Abstract
The effects of ions (i.e. Na+, Mg2+ and polyamines including spermidine and spermine) on the stability of various DNA oligonucleotides in solution were studied. These synthetic DNA molecules contained sequences that mimic various cellular DNA structures, such as duplexes, bulged loops, hairpins and/or mismatched base pairs. Melting temperature curves obtained from the ultraviolet spectroscopic experiments indicated that the effectiveness of the stabilization of cations on the duplex formation follows the order of spermine > spermidine > Mg2+ > Na+ > Tris–HCl buffer alone at pH 7.3. Circular dichroism spectra showed that salts and polyamines did not change the secondary structures of those DNA molecules under study. Surface plasmon resonance (SPR) observations suggested that the rates of duplex formation are independent of the kind of cations used or the structure of the duplexes. However, the rate constants of DNA duplex dissociation decrease in the same order when those cations are involved. The enhancement of the duplex stability by polyamines, especially spermine, can compensate for the instability caused by abnormal structures (e.g. bulged loops, hairpins or mismatches). The effects can be so great as to make the abnormal DNAs as stable as the perfect duplex, both kinetically and thermodynamically. Our results may suggest that the interconversion of various DNA structures can be accomplished readily in the presence of polyamine. This may be relevant in understanding the role of DNA polymorphism in cells.
INTRODUCTION
Polyamines are ubiquitous cellular constituents and they play important biological roles, such as in cell growth and differentiation (1,2). Intracellular concentrations of polyamines are in the high millimolar range (3). In some tumor cells, polyamines are regulated poorly, resulting in higher polyamine concentrations than those in normal cells (4). Therefore, inhibition of polyamine biosynthesis is a potential mechanism for cancer chemotherapy (5–7). Polyamines are aliphatic polycationic compounds, and they possess multiple positive charges (two for putrescine, three for spermidine and four for spermine) at physiological pH due to the protonations of their amine groups. They are almost exclusively bound to nucleic acids and are involved in many cellular processes that need nucleic acids (2). For example, polyamines affect the interactions of DNA-binding protein with nucleic acids (8–10), and modulate the transcription of various genes (11,12). Polyamines are essential materials for catenation of supercoiled DNA by topoisomerase (13,14). They also induce the transition of B-DNA to A-DNA or Z-DNA depending on the conditions (15–17).
Polyamines are flexible, positively charged molecules. Stabilization of DNA duplexes by polyamines has been studied for years. Intuitively, they should prefer to bind to the phosphate groups strongly in each strand of DNA duplex, and reduce the repulsion between the phosphate groups of DNA. However, the polyamine binding sites in DNA have been observed within both the minor and the major grooves of DNA (18–21). Theoretical modeling results suggested that polyamines bind to the major groove of GC-rich duplexes (22) and to the minor groove of AT-rich duplexes (23,24). Polyamines also have remarkable stabilizing effects on DNA–RNA hybrids (25), triple-helix DNA (26–28) as well as other double-helical secondary structures, such as stems and loops in rRNA, mRNA and tRNA (3,29). Polyamines can interact with DNA leading to the condensation of DNA (30,31) and chromatin (32,33), and protect DNA from denaturation (34,35). DNA lesions including bulged loops, hairpins or mismatched base pairs have significant biological consequence. Normally, perfectly base-paired DNA duplexes are considerably more stable than those with lesions. It would be of interest to know whether polyamines can stabilize DNAs with lesions, and if so, what biological roles they play.
In this study, we present a systematic study to reveal the role of salts, spermidine and spermine in the formation of various A,T-rich DNA duplexes. We synthesized six duplexes, as shown in Figure 1. Perfect duplex P1, containing no lesion, has 15 Watson–Crick base pairs (14 ATs and one GC). B1 and M1 contain a single base (G) bulge loop and an AG mismatched base pair in the center of P1, respectively. Finally, we synthesized three hairpins with either a seven or eight AT base-pair stem and a TGT loop. Using the ultraviolet (UV) melting and circular dichroism (CD) studies, we characterized the stabilizing and structural effects of polyamines on those DNA duplexes. In addition, we used surface plasmon resonance (SPR), a method to analyze biomolecular affinity interaction in real time (36), to study the kinetic behavior of those DNA duplexes.
Figure 1.
DNA duplexes that were used in this study include P1 (a perfect duplex), B1 (a bulged loop duplex), M1 (a mismatched duplex), H1, H2 and H3 (hairpin duplexes).
MATERIALS AND METHODS
Polyamine and oligodeoxyribonucleotide syntheses
Spermine tetrahydrochloride and spermidine trihydrochloride were purchased from Sigma Chemical Co. (St Louis, MO). Protected nucleotide phosphoramidites and controlled pore glass beads were obtained from Glen Research, Inc. (Sterling, VA). All oligodeoxyribonucleotides listed in Figure 1 were synthesized using an automated DNA synthesizer (Applied Biosystems Model 391) and purified by gel electrophoresis. The biotin-linked oligomers (5′-biotin-T7GT7 and 5′-biotin-T6CGCT8) were synthesized by incorporating the biotin synthon (Glen Research, Inc., Sterling, VA) at the 5′-end of the oligomers and can be immobilized to the streptavidin-coated biosensor chip used for the SPR experiments.
