ABSTRACT
Orientia tsutsugamushi is an etiologic agent of scrub typhus, a globally emerging rickettsiosis that can be fatal. The bacterium’s obligate intracellular lifestyle requires its interaction with host eukaryotic cellular pathways. The proteins it employs to do so and their functions during infection are understudied. Recombinant versions of the recently characterized O. tsutsugamushi deubiquitylase (OtDUB) exhibit high-affinity ubiquitin binding, mediate guanine nucleotide exchange to activate Rho GTPases, bind clathrin adaptor protein complexes 1 and 2, and bind the phospholipid phosphatidylserine. Whether OtDUB is expressed and its function during O. tsutsugamushi infection have yet to be explored. Here, OtDUB expression, location, and interactome during infection were examined. O. tsutsugamushi transcriptionally and translationally expresses OtDUB throughout infection of epithelial, monocytic, and endothelial cells. Results from structured illumination microscopy, surface trypsinization of intact bacteria, and acetic acid extraction of non-integral membrane proteins indicate that OtDUB peripherally associates with the O. tsutsugamushi cell wall and is at least partially present on the bacterial surface. Analyses of the proteins with which OtDUB associates during infection revealed several known O. tsutsugamushi cell wall proteins and others. It also forms an interactome with adapter protein complex 2 and other endosomal membrane traffic regulators. This study documents the first interactors of OtDUB during O. tsutsugamushi infection and establishes a strong link between OtDUB and the host endocytic pathway.
KEYWORDS: scrub typhus, Orientia, effector, clathrin, adaptor protein complexes, endocytosis, Golgi, membrane trafficking, obligate intracellular bacteria
INTRODUCTION
Orientia tsutsugamushi is a Gram negative obligate intracellular bacterium that causes scrub typhus, a febrile disease endemic to the Asia-Pacific where roughly one million cases are reported annually. Clinical presentation ranges from asymptomatic to severe and can include complications such as myocarditis, pneumonitis, meningitis, circulatory system collapse, organ failure, and death (1–4). Travel related scrub typhus occurs, and non-travel related cases in African, South American, and Middle Eastern countries indicate the endemic region is much larger than originally appreciated (5–23). O. tsutsugamushi is vectored by trombiculid mites (24, 25). Of all Rickettsiales members, it infects the broadest range of cell types in its mammalian reservoirs and dead-end human hosts (26). It invades dermal dendritic cells, monocytes, and macrophages at the mite feeding site (27, 28). Professional antigen-presenting cells harboring the bacterium traffic through the lymph and egress into the circulatory system, which leads to infection of vascular endothelial cells (27). The success of O. tsutsugamushi as an obligate endosymbiont that replicates to high numbers in the cytoplasm of arthropod and mammalian host cells indicates that it effectively interfaces with eukaryotic cellular processes. The proteins that it expresses during infection to do so and the host-microbe interactions that they facilitate are largely uncharacterized.
Obligate intracellular bacterial proteins have evolved to perform eukaryotic-like functions that are essential for symbiosis and, in the case of pathogens, virulence. Studying them can provide valuable insights into the molecular basis of disease, reveal novel examples of pathogen evolution, and yield new biochemical tools. A prime example of such a protein is the recently described O. tsutsugamushi deubiquitylase, OtDUB (OTT_1962). The name of this 1368-residue protein is derived from the biochemical activity that it was first described to exhibit (29). OtDUB also possesses a ubiquitin binding domain (OtDUBUBD), a guanine nucleotide exchange factor domain (OtDUBGEF), a domain that binds directly to clathrin adaptor protein complexes 1 and 2 (AP-1, AP-2), and a phosphatidylserine (PS)-binding domain (29–31). It is the first bacterial protein discovered to carry this combination of functional elements, and several of them have unique structural and biochemical features. For instance, its DUB domain bears a previously undescribed variable domain architecture and is capable of cleaving ubiquitin chains of different linkages. Unlike other members of the CE-clan of bacterial ubiquitin proteases that prefer K63-linked ubiquitin chains, OtDUB efficiently cleaves K33-, K48-, and K63-linked chains. OtDUBUBD regulates the activity of the DUB domain and has an extraordinary affinity for ubiquitin that makes it an attractive tool for enriching or detecting ubiquitin/ubiquitylated proteins (29, 32). OtDUBGEF preferentially exchanges GDP for GTP in the small GTPase Rac1 compared to the closely related CDC42 enzyme. Structurally, the OtDUBGEF is an excellent example of convergent evolution as it exhibits an entirely different topology compared to other bacterial GEFs and yet upon binding Rac1, adopts an overall V-shaped fold that is conserved among bacterial GEFs (30).
Rac1 functions at the inner leaflet of the plasma membrane, a membrane bilayer enriched for PS and the same subcellular site where AP-2-assisted clathrin-mediated endocytosis occurs. OtDUB binds to both AP complexes in cellular lysates and associates directly with recombinant AP proteins through its AP-1/2-binding domain (AP-1/2 BD; OtDUB275-675) (29–31). Notably, ectopic expression of OtDUB severely disrupts membrane trafficking, but can do so independently of the AP-1/2 BD. By contrast, the PS-binding domain (OtDUB760-1159) was found to disrupt membrane trafficking, and mutants deficient for PS-binding no longer caused trafficking pathway defects (31). All of these biochemical activities suggest OtDUB can function at the plasma membrane of the host cell if expressed and exposed to the host cytoplasm during infection.
To date, all data on OtDUB stem exclusively from structural and biochemical analyses of recombinant versions of the proteins or truncated portions thereof (29, 30). This represents a significant gap in the knowledge of OtDUB relevance to infection. In this study, we examined OtDUB transcriptional and translational expression, its location in the bacterial cell, and its interactome during O. tsutsugamushi infection of mammalian cells. OtDUB is expressed throughout infection of different host cell types, peripherally associates with the bacterial outer membrane, is at least partially surface-exposed, and interacts with both bacterial cell wall and other proteins. OtDUB also associates with numerous host cell membrane traffic regulators with an enrichment for AP complex subunits. Coimmunoprecipitation (CoIP) of AP-2 and AP-2 accessory proteins, but not AP-1, with endogenous OtDUB demonstrates the preferential binding of endocytic regulators during infection. Overall, this study establishes that OtDUB is expressed by O. tsutsugamushi during infection, suggests that it potentially functions at the bacterial cell surface, and establishes the AP-2 complex as a bona fide host interaction during infection.
RESULTS
OtDUB is conserved among O. tsutsugamushi clinical isolates.
OtDUB is encoded by a single-copy gene in the O. tsutsugamushi Ikeda strain, a clinical isolate from Japan that causes severe disease in humans and experimentally infected mice (33, 34). Candidatus Orientia chuto, which was isolated from a traveler that acquired scrub typhus in Dubai (35), encodes an OtDUB homolog, OcDUB (WP_052694629.1), that also binds and cleaves a variety of ubiquitin linkages (29). This protein’s conservation suggests its importance to Orientia pathobiology. Using Ikeda OtDUB nucleic acid (OTT_1962/OTT_RS09490) and protein (WP_012462337.1) sequences as queries, we sought to identify OtDUB homologs in other O. tsutsugamushi strains. Homology searches were conducted using the National Center for Biotechnology Information (NCBI) Nucleotide Basic Local Alignment Search Tool (BLASTN) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) and Protein BLAST (BLASTP) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), respectively. OtDUB homologs were identified as being encoded by single-copy genes in strains Kato (Japan), UT176 and UT76 (both from northeastern Thailand), Gilliam (Burma), Karp (New Guinea), Wuj/2014 (China), Boryong (South Korea), and TA686 (Central Thailand). Complete genomes for each of these strains recovered from scrub typhus patients are available in GenBank. These homologs displayed 96% to 98% nucleotide identity, 92% to 96% amino acid identity, and 95% to 98% amino acid similarity (Table 1). Hence, this protein is conserved among Candidatus O. chuto and multiple O. tsutsugamushi strains that are pathogenic to humans.
TABLE 1.
Comparison of full-length OtDUB homologs in other O. tsutsugamushi strains/isolates
| Strain/isolate | Geographic origin | Reference | Nucleotide length | % Nucleotide identity | Protein accession no. | AA length | % AA identity | % AA similarity |
|---|---|---|---|---|---|---|---|---|
| Ikeda | Japan | Tamura 1984 | 4107 | 100 | WP_012462337.1 | 1368 | 100 | 100 |
| Kato | Japan | Shishido 1958 | 4107 | 98 | WP_045915965.1 | 1368 | 96 | 98 |
| UT176 | NE Thailand | Blacksell 2008 |
4125 | 97 | WP_109489930.1 | 1374 | 96 | 97 |
| UT76 | NE Thailand | Luksameetanasan 2007 |
4122 | 97 | WP_109227152.1 | 1373 | 95 | 97 |
| Gilliam | Burma | Bengston 1945 | 4116 | 97 | WP_047220928.1 | 1371 | 94 | 97 |
| Karp | New Guinea | Rights 1948 | 3936 | 97 | WP_231967837.1 | 1311 | 95 | 97 |
| Wuj/2014 | China | 4113 | 97 | WP_150339717.1 | 1370 | 94 | 96 | |
| Boryong | South Korea | Chang 1990 | 4116 | 96 | WP_011944731.1 | 1371 | 93 | 96 |
| TA686 | Central Thailand | Elisberg 1968 | 4059 | 96 | SPR13742.1 | 1352 | 92 | 95 |
O. tsutsugamushi transcriptionally expresses otdub throughout mammalian cell infection.
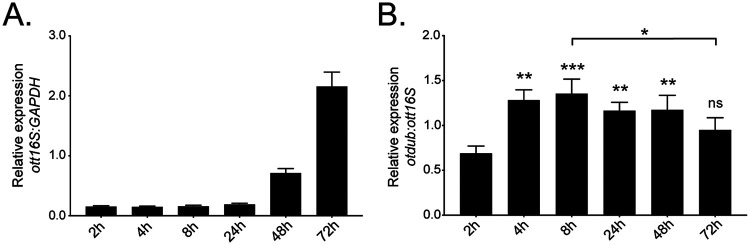
To determine if O. tsutsugamushi transcribes the otdub gene during infection of mammalian host cells, total RNA isolated from synchronously infected HeLa cells at 2, 4, 8, 24, 48, and 72 h was subjected to RT-qPCR analyses. Following entry into HeLa and other immortalized cell lines, O. tsutsugamushi undergoes minimal growth during the first 24 h after which it expands logarithmically until bacterial exit or host cell lysis (36–38). Consistent with this growth curve, normalized O. tsutsugamushi 16S rRNA (ott16S)-to-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were significantly elevated at 48 and 72 h (Fig. 1A). Monitoring relative otdub-to-ott16S expression detected otdub transcript at all time points, but at significantly higher levels at 4 h through 48 h (Fig. 1B). Thus, O. tsutsugamushi transcribes otdub throughout its intracellular life cycle in mammalian cells.
FIG 1.
O. tsutsugamushi transcribes otdub during infection of HeLa cells. HeLa cells were infected with O. tsutsugamushi at an MOI of 10. Total RNA was collected at 2, 4, 8, 24, 48, and 72 h. Quantitative reverse transcriptase PCR (RT-qPCR) was performed to assess expression of otdub, ott16S (O. tsutsugamushi 16S rRNA gene), or human GAPDH. Relative ott16S-to-human GAPDH (A) and otdub-to-ott16S expression (B) was determined using the 2−ΔΔCT method. Data are mean values ± SD from 3 independent experiments. One-way ANOVA with Tukey’s post hoc test was used to discern statistical significance of otdub levels across time points. Significance shown is compared to 2 h postinfection unless denoted otherwise (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).
