ABSTRACT
OTUD1 is a deubiquitinating enzyme involved in many cellular processes including cancer and innate, immune signaling pathways. Here, we perform a proximity labeling-based interactome study that identifies OTUD1 largely present in the translation and RNA metabolism protein complexes. Biochemical analysis validates OTUD1 association with ribosome subunits, elongation factors and the E3 ubiquitin ligase ZNF598 but not with the translation initiation machinery. OTUD1 catalytic activity suppresses polyA triggered ribosome stalling through inhibition of ZNF598-mediated RPS10 ubiquitination and stimulates formation of polysomes. Finally, analysis of gene expression suggests that OTUD1 regulates the stability of rare codon rich mRNAs by antagonizing ZNF598.
KEYWORDS: ribosome stalling, translation, ubiquitination
INTRODUCTION
Translation is a fundamental and highly sophisticated process required by cells to fulfill their protein needs. To ensure the quality of synthesized proteins, all phases of translation are tightly regulated (1–4). Events following protein synthesis such as folding can also have a direct impact on translation (5). Additionally, intrinsic features of every mRNA influence ribosome processivity. Encountering senseless, unreadable, or damaged sequence usually leads to translation arrest (6–9). Similarly, a high abundance of rare codons can limit the translation rate (10, 11).
Defects in mRNA can lead to the activation of specific rescue mechanisms generally termed as mRNA surveillance pathways (4). Currently, there are 3 well-established processes of mRNA-related translation control: i) nonstop mRNA decay (NSD) and ribosome-associated quality control (RQC) preventing readthrough polyA tail in the absence of stop codon (12, 13); ii) nonsense-mediated mRNA decay (NMD) for mRNA containing a premature stop codon (14); and iii) no-go decay (NGD) activated by secondary structures, truncated mRNA, or abundant rare codons (4, 15, 16). Ultimately, unrepairable mRNA undergoes exo- or endonucleolytic cleavage (17).
During translation, a single mRNA molecule can be occupied with multiple actively elongating ribosomes forming a structure known as polysome. Upon encountering senseless or unreadable sequence, translating ribosomes stall and eventually collide which induces the activation of the mRNA surveillance pathways. The E3 ubiquitin ligases ZNF598 and Makorin 1 (MKRN1) are the very first RQC proteins associated with the collided ribosomes formed on polyA stretches or sites with a high abundance of rare codons (18). ZNF598 can only bind to the interface between the 2 adjacent ribosomes (19), thus acting as a direct translation arrest sensor. It reversibly mono-ubiquitinates specific lysins in RPS10 and RPS20 proteins in the 40S ribosome subunit (18–21). These ubiquitination events trigger the RQC and mRNA decay pathways (22).
The ZNF598-mediated ubiquitination of stalled ribosomes on polyA stretches can be antagonized by deubiquitinating enzymes (DUBs) (23, 24). Recently, Garshott and colleagues (25) performed an overexpression DUBs screen and identified ovarian tumor (OTU) Domain Containing Protein 3 (OTUD3) and Ubiquitin Specific Peptidase 21 (USP21) as a negative regulators of ribosome stalling. Both DUBs seem to sequentially antagonize ZNF598 activity allowing ribosomes to read through the polyA sequence. The study uncovered several additional DUBs including OTUD1 with potential effect on ribosome stalling.
Currently described functions of OTUD1 include particularly its tumor suppressive role in multiple cancer types by promoting TGF-β, p53, yes-associated protein (YAP), iron transport and pro-apoptotic signaling pathways (26–30). Through its deubiqutinating activity, OTUD1 stimulates the inflammatory and innate immune responses (31). Recently, the oxidative sensor KEAP1 was identified as a direct interactor of OTUD1 providing a link to the redox signaling (32). Even though OTUD1 was found to mediate a response to premature polyA sequences in mRNA (25), the mechanistic details of how OTUD1 controls protein translation is not yet studied in detail.
In addition to premature polyA sequences, reduced ribosome processivity might be caused also by the presence of rare codons, lack of particular tRNA or extensive repeats of specific codons (4, 33, 34). Slowdown of ribosomes can lead to spontaneous mRNA decay or RQC that is dependent on ZNF598 (28). The mechanisms regulating the switch from mRNA decay to RQC and mRNA rescue pathways for rare codon-rich genes are not known.
Here, we revealed OTUD1 as a novel DUB associated with elongating ribosomes. We found that OTUD1 resolves the polyA-stalled ribosomes by antagonizing ZNF598-mediated mono-ubiquitination of RPS10 subsequently promoting the formation of polysomes. We also show that OTUD1 can mediate stability of mRNAs with abundant rare codons and direct a specific mRNA surveillance pathway. Thus, OTUD1 can be assigned as an important mediator of translation quality control.
RESULTS
OTUD1 interacts with elongating ribosomes.
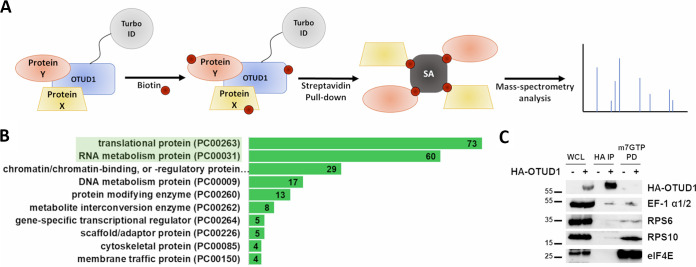
A recent study associated OTUD1 with ribosome stalling (25). However, the precise mechanism of how OTUD1 regulates translation machinery was not investigated. To validate the association of OTUD1 with the proteosynthetic apparatus, we performed TurboID proximity labeling (35) followed by mass spectrometry (MS) analysis (Fig. 1A). The obtained data set contained quantification of a relative protein abundance between the control (free TurboID) and test (TurboID-OTUD1) samples (Table S1).