Melting temperature measurement
The profiles of UV absorbance versus temperature were measured using a JASCO V560 UV/VIS spectrophotometer by monitoring the sample absorption (in OD) at 260 nm. The sample cell was equipped with a Peltier type cell holder (EHC-441), and the temperature regulated by a programmer (JASCO TPU-436). The concentration of the duplex DNAs in each sample was 4 µM in 20 mM Tris–HCl buffered solution, pH 7.3, containing 100 mM NaCl, 22 mM MgCl2, 5 mM spermidine or 5 mM spermine. The experiments were carried out by increasing the temperature at a rate of 0.5°C/min from 0 to 100°C and the temperature recorded every 30 s. The Tm values were determined from the polynomial fitting of the observed curves and taken as the temperatures corresponding to half-dissociation of the DNA duplexes. The first derivative of absorption with respect to temperature, dA/dT, of the melting curve was computer-generated and used for determining the Tm.
CD spectroscopy
CD spectra were obtained in a JASCO-720 CD spectropolarimeter. Temperature was controlled by water circulation in the cell jacket. The DNA samples were prepared under the conditions identical to those prepared for the Tm measurements. The CD spectra were performed in the region of 340–190 nm. The molar ellipticity [θ] was calculated from the equation [θ] = θ/Cl, where θ is the relative intensity, C is the molar concentration of oligonucleotides, and l is the path length of the cell in centimeters. Spectra were collected between 320 and 190 nm with bandwidth at 1-nm interval.
Affinity measurements
The affinity, association and dissociation of the duplexes, were measured in a BIAcore 2000A SPR instrument (Pharmacia, Uppsala, Sweden) with a SensorChip SA5 from Pharmacia by monitoring the refractive index change of the sensor chip surface. These changes are generally assumed to be proportional to the mass of the molecules bound to the chip and recorded in resonance unit (RU). The surface was first washed three times by injecting 10 µl 100 mM NaCl solution. To control the amount of the DNA bound to the SA chip surface, the biotinated oligomer was immobilized manually onto the surface of a streptavidin chip until 350 RU was reached. The chip surface was then washed to remove non-specific binding with 10 µl 10 mM HCl. The target strands were prepared in different buffers with appropriate concentrations (0.02–0.5 µM) and passed over the chip surface for 140 s at a flow rate of 30 µl/min to reach the equilibrium. The conditions of the solutions used for SPR were the same as those in UV melting curve studies. Blank buffer solution was then passed over the chip to initiate the dissociation reaction and this was continued for 600 s to complete the reaction. After 600 s, the surface was recovered by washing with 10 µl 10 mM HCl for each DNA duplex.
Kinetic analysis
Sensorgrams for the interactions of DNA duplex were analyzed with BIA evaluation software version 3. The association and dissociation constants of the binding reaction between the probe (A) and the target (B) can be measured as described below:
where ka and kd are association and dissociation constants, respectively. The formation rate of the complex can be described as following:
d[AB]/dt = ka[A][B] – kd[AB] 1
The above equation can be expressed in terms of resonance units as:
dR/dt = ka (Rmax – R)C – kdR
= kaRmaxC – (kaC + kd)R 2
where Rmax is the maximum binding capacity of the immobilized DNA probe, R is the amount of target DNA bound to the immobilized probe and C is the concentration of the flowing target DNA. In principle, the ka and kd can be calculated directly from a straight line obtained by plotting dR/dt versus R to yield a slope, ks, defined as
ks = kaC + kd. 3
Determination of ks was achieved at different concentrations of the target. The ka was obtained as the slope from a plot of ks versus C. However, kd was very close to the origin and an accurate determination of kd was difficult to establish. Therefore, an alternative method was devised. The target buffer pulse has traversed the chip surface and is replaced by a steady flow of buffer. The rate of dissociation of the formed complex, AB, is described by
dR/dt = –kdR 4
ln (R1/Rn) = kd(tn – t1) 5
where R1 is the response at t = 1 s and Rn is the response at time t = n s along the dissociation curve. Therefore, a plot of ln (R1/Rn) versus (tn – t1) should produce a straight line with the slope of kd.
