and (d) invasion were assessed using modified Boyden chambers following treatment with AMD3100 (0 to 30 μM) for 24 h. Untreated PIM3 OE cells exhibited significantly increased (c) migration and (d) invasion compared to untreated HuH6 EV cells (black bars). AMD3100 treatment of HuH6 EV and PIM3 OE cells significantly decreased (c) migration and (d) invasion in a concentration-dependent manner compared to untreated cells. (e) Following treatment with AMD3100 (30 μM), anchorage-independent growth was assessed using soft agar assays in HuH6 EV and PIM3 OE cells. Treatment with AMD3100 significantly decreased colony formation in both HuH6 EV and PIM3 OE cell lines. There was no significant difference in (c) migration, (d) invasion, or (e) anchorage-independent growth between PIM3 OE cells treated with AMD3100 and HuH6 EV cells treated with AMD3100 (light grey bars). Results reported as mean ± SEM. Experiments were repeated in biologic triplicates. Data analyzed with Student’s t-test. *p≤0.05, **p≤0.01, ***p≤0.001
CXCR4 Blockade Reduced the PIM3-Induced Lung Metastatic Burden In Vivo
To determine whether CXCR4 contributes to the PIM3-induced formation of lung metastasis in vivo, we injected PIM3 OELuc cells into tail veins of mice who were randomized to receive subcutaneous injections of either AMD3100 (5 mg/kg/day, n=7) or vehicle control (PBS, n=7) daily for 5 days a week. After 6 weeks, 3 of 7 control and 2 of 7 AMD3100 treated mice had evidence of gross lung metastasis (Fig. 6a,b). The metastatic burden was quantified with H&E. Lungs from animals treated with AMD3100 had significantly decreased microscopic pulmonary metastatic burden compared to vehicle treated animals (p≤0.05, Fig. 6c,d). These results indicate that treatment with AMD3100 diminished the PIM3-induced formation of pulmonary metastasis in vivo. The treatment did not significantly affect animal weight (Supplemental Information, Figure S3b).
Fig. 6. CXCR4 Blockade Reduced the PIM3-induced lung metastatic burden in vivo.
(a) Tail vein injections of 2 x 106 PIM3 OELuc cells were performed. Animals were randomized to receive subcutaneous injections of AMD3100 (5 mg/kg/day, n=7) or a vehicle control (PBS, n=7) daily for 5 days a week. Bioluminescence imaging was used to monitor the mice weekly for development of pulmonary metastasis. At 6 weeks, lungs (ex vivo) were examined with bioluminescence imaging. (b) At 6 weeks, gross metastases were detected in 3 out of 7 mice treated with vehicle and 2 out of 7 mice treated with AMD3100. (c) H&E-stained lung sections at three different levels from each mouse were imaged using light microscopy. Metastatic burden was quantified using ImageJ software by determining the total area of lung metastasis (in pixel squared). Treatment with AMD3100 significantly reduced pulmonary metastatic burden in animals injected with PIM3 OELuc. (d) Representative photomicrographs of H&E stained lung sections from both groups are shown with metastatic lesions marked (dotted lines); inset (right panel) shows tumor cells at higher magnification. Scale bars represent 300 μm. Results from each group were reported as mean ± SEM. Data evaluated with chi-square or Student’s t-test. * p≤0.05
DISCUSSION
Metastasis, the major cause of morbidity and mortality in most cancers [28], is a complex pathophysiologic process [29]. After traveling through the circulation, tumor cells extravasate, and attach and proliferate in their new location establishing metastatic tumors [30]. The difficulty in treating metastatic hepatoblastoma is the reliance on therapy to target both the primary tumor and metastatic lesions. While many of the molecular pathways that promote tumorigenesis also promote metastasis, some genes exert effects only on metastatic capability and colonization [31, 32]. The role of PIM3 kinase has not yet been delineated. To our knowledge, this study is novel in evaluating the role of PIM3 kinase in hepatoblastoma metastasis. With up to 20% of hepatoblastoma patients having pulmonary metastasis at presentation and survival rates as low as 25% [4], we believe PIM inhibition will be a useful tactic to treat children with metastatic disease.
