Abstract
Background
Presence of residual neurovascular activity within glioma lesions have been recently demonstrated via functional MRI (fMRI) along with active electrical synapses between glioma cells and healthy neurons that influence survival. In this study, we aimed to investigate whether gliomas demonstrate synchronized neurovascular activity with the rest of the brain, by measuring Blood Oxygen Level Dependent (BOLD) signal synchronization, that is, functional connectivity (FC), while also testing whether the strength of such connectivity might predict patients’ overall survival (OS).
Methods
Resting-state fMRI scans of patients who underwent pre-surgical brain mapping were analyzed (total sample, n = 54; newly diagnosed patients, n = 18; recurrent glioma group, n = 36). A seed-to-voxel analysis was conducted to estimate the FC signal profile of the tumor mass. A regression model was then built to investigate the potential correlation between tumor FC and individual OS. Finally, an unsupervised, cross-validated clustering analysis was performed including tumor FC and clinical OS predictors (e.g., Karnofsky Performance Status - KPS - score, tumor volume, and genetic profile) to verify the performance of tumor FC in predicting OS with respect to validated radiological, demographic, genetic and clinical prognostic factors.
Results
In both newly diagnosed and recurrent glioma patients a significant pattern of BOLD synchronization between the solid tumor and distant brain regions was found. Crucially, glioma-brain FC positively correlated with variance in individual survival in both newly diagnosed glioma group (r = 0.90–0.96; P < .001; R2 = 81–92%) and in the recurrent glioma group (r = 0.72; P < .001; R2 = 52%), outperforming standard clinical, radiological and genetic predictors.
Conclusions
Results suggest glioma’s synchronization with distant brain regions should be further explored as a possible diagnostic and prognostic biomarker.
Keywords: brain tumor, fMRI, functional connectivity, overall survival, predictor
Key Points.
Gliomas displayed organized functional connectivity with distant healthy brain regions.
Functional connectivity of solid tumor predicts patients’ survival.
Tumor functional connectivity outperformed classical clinical prognostic predictors.
Importance of the Study.
Neuroimaging research has previously focused on alterations of brain functional connectivity caused by the physical presence of gliomas, but has not considered the relevance of possible patterns of functional connectivity characterizing the tumor mass itself and its interplay with the rest of the brain. Recently, the presence of residual neurovascular activity within glioma lesions have been demonstrated via functional MRI (fMRI), and electrical synapses between gliomas and surroundings neurons have been discovered, along with preliminary evidences of their role in tumor aggressiveness and survival. Using resting-state fMRI in both newly diagnosed and recurrent glioma patients, we demonstrated the presence of solid tumor functional connectivity with local and distant healthy brain regions. Glioma-brain connectivity also strongly predicted patient survival, outperforming classical prognostic markers such as genetic and molecular features. Tumor connectivity analysis could help deepen the understanding of glioma pathophysiology and possibly optimize novel therapeutic approaches aimed at suppressing or manipulating such pathological crosstalk.
In the last few decades, functional neuroimaging has substantially increased our knowledge of large scale brain dynamics, shedding light on the functional role of various brain areas underlying specific behaviors, as well as on their correlation with clinical symptoms.1Resting-State Networks (RSNs) have been identified, indicating regions showing strong temporally correlated Blood Oxygen Level Dependent (BOLD) signal at rest (ie, functional connectivity, FC) and reflecting synchronous co-activation of distant regions supporting the execution of both fundamental (eg, Somato-motor and Language Networks2,3) and high-order cognitive abilities (ie, Fronto-Parietal Control and Dorsal Attention Networks4,5). Consequently, resting-state FC (rs-FC) alterations have been related to a wide range of neurological and psychiatric symptoms in patient populations, as well as to cognitive and psychological traits in healthy individuals.6–10 Interestingly, gliomas have recently been found to preferentially localize to key nodes (hubs) of association cortices, as well as in brain regions strongly expressing synaptic signaling genes.11 In addition, functional healthy brain tissue has been observed inside glioma lesions during intra-operative direct electrical stimulation performed to map eloquent regions.12,13 The presence and preservation of functioning neural populations within glioma is not surprising, considering its pathophysiological infiltrative behavior.14 However, only recently the existence of preserved functional fMRI connectivity within tumors has been documented, with preliminary evidence of its correlation with patients’ clinical profiles.15 Even though organized BOLD activity within the tumor lesion is a significant step towards new functional markers of tumor “activity”, equal attention should be focused on the potential interplay between glioma and healthy local and distant brain tissue that could represent a measure of tumor invasiveness and/or aggressiveness and therefore correlate with clinical status and overall survival.
On the other hand, recent electrophysiological data regarding molecular and cellular communications between gliomas and healthy neurons poses the basis to explore the potential influence of glioma cells on brain circuits, shifting the paradigm from gliomas exerting a passive disruption of brain structures to a more causal relationship between tumor activity and functional brain changes. Indeed, glioma cells have been recently found to establish electrically active glutamatergic synapses with healthy neurons mediated by AMPA ionotropic receptors,16,17 that allow their fast depolarization in response to presynaptic neuronal spiking, and ultimately promote the mitosis and migration of glioma cells.16,17 This newly discovered bidirectional neuron-to-glioma communication, on top of other tumor-promoting paracrine systems, seems to be an important biological mechanism contributing to the extreme aggressiveness of high-grade gliomas (HGG, WHO grade III and IV gliomas).18
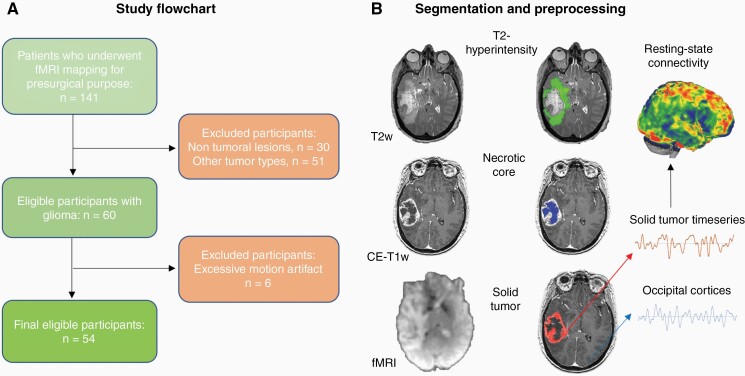
In light of the aforementioned recent evidence of glioma synaptic activity, as well as of preserved BOLD activity within gliomas, the evaluation of glioma behavior at micro-, meso- and macro-scale in humans could provide insight into its aggressiveness and potentially identify novel markers or therapeutic targets.19 In this study, we explored the macro-scale BOLD signal profile of gliomas via rs-fMRI data collected in patients with newly diagnosed or recurrent gliomas who required surgical intervention and functional imaging at the Brigham and Women’s Hospital (BWH) and Dana Farber Cancer Institute (DFCI) in Boston (MA, US; Figure 1A). First, we investigated whether the solid part of gliomas was characterized by significant BOLD synchronization (ie, functional connectivity—FC) with the grey matter of radiologically healthy brain areas. Due to location of the tumor in the left frontal or temporal lobe for the majority of cases, patients underwent pre-operative task-based fMRI assessment and functional mapping of eloquent brain regions for language/motor function were acquired a few days prior to surgery (see Supplementary Table S1). Rs-fMRI was collected in the same session for research purposes under IRB oversight. Manual segmentation of the solid part of the tumor as well as of its necrotic core and T2-hyperintense region was performed on contrast-enhanced T1w (CE-T1w) and T2w images following the Response Assessment in Neuro-Oncology (RANO) recommendations and previously applied procedures20 (Figure 1B). Rs-fMRI was analyzed via seed-to-voxel analysis using the solid tumor mask as the seed region (Figure 1B).