RESULTS
Effects of salts and polyamines on the melting temperature of DNA duplexes
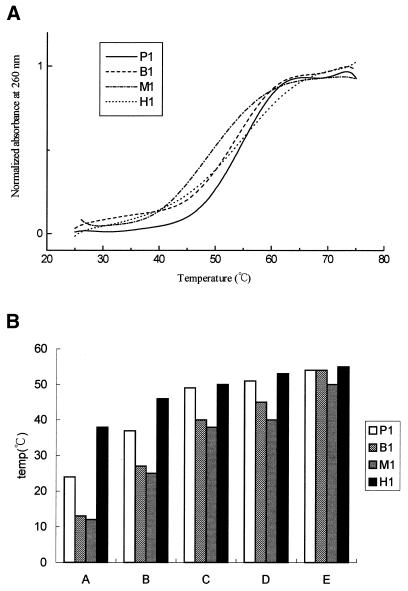
The Tm values of DNA duplexes listed in Figure 1 were determined by recording their A260 at a different temperature. The melting profile of various duplexes is depicted in Figure 2A showing different dissociation type in the presence of 5 mM spermine. The Tm values were graphed in Figure 2B. The P1 duplex is served as a reference for comparison with duplexes containing bulged loop, mismatch or hairpin. Figure 2B shows that in the absence of any salts and polyamines, regardless of the DNA sequences, hairpin (H1) shows the highest Tm, whereas both bulged loop (B1) and mismatched (M1) DNAs show lower Tm values than that of the perfect one (i.e. P1). This is to be expected. The Tm value of all oligonucleotides started to increase upon the addition of NaCl, MgCl2, spermidine and spermine, suggesting that electrostatic interactions play an important role in the stability of DNA duplexes. The Tm value of the duplex with a bulged base is ∼12°C lower than that without. However, the difference is reduced to 10°C when 100 mM NaCl is present. The difference drops sharply upon the addition of 20 mM MgCl2 or 5 mM spermidine. Notice that there is no difference in the stability between B1 and P1 duplexes when 5 mM spermine is present. Therefore, in terms of the melting of DNA duplexes, the two polyamines studied here can make imperfect DNA with abnormal structure behave like perfect ones as far as the Tm values are concerned.
Figure 2.
(A) Melting profile of the various duplexes including P1, B1, M1 and H1 in the presence 5 mM spermine and (B) the effect of salts and polyamines on the Tm value of duplexes with different structures in solution A, 20 mM Tris–HCl buffer, pH 7.3; solution B, 100 mM NaCl in solution A; solution C, 20 mM M MgCl2 in solution B; solution D, 5 mM spermidine in solution C; and solution E, 5 mM spermine in solution C. Tm values were taken as the temperatures corresponding to half-dissociation of the DNA duplexes. The first derivative of absorbance respect to temperature, dA/dT, of the melting curve was computer-generated and used for determining Tm.
We also studied the Tm value of hairpin duplexes with a stem of eight (H1) and seven base pairs (H2 and H3), all with a triplet TGT loop (Fig. 1). The Tm value of H1 is slightly higher than those of H2 and H3 (Table 1). In contrast to the bulged and mismatched DNA duplexes, the Tm values of all three H duplexes are on average 12°C higher than those of the perfect ones in 10 mM Tris–HCl buffer due to the uni-molecular formation characteristics of the hairpin duplexes. However, this gap is reduced to 8°C when 100 mM NaCl was present, and 1°C when 20 mM MgCl2 or 5 mM spermidine was present. There was virtually no difference among the Tm values between the hairpin and perfect duplexes when spermine was present.
Table 1. The melting temperatures (°C) of duplexes P1, B1, M1, H1, H2 and H3 at duplex concentration of 4 µM, pH 7.3, buffered by 20 mM Tris–HCl in the following conditions.
| Code | No salt | 100 mM NaCl | 20 mM MgCl2 | 5 mM Spermidine | 5 mM Spermine |
|---|---|---|---|---|---|
| P1 |
24 |
37 |
49 |
51 |
54 |
| B1 |
13 |
27 |
40 |
45 |
54 |
| M1 |
12 |
25 |
38 |
40 |
50 |
| H1 |
38 |
46 |
50 |
53 |
55 |
| H2 |
37 |
45 |
50 |
51 |
55 |
| H3 | 37 | 44 | 47 | 49 | 50 |
Effects of salts and polyamines on the conformation of DNA duplexes
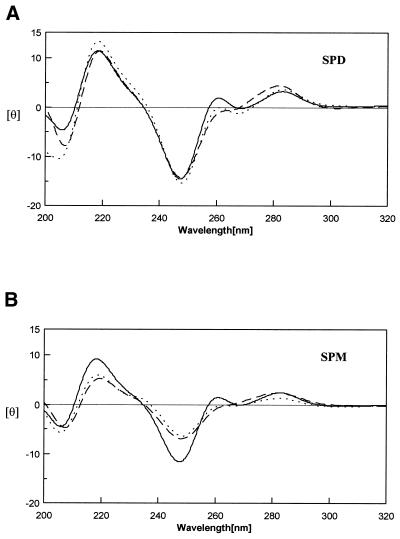
CD spectra are good indicators of DNA conformation. Figure 3A and B show the CD spectra of DNA duplexes with normal (P1) and abnormal (M1 and B1) sequences in the presence of spermidine and spermine, respectively. In the region of 260–290 nm of the spectrum, DNA duplexes (P1, M1 and B1) have a split peak which includes a large positive peak at 280 nm and a weak positive peak at 260 nm, as normally found for (A)n•(T)n sequence (37). No change was observed in the CD spectra of both perfect and imperfect DNA upon the addition of spermidine (Fig. 3A) and spermine (Fig. 3B) as compared with those of the perfect duplex in salt conditions (data not shown). In Figure 3A, the CD spectra of P1, M1 and B1 have a band with negative and positive peaks around 245 and 280 nm, typical of B-DNA (25). This suggests that the conformation of these DNA duplexes remains as B-DNA. However, spermine decreased the intensity of the CD spectra of both M1 and B1, and to a lesser extent P1 in the region of 240–250 nm.