A strength of the current study is the use of multiple methods to alter PIM3 expression. We genetically modified the PIM3 gene with both CRISPR/Cas9 knockout and overexpression plasmid as well as pharmacologic inhibition. Both PIM3 KO and PIM kinase inhibition decreased formation of pulmonary metastasis, while overexpression of PIM3 had the opposite result. These findings are congruent with our previous investigations demonstrating the effect of PIM kinase inhibition [5] and PIM3 KO [7] on hepatoblastoma cell migration and invasion in vitro. The findings in the current study are also consistent with those reported in other malignancies [15, 33-34]. For example, PIM3 overexpression enhanced the metastatic properties of prostate cancer xenografts [13], while shRNA PIM3 knockdown reduced pulmonary metastasis of melanoma cancer cells in vivo [34].
In addition to bioluminescence imaging to detect gross metastasis, we obtained histologic confirmation of microscopic pulmonary metastases in each of the in vivo studies. Using H&E staining, we visualized the secondary lesions invading the lung parenchyma (as opposed to tumor cell aggregations in the microvasculature). In the first in vivo study, one of the 7 mice injected with HuH6 WTLuc cells died immediately after the tail vein injection, attributed to a thromboembolic event. Sudden death due to thromboembolism and the lack of a gradual progression of tumor burden has previously led to criticism of the tail vein injection model of lung metastasis [35]; the limitations of which are expanded upon further in the discussion. These concerns highlight the importance of histologic examination which allowed for i) the identification of micrometastases that were undetected by bioluminescence and ii) the quantification of metastatic burden.
Utilizing publicly available hepatoblastoma databases, we demonstrated that PIM3 gene expression was significantly higher in patients with recurrent or progressive disease. Over half of those patients developed pulmonary metastasis. This analysis was consistent with our previous findings that higher PIM3 expression in hepatoblastoma tumor tissues correlated with worse patient survival [11]. We also demonstrated that PIM3 expression correlates positively with CXCR4 expression levels in primary hepatoblastoma tissues. The integration of comparative transcriptomic analysis of both PIM3 KO and PIM3 OE cells provided a unique opportunity to evaluate the genes differentially regulated by PIM3. For instance, we observed that the chemokine signaling pathway was significantly downregulated in PIM3 KO cells and upregulated in PIM3 OE cells. These observations corroborate the positive correlation between PIM3 and CXCR4 noted in database evaluation. The publicly available hepatoblastoma databases that span across various disease stages or risk groups are limited. Thus, whether PIM3 or CXCR4 expression is upregulated in metastatic lesions compared to primary hepatoblastoma tumor tissues and whether CXCR4 expression correlates with poor patient overall survival in hepatoblastoma remains unknown and an area of future investigations.
We postulated that PIM3 kinase enhances the migratory and invasive capabilities of hepatoblastoma cells through mediating CXCR4 surface expression. While the PIM3 and CXCR4 are structurally unrelated proteins, investigators have described a link between the two molecules in other malignancies. In both prostate cancer xenografts and acute myeloid leukemia (AML) cell lines, overexpression of PIM3 was associated with increased CXCR4 phosphorylation at Ser339 [13, 33]. Complementary to these findings, PIM kinase inhibition decreased CXCR4 phosphorylation (Ser339), impaired CXCR4-mediated migration of B-cell chronic lymphocytic leukemia (CLL), and downregulated CXCR4 cell surface expression [14]. Consistent with these previous reports, we demonstrated that PIM3 overexpression increased CXCR4 phosphorylation (Ser339) and cell surface expression, and increased CXCR4-mediated pulmonary metastasis of hepatoblastoma cells, therefore adding to the understanding of how PIM3 overexpression may promote hepatoblastoma metastasis.
