SUMMARY
Obesity comorbidities such as diabetes and cardiovascular disease are pressing public health concerns. Overconsumption of calories leads to weight gain; however, neural mechanisms underlying excessive food consumption are poorly understood. Here, we demonstrate that dopamine receptor D1 (Drd1) expressed in the agouti-related peptide/neuropeptide Y (AgRP/NPY) neurons of the arcuate hypothalamus is required for appropriate responses to a high-fat diet (HFD). Stimulation of Drd1 and AgRP/NPY co-expressing arcuate neurons is sufficient to induce voracious feeding. Delivery of a HFD after food deprivation acutely induces dopamine (DA) release in the ARC, whereas animals that lack Drd1 expression in ARCAgRP/NPY neurons (Drd1AgRP-KO) exhibit attenuated foraging and refeeding of HFD. These results define a role for the DA input to the ARC that encodes acute responses to food and position Drd1 signaling in the ARCAgRP/NPY neurons as an integrator of the hedonic and homeostatic neuronal feeding circuits.
Graphical Abstract
In brief
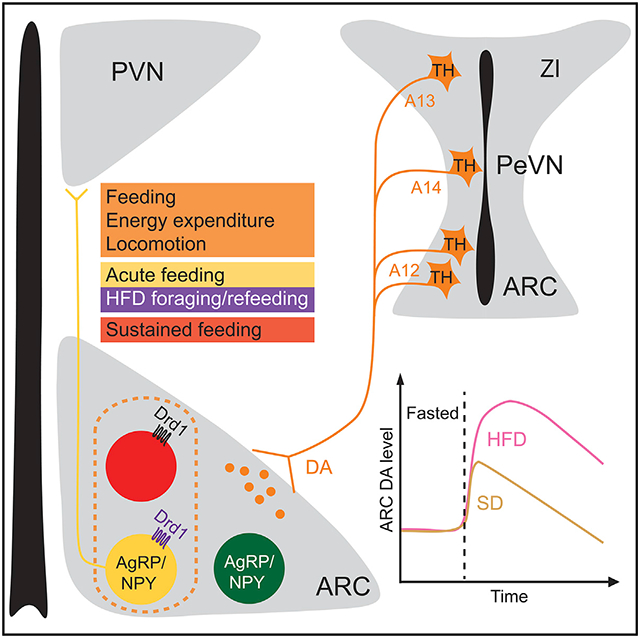
Zhang et al. show that DA release in the ARC is modulated by metabolic states and encodes the energy density of the available food. The Drd1-expressing neurons in the ARC including a subpopulation of ARCAgRP/NPY neurons induce feeding. Genetic ablation of Drd1 in ARCAgRP/NPY neurons attenuates HFD-induced hyperphagia.
INTRODUCTION
Obesity resulting from overconsumption and a sedentary lifestyle often leads to diabetes, cardiovascular disease, and the metabolic syndrome.1,2 Energy-dense foods increase appetite, leading to consumption even in the absence of homeostatic need.3 Mammals have evolved exquisite neural mechanisms to consume sufficient but not excessive calories, yet how a high-fat diet (HFD) short circuits these mechanisms remains unclear.
The drive to eat is determined by multiple neural circuits. The arcuate nucleus of the hypothalamus (ARC) integrates peripheral signals to regulate energy homeostasis.4 Neurons that synthesize both the agouti-related peptide (AgRP) and neuropeptide Y (NPY) are the predominant orexigenic population in this nucleus.5 During states of energy deficit, these ARCAgRP/NPY neurons are activated by hunger signals.5-9 The increased activity of ARCAgRP/NPY neurons promotes feeding and suppresses energy expenditure.10-13 Dopamine (DA) signaling is necessary for encoding feeding-associated behaviors, especially with the incentive and motivational aspects driving feeding.14-16 DA-deficient mice fail to eat and die of starvation.17 The mesolimbic DA system is well-established for reinforcing the value of rewarding food and facilitating motivated behaviors.16,18-22 However, counterintuitively, stimulation of the midbrain DA neurons fails to affect the amount of food consumed.23
Beyond the mesolimbic system, DA neurons in the ARC (A12 group) are primarily involved in prolactin secretion,24,25 and have been reported to promote feeding by inhibiting the anorexigenic proopiomelanocortin neurons (ARCPOMC neurons).26 Additionally, DA neurons in the zona incerta (ZI; A13 group) and in the periventricular nucleus (PeVN; A14 group) project to the ventral hypothalamus27,28; yet a gap remains as to whether or how these dopaminergic nodes contribute to the regulation of energy homeostasis. Notably, a potential role for DA receptor D1 (Drd1) in ARCAgRP/NPY neuron activity has been postulated26,29-31; however, the functional relevance of DA signaling in the ARC-dependent metabolic responses is unknown. Therefore, we evaluated the role of Drd1 signaling in the ARCAgRP/NPY neurons during feeding.
In this study, we show that the Drd1-expressing neurons in the ARC (ARCDrd1 neurons) comprise a heterogeneous population of cells, including a subpopulation that expresses AgRP. Optogenetic or chemogenetic stimulation of the ARCDrd1 neurons induces acute and reversible food consumption and promotes energy expenditure. These responses are attenuated by blocking neurotransmitter release from ARCAgRP/NPY neurons. In line with these findings, activation of the ARCDrd1 neurons that do not express the ARCAgRP/NPY neuronal marker, NPY, fails to initiate feeding, while stimulation of the neurons that co-express Drd1 and NPY increases food intake robustly. The ARC receives projections from a continuum of DA neurons in the hypothalamic and subthalamic areas. Food delivery acutely induces DA release in the ARC of fasted mice, whereas mice that lack Drd1 expression in ARCAgRP/NPY neurons (Drd1AgRP-knockout [KO]) exhibit attenuated foraging and refeeding of HFD. Our findings suggest that energy-dense foods increase DA signaling to the ARCAgRP/NPY neurons through Drd1 to promote feeding.
RESULTS
The ARCDrd1 neurons express genes of energy homeostasis
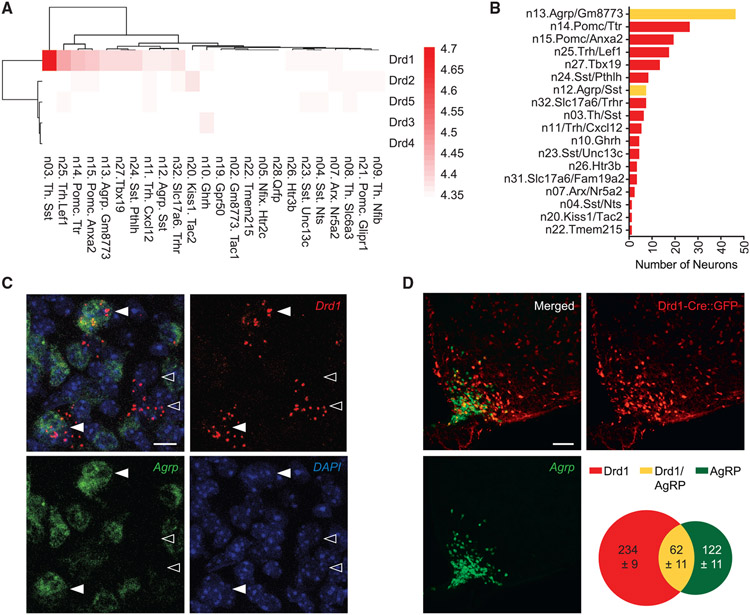
To identify neuronal populations that potentially receive dopaminergic input in the ARC, we examined the RNA levels of DA receptors (Drd1-5) using previously obtained single-cell RNA sequencing data.32 We found that Drd1 expression is enriched in multiple molecularly distinct neuronal groups, which is in contrast with the low expression levels of Drd2 to Drd5 in the same dataset (Figure 1A). This analysis revealed that 53 of the 173 Drd1-expressing neurons are from the ARCAgRP/NPY neuronal clusters (Agrp/Gm8773 and Agrp/Sst), while 26 of them express two other orexigenic markers, Sst and Th (Figures 1B and S1A). Juxtaposing this, 45 of the Drd1 -expressing cells are from two POMC neuronal clusters, Pomc/Ttr and Pomc/Anxa2 (Figures 1B and S1A). Because Drd1-dependent activation of ARCAgRP/NPY neurons has been previously observed,26 we confirmed that a subpopulation of ARCAgRP/NPY neurons expresses Drd1 messenger RNA (mRNA) using RNAscope (Figure 1C). To quantify the colocalization between the ARCDrd1 neurons and ARCAgRP/NPY neurons or ARCPOMC neurons, we probed Agrp or Pomc mRNA by RNAscope on brain sections from the Drd1tm1(cre)Rpa (Drd1-Cre) mice, which express Cre:GFP in the Drd1 positive cells.33 21.0 ± 2.2% of the Cre:GFP-expressing ARCDrd1 cells co-express Agrp, while 33.7 ± 5.9% of the Agrp-expressing cells co-express Drd1-Cre:GFP in the ARC (Figure 1D). In contrast, 9.0 ± 0.9% of the Cre:GFP-expressing ARCDrd1 cells co-express Pomc, while 18.2 ± 2.1% of the Pomc-expressing cells co-express Drd1-Cre:GFP in the ARC (Figure S1B).
Figure 1. The genetic profile of ARC neurons reveals an orexigenic neuronal population expressing Drd1.
(A) Batch-corrected, normalized expression values for Drd genes in each arcuate-median eminence (ARC-ME) neuron cluster from the Campbell et al., 2017 dataset.
(B) Number of Drd1 positive neurons from each cluster of ARC-ME neurons from A. Drd1 positive neurons from Agrp-expressing clusters are depicted in yellow.
(C) RNAscope images showing representative Drd1-expressing cells in the ARC with (▸) or without (▹) Agrp expression. Scale bar, 10 μm.
(D) Representative images showing the ARC of Drd1-Cre:GFP mice. Scale bar, 100 μm. Venn diagram indicates the average number of cells expressing only the Drd1-Cre:GFP (red), only Agrp (green), and both Drd1-Cre:GFP and Agrp (yellow). n = 3.
Optogenetic and chemogenetic activation of the ARCDrd1 neurons increases food intake
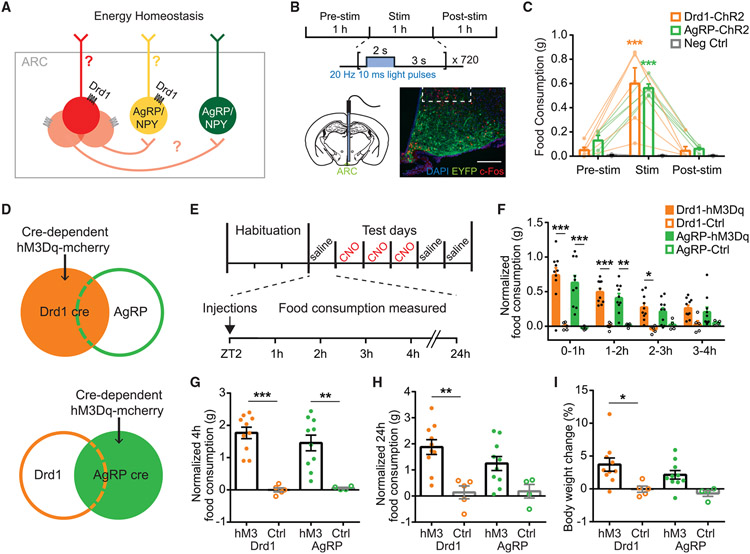
Given the genetic profile of the ARCDrd1 neurons, we sought to investigate the relative contribution of the AgRP/NPY-positive or -negative ARCDrd1 neurons on energy homeostasis and determine the role of Drd1 signaling in the ARCAgRP/NPY neurons (Figure 2A). To test whether increased activity of the ARCDrd1 neurons stimulates feeding, we targeted them by infusing an adeno-associated virus (AAV) into the ARC of adult Drd1-Cre mice. This AAV vector drives the expression of Channelrhodopsin-2 (ChR2; AAV2-DIO-ChR2-EYFP) in a Cre-dependent manner (Figure 2B). After at least 3 weeks of postoperative recovery, the ARCDrd1 neurons were activated by a 473-nm laser at 20 Hz with a 2-s-on/3-s-off pattern delivered via a fiber optic cannula implanted above the ARC. We measured food consumption in these mice before, during, and after the 1-h laser stimulation. Similar to optogenetic stimulation of AgRP neurons, activation of ARCDrd1 neurons induced rapid and reversible food intake, 1-h intake in Drd1 stimulated: 0.60 ± 0.13 g; AgRP stimulated: 0.56 ± 0.03 g; negative control stimulated: 0.005 ± 0.002 g; two-way ANOVA with Bonferroni post hoc comparison, Fgenotype (2, 14) = 14.56, p < 0.001 (Figure 2C).