Figure 1.
Study flowchart and MRI preprocessing. (A) A retrospective analysis was conducted on all patients who underwent rs-fMRI at BWH (Boston, MA, US) between September 1, 2012 and September 1, 2018 because of a suspected brain focal lesion. (B) Gliomas were manually segmented into their necrotic core/surgical cavity, solid region (mass) and T2-hyperintense area on the basis of T1w and T2w MRI scans. Average BOLD timeseries were extracted from the solid tumor mask of each patient and correlated with the rest of the brain at single voxel-level, using age and gender as covariates. CE-T1w, contrast-enhanced T1w; fMRI, functional Magnetic Resonance Imaging.
We hypothesized the presence of (1) significant tumor-brain connectivity expanding on previously recognized intra-tumoral BOLD activity,15 and (2) that the strength of tumor BOLD synchronization with extra-tumoral healthy brain tissue could inform on disease status/aggressiveness, therefore correlating with disease course and survival as observed in animal models.16,17
Materials and Methods
Patient Sample
A retrospective analysis was conducted on all patients who underwent rs-fMRI at BWH between September 1, 2012 and September 1, 2018 because of a suspected brain focal lesion (ie, tumor, vascular malformation, dysplasia; n = 141). The inclusion criteria were: (1) diagnosis of glioma (WHO I–II–III–IV), defined according to the 2016 World Health Organization Classification of Tumors of the Central Nervous System21; (2) no other brain lesion; (3) pre-surgical fMRI assessment including resting-state fMRI acquisition, (4) follow-up structural MRI scans at BWH. Patients with other types of tumor (ie, meningioma, metastasis) or lesion (ie, vascular malformation, cortical dysplasia) and not aligning to the aforementioned criteria (ie, without follow-up structural MRI scans) were excluded. Demographic data of the final eligible sample of patients are reported in Supplementary Table S1 (n = 54), including Overall Survival (OS) calculated from the day of the fMRI scan to the date of death. Karnofsky Performance Status Scale (KPS) score was retrieved from the closest neurooncological visit with respect to the fMRI date scan. Handedness was assessed via Edinburgh Handedness Inventory22 and clinical notes (Supplementary Table S1). HGG patients whose OS was not available in the clinical database were not included in the FC-OS analysis (Supplementary Table S2). Genetic profiling regarding IDH (isocitrate dehydrogenase) status and methylation of MGMT [O(6)-Methylguanine-DNA methyltransferase] gene is also reported in Supplementary Table S2. All the patients were native English speakers, and none were fluent in a second language. All procedures of the study were done in accordance with the Declaration of Helsinki, under HIPAA compliance and approved by the Partners Institutional Review Board. The study protocol was fully explained to the patients prior to the MRI data acquisition. Patients provided written informed consent for research use of their imaging and clinical data.
MRI Acquisition
Structural and functional MRI was performed on a 3.0 T Siemens scanner (Siemens Trio, Verio, Skyra, and Prisma Systems, Munich, Germany) with a 20-channel head coil. Participants were placed in a supine position with their head fixed by positioning cushions to minimize head motion artifacts. BOLD fMRI was acquired using single-shot T2*-weighted gradient echo-planar imaging (EPI) with the following parameters: repetition time (TR) = 2000 ms, echo time (TE) = 30 ms, flip angle = 85, Matrix = 64 × 64, field of view (FOV) = 220 mm × 220 mm, voxel size = 3.44 × 3.44 × (4.0 or 5.0) mm3, 24 or 32 axial slices, ascending interleaved sequence, 7 minutes duration. A high resolution T2-weighted image was also acquired for the clinical fMRI report. Structural MRI was performed for surgical planning as clinically indicated, including a high resolution T1-weighted anatomical image with contrast (gadolinium) administration (axial 1 mm slices). The structural images were used for spatial co-registration and normalization into standard MNI space of the fMRI. Task-fMRI data were also acquired but were not used for the present analyses.
Tumor Segmentation
The tumor masks (ie, solid part of the tumor, necrotic core/surgical cavity, and T2-hyperintense region) for each patient were created on the basis of the pre-surgical structural MRI assessment (CE-T1w and T2w images, Figure 1B) by a senior radiology resident, specifically trained in neuroradiology (GS) and reviewed by the senior author of the study (ES), following similar segmentation procedures applied in previous studies.20 Masks were manually segmented using MRIcro (https://people.cas.sc.edu/rorden/mricro/mricro.html) on the corresponding anatomical scan and normalized to the MNI (Montreal Neurological Institute) space, along with corresponding anatomical scans for group analysis, via SPM12 (https://www.fil.ion.ucl.ac.uk/spm/) and custom MATLAB (Matlab 2016b, The Mathworks, Natick, MA, USA) scripts, and visually inspected to verify the correct correspondence onto the normalized CE-T1w and T2w scans.
fMRI Preprocessing
Preprocessing steps were performed using custom MATLAB scripts leveraging libraries from SPM12 (https://www.fil.ion.ucl.ac.uk/spm/), CONN toolbox (https://web.conn-toolbox.org/) and FSL (https://fsl.fmrib.ox.ac.uk/fsl). The first five volumes of functional images were discarded to allow for steady-state magnetization. EPI (Echo-Planar Imaging) images were slice-time corrected using the interleaved ascending acquisition criteria, then realigned and resliced to correct for head motion using a mean functional volume. Subjects whose head motion exceeded 1.0 mm or rotation exceeded 1.0° during scanning were excluded (6 patients removed, Figure 1A). Anatomical data were segmented into grey matter, white matter, and CSF tissue classes using SPM12 unified segmentation procedure and normalized into standard MNI space using non-linear volume-based registration.23,24 Functional data were smoothed using spatial convolution with a Gaussian kernel of 6mm full width half maximum (FWHM).15 Additional smoothing parameters were also tested (2 and 8 mm FWHM) to test whether analyses were affected by blurring of BOLD signal across solid tumor, necrotic core and T2-hyperintense tissue. Results were consistent across FWHM applied, without significant differences in resulting topography of functional connectivity maps with differences only in the size of relevant clusters.