Recent research established that CXCR4 plays a central role in cancer metastasis in more than 75% of all cancers [36]. CXCR4 expressing cancer cells migrate into secondary organs where they proliferate, induce angiogenesis, and form metastatic tumors [37]. Given that complete disruption of the CXCR4 chemokine receptor gene is embryologic lethal in mice [38], current approaches to study CXCR4-mediated metastasis are based on the functional blockade of CXCR4 on cancer cell surface with CXCR4 antagonists [39] and CXCR4 neutralizing antibodies [40]. We chose to use pharmacologic blockade with AMD3100, a CXCR4 antagonist. We demonstrated that AMD3100 decreases the in vitro metastatic phenotype of PIM3 overexpressing hepatoblastoma cells and abrogated the effect of PIM3 OE. Further, AMD3100 treatment significantly decreased the pulmonary metastatic burden of mice injected with PIM3 OE hepatoblastoma cells. These findings suggest that CXCR4 mediates the PIM3-induced promotion of hepatoblastoma metastasis and CXCR4 inhibition could serve as therapeutic for preventing hepatoblastoma metastasis.
Other mechanisms may contribute to the PIM3-induced metastatic phenotype of hepatoblastoma cells tumors [35, 41]. Silencing of PIM3 with shRNA inhibited melanoma cell migration and invasion and decreased expression of matrix metalloproteinase 9 (MMP-9) [33]. PIM3 overexpression upregulated the intra-tumoral levels of vascular endothelial growth factor (VEGF) in melanoma leading to increased angiogenesis, an important step in metastasis [42]. In CLL, PIM kinases resulted in decreased phosphorylation of the CXCR4 receptor, enhanced internalization, and inhibited its function by reducing ERK phosphorylation, blocking the MAPK pathway [43]. Other investigators demonstrated that PIM inhibition blocked CXCR4 function by decreasing phosphorylation of mTOR pathway components and decreasing mTOR activation [14]. Given that CXCR4 was shown to promote metastasis through upregulation of both VEGF and MMP-9 [44], it is also possible that PIM3 kinase upregulates VEGF and MMP-9 downstream of CXCR4 or independently. Current and future investigations will explore signaling downstream of CXCR4 as well as CXCR4 independent mechanisms.
We utilized AMD3100, which is FDA approved and used to mobilize hematopoietic progenitor cells for autologous transplantation in patients with non-Hodgkin lymphoma and multiple myeloma [45]. At least nine other CXCR4 antagonists are in ongoing clinical trials for their effect on mobilizing hematopoietic stem cells [46] and have shown promising results in increasing chemosensitivity of hematopoietic malignancies [47, 48]. However, clinical trials have not investigated the effect of CXCR4 inhibition on the metastatic potential of solid tumors. One downside to CXCR4 antagonists is the induction of a counter-regulatory upregulation of CXCR4 cell surface expression, resulting in tolerance and reduced efficacy [49]. This effect may be partly explained by the fact that CXCR4 antagonists inhibit both G protein signaling by CXCR4 and β-arrestin-dependent receptor endocytosis with equal potency [27]. Targeting CXCR4 through PIM inhibition is an alternative that could mediate the issues with antagonist tolerance. PIM inhibitors block the function and externalization of the CXCR4 receptor [43, 50], so PIM inhibitors may represent a more efficient therapeutic strategy to target metastasis with less harmful side effects. To that extent, the PIM inhibitor used in this study, AZD 1208, is available for oral administration, safe in adults [51], and impairs formation of hepatoblastoma lung metastasis in vitro [5] and in vivo.
One of the limitations of the current study, and in fact with many studies involving hepatoblastoma, is the use of a single long-term passage cell line. Unfortunately, the HuH6 cell line remains the only commercially available, well described, fully characterized, established hepatoblastoma cell line, which is why we chose to utilize it as the basis of our experiments. By manipulating this cell line through the interventions we have described, including CRISPR knockout and formation of a metastatic cell line, we were better able to study the metastatic properties of hepatoblastoma as opposed to just analyzing the HuH6 cell line itself.