FIG 1.
OTUD1 Interacts with Elongating Ribosomes. (A) Schematic diagram of OTUD1-TurboID proximity labeling assay. (B) Gene ontology analysis of OTUD1 interactome identified by mass spectrometry. Enriched proteins with fold change more than 5 were selected for the analysis. Numbers in green bars correspond to the number of genes identified in the respective category. (C) Co-immunoprecipitation analysis of OTUD1 interaction with ribosomal proteins, initiation and elongation complex. The eluates were analyzed by Western blotting using indicated primary antibodies. WCL = whole cell lysate.
Further, we used the Panther classification system (36) to analyze the gene ontology enrichment of proteins present in the OTUD1-TurboID samples with a fold change greater than 5 compared to the TurboID control. The analysis identified 228 protein class hits. The relative abundances of the most enriched protein classes were: 25.5% translation (PC00263), 21% RNA metabolism (PC00031), and 10.1% chromatin/chromatin-binding, or -regulatory proteins (PC00077). In the translation protein class, 97.3% of proteins belonged to the ribosomal subunits (PC00202), supporting the presence of OTUD1 in the ribosomal complexes and translation machinery (Fig. 1B).
Immunoprecipitation confirmed association of OTUD1 with endogenous ribosomal proteins (RPS10, RPS6) (Fig. 1C). Importantly, the OTUD1 pulled down elongation factor-1 α1/2 (EF-1 α1/2) but not eukaryotic translation initiation factor 4E (eIF4E). In agreement, OTUD1 was not present in the m7GTP pull-down which was used to isolate the initiation complex. These results indicate that OTUD1 is excluded from the partially assembled ribosomes in the 5′-UTR but it is likely present on the mRNA-associated elongating ribosomes.
OTUD1 counteracts ribosome stalling in a catalytically dependent way.
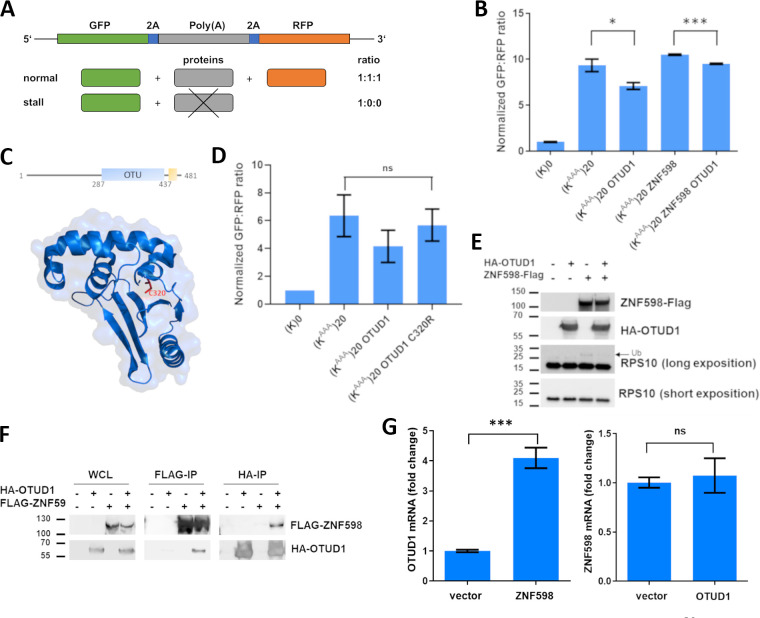
Based on our and recently published (25) results, we speculated that OTUD1 might be involved in polyA-mediated ribosome stalling. Therefore, we utilized a polyA-containing reporter (12) that consists of GFP and RFP fluorescent cassettes separated by a polyA sequence (KAAA)20. Complete reporter translation without any stalling event would lead to production of equal amounts of GFP and RFP, while the presence of a polyA stretch should induce ribosome stalling and move the fluorescence ratio toward more GFP (Fig. 2A).
FIG 2.
OTUD1 Suppresses polyA-induced Ribosome Stalling. (A) Schematic representation of the polyA ribosome stalling reporter system. (B) Normalized GFP:RFP fluorescence ratio of the polyA ribosome stalling reporter system coexpressed with OTUD1 WT and/or ZNF598. Significance was compared using the two-tailed Student's t test, * = P < 0.05; *** = P < 0.001. Error bars represent the mean ± SD. (C) OTUD1 structure (pdb code: 4BOP) with the catalytic cysteine (C320) marked in red. (D) Normalized GFP:RFP fluorescence ratio of the polyA ribosome stalling reporter system coexpressed with OTUD1 WT and OTUD1 C320R. Significance was compared using the two-tailed Student's t test. Error bars represent the mean ± SD. (E) The Western blot analysis of ribosomal protein S10 ubiquitination in HEK293 cell lysates with overexpression of OTUD1 WT or/and ZNF598. (F) Co-immunoprecipitation analysis of OTUD1 interaction with ZNF598. The eluates were analyzed by Western blotting using the indicated primary antibodies. (G) Amount of endogenous ZNF598 (left) or OTUD1 (right) mRNA in HEK293 cells upon overexpression of OTUD1 or ZNF598, respectively, were analyzed by RT-PCR. Significance was compared using the two-tailed Student's t test, *** = P < 0.001. Error bars represent the mean ± SD.