Figure 2. Optogenetic or chemogenetic stimulation of ARCDrd1 neurons promote feeding.
(A) Hypothetical model of the ARCDrd1 neurons involved in inducing orexigenic responses.
(B) Schematic depicting optogenetic experiment schedule. Photostimulation was conducted for 1 h with 20 Hz 10 msec light pulses applied for 2 s followed by a 3-s break. Representative images showing the expression of ChR2-EYFP in the ARC of Drd1-Cre mice. Scale bar, 100 μm.
(C) Food consumption before, during, and after the 1-h photostimulation in Drd1-Cre (orange), AgRP-Cre (green), or wild-type (WT) negative control (gray) mice with Cre-dependent ChR2-EYFP delivered to the ARC. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 5–6/group; Fgenotype (2, 14) = 14.56, p < 0.001; Ftreatment (2, 28) = 58.52, p < 0.001.
(D) Schematic depicting Cre-dependent hM3Dq-mCherry expression in the ARC of Drd1-Cre or AgRP-Cre mice.
(E) Food intake experiment schedule. Mice were individually caged for at least 3 days and habituated to intraperitoneal injections on the last day. During test days, mice were injected with saline on days 1, 5, and 6, and with CNO on days 2 through 4 at ZT 2. Food intake was measured 1, 2, 3, 4, and 24 h after the injections.
(F) Hourly food consumption after CNO administration in Drd1-hM3Dq, AgRP-hM3Dq mice, or control mice (Ctrl). Control mice are Drd1-Cre or AgRP-Cre mice with cre-dependent mCherry targeted to the ARC. Responses were normalized by subtracting the average food consumption on days 1, 5, and 6 after saline injections. Repeated-measures three-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Fgenotype (1,25) = 0.2733, p = 0.6057; Ftransgene (1,25) = 46.94, p < 0.001; Ftime (3, 75) = 9.785, p < 0.001.
(G) Normalized 4-h food consumption after CNO administration in Drd1-hM3Dq, AgRP-hM3Dq, or Ctrl mice. Two-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Fgenotype (1, 25) = 0.2998, p = 0.5889; Ftransgene (1, 25) = 46.38, p < 0.001.
(H) Normalized 24-h food consumption after CNO administration in Drd1-hM3Dq, AgRP-hM3Dq, or control mice. two-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Fgenotype (1, 25) = 0.8396, p = 0.3683; Ftransgene (1, 25) = 19.20, p < 0.001.
(I) Percentage body weight change after 3 consecutive days of CNO administrations in Drd1-hM3Dq or AgRP-hM3Dq mice. Two-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Fgenotype (1,25) = 1.377, p = 0.2517; Ftransgene (1,25) = 12.25, p = 0.0018. Data are represented as mean ± standard error of the mean. *p < 0.05; **p < 0.01; ***p < 0.001.
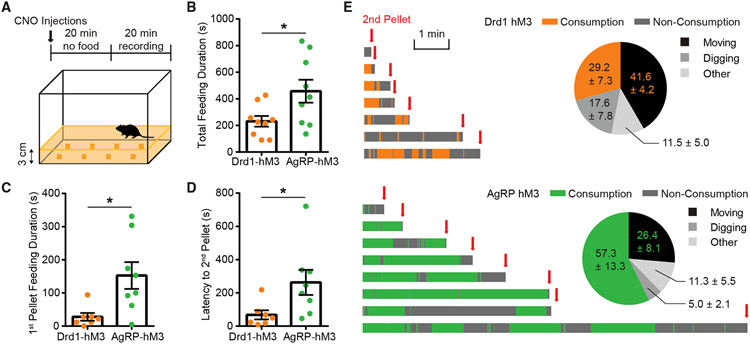
To determine the effect of extended stimulation of the ARCDrd1 neurons, we similarly expressed the designer receptor hM3Dq (AAV8-DIO-hM3Dq-mCherry) in the ARC of Drd1-Cre mice (Drd1-hM3Dq) (Figure 2D). We activated the ARCDrd1 neurons with an intraperitoneal injection of 0.3 mg/kg clozapine N-oxide (CNO). At the beginning of the light phase, when mice normally refrain from eating, this stimulation significantly increased food consumption in the Drd1-hM3Dq mice (4-h food consumption, saline: 0.24 ± 0.033 g vs. CNO: 1.14 ± 1.127 g; repeated-measures three-way ANOVA with Bonferroni post hoc comparison;, p < 0.001) (Figure S2C). Reflecting the increased daily food intake, 3 consecutive days of CNO injections led to a significant amount of body weight gain in these animals (body weight change, Drd1-hM3Dq: 3.17 ± 0.677% vs. Drd1-mCherry controls: −1.29 ± 0.487%; Student two-tailed t test; p < 0.001) (Figure S2D). The temporal pattern of food consumption over 24 h (Figures 2F-2H and S2G) and the body weight gain across three days of CNO treatment (Figure 2I) were similar between chemogenetic stimulation of the ARCAgRP/NPY (AgRP-hM3Dq) and the ARCDrd1 neurons. Next, we evaluated the food-seeking behavior of AgRP-hM3Dq and Drd1-hM3Dq mice in an open field where eight pellets of standard chow (PicoLab Rodent Diet 20 5053) were buried under the corn bedding (Figure 3A). Stimulated AgRP-hM3Dq mice spent significantly more time eating overall than Drd1-hM3Dq mice (Figure 3B). Specifically, compared with AgRP-hM3Dq mice, Drd1-hM3Dq mice stopped consuming the initially acquired pellet more rapidly and instead searched for the next pellet (Figures 3C-3E). These results suggest that the ARCDrd1 and ARCAgRP/NPY neurons differentially regulate food consumption and activation of ARCDrd1 neurons promotes exploration even after a food source has already been acquired.
Figure 3. Activation of ARCDrd1 neurons promotes food exploration.
(A) Foraging paradigm: Habituated mice were injected with CNO for 20 min without food, and then placed in an arena with eight pellets of SD buried under 3-cm-thick corn bedding on the cage floor and video recorded for 20 min.
(B) Total feeding duration during the 20-min test. Student two-tailed t test; n = 9/group.
(C) First pellet feeding duration. Student two-tailed t test; n = 7–8/group.
(D) Latency to the acquisition of the second pellet after the acquisition of the first pellet. Student two-tailed t test; n = 7–8/group.
(E) Annotated consumption or non-consummatory behaviors of each mouse during the test after the acquisition of the first pellet and before the acquisition of the second pellet. Consumption time of Drd1-hM3Dq and AgRP-hM3Dq mice is shown as orange and green, respectively. Time spent performing non-consummatory behaviors is shown as dark gray. Pie charts indicated the percentage time spent consuming food (orange or green), moving (black), digging (gray), and other non-consummatory behaviors (light gray). Data are represented as mean ± standard error of the mean. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
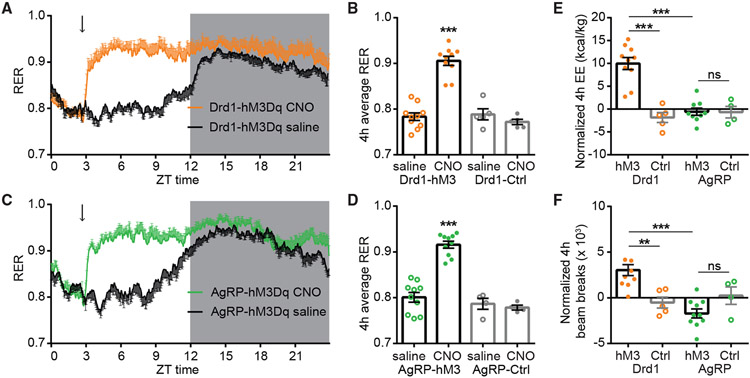
The ARCDrd1 neurons promote energy expenditure and locomotion
To gain insight into how energy homeostasis is regulated by ARCDrd1 neurons, we monitored the metabolic activity during their chemogenetic activation using the Homecage Comprehensive Lab Animal Monitoring System (CLAMS). Paralleling the increased food consumption, we observed increased respiratory exchange rates (RER) in Drd1-hM3Dq and AgRP-hM3Dq mice after CNO administration (Figures 4A-4D). This reflects the rapid switch of substrate utilization from fats to carbohydrates as the animals begin to consume food. Interestingly, stimulation of Drd1-hM3Dq and AgRP-hM3Dq neurons even in the absence of food still increased RER (Figures S2K and S2L), indicating that, like ARCAgRP/NPY neurons, ARCDrd1 neurons regulate substrate utilization independent of food consumption. Consistent with previous reports,34 we did not observe increased energy expenditure (EE) after ARCAgRP/NPY neuron stimulation (Figure 4E). However, stimulation of the ARCDrd1 neurons significantly increased EE (Figure 4E) and locomotor activity (Figure 4F), suggesting a distinct role for these neurons in energy homeostasis.
Figure 4. ARCDrd1 neurons regulate energy homeostasis.
(A and C) A 24-h RER in Drd1-hM3Dq (A) or AgRP-hM3Dq mice (C) after saline (black traces) or CNO administration (orange and green traces, respectively). The dark portion of the light cycle is indicated by gray shading. Arrows indicate the time of injections.
(B) Average 4-h RER in Drd1-hM3Dq and control mice after saline or CNO administration. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 5–10/group; Ftransgene (1, 13) = 48.61, p < 0.001; Ftreatment (1, 13) = 21.21, p < 0.001.
(D) Average 4-h RER AgRP-hM3Dq and control mice following saline or CNO administration. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Ftransgene (1, 12) = 60.55, p < 0.001; Ftreatment (1, 12) = 17.85, p = 0.0012.
(E) Normalized 4-h EE in Drd1-hM3Dq, AgRP-hM3Dq, or control mice. Responses were normalized by subtracting the EE after saline injections. Two-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Fgenotype (1, 24) = 13.11, p = 0.0014; Ftransgene (1, 24) = 21.19, p < 0.001.
(F) Normalized 4-h beam breaks in Drd1-hM3Dq, AgRP-hM3Dq, or control mice. Responses were normalized by subtracting the beam breaks after saline injections. Two-way ANOVA with Bonferroni post hoc comparison; n = 4–10/group; Fgenotype (1, 25) = 8.372, p = 0.0078; Ftransgene (1, 25) = 1.353, p = 0.2557. Data are represented as mean ± standard error of the mean. *p < 0.05; **p < 0.01; ***p < 0.001.
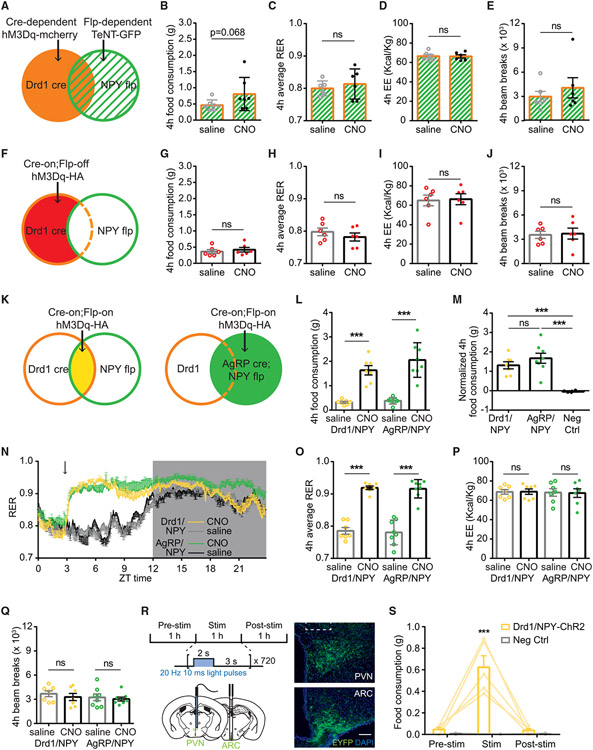
Inhibition of ARCAgRP/NPY neuron neurotransmitter release dampens ARCDrd1 neuron-induced food consumption
We sought to elucidate whether the ARCDrd1 neuron-induced feeding depends on the ARCAgRP/NPY neurons. To this end, we used the Npytm1.1(flpo)Hze (NPY-flp) mice, which express Flp recombinase in the ARCAgRP/NPY neurons, and crossed them with the Drd1-Cre mice to generate Drd1-Cre;NPY-Flp mice. This allowed us to express two independent transgenes in these two neuron populations simultaneously. By expressing Cre-dependent AAV8-DIO-hM3Dq-mCherry in the ARCDrd1 neurons and Flp-dependent tetanus neurotoxin (AAV1-fDIO-TeNT-GFP) in the ARCAgRP/NPY of the Drd1-Cre;NPY-Flp mice, we blocked neurotransmitter release in the ARCAgRP/NPY neurons while being able to chemogeneticaly stimulate ARCDrd1 neurons (Figure 5A). We found that blocking the ARCAgRP/NPY-neuron neurotransmission dramatically dampened the increased food consumption induced by stimulation of the ARCDrd1 neurons (Figures 5B and S3A), although a trend toward increased 4-h food consumption was still observed compared to the saline control group (4-h food consumption, saline injected: 0.46 ± 0.06 g vs. CNO injected: 0.81 ± 0.19 g; Student two-tailed t test; p = 0.0677) (Figure 5B). Likewise, after the CNO injection, we did not observe an increase in the RER, EE, or locomotion in these animals (Figures 5C-5E), unlike stimulation of ARCDrd1 neurons in the absence of ARCAgRP/NPY neurotransmission blockade. Taken together, these results demonstrate that intact ARCAgRP/NPY neuron signaling is required for the ARCDrd1 neuron-induced changes in energy homeostasis.