Generation and validation of OtDUB-specific antisera.
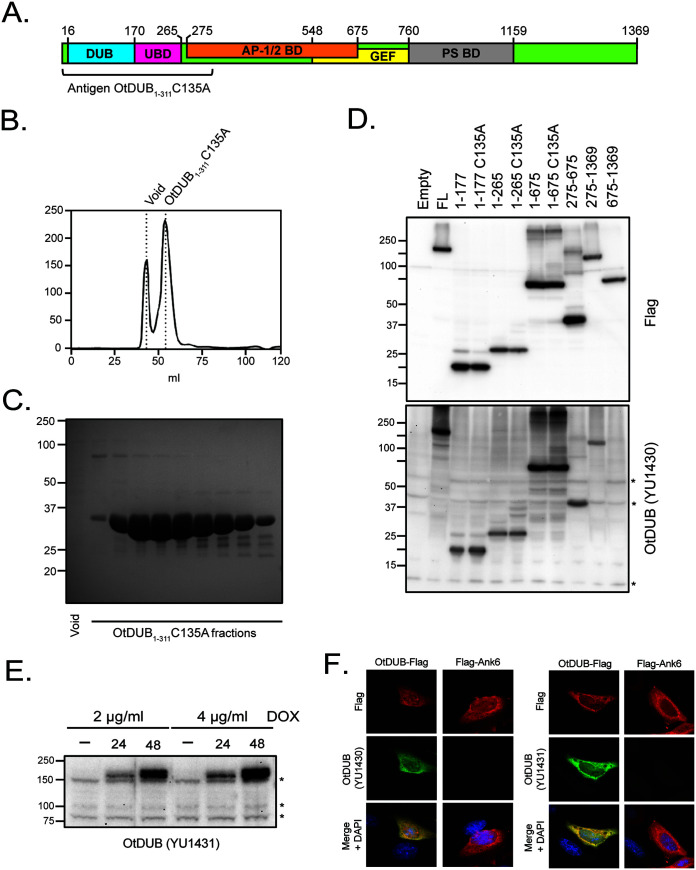
To assess O. tsutsugamushi OtDUB translational expression, we first had to raise antiserum against this protein and confirm its specificity. Figure 2A presents a schematic of Ikeda OtDUB with its DUB, UBD, AP-1/2 binding, GEF, and PS-binding domains color-coded. Two polyclonal antisera were generated against the previously characterized N-terminal fragment containing a catalytically dead DUB domain, OtDUB1-311-C135A (29). Recombinantly expressed glutathione S-transferase (GST)-OtDUB1-311-C135A was affinity purified followed by cleavage and depletion of the GST-tag, and further purified via size exclusion chromatography (Fig. 2B). Fractions from the main protein peak were devoid of major contaminants or breakdown products, and the purified protein resolved at the expected size (calculated molecular weight [MW] 35.3 kDa) (Fig. 2C). Purified OtDUB1-311-C135A was inoculated into 2 rabbits to yield antisera, Yale University (YU)1430 and YU1431. Immunoblotted lysates from Saccharomyces cerevisiae expressing various Flag-tagged OtDUB fragments were probed with YU1430 serum or Flag-tag antibody as a control. YU1430 detected full-length OtDUB-Flag and fragments thereof that included all or some of the antigen but failed to recognize OtDUB675-1369-Flag (Fig. 2D).
FIG 2.
Production and evaluation of OtDUB antisera YU1430 and YU1431. (A) Cartoon diagram of full-length OtDUB with known functional domains: DUB (deubiquitylase; blue), UBD (ubiquitin binding domain; magenta), AP-1/2 BD (adaptor protein complex 1/2 binding domain; orange), GEF (guanine nucleotide exchange factor; yellow), and PS BD (phosphatidylserine binding domain; gray). The antigen used to produce both antisera was OtDUB1-311-C135A (bracket). (B) Superdex 75 gel filtration chromatogram of the OtDUB1-311-C135A preparation used for the antigen. (C) Representative SDS-PAGE gel of OtDUB1-311-C135A fractions stained with Coomassie brilliant blue. (D) Equivalent OD600 units of protein lysates from galactose-induced yeast expressing various OtDUB fragments, catalytically dead versions (C135A) thereof, or an empty vector-transformed control were resolved on parallel SDS-PAGE gels and immunoblotted with Flag antibody or YU1430 OtDUB antiserum. Asterisks denote nonspecific bands that served as a loading control. (E) Western-blotted whole cell lysates from OtDUB Flp-In HeLa cells incubated for 24 or 48 h in the presence or absence of doxycycline (DOX) were probed with YU1431 OtDUB antiserum. Asterisks indicate nonspecific signals, which serve as loading controls. (F) Representative immunofluorescence images of HeLa cells ectopically expressing either Flag-Ank6 or OtDUB-Flag. Fixed cells were immunolabeled with anti-Flag antibody together with YU1430 or YU1431 OtDUB antiserum and stained with DAPI.
A stably transfected cell line was generated expressing OtDUB-Flag under the control of a doxycycline-inducible promoter (HeLa Flp-In OtDUB-Flag). Lysates from HeLa Flp-In OtDUB-Flag cells incubated with doxycycline or vehicle were collected at 24 or 48 h, immunoblotted, and screened using YU1431 serum. Only weak, nonspecific bands were detected in vehicle-treated controls (Fig. 2E). In the presence of doxycycline, YU1431 detected robust expression of a band corresponding to the expected size of OtDUB-Flag (approximately 150 kDa) that increased in abundance over the duration of induction and in a dose-dependent manner. Both antisera were also assayed for the ability to immunolabel OtDUB in fixed cells. HeLa cells transfected to ectopically express OtDUB-Flag were fixed, probed with anti-Flag antibody together with either YU1430 or YU1431 antiserum, and stained with DAPI. To confirm specificity, HeLa cells ectopically expressing a Flag-tagged version of the O. tsutsugamushi Ank6 effector (39) were also screened. Whereas Flag antibody immunolabeled both OtDUB-Flag and Flag-Ank6, YU1430 and YU1431 recognized only OtDUB-Flag (Fig. 2F). Overall, these results demonstrate that both OtDUB antisera specifically detect the antigen.
O. tsutsugamushi translationally expresses OtDUB during infection of mammalian cell lines.
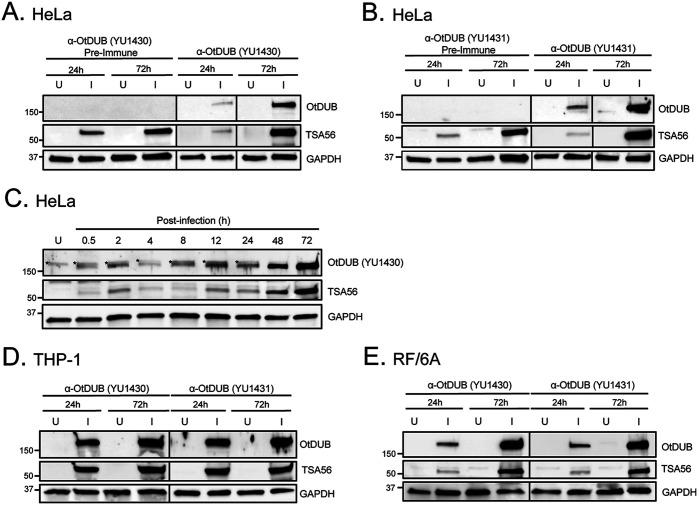
To confirm if O. tsutsugamushi expresses OtDUB protein during infection, immunoblots of whole cell lysates recovered from synchronously infected HeLa cells at 24 and 72 h were probed with YU1430, YU1431, or preimmune serum from the respective rabbits. Antiserum against TSA56 (56-kDa type-specific antigen), which is an outer membrane protein (OMP) that O. tsutsugamushi abundantly expresses throughout infection (40), was a positive control. YU1430 and YU1431 detected OtDUB in infected cell lysates, while the preimmune sera did not (Fig. 3A and B). The OtDUB immunosignal increased with bacterial load. When OtDUB expression in infected HeLa cells between 30 min and 72 h was examined, the signal intensity of the OtDUB band detected by YU1430 mirrored that of TSA56 at every time point (Fig. 3C). OtDUB was detected at 0.5 h and 2 h, time points that correspond to when bound O. tsutsugamushi organisms are initiating the internalization process or are newly internalized, respectively (41, 42). Lower detection of both OtDUB and TSA56 at 4 h likely reflects a reduction in the overall number of cell-associated bacteria due to the host cells having been washed just after 2 h to remove unbound and/or non-internalized bacteria that remained weakly associated at the cell surface. The OtDUB immunosignal was greater than the TSA56 signal at 8 h, suggesting that O. tsutsugamushi potentially begins to upregulate OtDUB expression prior to that of TSA56. A nonspecific host cell protein was faintly detected by YU1430 in both uninfected cells and infected cells. Importantly, both YU1430 and YU1431 also recognized OtDUB in lysates of infected THP-1 monocytic cells and RF/6A endothelial cells (Fig. 3D and E). Thus, O. tsutsugamushi expresses OtDUB during infection of multiple mammalian cell types including ones that are representative of those that it colonizes in animal reservoirs and humans.
FIG 3.
O. tsutsugamushi expresses OtDUB during infection of HeLa cells as well as monocytic and endothelial cell lines. (A and B) Lysates from uninfected [U] and O. tsutsugamushi-infected [I] HeLa cells (MOI 10) were resolved by SDS-PAGE and immunoblotted with TSA56 antiserum as an infection control, anti-human GAPDH for a loading control, YU1430 (A) or YU1431 (B), or the respective preimmune serum. (C) Infected HeLa cells were harvested at various time points postinfection, resolved by SDS-PAGE, and immunoblotted with antibodies against TSA56, GAPDH, and OtDUB (YU1430). Asterisks denote nonspecific signals. (D and E) Lysates from U and I THP-1 monocytic (D) and RF/6A endothelial (E) cells (MOI 10) were resolved by SDS-PAGE and immunoblotted with antibody against TSA56, GAPDH, and either YU1430 or YU1431. Data are representative of two or three independent experiments.
OtDUB peripherally associates with the bacterial cell wall and is at least partially surface-exposed.
