ABSTRACT
This report describes genome sequences for nine Listeria innocua strains that varied in hemolytic phenotypes on sheep blood agar. All strains were sequenced using Pacific Biosciences (PacBio) single-molecule real-time (SMRT) chemistry; overall, the average read length of these sequences was 2,869,880 bp, with an average GC content of 37%.
ANNOUNCEMENT
The Listeria genus represents a diverse group of Gram-positive organisms that are commonly found in environmental reservoirs. Public health focuses on Listeria monocytogenes, which can cause fatal infections in vulnerable human populations (1, 2). Unlike L. monocytogenes, Listeria innocua typically is nonpathogenic but in rare cases has been linked to human infections (3, 4). Genotypic characterization has identified L. innocua as a close relative of L. monocytogenes (2, 4–6). Although L. innocua is typically nonhemolytic, some isolates within the species exhibit β-hemolysis on blood agar (2, 5, 7, 8). These atypical strains also share virulence factors with L. monocytogenes (2, 5, 7). A genomic assessment was performed with nine L. innocua strains that had been isolated from various sources and were archived at the National Listeria Reference Laboratory at the Centers for Disease Control and Prevention (CDC). Here, we present Pacific Biosciences (PacBio) sequences of nine L. innocua isolates that were received between 1987 and 2015.
Genomic DNA was extracted from single-colony isolates using the DNeasy blood and tissue kit (Qiagen, Germany) after overnight growth on Trypticase soy agar II with 5% sheep blood (BD, Germany) at 36°C. Samples were assessed for quality (5 to 10 μg DNA in 100 μL buffer) for submission for PacBio sequencing. The Biotechnology Core Facility Branch at the CDC performed sequencing and raw read analysis.
Genomic DNA was sheared to 10 kb or 20 kb using needle shearing. Plasmids were identified as contigs of <0.5 MB, consistent with standard threshold cutoff values. Contigs from strains H0996, 2010L-1951, and 2010L-2059 were linear; all other contigs were circularized. Default parameters were used for all software unless otherwise specified. Libraries were generated with the SMRTbell template preparation kit v1.0 (PacBio, Menlo Park, CA). The 2012L-5520 and 2015L-6726 sample libraries were size selected to 8 kb with the BluePippin system (Sage Science, Beverly, MA). All libraries were bound to polymerase using the DNA/polymerase binding kit P6 v2 (PacBio), loaded on one single-molecule real-time (SMRT) Cell (PacBio), and sequenced with C4 v2 chemistry (PacBio) for 270- or 360-min movies, depending on the length, with an RS II instrument (PacBio). Sequence assembly was performed on the HGAP v3 platform (9), with an average yield of 2,869,880 bp.
A phylogenetic tree was inferred on the BioLinux command line using Parsnp v1.2 (10); it included 30 published sequences from the NCBI database annotated as Listeria innocua and was visualized in MEGA7 (6) (Fig. 1). The level of base alignment with the reference genome (H0996) was 73.8%, as assessed with Gingr v1.3 (10).
FIG 1.