Figure 5. Stimulation of ARCDrd1 neurons expressing AgRP/NPY induces feeding acutely.
(A, F, and K) Schematic depicting Cre-dependent hM3Dq-mCherry and Flp-dependent TeNT-GFP expression (A), Cre-on;Flp-off hM3Dq-HA expression (F), Cre-on;Flp-on hM3Dq-HA expression (K) in the ARC of Drd1-Cre;NPY-Flp mice.
(B–E) Four-hour food consumption (B), RER (C), EE (D), or beam breaks (E) in mice indicated in (A) after saline or CNO administration. Student two-tailed t test; n = 6–7/group.
(G–J) Four-hour food consumption (G), RER (H), EE (I), or beam breaks (J) in mice indicated in (F) after saline or CNO administration. Student two-tailed t test; n = 6/group.
(L) Four-hour food consumption in Drd1/NPY-hM3Dq or AgRP/NPY-hM3Dq mice after saline or CNO administration. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 7–8/group; Fgenotype (1, 26) = 2.140, p = 0.1555; Ftreatment (1, 26) = 82.60, p < 0.001.
(M) Normalized 4-h food consumption in Drd1/NPY-hM3Dq, AgRP/NPY-hM3Dq, or negative control mice (Neg Ctrl). Negative control mice are Drd1-Cre or NPY-Flp mice with Cre-on; Flp-on hM3Dq-HA delivered to the ARC. Responses were normalized by subtracting the average food consumption on day 1, 5, 6 after saline injections. One-way ANOVA with Bonferroni post hoc comparison; n = 6–8/group; Fgenotype (2, 18) = 18.92, p < 0.001.
(N) 24-h RER in the Drd1/NPY-hM3Dq mice following CNO (yellow trace) or saline (gray trace) administration, or the AgRP/NPY-hM3Dq mice following CNO (green trace) or saline (black trace) administration. Dark portion of the light cycle is indicated by gray shading. Arrows indicate the time of injections.
(O) Average 4-h RER in Drd1/NPY-hM3Dq or AgRP/NPY-hM3Dq mice following saline or CNO administration. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 7–8/group; Fgenotype (1, 13) = 0.09766, p = 0.7596; Ftreatment (1, 13) = 267.4, p < 0.001.
(P) Average 4-h EE n Drd1/NPY-hM3Dq or AgRP/NPY-hM3Dq mice following saline or CNO administration. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 7/group; Fgenotype (1, 12) = 0.04569, p = 0.8343; Ftreatment (1, 12) = 0.03815, p = 0.8484.
(Q) Average 4-h beam breaks in Drd1/NPY-hM3Dq or AgRP/NPY-hM3Dq mice following saline or CNO administration. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 7–8/group; Fgenotype (1, 13) = 0.5810, p = 0.4595; Ftreatment (1, 26) = 1.331, p = 0.2693.
(R) Schematic and representative image showing the Cre-on; Flp-on ChR2-EYFP expression in the ARC of Drd1-Cre; NPY-Flp mice and optogenetic stimulation of the ChR2-EYFP expressing-neuron projections in the PVN. Scale bar represents 100 μm.
(S) Food consumption before, during, and after the 1-h photostimulation in Drd1-Cre; NPY-Flp mice or WT mice with Cre-on; Flp-on ChR2-EYFP delivered to the ARC. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 5/group; Fgenotype (1, 8) = 44.51, p < 0.001; Ftreatment (1, 16) = 26.85, p < 0.001. Data are represented as mean ± standard error of the mean. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
The subpopulation of ARCDrd1 neurons expressing AgRP/NPY is sufficient to induce voracious food consumption
Because ARCDrd1 neuron-induced food consumption depends on the ARCAgRP/NPY neurons, either the ARCDrd1 neurons increase ARCAgRP/NPY neuron activity to induce feeding or activation of a subpopulation of the ARCDrd1 neurons that express AgRP/NPY are sufficient for this behavior. To distinguish between these two possibilities, we delivered an AAV in which hM3Dq expression is turned on by Cre recombinase but turned off by Flp recombinase (AAV1-Con/Foff-hM3Dq), to the ARC of Drd1-Cre;NPY-Flp mice (Figure 5F). Activation of the non-AgRP/NPY ARCDrd1 neurons failed to induce feeding or metabolic changes in RER, EE, and locomotion that were observed during activation of all ARCDrd1 neurons within 4 h (Figures 5G-5J). Interestingly, activation of this subpopulation of ARCDrd1 neurons still increased 24-h food consumption after CNO injections (24-h food consumption: saline injected: 5.79 ± 0.24 g; CNO injected: 6.33 ± 0.29 g; Student two-tailed t test; p = 0.003 (Figure S3B)), suggesting a role for these neurons in sustained feeding response. To test the possibility that ARCDrd1 neurons expressing AgRP/NPY are sufficient to induce food intake, we targeted them by infusing an AAV that expresses hM3Dq only after Cre- and Flp-dependent recombination (AAV1-Con/Fon-hM3Dq) into the ARC of the Drd1-Cre;NPY-Flp (Drd1/NPY-hM3Dq) mice (Figure 5K). As a positive control, we delivered the same virus to the ARC of the AgRP-Cre;NPY-Flp (AgRP/NPY-hM3Dq) mice (Figure 5K). CNO-stimulated Drd1/NPY-hM3Dq mice rapidly initiated voracious feeding within 1 h (Figure S3D), which lasted at least for 4 h, similar to the AgRP/NPY-hM3Dq mice (Figures 5L and 5M). However, 24-h food consumption in Drd1/NPY-hM3Dq mice was not as robust as AgRP/NPY-hM3Dq mice (normalized 24-h food consumption: Drd1/NPY-hM3Dq: 0.73 ± 0.26 g; AgRP/NPY-hM3Dq: 2.30 ± 0.53 g; one-way ANOVA with Bonferroni post hoc comparison, p = 0.0251) (Figure S3F). This suggests that the subset of ARC neurons that express AgRP, NPY, and Drd1 (ARCAgRP/NPY/Drd1) are engaged during acute feeding response. Consistent with this finding, Drd1/NPY-hM3Dq mice did not gain body weight after 3 consecutive days of CNO injections (body weight change: Drd1/NPY-hM3Dq: 0.62 ± 0.60%; AgRP/NPY-hM3Dq: 6.38 ± 1.59%; one-way ANOVA with Bonferroni post hoc comparison, p = 0.0079) (Figure S3G). We next determined the role of the ARCAgRP/NPY/Drd1 neurons in metabolic responses using CLAMS. Similar to AgRP/NPY-hM3Dq mice, Drd1/NPY-hM3Dq mice exhibited a rapid increase in their RER after CNO injections in both ad lib-fed, 4-h average RER: saline injected: 0.79 ± 0.01; CNO injected: 0.92 ± 0.01; two-way ANOVA with Bonferroni post hoc comparison, Ftreatment (1, 13) = 267.4, p < 0.001 (Figures 5N and 5O), or food-deprived states, 4-h average RER with no food: saline injected: 0.75 ± 0.01; CNO injected: 0.92 ± 0.01; Ftreatment (1, 13) = 371.5, p < 0.001 (Figure S3H). Neither Drd1/NPY-hM3Dq nor AgRP/NPY-hM3Dq mice showed changes in EE (Figure 5P) or locomotion (Figure 5Q).
Optogenetic stimulation of the ARCAgRP/NPY/Drd1 neuron projections in the paraventricular nucleus induces robust food consumption
To investigate the downstream brain regions that receive projections from the ARCAgRP/NPY/Drd1 neurons, we examined the distribution of hM3Dq-HA-labeled axons in brain regions of Drd1/NPY-hM3Dq mice. We observed that the paraventricular nucleus (PVN) is densely innervated by the axons of the HA-expressing Drd1/NPY neurons (Figure S3C). Therefore, we sought to evaluate the functional role of the Drd1/NPY neuron projections in the PVN. To this end, we targeted the ARC of Drd1-cre; NPY-flp mice with an AAV that expresses ChR2-EYFP after Cre- and Flp-dependent recombination (AAV8-Con/Fon-ChR2-EYFP). After at least 3 weeks of postoperative recovery, the axon terminals from the Drd1/NPY neurons in the PVN were activated by a 473-nm laser at 20 Hz with a 2-s-on/3-s-off pattern delivered via a fiber optic cannula (Figure 5R). We measured food consumption in these mice before, during, and after 1-h laser stimulation. Stimulation of the Drd1/NPY neuron projections in the PVN robustly increased food consumption, indicating that the ARCAgRP/NPY/Drd1 → PVN circuit is sufficient to drive feeding, 1-h intake in Drd1/NPY stimulated: 0.624 ± 0.107 g vs. in negative control stimulated: 0 ± 0 g; two-way ANOVA with Bonferroni post hoc comparison, Fgenotype (1, 8) = 44.51; p < 0.001) (Figure 5S).
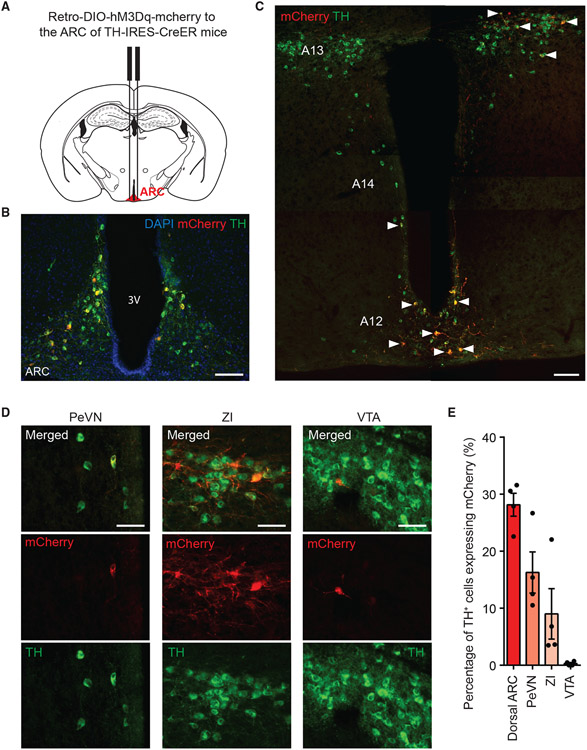
The ARC receives anatomic inputs from multiple dopaminergic neuronal populations
To gain insight into the potential sources of DA inputs to the ARC, we targeted the ARC of adult wild-type mice with retrogradely transported red fluorescent beads (retrobeads) (Figures S4A and S4B). One week after surgery, we examined the distribution of retrobeads in brain regions inhabited by dopaminergic neurons. We observed retrobeads in the ARC, ZI, and ventral tegmental area (VTA). There was notable co-localization of retrobeads and tyrosine hydroxylase (TH) (the rate-limiting enzyme in DA synthesis) immunoreactivity in the ARC and ZI, but relatively lower in the VTA (Figure S4C). This suggests that the projections from the VTA to the ARC are non-dopaminergic and dopaminergic input to the ARC originates in the hypothalamic and subthalamic areas. To identify the dopaminergic neurons that make synaptic contacts within the ARC, we delivered a retrogradely transported AAV that Cre recombinase dependently expresses hM3Dq-mCherry (retrograde AAV-DIO-hM3Dq-mCherry) to the ARC of Thtm1(cre/Esr1)Nat (TH-IRES-CreER) mice, which express Cre in TH-positive cells after 5 consecutive days of 75 mg/kg tamoxifen injections (Figures 6A and 6B). In these mice, we observed robust mCherry and TH colocalization in the dorsal ARC, PeVN, and ZI (Figures 6C-6E), but relatively sparsely in the VTA (Figures 6D and 6E). This indicates that the dopaminergic neurons in the dorsal ARC, PeVN, and ZI are the likely sources of DA in the ARC.