Functional Connectivity Analysis
Analysis of fMRI data included preprocessing and analysis steps, performed using multiple toolbox/packages within the MATLAB environment: SPM12, CONN toolbox, as well as Connectome Workbench (https://www.humanconnectome.org/software/connectome-workbench), SPSS 12 and SPSS modeler (IBM Corp. Armonk, NY).
First, a seed-based connectivity analysis was performed to characterize the connectivity profile of the solid tumor in each patient (Figure 2). Clusters of significant cortical and subcortical connectivity were mapped onto known RSNs to establish network-level connectivity of solid tumor (“network mapping”) according to published methods based on Dice coefficient25 (Figure 3). Then, individual connectivity profiles were used to predict OS by means of a voxel-wise whole-brain multiple regression model (Figures 4 and 5). All the analyses were performed separately on newly diagnosed and recurrent glioma samples. Below we report details on each analysis.
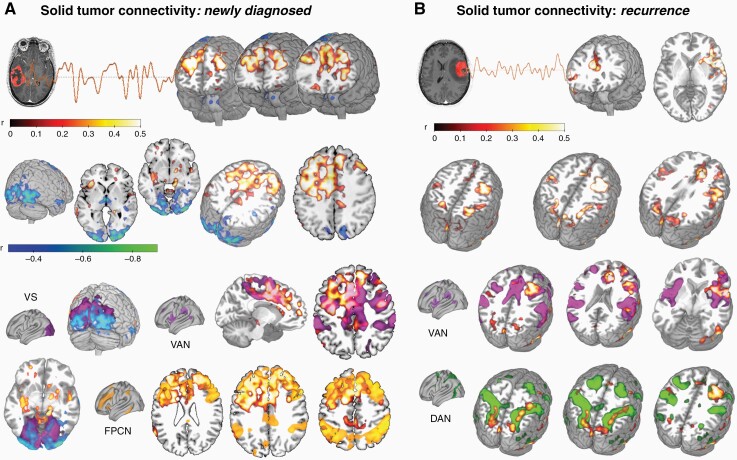
Figure 2.
Solid tumor connectivity profile. (A) Seed-to-voxel analysis revealed significant glioma BOLD signal synchronization (ie, functional connectivity, FC) with bifrontal regions and occipital cortices in newly diagnosed patients (P < .05; n = 18), roughly overlapping with the known resting-state networks such as the FPCN, VAN and VS. (B) Seed-to-voxel analysis performed on patients at recurrence (n = 36) revealed significant FC between the tumor mass and bilateral fronto-parietal regions (P < .05), resembling the VAN and DAN RSNs. DAN, dorsal attention network; FPCN, fronto-parietal control network; VAN, ventral attention network; VS, visual network.
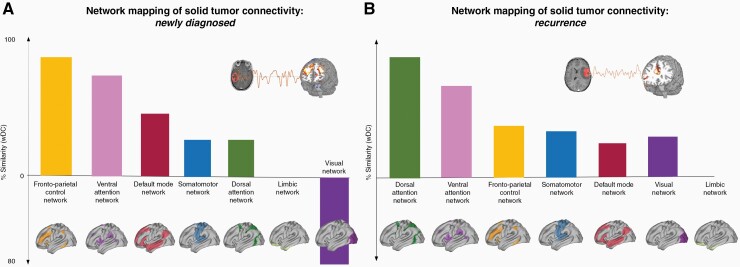
Figure 3.
Network mapping of solid tumor connectivity. Newly diagnosed (A) and recurrent glioma (B) functional connectivity profile revealed overlap with several networks, with a preference for distributed fronto-parietal networks such the FPCN, DAN and VAN. DAN, dorsal attention network; FPCN, fronto-parietal control network; VAN, ventral attention network, wDC, weighted dice coefficient.
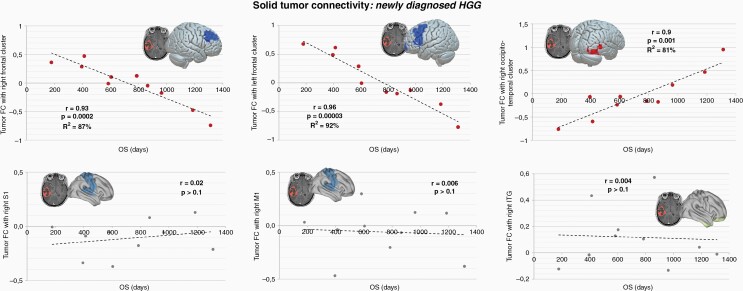
Figure 4.
Connectivity and overall survival in newly diagnosed HGG. In newly diagnosed HGG patients, three significant cortical clusters whose connectivity with the tumor mass displayed predictive power over OS were identified: two clusters located in the right and left frontal lobes, and the right occipito-temporal cortex (the correlation analyses revealed a variance in explained OS ranging from R2 = 81–92%; P < .001). FC of solid tumor and three control brain regions were extracted and correlated with OS, showing no significant correlation with overall survival (lower panels; r < 0.1, P > .1; for additional control regions see Supplementary Figure S2). Regression model parameters are reported in the Results section. FC, functional connectivity; OS, overall survival.
Figure 5.
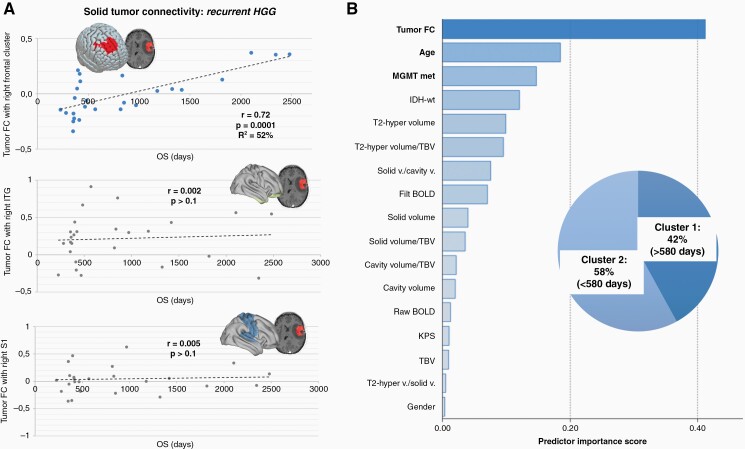
Overall survival in recurrent HGG and patients’ classification. (A) In HGG at recurrence, a single cluster in the right frontal lobe showing FC with the tumor mass was found to significantly predict OS variability across patients. Connectivity with control regions located in right ITG and right S1 showed no significant link with OS (for additional control regions please see Supplementary Figure S2). Regression model parameters are reported in the Results section. (B) A clustering algorithm was used to classify recurrent HGG patients based on OS, producing two groups: (1) long-term survivor—Cluster 1, and (2) short term survivor—Cluster 2. Among available predictors, including clinical, molecular, demographic and radiological parameters (ie, volume of the T2-hyperintense region, solid part, IDH status; see Supplementary Table S2 and Figure S3), tumor functional connectivity resulted as the best predictor of OS. FC, functional connectivity; filt BOLD, filtered BOLD; IDH-wt, IDH-wild type; ITG, inferior temporal gyrus; KPS, Karnofsky Performance Status score; MGMT met, methylation status of MGMT promoter; OS, overall survival; S1, primary sensory cortex; T2-hyper, T2-hyperintense region; TBV, total brain volume; v., volume.