In our studies, we used an experimental, as opposed to spontaneous, in vivo model of metastasis. One may argue that this model does not recapitulate the first steps of the metastatic cascade and only reflects homing of circulating tumor cells in the bloodstream to a limited set of secondary organs [52]. Sites of vascular injection in experimental models define the site of colonization [53]. For example, injection of tumor cells into the tail vein predominantly results in pulmonary metastasis, because the lung capillary bed is the first the injected cells encounter [54]. The selection of the injection site must consider the usual distribution of metastases for the human cancer being investigated. For example, human prostate and breast cancers commonly metastasize to bone, therefore, intra-cardiac injections are employed to model their metastases [55, 56]. The lungs are the most common site of metastasis in hepatoblastoma [57], therefore we elected to employ tail vein injections [54]. Another limitation of experimental metastatic models is that they bypass the formation of a primary tumor [52], excluding the systemic influence of primary tumor-derived cytokines and growth factors that may promote distal metastatic tumors [58] or may inhibit metastatic seeding [59]. Despite these issues, experimental metastatic models have been instrumental in evaluating the capacity of cancer cells to arrest, extravasate, and grow in ectopic sites following intravascular injection [59]. With consideration for the complex series of steps required for successful metastasis, extravasation from blood vessels in target organs is regarded as a rate-limiting step and critical process [60].
In conclusion, our results show a role for PIM3 kinase in promoting hepatoblastoma metastasis in vivo and provide evidence for targeting PIM3 as a potential novel therapeutic approach for metastatic disease. Using functional and mechanistic studies, as well as comparative transcriptomic analyses, we identified CXCR4 as a plausible mechanism that mediates PIM3’s promotion of hepatoblastoma metastasis. These findings provide critical knowledge to developing better therapies for hepatoblastoma.
Supplementary Material
Acknowledgments
The authors wish to thank Vidya Sagar Hanumanthu from the UAB Comprehensive Flow Cytometry Core and Dr. Michael Crowley from the UAB Genomics Core.
Financial Support:
This work was supported in part by funding from National Institutes of Health (NIH) T32 CA091078 Surgical Oncology Research Training Program (LLS); NIH T32 CA229102 Surgical Oncology Research Training Program (JRJ, LVB); NIH 5T32GM008361 MSTP Training Program (CHQ); Vince Lombardi Cancer Research Fund Bart Starr Award, Hyundai Hope on Wheels, Kaul Pediatric Research Foundation, Sid Strong Foundation, Elaine Roberts Foundation, and Open Hands Overflowing Hearts (EAB). This work utilized the University of Alabama at Birmingham (UAB) Comprehensive Flow Cytometry Core (supported by NIH P30 AR048311 and NIH P30 AI27667), and the Comprehensive Genomics and Preclinical Imaging Cores, shared facilities which are supported by the O’Neal Comprehensive Cancer Center (P30 CA013148).
Footnotes
Competing Interests: The authors declare no financial conflicts of interest.