Figure 3.
CD spectra of P1 (solid line), B1 (dotted line) and M1 (broken line) in solution of 20 mM Tris–HCl buffer at pH 7.3 with 5 mM (A) spermidine at (B) spermine at 10°C. The protocols for CD data collection are described in the Materials and Methods section.
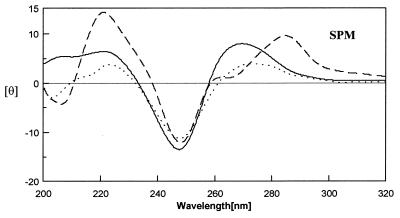
Except for the hairpin duplex of H2, which shows a CD spectral character of oligo-(A)n•(T)n in the presence of spermine, the DNA hairpins H1 and H3 with a (dA•dT)n duplex stem have positive bands at 265 nm characteristics of the CD spectra of A-DNA (38) (Fig. 4). Hence, the conformations of H1 and H3 may be an intermediate between B-DNA and A-DNA. Spermidine and salts had similar effects on the conformation of these DNA hairpins to those caused by spermine (data not shown).
Figure 4.
CD spectra of H1 (solid line), H2 (broken line) and H3 (dotted line) in 20 mM Tris–HCl buffer at pH 7.3 with 5 mM spermine at 10°C. The protocols for CD data collection are described in the Materials and Methods section.
Binding affinity of DNA duplex with normal and abnormal structures
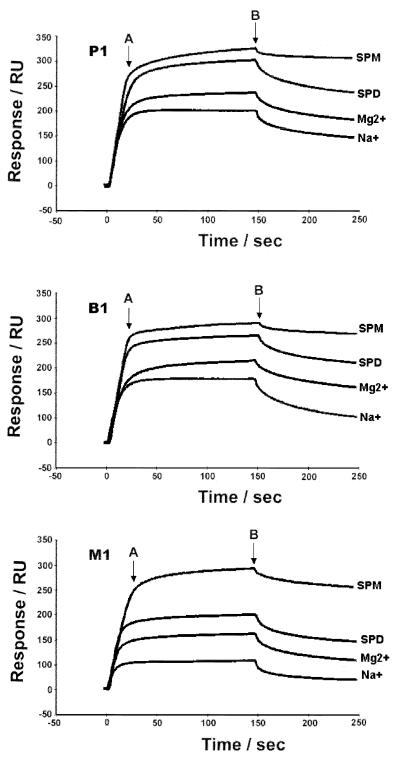
In order to study the effects of salts and polyamines on the binding affinity of DNA–DNA interaction including P1, B1 and M1, the maximum binding capacity (Rmax) (in RU) between the two strands of these DNA molecules was measured using SPR in the presence of salts and polyamines. The 5′-biotin-T7GT7 was used as the probe for 0.1 µM A7CA7 and A15 to form duplexes with perfect (P1) and mismatched (M1) base pairs, respectively, while the 5′-biotin-T7CGCT6 was used for A6G2A7 to form a bulged G loop (B1). In Figure 5, at certain concentrations of salts and polyamines, regardless of DNA sequences, the maximum binding capacity of each DNA duplex increased as shown by their RU values from Na+ to spermine, consistent with the increasing number of positive charges of these cations. Spermine caused the largest effect on DNA-binding affinity, presumably by holding together the two strands of DNA. Sodium ion had little effect on the binding affinity of DNA containing a mismatch, but binding affinity of the bulged G loop (B1) was close to the perfect one (P1) in the presence of Na+ and Mg2+. For P1, B1 and M1, the highest RU values caused by spermine are nearly the same. Spermine appears to be a much better species than any other cations in terms of the binding affinity of mismatched DNA (M1).
Figure 5.
Binding capacity sensorgrams of DNA–DNA interaction of immobilized 5′-T7GT7 and the target of 5′-A7CA7 (P1), 5′-A15 (M1) and immobilized 5′-T7G2T6 and the target 5′- A6CGCA7 (B1). The concentration of each target DNA strand is 0.1 µM, in 100 mM NaCl (Na+), 20 mM MgCl2 (Mg2+), 5 mM spermidine (SPD), or 5 mM spermine (SPM), buffered by 20 mM Tris–HCl at pH 7.3. The end point of the association (A) and the starting point of dissociation (B) are indicated by arrows.
Kinetics study of DNA duplexes with normal and abnormal structures
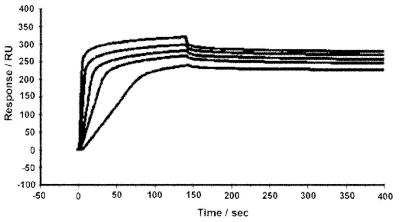
To study the kinetics of molecular interactions, the SPR experiment is superior to the conventional stop-flow technique because SPR can be used to study both the association and dissociation of DNA strands. The kinetic experiments were carried out by measuring the binding affinity between one DNA strand and its complementary target strand. Five concentrations of the target DNA were used for the determination of both ka and kd in different cation solutions at 20°C. In Figure 6, the Biacore SPR traces of P1 in the presence of spermine are shown.