To test our hypothesis, we used the control (K)0 and stalling (KAAA)20 constructs together with overexpression of OTUD1 wild type (WT) and ZNF598. As expected, expression of OTUD1 WT significantly decreased the GFP/RFP ratio in normal genetic background and also upon co-expression with ZNF598 (Fig. 2B). In contrast to the previous report (25), the effect of OTUD1 on ribosome stalling was fully dependent on the presence of the catalytic cysteine (C320) (Fig. 2C) as the catalytically inactive OTUD1 mutant (C320R) (37) was unable to revert ribosome stalling (Fig. 2D).
To validate and mechanistically explain these results, we expressed OTUD1 and ZNF598 separately or in combination. Biochemical analysis revealed that OTUD1 can revert ZNF598-mediated mono-ubiquitination of the ribosomal subunit RPS10 (Fig. 2E). Additionally, we performed reciprocal co-immunoprecipitation of OTUD1 and ZNF598 that confirmed association of both proteins (Fig. 2F). Interestingly, overexpression of OTUD1 did not alter amount of ZNF598 mRNA (Fig. 2G, left). On the other hand, ZNF598 potentiated expression of OTUD1 (Fig. 2G, right) providing a potential regulatory feedback loop. Together, these results support the model where OTUD1 antagonizes E3 ubiquitin ligase activity induced by stalled ribosomes.
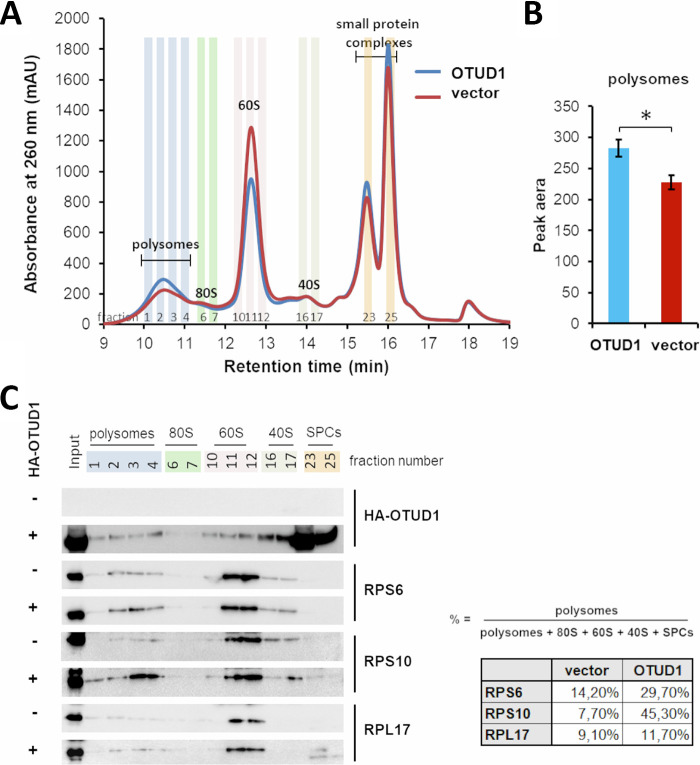
Polysome formation is promoted by OTUD1.
ZNF598 is a well-established regulator of the RQC pathway particularly in prematurely polyadenylated mRNA (12, 22, 38) which appears in about 1% of all transcripts (39, 40). Our data indicate that OTUD1 counteracts ZNF598 by deubiquitinating RPS10. Given the relatively large abundance of non-terminal polyA sequences in human transcripts, we speculated that overexpression of OTUD1 might alter the overall polysome profile. To explore this hypothesis, we utilized an optimized protocol for polysome profiling based on the Ribo Mega-SEC method (41). As expected, expression of OTUD1 significantly increased the relative amount of polysomes in the cell lysates together with a corresponding drop in the 60S ribosome subunit (Fig. 3A). The quantitative analysis of the polysome fractions from two independent experiments validated a strong positive effect of OTUD1 on polysome formation (Fig. 3B). The Western blot analysis and densitometry analysis of particular fractions from the Ribo Mega-SEC analysis confirmed the presence of OTUD1 in polysome fractions and detected an increased abundance of ribosome subunits in polysomes upon OTUD1 expression (Fig. 3C).
FIG 3.
OTUD1 Promotes Formation of Polysomes. (A) Polysome analysis using Ribo Mega-SEC. Cell lysates from HEK293 cells transfected with empty vector or WT OTUD1 were separated using Agilent Bio SEC-5 2000 Å column. The retention time is indicated on the x axis and the UV absorbance at 260 nm is shown on the y axis. (B). Quantification of the area under the polysomal peak. The values represent a mean ± SD from two biological replicates. (C) The highlighted fractions from (A) were analyzed by Western blotting with the indicated primary antibodies. SPCs = small protein complexes (left panel). The relative abundance of ribosomal proteins (RPS6, RPS10 and RPL17) present in the polysomal fractions (1–4) was quantified by densitometry analysis of band intensities using the indicated formula (right panel).
The stability of rare codon-rich mRNA is regulated by OTUD1.