Figure 6. The ARC receives anatomic inputs from multiple dopaminergic neuronal populations.
(A) ARC retrograde tracing paradigms depicting retrograde AAV-DIO-hM3Dq-mCherry delivered to the ARC of TH-IRES-CreER mice.
(B) Representative images showing hM3Dq-mCherry expression in the ARC. Scale bar, 100 μm.
(C) Representative stitched images showing expression of hM3Dq-mCherry and TH marking the A12-14 groups of DA neurons. Arrows indicate cells co-expressing hM3Dq-mCherry and TH. Scale bar, 100 μm.
(D) Representative images of hM3Dq-mCherry- and TH-expressing cells in the PeVN (left), the ZI (middle) and the VTA (right). Scale bar, 50 μm.
(E) Percentage of TH-expressing cells with hM3Dq-mCherry expression, n = 4. Data are represented as mean ± standard error of the mean.
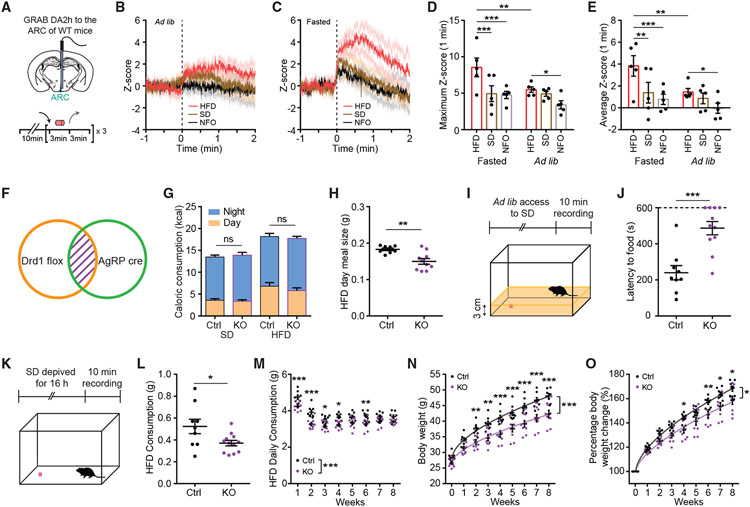
DA is released in the ARC in response to food
To investigate the DA levels in the ARC during changing metabolic states, we virally delivered the G-protein-coupled receptor-based DA sensor, GRAB DA2h (AAV9-GRAB-DA2h),35 to the ARC of adult WT mice (Figure 7A). We monitored GRAB DA2h signal during repeated delivery and withdrawal of a standard diet (SD), HFD, or non-food object (NFO) in ad libitum or overnight fasted mice (Figure 7A). Delivery of HFD mildly increased DA levels in the ARC of ad libitum mice, while SD or NFO failed to alter DA levels significantly (Figures 7B and S5A-S5E). Surprisingly, SD presentation induced immediate DA releases in the ARC of overnight-fasted mice (Figures 7C and S5G), suggesting that energy deficiency enhances the DA response to regular food. However, delivery of HFD evoked rapid and more robust DA release in these mice (Figure 7C and S5F-S5J; Video S1) when compared with SD or NFO delivery and when compared with HFD delivery to ad libitum mice (Figures 7D, 7E, and S5K-S5M). These findings indicate that DA release in the ARC is modulated by metabolic states and encodes the energy density of the available food.
Figure 7. Food-induced DA signaling in ARCAgRP/NPY neurons promotes feeding.
(A) Schematic depicting fiber photometry experiment schedule. WT mice with GRAB DA2h expression in the ARC were ad libitum or overnight fasted before the test day. Black arrow indicates the delivery of HFD, SD, or NFO; gray arrow indicates the withdrawal of HFD, SD, or NFO. During test days, HFD, SD, or NFO were repeatedly delivered 3 times for 3 min with 3-min intervals.
(B) Fiber photometry monitoring of GRAB DA2h activity in the ARC in response to HFD (red trace), SD (brown trace), or NFO (black trace) delivery in ad libitum mice. Responses from three trials were averaged for each mouse, n = 5.
(C) Fiber photometry monitoring of GRAB DA2h activity in the ARC in response to HFD (red trace), SD (brown trace), or NFO (black trace) delivery in overnight-fasted mice. Responses from three trials were averaged for each mouse, n = 5.
(D) Maximum Z score during the 1-min period following HFD (red), SD (brown), or NFO (gray) delivery in overnight-fasted or ad libitum mice. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 5/group; Fcondition (1, 4) = 4.796, p = 0.0937; Ftreatment (2, 8) = 11.44, p = 0.0045.
(E) Average Z score during the 1-min period after HFD (red), SD (brown), or NFO (gray) delivery in overnight-fasted or ad libitum mice. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 5/group; Fcondition (1, 4) = 7.002, p = 0.0572; Ftreatment (2, 8) = 8.531, p = 0.0104.
(F) Schematic indicating the genetic ablation of Drd1 from ARCAgRP/NPY ceils in AgRP-Crecre/+;Drd1fl/fl mice (Drd1AgRP-KO).
(G) Average day and night kilocalories (kcal) consumed by control or Drd1AgRP-KO mice during 2 days when provided with ad libitum access to SD and the first 2 days when switched to ad libitum access HFD. Repeated-measures three-way ANOVA with Bonferroni post hoc comparison; n = 8–10/group; Ftime (1, 16) = 198.7, p < 0.001; Fgenotype (1, 16) = 0.0007562, p = 0.9784; Fdiet (1, 16) = 34.43, p < 0.001.
(H) Average of day-time HFD meal size of control or Drd1AgRP-KO mice during the first 2 days when switched to ad libitum access HFD. Student two-tailed t test; n = 8–10/group.
(I) Open field foraging paradigm.
(J) Latency to food of control or Drd1AgRP-KO mice during the 10-min recording. Mice that failed to find food are represented with latency of 10 min. Student two-tailed t test; n = 9–11/group.
(K) Open field refeeding paradigm.
(L) HFD consumption of control or Drd1AgRP-KO mice during the 10-min recording. Student two-tailed t test; n = 9–12/group.
(M) Daily HFD consumption of control or Drd1AgRP-KO mice through eight weeks of ad libitum access to the HFD. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 8–9/group; Fgenotype (1,15) = 25.62, p < 0.001; Ftime (7,105) = 58.96, p < 0.001.
(N) Body weights of control or Drd1AgRP-KO mice through 8 weeks of ad libitum access to the HFD. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 8–9/group; Fgenotype (1, 15) = 16.64, p = 0.001; Ftime (8, 120) = 448.9, p < 0.001.
(O) Percentage body weight change of control or Drd1AgRP-KO mice through eight weeks of ad libitum access to the HFD. Repeated-measures two-way ANOVA with Bonferroni post hoc comparison; n = 8–9/group; Fgenotype (1, 15) = 8.5758, p = 0.0104; Ftime (8, 120) = 460.7, p < 0.001. Data are represented as mean ± standard error of the mean. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
Genetic ablation of Drd1 in the ARCAgRP/NPY neurons attenuates HFD-associated feeding behaviors
To investigate the function of Drd1 in the ARCAgRP/NPY neurons, we genetically ablated the Drd1 expression from these neurons by crossing the mice that carry the floxed Drd1 allele (Drd1tm2.1Stl) with the AgRP-Cre mice (Drd1AgRP-KO) (Figures 7F and S6A-S6C). Unlike the germline Drd1 KO mice,36 at 8 weeks of age, the Drd1AgRP-KO mice had a similar body weight compared with the control littermates (Figure S6D). To test whether genetic ablation of the Drd1 expression from the ARCAgRP/NPY neurons affects feeding, we first monitored the gross changes of feeding structure in Drd1AgRP-KO and control groups when switched from a SD to a HFD during the initial 2-day period. We did not observe any difference in consumption amounts between the Drd1AgRP-KO and control groups during the day or night periods (Figures 7G and S6E). Next, we determined the meal microstructure of the Drd1AgRP-KO and control mice fed on either of these diets. Meals were defined as feeding bouts clustering within 5-min intervals,37 with a consumption amount of no less than 0.05 g and a duration of no less than 5 s. On SD, Drd1AgRP-KO and control mice had similar feeding microstructure (Figures S6F-S6I). However, on HFD, the Drd1AgRP-KO mice had significantly smaller meals during the daytime (HFD meal size in day time: Drd1AgRP-KO: 0.15 ± 0.008 g; control: 0.18 ± 0.004 g; Student two-tailed t test, p = 0.0026) (Figure 7H). Therefore, we investigated the role of Drd1 in ARCAgRP/NPY neurons in HFD-associated foraging behaviors by assessing food seeking in Drd1AgRP-KO mice and controls. For this, we placed habituated mice in an open field with one pellet of the HFD buried under the bedding (Figure 7I).36 The latency to find the pellet was significantly longer in the Drd1AgRP-KO mice compared with controls (latency to food: Drd1AgRP-KO: 486.9 ± 36.83 s; control: 239.9 ± 39.60 s; Student two-tailed t test, p < 0.001) (Figure 7J), which implicates a role for Drd1AgRP signaling in foraging of palatable food. To examine whether the refeeding of HFD after food deprivation is affected in the Drd1AgRP-KO mice, we placed overnight-fasted mice in an open field where one pellet of HFD was provided during the 10-min recording period (Figure 7K). The Drd1AgRP-KO mice consumed significantly less HFD compared with controls (HFD refeeding consumption: Drd1AgRP-KO: 0.37 ± 0.03 g; control: 0.52 ± 0.07 g; Student two-tailed t test, p = 0.0270) (Figure 7L). This is in support of the notion that DA signaling via Drd1 in ARCAgRP/NPY cells is necessary for promotion of HFD consumption during an energy-deprived state. To investigate the long-term consequences of this response, we put Drd1AgRP-KO mice and controls on the HFD for 8 weeks and measured their food consumption and body weight weekly. Although we did not observe a difference in consumption amount during the initial 2-day period (Figure 7G), the Drd1AgRP-KO mice exhibited reduced food consumption (Figure 7M) and body weight gain (Figures 7N and 7O) compared with the controls during the long-term exposure to HFD. These observations confirm that Drd1 signaling in the ARCAgRP/NPY neurons is a contributor to HFD-induced hyperphagia and body weight gain.
DISCUSSION
The decision to eat is informed by many factors, including peripheral energy balance signals and sensory inputs relaying the palatability of the food.38,39 Neuronal circuits that originate in the ARC process these inputs to initiate or terminate feeding.10,26,40-43 The nature of how ARC neurons monitor and respond to blood-borne signals are well described44; however, the contribution of the central projections carrying sensory information to the ARC during feeding-associated behaviors remains obscure.45
Dopaminergic signaling in the brain is involved in many aspects of feeding-associated behaviors.14-16,46 The ARC receives projections from an array of DA neurons in the hypothalamic and subthalamic areas (Figures 6 and S4).26-28 In this study, we show that presentation of HFD to a food-deprived animal induces robust DA release in the ARC. The hypothalamic engagement of this dopaminergic signaling is likely multi-faceted, with Drd1 expressing ARC neurons being one of the major targets.26,29,31 Drd1, as a predominantly Gs-coupled G protein-coupled receptor (GPCR), could trigger sustained increase in food intake similar to exogenously expressed designer Gs-coupled GPCRs in ARCAgRP/NPY neurons.47 This is unlike Gq-coupled activation of ARCAgRP/NPY neurons, which increases food consumption only acutely and minimally contributes to weight gain.48 Our findings provide a plausible mechanism for the HFD-induced modulation of the ARC cells, where an HFD increases the intrinsic excitability of ARCDrd1 neurons (including a subpopulation of ARCAgRP/NPY neurons) through Drd1-dependent DA signaling. This also brings up an interesting potential feedforward mechanism in which intra-hypothalamic DA can regulate the metabolic state-dependent ARCAgRP/NPY neuron-mediated mesolimbic responses.29,49,50 Therefore, this node represents a significant integration point for DA neurotransmission in energy homeostasis.
A significant proportion of the ARCDrd1 neurons that express AgRP/NPY (ARCAgRP/NPY/Drd1 neurons) are within the Agrp/Gm8773 neuronal cluster. Although a second subset of the ARCAgRP/NPY neurons mediate glucoregulation and metabolism (i.e., the somatostatinergic Agrp/Sst neurons),32 whether Agrp/Gm8773 neurons have a unique role in energy homeostasis is unclear. We established that the stimulation of ARCAgRP/NPY/Drd1 neurons is sufficient to initiate robust feeding (Figures 5L and 5M) and that genetic ablation of Drd1 in these neurons disturbs food-seeking and reduces refeeding of HFD (Figures 7J and 7L). These data suggest that Drd1-expressing Agrp/Gm8773 neurons are potent drivers of orexigenic responses and can be modulated by DA in response to availability of energy-dense foods, making them a target of interest for therapeutic intervention for eating disorders.