Figure 6.
Sensorgrams of DNA–DNA interaction between immobilized 5′-T7GT7 and the target DNA of 5′-A7CA7 at 0.5, 0.2, 0.1, 0.05 and 0.02 µM, respectively, from top to bottom in 5 mM spermine, buffered by 20 mM Tris–HCl at pH 7.3. The association rate was increased along with the concentrations of target strands and the dissociation rate was independent of concentration.
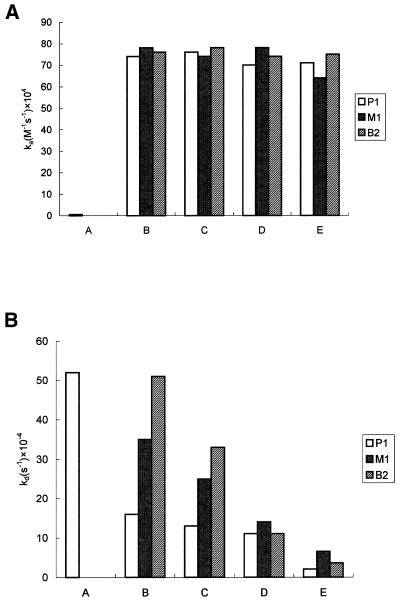
The ka and kd of P1, M1 and B1, calculated according to equations 3–5, are shown in Figure 7A and B. The ka and kd of P1 in Tris–HCl buffer are ∼6.84 × 103 M–1 s–1 and 5.33 × 10–3 s–1, respectively, whereas no duplex interaction was detected for M1 and B1. This is because the temperature (20°C), where kinetic experiments were carried out, was above the Tm (∼14°C) of both M1 and B1. In the presence of salts and polyamines, the ka of P1 was elevated ~100-fold as compared with the controls. Furthermore, under the same conditions (salts and polyamines), the ka values of M1 and B1 were not only measurable, but also increased to approximately the same level as those of P1 (Fig. 7A). So the stabilizing effect on the association of DNA duplexes is neither dependent of the type of cations nor the structure of the duplex. On the other hand, the kd values of P1, M1 and B1 were decreased in the presence of salts and, particularly, polyamines. As shown in Figure 7B, the kd of both M1 and B1 are higher than that of P1. The kd of P1 was decreased by spermine to a much lower extent than that by salts and spermidine. The kd of B1 are higher than those of M1 in the presence of salts. However, the kd of B1 are lower than those of M1 in the presence of polyamines, suggesting that polyamines interact with these imperfect DNAs using a different mode as compared with salts.
Figure 7.
(A) The ka and (B) the kd values derived from the DNA–DNA-binding sensorgrams between the immobilized oligonucleotide and its corresponding target strand of P1, M1 and B1 as described in Figure 5. Kinetic measurements were carried out under the following conditions A–E as described in Figure 2. All binding experiments were completed with at least five different concentrations for each target strand. The methods for determining ka and kd are described in the Materials and Methods section. Typical error levels for the ka and kd values are ±20 and ±10%, respectively.
DISCUSSION
The purpose of this study was to examine the ability of spermine and spermidine to stabilize various duplexes including perfect, mismatched, bulged loop and hairpin duplexes. Polyamines may participate in the secondary structure formation of DNA in cells, e.g. in the stabilization of stem and loop of RNA in vivo (2). Our results show that salts and particularly polyamines stabilize DNA duplexes with abnormal sequences by elevating their Tm values. Among the biological functions of polyamines, one notable feature is their ability to condense DNA (31). Polyamine-induced DNA condensation is likely driven by non-specific electrostatic interactions. In fact, some reports applied the counterion condensation theory to DNA condensation and proposed that polyamines formed a positive ionic cloud around DNA and neutralized the negative charges of the phosphate groups of DNA (31,39). DNA condensation is caused by lowering the electrostatic repulsion between negative charges of the phosphate groups.
By examining the thermodynamic properties of DNA duplexes, it was found recently by Rouzina and Bloomfield (40) and Chalikian et al. (41) that melting of DNA duplexes is accompanied with positive heat capacity (ΔCp) change depending on base composition and base sequence, whereas Tm is only weakly affected by ΔCp. They also suggest the transition enthalpy (ΔH) at a common reference temperature is less affected by duplex type, base composition, or base sequence than previously believed on the basis of the conventional assumption of a near-zero value for ΔCp. However, the changes in the thermodynamic properties of DNA duplexes were not investigated in our study. It would be interesting to study the effect of structures on the heat capacity of various duplexes in the presence of salts and polyamine in the future.