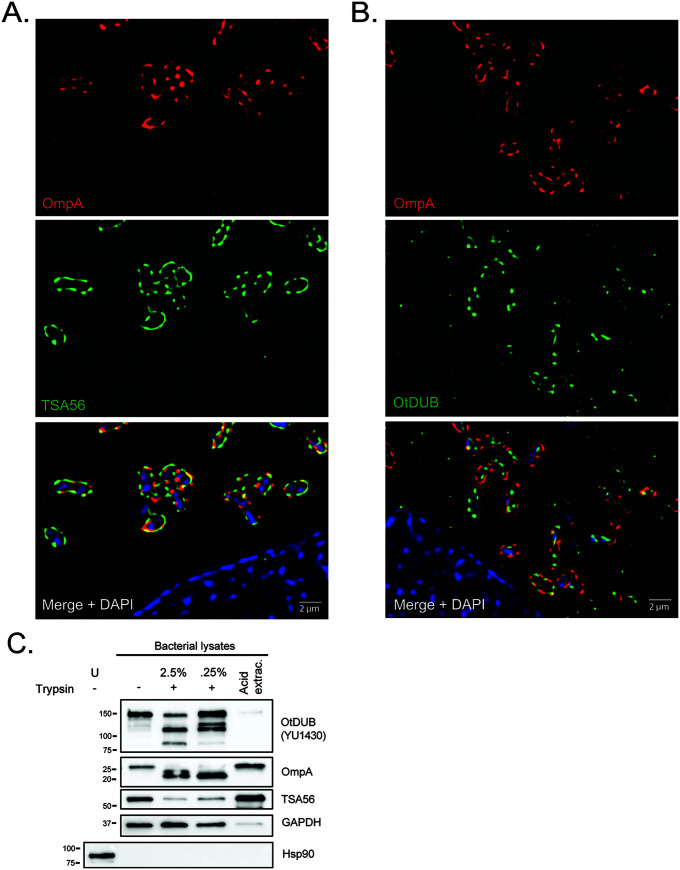
Studies of OtDUB in recombinant form demonstrated that it performs at least 4 eukaryotic-like activities that enable it to interact with signaling pathways present in eukaryotes, but not bacteria (29–31). These observations suggest that it is exposed to the host cellular milieu during O. tsutsugamushi infection. To assess potential OtDUB association with the bacterial cell wall, infected HeLa cells were fixed, co-immunolabeled with YU1430 and antisera specific for O. tsutsugamushi outer membrane protein A (OmpA), followed by visualization using structured illumination microscopy (SIM). This form of super-resolution microscopy has a lateral resolution of ~110 nm (43). As a control, duplicate samples labeled with antisera against OmpA and TSA56 were also examined. OmpA and TSA56 immunosignals exhibited punctate patterns that encircled the DAPI-stained nucleoids of individual O. tsutsugamushi bacteria (Fig. 4A). Consistent with both being OMPs, several instances of OmpA and TSA56 immunosignal colocalization were apparent. By comparison, there were fewer instances of OmpA and OtDUB immunosignal colocalization (Fig. 4B). OtDUB immunolabeling resulted in a punctate pattern that encircled DAPI-stained bacterial nucleoids and infrequently colocalized with OmpA immunosignal. Numerous instances of punctate OtDUB immunosignal were also detected within the cytoplasm of infected cells independent of intracellular bacteria, a trend that was not observed for TSA56 immunosignal.
FIG 4.
OtDUB peripherally associates with the bacterial cell wall and is at least partially surface-exposed. (A and B) HeLa cells infected with O. tsutsugamushi (MOI 10) for 24 h were fixed, immunolabelled for OmpA (red) and TSA56 (green) (A) or OmpA (red) and OtDUB (green) (B), stained with DAPI, and subjected to structured illumination microscopy. Representative fluorescence images are shown. Scale bar is 2 μm. (C) Representative blot of uninfected HeLa cells or host cell-free O. tsutsugamushi bacteria subjected to no treatment (-), varying concentrations of trypsin (+), or extraction with 50 mM acetic acid (Acid extrac.). Lysates were resolved by SDS-PAGE and immunoblotted for OtDUB (YU1430), OmpA or TSA56 as bacterial transmembrane protein controls, GAPDH as a bacterial cytosolic protein control, and Hsp90 as a host cell protein control. Data are representative of five experiments with similar results.
As a complementary approach, O. tsutsugamushi bacteria were isolated from highly infected HeLa cells at 72 h postinfection and treated with trypsin or acetic acid. Surface trypsinolysis has been used to confirm surface localization of proteins of other obligate intracellular bacteria including Anaplasma phagocytophilum (44–46) and Chlamydia trachomatis (47). Acid hydrolysis of intact bacteria will solubilize proteins that are intracellular or peripherally associated with the outer membrane but, in the absence of a protease is insufficient to solubilize integral membrane proteins (48, 49). Intact host cell-free O. tsutsugamushi were incubated with 2.5% or 0.25% trypsin, 50 mM acetic acid, or vehicle prior to lysis and immunoblot analyses. Lysates of vehicle-treated bacteria were confirmed to be devoid of host cellular proteins based on the absence of human-specific heat shock protein 90 (Hsp90) (Fig. 4C). Treatment efficacy was verified by screening with antisera against bacterial OmpA and TSA56, both of which are surface-exposed transmembrane proteins (34, 50), and GAPDH, which resides in the bacterial cytoplasm (33). Signal corresponding to intact versions of OmpA and TSA56 were each considerably reduced and smaller-sized anti-OmpA immunoreactive bands were detected in lysates of trypsin-treated bacteria. Acid extraction had no effect on either protein. GAPDH was unaffected by surface trypsinolysis but was barely detectable following incubation with acetic acid. Trypsin treatment of intact O. tsutsugamushi bacteria reduced levels of and/or yielded cleavage products of OtDUB, whereas acid extraction nearly abolished the ability to immunodetect the protein. Taken together, these data indicate that OtDUB is not a membrane-spanning protein, is at least proximally associated with the cell wall, and is at least partially presented on the O. tsutsugamushi surface.
The OtDUB interactome during infection is enriched for AP complexes.
As a first step toward elucidating the role(s) of OtDUB during O. tsutsugamushi infection, we sought to identify the interacting partners of OtDUB endogenously expressed by the bacterium during infection of mammalian host cells using coimmunoprecipitation coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS). HeLa cells were synchronously infected and lysed 72 h later. The lysate was split evenly and incubated with either YU1431 or rabbit preimmune serum in the presence of Protein A/G beads followed by elution, trypsin digestion, and identification of the tryptic peptides by LC-MS/MS. Specific metrics were applied to sort through the data and isolate high confidence hits that were likely to be bona fide interactors.
After removing OtDUB peptide hits, the remaining peptides mapped to 16 O. tsutsugamushi proteins (Table 2) and 88 human proteins (Data set S1). Five of the identified O. tsutsugamushi proteins associate with the outer membrane (TSA56, TSA47) or inner membrane (HflC, HflK, FtsH) (33, 51–53), while the remaining 11 are annotated as functioning in the bacterial cytoplasm with roles tied to stress responses, metabolism, transcription, and translation (33, 54). Of the 88 potential host hits, 19 localize to subcellular compartments or have roles in DNA replication, transcription, or mRNA processing. Given that OtDUB associates with the O. tsutsugamushi cell wall and the coimmunoprecipitation was performed at a time point during infection when the bacteria were in the host cell cytoplasm, these 19 were considered lower-confidence hits.
TABLE 2.
Proteomics based screen identifies OtDUB bacterial interactions during infection
| Accession no. | Description | Role/location | MW (kDa) | IgG spectral counts | OtDUB spectral counts | F-scorea |
|---|---|---|---|---|---|---|
| BAG41086.1 | ATP-dependent protease La | Stress response | 88 | 0 | 33 | 1 |
| BAG40771.1 (+8) | Hypothetical protein OTT_1313 | Stress response | 35 | 0 | 9 | 1 |
| BAG40249.1 | Heat shock protein HtpG | Stress response | 72 | 0 | 7 | 1 |
| BAG40032.1 | NifS protein homolog | Metabolism | 48 | 0 | 6 | 1 |
| BAG41073.1 (+7) | 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase | Metabolism | 32 | 0 | 5 | 1 |
| BAG40289.1 (+6) | Acetylglutamate kinase | Metabolism | 34 | 0 | 4 | 1 |
| BAG40388.1 (+2) | 30S ribosomal protein S9 | Protein synthesis | 18 | 0 | 2 | 1 |
| BAG40534.1 | Thy1 | Metabolism | 35 | 0 | 2 | 1 |
| BAG40292.1 | Cell surface antigen | Outer membrane protein | 105 | 0 | 2 | 1 |
| BAG40526.1 (+1) | DNA-directed RNA polymerase alpha chain | Transcription | 38 | 0 | 1 | 1 |
| BAG41326.1 | NAD-glutamate dehydrogenase | Metabolism | 187 | 3 | 18 | 0.86 |
| BAG40776.1 (+5) | HflC | Inner membrane | 33 | 1 | 6 | 0.86 |
| BAG40775.1 (+9) | HflK | Inner membrane | 40 | 1 | 4 | 0.8 |
| BAG40403.1 | 56 kDa type-specific antigen | Outer membrane protein | 56 | 2 | 7 | 0.83 |
| SPR13391.1 | Type surface antigen 47 (47 kDa/HtrA) | Outer membrane protein | 51 | 5 | 27 | 0.81 |
| BAG41041.1 (+3) | FtsH | Cytoplasmic membrane protease | 69 | 4 | 13 | 0.80 |
F-score, ratio of peptides from the OtDUB IP divided by the sum of peptides from both IgG and OtDUB IPs.
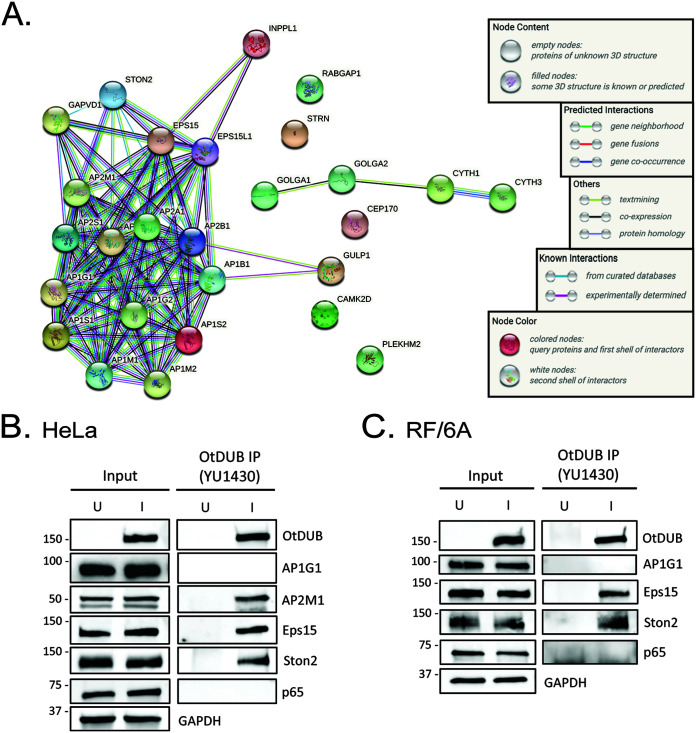
After categorizing the remaining 69 hits based on their cellular roles, 27 grouped into membrane trafficking-related activities (Table 3). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis (55) on these 27 proteins revealed a functional enrichment for components of AP-1 and AP-2 complexes (Fig. 5A), which is consistent with published results for recombinant OtDUB (31). AP-1/AP-2 complexes interact with clathrin to mediate different membrane trafficking pathways. Specifically, AP-1 is required for trafficking from the trans-Golgi network-to-early endosomes and from the endosomal system to other destinations, whereas AP-2 is essential for endocytosis (56). Additional identified OtDUB interacting partners were involved in endocytosis, maintenance of Golgi structural integrity, calcium signaling, microtubule-organization, and other aspects of membrane traffic (Table 3).
TABLE 3.