The ARCAgRP/NPY/Drd1 neuron stimulation-induced 24-h food consumption response (Figure S3E) is not as robust as the stimulation of the entire ARCAgRP/NPY neuron population (Figure S3F). Notably, the stimulation of the ARCDrd1 neurons that lack NPY expression increases 24-h food consumption without significant induction during the acute phase of the feeding response, while blocking neurotransmitter release from ARCAgRP/NPY neurons attenuates the ARCDrd1 neuron-induced feeding responses. These findings suggest that the non-AgRP/NPY ARCDrd1 orexigenic neurons (e.g., ARCSst/Drd1 or ARCTh/Drd1 neurons) contribute to the sustained food consumption after the acquisition of food when the activity of the ARCAgRP/NPY neurons are dampened.45,51,52 Interestingly, chemogenetic activation of the ARCDrd1 neurons induces distinct metabolic phenotypes from the stimulation of the ARCAgRP/NPY neurons, including increased EE and locomotion (Figures 4E and 4F). These responses may be associated with the simultaneous activation of multiple ARC dependent circuits, including the neurons coexpressing POMC, which are known to promote EE and ambulatory movements. Expression of Drd1 in both ARCAgRP/NPY and ARCPOMC neurons could be a reflection of their shared hypothalamic progenitor origin53; however, the role of Drd1 signaling in ARCPOMC neurons remains unclear.
The daily rhythm and the microstructure of feeding are two critical parameters that are required for maintaining energy balance. Energy-dense foods increase appetite and encourage snacking outside of regular mealtimes.36 Mice fed an ad libitum HFD lose circadian rhythmicity of their food consumption, which contributes to obesity and is reversible by time-restricted feeding.54 Larger portion sizes of food lead to significantly higher energy intake and weight gain,55-57 while a greater number of smaller meals is associated with a lower risk of obesity.58 Wild-type mice rapidly increase their meal size during the resting phase when presented with an HFD (Figures 7H and S6F). Surprisingly, this diet-induced meal size change is reduced in the AgRPDrd1-KO mice. Thus, Drd1 signaling in the ARCAgRP/NPY neurons contributes to the diet-induced disturbance of temporal feeding microstructure. Because A12 and A14 DA neurons exhibit circadian rhythmicity in their activity,59 DA input from these neurons are a potential source for time of day signal. Consumption of the HFD might be disrupting the temporal pattern of DA release in the ARC from the A12 or A14 group of DA neurons and therefore promoting feeding during the resting phase. In future studies, we will delineate the mechanism by which the ARC-projecting DA neurons get modulated and contribute to consummatory behaviors.
Limitations of the study
Extra-synaptic DA transmission in dopaminergic neurons has been well defined.60,61 This brings challenges in the interpretation of retrograde tracing experiments seeking to determine the source of DA for ARCDrd1 neurons. The proximity of dorsal and ventral ARC is another confound due to the potential somatic uptake of retrobeads or retrograde AAV. Therefore, although we demonstrated that the ARC receives DA projections from the dorsal ARC, PeVN, and ZI, we cannot rule out a possible contribution of DA from other dopaminergic neurons (i.e., midbrain). To monitor ARC DA, we used GRAB DA2h, which preferentially detects DA over other catecholamines35; however, it is formally possible that other catecholamines could be contributing to the observed responses. Last, previous findings have demonstrated that long-term access to HFD diminishes the capacity of SD to suppress the activity of ARCAgRP/NPY neurons.49,62 This indicates that ARCAgRP/NPY neurons are dysregulated during diet-induced obesity. Although our results demonstrate that the DA-ARC axis is engaged during HFD feeding, we did not investigate the precise role it plays during the development of obesity or an HFD-induced neuroplasticity, which will be the focus of future studies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ali D. Güler (aguler@virginia.edu).
Materials availability
AAV1-hSyn-FRT-TeNT-GFP and AAV1-Ef1a-ConFoff-hM3-HA generated in this study will be made available on request, but we may require a payment and/or a completed materials transfer agreement if there is potential for commercial application.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-cFos | Santa Cruz Biotechnology | Cat# sc-52; AB_2106783 (production is discontinued) |
| Rabbit polyclonal anti-dsRed | Clontech Laboratories | Cat#632496; RRID: AB_10013483 |
| Rabbit monoclonal anti-HA | Cell Signaling Technology | Cat#3724S; RRID: AB_1549585 |
| Chicken polyclonal anti-Tyrosine Hydroxylase (TH) | EMD Millipore Corporation | Cat# AB9702; RRID: AB_570923 |
| Chicken polyclonal anti-mCherry | Novus Biologicals | Cat# NBP2-25158, RRID:AB_2636881 |
| Goat polyclonal anti-GFP | Rockland Immunochemicals | Cat# 600-101-215, RRID:AB_218182 |
| Donkey polyclonal anti-rabbit Cy2 | Jackson ImmunoResearch | Cat# 711-225-152; RRID: AB_2340612 |
| Donkey polyclonal anti-rabbit Cy3 | Jackson ImmunoResearch | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey polyclonal anti-chicken Cy2 | Jackson ImmunoResearch | Cat# 703-225-155; RRID:AB_2340370 |
| Donkey polyclonal anti-chicken Cy3 | Jackson ImmunoResearch | Cat# 703-165-155; RRID:AB_2340363 |
| Donkey polyclonal anti-goat Alexa Fluor 488 | Jackson ImmunoResearch | Cat# 705-545-003; RRID:AB_2340428 |
| Bacterial and virus strains | ||
| AAV8-hSyn-DIO-hM3Dq-mCherry | Addgene | Cat# 44361-AAV8; RRID:Addgene_44361 |
| AAV8-hSyn-DIO-mCherry | Addgene | Cat# 50459-AAV8; RRID:Addgene_50459 |
| AAV2-Ef1a-DIO-hChR2(H134R)-EYFP-WPRE-pA | UNC vector core | Cat# VB4652; RRID:Addgene_55639 |
| AAV1-hSyn-FRT-TeNT-GFP | Lab of L.S. Zweifel | N/A |
| AAV1-Ef1a-ConFon-hM3-HA | https://vvf.ethz.ch | N/A |
| AAV1-Ef1a-ConFoff-hM3-HA | Lab of A.D. Güler | N/A |
| AAV8-hSyn-Con/Fon-ChR2-EYFP | Addgene | Cat# 55645-AAV8; RRID:Addgene_55645 |
| retrograde AAV-hSyn-DIO-hM3Dq-mCherry | Addgene | Cat# 44361-AAVrg; RRID:Addgene_44361) |
| AAV9-hSyn-GRAB-DA2h | Addgene | Cat# 140554-AAV9; RRID:Addgene_140554 |
| Chemicals, peptides, and recombinant proteins | ||
| Clozapine-N-oxide (CNO) | Sigma Aldrich | Cat# C0832 |
| Critical commercial assays | ||
| RNAscope® Multiplex Fluorescent Reagent Kit v2 Assay | Advanced cell diagnostics | Cat# 323100 |
| RNAscope® Probe- Mm-Drd1-C2 | Advanced cell diagnostics | Cat# 461901-C2 |
| RNAscope® Probe- Mm-AgRP | Advanced cell diagnostics | Cat# 400711 |
| RNAscope® Probe- Mm-Pomc-C3 | Advanced cell diagnostics | Cat# 314081-C3 |
| Deposited data | ||
| Custom MATLAB script for fiber photometry analysis | This paper | https://doi.org/10.5281/zenodo.7222038 |
| Experimental models: Organisms/strains | ||
| Mouse; C57BL6/J | Jackson Laboratory | Cat# JAX#000664; RRID: IMSR_JAX:000,664 |
| Drd1-Cre | Palmiter Lab, University of Washington | N/A |
| Drd1tm2.1Stl | Jackson Laboratory | Cat# JAX#025700; RRID: IMSR_JAX:025,700 |
| Agrptm1(cre)Lowl | Jackson Laboratory | Cat# JAX#012899; RRID: IMSR_JAX:012,899 |
| Npytm1.1(flpo)Hze | Jackson Laboratory | Cat# JAX#030211; RRID: IMSR_JAX:030,211 |
| Thtm1(cre/Esr1)Nat | Jackson Laboratory | Cat# JAX#008532; RRID: IMSR_JAX:008,532 |
| Software and algorithms | ||
| MATLAB R2019b | MathWorks | https://www.mathworks.com/products/matlab.html |
| Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Axiovision 4.6 software | Zeiss | https://www.zeiss.com/microscopy/us/downloads.html?vaURL=www.zeiss.com/microscopy/us/downloads/axiovision-downloads.html |
| ImageJ | NIH | https://ImageJ.nih.gov/ij/RRID:SCR_003070 |
| CLAMS-HC | Columbus instruments | https://www.colinst.com/product-detail?pid=35 |
| Other | ||
| Dapi-Fluoromount-G | Southern Biotechnology | Cat# OB010020 |
| Red Retrobeads IX | Lumafluor Inc | Cat# R180 |
| Stereotaxic apparatus | David Kopf Instruments | Model:1900 |
| Standard chow diet (Teklad F6 Rodent Diet) | Envigo | 8664 (production is terminated) |
| Standard chow diet (PicoLab Rodent Diet 20 5053) | LabDiet | Cat# 0007688 |
| High-fat high-sugar diet (Rodent Diet with 45% kcal% fat) | OpenSource Diets | D12451 |
| 4 valve solineoid perfusion exchange system, with hardware controller and 4 channel manifold | Automate Scientific | Economy Vlave Pinch System |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal care experiments were conducted in accordance with the University of Virginia Institutional Animal Care and Committee. Mice were housed in a temperature and humidity controlled vivarium at 22–24°C and ~40% humidity with a 12h/12h light/dark cycle. Adult male and female animals older than 8 weeks were used in all behavioral experiments with the following genotypes: C57BL6 (The Jackson Laboratory Cat# JAX#000664; RRID: IMSR_JAX:000,664), Drd1-Cre (Palmiter Lab, University of Washington), Drd1tm2.1Stl (The Jackson Laboratory Cat# JAX#025700; RRID: IMSR_JAX:025,700), Agrptm1(cre)Lowl (The Jackson Laboratory Cat# JAX#012899; RRID: IMSR_JAX:012,899), Npytm1.1(flpo)Hze (The Jackson Laboratory Cat# JAX#030211; RRID: IMSR_JAX:030,211), Thtm1(cre/Esr1)Nat (The Jackson Laboratory Cat# JAX#008532; RRID: IMSR_JAX:008,532).
METHOD DETAILS
Mouse diets
Standard diet (SD): Teklad 8664 (Envigo, United Kingdom: 3.07 kcal/gram; 19% fat, 31% protein, 50% carbohydrates. PicoLab Rodent Diet 20 5053 (3.07 kcal/gram; 13% fat, 24% protein, 62% carbohydrates; 3.2% sucrose). High-fat diet (HFD): Open Source D12451 (4.73 kcal/gram; 45% fat, 20% protein, 35% carbohydrates; 17% sucrose).
RNAseq analysis
The batch-corrected and normalized cluster-level expression values for Drd1, Drd2, Drd3, and Drd5 were downloaded from the Broad Single Cell Atlas and read into the R package to generate a heatmap with the following code:
expr < - read.csv(file = "ForAli_v4_heatmap.csv", header = TRUE, row.names = 1)
expr_mat < - data.matrix(expr)cols = colorRampPalette(c("white", "red")) (30)
pheatmap(expr_mat,border_color = "NA", scale = "none", show_rownames = TRUE, main = "Dopamine Receptor Isoforms Expressed by Arc-ME Neurons",cluster_rows = T, cluster_cols = T, legend = T, color = cols)
The number of Drd1 positive neurons from each molecular cluster according to the Broad Single Cell Atlas was quantified to make the bar graph.