In polyamines, the distance separating the charge is a function of the length of the carbon chain between amino groups and is important for the induction of conformational changes in DNA (42). It was shown previously that polyamines can promote the B to Z conversion of poly(dG–dC)•poly(dG–dC) (15,43). In addition, G/C-rich B-DNA may be converted to A-DNA reversibly in the presence of polyamine (16,44). The sequences of DNA duplexes studied here were mainly (A)n•(T)n. According to the results from CD studies, both the perfect and imperfect DNAs retained their B-type conformation in the presence of high concentration polyamines. The results indicated that the spermine-induced change of conformations of oligonucleotides is sequence dependent. Furthermore, several X-ray structures have reported evidence for the polyamines in B-DNA stabilization with different a mechanism of polyamine–DNA interaction. In the previous work, Drew and Dickerson (45) have shown a spermine molecule that spans the upper end of the major groove in the B-DNA crystal structure of dodecamer d(CGCGAATTCGCG)2. In contrast, Tari and Secco (21) have found a spermine molecule in the narrow minor groove of a distorted B-DNA. In addition, Gao et al. (46) found a spermine molecule bridging two tandem GpA steps from two symmetry-related duplexes by making contacts with four guanines of the G•A mismatches. This may also suggest a possible mode of stabilization by spermine in M1 duplex.
The biosensor method is sensitive not only to discriminate the gross difference caused by lesions in a DNA duplex, but also to detect the kinetic effects under different conditions. Here we report the first example of the analysis on the association and dissociation kinetics of DNA containing abnormal sequences using SPR technique in the presence of different cations. A previous SPR study has shown that the kinetic parameters of DNA hybridization can be significantly affected by the number and location of mismatches in a DNA duplex of less than 20 bases (47). Jensen et al. (48) have shown that the binding of pentadecamer DNA and RNA to peptide nucleic acid, DNA and RNA sequences containing single mismatches at various positions in the center resulted in increased kd values and decreased ka values. They also suggested that the most stable duplexes (the highest Tm) exhibited the slowest dissociation rates and the fastest association rates. In the SPR study of this report, we observed that high concentration of salts or polyamine increased the association rate constant of M1, B1 and P1 to nearly the same level in the high positive ionic strength environment. But they also caused different binding capacity on the duplex formation in the order of spermine > spermidine > Mg2+ > Na+. Particularly notable is that polyamines can stabilize DNA containing mismatched and bulged base pairs as shown by their decreased kd values which generally correlate with increased Tm.
We conclude that the binding of polyamine to DNA duplexes removes the stability differences among various DNA structures whose inter-conversion may be carried out with minimal energy barriers in the presence of polyamine. This may have relevance in the possible biological functions associated with DNA polymorphism. In addition, some studies suggest that mismatched base pair stacking site is a preferred recognition position for protein–DNA interaction (49), especially for repair enzyme (50). Therefore, a high concentration of polyamines, often found in tumor cells, may be able to protect a mismatched base pair in DNA from repair enzymes, leading to deleterious mutations.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the National Health Research Insitute for use of their BIAcore 2000 SPR instrument. This work is supported by NSC grants 89-2113-M-001-093 (Taiwan, ROC) to L.-S.K and NSC grants 90-2311-B-001-099 (Taiwan, ROC) to A.H.-J.W.
REFERENCES
- 1.Cohen S.S. (1998) A Guide to the Polyamines. Oxford University Press, NY.
- 2.Igarashi K. and Kashiwagi,K. (2000) Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun., 271, 559–564. [DOI] [PubMed] [Google Scholar]
- 3.Tabor C.W. and Tabor,H. (1984) Polyamines. Annu. Rev. Biochem., 53, 749–790. [DOI] [PubMed] [Google Scholar]
- 4.Pegg A.E. (1988) Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res., 48, 759–774. [PubMed] [Google Scholar]
- 5.Davidson N.E., Hahm,H.A., McCloskey,D.E., Woster,P.M. and Casero,R.A. (1999) Clinical aspects of cell death in breast cancer: the polyamine pathway as a new target for treatment. Endocrinol. Relat. Cancer, 6, 69–73. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi Y., Mai,M. and Nishioka,K. (2000) Alpha-difluoromethylornithine induces apoptosis as well as anti-angiogenesis in the inhibition of tumor growth and metastasis in a human gastric cancer model. Int. J. Cancer, 85, 243–247. [PubMed] [Google Scholar]