Proteomics based screen identifies OtDUB host interactions during infection
| Gene | Description | Pathway involvement | MW (kDa) | IgG spectral counts | OtDUB spectral counts | F-scorea |
|---|---|---|---|---|---|---|
| EPS15 | Epidermal growth factor receptor substrate 15 | Endocytosis | 99 | 0 | 65 | 1 |
| GOLGA1 | Golgin subfamily A member 1 | Golgi adhesion | 88 | 0 | 48 | 1 |
| EPS15L1 | Epidermal growth factor receptor substrate 15-like 1 | Endocytosis | 94 | 0 | 39 | 1 |
| CYTH1 | Cytohesin-1 | GEF; membrane trafficking | 46 | 0 | 29 | 1 |
| INPPL1/SHIP2 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | Endocytosis | 139 | 0 | 26 | 1 |
| RABGAP1 | Rab GTPase-activating protein 1 | GAP; Rab6A vesicle transport | 122 | 0 | 22 | 1 |
| CEP170 | Centrosomal protein of 170 kDa | Microtubule | 175 | 0 | 19 | 1 |
| CYTH3 | Cytohesin-3 | GEF; membrane trafficking | 46 | 0 | 18 | 1 |
| PLEKHM2 | Pleckstrin homology domain-containing family M member 2 | Golgi/microtubules | 113 | 0 | 18 | 1 |
| GOLGA2 | Golgin subfamily A member 2 | Golgi adhesion | 113 | 0 | 18 | 1 |
| GULP1 | PTB domain-containing engulfment adapter protein 1 | Endocytosis/trafficking | 34 | 0 | 16 | 1 |
| STON2 | Stonin-2 | Endocytosis | 101 | 0 | 15 | 1 |
| CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Ca2+ signaling | 56 | 0 | 13 | 1 |
| STRN | Striatin | Ca2+ signaling | 86 | 0 | 13 | 1 |
| GAPVD1 | GTPase-activating protein and VPS9 domain-containing protein 1 | Endocytosis | 165 | 0 | 10 | 1 |
| AP1B1 | AP-1 complex subunit beta-1 | Golgi trafficking | 105 | 0 | 84 | 1 |
| AP1G1 | AP-1 complex subunit gamma-1 | Golgi trafficking | 91 | 0 | 13 | 1 |
| AP1M1 | AP-1 complex subunit mu-1 | Golgi trafficking | 49 | 0 | 8 | 1 |
| AP1G2 | AP-1 complex subunit gamma-like 2 | Golgi trafficking | 87 | 0 | 6 | 1 |
| AP1S1 | AP-1 complex subunit sigma-1A | Golgi trafficking | 19 | 0 | 6 | 1 |
| AP1M2 | AP-1 complex subunit mu-2 | Golgi trafficking | 48 | 0 | 2 | 1 |
| AP1S2 | AP-1 complex subunit sigma-2 | Golgi trafficking | 19 | 0 | 2 | 1 |
| AP2A2 | AP-2 complex subunit alpha-2 | Endocytosis | 104 | 0 | 82 | 1 |
| AP2M1 | AP-2 complex subunit mu | Endocytosis | 50 | 0 | 38 | 1 |
| AP2S1 | AP-2 complex subunit sigma | Endocytosis | 17 | 0 | 18 | 1 |
| AP2A1 | AP-2 complex subunit alpha-1 | Endocytosis | 108 | 1 | 154 | 0.99 |
| AP2B1 | AP-2 complex subunit beta | Endocytosis | 105 kDa | 3 | 158 | 0.98 |
F-score, ratio of peptides from the OtDUB IP divided by the sum of peptides from both IgG and OtDUB IPs.
FIG 5.
OtDUB interacts with AP complexes during infection. (A) The 27 host proteins that coimmunoprecipitated with endogenous OtDUB from O. tsutsugamushi-infected HeLa cells and are functionally enriched for membrane trafficking pathways were subjected to STRING analysis. (B and C) OtDUB preferentially interacts with AP-2 complex proteins. HeLa (B) and RF/6A (C) cells were uninfected (U) or infected with O. tsutsugamushi for 72 h (I), lysed, and subjected to OtDUB precipitation using YU1430 antiserum. Input lysates and eluates were resolved by SDS-PAGE and immunoblotted for OtDUB, AP1G1, AP2M1, Eps15, Ston2, p65, and GAPDH. Data are representative of three independent experiments.
By spectral counts, there were 4-fold more AP-2 than AP-1 peptides. In agreement, OtDUB coimmunoprecipitated AP2M1, but not AP1G1 from infected HeLa cells (Fig. 5B). Immunoprecipitation of OtDUB from infected RF/6A cells also failed to recover AP1G1 (Fig. 5C). As further validation that OtDUB interacts with AP-2 complexes, it coimmunoprecipitated AP-2 accessory proteins Eps15 (epidermal growth factor receptor substrate 15) and Ston2 (stonin 2) from infected HeLa and RF/6A cells (Fig. 5B and C). AP2M1 antibodies from 2 different commercial sources detected multiple background bands in infected and uninfected RF/6A cells lysates, which prevented direct validation that OtDUB interacts with AP-2 during infection of these cells. Screening OtDUB coimmunoprecipitated proteins with antibody against p65, an NF-κB component that was not detected by LC-MS/MS as an OtDUB interacting partner, validated that immunoprecipitation conditions did not recover nonspecific interactions. Overall, however, these results collectively indicate that OtDUB preferentially interacts with the endocytic AP-2 clathrin adaptor during infection.
DISCUSSION
This report establishes that the multifunctional OtDUB protein is expressed during infection and reveals its first bona fide interactions in infection. Consistent with recently published data for the protein in recombinant form, OtDUB expressed by O. tsutsugamushi interacts with proteins of membrane trafficking systems with a preference for the AP-2 complex and AP-2 associated proteins (31). This capability, along with its ubiquitin binding, deubiquitylation, and GEF activities would require OtDUB exposure to the host cell cytoplasmic milieu. Multiple lines of evidence support this premise. The otdub gene is transcriptionally upregulated beginning between 2 and 4 h postinfection, a time period that corresponds with when the pathogen exits the endosome and initiates its cytoplasmic phase of infection (30). This leads to OtDUB protein levels increasing with bacterial load in HeLa epithelial, THP-1 monocytic, and RF/6A endothelial cells, a trend that implies the protein’s importance for O. tsutsugamushi fitness during mammalian cell infection. Results obtained using SIM, bacterial surface trypsinization, and acid extraction indicate that OtDUB peripherally associates with the cell wall. Although OtDUB cleavage products were detected following trypsin treatment, a substantial amount of full-length OtDUB remained, indicating that only portions of it are accessible to trypsin on the O. tsutsugamushi surface. OtDUB trypsin-inaccessible regions could be periplasmic or intracellular, as is consistent with its immunofluorescence signal being interior relative to that of OmpA. Alternatively, portions of it could be occluded by its proximity to/direct interaction with bacterial OMPs or host cytoplasmic proteins, possibilities that are supported by our coimmunoprecipitation-proteomics results.
OtDUB peripherally associates with the O. tsutsugamushi cell wall. The additional detection of OtDUB punctate immunosignal in the cytoplasm of infected cells could be due to nonspecific antibody binding of a host protein or because OtDUB is a secreted effector. If the latter, then its peripheral association with the O. tsutsugamushi cell wall would be also due to it being translocated through the cell wall. Other Rickettsiales effectors are both secreted into the host cell milieu and associate with membrane-bound proteins on the bacterial surface. These multifunctional effectors perform different roles based on their spatiotemporal location. The Ehrlichia chaffeensis tandem repeat protein effector, TRP120, is an invasin when localized at the bacterial surface. Post entry, E. chaffeensis translocates TRP120 using the Type 1 secretion system into its host cell where it interfaces with multiple eukaryotic cellular pathways to become both ubiquitylated and sumoylated, interact with the cytoskeleton, co-opt Notch signaling, and modulate host cell gene expression (57–62). A. phagocytophilum utilizes heat shock protein 70, a predicted Type 4 secretion system effector, to interact with major surface protein 4 on the bacterial surface and mediate tick cell invasion (63). It will be important to determine whether OtDUB is a secreted effector once inside its host cell and/or if it mediates early host-pathogen interactions at the O. tsutsugamushi cell surface.
OtDUB interfaces with the endosomal system during infection and does so through its preferential interaction with AP-2 complexes. Our inability to detect AP1G1 via Western blot among endogenous OtDUB interacting partners agrees with the overall lower spectral counts of AP-1 complex subunit peptides detected by LC-MS/MS, but is in contrast to the ability of recombinant OtDUB to recover AP-1 and AP-2 subunits with comparable efficiency via gel filtration chromatography (31). The difference in AP-1 recovery presumably lies in the 2 assays. Recombinant OtDUB immobilized in abundance on an affinity matrix column is likely able to better retain lower-affinity interactions with the AP-1 complex versus endogenous OtDUB expressed at lower levels by O. tsutsugamushi. Eps15 and Ston2 interact with each other and, via Eps15, complex with AP-2 (64). Coimmunoprecipitation of both proteins by OtDUB from infected HeLa and RF/6A cells further supports that OtDUB interacts with the AP-2 complex during infection.
While the AP-1/2 BD of OtDUB has been mapped to residues 275–675 based on direct protein:protein interaction assays performed using recombinant versions of each partner (31), its relevance to O. tsutsugamushi infection remains unknown. Coxiella burnetii hijacks AP-2 vesicular complexes to support its intracellular growth and development of its parasitophorous vacuole (65). Although not a vacuole-adapted pathogen, perhaps O. tsutsugamushi uses OtDUB to harness cargo-containing AP-2 vesicles for optimal intracellular fitness. Notably, Eps15 and Eps15L1 have ubiquitin-interacting motifs (UIMs) that bind ubiquitin to recruit them to ubiquitinated receptors and initiate endocytosis (66, 67). The Eps15 and Eps15L1 UIMs also induce coupled monoubiquitination of both proteins (68), which could be potential sites for OtDUBUBD interactions. OtDUB-AP-2 interactions might negatively regulate endocytosis, which in the context of dendritic cells could prevent cytokine receptor endocytosis, a phenotype of O. tsutsugamushi infection (69). OtDUBGEF activation of Rac1 and other Rho GTPases is yet another means by which OtDUB could co-opt or modulate endocytic and other membrane trafficking pathways at the plasma membrane. Further, the PS-binding activity of OtDUB helps it associate with the host plasma membrane (31), which may also help alter host membrane trafficking.
The OtDUB interactome is enriched as well for microtubule- and Golgi-associated proteins, consistent with prior observations that following endosomal escape, O. tsutsugamushi bacteria mobilize themselves via microtubules to the microtubule organizing center and thereafter remain juxtaposed to the Golgi throughout infection until exiting the host cell. OtDUB interactions with GOLGA1 and GOLGA2, which are involved in endosome-to-Golgi trafficking and maintaining Golgi integrity, are conspicuous because O. tsutsugamushi perturbs Golgi structure and disrupts the secretory pathway during infection (70, 71). ChlaDUB1, a C. trachomatis acetyltransferase and deubiquitylase of K63 chains, fragments the Golgi to promote redistribution of the stacks around the chlamydial inclusion (72). In biochemical assays, OtDUB cleaves K63 polyubiquitin types but against diubiquitin substrates demonstrates even stronger activity toward K33 linkages, an interesting attribute as no other DUB has a reported preference for this linkage type (29, 73). K33 linkages are associated with post-Golgi protein trafficking (74). It will be useful to investigate if OtDUB contributes to O. tsutsugamushi-Golgi interactions and the bacterium’s abilities to alter the organelle’s structure and function. Overall, based on OtDUB interactions with AP complexes, associated endosomal and Golgi network proteins, and potentially related roles of Rac1 and PS, it is likely that this protein plays one or more key roles in O. tsutsugamushi modulation of vesicular traffic between the Golgi and plasma membrane using its DUB, ubiquitin binding, GEF, AP1/2-binding, and/or PS-binding domains.