RNAscope
Mice were anesthetized with ketamine and xylazine (i.p.) and perfused transcardially with PBS followed by 4% paraformaldehyde. Brains were harvested and fixed overnight in 4% paraformaldehyde at 4°C. Coronal sections were cut at 30 μm on a vibratome and dried on slides overnight in the dark and a hydrophobic barrier was applied to the slides. In situ hybridization was performed using the RNAscope Multiplex Fluorescent Reagent Kit v2 Assay (ACD) in accordance with the manufacturer’s instruction. Probes were used against mouse Drd1 (RNAscope Probe- Mm-Drd1-C2, Cat No. 461901), mouse AgRP (RNAscope Probe- Mm-AgRP, Cat No. 400711), and mouse POMC (RNAscope Probe- Mm-POMC-C3 Cat No. 314081). Sections were immersed with RNAscope hydrogen peroxide to block the activity of endogenous peroxidases. After a wash in distilled water, sections were permeabilized with RNAscope protease IV for 30 min at 40°C. Sections were hybridized with the Drd1, AgRP or POMC probe at 40°C for 2 h, followed by amplification incubation steps: Amp 1, 30 min at 40°C; Amp 2, 30 min at 40°C; Amp 3, 15 min at 40°C. HRP signals were developed with RNAscope Multiplex FL v2 HRP and TSA Plus fluorophores (1:750). Sections were washed with the provided washing buffer 2 × 2 min in between each step. Sections were then coverslipped with DAPI Fluoromount-G (Southern Biotech). For Figures 1D and S1B, sections were incubated with goat anti-GFP primary antibodies overnight and donkey anti-goat Alexa Fluor 488 secondary antibodies for 2 h at room temperature following the amplification incubation steps. Images were captured on a Zeiss Axioplan 2 imaging microscope equipped with an AxioCam MRm camera using AxioVision 4.6 software (Zeiss), or with a confocal Leica SP5 X imaging system in W.M. Keck Center for Cellular Imaging, University of Virginia. Brightness and contrast were corrected with ImageJ. The manipulation of the images between control and experimental conditions were kept consistent.
Immunohistochemistry
Mice were anesthetized with ketamine and xylazine (i.p.) and perfused transcardially with PBS followed by 4% paraformaldehyde. Brains were harvested and fixed overnight in 4% paraformaldehyde at 4°C and then were then placed into 30% sucrose for around 24h until they sank to the bottom of the container. The tissue was sectioned coronally at 30 μm on a cryostat and immersed in PBS with 0.004% sodium azide. Sections were treated with 3% donkey serum in 0.3% Triton X-100 in PBS for 30 min, and then incubated with primary antibodies overnight at 4°C. After washing with PBS (5 min × 2), brain sections were incubated with secondary antibodies for 2 h at room temperature. Sections were washed in PBS (5 min × 2) and mounted using DAPI Fluoromount-G (Southern Biotech). Images were captured on a Zeiss Axioplan 2 Imaging microscope equipped with an AxioCam MRm camera using AxioVision 4.6 software (Zeiss). Brightness and contrast were corrected with ImageJ. The manipulation of the images between control and experimental conditions were kept consistent.
For c-Fos staining, Drd1-ChR2 mice were treated with a 20-min, 20-Hz, and 2s-on-3s-off 473 nm laser above the ARC 90 min before anesthesia; Drd1-hM3Dq and Drd1-Ctrl mice were i.p. injected with 0.3 mg/kg CNO two hours before anesthesia. Brain sections were treated with 5% donkey serum in 0.3% Triton X-100 in PBS for 1 h, and then incubated with rabbit anti-c-Fos (1:200) for 20h at room temperature. After washing with PBS, brain sections were incubated with secondary donkey anti-rabbit Cy2 (1:250) for 3h at room temperature.
The following primary antibodies were used for fluorescent labeling: rabbit anti-c-Fos (1:200, Santa Cruz Sc-52), rabbit anti-DsRed (1:1000, Clontech 632,496), rabbit anti-HA (1:500, Cell Signaling Technology C29F4), chicken anti-TH (1:500, Millipore AB9702), chicken anti-mCherry (1:500, NOVUSBIO NBP2-25158), goat anti-GFP (1:500, Rockland Immunochemicals Inc 600-101-215). The secondary antibodies (Jackson ImmunoResearch) used were Cy2-or Cy3-conjugated donkey anti-rabbit (1:250), donkey anti-chicken (1:250) and donkey anti-goat Alexa Fluor 488 (1:250).
Stereotaxic surgery
For stereotaxic injections of virus vectors, mice older than 8 weeks were anesthetized with 5% gaseous isoflurane in a closed container, and then placed into the Kopf Small Animal Stereotaxic Frame on a heating pad to maintain body temperature. The isoflurane level was lowered to 2–3% when breathing rhythms steadily reached one breath per 1–2 s. The skull was exposed via a small incision after 0.8 mL bupivacaine injection under the scalp. A small hole in the skull was drilled above the targeted injection site. A 26 gauge Hamilton syringe was inserted into the ARC (coordinates: bregma: AP: −1.40 mm, DV: −5.90 mm, L: 0.30 mm) and 500 nL of the virus was bilaterally injected at a flow rate of 100 nL/min using an automated delivery system (World Precision Instruments microsyringe controller). For optogenetic surgeries, a fiber optic cannula was implanted above the ARC following the injection of the virus (coordinates: bregma: AP: −1.40 mm, DV: −5.65 mm, L: 0.30 mm). The exposed end of the fiber optic cannula was fixed to the skull with Metabond. Mice were allowed to recover for 3 weeks before any behavioral tests. All surgical procedures were performed in sterile conditions and in accordance with University of Virginia IACUC guidelines.
Viral constructs
AAV8-hSyn-DIO-hM3Dq-mCherry (Addgene 44,361-AAV8; 9 × 10^12 viral genomes/mL), AAV8-hSyn-DIO-mCherry (Addgene 50,459-AAV8; 1.2 × 10^13 viral genomes/mL), AAV2-Ef1a-DIO-hChR2(H134R)-EYFP-WPRE-pA (University of North Carolina at Chapel Hill Gene Therapy Center Vector Core Services; 2.1 × 10^13 viral genomes/mL), AAV1-hSyn-FRT-TeNT-GFP (generated and produced in the Zweifel lab; 10^12 viral genomes/mL), AAV1-Ef1a-ConFon-hM3-HA (ssAAV-1/2-hEf1alpha/hTLV1-Fon/Con [dFRT-HA_hM3D(Gq) (rev)-I-dlox-I-HA_hM3D(Gq)-I-dlox-I-HA_hM3D(Gq) (rev)-dFRT]-WPRE-hGHp(A); entry v605 at https://vvf.ethz.ch; 4.6 × 10^12 viral genomes/mL), AAV1-Ef1a-ConFoff-hM3-HA (generated in the Güler lab, produced at the University of North Carolina at Chapel Hill Gene Therapy Center Vector Core Services; 2.2 × 10^13 viral genomes/mL), AAV8-hSyn-Con/Fon-ChR2-EYFP (Addgene 55,645-AAV8; 2.1 × 10^13 viral genomes/mL), retrograde AAV-hSyn-DIO-hM3Dq-mCherry (Addgene 44361-AAVrg; 7 × 10^12 viral genomes/mL), AAV9-hSyn-GRAB-DA2h (Addgene 140,554-AAV9; 2.5 × 10^13 viral genomes/mL) were injected to the ARC (ML: ± 0.30 mm, AP: − 1.40 mm, DV: −5.90 mm). All coordinates are relative to bregma (George Paxinos and Keith B. J. Franklin).
Food intake analysis
For optogenetic food intake analysis, adult male Drd1-Cre and AgRP-Cre mice were single-housed for at least 3 days and habituated to the head-tether before the test. During the test day, after a brief habituation, mice were given no laser stimulation for 1 h, laser stimulation for 1 h, and no laser stimulation for another hr. One pellet of the SD around 4 g was provided. The amount of food consumption within each hour was manually measured. The laser stimulation was 20 Hz 473 nm blue light with a 2s-on-3s-off pattern. The light power exiting the fiber optic cable measured by an optical power meter (Thorlabs) was 7–8 mW in all experiments.
For chemogenetic food intake analysis, adult male Drd1-Cre and AgRP-Cre mice and adult male and female Drd1-Cre; NPY-flp mice were single-housed for at least 3 days and habituated to i.p. injection of saline before the test. During the test, mice were i.p. injected with saline on days 1, 5, and 6, and i.p. injected with 0.3 mg/kg CNO on days 2, 3 and 4. The injections occur at the beginning of the light cycle, from ZT2 to ZT3. Three pellets of SD were placed on the floor of the home cages with the total weight of food around 13 g. Food consumption was manually measured at 1h, 2h, 3h, 4h, and 24h after the injections. Body weights were measured 24h after injections.
For HFD intake and body weight measurements, 16-week to 24-week male control and AgRPDrd1–KO littermates were provided with ad lib access to HFD for 8 weeks. 10 pellets of pre-weighed HFD placed on the cage floor were weighed and refreshed weekly. Body weights were measured weekly.
Open field 8-pellet SD foraging analysis
All the open field 8-pellet SD foraging experiments were conducted after the food intake analysis (Figures 3D-3I). Adult male Drd1-hM3Dq mice and AgRP-hM3Dq mice were habituated to the test conditions on day 1. On day 5, mice were placed in the open field before the test for 20 min. During the test, Drd1-hM3Dq mice and AgRP-hM3Dq mice were injected with 0.3 mg/kg CNO and placed into the open field without food for 20 min for CNO to take effect. Mice were put into their home cage without food when 8 pellets of the SD were buried under the 3-cm-thick corn bedding in the open field, and then placed back into the open field. Behavior was recorded with a camera above the open-field chamber for 20 min. The total number of food pellets found within 20 min, the latency to the first and second food pellets, the feeding time of the first food pellet, and the total feeding time were quantified offline. Mice that failed to find the second food pellet were not included in the analysis of the latency to the second food pellet (Drd1-hM3Dq: n = 2; AgRP-hM3Dq: n = 1).
Comprehensive lab animal monitoring system (CLAMS)
The CLAMS system (Columbus Instruments) was used for indirect calorimeter and ambulatory locomotor activity measurements during ad lib access to SCD or HFD. For chemogenetic activation experiments, mice previously used in the food intake analysis were acclimated to metabolic cages for 3 days and habituated to i.p. injection on the second day. During test days, mice were injected with saline or CNO between ZT 3 to ZT 4 and then monitored for 24 h following the manufacturer’s instructions. Adult male control and AgRPDrd1-KO mice were acclimated to metabolic cages for 2 days, and then monitored for 48 h with ad lib access to SD. Mice were switched to HFD afterward and then monitored for another 48 h with ad lib access to HFD.
Energy expenditure (EE) in watts per kilogram of lean mass [W/kg] was calculated with the following formula63:
EE[W/kg] = 1/60*((0.2716[W*min/mL]*VO2[mL/kg/h])+(0.07616[W*min/mL]*VCO2[mL/kg/h])
The unit Watts was converted to kcal/h by multiplying factor of 0.86 to report EE as kcal/h/kg of lean mass.
Locomotor activity was reported as the sum of ambulatory beam breaks in x and y directions.
Retrograde tracing
Retro-beads tracing: 300 nL of fluorescent retrobeads (Lumafluor) were bilaterally injected to the ARC of adult male WT mice. Brains were processed 3 days after retrobead delivery.
Retrograde AAV tracing: 500nL retrograde AAV-Con-hM3Dq-mCherry was bilaterally injected to the ARC of adult male TH-IRES-CreER mice. Brains were processed three weeks after AAV delivery for immunohistochemistry as described above.
Open field HFD foraging analysis
Open field HFD foraging analysis was conducted as described previously.36 8-week to 9-week old male control and AgRPDrd1-KO littermates were single-housed for at least one week and habituated to the HFD on day 1 and day 2 with 1-h access to one pellet of the HFD provided each day. On day 3, mice were habituated to the open field for 10 min without food. On day 5, control mice and AgRPDrd1-KO mice were placed into the open field with one pellet of the HFD buried under the 3cm-thick corn bedding. Behavior was recorded with a camera above the open-field chamber for 10 min and latency to food was quantified offline. The mice that failed to find the food pellet after 10 min were given a latency score of 10 min. One outlier in the AgRPDrd1-KO group was detected by ROUT outlier test (Q = 1%) and excluded for analysis and data presentation.
Open field HFD refeeding analysis
The same cohort of control and AgRPDrd1-KO littermates previously used in the open field HFD foraging experiment were used in the open field HFD foraging experiment when 12-week to 15-week old. Mice were overnight fasted for 16 h before the test day. During the test day, control mice and AgRPDrd1-KO mice were placed into the open field with one pellet of the HFD provided. Behavior was recorded with a camera above the open-field chamber for 10 min and consumption of the HFD was measured manually.