REFERENCES
- 1.Feng J, Polychronidis G, Heger U, Frongia G, Mehrabi A, Hoffmann K. Incidence trends and survival prediction of hepatoblastoma in children: a population-based study. Cancer Commun (Lond). 2019;39(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyers RL, Tiao G, de Ville de Goyet J, Superina R, Aronson DC. Hepatoblastoma state of the art: pre-treatment extent of disease, surgical resection guidelines and the role of liver transplantation. Curr Opin Pediatr. 2014;26(1):29–36. [DOI] [PubMed] [Google Scholar]
- 3.Katzenstein HM, London WB, Douglass EC, Reynolds M, Plaschkes J, Finegold MJ, et al. Treatment of unresectable and metastatic hepatoblastoma: a pediatric oncology group phase II study. J Clin Oncol. 2002;20(16):3438–44. [DOI] [PubMed] [Google Scholar]
- 4.Perilongo G, Brown J, Shafford E, Brock P, De Camargo B, Keeling JW, et al. Hepatoblastoma presenting with lung metastases: treatment results of the first cooperative, prospective study of the International Society of Paediatric Oncology on childhood liver tumors. Cancer. 2000;89(8):1845–53. [DOI] [PubMed] [Google Scholar]
- 5.Stafman LL, Mruthyunjayappa S, Waters AM, Garner EF, Aye JM, Stewart JE, et al. Targeting PIM kinase as a therapeutic strategy in human hepatoblastoma. Oncotarget. 2018;9(32):22665–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maryati R, Julson JR, Bownes LV, Quinn CH, Hutchins SC, Williams AP, Markert HR, Beierle AM, Stewart JE, Hjelmeland AB, Mroczek-Musulman E, Beierle EA. Metastatic human hepatoblastoma cells exhibit enhanced tumorigenicity, invasiveness and a stem-cell like phenotype. J of Pediatr Surg. 2022; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marayati R, Stafman LL, Williams AP, Bownes LV, Quinn CH, Markert HR, et al. CRISPR/Cas9-mediated knockout of PIM3 suppresses tumorigenesis and cancer cell stemness in human hepatoblastoma cells. Cancer Gene Ther. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng HC, Tsuneyama K, Takahashi H, Miwa S, Sugiyama T, Popivanova BK, et al. Aberrant Pim-3 expression is involved in gastric adenoma-adenocarcinoma sequence and cancer progression. J Cancer Res Clin Oncol. 2008;134(4):481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang H, Zhao MY, Hei KW, Yang BC, Sun L, Du X, et al. Aberrant expression of pim-3 promotes proliferation and migration of ovarian cancer cells. Asian Pac J Cancer Prev. 2015;16(8):3325–31. [DOI] [PubMed] [Google Scholar]
- 10.Qu Y, Zhang C, Du E, Wang A, Yang Y, Guo J, et al. Pim-3 is a Critical Risk Factor in Development and Prognosis of Prostate Cancer. Med Sci Monit. 2016;22:4254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stafman LL, Waldrop MG, Williams AP, Aye JM, Stewart JE, Mroczek-Musulman E, et al. The presence of PIM3 increases hepatoblastoma tumorigenesis and tumor initiating cell phenotype and is associated with decreased patient survival. J Pediatr Surg. 2019;54(6):1206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Q, Gao BL, Zhang XJ, Liu GC, Xu F, Fan QY, et al. CXCL12-CXCR4 Axis Promotes Proliferation, Migration, Invasion, and Metastasis of Ovarian Cancer. Oncol Res. 2014;22(5-6):247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santio NM, Eerola SK, Paatero I, Yli-Kauhaluoma J, Anizon F, Moreau P, et al. Pim Kinases Promote Migration and Metastatic Growth of Prostate Cancer Xenografts. PLoS One. 2015;10(6):e0130340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bialopiotrowicz E, Gorniak P, Noyszewska-Kania M, Pula B, Makuch-Lasica H, Nowak G, et al. Microenvironment-induced PIM kinases promote CXCR4-triggered mTOR pathway required for chronic lymphocytic leukaemia cell migration. J Cell Mol Med. 2018;22(7):3548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillory LA, Stewart JE, Megison ML, Nabers HC, Mroczek-Musulman E, Beierle EA. FAK Inhibition Decreases Hepatoblastoma Survival Both In Vitro and In Vivo. Transl Oncol. 2013;6(2):206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Ren X, Ren R, Wang Y, Shang J. The combination of G-CSF and AMD3100 mobilizes bone marrow-derived stem cells to protect against cisplatin-induced acute kidney injury in mice. Stem Cell Research & Therapy. 2021;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiyama E, Ueda Y, Kurihara S, Kawashima K, Ikeda K, Morihara N, Fukazawa T, Kanawa M, Hiyama K Gene expression profiling in hepatoblastoma cases of the Japanese Study Group for Pediatric Liver Tumors-2 (JPLT-2) trial. Eur J Mol Cancer. 2019. [Google Scholar]