OtDUB is a unique protein capable of facilitating host-pathogen interactions by executing at least 4 distinct eukaryotic-like activities – high-affinity ubiquitin binding, deubiquitylation, promoting GTPase guanine nucleotide exchange, and binding the AP-2 complex – and is robustly expressed throughout O. tsutsugamushi infection of different mammalian cell types, including those of monocytic and endothelial lineages. Additionally, otdub is conserved in the genome of every O. tsutsugamushi strain assessed. Collectively, these findings suggest the importance of OtDUB for the pathogen’s success as an endosymbiont. Discerning the specific role that each OtDUB activity plays in O. tsutsugamushi pathogenesis will serve as an intriguing model of host-intracellular microbe interactions and will provide insights into how this single multifunctional bacterial protein contributes to scrub typhus progression.
MATERIALS AND METHODS
Mammalian cell culture.
HeLa cells (CCL-2; American Type Culture Collection [ATCC], Manassas, VA) were cultured in complete medium: Dulbecco’s Modified Eagle Medium (ThermoFisher) supplemented with 10% (vol/vol) antibiotic-free fetal bovine serum (Gemini Bio-Products, Sacramento, CA) and 1% (vol/vol) penicillin/streptomycin in a 37°C humidified incubator containing 5% CO2. THP-1 cells (TIB-202; ATCC) were cultured as previously described (75). RF/6A rhesus monkey choroidal endothelial cells (CRL-1780; ATCC) were cultured as previously described (76).
Generation of stable HeLa Flp-In OtDUB-Flag cell lines.
HeLa Flp-In T-Rex cells were plated at 2.2x106 cells per 10 cm plate and 24 h later transfected with X-tremeGENE 9 (Roche) according to the manufacturer’s protocol. In brief, 1 μg of pCDNA5FRT/TO OtDUB-Flag variants was mixed with 9 μg pOG44 Flp Recombinase vector in 500 μL Opti-MEM and 10 μL transfection reagent. The mixture was incubated for 15 min at room temperature before being added dropwise to cells in antibiotic-free media. Forty-eight hours after plating, the media was replaced with complete medium containing 200 μg/mL hygromycin B. After selection, polyclonal cell lines were expanded and maintained in complete medium containing 150 μg/mL hygromycin B. HeLa Flp-In T-Rex cells (ThermoFisher) were maintained in complete media supplemented with 100 μg/mL zeocin.
O. tsutsugamushi culturing and infection.
O. tsutsugamushi str. Ikeda was maintained and passaged in HeLa cells as previously described (39). For infection experiments, O. tsutsugamushi organisms were obtained from highly infected (≥90%) HeLa cells and then introduced at a multiplicity of infection (MOI) of 10 into cell types of interest (39). RF/6A (77) and THP-1 cells (75) were infected as previously described. Two hours after infection, the medium was replaced with fresh media containing 1% (vol/vol) fetal bovine serum (Gemini BioSciences). Achievement of the desired MOI was confirmed by examining an aliquot of the infected culture at 2 to 3 h via immunofluorescence microscopy using TSA56 antibody as previously described (77).
RNA isolation and RT-qPCR.
Total RNA collected from HeLa cells was isolated using a Rneasy minikit (Qiagen). Samples were normalized, Dnase treated, synthesized to cDNA, and verified genomic DNA-free as described previously (36). qPCR was performed with cDNA as template using SsoFast EvaGreen supermix (Bio-Rad) and primers (5′-TTGATTGTGCAGTCCCGCTA-3′ and 5′-TGCAAAGTGTTCAGCGCAAG-3′, which are specific for the sense and antisense strands, respectively, of O. tsutsugamushi str. Ikeda otdub. Primers specific for human GAPDH (75) and O. tsutsugamushi ott16S (78) have been previously described. Thermal cycling conditions used were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 5 s. Bacterial load was assessed by normalizing ott16S gene levels to gapdh. The otdub expression profile was developed by normalizing relative otdub gene levels to ott16S using the 2−ΔΔCT method (79) in CFX Maestro for Mac 1.0 software package (Bio-Rad).
Antigen preparation.
Rosetta DE3 Escherichia coli transformed with pGEX6P-1 OtDUB1-311-C135A (29) were back diluted in Luria-Bertani (LB) medium supplemented with 100 μg/mL ampicillin and grown at 37°C to an OD600 of 0.5 to 0.6. Protein expression was induced with 300 μM isopropyl β-d-1-thiogalactopyranoside for 16 h at 18°C. The culture was then centrifuged, and the resulting bacterial pellet resuspended in ice-cold phosphate-buffered saline (PBS; 1.05 mM KH2PO4, 155 mM NaCl, 2.96 mM Na2HPO4, pH 7.4) + 400 mM KCl, 1 mM DTT, 2 mM PMSF, lysozyme, and Dnase. After incubating on ice for 1 h, bacteria were mechanically disrupted in a French press and the insoluble material pelleted by high-speed centrifugation. The resulting lysates were pumped onto a glutathione resin gravity column pre-equilibrated with PBS + 400 mM KCl. The column was washed thoroughly with PBS + 400 mM KCl before proteins were eluted with 250 mM Tris-HCl pH 8, 0.5 M KCl, 10 mM reduced glutathione. The N-terminal GST-tag was then cleaved with GST-HRV 3C during extensive dialysis in PBS + 400 mM KCl, 1 mM DTT. The free GST and GST-HRV 3C were captured over glutathione resin and the flow-through containing OtDUB1-311-C135A was concentrated by centrifugation through an Amicon Ultra with a 10 kDa cutoff (MilliporeSigma). The protein was further purified by size exclusion fast protein liquid chromatography (Akta, GE Amersham Pharmacia) on a Superdex 75 HiLoad 16/600 that was equilibrated with 50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT. OtDUB1-311-C135A main peak fractions were pooled and concentrated to 3 mg/mL as determined by A280. The protein was aliquoted, flash frozen in liquid N2, and stored at −80°C.
Antibody production.
OtDUB1-311-C135A antigen aliquots were shipped to Cocalico Biologicals (Stevens, PA) for their standard custom antibody protocol using Freund’s adjuvant to generate 2 polyclonal rabbit antisera, YU1430 and YU1431.
Antigen-based affinity purification of OtDUB antibodies.
Additional antigen was affinity purified as described above but the GST-tag was left intact to increase the number of available cysteine residues for conjugating to Sulfolink resin (ThermoFisher) according to the manufacturer’s protocol. In brief, 10 mg of GST-OtDUB1-311-C135A was diluted into coupling buffer (50 mM Tris-HCl pH 8.5, 5 mM EDTA) + 25 mM TCEP (tris[2-carboxyethyl]phosphine). After binding to 2 mL of resin, the column was washed with coupling buffer and unreacted iodoacetyl groups were quenched with free cysteine. The column was then stringently washed with 50 mM Tris-HCl, 1M NaCl followed by 0.1 M glycine pH 3.5 before washing and storing in PBS. For YU1430 and YU1431 each, bleeds 1 and 2 were blended and 20 mL was mixed with 1 mL of 20X PBS. The sera were then centrifuged to pellet any protein aggregates before being added to PBS-equilibrated GST-OtDUB1-311-C135A-Sulfolink columns. After binding, the columns were washed with PBS and the affinity purified antibodies were eluted with 1 mL fractions of 0.1 M glycine, pH 3.5. Eluates were immediately neutralized with 25 μL 1M Tris-HCl pH 8.0. Antibodies were buffer exchanged into PBS + 50% vol/vol glycerol in a 30 kDa cutoff concentrator (Amicon Ultra) and concentrated to ~1 mg/mL by A280. The antibody was aliquoted, flash frozen in liquid N2 and stored at −80°C.
Antibodies.
Generation of YU1430 and YU1431 are described above. Rabbit TSA56 antiserum (70) and rat OmpA antiserum (80) were described previously. Commercially available antibodies used in this study were mouse anti-Flag (Sigma-Aldrich; [F1804]), mouse anti-Hsp90 (Santa Cruz; [sc-13119]), mouse anti-GAPDH (Santa Cruz; [sc-365062]), mouse anti-AP1G1 (BD Bioscience; [610385]), mouse anti-AP2M1 (BD Bioscience; [611350]), mouse anti-Eps15 (Santa Cruz; [sc-390259]), rabbit anti-Eps15 (Cell Signaling Technology; [12460]), rabbit anti-Ston2 (Invitrogen; [PA5-51821]), mouse anti-Ston2 (Santa Cruz; [sc-514542]), and mouse anti-p65 (Santa Cruz; [sc-8008]).
Yeast culturing.
S. cerevisiae background strain W303 (MHY2416) (81) was grown using standard conditions; incubating at 30°C using rich (yeast extract-peptone-dextrose) or minimal (SD) media (82). S. cerevisiae was transformed with various p416GAL1-OtDUB variants (83) or empty p416GAL1. Individual isolated transformants were grown overnight in SD media-URA supplemented with Casamino Acids. Cultures were then back diluted into SD media-URA with 2% raffinose and grown overnight. Finally, the SD media-URA raffinose cultures were back diluted to an OD600 absorbance of 0.2 in SD media-URA + 2% galactose and induced for 8 h. The final OD600 absorbances ranged from 0.5 to 1.5 and culture volumes normalized to OD600 2.0 were processed by alkaline lysis, resuspended in Laemmli sample buffer + 4% β-mercaptoethanol (βME), and analyzed by immunoblot.
Immunoblotting.
For analyses of ectopically expressed proteins, samples were resolved by SDS-PAGE and transferred to PVDF membranes (Immobilon P; MilliporeSigma). Membranes were blocked in either 3% wt/vol bovine serum albumin (BSA) or 5% vol/vol non-fat milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T-0.1%). After blocking, blots were incubated in primary antibodies diluted in TBS-T-0.1% (1:5000 for YU1430, YU1431, and anti-Flag). Blots were thoroughly washed with TBS-T before incubation with goat anti-mouse or goat anti-rabbit horseradish-peroxidase (HRP)-conjugated secondary antibody (NA931V or NA934V, respectively, GE Healthcare), which was diluted 1:5000 in the same blocking buffer as the primary antibody for 1 to 2 h at room temperature. Blots were visualized by enhanced chemiluminescence on a G:Box imaging system (Syngene).
For analyses of infection experiments, uninfected and infected HeLa, THP-1, and RF/6A cells were lysed in modified radioimmunoprecipitation assay buffer (RIPA; 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA [pH 8]) containing Halt protease and phosphatase inhibitor cocktail. Protein lysates were normalized via Bradford Assay (Bio-Rad). Normalized lysates were resolved and transferred onto nitrocellulose as described previously (36). Resulting blots were blocked in 5% non-fat milk in Tris-buffered saline with 0.05% Tween 20 (TBS-T-0.05%) and screened with YU1430 (1:5000), YU1431 (1:2500), preimmune sera from each respective rabbit at either 1:5000 or 1:2500, TSA56 antiserum (1:1000), OmpA antiserum (1:750), anti-Hsp90 (1:200), and anti-GAPDH (1:750). Blots were washed with TBS-T-0.05% before incubation with HRP-conjugated horse anti-mouse, anti-rabbit, or anti-rat IgG secondary antibody diluted 1:10000 (Cell Signaling Technology) in 5% milk in TBS-T-0.05%. Blots were incubated with either SuperSignal West Pico PLUS, SuperSignal West Dura, or SuperSignal West Femto chemiluminescent substrate (ThermoFisher) prior to imaging in a ChemiDoc Touch Imaging System (Bio-Rad).