Fiber photometry recording
Adult male and female WT mice were previously habituated to HFD, NFO (a lego brick), and experimental conditions and were ad lib fed on SD or overnight fasted for 16 h before the test day. During the test day, mice were transferred to the recording room and individually housed in their home cages with the cage top left open. Implanted fiber-optic cannula (Thorlabs, 0.39 NA, ∅200 μm Core) was connected to fiber-optic cable (Doric Lenses, 0.37 NA, ∅200 μm Core) with the use of a zirconia mating sleeve (Doric Lenses). The fiber-optic cables were wrapped with metal sheath (Doric Lenses) to prevent breakage and connected to rotary joints (Doric Lenses) to allow free-movement. Mice were allowed to acclimate to the experiment environment and fiber-optic tethering 10 min before recording. The fiber photometry system used in this work records the fluorescent signal from both DA-dependent (465 nm) and DA-independent isosbestic (405 nm) excitation light wavelengths, in which the isosbestic wavelength excitation signal can be used to control for the artifacts from animal movement, fluorescent reporter expression and photobleaching. The light power exiting the fiber optic cable measured by an optical power meter (Thorlabs) was 20–30 μW for 465 nm and 4–5 μW for 405 nm in all experiments. Signals were collected at 120Hz sampling rate, with manually entered timestamps during the experiment tracked automatically by the software. During the test, 10 min of baseline signal was recorded prior to first delivery of food/object. 3 pellets of HFD, SD, or NFO were delivered and withdrawn at the set time of the recording session (delivery: 600s, 960s, 1320s; withdrawal: 780s, 1140s, 1500s).
Fiber photometry data analysis
Data were processed in MATLAB R2019b with custom scripts. Recordings from 90s before to 180 s after treatments (food delivery or withdrawal) were extracted as individual sessions for further analysis. To eliminate the artifact from motion or protein expression, the DA-independent isosbestic control (405nm induced) signal was fitted to the DA-dependent (465nm induced) signal with a linear least-squares fit. To normalize the variability caused by signal strength, Z score ((signal-average of baseline signal)/standard deviation of baseline signal) is calculated to present the data where the first 15 s of the 270 s sessions were used as baseline. To account for different photobleaching dynamics, each analyzed curve was further detrended by an exponential fit where the first 60 s were used to calculate the coefficients.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of histological images
30 μm thick brain sections containing brain regions of interest were collected from adult male mice. The determination of the brain region borders were guided by DAPI nuclear staining. Neurons labeled by fluorescent markers were blindly counted by trained observers. Cell counts of multiple sections were averaged for each animal for further statistical analysis.
Statistical analysis
When comparing two groups of normally distributed data, a Student’s two tailed t test was used. In experiments with a single variable across more than two groups, an one-way ANOVA was performed. To compare the effects of genotypes and treatments within 4 groups, a two-way ANOVA test was used. To compare the effects of genotype and treatment within 4 groups at multiple time points, a three-way ANOVA test was performed. Following a significant effect in the ANOVA test, Bonferroni’s post hoc comparison was used to determine differences between individual data points. Analyses were conducted using the GraphPad Prism 8 statistical software for Windows. All data are presented as means ± standard error of the mean with p < 0.05 considered statistically significant.
Supplementary Material
Highlights.
Dopamine is released in the ARC in response to food
ARCDrd1 neurons regulate feeding and energy homeostasis
Stimulation of ARCDrd1/AgRP/NPY neurons is sufficient to induce feeding
Drd1 signaling in the ARCAgRP/NPY neurons promotes HFD consumption
ACKNOWLEDGMENTS
Research was supported by NIH National Institute of General Medicinal Sciences R01GM121937 and R35GM140854 (A.D.G.), UVA Faculty Start-up Funds (A.D.G.), UVA Brain Institute 2018 Seed Funding Award (A.D.G.), and UVA Presidential Fellowship for Collaborative Neuroscience (Q.Z.). The authors thank all current and previous members of Dr. Güler’s and Dr. Deppmann’s labs for their technical and mental support. We thank Dr. Campbell’s lab for their technical support for RNAscope experiments. We thank Hanns Ulrich Zeilhofer and Hendrik Wildner for the AAV1-ConFon-hM3Dq construct which was produced by the University of Zurich Viral Vector Facility. We thank Aundrea Rainwater and Ricardo Salinas for their help with animal husbandry. We would also like to thank Dr. Calhan, Dr. Deppmann, and Dr. Provencio for their input on the manuscript. We acknowledge the Keck Center for the use of the Leica confocal microscopy system (RR025616).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111718.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Piché ME, Tchernof A, and Després JP (2020). Obesity phenotypes, diabetes, and cardiovascular diseases. Circ. Res 126, 1477–1500. 10.1161/CIRCRESAHA.120.316101. [DOI] [PubMed] [Google Scholar]
- 2.Chaput JP, Klingenberg L, Astrup A, and Sjödin AM (2011). Modern sedentary activities promote overconsumption of food in our current obesogenic environment. Obes. Rev 12, e12–e20. 10.1111/j.1467-789X.2010.00772.x. [DOI] [PubMed] [Google Scholar]
- 3.Ferrario CR, Labouèbe G, Liu S, Nieh EH, Routh VH, Xu S, and O’Connor EC (2016). Homeostasis meets motivation in the battle to control food intake. J. Neurosci 36, 11469–11481. 10.1523/JNEUROSCI.2338-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joly-Amado A, Cansell C, Denis RGP, Delbes A-S, Castel J, Martinez S, and Luquet S (2014). The hypothalamic arcuate nucleus and the control of peripheral substrates. Best Pract. Res. Clin. Endocrinol. Metab 28, 725–737. 10.1016/j.beem.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Hahn TM, Breininger JF, Baskin DG, and Schwartz MW (1998). Co-expression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat. Neurosci 1, 271–272. 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- 6.Liu T, Kong D, Shah BP, Ye C, Koda S, Saunders A, Ding JB, Yang Z, Sabatini BL, and Lowell BB (2012). Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron 73, 511–522. 10.1016/j.neuron.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi KA, and Cone RD (2005). Fasting induces a large, leptin-dependent increase in the intrinsic action potential frequency of orexigenic arcuate nucleus neuropeptide Y/Agouti-related protein neurons. Endocrinology 146, 1043–1047. 10.1210/en.2004-1397. [DOI] [PubMed] [Google Scholar]
- 8.Andrews ZB, Liu Z-W, Walllingford N, Erion DM, Borok E, Friedman JM, Tschöp MH, Shanabrough M, Cline G, Shulman GI, et al. (2008). UCP2 mediates ghrelin’s action on NPY/AgRP neurons by lowering free radicals. Nature 454, 846–851. 10.1038/nature07181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HY, Trumbauer ME, Chen AS, Weingarth DT, Adams JR, Frazier EG, Shen Z, Marsh DJ, Feighner SD, Guan XM, et al. (2004). Orexigenic action of peripheral ghrelin is mediated by neuropeptide Y and agouti-related protein. Endocrinology 145, 2607–2612. 10.1210/en.2003-1596. [DOI] [PubMed] [Google Scholar]
- 10.Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL, and Lowell BB (2011). Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest 121, 1424–1428. 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Small CJ, Kim MS, Stanley SA, Mitchell JR, Murphy K, Morgan DG, Ghatei MA, and Bloom SR (2001). Effects of chronic central nervous system administration of agouti-related protein in pair-fed animals. Diabetes 50, 248–254. 10.2337/diabetes.50.2.248. [DOI] [PubMed] [Google Scholar]
- 12.Ruan H-B, Dietrich MO, Liu Z-W, Zimmer MR, Li M-D, Singh JP, Zhang K, Yin R, Wu J, Horvath TL, and Yang X (2014). O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell 159, 306–317. 10.1016/j.cell.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aponte Y, Atasoy D, and Sternson SM (2011). AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat. Neurosci 14, 351–355. 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmiter RD (2007). Is dopamine a physiologically relevant mediator of feeding behavior? Trends Neurosci. 30, 375–381. 10.1016/j.tins.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Narayanan NS, Guarnieri DJ, and DiLeone RJ (2010). Metabolic hormones, dopamine circuits, and feeding. Front. Neuroendocrinol 31, 104–112. 10.1016/j.yfrne.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coccurello R, and Maccarrone M (2018). Hedonic eating and the “delicious circle”: from lipid-derived mediators to brain dopamine and back. Front. Neurosci 12, 271. 10.3389/fnins.2018.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szczypka MS, Rainey MA, Kim DS, Alaynick WA, Marck BT, Matsumoto AM, and Palmiter RD (1999). Feeding behavior in dopamine-deficient mice. Proc. Natl. Acad. Sci. USA 96, 12138–12143. 10.1073/pnas.96.21.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salamone JD, and Correa M (2012). The mysterious motivational functions of mesolimbic dopamine. Neuron 76, 470–485. 10.1016/j.neuron.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wise RA (2004). Dopamine, learning and motivation. Nat. Rev. Neurosci 5, 483–494. 10.1038/nrn1406. [DOI] [PubMed] [Google Scholar]
- 20.Salamone JD, Correa M, Mingote S, and Weber SM (2003). Nucleus accumbens dopamine and the regulation of effort in food-seeking behavior: implications for studies of natural motivation, psychiatry, and drug abuse. J. Pharmacol. Exp. Ther 305, 1–8. 10.1124/jpet.102.035063. [DOI] [PubMed] [Google Scholar]
- 21.Fields HL, Hjelmstad GO, Margolis EB, and Nicola SM (2007). Ventral tegmental area neurons in learned appetitive behavior and positive reinforcement. Annu. Rev. Neurosci 30, 289–316. 10.1146/annurev.neuro.30.051606.094341. [DOI] [PubMed] [Google Scholar]
- 22.Berridge KC (2007). The debate over dopamine’s role in reward: the case for incentive salience. Psychopharmacology (Berl) 191, 391–431. 10.1007/s00213-006-0578-x. [DOI] [PubMed] [Google Scholar]
- 23.Boekhoudt L, Roelofs TJM, de Jong JW, de Leeuw AE, Luijendijk MCM, Wolterink-Donselaar IG, van der Plasse G, and Adan RAH (2017). Does activation of midbrain dopamine neurons promote or reduce feeding? Int. J. Obes 41, 1131–1140. 10.1038/ijo.2017.74. [DOI] [PubMed] [Google Scholar]
- 24.Gudelsky GA (1981). Tuberoinfundibular dopamine neurons and the regulation of prolactin secretion. Psychoneuroendocrinology 6, 3–16. 10.1016/0306-4530(81)90044-5. [DOI] [PubMed] [Google Scholar]