- 22.Carrillo-Reixach J, Torrens L, Simon-Coma M, Royo L, Domingo-Sabat M, Abril-Fornaguera J, et al. Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol. 2020;73(2):328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. [DOI] [PubMed] [Google Scholar]
- 24.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270(1):41–9. [DOI] [PubMed] [Google Scholar]
- 25.Tang L, Zhang H, Zhang B. A note on error bars as a graphical representation of the variability of data in biomedical research: Choosing between standard deviation and standard error of the mean. J Pancreatol. 2019;2(3):69–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. J Vis Exp. 2014(92):e51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatse S, Princen K, Bridger G, De Clercq E, Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002;527(1-3):255–62. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 29.Welch DR, Hurst DR. Defining the Hallmarks of Metastasis. Cancer Res. 2019;79(12):3011–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–72. [DOI] [PubMed] [Google Scholar]
- 31.Nagle JA, Ma Z, Byrne MA, White MF, Shaw LM. Involvement of insulin receptor substrate 2 in mammary tumor metastasis. Mol Cell Biol. 2004;24(22):9726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin SS, Ridgeway AG, Pinkas J, Lu Y, Reginato MJ, Koh EY, et al. A cytoskeleton- based functional genetic screen identifies Bcl-xL as an enhancer of metastasis, but not primary tumor growth. Oncogene. 2004;23(26):4641–5. [DOI] [PubMed] [Google Scholar]
- 33.Luo H, Sun R, Zheng Y, Huang J, Wang F, Long D, et al. PIM3 Promotes the Proliferation and Migration of Acute Myeloid Leukemia Cells. Onco Targets Ther. 2020;13:6897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Qu X, Shao L, Hu Y, Yu X, Lan P, et al. Pim-3 enhances melanoma cell migration and invasion by promoting STAT3 phosphorylation. Cancer Biol Ther. 2018;19(3):160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashid OM, Nagahashi M, Ramachandran S, Dumur CI, Schaum JC, Yamada A, et al. Is tail vein injection a relevant breast cancer lung metastasis model? J Thorac Dis. 2013;5(4):385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther. 2013;6:1347–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. [DOI] [PubMed] [Google Scholar]
- 38.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393(6685):595–9. [DOI] [PubMed] [Google Scholar]
- 39.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23(1):43–52. [DOI] [PubMed] [Google Scholar]
- 40.Sun YX, Wang J, Shelburne CE, Lopatin DE, Chinnaiyan AM, Rubin MA, et al. Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem. 2003;89(3):462–73. [DOI] [PubMed] [Google Scholar]
- 41.Dang Y, Jiang N, Wang H, Chen X, Gao Y, Zhang X, et al. Proto-Oncogene Serine/Threonine Kinase PIM3 Promotes Cell Migration via Modulating Rho GTPase Signaling. J Proteome Res. 2020;19(3):1298–309. [DOI] [PubMed] [Google Scholar]
- 42.Liu B, Wang Z, Li HY, Zhang B, Ping B, Li YY. Pim-3 promotes human pancreatic cancer growth by regulating tumor vasculogenesis. Oncol Rep. 2014;31(6):2625–34. [DOI] [PubMed] [Google Scholar]
- 43.Decker S, Finter J, Forde AJ, Kissel S, Schwaller J, Mack TS, et al. PIM kinases are essential for chronic lymphocytic leukemia cell survival (PIM2/3) and CXCR4-mediated microenvironmental interactions (PIM1). Mol Cancer Ther. 2014;13(5):1231–45. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Diao X, Sun J, Chen Z. Regulation of VEGF, MMP-9 and metastasis by CXCR4 in a prostate cancer cell line. Cell Biol Int. 2011;35(9):897–904. [DOI] [PubMed] [Google Scholar]