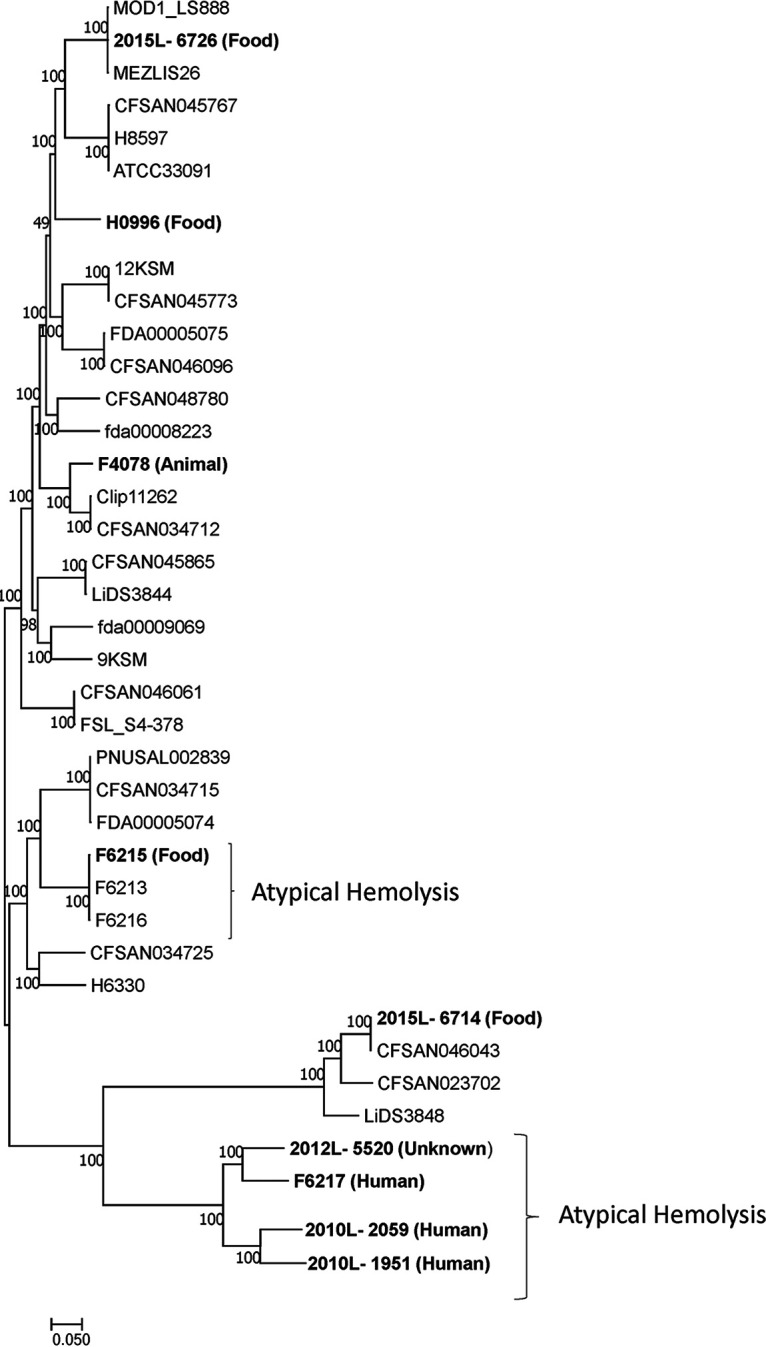
Phylogenetic tree based on the full genome sequences for 39 L. innocua isolates. PacBio sequences for the 9 isolates featured in this study are shown in bold, and their sources are listed in parentheses. Strains exhibiting the atypical hemolysis phenotype are indicated by brackets. The phylogeny was constructed by Parsnp using H0996 as the reference genome. Bootstrap branch support values are depicted beside the root and branches.
All isolates that exhibited β-hemolysis were in two distinct clades (Fig. 1). Isolate 2012L-5520 (FSL J1-023) is of unknown origin, F6215 from a food source, and F6217, 2010L-2059, and 2010L-1951 from human infections (Table 1). The description of these atypical strains confirms that L. innocua can contribute to human illness. These genome sequences serve as useful resources for additional characterization of the evolutionary mechanisms of Listeria.
TABLE 1.