- 25.Stagkourakis S, Dunevall J, Taleat Z, Ewing AG, and Broberger C (2019). Dopamine release dynamics in the tuberoinfundibular dopamine system. J. Neurosci 39, 4009–4022. 10.1523/JNEUROSCI.2339-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, and van den Pol AN (2016). Hypothalamic arcuate nucleus tyrosine hydroxylase neurons play orexigenic role in energy homeostasis. Nat. Neurosci 19, 1341–1347. 10.1038/nn.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriya S, and Kuwaki T (2021). A13 dopamine cell group in the zona incerta is a key neuronal nucleus in nociceptive processing. Neural Regen. Res 16, 1415–1416. 10.4103/1673-5374.300991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, Alpár A, Mulder J, Clotman F, Keimpema E, et al. (2017). Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci 20, 176–188. 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alhadeff AL, Goldstein N, Park O, Klima ML, Vargas A, and Betley JN (2019). Natural and drug rewards engage distinct pathways that converge on coordinated hypothalamic and reward circuits. Neuron 103, 891–908, e6. 10.1016/j.neuron.2019.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero-Fernandez W, Borroto-Escuela DO, Vargas-Barroso V, Narváez M, Di Palma M, Agnati LF, Larriva Sahd J, and Fuxe K (2014). Dopamine D1 and D2 receptor immunoreactivities in the arcuate-median eminence complex and their link to the tubero-infundibular dopamine neurons. Eur. J. Histochem 58, 2400. 10.4081/ejh.2014.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chadwick SR, and Güler AD (2022). Local Drd1-neurons input to subgroups of arcuate AgRP/NPY-neurons. iScience 25, 104605. 10.1016/j.isci.2022.104605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AMJ, Resch JM, McCarroll SA, et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci 20, 484–496. 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heusner CL, Beutler LR, Houser CR, and Palmiter RD (2008). Deletion of GAD67 in dopamine receptor-1 expressing cells causes specific motor deficits. Genesis 46, 357–367. 10.1002/dvg.20405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavalcanti-de-Albuquerque JP, Bober J, Zimmer MR, and Dietrich MO (2019). Regulation of substrate utilization and adiposity by Agrp neurons. Nat. Commun 10, 311. 10.1038/s41467-018-08239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun F, Zhou J, Dai B, Qian T, Zeng J, Li X, Zhuo Y, Zhang Y, Wang Y, Qian C, et al. (2020). Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nat. Methods 17, 1156–1166. 10.1038/s41592-020-00981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grippo RM, Tang Q, Zhang Q, Chadwick SR, Gao Y, Altherr EB, Sipe L, Purohit AM, Purohit NM, Sunkara MD, et al. (2020). Dopamine signaling in the suprachiasmatic nucleus enables weight gain associated with hedonic feeding. Curr. Biol 30, 196–208, e8. 10.1016/j.cub.2019.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathod YD, and Di Fulvio M (2021).The feeding microstructure of male and female mice. PLoS One 16, e0246569. 10.1371/journal.pone.0246569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulton S (2010). Appetite and reward. Front. Neuroendocrinol 31, 85–103. 10.1016/j.yfrne.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Kim ER, Xu Y, Cassidy RM, Lu Y, Yang Y, Tian J, Li D-P, Van Drunen R, Ribas-Latre A, Cai Z-L, et al. (2020). Paraventricular hypothalamus mediates diurnal rhythm of metabolism. Nat. Commun 11, 3794. 10.1038/s41467-020-17578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo SX, Huang J, Li Q, Mohammad H, Lee C-Y, Krishna K, Kok AM-Y, Tan YL, Lim JY, Li H, et al. (2018). Regulation of feeding by somatostatin neurons in the tuberal nucleus. Science 361, 76–81. 10.1126/science.aar4983. [DOI] [PubMed] [Google Scholar]
- 41.Jais A, Paeger L, Sotelo-Hitschfeld T, Bremser S, Prinzensteiner M, Klemm P, Mykytiuk V, Widdershooven PJM, Vesting AJ, Grzelka K, et al. (2020). PNOCARC neurons promote hyperphagia and obesity upon high-fat-diet feeding. Neuron 106, 1009–1025, e10. 10.1016/j.neuron.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan C, Zhou J, Feng Q, Zhang J-E, Lin S, Bao J, Wu P, and Luo M (2013). Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J. Neurosci 33, 3624–3632. 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fenselau H, Campbell JN, Verstegen AMJ, Madara JC, Xu J, Shah BP, Resch JM, Yang Z, Mandelblat-Cerf Y, Livneh Y, and Lowell BB (2017). A rapidly acting glutamatergic ARC→PVH satiety circuit postsynaptically regulated by α-MSH. Nat. Neurosci 20, 42–51. 10.1038/nn.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jais A, and Brüning JC (2022). Arcuate nucleus-dependent regulation of metabolism - pathways to obesity and diabetes mellitus. Endocr. Rev 43, 314–328. 10.1210/endrev/bnab025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Lin Y-C, Kuo T-W, and Knight ZA (2015). Sensory detection of food rapidly modulates arcuate feeding circuits. Cell 160, 829–841. 10.1016/j.cell.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denis RGP, Joly-Amado A, Webber E, Langlet F, Schaeffer M, Padilla SL, Cansell C, Dehouck B, Castel J, Delbès AS, et al. (2015). Palatability can drive feeding independent of agrp neurons. Cell Metab. 22, 646–657. 10.1016/j.cmet.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakajima K.i., Cui Z, Li C, Meister J, Cui Y, Fu O, Smith AS, Jain S, Lowell BB, Krashes MJ, and Wess J (2016). Gs-coupled GPCR signalling in AgRP neurons triggers sustained increase in food intake. Nat. Commun 7, 10268. 10.1038/ncomms10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ewbank SN, Campos CA, Chen JY, Bowen AJ, Padilla SL, Dempsey JL, Cui JY, and Palmiter RD (2020). Chronic Gq signaling in AgRP neurons does not cause obesity. Proc. Natl. Acad. Sci. USA 117, 20874–20880. 10.1073/pnas.2004941117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzone CM, Liang-Guallpa J, Li C, Wolcott NS, Boone MH, Southern M, Kobzar NP, Salgado I.d.A., Reddy DM, Sun F, et al. (2020). High-fat food biases hypothalamic and mesolimbic expression of consummatory drives. Nat. Neurosci 23, 1253–1266. 10.1038/s41593-020-0684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reichenbach A, Clarke RE, Stark R, Lockie SH, Mequinion M, Dempsey H, Rawlinson S, Reed F, Sepehrizadeh T, DeVeer M, et al. (2022). Metabolic sensing in AgRP neurons integrates homeostatic state with dopamine signalling in the striatum. Elife 11, e72668. 10.7554/eLife.72668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Betley JN, Xu S, Cao ZFH, Gong R, Magnus CJ, Yu Y, and Sternson SM (2015). Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature 521, 180–185. 10.1038/nature14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beutler LR, Chen Y, Ahn JS, Lin Y-C, Essner RA, and Knight ZA (2017). Dynamics of gut-brain communication underlying hunger. Neuron 96, 461–475, e5. 10.1016/j.neuron.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Padilla SL, Carmody JS, and Zeltser LM (2010). Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat. Med 16, 403–405. 10.1038/nm.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JAJ, et al. (2012). Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860. 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rolls BJ, Roe LS, Meengs JS, and Wall DE (2004). Increasing the portion size of a sandwich increases energy intake. J. Am. Diet Assoc 104, 367–372. 10.1016/j.jada.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 56.Rolls BJ, Morris EL, and Roe LS (2002). Portion size of food affects energy intake in normal-weight and overweight men and women. Am. J. Clin. Nutr 76, 1207–1213. 10.1093/ajcn/76.6.1207. [DOI] [PubMed] [Google Scholar]
- 57.Rolls BJ, Roe LS, Kral TVE, Meengs JS, and Wall DE (2004). Increasing the portion size of a packaged snack increases energy intake in men and women. Appetite 42, 63–69. 10.1016/S0195-6663(03)00117-X. [DOI] [PubMed] [Google Scholar]
- 58.Ma Y, Bertone ER, Stanek EJ, Reed GW, Hebert JR, Cohen NL, Merriam PA, and Ockene IS (2003). Association between eating patterns and obesity in a free-living US adult population. Am. J. Epidemiol 158, 85–92. 10.1093/aje/kwg117. [DOI] [PubMed] [Google Scholar]
- 59.Lerant A, Herman ME, and Freeman ME (1996). Dopaminergic neurons of periventricular and arcuate nuclei of pseudopregnant rats: semicircadian rhythm in Fos-related antigens immunoreactivities and in dopamine concentration. Endocrinology 137, 3621–3628. 10.1210/endo.137.9.8756525. [DOI] [PubMed] [Google Scholar]
- 60.Ford CP, Gantz SC, Phillips PEM, and Williams JT (2010). Control of extracellular dopamine at dendrite and axon terminals. J. Neurosci 30, 6975–6983. 10.1523/JNEUROSCI.1020-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C, Goel P, and Kaeser PS (2021). Spatial and temporal scales of dopamine transmission. Nat. Rev. Neurosci 22, 345–358. 10.1038/s41583-021-00455-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beutler LR, Corpuz TV, Ahn JS, Kosar S, Song W, Chen Y, and Knight ZA (2020). Obesity causes selective and long-lasting desensitization of AgRP neurons to dietary fat. Elife 9, e55909. 10.7554/eLife.55909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer AW, Cannon B, and Nedergaard J (2018). Optimal housing temperatures for mice to mimic the thermal environment of humans: an experimental study. Mol. Metab 7, 161–170. 10.1016/j.molmet.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-cFos | Santa Cruz Biotechnology | Cat# sc-52; AB_2106783 (production is discontinued) |
| Rabbit polyclonal anti-dsRed | Clontech Laboratories | Cat#632496; RRID: AB_10013483 |
| Rabbit monoclonal anti-HA | Cell Signaling Technology | Cat#3724S; RRID: AB_1549585 |
| Chicken polyclonal anti-Tyrosine Hydroxylase (TH) | EMD Millipore Corporation | Cat# AB9702; RRID: AB_570923 |
| Chicken polyclonal anti-mCherry | Novus Biologicals | Cat# NBP2-25158, RRID:AB_2636881 |
| Goat polyclonal anti-GFP | Rockland Immunochemicals | Cat# 600-101-215, RRID:AB_218182 |
| Donkey polyclonal anti-rabbit Cy2 | Jackson ImmunoResearch | Cat# 711-225-152; RRID: AB_2340612 |
| Donkey polyclonal anti-rabbit Cy3 | Jackson ImmunoResearch | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey polyclonal anti-chicken Cy2 | Jackson ImmunoResearch | Cat# 703-225-155; RRID:AB_2340370 |
| Donkey polyclonal anti-chicken Cy3 | Jackson ImmunoResearch | Cat# 703-165-155; RRID:AB_2340363 |
| Donkey polyclonal anti-goat Alexa Fluor 488 | Jackson ImmunoResearch | Cat# 705-545-003; RRID:AB_2340428 |
| Bacterial and virus strains | ||
| AAV8-hSyn-DIO-hM3Dq-mCherry | Addgene | Cat# 44361-AAV8; RRID:Addgene_44361 |
| AAV8-hSyn-DIO-mCherry | Addgene | Cat# 50459-AAV8; RRID:Addgene_50459 |
| AAV2-Ef1a-DIO-hChR2(H134R)-EYFP-WPRE-pA | UNC vector core | Cat# VB4652; RRID:Addgene_55639 |
| AAV1-hSyn-FRT-TeNT-GFP | Lab of L.S. Zweifel | N/A |
| AAV1-Ef1a-ConFon-hM3-HA | https://vvf.ethz.ch | N/A |
| AAV1-Ef1a-ConFoff-hM3-HA | Lab of A.D. Güler | N/A |
| AAV8-hSyn-Con/Fon-ChR2-EYFP | Addgene | Cat# 55645-AAV8; RRID:Addgene_55645 |
| retrograde AAV-hSyn-DIO-hM3Dq-mCherry | Addgene | Cat# 44361-AAVrg; RRID:Addgene_44361) |
| AAV9-hSyn-GRAB-DA2h | Addgene | Cat# 140554-AAV9; RRID:Addgene_140554 |
| Chemicals, peptides, and recombinant proteins | ||
| Clozapine-N-oxide (CNO) | Sigma Aldrich | Cat# C0832 |
| Critical commercial assays | ||
| RNAscope® Multiplex Fluorescent Reagent Kit v2 Assay | Advanced cell diagnostics | Cat# 323100 |
| RNAscope® Probe- Mm-Drd1-C2 | Advanced cell diagnostics | Cat# 461901-C2 |
| RNAscope® Probe- Mm-AgRP | Advanced cell diagnostics | Cat# 400711 |
| RNAscope® Probe- Mm-Pomc-C3 | Advanced cell diagnostics | Cat# 314081-C3 |
| Deposited data | ||
| Custom MATLAB script for fiber photometry analysis | This paper | https://doi.org/10.5281/zenodo.7222038 |
| Experimental models: Organisms/strains | ||
| Mouse; C57BL6/J | Jackson Laboratory | Cat# JAX#000664; RRID: IMSR_JAX:000,664 |
| Drd1-Cre | Palmiter Lab, University of Washington | N/A |
| Drd1tm2.1Stl | Jackson Laboratory | Cat# JAX#025700; RRID: IMSR_JAX:025,700 |
| Agrptm1(cre)Lowl | Jackson Laboratory | Cat# JAX#012899; RRID: IMSR_JAX:012,899 |
| Npytm1.1(flpo)Hze | Jackson Laboratory | Cat# JAX#030211; RRID: IMSR_JAX:030,211 |
| Thtm1(cre/Esr1)Nat | Jackson Laboratory | Cat# JAX#008532; RRID: IMSR_JAX:008,532 |
| Software and algorithms | ||
| MATLAB R2019b | MathWorks | https://www.mathworks.com/products/matlab.html |
| Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Axiovision 4.6 software | Zeiss | https://www.zeiss.com/microscopy/us/downloads.html?vaURL=www.zeiss.com/microscopy/us/downloads/axiovision-downloads.html |
| ImageJ | NIH | https://ImageJ.nih.gov/ij/RRID:SCR_003070 |
| CLAMS-HC | Columbus instruments | https://www.colinst.com/product-detail?pid=35 |
| Other | ||
| Dapi-Fluoromount-G | Southern Biotechnology | Cat# OB010020 |
| Red Retrobeads IX | Lumafluor Inc | Cat# R180 |
| Stereotaxic apparatus | David Kopf Instruments | Model:1900 |
| Standard chow diet (Teklad F6 Rodent Diet) | Envigo | 8664 (production is terminated) |
| Standard chow diet (PicoLab Rodent Diet 20 5053) | LabDiet | Cat# 0007688 |
| High-fat high-sugar diet (Rodent Diet with 45% kcal% fat) | OpenSource Diets | D12451 |
| 4 valve solineoid perfusion exchange system, with hardware controller and 4 channel manifold | Automate Scientific | Economy Vlave Pinch System |