- 45.Calandra G, McCarty J, McGuirk J, Tricot G, Crocker SA, Badel K, et al. AMD3100 plus G-CSF can successfully mobilize CD34+ cells from non-Hodgkin's lymphoma, Hodgkin's disease and multiple myeloma patients previously failing mobilization with chemotherapy and/or cytokine treatment: compassionate use data. Bone Marrow Transplant. 2008;41(4):331–8. [DOI] [PubMed] [Google Scholar]
- 46.Hira VVV, Van Noorden CJF, Molenaar RJ. CXCR4 Antagonists as Stem Cell Mobilizers and Therapy Sensitizers for Acute Myeloid Leukemia and Glioblastoma? Biology (Basel). 2020;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraham M, Biyder K, Begin M, Wald H, Weiss ID, Galun E, et al. Enhanced unique pattern of hematopoietic cell mobilization induced by the CXCR4 antagonist 4F-benzoyl-TN14003. Stem Cells. 2007;25(9):2158–66. [DOI] [PubMed] [Google Scholar]
- 48.Abraham M, Pereg Y, Bulvik B, Klein S, Mishalian I, Wald H, et al. Single Dose of the CXCR4 Antagonist BL-8040 Induces Rapid Mobilization for the Collection of Human CD34(+) Cells in Healthy Volunteers. Clin Cancer Res. 2017;23(22):6790–801. [DOI] [PubMed] [Google Scholar]
- 49.Sison EA, Magoon D, Li L, Annesley CE, Rau RE, Small D, et al. Plerixafor as a chemosensitizing agent in pediatric acute lymphoblastic leukemia: efficacy and potential mechanisms of resistance to CXCR4 inhibition. Oncotarget. 2014;5(19):8947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grundler R, Brault L, Gasser C, Bullock AN, Dechow T, Woetzel S, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD–induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4–mediated homing and migration. J Exp Med 2009;206:1957–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortes J, Tamura K, DeAngelo DJ, de Bono J, Lorente D, Minden M, et al. Phase I studies of AZD1208, a proviral integration Moloney virus kinase inhibitor in solid and haematological cancers. Br J Cancer. 2018;118(11):1425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez-Cuadrado L, Tracey N, Ma R, Qian B, Brunton VG. Mouse models of metastasis: progress and prospects. Dis Model Mech. 2017;10(9):1061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26(3):513– 23. [DOI] [PubMed] [Google Scholar]
- 54.Elkin M, Vlodavsky I. Tail vein assay of cancer metastasis. Curr Protoc Cell Biol. 2001;Chapter 19:1921–27. [DOI] [PubMed] [Google Scholar]
- 55.Harms JF, Welch DR. MDA-MB-435 human breast carcinoma metastasis to bone. Clin Exp Metastasis. 2003;20(4):327–34. [DOI] [PubMed] [Google Scholar]
- 56.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, et al. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2(9):1627–36. [PubMed] [Google Scholar]
- 57.Meyers RL, Katzenstein HM, Krailo M, McGahren ED 3rd, Malogolowkin MH. Surgical resection of pulmonary metastatic lesions in children with hepatoblastoma. J Pediatr Surg. 2007;42(12):2050–6. [DOI] [PubMed] [Google Scholar]
- 58.Kuznetsov HS, Marsh T, Markens BA, Castano Z, Greene-Colozzi A, Hay SA, et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov. 2012;2(12):1150–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang SY, Halvorsen OJ, Gravdal K, Bhattacharya N, Lee JM, Liu NW, et al. Prosaposin inhibits tumor metastasis via paracrine and endocrine stimulation of stromal p53 and Tsp-1. Proc Natl Acad Sci U S A. 2009;106(29):12115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart IR, Fidler IJ. Cancer invasion and metastasis. Q Rev Biol. 1980;55(2):121–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Expression profile data analyzed in this study were obtained from GEO GSE131329 and GSE133039. The sequence data generated in this study are publicly available in GEO at GSE164082 and GSE176152.