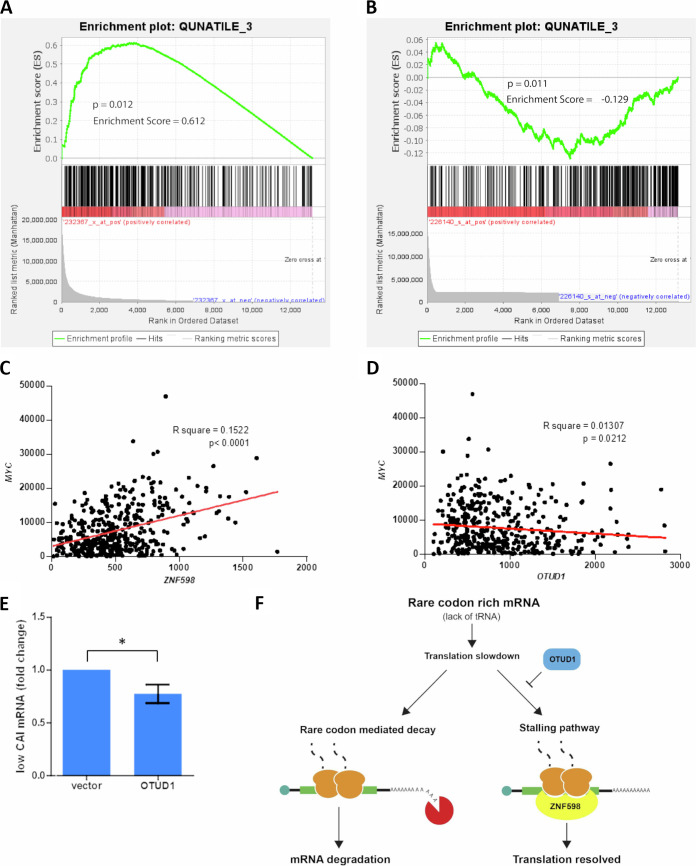
PolyA sequence is only one of many signals known to induce ribosome stalling. It was shown that tRNA starvation and the presence of rare codons in mRNA can also lead to ribosome collisions (42). Therefore, we speculated that ZNF598 and OTUD1 might be involved in the regulation of the surveillance pathways for mRNA with a low codon adaptation index (CAI). Our previous analysis identified the highest expression of OTUD1 in the hematopoietic system, particularly in the immunoglobulin-producing plasma cells (43). Therefore, we decided to use the relevant expression data set GSE4581.
To reveal the potential effect of the ribosome stalling pathway on mRNA with CAI (<50% to 1%), we performed a gene set enrichment analysis (GSEA) using ZNF598 and OTUD1 expression as a continuous label. Surprisingly, we observed a strong positive correlation between ZNF598 expression and mRNAs with CAI lower than median (Fig. 4A). GSEA using OTUD1 revealed the opposite trend for the mRNAs with the lowest CAI (lower 3%) (Fig. 4B). This indicates that the ribosome stalling pathway might be required for the stability of transcripts rich in rare codons.
FIG 4.
OTUD1 and ZNF598 affect stability of rare codon rich mRNAs. (A) and (B) Gene set enrichment analysis of genes with low CAI using ZNF598 (A) and OTUD1 (B) expression as a continuous label. The y axis represents enrichment score (ES) and on the x axis are genes (vertical black lines) represented in the gene set. The green line connects points of ES and genes. ES is the maximum deviation from zero as calculated for each gene going down the ranked list and represents the degree of over-representation of a gene set at the top or the bottom of the ranked gene list. The colored band at the bottom represents the degree of correlation of genes with ZNF598 (A) and OTUD1 (B) expression. (C) and (D) Correlation of ZNF598 (C) and OTUD1 (D) and C-MYC mRNA levels. (E) Expression of 7 genes from the bottom 1% (those with the lowest CAI) which expression had the strongest negative correlation with OTUD1 (B), was analyzed by RT-PCR HEK293 cell lines. Significance was compared using the two-tailed Student's t test, * = P < 0.05. Error bars represent the mean ± SD. (F) Schematic model of OTUD1-mediated translation control. Abundance of rare codons causes translation slowdown eventually triggering mRNA decay (left). Alternatively in response to ribosome collision ZNF598 may activate a stalling pathway that leads to reversible translation pause; depending on presence of cognate tRNA, translation can be resumed or no-go decay is initiated. OTUD1 acts as an antagonist to ZNF598 and by suppressing stalling pathway facilitates rare codon-mediated decay (right).
On the other hand, it was recently shown (44) that the presence of rare codons can induce ribosome slowdown and mRNA decay. In particular, c-MYC mRNA contains stretches of rare codons making it prone to a rare codon-mediated decay (45). Similarly, as in GSEA with a global analysis of the low CAI genes, we observed a positive correlation between the amount of ZNF598 and MYC transcripts, while comparing OTUD1 and MYC mRNA revealed an opposite trend (Fig. 4C and D). Based on these data, we suggest ZNF598 might protect rare codon rich mRNAs from degradation, possibly by triggering the ribosome stalling pathway. In reverse, OTUD1 negatively affects stability of mRNAs with low CAI supposedly by antagonizing ZNF598-mediated ribosome ubiquitination.
For experimental validation of our results obtained in the bioinformatic analysis, we initially sorted the genes from the GSE4581 expression data set based on their CAI. Then, we selected the 10 genes from the bottom 1% (those with the lowest CAI) which expression had the strongest negative correlation with OTUD1. From the 10 genes, 7 were expressed in the cell line used. In our model, OTUD1 affects mRNA stability by suppressing the ZNF598-mediated ribosome stalling pathway. Therefore, we used the cells with ZNF598 overexpression in which we analyzed the effect of OTUD1 on the stability of the above-mentioned gene products (low CAI mRNAs). As expected, the amount of these mRNAs significantly decreased upon elevated expression of OTUD1 (Fig. 4E).
Altogether, based on our results, we propose a model where the activation of the ZNF598-mediated ribosome stalling pathway prevents a decay of mRNAs with low CAI and deubiquitinase OTUD1 acts as a negative regulator of this process (Fig. 4F).