Indirect immunofluorescence microscopy.
HeLa cells were seeded onto glass coverslips in 24-well plates. For ectopic expression studies, cells were transiently transfected with 1 μg of pFlag-Ank6 (78) or pCDNA3.1(+)OtDUB-Flag using Lipofectamine 2000 (Invitrogen) for 18 h. At the time of collection, cells were fixed in −20°C methanol for 5 min, quenched with PBS, and blocked in 5% BSA in PBS. Coverslips of ectopic protein-expressing cells were incubated with mouse anti-Flag (1:1000) and rabbit YU1430 or YU1431 (1:500) in 5% BSA in PBS, washed three times in PBS, and incubated for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-rabbit IgG or Alexa Fluor 564-conjugated goat anti-mouse IgG secondary antibody in 5% BSA (ThermoFisher; 1:10,000). Coverslips of cells ectopically expressing fusion proteins were imaged with an Olympus BX51 spinning disc confocal microscope (Olympus).
For OtDUB localization experiments, cells were infected with O. tsutsugamushi at an MOI of 10 for 24 h. At the time of collection, cells were fixed in −20°C methanol for 5 min, quenched with PBS, and blocked in 5% BSA in PBS. Coverslips were incubated with anti-TSA56 (1:1000), OmpA antiserum (1:500), or YU1430 (1:500) in 5% BSA in PBS, washed three times in PBS, and incubated for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-rabbit IgG or Alexa Fluor 564-conjugated goat anti-mouse IgG secondary antibody in 5% BSA (ThermoFisher; 1:10,000). Series of optical sections were acquired of infected cells using a structured illumination microscope (Nikon N-SIM controlled with NIS-Elements AR), equipped with a 100x Plan Apo oil immersion objective (NA 1.45) and an Andor iXon EMCCD camera (Oxford Instruments). Fluorescence of DAPI was excited with 405 nm (diode laser) whereas that of Alexa Fluor fluorophores with 488 nm diode and 561 nm Diode Pumped Solid State (DPSS) lasers, respectively. Fluorescence emission was detected sequentially in 420 – 475 nm, 500 – 550 nm and 575 – 620 nm range, respectively. Acquisition time was set between 300 and 450 ms and EM gain between 250 and 285, depending on the fluorophore. Pixel size of the raw data (15 images per section) was 64 nm, whereas the z-step was 100 nm. The 3D images were reconstructed with default settings (linear weighting of spatial frequencies). The same acquisition and processing settings were used to produce images of 110 nm beads (3 color channels), then in turn were used to reconstruct the Point spread Function (PSF) of the system with PSF distiller function of Scientific Volume Imaging (SVI) Huygens. This experimental PSF was then used for deconvolution of the reconstructed 3D SIM images with Classic Maximum Likelihood Estimation (CMLE) algorithm, signal-to-noise ratio of 12 – 16 and 15 – 20 iterations, depending on the fluorophore. Maximum intensity projections (nominal depth of 300 nm) were generated with SVI Huygens. The images were equally adjusted for brightness and contrast across all conditions based on visual signal intensity in ImageJ-Fiji 565 (NIH) (84). Line-profile analyses were performed on a single z-section using ImageJ and relative signal intensity was generated for TSA56, OmpA, and OtDUB for selected bacteria by normalizing values to the highest fluorescence intensity for each individual channel.
Treatment of host cell-free O. tsutsugamushi with trypsin or acetic acid.
HeLa cells infected with O. tsutsugamushi were mechanically disrupted using glass beads. Differential centrifugation was used to isolate the bacteria from host cells and debris (39). Isolated bacteria were washed with PBS twice and subjected to one of the following conditions: 2.5% (vol/vol) HyClone Trypsin Protease (Sigma-Aldrich) in PBS, 0.25% (vol/vol) HyClone Trypsin Protease in PBS, or 50 mM acetic acid. Treatment was performed at 35°C for 30 min with intermittent shaking. Reactions were quenched with RPMI media containing 10% (vol/vol) FBS, spun down at 10000 × g for 10 min, washed with PBS twice, and lysed in RIPA containing Halt protease and phosphatase inhibitor cocktail. Uninfected cells were processed in parallel with PBS as a control for verifying the absence of host cell debris in the bacterial lysates. Protein lysates were normalized, resolved via SDS-PAGE, and Western-blotted as described above. Blots were blocked in 5% milk in TBS-T-0.05% prior to being screened with YU1430 (1:5000), TSA56 antiserum (1:1000), OmpA antiserum (1:750), anti-Hsp90 (1:200), or anti-GAPDH (1:750). Blots were probed with secondary antibodies and imaged as described above.
Sample preparation for mass spectrometry.
HeLa cells infected with O. tsutsugamushi at an MOI of 10 were harvested by tryptic detachment at 72 h. Cells were pelleted and washed once in PBS. The washed pellet was resuspended in 20 mL lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Triton X-100) containing Halt protease and phosphatase inhibitor cocktail. After lysing on ice for 1 h with intermittent vortexing, the insoluble fraction was pelleted by high-speed centrifugation (16000 × g, 10 min, 4°C). The clarified lysates were quantified by Bradford analysis to be 5.5 mg mL−1. The sample was split in two with 9.5 mL of lysate added to 15 mL conical tubes containing 100 μL of packed Protein A/G resin (ThermoFisher) and either 40 μL of 1 mg mL−1 rabbit IgG or combined YU1431 serum bleeds 1 and 2. Samples were rotated overnight at 4°C. After pelleting, the supernatants were aspirated, and the resin pellets resuspended in 1 mL of lysis buffer and transferred to 1.5-mL microfuge tubes. The resin was washed three more times with 1 mL of lysis buffer. Proteins were eluted from the resin with 0.3 mL of 0.2 M glycine pH 2.6, 300 mM NaCl by rotating for 10 min at room temperature. After centrifugation, the eluates were carefully removed from the pelleted resin and neutralized with 50 μL of 1M Tris-HCl pH 8.5. Samples were stored at −80°C until processed for mass spectrometry.
In-solution protein digestion.
Eluted proteins were precipitated by the addition of 4 volumes of ice-cold acetone and storage at −20°C overnight, followed by centrifugation at 14,600 rpm for 10 min at 4°C. The supernatant was discarded, and the pellets allowed to air dry for 5 min. Protein pellets were dissolved and denatured in 8 M urea, 0.4 M ammonium bicarbonate, pH 8. The proteins were reduced by the addition of 1/10 volume of 45 mM dithiothreitol and incubated at 37°C for 30 min, then alkylated with the addition of 1/10 volume of 100 mM iodoacetamide following incubation in the dark at room temperature for 30 min. The urea concentration was adjusted to 2 M by the addition of water prior to enzymatic digestion at 37°C with trypsin for 16 h. Protease:protein ratios were estimated at 1:50. Samples were acidified by the addition of 1/40 volume of 20% trifluoroacetic acid, then desalted using BioPureSPN PROTO 300 C18 Macro spin columns (The Nest Group [#HMM S18V]) following the manufacturer’s directions to elute peptides with 0.1% TFA (vol/vol), 80% acetonitrile (vol/vol) into water. Eluted peptides were lyophilized in a Speed Vac and dissolved in MS loading buffer (2% acetonitrile, 0.2% trifluoroacetic acid in water). Protein concentrations were determined via NanoDrop measurement (A260/A280). Each sample was then further diluted with MS loading buffer to 0.05 μg/μL, with 0.25 μg (5 μL) injected for LC-MS/MS analysis.
LC-MS/MS analysis and peptide identification.
LC-MS/MS analysis was performed on a Thermo Scientific Q Exactive Plus equipped with a Waters nanoAcquity UPLC system utilizing a binary solvent system (A: 100% water, 0.1% formic acid; B: 100% acetonitrile, 0.1% formic acid). Trapping was performed at 5 μL min−1, 99.5% Buffer A for 3 min using a Waters ACQUITY UPLC M-Class Symmetry C18 Trap Column (100 Å, 5 μm, 180 μm × 20 mm, 2G, V/M). Peptides were separated at 37°C using a Waters ACQUITY UPLC M-Class Peptide BEH C18 Column (130 Å, 1.7 μm, 75 μm × 250 mm) and eluted at 300 NI min−1 with the following gradient: 3% buffer B at initial conditions; 5% B at 1 min; 25% B at 90 min; 50% B at 110 min; 90% B at 115 min; 90% B at 120 min; return to initial conditions at 125 min. MS was acquired in profile mode over the 300-1,700 m/z range using 1 microscan, 70,000 resolution, AGC target of 3E6, and a maximum injection time of 45 ms. Data dependent MS/MS were acquired in centroid mode on the top 20 precursors per MS scan using 1 microscan, 17,500 resolution, AGC target of 1E5, maximum injection time of 100 ms, and an isolation window of 1.7 m/z. Precursors were fragmented by higher energy collisional dissociation activation with a collision energy of 28%. MS/MS were collected on species with an intensity threshold of 1E4, charge states 2 to 6, and peptide match preferred. Dynamic exclusion was set to 20 s.
Data were analyzed using Proteome Discoverer software (version 2.2.0.388, Thermo Scientific). Data searching was performed using the Mascot algorithm (version 2.6.1, Matrix Science) against a custom database containing protein sequences for the OtDUB constructs of interest, the O. tsutsugamushi str. Ikeda proteome ([AP008981] derived from the NCBI-nr database), and E. coli and human proteomes (derived from the Swissprotein database). The search parameters included tryptic digestion with up to 2 missed cleavages, 10 ppm precursor mass tolerance and 0.02 Da fragment mass tolerance, and variable (dynamic) modifications of methionine oxidation and carbamidomethylated cysteine. Normal and decoy database searches were run with the confidence level set to 95% (P < 0.05). Scaffold (version 4.11.0, Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides.
Mass spectrometry data analysis.
Recovered MS data was sorted using a scoring system (F-score), which is the number of peptides from the OtDUB IP divided by the sum of peptides from both IgG and OtDUB IP (85). The resulting F-score is a ratio from 0 to 1, where 1 represents a protein only found in the OtDUB IP and 0 is a protein only found in the IgG IP. To identify potential interactors, the F-score threshold was set to ≥ 0.75. To further increase the stringency of our putative interactor list, only proteins with ≥ 4 identified peptides being enriched in the OtDUB IP over the IgG IP sample were included. STRING (55) was used to analyze protein-protein interaction networks among the high potential OtDUB interacting partners (https://string-db.org/cgi/input.pl).
Immunoprecipitation.
Uninfected and O. tsutsugamushi-infected HeLa or RF/6A cells were scraped down in ice-cold PBS and subsequently lysed in Flag IP Buffer (50 mM Tris-HCl pH 7.4, 400 mM NaCl, 1 mM EDTA, 1.0% Triton X-100) containing Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). After lysis on ice, the insoluble fraction was pelleted by centrifugation at 2793 × g for 10 min at 4°C. The clarified lysates were quantified by the Bradford assay (Bio-Rad). Normalized HeLa lysates (0.8 mL at 1.0 mg/mL with 4 μL of affinity purified YU1430) and RF/6A lysates (0.4 mL at 1.0 mg/mL with 2 μL of affinity purified YU1430) were incubated with Protein A/G resin (ThermoFisher). Samples were rotated overnight at 4°C before being washed in Flag IP buffer. Proteins were eluted with Laemmli buffer containing 4% βME and resolved by SDS-PAGE next to input lysates and immunoblotted as described above. Resulting blots were blocked in 3% BSA in TBS-T-0.1% and screened with anti-AP1G1 (1:500), anti-AP2M1 (1:500), or blocked in 5% non-fat milk in TBS-T-0.05% and screened with anti-Eps15 (mouse 1:500; rabbit 1:1000), anti-Ston2 (mouse 1:100; rabbit 1:1000), or anti-p65 (1:200).