Seed-Based Connectivity Profiling
Seed-to-voxel analyses were conducted to identify FC of solid tumor (seed) with the rest of the brain (Figure 2), using SPM and the CONN toolbox. The average BOLD signal inside the solid tumor mask of each patient was correlated with remaining voxels in the brain by means of Pearson product-moment correlation,26 using a voxel-wise threshold = P < .05 (two sided) and a cluster-level threshold = P < .05 FDR corrected. Age and gender were entered as covariates. This analysis resulted in voxel-wise maps representing positive and negative connectivity of the brain with the tumor mass, for each patient.
Network Mapping
In order to characterize the functional profile of each resulting network, functional labeling was performed by looking at the spatial similarity of each network map and those of known RSNs using a weighted variant of the original Dice coefficient (weighted dice coefficient, wDC; Figure 3).25 RSNs were defined following the brain parcellation scheme by Yeo et al5 reporting 7 non-overlapping maps of different networks: Default Mode Network (DMN), Fronto-Parietal Control Network (FPCN), Dorsal Attention Network (DAN), Ventral Attention Network (VAN), Limbic Network (LIM), Visual network (VS), and Somato-motor network (SM).5 RSNs maps were computed for 1000 healthy control subjects using the Human Connectome Project (HCP) dataset, resulting in weighted maps reporting both positive and negative connectivity patterns that were morphed into each patient’s individual MRI space. Therefore, the comparison of weighted, unthresholded connectivity maps for each solid tumor mask and RSNs map at voxel-level requires considering not only spatial similarity, but also similarity of the connectivity sign (ie, positive and negative connectivity). The wDC was obtained by computing the product of each voxel’s value across two maps (eg, voxel j located at x = 15, y = 32, z = 24 in both solid tumor and DMN connectivity maps), resulting in a map where positive values represent voxels with the same sign in both maps (ie, positive connectivity in both solid tumor and DMN), while negative ones represent opposite signs (ie, positive connectivity value in voxel j in solid tumor, negative in DMN). As a result, the magnitude of the similarity index represents the similarity of connectivity strength in any two given maps,27,28 therefore identifying RSNs with higher or lower similarity to glioma’s connectivity map (Figure 3). The analysis was performed to gain insight on possible tumor associations with specific RSNs, and did not inform subsequent clinical analyses related to OS.
Correlation with OS
In order to verify whether the existence and strength of glioma’s BOLD synchronization with the rest of the brain could be predictive of disease progression/survival, a whole-brain voxel-wise regression model was built using the solid tumor mask as a seed region (independent variable) and individual OS (expressed in days) as dependent variable in the CONN software (Figures 4 and 5). The analysis was conducted separately for newly diagnosed and recurrent HGG with age and gender as covariates (voxel-level threshold = P < .05, cluster-level threshold = P < .001, FDR corrected to refine the spatial specificity of the results). Given the inhomogeneity of HGG and LGG in terms of OS, the association between FC and OS was built separately for the two groups. Even though both analyses resulted in significant results, in order to facilitate both data visualization and interpretation findings related to HGG are reported in the main manuscript, while those for LGG are reported as part of the Supplementary Material (Figure S2). Significant clusters resulting from the model were labeled according to their anatomical location. After this step, connectivity values between each seed region (solid tumor) and each cluster of significant FC were extracted and visualized against days of survival (scatterplot) to check for the correlation between these brain-tumor FC values and OS, whose values are reported in Figures 4 and 5. Only patients for whom OS was available were included into the analysis (Supplementary Table S1). To further ensure that the observed association between FC and OS were not due to spurious correlations in the data, the same analysis and visualization were computed for control regions located in different lobes/regions/hemispheres. Specifically, we analyzed the connectivity between solid tumor and right primary motor and sensory cortices (M1 and S1), as well as right inferior temporal gyrus (ITG), given that the majority of the lesions were located in analogous areas in the left hemisphere (Supplementary Table S1). We also selected left hemisphere regions in the temporal lobe not overlapping with the tumor masks, such as left parietal operculum and left planum temporale (see Figures 4, 5 and Supplementary Figure S2). Regions were identified based on the Desikan atlas.29
Comparison with Classical Predictors of OS
Significant connectivity clusters were also compared in terms of their predictive power with canonical predictors of survival used in clinical practice. The validation included a two-step unsupervised clustering approach of recurrent HGG patients based on individual survival. Clustering was based on Akaike Information Criterion,30 no restriction on number of clusters, log-likelihood distance estimation and noise handling 25% for outlier removal. The predictive value of solid tumor’s connectivity with the rest of the brain was included as a predictor together with established markers of OS, including demographic (eg, age, gender),31 clinical (eg, KPS),31 genetic (eg, IDH mutation and MGMT methylation status32,33), and neuroimaging markers34,35 (see Supplementary Table S2 and Figure S1, S3). Neuroimaging predictors were: (1) the volume of the different tumor components, for example, T2-hyperintense region, solid part, necrotic core (or surgical cavity for recurrent gliomas); (2) the average amplitude of raw BOLD as well as filtered BOLD (pass band 0.008–0.09 Hz) time series extracted from the solid tumor mask, (3) total brain volume (TBV), extracted by combining grey and white matter segmentation masks for each patient, as control neuroimaging marker (not expected to be relevant for OS). The model produced a Predictor Importance score related to the independent (non-cumulative) amount of influence each predictor holds on the proposed sample portioning (range 0–1), that is, the residual sum of squares with the predictor removed from the model, normalized so that the importance values sum to 136 (Figure 5B).
Finally, a k-fold cross-validation analyses on recurrent HGG sample was performed to verify the replicability of results despite the relatively small sample, along with Cox regression analyses (see Supplementary Materials).
Results
In total, 60 patients with gliomas met criteria for inclusion in the study. Extraction of FC maps from resting-state scans were not possible in six patients due to movement artifacts, therefore the final sample consisted of 54 patients (male = 34, mean age = 50.83 years, SD = 14.76; Supplementary Table S1; Figure 1A). Eighteen (18) patients were scanned at first presentation of the tumor (newly diagnosed HGG = 12, male = 11, mean age = 54.5; SD = 16.5; mean OS = 951 days; Supplementary Table S1), and 36 patients were scanned at tumor recurrence (recurrent HGG = 26, male = 23, mean age = 47.1, SD = 13.4; mean OS = 289 days; Supplementary Table S1).