DISCUSSION
Translation quality control is a crucial step to avoid production of dysfunctional or harmful proteins. In cases when ribosome encounters damaged or difficult-to-read mRNA sequence, translation is slowed down eventually leading to ribosome collisions. Induction of the RQC pathway is required to resolve the stalled ribosomes. In this study, we provide evidence of the direct involvement of deubiquitinating enzyme OTUD1 in the regulation of ribosome stalling. Proteomic analysis revealed the presence of OTUD1 in the translation complex, specifically in elongating ribosomes. Using a fluorescent reporter system, we found that OTUD1 enables ribosome readthrough polyA-containing mRNA.
Translation of polyA would add a long stretch of lysins to the N-terminus of protein which could negatively affect its function. Ubiquitination of small ribosomal subunits by the E3 ligases ZNF598 and MRKN1 is well-established as one of the initial signals driving the RQC pathway when ribosome reaches polyA sequence (12). In contrast to a study (25), we found the enzymatic activity of OTUD1 is required for its role in rescuing polyA-stalled ribosomes. Specifically, our results show that OTUD1 is antagonizing ZNF598-mediated RPS10 ubiquitination. Because ZNF598 recognizes the interface between 2 stalled ribosomes (19) and associates with OTUD1, it might act as an adaptor directing OTUD1 for RPS10 deubiquitination. Termination of ZNF598 signals would inhibit further steps in the RQC and enable continuous translation. This observation is supported by the enhanced formation of polysomes in cells with high expression of OTUD1.
The polyA reporters are useful tool in studying ribosome stalling. However, they represent an artificial approximation of the relatively rare situations, when ribosomes skip the stop codon. Another potentially deleterious event associated with translation is the presence of rare codon rich sequences. Multiple reports indicate ribosomes pausing at rare codons is directly linked to frame shifts (46) and decreased mRNA half-life (47, 48). At these sites, ribosomes tend to stop either due to a limited amount of cognate tRNA or due to complex secondary mRNA structures (49). Following translation slowdown could lead to ribosome collisions that might resemble features of a damage-associated ribosome stalling.
It was shown that the rare codon rich mRNAs undergo mRNA decay independently of the canonical RQC pathway (44). We found a strong positive correlation between ZNF598 expression and rare codon rich mRNAs, while OTUD1 negatively affected the amount of mRNAs with abundant rare codons. This suggests a new role of the ribosome stalling pathway regulators in the mRNA homeostasis. Further experimental work is required to delineate the precise mechanistic involvement of ZNF598 and OTUD1 in the control of the stability of rare codon rich mRNAs.
MATERIALS AND METHODS
Cell lines, culture conditions, and transfections.
HEK293 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM; high glucose, pyruvate, and l-glutamine) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained in a 5% CO2 humidified incubator. Cells were transiently transfected with the respective plasmids using polyethyleneimine (PEI; 1 mg/mL) and serum-free media (Opti-MEM), mixed, and incubated 15 min at RT when cells were at 60% confluence 24 h after seeding. Stable doxycycline-inducible cell lines were treated with doxycycline hyclate (2 μg/mL) for 2 to 3 days prior analysis.
Quantitative real-time PCR.
The RNeasy minikit (Qiagen) was used to extract total RNA. The aliquots of RNA were stored at −80°C. The RNA samples’ quality (purity and integrity) was determined utilizing the Agilent 2100 Bioanalyser with the RNA 600 NanoLabChip reagent set (Agilent Technologies). The RNA was quantified by Qubit fluorometry (Thermo Scientific). The RevertAid First Strand cDNA Synthesis Kit (Thermo 656 Scientific) was used for Complementary DNA (cDNA) synthesis according to the manufacturer's instructions. Quantitative PCR was conducted using PowerUpTMSYBRTM Green Master Mix (Applied Biosystems) on StrepOnePlus real-time PCR system (Applied Biosystems). Relative mRNA expression was calculated by 2-ΔΔCt method and normalized to GAPDH transcripts.
Flow cytometry analysis.
Ribosome stalling analysis employing flow cytometry was performed as described previously (12). Briefly, 106 HEK293 cells transfected with a dual-fluorescent reporter containing K0 or K20 stalling cassettes or co-transfected with K20 reporter together with full length OTUD1 wild type (WT), OTUD1 C320R, or ZNF598, were trypsinized after 72 h transfection using TrypleX, sedimented (600 g for 3 min), washed in PBS, sedimented again and resuspended in PBS containing SYTOX blue and analyzed using a Beckman Coulter's CytoFLEX S instrument. Approximately 104 events were collected in SYTOX negative gate for each sample, geometric mean statistics were used to analyze MFI in the FlowJo software.
TurboID proximity labeling assay.
A total of 2.5*107 HEK293 cells expressing doxycycline-inducible OTUD1 WT N-terminally fused to TurboID or free TurboID were treated with biotin (50 μM) for 1 h before lysis in Urea buffer (8 M Urea, 1 mM DTT, 50 mM TRIS, pH 7.6), complemented with protease inhibitor cocktail (Thermo Fisher Scientific) and 1% Triton X-100. After 1 h incubation on ice, the lysates were diluted with Urea buffer to lower the concentration of Triton X-100 to 0.5%. The lysates were further sonicated and cleared by centrifugation at 17,000 g for 15 min at 4°C. Protein concentration supernatant was determined by BCA protein assay (Thermo Fisher Scientific). A total of 25 μL Streptavidin Sepharose high-performance resin (Cytiva) was added to lysates and incubated on a rotator overnight at 4°C. Beads extensively washed with Urea buffer were transferred to new tubes and washed with ammonium bicarbonate buffer (1 mM biotin, 50 mM ammonium bicarbonate). On-bead trypsin digestion was performed on enriched biotinylated proteins (1 μg trypsin overnight at 37°C). Collected digested peptides were desalted using in-house-made StageTips packed with C18 disks (Empire) before MS analysis.