Summary of genomic characteristics from PacBio sequencing for the L. innocua isolates
| Isolate | Host (source) | Depth of coverage (×) | No. of reads | Avg read length (bp) | No. of contigs (no. of plasmids) | Contig N50 (bp) | GC content (%) | Total genome size (bp) | SRA accession no. | BioSample accession no. | Nucleotide accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F4078 (ATCC 33090) | Animal (cow brain) | 79.51 | 15,820 | 16,676 | 2a | 2,794,052 | 37.6 | 2,782,538 | SRS12596339 | SAMN13941777 | NZ_JABXLC000000000 |
| F6215 | Food (meat) | 178.65 | 35,563 | 18,169 | 2a | 2,901,888 | 37.5 | 2,939,591 | SRS12596348 | SAMN13941779 | NZ_JABXLE000000000 |
| F6217 | Human (peritoneum) | 353.39 | 79,416 | 17,132 | 1a | 2,884,312 | 37.4 | 2,855,969 | SRS952935 | SAMN03761682 | NZ_CP095724.1 |
| 2015L-6714 | Food (meat) | 223.30 | 49,392 | 16,497 | 2a | 2,930,612 | 37.38 | 2,984,334 | SRR21942761 | SAMN27548027 | NZ_CP095723.1 |
| H0996 (ATCC 51742) | Food (cabbage) | 394.95 | 84,980 | 19,213 | 3 (1) | 2,801,715 | 37.5 | 2,795,487 | SRR21942762 | SAMN10869156 | NZ_CP095730.1 |
| 2010L-1951 | Human (abscess [armpit]) | 431.68 | 72,490 | 21,319 | 3 (1) | 2,818,897 | 37.4 | 2,810,256 | SRS952976 | SAMN03761736 | NZ_CP095727.1 |
| 2010L-2059 (PNUSAL000003) | Human (knee fluid) | 322.32 | 81,774 | 18,681 | 15 (2) | 2,340,577 | 37.4 | 2,816,380 | SRR21982767 | SAMN10869157 | NZ_CP095726.1 |
| 2012L-5520 (FSL J1-023) | Unknown | 337.92 | 77,923 | 16,424 | 1a | 2,826,835 | 37.5 | 2,826,835 | SRR2157026 | SAMN13941775 | NZ_JABXLA000000000 |
| 2015L-6726 | Food (meat) | 249.48 | 66,858 | 12,899 | 2a | 2,857,050 | 37.4 | 2,926,353 | SRS1160884 | SAMN04263682 | NZ_CP095728.1 |
Circularized contig.
Data availability.
The GenBank accession numbers for all strains are provided in Table 1. PacBio sequences are available under BioProject accession number PRJNA212117. All isolates are available upon request.
ACKNOWLEDGMENT
We thank Jasmine Huffman of the Enteric Diseases Laboratory Branch for her assistance with uploading sequences to NCBI.
Contributor Information
Zuzana Kucerova, Email: zik0@cdc.gov.
Christine Lee, Email: clee13@cdc.gov.
Vanja Klepac-Ceraj, Wellesley College.
REFERENCES
- 1.Stea EC, Purdue LM, Jamieson RC, Yost CK, Truelstrup Hansen L. 2015. Comparison of the prevalences and diversities of Listeria species and Listeria monocytogenes in an urban and a rural agricultural watershed. Appl Environ Microbiol 81:3812–3822. doi: 10.1128/AEM.00416-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson J, Jinneman K, Stelma G, Smith BG, Lye D, Messer J, Ulaszek J, Evsen L, Gendel S, Bennett RW, Swaminathan B, Pruckler J, Steigerwalt A, Kathariou S, Yildirim S, Volokhov D, Rasooly A, Chizhikov V, Wiedmann M, Fortes E, Duvall RE, Hitchins AD. 2004. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl Environ Microbiol 70:4256–4266. doi: 10.1128/AEM.70.7.4256-4266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favaro M, Sarmati L, Sancesario G, Fontana C. 2014. First case of Listeria innocua meningitis in a patient on steroids and eternecept. JMM Case Rep doi: 10.1099/jmmcr.0.003103. [DOI] [Google Scholar]
- 4.Perrin M, Bemer M, Delamare C. 2003. Fatal case of Listeria innocua bacteremia. J Clin Microbiol 41:5308–5309. doi: 10.1128/JCM.41.11.5308-5309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moura A, Disson O, Lavina M, Thouvenot P, Huang L, Leclercq A, Fredriksson-Ahomaa M, Eshwar AK, Stephan R, Lecuit M. 2019. Atypical hemolytic Listeria innocua isolates are virulent, albeit less than Listeria monocytogenes. Infect Immun 87:e00758-18. doi: 10.1128/IAI.00758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volokhov DV, Duperrier S, Neverov AA, George J, Buchrieser C, Hitchins AD. 2007. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl Environ Microbiol 73:1928–1939. doi: 10.1128/AEM.01796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton EM, Daly KM, Guinane CM, Hill C, Cotter PD, Ross PR. 2014. Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol 14:58. doi: 10.1186/1471-2180-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 10.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GenBank accession numbers for all strains are provided in Table 1. PacBio sequences are available under BioProject accession number PRJNA212117. All isolates are available upon request.