Seed-Based Connectivity and Network Mapping
In patients with newly diagnosed gliomas (n = 18; Figure 2A), solid tumor revealed a pattern of significant BOLD synchronization (or FC) (P < .05, t(15) = 2.13) with non-tumor bearing cortical regions; specifically, positive functional connectivity was demonstrated with bifrontal areas (middle frontal gyri, superior frontal gyri, anterior cingulate gyrus, paracingulate gyri, frontal poles; MNI coordinates of cluster centroid x = −12, y = +36, z = +16), while negative FC was observed with the occipital poles (MNI coordinates x = −38, y = −88, z = −04; Figure 2A). Those regions were noticed to overlap with known RSNs such as the FPCN, VAN and the Visual Network (Figure 3A).
In patients with recurrent gliomas (n = 36), solid tumor revealed a significant positive BOLD synchronization (P < .05; t(33) = 2.03) with bilateral fronto-parietal areas (superior parietal lobules, intraparietal sulci, anterior cingulate gyrus, right insular cortex, right frontal eye field; MNI coordinates of cluster centroids: x = +32, y = +00, z = +32; x = +32, y = −48, z = +58; x = +10, y = +28, z = +24; Figure 2B), again overlapping with the VAN and DAN (Figure 3B).
Connectivity Association with OS
Given the link between electrical integration of gliomas and survival in mouse models,16,17 we explored whether a relationship between solid tumor connectivity and OS among patients with gliomas could be also observed. Based on the differential clinical prognosis of HGG with respect to LGG, below we specifically focus on HGG patients (for results on LGG patients see Supplementary Resulst and Figure S2). A multiple regression model between OS and solid tumor functional connectivity with respect to the whole-brain was performed.37 We found that functional connectivity of solid tumor and specific brain areas significantly explained variance in OS for both newly diagnosed and recurrent HGG patients (newly diagnosed HGG, t(6) = 2.45, P < .05; recurrent HGG, t(24) = 2.06, P < .05) (Figures 4 and 5). Specifically, in newly diagnosed HGG (n = 10) three brain cluster regions whose FC with the tumor is related to OS were revealed by the regression analysis: right frontal lobe (mapped on middle frontal gyrus, frontal pole and paracingulate gyrus; MNI cluster centroid: x = −24, y = +26, z = +38), left frontal lobe (ie, middle frontal gyrus, superior frontal gyrus, frontal pole, inferior frontal gyrus; MNI cluster centroid x = −14, y = −08, z = +06) and right occipito-temporal regions (ie, temporal occipital fusiform cortex, lateral occipital cortex, middle temporal gyrus, occipital fusiform gyrus; MNI cluster centroid x = +44, y = −36, z = +04).
Then, we extracted the brain-tumor FC values of these relevant regions and performed a correlation analysis with OS revealing strong and significant correlations with patient survival (Figure 4): (1) right frontal lobe, r = 0.93, F(28) = 28.57, P = .0002, variance in OS explained by FC R2 = 87%; left frontal lobe, r = 0.96, F(28) = 49.54, P = .00003, variance in OS explained by FC R2 = 92%; and right occipito-temporal regions, r = 0.90, F(28) = 17.89, P = .001, variance in OS explained by FC R2 = 81%, (Figure 4). The regression model parameters testing the predictive power of OS based on the three FC clusters were as follows: (1) [right frontal lobe] Constant = 719.302, tumor FC beta = −902.402, t = −7.555, adjusted R2 = 86.2%, P < .001; (2) [left frontal lobe] Constant = 761.884, tumor FC beta = −745.748, t = −9.899, adjusted R2 = 91.5%, P < .001; (3) [right occipito-temporal regions] Constant = 758.332, tumor FC beta = 662.231, t = 5.948, adjusted R2 = 79.3%, P < .001.
Similarly, in patients with recurrent HGG (n = 26) BOLD synchronization between solid tumor and right frontal regions (superior and middle frontal gyri, frontal pole; MNI cluster centroid x = +22, y = +42, z = +44) was found to be relevant for OS by the regression analysis. Exploring the correlation between the cluster brain region-tumor FC with OS revealed a strong and significant correlation: r = 0.72, F(2 24) = 13.27; P = .0001, variance in OS explained by FC R2 = 52% (Figure 5A). The regression model parameters testing the predictive power of OS based on FC were as follows: Constant = 867.684, tumor FC beta = 2479.709, t = 5.142, adjusted R2 = 50.4%, P < .001. Given the bimodal distribution of OS in patients with recurrent HGG, with two clusters of survival above or below 580 days (see following paragraph), a model solely based on patients with >580 days of survival was built as well: Constant = 1142.662, tumor FC beta = 3008.263, t = 6.280, adjusted R2 = 77.8%, P < .001.
In LGG patients, functional connectivity of solid tumor with the same brain regions still predicted OS but with lower strength (R2 across regions = 13–79%; for additional LGG results see Supplementary Materials, Figure S2). Importantly, as a control analysis, the connectivity between solid tumor and multiple ipsi/homolateral control cortical regions were also extracted (eg, motor cortex—M1, sensory cortex—S1, see Methods section for details about control regions selection), revealing no significant correlation with OS across all groups (r < 0.1; P > .1; for newly diagnosed gliomas see Figure 4 and Supplementary Figure S2A; for the recurrent group see Figure 5 and Supplementary Figure S2B).
Comparison with Classical Predictors of Survival
To further confirm the potential relevance of FC association with OS when compared with standard clinical predictors, patients at HGG recurrence (ie, the largest group) were clustered based on their OS via a two-step clustering algorithm,30 leading to the identification of two subgroups: (1) “long-term survival group” with survival over 580 days (42%) and (2) “short-term survival group” with survival lower than 580 days (58%) (Figure 5B). The strength of the connectivity between solid tumor and right frontal lobe (Figure 5B)—identified as the most significant cluster in the seed-based regression model—outperformed gold standard clinical (KPS, IDH-wt, MGMT methylation), demographic (age, gender) and radiological markers (T2-hypertense volume, solid tumor volume, average raw and filtered BOLD amplitude—band pass 0.008 Hz/0.09 Hz; Figure 5B and Supplementary Figure S3), with a predictive power over OS for glioma FC (Predictor Importance score = 0.42, R2 = 54% for tumor FC) roughly two times higher than the second and third best predictors (ie, age, Predictor Importance score = 0.18, R2 = 24%; and genetic profile, Predictor Importance score = 0.16, R2 = 23%; for MGMT methylations status; Figure 5B and Supplementary Figure S3). Genetic and age performances in predicting OS were in line with literature.38,39 For k-fold cross-validation analyses on recurrent HGG sample see Supplementary Materials and Figure S4.