Mass spectrometry and proteomic data analysis.
Separated peptides were analyzed on an UltiMate 3000 RSLCnano system coupled to an Orbitrap Fusion Tribrid mass spectrometer (both from Thermo Fisher Scientific). Acclaim PepMap300 trap column (300 μm × 5 mm) packed with C18 (5 μm, 300 Å, Thermo Fisher Scientific) were the first loaded with peptides in loading buffer (0.1% trifluoroacetic acid in 2% acetonitrile) for 4 min at 15 μL/min and then the peptides were separated in an EASY-Spray column (75 μm × 50 cm) packed with C18 (2 μm, 100 Å, Thermo Fisher Scientific) at a flow rate of 300 nl/min. To establish a 60-min gradient from 4% to 35% B, mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) were used. Peptides were ionized by electrospray after elution. A full MS spectrum (350-1400 m/z range) was obtained at a resolution of 120,000 at m/z 200 and a 100 ms maximum ion accumulation time. Dynamic exclusion was set to 60 s. Higher-energy collisional dissociation (HCD) MS/MS spectra were acquired in an ion-trap in rapid mode and the normalized collision energy was set to 30% with a maximum ion accumulation time of 35 ms. The MS and MS2 had set up automatic gain control at 1E 6 and 5E4, respectively. Top speed mode with 2 s cycle time and lower intensity threshold 5E3 were selected. An isolation width of 1.6 m/z units was used for MS. MaxQuant 1.6.3.4 was used to process and search through all raw data with the UniProtKB reviewed human protein database (release 2020_07; 20,381 sequences). Trypsin specificity was set C-terminally to arginine and lysine residues, also allowing the cleavage of proline bonds. Two missed cleavage sites of trypsin were allowed. Carbamidomethylation of cysteine was selected as a fixed modification and N-terminal protein acetylation and methionine oxidation as variable modifications. The false discovery rate of both peptide identification and protein identification was set as 1%. The options of “Second peptides” and “Match between runs” were enabled. Label-free quantification was used to quantify the difference in protein abundance between different samples. Data analysis was performed using Perseus 1.6.1.3 software. The final data set contains relative fold change of spectra counts from control (free TurboID) and test (OTUD1-TurboID) samples.
Pathways analysis and gene ontology.
Proteins identified as OTUD1 interactors in the TurboID proximity labeling assay (fold change >5 compared to free TurboID control in the MS analysis) underwent the gene ontology (GO) classification ontology analysis using the GO PANTHER software version 17.0 (http://pantherdb.org/).
Preparation of cell lysates for Ribo Mega-SEC.
A total of 2*107 HEK293 cells transfected with either OTUD1 WT or empty vector (pcDNA3.1) were treated with 50 μg/mL cycloheximide to maintain polysome stability for 5 min under 37°C and 5% CO2 before harvest. Cells were then washed with ice-cold PBS containing 50 μg/mL cycloheximide, lysed by vortexing for 10 s in 400 μL of polysome extraction buffer (20 mM HEPES-NaOH, pH 7.4, 130 mM NaCl, 10 mM MgCl2, 1% CHAPS, 2.5 mM DTT, 50 μg/mL cycloheximide, 20 U RNase inhibitor murine, New England BioLabs, EDTA-free protease inhibitor cocktail, Thermo Scientific) and incubated for 30 min on ice. Insoluble material was pelleted by centrifugation at 17,000 g for 10 min at 4°C. The supernatant was filtered through 0.45 μm Ultrafree-MC HV centrifugal filter units by 12,000 g for 2 min, and the total RNA amount in the filtrate was quantified by Qubit fluorometry (Thermo Scientific).
Polysome separation by Ribo Mega-SEC.
Agilent Bio SEC-5 column (2000 Å, 7.8 × 300 mm, 5 μm particles) together with Agilent Bio SEC-5 guard column (2000 Å, 7.8 × 50 mm, 5 μm particles) was connected to the Agilent 1200 HPLC system (Agilent Technologies) and equilibrated with 2 column volumes (CV) of filtered SEC buffer (20 mM HEPES-NaOH, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 0.3% CHAPS, 2.5 mM DTT). 100 μL of 10 mg/mL of filtered bovine serum albumin (BSA) solution diluted by SEC buffer was injected once to block the sites for nonspecific interactions. After monitoring the column condition by injecting 25 μL of GeneRuler 1 kb DNA Ladder (Thermo Scientific), 100 μL of cell lysate containing 70 μg of RNA was loaded on the column. All column conditioning and separation were done at 12°C. The chromatogram was monitored by measuring UV absorbance at 215, 260, and 280 nm by the diode array detector. The flow rate was 0.8 mL/min and 26 × 250 μL fractions were collected from 10 min to 16.5 min and analyzed by Western blotting.
Co-immunoprecipitation assay.
HEK293 cells expressing N-terminally HA-tagged OTUD1 and/or C-terminally FLAG-tagged ZNF598 were harvested, washed with ice-cold PBS, and lysed in polysome extraction buffer for 30 min on ice. The lysates were clarified by centrifugation at 17,000 g for 10 min at 4°C, and mixed with anti-HA agarose, anti-FLAG M2 agarose (both from Sigma-Aldrich), or γ-aminophenyl-m7GTP agarose (Jena Bioscience). After 2 h incubation on a rotator at 4°C, beads were washed extensively with polysome extraction buffer. Bound proteins were eluted by HA peptide, FLAG peptide (0.4 mg/mL, both from Sigma-Aldrich), or by boiling the γ-aminophenyl-m7GTP agarose beads and analyzed by Western blotting.