- 7.Gupta S., Ahmad,N., Marengo,S.R., Mac Lennan,G.T., Greenberg,N.M. and Mukhtar,H. (2000) Chemoprevention of prostate carcinogenesis by alpha-difluoromethylornithine in TRAMP mice. Cancer Res., 60, 5125–5133. [PubMed] [Google Scholar]
- 8.Oller A.R., Van den Broek,W., Conrad,M. and Topal,M.D. (1991) Ability of DNA and spermidine to affect the activity of restriction endonucleases from several bacterial species. Biochemistry, 30, 2543–2549. [DOI] [PubMed] [Google Scholar]
- 9.Panagiotidis C.A., Artandi,S., Calame,K. and Silverstein,S.J. (1995) Polyamines alter sequence-specific DNA-protein interactions. Nucleic Acids Res., 23, 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel A.R. and Wang,J.Y. (1999) Polyamine depletion is associated with an increase in JunD/AP-1 activity in small intestinal crypt cells. Am. J. Physiol., 276, G441–450. [DOI] [PubMed] [Google Scholar]
- 11.Frugier M., Florentz,C., Hosseini,M.W., Lehn,J.M. and Giege,R. (1994) Synthetic polyamines stimulate in vitro transcription by T7 RNA polymerase. Nucleic Acids Res., 22, 2784–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Devereux,W., Stewart,T.M. and Casero,R.A. (2001) Characterization of the interaction between the transcription factors human polyamine modulated factor (PMF-1) and NF-E2-related factor 2 (Nrf-2) in the transcriptional regulation of the spermidine/spermine N1-acetyltransferase (SSAT) gene. Biochem. J., 355, 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westerhoff H.V., O’Dea,M.H., Maxwell,A. and Gellert,M. (1988) DNA supercoiling by DNA gyrase. A static head analysis. Cell Biophys., 12, 157–181. [DOI] [PubMed] [Google Scholar]
- 14.Pommier Y., Kerrigan,D. and Kohn,K. (1989) Topological complexes between DNA and topoisomerase II and effects of polyamines. Biochemistry, 28, 995–1002. [DOI] [PubMed] [Google Scholar]
- 15.Ohishi H., Terasoma,N., Nakanishi,I., van der Marel,G., van Boom,J.H., Rich,A., Wang,A.H.-J., Hakoshima,T. and Tomita,K. (1996) Interaction between left-handed Z-DNA and polyamine-3. The crystal structure of the d(CG)3 and thermospermine complex. FEBS Lett., 398, 291–296. [DOI] [PubMed] [Google Scholar]
- 16.Gao Y.G., Robinson,H. and Wang,A.H.-J. (1999) High-resolution A-DNA crystal structures of d(AGGGGCCCCT). An A-DNA model of poly(dG)•poly(dC). Eur. J. Biochem., 261, 413–420. [PubMed] [Google Scholar]
- 17.Thomas T.J. and Messner,R.P. (1986) A left-handed (Z) conformation of poly(dA–dC)•poly(dG–dT) induced by polyamines. Nucleic Acids Res., 14, 6721–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egli M., Tereshko,V., Teplova,M., Minasov,G., Joachimiak,A., Sanishvili,R., Weeks,C.M., Miller,R., Maier,M.A., An,H., Dan Cook,P. and Manoharan,M. (1998) X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers, 48, 234–252. [DOI] [PubMed] [Google Scholar]
- 19.Shui X., McFail-Isom,L., Hu,G.G. and Williams,L.D. (1998) The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry, 37, 8341–8355. [DOI] [PubMed] [Google Scholar]
- 20.Jain S., Zon,G. and Sundaralingam,M. (1989) Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC). Biochemistry, 28, 2360–2364. [DOI] [PubMed] [Google Scholar]
- 21.Tari L.W. and Secco,A.S. (1995) Base-pair opening and spermine binding B-DNA features displayed in the crystal structure of a gal operon fragment: implications for protein-DNA recognition. Nucleic Acids Res., 23, 2065–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feuerstein B.G., Pattabiraman,N. and Marton,L.J. (1990) Molecular mechanics of the interactions of spermine with DNA: DNA bending as a result of ligand binding. Nucleic Acids Res., 18, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feuerstein B.G., Pattabiraman,N. and Marton,L.J. (1986) Spermine-DNA interactions: a theoretical study. Proc. Natl Acad. Sci. USA, 83, 5948–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zakrzewska K. and Pullman,B. (1986) Spermine-nucleic acid interactions: a theoretical study. Biopolymers, 25, 375–392. [DOI] [PubMed] [Google Scholar]
- 25.Antony T., Thomas,T., Shirahata,A. and Thomas,T.J. (1999) Selectivity of polyamines on the stability of RNA-DNA hybrids containing phosphodiester and phosphorothioate oligodeoxyribonucleotides. Biochemistry, 38, 10775–10784. [DOI] [PubMed] [Google Scholar]
- 26.Musso M., Thomas,T., Shirahata,A., Sigal,L.H., Van Dyke,M.W. and Thomas,T.J. (1997) Effects of chain length modification and bis(ethyl) substitution of spermine analogs on purine-purine-pyrimidine triplex DNA stabilization, aggregation, and conformational transitions. Biochemistry, 36, 1441–1449. [DOI] [PubMed] [Google Scholar]
- 27.Saminathan M., Antony,T., Shirahata,A., Sigal,L.H., Thomas,T. and Thomas,T.J. (1999) Ionic and structural specificity effects of natural and synthetic polyamines on the aggregation and resolubilization of single-, double-, and triple-stranded DNA. Biochemistry, 38, 3821–3830. [DOI] [PubMed] [Google Scholar]