Discussion
Results show that the solid component of gliomas displays significant BOLD synchronization with distant brain regions (Figure 2), expanding on recent findings of functional activation detected within GBM15 and possibly recapitulating the macro-scale functional communication of gliomas recently observed at micro-scale level in mouse models.16,17 In previous studies on smaller samples, gliomas have been related to both local and widespread functional changes,40 with HGG inducing more diffuse functional connectivity alterations than LGG. However, the BOLD synchronization (and thus the functional connectivity) between the solid tumor and the rest of the brain in humans has not been explored yet, instead limiting observations to tissue within or closely surrounding gliomas.15,40 Interestingly, the solid tumor functional connectivity pattern with the healthy brain seems to mirror the distribution of some RSNs (Figure 3), validating the previous detection of functional neural activity within the infiltrating tumor lesion15 and their preferential localization in functional hubs,11 thus providing further support for the non-randomness of the observed tumor functional connectivity pattern. Our sample however, was constituted mainly by frontal and temporal lobe gliomas, with a preferential left-sided location in the newly diagnosed group. Even if those represent the predominant locations for adult gliomas in general, present results should be not generalized to the entire class of gliomas, especially in term of the specific brain regions whose functional connectivity seems to be related to OS that could be dependent on tumor location. Results require further validation in other glioma cohorts, also considering the limited sample size.
The most clinically relevant finding is that this solid tumor connectivity with distant brain regions is significantly associated with individual OS of both newly diagnosed and recurrent glioma patients, surpassing the performance obtained with classical predictors usually applied in clinical settings such as age, genetic profile and KPS, on their turn fitting with the performance reported in literature, thus validating our method (Figure 5). Specifically, association between FC and patient survival based on tumor connectivity aligns with the aforementioned preclinical observations in glioma in which post-synaptic electrical signaling induced by glutamatergic neurons drives tumor progression and influences mouse survival (in patient-derived GBM xenograft models),16,17 while also expanding on the recent observation of intratumor FC correlation with clinical patient status.15 Moreover, results fit to the recent demonstration of glioma preferential localization in functional hubs with high connectivity and centrality nodes, linking different cognitive subsystems with one another, thus in regions presenting long-distance connections.11 Finally, glioma-infiltrated cortex has been found to recruit a diffuse, atypical network of frontal cortical regions during speech planning,41 partially overlapping with the frontal cortical regions whose connectivity with the tumor strongly relate to OS (ie, middle frontal gyrus) found in the present study. The authors provide evidences for engagement of glioma-infiltrated areas in neural response to speech production and their functional integrations with long-range task-relevant brain circuits, even if the compensatory or pathologic nature of this plastic functional adaptation remains to be elucidated.41 Therefore, present results in term of solid tumor-frontal BOLD synchronization (Figures 2–4) could also represent a sign of functional, long-range modeling of brain circuits present even at rest.
In recurrent HGG, FC of the solid tumor with specific right frontal lobe clusters showed a slightly lower performance in explaining OS compared to the newly diagnosed group. A possible explanation could be that patients with recurrent gliomas had already undergone multiple treatments including radiation therapy and, in many cases, other interventions as well (ie, re-resection or investigational drugs), potentially representing a much more inhomogeneous sample with multiple factors influencing brain connectivity by the time of fMRI acquisition. Moreover, the heterogeneity in the location of the tumors across the recurrent cohort compared to the more homogeneous localization of the lesion in the newly diagnosed group (mostly involving the left temporal lobe) could have limited the significance of the results in this group (Supplementary Table S1), along with the slightly higher prevalence of grade III tumors respect to the newly diagnosed group patients. Also, tumor FC seems to be a better predictor in patients at recurrence with longer OS (>580 days); we can speculate that, in patients with shorter survival, the tumor could have already microscopically but significantly spread beyond its primary site, making our estimate of “tumor BOLD signal” less accurate and less clinically meaningful. For instance, midbrain invasion has been commonly found in patients with end-stage disease,42 thus a microscopic but not radiologically evident spread seems plausible in the lower survival subgroup. Also, the weaker correlation of solid tumor FC and OS in the LGG group is congruent with LGG lower clinical aggressiveness, possibly reflecting the reported lower upregulation of synaptic genes with respect to HGG16 and supporting BOLD alteration as a potential disease severity index. Along this line, future investigation should also explore if tumor FC is grade dependent even between grade III and grade IV tumors, and if it could represent a marker of infiltration/residual neuronal population, (ie, higher FC in grade III gliomas correlates with less parenchymal infiltration).
Another possibility is that the prominence of neurovascular uncoupling (NVU) could have altered BOLD signal and affected our functional connectivity estimation.43 The progression of NVU—ie, the alteration of hemodynamic response to neural activation—is concomitant with tumor progression, related to the tumor grade and can lead to significant attenuation of neurovascular response in tumor regions,44 thus could have exert a strong influence on the recurrent HGG sample. Of note, a complete loss of fMRI signal due to NVU has not been observed in the literature, but rather a decrease in the strength and topography of functional connectivity in both murine models and patients.40,45 Additionally, antiepileptic drugs (AED) could have exerted a significant effect on the functional connectivity observed, potentially lowering the FC magnitude, since AED usually decrease the neuronal spiking and task-based fMRI studies have shown a decrease of BOLD signal response both at local and network level46 in patients undergoing medications. Finally, our findings do not bypass the more general concern about the nature of BOLD signal extracted from tumor tissue. The most recent hypotheses suggest, on one hand, that BOLD signal indexes bulk blood flow in the pathological vasculature of the tumor, with an expected more altered hemodynamic response in HGG with respect to LGG.47 On the other hand, BOLD could also capture the activity of residual active neuronal populations within gliomas’ infiltrative nature.15 Also, the presence of both positive and negative connectivity between the tumor and distant brain regions, that partially overlap with defined RSNs, seems to align with the observed engagement of glioma-infiltrated areas in neural response via recruitment of long-range task-relevant brain regions, that mirror the physiological recruitment of normal appearing cortex.41 One or more of all these factors might also contribute to explain the different pattern of connectivity found in recurrent glioma group in comparison to newly diagnosed one, as well as the presence of solid tumor BOLD signal and synchronization per se.
Of note, in the present manuscript we indeed refer to the tumor-brain functional connectivity also as “BOLD synchronization”, given the fact that at the present time it is impossible to univocally understand the nature of this signal and correlation, and uncritically talk of “functional connectivity”, as commonly accepted in literature when considering brain-to-brain BOLD signal. Even though the resolution of functional MRI data used in the present investigation, as well as the new and limited knowledge in the arising field of the so called Cancer Neuroscience,48 does not allow us to disentangle the contribution of the neural/tumor vascular effect to the BOLD signal, results point toward a potentially valuable clinical marker regardless of its definitive mechanistic explanation(s), beyond the scope of the present investigation. Future studies with high-resolution perfusion, BOLD, SWI (Susceptibility Weighted Imaging) and diffusion MRI as well as cellular imaging and electrophysiology in preclinical models, will allow to disentangle the nature of BOLD signal, the exact biological significance of the observed tumor-brain BOLD synchronization and possibly further improve OS estimation.