Statistical analyses.
Statistical analyses were performed using the Prism 8.0 software package (GraphPad). One-way ANOVA with Tukey’s post hoc test was used to test for significant differences between groups. Statistical significance was set at P-values of < 0.05.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases (https://www.niaid.nih.gov/) grants 1R01 AI123346, 2R56 AI123346, 1R01 AI167857, and R21 AI152513 to J.A.C., GM136325 to M.H., and American Heart Association grant 20PRE35210610 to H.E.A. Mass spectrometry data collection and analysis was performed by the Mass Spectrometry (MS) & Proteomics Resource of the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University, supported by NIH S10 (SIG) OD018034. Microscopy of infected samples was performed at the VCU Microscopy Facility, which is supported, in part, by funding from NIH-NCI Cancer Center Support Grant P30 CA016059.
Footnotes
This article is a direct contribution from Jason A. Carlyon, a member of the Infection and Immunity Editorial Board, who arranged for and secured reviews by Jere McBride, University of Texas Medical Branch at Galveston, and Alison Luce-Fedrow, Shippensburg University.
Supplemental material is available online only.
Contributor Information
Mark Hochstrasser, Email: mark.hochstrasser@yale.edu.
Jason A. Carlyon, Email: jason.carlyon@vcuhealth.org.
Andreas J. Bäumler, University of California, Davis
REFERENCES
- 1.Rapmund G. 1984. Rickettsial diseases of the Far East: new perspectives. J Infect Dis 149:330–338. 10.1093/infdis/149.3.330. [DOI] [PubMed] [Google Scholar]
- 2.Richards AL, Soeatmadji DW, Widodo MA, Sardjono TW, Yanuwiadi B, Hernowati TE, Baskoro AD, Roebiyoso Hakim L, Soendoro M, Rahardjo E, Putri MP, Saragih JM, Strickman D, Kelly DJ, Dasch GA, Olson JG, Church CJ, Corwin AL. 1997. Seroepidemiologic evidence for murine and scrub typhus in Malang, Indonesia. Am J Trop Med Hyg 57:91–95. 10.4269/ajtmh.1997.57.91. [DOI] [PubMed] [Google Scholar]
- 3.Kelly DJ, Richards AL, Temenak J, Strickman D, Dasch GA. 2002. The past and present threat of rickettsial diseases to military medicine and international public health. Clin Infect Dis 34:S145–S169. 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AJ, Paris DH, Newton PN. 2015. A systematic review of mortality from untreated scrub typhus (Orientia tsutsugamushi). PloS Negl Trop Dis 9:e0003971. 10.1371/journal.pntd.0003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly DJ, Fuerst PA, Ching W-M, Richards AL. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. Clin Infect Dis 48 Suppl 3:S203–S230. 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 6.Abarca K, Weitzel T, Martinez-Valdebenito C, Acosta-Jamett G. 2018. Scrub typhus, an emerging infectious disease in Chile. Rev Chilena Infectol 35:696–699. 10.4067/S0716-10182018000600696. [DOI] [PubMed] [Google Scholar]
- 7.Balcells ME, Rabagliati R, García P, Poggi H, Oddó D, Concha M, Abarca K, Jiang J, Kelly DJ, Richards AL, Fuerst PA. 2011. Endemic scrub typhus-like illness, Chile. Emerg Infect Dis 17:1659–1663. 10.3201/eid1709.100960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghorbani RP, Ghorbani AJ, Jain MK, Walker DH. 1997. A case of scrub typhus probably acquired in Africa. Clin Infect Dis 25:1473–1474. 10.1086/516990. [DOI] [PubMed] [Google Scholar]
- 9.Osuga K, Kimura M, Goto H, Shimada K, Suto T. 1991. A case of Tsutsugamushi disease probably contracted in Africa. Eur J Clin Microbiol Infect Dis 10:95–96. 10.1007/BF01964418. [DOI] [PubMed] [Google Scholar]
- 10.Weitzel T, Martinez-Valdebenito C, Acosta-Jamett G, Jiang J, Richards AL, Abarca K. 2019. Scrub typhus in Continental Chile, 2016-2018(1). Emerg Infect Dis 25:1214–1217. 10.3201/eid2506.181860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciceroni L, Pinto A, Ciarrocchi S, Ciervo A. 2006. Current knowledge of rickettsial diseases in Italy. Ann N Y Acad Sci 1078:143–149. 10.1196/annals.1374.024. [DOI] [PubMed] [Google Scholar]
- 12.Cracco C, Delafosse C, Baril L, Lefort Y, Morelot C, Derenne JP, Bricaire F, Similowski T. 2000. Multiple organ failure complicating probable scrub typhus. Clin Infect Dis 31:191–192. 10.1086/313906. [DOI] [PubMed] [Google Scholar]
- 13.Dupon M, Rogues AM, Malou M, d’Ivernois C, Lacut JY. 1992. Scrub typhus: an imported rickettsial disease. Infection 20:153–154. 10.1007/BF01704608. [DOI] [PubMed] [Google Scholar]
- 14.Edlinger E, Navarro P. 1983. [Rickettsioses. A disease of tourism]. Sem Hop 59:2053–2054. [PubMed] [Google Scholar]
- 15.Eisermann P, Rauch J, Reuter S, Eberwein L, Mehlhoop U, Allartz P, Muntau B, Tappe D. 2020. Complex cytokine responses in imported scrub typhus cases, Germany, 2010-2018. Am J Trop Med Hyg 102:63–68. 10.4269/ajtmh.19-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensenius M, Montelius R, Berild D, Vene S. 2006. Scrub typhus imported to Scandinavia. Scand J Infect Dis 38:200–202. 10.1080/00365540500277342. [DOI] [PubMed] [Google Scholar]
- 17.Mahdi AS, Al-Khalili SM, Chung CC, Molai M, Ibrahim H, Eskild P, Khamis F, Nenad P. 2019. Scrub typhus complicated by ARDS, myocarditis, and encephalitis imported to Oman from Nepal. Oman Med J 34:254–256. 10.5001/omj.2019.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maher PH. 1976. Imported scrub typhus. Conn Med 40:377–378. [PubMed] [Google Scholar]
- 19.Marschang A, Nothdurft HD, Kumlien S, von Sonnenburg F. 1995. Imported rickettsioses in German travelers. Infection 23:94–97. 10.1007/BF01833873. [DOI] [PubMed] [Google Scholar]
- 20.McDonald JC, MacLean JD, McDade JE. 1988. Imported rickettsial disease: clinical and epidemiologic features. Am J Med 85:799–805. 10.1016/s0002-9343(88)80024-x. [DOI] [PubMed] [Google Scholar]
- 21.Nachega JB, Bottieau E, Zech F, Van Gompel A. 2007. Travel-acquired scrub typhus: emphasis on the differential diagnosis, treatment, and prevention strategies. J Travel Med 14:352–355. 10.1111/j.1708-8305.2007.00151.x. [DOI] [PubMed] [Google Scholar]
- 22.Vliegenthart-Jongbloed K, de Mendonca Melo M, Slobbe L, Beersma MF, van Genderen PJ. 2013. Imported scrub typhus in The Netherlands. Travel Med Infect Dis 11:197–199. 10.1016/j.tmaid.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Watt G, Strickman D. 1994. Life-threatening scrub typhus in a traveler returning from Thailand. Clin Infect Dis 18:624–626. 10.1093/clinids/18.4.624. [DOI] [PubMed] [Google Scholar]
- 24.Pen SJ, X M. 1948. Characterization of Rickettsia tsutsugamushi from scrub typhus patients in Guangzhou isolated in mice. Acta Medicinae Zhongsh 35. [Google Scholar]
- 25.Bl L. 1952. Larvae of Leptotrombicula deliensis in Guangzhou and report of a simple method of rearing chiggers. Chin Med J 38:759–765. [Google Scholar]
- 26.Salje J. 2021. Cells within cells: rickettsiales and the obligate intracellular bacterial lifestyle. Nat Rev Microbiol 19:375–390. 10.1038/s41579-020-00507-2. [DOI] [PubMed] [Google Scholar]
- 27.Paris DH, Phetsouvanh R, Tanganuchitcharnchai A, Jones M, Jenjaroen K, Vongsouvath M, Ferguson DP, Blacksell SD, Newton PN, Day NP, Turner GD. 2012. Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PloS Negl Trop Dis 6:e1466. 10.1371/journal.pntd.0001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tantibhedhyangkul W, Prachason T, Waywa D, El Filali A, Ghigo E, Thongnoppakhun W, Raoult D, Suputtamongkol Y, Capo C, Limwongse C, Mege J-L. 2011. Orientia tsutsugamushi stimulates an original gene expression program in monocytes: relationship with gene expression in patients with scrub typhus. PloS Negl Trop Dis 5:e1028. 10.1371/journal.pntd.0001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berk JM, Lim C, Ronau JA, Chaudhuri A, Chen H, Beckmann JF, Loria JP, Xiong Y, Hochstrasser M. 2020. A deubiquitylase with an unusually high-affinity ubiquitin-binding domain from the scrub typhus pathogen Orientia tsutsugamushi. Nat Commun 11:2343. 10.1038/s41467-020-15985-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim C, Berk JM, Blaise A, Bircher J, Koleske AJ, Hochstrasser M, Xiong Y. 2020. Crystal structure of a guanine nucleotide exchange factor encoded by the scrub typhus pathogen Orientia tsutsugamushi. Proc Natl Acad Sci USA 117:30380–30390. 10.1073/pnas.2018163117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berk JM, Lee MJ, Zhang M, Lim C, Hochstrasser M. 2022. OtDUB from the human pathogen Orientia tsutsugamushi modulates host membrane trafficking by multiple mechanisms. Mol Cell Biol 42:e0007122. 10.1128/mcb.00071-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Berk JM, Mehrtash AB, Kanyo J, Hochstrasser M. 2022. A versatile new tool derived from a bacterial deubiquitylase to detect and purify ubiquitylated substrates and their interacting proteins. PloS Biol 20:e3001501. 10.1371/journal.pbio.3001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, Ooka T, Oshima K, Tamura A, Hattori M, Hayashi T. 2008. The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res 15:185–199. 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura A, Takahashi K, Tsuruhara T, Urakami H, Miyamura S, Sekikawa H, Kenmotsu M, Shibata M, Abe S, Nezu H. 1984. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol Immunol 28:873–882. 10.1111/j.1348-0421.1984.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 35.Izzard L, Fuller A, Blacksell SD, Paris DH, Richards AL, Aukkanit N, Nguyen C, Jiang J, Fenwick S, Day NPJ, Graves S, Stenos J. 2010. Isolation of a novel Orientia species (O. chuto sp. Nov.) from a patient infected in Dubai. J Clin Microbiol 48:4404–4409. 10.1128/JCM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adcox HE, Hatke AL, Andersen SE, Gupta S, Otto NB, Weber MM, Marconi RT, Carlyon JA. 2021. Orientia tsutsugamushi nucleomodulin Ank13 exploits the RaDAR nuclear import pathway to modulate host cell transcription. mBio 12:e01816-21. 10.1128/mBio.01816-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodino KG, VieBrock L, Evans SM, Ge H, Richards AL, Carlyon JA. 2018. Orientia tsutsugamushi modulates endoplasmic reticulum-associated degradation to benefit its growth. Infect Immun 86:e00596-17. 10.1128/IAI.00596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giengkam S, Blakes A, Utsahajit P, Chaemchuen S, Atwal S, Blacksell SD, Paris DH, Day NPJ, Salje J. 2015. Improved quantification, propagation, purification and storage of the obligate intracellular human pathogen Orientia tsutsugamushi. PloS Negl Trop Dis 9:e0004009. 10.1371/journal.pntd.0004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans SM, Rodino KG, Adcox HE, Carlyon JA. 2018. Orientia tsutsugamushi uses two Ank effectors to modulate NF-κB p65 nuclear transport and inhibit NF-κB transcriptional activation. PloS Pathog 14:e1007023. 10.1371/journal.ppat.1007023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen HW, Zhang Z, Huber E, Mutumanje E, Chao CC, Ching WM. 2011. Kinetics and magnitude of antibody responses against the conserved 47-kilodalton antigen and the variable 56-kilodalton antigen in scrub typhus patients. Clin Vaccine Immunol 18:1021–1027. 10.1128/CVI.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H, Lee J-H, Han S-H, Kim S-Y, Cho N-H, Kim I-S, Choi M-S. 2006. Exploitation of the endocytic pathway by Orientia tsutsugamushi in nonprofessional phagocytes. Infect Immun 74:4246–4253. 10.1128/IAI.01620-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge Y, Rikihisa Y. 2011. Subversion of host cell signaling by Orientia tsutsugamushi. Microbes Infect 13:638–648. 10.1016/j.micinf.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Allen JR, Ross ST, Davidson MW. 2014. Structured illumination microscopy for superresolution. Chemphyschem 15:566–576. 10.1002/cphc.201301086. [DOI] [PubMed] [Google Scholar]