SDS-PAGE and Western blotting.
The immunoprecipitates and cell lysates were resolved by SDS-PAGE and transferred to the polyvinylidene difluoride (PVDF) membrane. Membranes were blocked in 5% (wt/vol) nonfat milk (Roth) in PBS-T (phosphate buffer saline, 0.05% Tween 20) and incubated with the appropriate primary antibodies overnight at 4°C in 1% (wt/vol) BSA/PBS-T. Primary antibodies used at indicated dilutions include: anti-FLAG M2 (F1804, Sigma-Aldrich, 1:1,000), anti-HA (11867423001, Roche, 1:1000), anti-OTUD1 (ab122481, Abcam, 1:200), anti-EF-1 α1/2 (sc-377439, SantaCruz, 1:1,000), anti-RPS6 (sc-74459, SantaCruz, 1:1,000), anti-RPS10 (sc-515655, SantaCruz, 1:1,000), anti-eIF4E (sc-9976, SantaCruz, 1:1,000), and anti-RPL17 (sc-515904, SantaCruz, 1:1,000). Membranes were subsequently washed with PBS-T and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. HRP-coupled secondary antibodies used at indicated dilutions include: goat anti-rabbit-IgG (111-035-144, Jackson ImmunoResearch, 1:2,000), goat anti-mouse-IgG (115-035-146, Jackson ImmunoResearch, 1:2,000), and goat anti-rat-IgG (NA935, Cytiva, 1:2,000). Signal detection was performed using ECL (Thermo Fisher Scientific) and ChemiDoc MP System (Bio-Rad). Protein bands analysis by densitometry utilized ImageJ v1.49 (National Institutes of Health).
GSEA.
GSEA was performed using GSEA 4.2.3 software (50, 51) and expression data set GSE4581. Codon adaptation index was calculated as in Jansen et al. (52). Gene lists (14) for analysis included genes with low CAI (lower 50, 40, 30, 20, 10, 9-1%). ZNF598 and OTUD1 expression were used as continuous labels.
Statistical analysis.
The statistical significance of differences between various groups was calculated with the two-tailed unpaired t test. Error bars represent the standard deviation of the mean (SD). Statistical analyses, unless otherwise indicated, were performed using GraphPad Prism 5. Data are shown as mean ± SD.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bonderoff JM, Lloyd RE. 2010. Time-dependent increase in ribosome processivity. Nucleic Acids Res 38:7054–7067. 10.1093/nar/gkq566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux PP, Topisirovic I. 2018. Signaling pathways involved in the regulation of mRNA translation. Mol Cell Biol 38:e00070-18. 10.1128/MCB.00070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker CJ, Green R. 2012. Translation drives mRNA quality control. Nat Struct Mol Biol 19:594–601. 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Döring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O'Brien EP, Kramer G, Bukau B. 2017. Profiling ssb-nascent chain interactions reveals principles of hsp70-assisted folding. Cell 170:298–311. 10.1016/j.cell.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson BH, Zaher HS. 2015. O 6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity. RNA 21:1648–1659. 10.1261/rna.052464.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrasekaran V, Juszkiewicz S, Choi J, Puglisi JD, Brown A, Shao S, Ramakrishnan V, Hegde RS. 2019. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat Struct Mol Biol 26:1132–1140. 10.1038/s41594-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arpat AB, Liechti A, de Matos M, Dreos R, Janich P, Gatfield D. 2020. Transcriptome-wide sites of collided ribosomes reveal principles of translational pausing. Genome Res 30:985–999. 10.1101/gr.257741.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan LL, Zaher HS. 2021. Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol Cell 81:614–628. 10.1016/j.molcel.2020.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q, Yu C-H, Zhao F, Dang Y, Wu C, Xie P, Sachs MS, Liu Y. 2019. eRF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons. Nucleic Acids Res 47:9243–9258. 10.1093/nar/gkz710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeuchi K, Izawa T, Inada T. 2018. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front Genet 9:743. 10.3389/fgene.2018.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juszkiewicz S, Hegde RS. 2017. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol Cell 65:743–750. 10.1016/j.molcel.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262–2264. 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 14.Kurosaki T, Popp MW, Maquat LE. 2019. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20:406–420. 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontos H, Napthine S, Brierley I. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol Cell Biol 21:8657–8670. 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doma MK, Parker R. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440:561–564. 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karamyshev AL, Karamysheva ZN. 2018. Lost in translation: ribosome-associated mRNA and protein quality controls. Front Genet 9:431. 10.3389/fgene.2018.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris C, Cluet D, Ricci EP. 2021. Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny. Wiley Interdiscip Rev RNA 12. 10.1002/wrna.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS. 2018. ZNF598 is a quality control sensor of collided ribosomes. Mol Cell 72:469–481. 10.1016/j.molcel.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer C, Garzia A, Morozov P, Molina H, Tuschl T. 2020. The G3BP1-family-USP10 deubiquitinase complex rescues ubiquitinated 40S subunits of ribosomes stalled in translation from lysosomal degradation. Mol Cell 77:1193–1205. 10.1016/j.molcel.2019.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Shao S, Brown A, Santhanam B, Hegde RS. 2015. Structure and assembly pathway of the ribosome quality control complex. Mol Cell 57:433–444. 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, Sonenberg N. 2017. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8:16056. 10.1038/ncomms16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joazeiro CAP. 2017. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu Rev Cell Dev Biol 33:343–368. 10.1146/annurev-cellbio-111315-125249. [DOI] [PubMed] [Google Scholar]