Importantly, the comparison of tumor BOLD signal with gold standard clinical predictors of OS (ie, genetic profile, demographics and tumor size) in the recurrent HGG sample—the largest group but also the one where FC demonstrated lower correlation with survival, still suggests FC as a new potential prognostic biomarker in glioma management, with FC-based predictions matching and surpassing those based on age, MGMT methylation and IDH status, the classical clinical predictors. Longitudinal studies exploring the status of tumor FC at different stage of the disease (ie, newly-diagnosed mass, post-surgical residual tumor, recurrent tumor) would be important to clearly define the prognostic impact of tumor-brain BOLD signal.
Altogether, observations support a possible pathophysiological relevance of tumor-healthy brain BOLD synchronization profile. A network-oriented view, considering the impact of long-range FC patterns linking tumor activity and FC fluctuations, could further detail the pathophysiology of gliomas and its impact on brain function, also in line with recent successes obtained via similar connectivity-based methods in identifying brain-symptoms relationships (eg, “lesion-network mapping” approach49). The present manuscript offers a first preliminary evidence of the existence of a BOLD signal synchronization between glioma lesion and the healthy brain in a sample of gliomas mostly involving the fronto-temporal lobes, also identifying patterns of connectivity with cortical areas linked to patients’ clinical status, while did not aim to offer a definitive answer in terms of its biological substrate, significance and definite relevance, but rather hope to promote a fruitful scientific discussion and extensive investigation among the community.
Multiple limitations of the present design must be considered. Specifically, the limited sample size—due to the unique and well characterized patient population—should be addressed and cross-validation via multiple independent datasets should be prioritized. Our sample is over-represented by left-sided lesions leading to pre-surgical fMRI assessment to characterize the involvement of eloquent cortices, therefore future studies should validate the same analysis in a more balanced glioma sample, or focusing on lesion specifically involving the same lobe. Also, we did not select patients on the basis of handedness, more homogenous samples should be created in the future. To this aim, comprehensive and uniform data collection as well as data sharing should be implemented to also disentangle potential BOLD synchronization differences due to the different tumor location. Also, the length of the resting-state scan, limited to 7 minutes in our study, could have affected the reliability of our findings.50 Finally, analyses linking FC to patients’ longitudinal cognitive changes and to trajectories of radiological recurrence are fundamental aspects to implement in future studies to fully understand the role of FC in glioma’s prognosis. Nevertheless, we still consider this preliminary evidence of interest to both the clinical and research neurooncological community, especially in the light of the constant redefinition of glioma landscape via new molecular and biological features, as recently presented with the 2021 WHO CNS tumor classification.51
In conclusion, gliomas seem to display functional BOLD correlation with distant healthy brain regions in humans, that also correlates with patients’ overall survival. Tumor connectivity should be further explored as a possible diagnostic and prognostic biomarker possibly reflecting tumor aggressiveness and infiltration trajectory, as well as to guide therapeutic solutions aimed at inhibiting tumor-brain communication.52,53
Supplementary Material
Acknowledgments
We would like to thank the study participants and their families, as well as the radiology technicians for their essential support during the MRI acquisitions.
Contributor Information
Giulia Sprugnoli, Precision Neuroscience & Neuromodulation Program and Network Control Laboratory, Gordon Center for Medical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA; Radiology Unit, Department of Medicine and Surgery, University of Parma, Parma, Italy; Image Guided Neurosurgery Laboratory, Department of Neurosurgery and Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Laura Rigolo, Image Guided Neurosurgery Laboratory, Department of Neurosurgery and Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Meghan Faria, Image Guided Neurosurgery Laboratory, Department of Neurosurgery and Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Parikshit Juvekar, Image Guided Neurosurgery Laboratory, Department of Neurosurgery and Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Yanmei Tie, Image Guided Neurosurgery Laboratory, Department of Neurosurgery and Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Simone Rossi, Department of Medicine, Surgery and Neuroscience, Unit of Neurology and Clinical Neurophysiology, Siena Brain Investigation and Neuromodulation Lab (Si-BIN Lab), University of Siena, Italy.
Nicola Sverzellati, Radiology Unit, Department of Medicine and Surgery, University of Parma, Parma, Italy.
Alexandra J Golby, Image Guided Neurosurgery Laboratory, Department of Neurosurgery and Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Emiliano Santarnecchi, Precision Neuroscience & Neuromodulation Program and Network Control Laboratory, Gordon Center for Medical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Funding
Dr. Santarnecchi is supported by National Institutes of Health (R01 AG060981-01) and the Alzheimer’s Drug Discovery Foundation (ADDF) (ADDF-FTD GA201902–2017902). A.J.G. is supported by National Institutes of Health via 5P41EB028741, the Haley Distinguished Chair in the Neurosciences, and the Jennifer Oppenheimer Cancer Research Initiative. A.J.G. and Y.T. are supported by the National Institutes of Health (NIH) via R21CA198740.
Author contributions
Conceptualization: G.S., E.S., A.G. Methodology: G.S., E.S., A.G. Investigation: L.R., P.J., Y.T., M.F. Software: E.S. and G.S. Formal analysis: E.S and G.S. Writing: Original draft, G.S. and E.S. Review and editing: A.G, S.R., Y.T., P.J., N.S., L.R., M.F. Resources: A.G., E.S. and Y.T. Supervision: A.G., S.R., N.S. and E.S.
Conflict of interests:
The authors declare that they have no competing interests.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data may be requested from the authors under the BWH/Harvard policy.