- 44.Ojogun N, Kahlon A, Ragland SA, Troese MJ, Mastronunzio JE, Walker NJ, Viebrock L, Thomas RJ, Borjesson DL, Fikrig E, Carlyon JA. 2012. Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect Immun 80:3748–3760. 10.1128/IAI.00654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidman D, Ojogun N, Walker NJ, Mastronunzio J, Kahlon A, Hebert KS, Karandashova S, Miller DP, Tegels BK, Marconi RT, Fikrig E, Borjesson DL, Carlyon JA. 2014. Anaplasma phagocytophilum surface protein AipA mediates invasion of mammalian host cells. Cell Microbiol 16:1133–1145. 10.1111/cmi.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahlon A, Ojogun N, Ragland SA, Seidman D, Troese MJ, Ottens AK, Mastronunzio JE, Truchan HK, Walker NJ, Borjesson DL, Fikrig E, Carlyon JA. 2013. Anaplasma phagocytophilum Asp14 is an invasin that interacts with mammalian host cells via its C terminus to facilitate infection. Infect Immun 81:65–79. 10.1128/IAI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX. 2006. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci 15:122–134. 10.1110/ps.051616206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabilloud T. 2003. Membrane proteins ride shotgun. Nat Biotechnol 21:508–510. 10.1038/nbt0503-508. [DOI] [PubMed] [Google Scholar]
- 49.Zhong H, Marcus SL, Li L. 2005. Microwave-assisted acid hydrolysis of proteins combined with liquid chromatography MALDI MS/MS for protein identification. J Am Soc Mass Spectrom 16:471–481. 10.1016/j.jasms.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 50.Hillman RD, Jr, Baktash YM, Martinez JJ. 2013. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with alpha2beta1 integrin. Cell Microbiol 15:727–741. 10.1111/cmi.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ha NY, Kim Y, Min CK, Kim HI, Yen NTH, Choi MS, Kang JS, Kim YS, Cho NH. 2017. Longevity of antibody and T-cell responses against outer membrane antigens of Orientia tsutsugamushi in scrub typhus patients. Emerg Microbes Infect 6:e116. 10.1038/emi.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CC, Chou CH, Lin TC, Yang MC, Cho CL, Chang CH, Yu HS, Lai CH, Chen LK, Hong YR. 2012. Molecular characterization of three major outer membrane proteins, TSA56, TSA47 and TSA22, in Orientia tsutsugamushi. Int J Mol Med 30:75–84. 10.3892/ijmm.2012.967. [DOI] [PubMed] [Google Scholar]
- 53.Sharma D, Sharma A, Singh B, Verma SK. 2019. Bioinformatic exploration of metal-binding proteome of zoonotic pathogen Orientia tsutsugamushi. Front Genet 10:797. 10.3389/fgene.2019.00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma D, Kumar S, Sharma A, Kumar R, Kumar R, Kulharia M, Kumar M, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, District Kangra, Himachal Pradesh, India — 176206 . 2022. Functional assignment to hypothetical proteins in Orientia tsutsugamushi strain Ikeda. Biomedical Informatics 18:188–195. 10.6026/97320630018188. [DOI] [Google Scholar]
- 55.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beacham GM, Partlow EA, Hollopeter G. 2019. Conformational regulation of AP1 and AP2 clathrin adaptor complexes. Traffic 20:741–751. 10.1111/tra.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo T, Dunphy PS, McBride JW. 2017. Ehrlichia chaffeensis tandem repeat effector targets differentially influence infection. Front Cell Infect Microbiol 7:178. 10.3389/fcimb.2017.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakeel A, den Dulk-Ras A, Hooykaas PJ, McBride JW. 2011. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front Cell Infect Microbiol 1:22. 10.3389/fcimb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunphy PS, Luo T, McBride JW. 2014. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun 82:4154–4168. 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitra S, Dunphy PS, Das S, Zhu B, Luo T, McBride JW. 2018. Ehrlichia chaffeensis TRP120 effector targets and recruits host polycomb group proteins for degradation to promote intracellular infection. Infect Immun 86:e00845-17. 10.1128/IAI.00845-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JY, Zhu B, Patterson LL, Rogan MR, Kibler CE, McBride JW. 2020. Ehrlichia chaffeensis TRP120-mediated ubiquitination and proteasomal degradation of tumor suppressor FBW7 increases oncoprotein stability and promotes infection. PloS Pathog 16:e1008541. 10.1371/journal.ppat.1008541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popov VL, Yu X, Walker DH. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28:71–80. 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 63.Villar M, Ayllon N, Kocan KM, Bonzon-Kulichenko E, Alberdi P, Blouin EF, Weisheit S, Mateos-Hernandez L, Cabezas-Cruz A, Bell-Sakyi L, Vancova M, Bily T, Meyer DF, Sterba J, Contreras M, Rudenko N, Grubhoffer L, Vazquez J, de la Fuente J. 2015. Identification and characterization of Anaplasma phagocytophilum proteins involved in infection of the tick vector, Ixodes scapularis. PloS One 10:e0137237. 10.1371/journal.pone.0137237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martina JA, Bonangelino CJ, Aguilar RC, Bonifacino JS. 2001. Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J Cell Biol 153:1111–1120. 10.1083/jcb.153.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci USA 110:E4770–E4779. 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klapisz E, Sorokina I, Lemeer S, Pijnenburg M, Verkleij AJ, van Bergen En Henegouwen PM. 2002. A ubiquitin-interacting motif (UIM) is essential for Eps15 and Eps15R ubiquitination. J Biol Chem 277:30746–30753. 10.1074/jbc.M203004200. [DOI] [PubMed] [Google Scholar]
- 67.van Bergen En Henegouwen PM. 2009. Eps15: a multifunctional adaptor protein regulating intracellular trafficking. Cell Commun Signal 7:24. 10.1186/1478-811X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451–455. 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 69.Choi JH, Cheong TC, Ha NY, Ko Y, Cho CH, Jeon JH, So I, Kim IK, Choi MS, Kim IS, Cho NH. 2013. Orientia tsutsugamushi subverts dendritic cell functions by escaping from autophagy and impairing their migration. PloS Negl Trop Dis 7:e1981. 10.1371/journal.pntd.0001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beyer AR, Rodino KG, VieBrock L, Green RS, Tegels BK, Oliver LD, Marconi RT, Carlyon JA. 2017. Orientia tsutsugamushi Ank9 is a multifunctional effector that utilizes a novel GRIP-like Golgi localization domain for Golgi-to-endoplasmic reticulum trafficking and interacts with host COPB2. Cell Microbiol 19:e12727. 10.1111/cmi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barr FA. 1999. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol 9:381–384. 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- 72.Pruneda JN, Bastidas RJ, Bertsoulaki E, Swatek KN, Santhanam B, Clague MJ, Valdivia RH, Urbe S, Komander D. 2018. A chlamydia effector combining deubiquitination and acetylation activities induces Golgi fragmentation. Nat Microbiol 3:1377–1384. 10.1038/s41564-018-0271-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hermanns T, Hofmann K. 2019. Bacterial DUBs: deubiquitination beyond the seven classes. Biochem Soc Trans 47:1857–1866. 10.1042/BST20190526. [DOI] [PubMed] [Google Scholar]
- 74.Baumann K. 2014. Post-translational modifications: Lys33-linked ubiquitin in post-Golgi transport. Nat Rev Mol Cell Biol 15:365. 10.1038/nrm3812. [DOI] [PubMed] [Google Scholar]
- 75.Rodino KG, Adcox HE, Martin RK, Patel V, Conrad DH, Carlyon JA. 2019. The obligate intracellular bacterium Orientia tsutsugamushi targets NLRC5 to modulate the major histocompatibility complex Class I pathway. Infect Immun 87:e00876-18. 10.1128/IAI.00876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, Borjesson DL, Carlyon JA. 2010. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb Pathog 49:273–284. 10.1016/j.micpath.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wangsanut T, Brann KR, Adcox HE, Carlyon JA. 2021. Orientia tsutsugamushi modulates cellular levels of NF-kappaB inhibitor p105. PloS Negl Trop Dis 15:e0009339. 10.1371/journal.pntd.0009339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, Singh S, Rodino KG, Heinzen RA, Richards AL, Carlyon JA. 2015. Orientia tsutsugamushi ankyrin repeat-containing protein family members are Type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol 4:186. 10.3389/fcimb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 80.Beyer AR, VieBrock L, Rodino KG, Miller DP, Tegels BK, Marconi RT, Carlyon JA. 2015. Orientia tsutsugamushi strain Ikeda ankyrin repeat-containing proteins recruit SCF1 ubiquitin ligase machinery via poxvirus-like F-box motifs. J Bacteriol 197:3097–3109. 10.1128/JB.00276-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas BJ, Rothstein R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619–630. 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 82.Guthrie C, Fink GR. 1991. Guide to Yeast Genetics and Molecular Biology: Methods in Enzymology, vol 194. Academic Press, San Diego. [PubMed] [Google Scholar]
- 83.Mumberg D, Muller R, Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768. 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beckmann JF, Sharma GD, Mendez L, Chen H, Hochstrasser M. 2019. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. Elife 8:e50026. 10.7554/eLife.50026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download iai.00469-22-s0001.xlsx, XLSX file, 0.1 MB (61.1KB, xlsx)