- 24.Clague MJ, Urbé S, Komander D. 2019. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol 20:338–352. 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 25.Garshott DM, Sundaramoorthy E, Leonard M, Bennett EJ. 2020. Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. Elife 9:e54023. 10.7554/eLife.54023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Fan Y, Xie F, Zhou H, Jin K, Shao L, Shi W, Fang P, Yang B, van Dam H, ten Dijke P, Zheng X, Yan X, Jia J, Zheng M, Jin J, Ding C, Ye S, Zhou F, Zhang L. 2017. Breast cancer metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat Commun 8. 10.1038/s41467-017-02029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carneiro AP, Reis CF, Morari EC, Maia YCP, Nascimento R, Bonatto JMC, de Souza MA, Goulart LR, Ward LS. 2014. A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumours. Br J Cancer 111:551–558. 10.1038/bjc.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piao S, Pei HZ, Huang B, Baek S-H. 2017. Ovarian tumor domain-containing protein 1 deubiquitinates and stabilizes p53. Cell Signal 33:22–29. 10.1016/j.cellsig.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Yao F, Zhou Z, Kim J, Hang Q, Xiao Z, Ton BN, Chang L, Liu N, Zeng L, Wang W, Wang Y, Zhang P, Hu X, Su X, Liang H, Sun Y, Ma L. 2018. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat Commun 9. 10.1038/s41467-018-04620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song J, Liu T, Yin Y, Zhao W, Lin Z, Yin Y, Lu D, You F. 2021. The deubiquitinase OTUD1 enhances iron transport and potentiates host antitumor immunity. EMBO Rep 22. 10.15252/embr.202051162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Zhang H, Wang X, Shao Z, Li Y, Zhao G, Liu F, Liu B, Zheng Y, Chen T, Zheng H, Zhang L, Gao C. 2021. OTUD1 regulates antifungal innate immunity through deubiquitination of CARD9. J Immunol 206:1832–1843. 10.4049/jimmunol.2001253. [DOI] [PubMed] [Google Scholar]
- 32.Oikawa D, Gi M, Kosako H, Shimizu K, Takahashi H, Shiota M, Hosomi S, Komakura K, Wanibuchi H, Tsuruta D, Sawasaki T, Tokunaga F. 2022. OTUD1 deubiquitinase regulates NF-κB- and KEAP1-mediated inflammatory responses and reactive oxygen species-associated cell death pathways. Cell Death & Dis 13. 10.1038/s41419-022-05145-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varenne S, Buc J, Lloubes R, Lazdunski C. 1984. Translation is a non-uniform process. J Mol Biol 180:549–576. 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 34.Cardinaud S, Starck SR, Chandra P, Shastri N. 2010. The synthesis of truncated polypeptides for immune surveillance and viral evasion. PLoS One 5:e8692. 10.1371/journal.pone.0008692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho KF, Branon TC, Udeshi ND, Myers SA, Carr SA, Ting AY. 2020. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat Protoc 15:3971–3999. 10.1038/s41596-020-0399-0. [DOI] [PubMed] [Google Scholar]
- 36.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13:2129–2141. 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrow ME, Morgan MT, Clerici M, Growkova K, Yan M, Komander D, Sixma TK, Simicek M, Wolberger C. 2018. Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins. EMBO Rep 19. 10.15252/embr.201745680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. 2017. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell 65:751–760. 10.1016/j.molcel.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258–2261. 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 40.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143:1018–1029. 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshikawa H, Larance M, Harney DJ, Sundaramoorthy R, Ly T, Owen-Hughes T, Lamond AI. 2018. Efficient analysis of mammalian polysomes in cells and tissues using Ribo Mega-SEC. Elife 7:e36530. 10.7554/eLife.36530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Hirano R, Tagami H, Aiba H. 2006. Protein tagging at rare codons is caused by tmRNA action at the 3′ end of nonstop mRNA generated in response to ribosome stalling. RNA 12:248–255. 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vdovin A, Jelinek T, Hrdinka M, Bago JR, Sevcikova T, Hajek R, Simicek M. 2019. Identification of deubiquitinase OTUD1 as a novel player in resistance of multiple myeloma to bortezomib. Blood 134:5526–5526. 10.1182/blood-2019-127930. [DOI] [Google Scholar]
- 44.Mishima Y, Han P, Ishibashi K, Kimura S, Iwasaki S. 2022. Ribosome slowdown triggers codon-mediated mRNA decay independently of ribosome quality control. EMBO J 41:e109256. 10.15252/embj.2021109256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemm I, Ross J. 2002. Regulation of c- myc mRNA decay by translational pausing in a coding region instability determinant. Mol Cell Biol 22:3959–3969. 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simms CL, Yan LL, Qiu JK, Zaher HS. 2019. Ribosome collisions result in +1 frameshifting in the absence of no-go decay. Cell Rep 28:1679–1689. 10.1016/j.celrep.2019.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Presnyak V, Alhusaini N, Chen Y-H, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160:1111–1124. 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radhakrishnan A, Chen Y-H, Martin S, Alhusaini N, Green R, Coller J. 2016. The DEAD-box protein dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167:122–132. 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, Link K, Khatwani N, Reynders J, Moore MJ, McFadyen IJ. 2019. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci USA 116:24075–24083. 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 52.Jansen R, Bussemaker HJ, Gerstein M. 2003. Revisiting the codon adaptation index from a whole-genome perspective: analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res 31:2242–2251. 10.1093/nar/gkg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download mcb.00265-22-s0001.xlsx, XLSX file, 0.9 MB (935KB, xlsx)