- 28.Musso M. and Van Dyke,M.W. (1995) Polyamine effects on purine-purine-pyrimidine triple helix formation by phosphodiester and phosphorothioate oligodeoxyribonucleotides. Nucleic Acids Res., 23, 2320–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusama-Eguchi K., Irisawa,M., Watanabe,S., Watanabe,K. and Igarashi,K. (1991) Increase in fidelity of rat liver Ile-tRNA formation by both spermine and the aminoacyl-tRNA synthetase complex. Arch. Biochem. Biophys., 288, 495–499. [DOI] [PubMed] [Google Scholar]
- 30.Deng H., Bloomfield,V.A., Benevides,J.M. and Thomas,G.J. (2000) Structural basis of polyamine-DNA recognition: spermidine and spermine interactions with genomic B-DNAs of different GC content probed by Raman spectroscopy. Nucleic Acids Res., 28, 3379–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelta J., Livolant,F. and Sikorav,J.L. (1996) DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem., 271, 5656–5662. [DOI] [PubMed] [Google Scholar]
- 32.Smirnov I.V., Dimitrov,S.I. and Makarov,V.L. (1987) Interaction of polyamines with chromatin and DNA: formation of compact structures. Mol. Biol., 21, 1411–1421. [PubMed] [Google Scholar]
- 33.Smirnov I.V., Dimitrov,S.I. and Makarov,V.L. (1988) Polyamine-DNA interactions. Condensation of chromatin and naked DNA. J. Biomol. Struct. Dyn., 5, 1149–1161. [DOI] [PubMed] [Google Scholar]
- 34.Rodger A., Taylor,S., Adlam,G., Blagbrough,I.S. and Haworth,I.S. (1995) Multiple DNA binding modes of anthracene-9-carbonyl-N1-spermine. Bioorg. Med. Chem., 3, 861–872. [DOI] [PubMed] [Google Scholar]
- 35.Tabor H. (1962) The protective effect of spermine and other polyamines against heat denaturation of deoxyribonucleic acid. Biochemistry, 1, 496–500. [DOI] [PubMed] [Google Scholar]
- 36.Green R.J., Frazier,R.A., Shakesheff,K.M., Davies,M.C., Roberts,C.J. and Tendler,S.J. (2000) Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials, 21, 1823–1835. [DOI] [PubMed] [Google Scholar]
- 37.Marky L.A., Snyder,J.G. and Breslauer,K.J. (1983) Calorimetric and spectroscopic investigation of drug-DNA interactions: II. Dipyrandium binding to poly d(AT). Nucleic Acids Res., 11, 5701–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyi J.I., Conn,G.L., Lane,A.N. and Brown,T. (1996) Comparison of the thermodynamic stabilities and solution conformations of DNA.RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry, 35, 12538–12548. [DOI] [PubMed] [Google Scholar]
- 39.Bloomfield V.A. (1996) DNA condensation. Curr. Opin. Struct. Biol., 6, 334–341. [DOI] [PubMed] [Google Scholar]
- 40.Rouzina I. and Bloomfield,V.A. (1999) Heat capacity effects on the melting of DNA. 1. General aspects. Biophys. J., 77, 3242–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chalikian T.V., Volker,J., Plum,G.E. and Breslauer,K.J. (1999) A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques. Proc. Natl Acad. Sci. USA, 96, 7853–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basu H.S. and Marton,L.J. (1987) The interaction of spermine and pentamines with DNA. Biochem. J., 244, 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Behe M., Zimmerman,S. and Felsenfeld,G. (1981) Changes in the helical repeat of poly(dG–m5dC)•poly(dG–m5dC) and poly(dG–dC)•poly (dG–dC) associated with the B-Z transition. Nature, 293, 233–235. [DOI] [PubMed] [Google Scholar]
- 44.Robinson H. and Wang,A.H.-J. (1996) Neomycin, spermine and hexaamminecobalt (III) share common structural motifs in converting B- to A-DNA. Nucleic Acids Res., 24, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drew H.R. and Dickerson,R.E. (1981) Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol., 151, 535–556. [DOI] [PubMed] [Google Scholar]
- 46.Gao Y.G., Robinson,H., Sanishvili,R., Joachimiak,A. and Wang,A.H.-J. (1999) Structure and recognition of sheared tandem G × A base pairs associated with human centromere DNA sequence at atomic resolution. Biochemistry, 38, 16452–16460. [DOI] [PubMed] [Google Scholar]
- 47.Gotoh M., Hasegawa,Y., Shinohara,Y., Shimizu,M. and Tosu,M. (1995) A new approach to determine the effect of mismatches on kinetic parameters in DNA hybridization using an optical biosensor. DNA Res., 2, 285–293. [DOI] [PubMed] [Google Scholar]
- 48.Jensen K.K., Orum,H., Nielsen,P.E. and Norden,B. (1997) Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry, 36, 5072–5077. [DOI] [PubMed] [Google Scholar]
- 49.Su S., Gao,Y.G., Robinson,H., Liaw,Y.C., Edmondson,S.P., Shriver,J.W. and Wang,A.H.-J. (2000) Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs. J. Mol. Biol., 303, 395–403. [DOI] [PubMed] [Google Scholar]
- 50.Chepanoske C.L., Porello,S.L., Fujiwara,T., Sugiyama,H. and David,S.S. (1999) Substrate recognition by Escherichia coli MutY using substrate analogs. Nucleic Acids Res., 27, 3197–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]