References
- 1. Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011;1224:109–125. [DOI] [PubMed] [Google Scholar]
- 2. Tie Y, Rigolo L, Norton IH, et al. Defining language networks from resting-state fMRI for surgical planning—a feasibility study. Hum Brain Mapp. 2014;35(3):1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Unadkat P, Fumagalli L, Rigolo L, et al. Functional MRI task comparison for language mapping in neurosurgical patients. J Neuroimaging. 2019;29(3):348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. [DOI] [PubMed] [Google Scholar]
- 5. Yeo BTT, Krienen FM, Sepulcre J, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van den Heuvel MP, Scholtens LH, Kahn RS. Multiscale neuroscience of psychiatric disorders. Biol Psychiatry. 2019;86(7):512–522. [DOI] [PubMed] [Google Scholar]
- 7. Bassett DS, Sporns O. Network neuroscience. Nat Neurosci. 2017;20(3):353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruffini G, Wendling F, Sanchez-Todo R, Santarnecchi E. Targeting brain networks with multichannel transcranial current stimulation (tCS). Curr Opin Biomed Eng. 2018;8:70–77. [Google Scholar]
- 9. Santarnecchi E, Momi D, Sprugnoli G, et al. Modulation of network-to-network connectivity via spike-timing-dependent noninvasive brain stimulation. Hum Brain Mapp. 2018;39(12):4870–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Santarnecchi E, Sprugnoli G, Tatti E, et al. Brain functional connectivity correlates of coping styles. Cogn Affect Behav Neurosci. 2018;18(3):495–508. [DOI] [PubMed] [Google Scholar]
- 11. Mandal AS, Romero-Garcia R, Hart MG, Suckling J. Genetic, cellular, and connectomic characterization of the brain regions commonly plagued by glioma. Brain J Neurol. 2020;143(11):3294–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ojemann JG, John WM, Daniel LS. Preserved function in brain invaded by tumor. Neurosurgery. 1996;39(2):253–259. [DOI] [PubMed] [Google Scholar]
- 13. Skirboll SS, Ojemann GA, Berger MS, Lettich E, Winn HR. Functional cortex and subcortical white matter located within gliomas. Neurosurgery. 1996;38(4):678–84; discussion 684. [PubMed] [Google Scholar]
- 14. Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15(7):455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daniel AGS, Park KY, Roland JL, et al. Functional connectivity within glioblastoma impacts overall survival. Neuro-Oncology. 2020. doi: 10.1093/neuonc/noaa189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venkataramani V, Tanev DI, Strahle C, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573(7775):532–538. [DOI] [PubMed] [Google Scholar]
- 17. Venkatesh HS, Morishita W, Geraghty AC, et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573(7775):539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venkatesh HS. The neural regulation of cancer. Science. 2019;366(6468):965. [DOI] [PubMed] [Google Scholar]
- 19. Winkler F, Wick W. Harmful networks in the brain and beyond. Science. 2018;359(6380):1100–1101. [DOI] [PubMed] [Google Scholar]
- 20. Sprugnoli G, Monti L, Lippa L, et al. Reduction of intratumoral brain perfusion by noninvasive transcranial electrical stimulation. Sci Adv. 2019;5(8):eaau9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol (Berl). 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 22. Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. [DOI] [PubMed] [Google Scholar]
- 23. Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38(1):95–113. [DOI] [PubMed] [Google Scholar]
- 24. Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26(1053–8119 (Print):839–851. [DOI] [PubMed] [Google Scholar]
- 25. Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 26. Fox MD, Snyder AZ, Vincent JL, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci. 2005;102(27):9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santarnecchi E, Emmendorfer A, Pascual-Leone A. Dissecting the parieto-frontal correlates of fluid intelligence: a comprehensive ALE meta-analysis study. Intelligence. 2017;63:9–28. [Google Scholar]
- 28. Mencarelli L, Biagi MC, Salvador R, et al. Network mapping of connectivity alterations in disorder of consciousness: towards targeted neuromodulation. J Clin Med. 2020;9(3):828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. [DOI] [PubMed] [Google Scholar]
- 30. Akaike H. Prediction and entropy. In: Atkinson AC, Fienberg SE, eds. A Celebration of Statistics. New York, NY: Springer; 1985:1–24. doi: 10.1007/978-1-4613-8560-8_1. [DOI] [Google Scholar]
- 31. Chaudhry NS, Shah AH, Ferraro N, et al. Predictors of long-term survival in patients with glioblastoma multiforme: advancements from the last quarter century. Cancer Invest. 2013;31(5):287–308. [DOI] [PubMed] [Google Scholar]
- 32. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. [DOI] [PubMed] [Google Scholar]
- 33. Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 34. Iliadis G, Kotoula V, Chatzisotiriou A, et al. Volumetric and MGMT parameters in glioblastoma patients: survival analysis. BMC Cancer. 2012;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. Am J Neuroradiol. 2005;26(10):2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 36. Kent P, Jensen RK, Kongsted A. A comparison of three clustering methods for finding subgroups in MRI, SMS or clinical data: SPSS TwoStep cluster analysis, latent gold and SNOB. BMC Med Res Methodol. 2014;14:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ranganathan P, Pramesh CS, Aggarwal R. Common pitfalls in statistical analysis: logistic regression. Perspect Clin Res. 2017;8(3):148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thuy MNT, Kam JKT, Lee GCY, et al. A novel literature-based approach to identify genetic and molecular predictors of survival in glioblastoma multiforme: analysis of 14,678 patients using systematic review and meta-analytical tools. J Clin Neurosci. 2015;22(5):785–799. [DOI] [PubMed] [Google Scholar]
- 39. Verhaak RGW. Moving the needle: optimizing classification for glioma. Sci Transl Med. 2016;8(350):350fs14. [DOI] [PubMed] [Google Scholar]
- 40. Ghinda DC, Wu JS, Duncan NW, Northoff G. How much is enough-Can resting state fMRI provide a demarcation for neurosurgical resection in glioma? Neurosci Biobehav Rev. 2018;84:245–261. [DOI] [PubMed] [Google Scholar]
- 41. Aabedi AA, Lipkin B, Kaur J, et al. Functional alterations in cortical processing of speech in glioma-infiltrated cortex. Proc Natl Acad Sci USA. 2021;118(46):e2108959118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drumm MR, Dixit KS, Grimm S, et al. Extensive brainstem infiltration, not mass effect, is a common feature of end-stage cerebral glioblastomas. Neuro-Oncology. 2020;22(4):470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pak RW, Hadjiabadi DH, Senarathna J, et al. Implications of neurovascular uncoupling in functional magnetic resonance imaging (fMRI) of brain tumors. J Cereb Blood Flow Metab. 2017;37(11):3475–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Montgomery MK, Kim SH, Dovas A, et al. Glioma-induced alterations in neuronal activity and neurovascular coupling during disease progression. Cell Rep. 2020;31(2):107500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hadjiabadi DH, Pung L, Zhang J, et al. Brain tumors disrupt the resting-state connectome. NeuroImage Clin. 2018;18:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wandschneider B, Koepp MJ. Pharmaco fMRI: determining the functional anatomy of the effects of medication. NeuroImage Clin. 2016;12:691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta L, Gupta RK, Postma AA, et al. Advanced and amplified BOLD fluctuations in high-grade gliomas. J Magn Reson Imaging JMRI. 2017. doi: 10.1002/jmri.25869. [DOI] [PubMed] [Google Scholar]
- 48. Monje M, Borniger JC, D’Silva NJ, et al. Roadmap for the emerging field of cancer neuroscience. Cell. 2020;181(2):219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fox MD. Mapping symptoms to brain networks with the human connectome. N Engl J Med. 2018;379(23):2237–2245. [DOI] [PubMed] [Google Scholar]
- 50. Birn RM, Molloy EK, Patriat R, et al. The effect of scan length on the reliability of resting-state fMRI connectivity estimates. NeuroImage. 2013;83:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncology. 2021;( noab106). doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sprugnoli G, Golby AJ, Santarnecchi E. Newly discovered neuron-to-glioma communication: new noninvasive therapeutic opportunities on the horizon? Neuro-Oncol Adv. 2021;3(1):vdab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sprugnoli G, Rossi S, Rotenberg A, et al. Personalised, image-guided, noninvasive brain stimulation in gliomas: Rationale, challenges and opportunities. EBioMedicine. 2021;70:103514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.